Abstract
In this study, we developed a single helper-dependent adenovirus (HDAd) to deliver all of the components (donor DNA, CRISPR-associated protein 9 [Cas9], and guide RNA [gRNA]) needed to achieve high-efficiency gene targeting and homology-directed repair in transduced cells. We show that these “all-in-one” HDAds are up to 117-fold more efficient at gene targeting than donor HDAds that do not express CRISPR/Cas9 in human induced pluripotent stem cells (iPSCs). The vast majority (>90%) of targeted recombinants had only one allele targeted, and this was accompanied by high-frequency indel formation in the non-targeted allele at the site of Cas9 cleavage. These indels varied in size and nature, and included large deletions of ∼8 kb. The remaining minority of recombinants had both alleles targeted (so-called bi-allelic targeting). These all-in-one HDAds represent an important platform for accomplishing and expanding the utility of homology-directed repair, especially for difficult-to-transfect cells and for in vivo applications.
Keywords: homology-directed repair, bi-allelic, CFTR, cystic fibrosis, helper-dependent adenovirus, homologous recombination, gene targeting, gene editing, CRISPR, Cas9, indels
Introduction
We and others have shown that helper-dependent adenoviruses (HDAds) can efficiently deliver donor DNA to achieve homology-directed repair (HDR) by spontaneous homologous recombination (HR).1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 The HDAd offers a number of advantages as a gene-targeting vector. First, HDAds can very efficiently transduce many cell types from many species, rendering them an excellent platform for achieving gene targeting in hard-to-transfect cells such as primary cells and cells in vivo. Second, because of the HDAd’s tremendous cloning capacity of 37 kb, they can accommodate very long homology arms, and consequently we have shown that multiple genetic alterations up to 22.2 kb apart can be efficiently introduced simultaneously into the chromosomal locus using a single HDAd.14 This also means that a single HDAd can be used to correct many different mutations from many different individuals. However, the rate-limiting step in HDAd-mediated gene targeting is the reliance on rare, stochastic double-strand breaks (DSBs) in their homology arms to initiate HR.14 In this study, we develop a single HDAd to deliver the donor DNA, CRISPR-associated protein 9 (Cas9), and guide RNA (gRNA) into the nucleus of transduced cells. This all-in-one HDAd substantially improves the efficiency of gene targeting because it no longer relies on the occurrence of rare spontaneous DSBs in the vector’s homology arms to initiate HR. Instead HR is efficiently initiated by a DSB at the chromosomal target by the HDAd-encoded Cas9 and gRNA.
Results
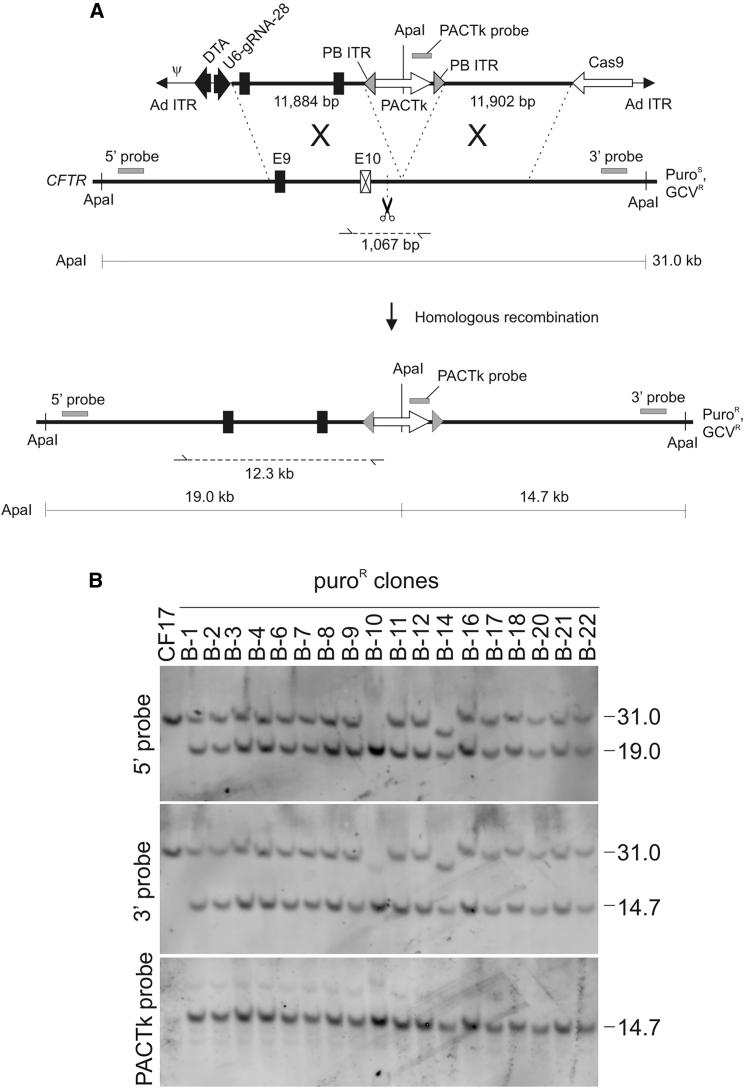
The objective of this study was to develop a single, all-in-one HDAd to efficiently deliver the three components (donor DNA, Cas9, and gRNA) needed to achieve high-efficiency gene targeting and homology-directed repair (HDR) into the nucleus of transduced cells. As a model system, we targeted the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) gene in a human induced pluripotent stem cell (iPSC) line, called CF17, which is compound heterozygous at the CFTR locus, with one allele bearing the ΔF508 mutation and the other allele bearing the ΔI507 mutation in exon 10.16 To accomplish this, we started with HD-23.8-CFTRm-PACTk-DTA, an HDAd that we previously used for footprintless HDR at the CFTR locus of CF17 iPSCs by spontaneous HR,14 and modified it to create the all-in-one HDAd. This was accomplished by replacing the LacZ expression cassette with a Cas9 expression cassette and by inserting the gRNA expression cassette into the vector. Two different all-in-one HDAds were made in this way, differing in the gRNA expressed; the first expressed a gRNA that directed Cas9 cleavage 28 bp upstream of the site of PACTk insertion, and this all-in one HDAd was named HD-23.8-CFTRm-PACTk-DTA+Cas9−28 (the superscript −28 indicates the position of the DSB with respect to PACTk) (Figure 1A). To prevent self-cleavage, we changed the 5′-CCA-3′ protospacer adjacent motif (PAM) in the donor to 5′-TCA-3′. The second all-in-one HDAd expressed a gRNA that directed Cas9 cleavage exactly at the site of the PACTk insertion, and this all-in one HDAd was named HD-23.8-CFTRm-PACTk-DTA+Cas9+0 (the superscript +0 denotes the location of the DSB with respect to PACTk) (Figure 2A). Both all-in-one HDAds bore the positive/negative selectable PACTk marker flanked by piggyBac (PB) inverted terminal repeats (ITRs) to permit its footprintless removal by PB transposase.
Figure 1.
Gene Targeting with All-in-One HDAd
(A) The all-in-one HD-23.8-CFTRm-PACTk-DTA+Cas9−28 possesses the donor DNA bearing a wild-type exon 10, a Cas9 expression cassette, and a gRNA expression cassette. The gRNA directs Cas9 cleavage 28 bp upstream of the PACTk insertion site indicated by the scissor. The 5′-CCA-3′ PAM in the donor was modified to 5′-TCA-3′ to prevent self-cleavage. PB inverted terminal repeats (ITRs) flank the PACTk cassette to permit its footprintless excision in the presence of PB transposase. Sizes of the diagnostic ApaI fragments and the locations of the 5′ external probe, 3′ external probe, and PACTk probe used for Southern blot analyses are shown. The 1,067-bp PCR product from the non-targeted allele is shown and was sequenced to determine its identity and the presence of indels. The 12.3-kb PCR product from the targeted allele is shown and was sequenced to determine whether the wild-type sequence was introduced into the target locus after gene targeting. The ΔI507 and ΔF508 mutations are 208 bp from the site of PACTk insertion. The position of the adenoviral packaging signal (ψ), adenoviral (Ad) ITR, and the diphtheria toxin A-fragment gene (DTA) for counter-selection against random integrations are shown for the HDAd. (B) Representative Southern blots of genomic DNA extracted from puroR colonies analyzed with the 5′ external probe, the 3′ external probe, and the PACTk probe.
Figure 2.
Gene Targeting with All-in-One HDAd
(A) The all-in-one HD-23.8-CFTRm-PACTk-DTA+Cas9+0 possesses the donor DNA bearing a wild-type exon 10, a Cas9 expression cassette, and a gRNA expression cassette. The gRNA directs Cas9 cleavage precisely at the PACTk insertion site indicated by the scissor. The 1,912-bp PCR product from the unmodified allele is shown, which was sequenced to determine its identity and presence of indels. The 15.0-kb PCR product from the targeted allele is shown, which was sequenced to determine whether the wild-type exon 10 had been introduced into the target locus after gene targeting. The ΔI507 and ΔF508 mutations are 1.8 kb from the site of PACTk insertion. All other elements are as described in Figure 1. (B) Representative Southern blots of genomic DNA extracted from puroR clones analyzed with the 5′ external probe, the 3′ external probe, and the PACTk probe.
To access gene-targeting efficiency, 2 × 106 CF17 cells were transduced with the two all-in-one HD-23.8-CFTRm-PACTk-DTA-Cas9+28 and HD-23.8-CFTRm-PACTk-DTA-Cas9+0 at an MOI of 350 viral particles (vp)/cell. A total of 2 × 106 CF17 cells were also transduced with the parental HD-23.8-CFTRm-PACTk-DTA at an MOI of 350 vp/cell to serve as a control. As shown in Table 1, transduction with the parental HD-23.8-CFTRm-PACTk-DTA yielded a total of 30 puromycin-resistant (puroR) colonies for a puroR frequency of 1.5 × 10−5. In contrast, the all-in-one HD-23.8-CFTRm-PACTk-DTA+Cas9−28 yielded a total of 1,937 puroR colonies for a puroR frequency of 9.7 × 10−4, and the all-in-one HD-23.8-CFTRm-PACTk-DTA+Cas9+0 yielded a total of 3,501 colonies for a puroR frequency of 1.8 × 10−3. Therefore, the all-in-one HDAds improved puroR frequencies by 65-fold and 117-fold, respectively.
Table 1.
Gene-Targeting Frequencies
| HDAd | No. of Cells Transduced | No. of puroR Colonies | Freq puroR | Correct Gene Targeting | Gene Correction | One Allele Targeted | Both Alleles Targeted | Indels in Non-targeted Allele |
|---|---|---|---|---|---|---|---|---|
| HD-23.8-CFTRm-PACTk-DTA | 2 × 106 | 30 | 1.5 × 10−5 | NDa | NDb | NDc | NDd | NA |
| HD-23.8-CFTRm-PACTk-DTA+Cas9−28 | 2 × 106 | 1,937 | 9.7 × 10−4 | 100% (21/21) | 93.8% (15/16) | 100% (21/21) | 0% (0/21) | 75% (15/20) |
| HD-23.8-CFTRm-PACTk-DTA+Cas9+0 | 2 × 106 | 3,501 | 1.8 × 10−3 | 95.7% (22/23) | 60% (12/20) | 90.9% (20/22) | 9.1% (2/22) | 100% (21/21) |
Freq, Frequency; NA, not applicable; ND, not determined.
ND in this study but was 89.5% in a prior study with the same HDAd.14
ND in this study but was 100% in a prior study with the same HDAd.14
ND in this study but was 100% in a prior study with the same HDAd.14
ND in this study but was 0% in a prior study with the same HDAd.14
To verify correct gene targeting by the all-in-one HDAds, DNA was extracted from 21 and 23 puroR colonies generated with HD-23.8-CFTRm-PACTk-DTA+Cas9−28 and HD-23.8-CFTRm-PACTk-DTA+Cas9+0, respectively, and analyzed by Southern blot hybridization. As shown in Figure 1A, in the case of HD-23.8-CFTRm-PACTk-DTA+Cas9−28, gene targeting converts the 31-kb endogenous ApaI fragment (revealed by the 5′ external probe and 3′ external probe) to a 19.0-kb ApaI fragment (revealed by the 5′ external probe) and a 14.5-kb ApaI fragment (revealed by the external 3′ probe and internal PACTk probe). The results of the Southern blot analyses are summarized in Table 1, and representative Southern blots are shown in Figure 1B. These analyses revealed that targeted vector integration into the CFTR locus had occurred in 100% (21/21) of the puroR colonies as evident by the presence of the 19.0- and 14.5-kb ApaI fragments when analyzed with the 5′ external probe and the 3′ external probe, respectively. To determine whether gene targeting resulted in gene correction (introduction of the wild-type sequence into the chromosomal target), we used PCR to specifically amplify a unique 12.3-kb fragment encompassing the 5′ region of homology from the targeted allele, which was then sequenced (Figure 1A). We performed this analysis on 16 representative targeted clones and found that 93.8% (15/16) had introduced the wild-type sequence, with the remainder 3.6% (1/16) having retained the mutant sequence. Of the 21 targeted clones, 19 (90.5%) have the 31.0-kb ApaI fragment when analyzed with either the 5′ external or 3′ external probes indicating that in these clones, only one CFTR allele was targeted (Figure 1B). The remaining 2/21 possessed, instead of the expected 31-kb band, a smaller band of ∼23 kb (clones B-10 and B-14 in Figure 1B), indicating that the non-targeted allele had suffered a deletion of ∼8 kb. Significantly, such a large deletion of the non-targeted allele was not observed previously in all 287 targeted recombinants obtained by HDAd-mediated spontaneous HR.13,14 Therefore, we believe that the large deletion arose as a consequence of Cas9 cleavage at the non-targeted allele. This prompted us to determine whether more subtle indels were present at the Cas9 cleavage site in the non-targeted allele of the 18 puroR colonies (not 19 because 1 puroR colony was lost before we could do this analysis) with the 31-kb ApaI fragment. To investigate this, we performed PCR to amplify a 1,067-bp fragment specifically from the non-targeted allele encompassing the Cas9 cleavage site (Figure 1A) of the 18 puroR colonies for sequencing. The results revealed that 2/18 yielded no PCR product, and we interpreted this to mean Cas9 induced deletion involving at least a portion of the template and/or PCR primer binding site(s). Of the 16 colonies that yielded a ∼1-kb PCR product, sequencing revealed that 5 did not bear indels, having the expected sequence around the Cas9 cleavage site, whereas the remaining 11 harbored various indels at the Cas9 cleavage site, and representative examples are shown in Figure 3A. Thus, of the 20 colonies analyzed, 75% (15/20) had indels at the Cas9 cleavage site in the non-targeted allele.
Figure 3.
Indels in the Non-targeted Allele of Targeted Recombinants
Indels in the non-targeted allele of representative targeted recombinants generated with (A) HD-23.8-CFTRm-PACTk-DTA+Cas9−28 and (B) HD-23.8-CFTRm-PACTk-DTA+Cas9+0. Boxed sequence indicates gRNA sequence.
As shown in Figure 2A, in the case of HD-23.8-CFTRm-PACTk-DTA+Cas9+0, gene targeting would convert the 31-kb endogenous ApaI fragment (revealed by the 5′ external probe and 3′ external probe) into a 17.0-kb fragment (revealed by the external 5′ probe) and a 16.7-kb ApaI fragment (revealed by the external 3′ probe and internal PACTk probe). The results of the Southern blot analyses are summarized in Table 1, and representative Southern blots are shown in Figure 2B. These analyses revealed that correct targeted vector integration into the CFTR gene had occurred in 95.7% (22/23) of the puroR colonies. The remaining (1/23) puroR colony had an aberrantly targeted vector integration with correct HR in the 5′ homology (as evident by the presence of the 17.0-kb band when analyzed with the 5′ external probe; see clone C-22 in Figure 2B), but not the 3′ homology arm (as evident by the absence of the 16.7-kb band when analyzed with the 3′ external probe and PACTk probe; see clone C-22 in Figure 2B). Of the 22 correctly targeted colonies, 20 (90.9%) had targeting into one allele as evident by the continued presence of the endogenous 31-kb ApaI fragment when analyzed with the external 5′ and 3′ probes. The other 2 (9.1% or 2/22) had targeted vector integration into both alleles (so-called bi-allelic targeting) (see, for example, clone C-11 in Figure 2B) as evident by the absence of the endogenous 31-kb ApaI fragment when analyzed with the 5′ external probe and 3′ external probe. To determine whether vector integration resulted in gene correction (introduction of the wild-type sequence into the chromosomal target), we used PCR to specifically amplify a unique 15-kb fragment encompassing the 3′ region of homology and sequenced it. We performed this analysis on 20 correctly targeted clones, and 60% (12/20) had introduced the wild-type sequence, with the remaining 40% (8/20) having retained the mutant sequence. Finally, for the 20 puroR colonies with targeted vector integration into one CFTR allele, we next sought to determine whether the non-targeted allele acquired indels as a consequence of Cas9 cleavage. To accomplish this, we used PCR to amplify a 1,912-bp region specifically from the non-targeted allele and sequenced it. As expected, all clones yielded a band at ∼1.9 kb, except clones C-15 and C-18, which yielded bands slightly smaller than 1.9 kb (data not shown). The PCR products were sequenced and the results revealed that 100% (21/21, including the one aberrantly targeted clone) had acquired indels at the Cas9 cleavage site in the non-targeted allele. Representative examples are shown in Figure 3B and as shown, clones C-15 and C-18 bore 184- and 429-bp deletions, respectively, encompassing the gRNA sequence consistent with the smaller-size PCR product described above.
In summary, gene targeting is improved up to 117-fold with all-in-one donor HDAd expressing Cas9 and gRNA to create a DSB at the chromosomal target compared with HDAd relying on spontaneous HR. Furthermore, bi-allelic targeting can be achieved relatively easily with an all-in-one HDAd. However, most recombinants have only one allele targeted, and the non-targeted allele often acquires indels, which can be large deletions of up to ∼8 kb.
Discussion
In this study, we developed a single, all-in-one HDAd to transduce the three components (donor DNA, Cas9, and gRNA) needed to achieve high-efficiency gene targeting and HDR. We assessed two such all-in-one HDAds named HD-23.8-CFTRm-PACTk-DTA+Cas9−28 and HD-23.8-CFTRm-PACTk-DTA+Cas9+0. The gRNA expressed by HD-23.8-CFTRm-PACTk-DTA+Cas9−28 directs Cas9 to cleave the chromosomal target 28 bp upstream of the selectable marker insertion site, whereas the gRNA expressed by HD-23.8-CFTRm-PACTk-DTA+Cas9+0 directs Cas9 to cleave the chromosomal target precisely at the selectable marker insertion site. We found that these all-in-one HDAds improved gene-targeting efficiencies by 65-fold and 117-fold, respectively, compared with donor HDAd that relied on spontaneous HR.
Our data show that HD-23.8-CFTRm-PACTk-DTA+Cas9+0 was 1.8-fold more efficient at gene targeting than HD-23.8-CFTRm-PACTk-DTA+Cas9−28. Bi-allelic targeting, at a relatively high 8.7%, was also obtained with HD-23.8-CFTRm-PACTk-DTA+Cas9+0, but not HD-23.8-CFTRm-PACTk-DTA+Cas9−28. Lastly, HD-23.8-CFTRm-PACTk-DTA+Cas9+0 was also more efficient at producing indels in the non-targeted allele (100%) than HD-23.8-CFTRm-PACTk-DTA+Cas9−28 (75%). Together, these results suggest that the gRNA expressed by HD-23.8-CFTRm-PACTk-DTA+Cas9+0 is more efficient at directing Cas9 cleavage than the gRNA expressed by HD-23.8-CFTRm-PACTk-DTA+Cas9+0. However, other factors such as location of Cas9-mediated DSB may have contributed to some of these differences and cannot be ruled out.
HD-23.8-CFTRm-PACTk-DTA+Cas9+0 has the advantage that the target gRNA sequence is not present in the donor because it is interrupted by the selectable marker. Therefore, mutating the PAM in the donor is not necessary to avoid vector self-cleavage, unlike with HD-23.8-CFTRm-PACTk-DTA+Cas9−28. Furthermore, in the case of HD-23.8-CFTRm-PACTk-DTA+Cas9−28, we have found that mutating the PAM was not sufficient to prevent self-cleavage during vector production,17 and that this vector could be produced only by downregulating Cas9 mRNA and inhibiting Cas9 protein activity during vector production.17
However, a disadvantage of the strategy employed by HD-23.8-CFTRm-PACTk-DTA+Cas9+0 is that the gRNA sequence must be 5′-CCNNTTAAN15-3′ to accommodate the 5′-TTAA-3′ piggbac insertion site, and this greatly limits where the selectable marker can be placed relative to the site of the intended modification. This is important because we have previously shown that there is an inverse relationship between the efficiency of incorporating a genetic modification present in the donor and its distance from the selectable marker.14 In this case, the closest 5′-CCNNTTAAN15-3′ motif for selectable marker insertion was located 1,776 bp upstream of the ΔF508/ΔI507 mutation. This is in contrast with HD-23.8-CFTRm-PACTk-DTA+Cas9−28, where the site of selectable marker insertion is only 208 bp upstream of the ΔF508/ΔI507 mutation, and this explains why the wild-type sequence was introduced into the target locus in 93.8% of the targeted recombinants generated with HD-23.8-CFTRm-PACTk-DTA+Cas9−28, whereas only 60% of the targeted recombinants generated with HD-23.8-CFTRm-PACTk-DTA+Cas9+0 had introduced the wild-type sequence into the target locus.
As mentioned above, bi-allelic targeting can be efficiently achieved using an all-in-one HDAd. This is in contrast with gene targeting with conventional HDAd, which relies on spontaneous HR in which bi-allelic targeting could be achieved only by targeting each allele one at a time by sequential vector transductions.11,15 Nevertheless, gene targeting by the all-in-one HDAd occurred in only one allele in the majority of the targeted recombinants, and in these clones, the non-targeted allele acquired indels at high frequency (up to 100%). Although the consequence of this may depend on the target gene and the nature of the indels, it should be emphasized that we observed deletions of ∼8 kb, and such large deletions might not be benign.
Finally, for HDR, should the all-in-one HDAd be preferred over conventional HDAds that rely on spontaneous HR because of their significantly higher efficiency? The answer may be yes if the application requires the highest absolute efficiency. However, for applications where lower efficiencies are acceptable, then conventional HDAds might be preferred to avoid concerns related to indel formation in the non-targeted allele and off-target effects. However, Xia et al.18 recently showed that Cas9 expressed from an HDAd is quickly lost due to rapid degradation of the HDAd genome as a consequence of recombining with the chromosomal target, thus limiting potential off-targeting and toxic effects of Cas9. In any case, both options are now available, and the choice will be based on the application’s unique requirements.
Materials and Methods
HDAds
The all-in-one HDAd, HD-23.8-CFTRm-PACTk-DTA+Cas9−28, was derived from HD-23.8 m-PACTk-DTA14 by replacing the 4,487-bp AscI fragment containing the LacZ expression cassette with the 5,896-bp AscI fragment containing the Cas9 expression cassette, and then by inserting the gRNA expression cassette into the unique SbfI site, and finally by changing the 5′-CCA-3′ PAM to 5′-TCA-3′ to prevent self-cleavage. However, even with the mutated PAM, HD-23.8-CFTRm-PACTk-DTA+Cas9−28 had to be produced in 116Acr3 cells with the helper virus AdNG163Acr to prevent self-cleavage during production.17 The all-in-one HDAd, HD-23.8-CFTRm-PACTk-DTA+Cas9+0, was derived from HD-23.8 m-PACTk-DTA14 by first moving the PACTk upstream 1,988 bp, which was necessary for having the Cas9 cleavage site located precisely at the PACTk insertion as depicted in Figure 2A. Next, the 4,487-bp AscI fragment containing the LacZ expression cassette was replaced with the 5,896-bp AscI fragment containing the Cas9 expression cassette. Finally, the gRNA expression cassette was inserted into the unique SbfI site. The Cas9 expression cassette used in both all-in-one HDAds possessed the target sequences for the helper-virus-encoded microRNA (miRNA), mivaRNAI, and producer-cell-encoded miRNAs, hsa-miR183-5p and hsa-miR218-5p, into the 3′ untranslated region to downregulate Cas9 mRNA expression during vector production.17 The all-in-one HDAds were produced in 116Acr3 cells with the helper virus AdNG163Acr as described previously.17
Transduction of iPSCs
CF17, the feeder free human CF iPSC line used in this study, is described elsewhere16 and was maintained in mTeSR 1 (STEMCELL Technologies, Vancouver, BC, Canada) on Matrigel (Corning, Tewksbury, MA, USA)-coated plates. Transduction of iPSCs for gene targeting was performed as follows: 2 × 106 cells were resuspended in 1 mL mTeSR 1 supplemented with Y27632 (Reagents Direct, Encinitas, CA, USA) to 10 μM in a 1.5-mL microfuge tube and transduced with HDAd at an MOI of 350 vp/cell for 1 h at 37°C with gentle rocking. Following transduction, cells were washed twice with 1 mL mTeSR 1 supplemented with Y27632 to 10 μM and plated onto six-well plates at a density of 7.7 × 104 cells/well in nonselective media. Forty-eight hours later, the media were replaced with media supplemented with puromycin to a final concentration of 0.5 μg/mL. Well-isolated puroR colonies were picked and DNA extracted for Southern blot analysis and PCR.
DNA Analyses
DNA extraction and non-radioactive digoxigenin-based Southern blot hybridization were performed as previously described.13 The 5′ region of homology was amplified from the targeted clone generated with HD-23.8-CFTRm-PACTk-DTA+Cas9−28 using primers 5′-atgagggaaggactcatgagagggaagtag-3′ and 5′-atgctccagactgccttgggaaaagcg-3′. The latter primer was also used to sequence the 5′ PCR product to determine whether exon 10 bore the wild-type or ΔF508 or ΔI507 sequence. The 3′ region of homology was amplified from the targeted clones generated with HD-23.8-CFTRm-PACTk-DTA+Cas9+0 using primers 5′-tctatggcttctgaggcggaaagaaccag-3′ and 5′-acgtgtatctgagagtgttacatggccctg-3′. The latter primer was also used to sequence the 5′ PCR product to determine whether exon 10 bore the wild-type or ΔF508 or ΔI507 sequence. The above PCR amplifications were performed with PrimeSTAR GXL DNA polymerase (Takara/Clontech, Mountain View, CA, USA) with final concentrations of 0.2 mM dNTP and 0.2 μM of each primer. Thermocycling conditions were as follows: 1 min at 94°C, followed by 30 cycles of 98°C for 10 s and 72°C for 10 minutes, and a final extension of 10 min at 72°C.
The non-targeted allele from targeted clones generated with HD-23.8-CFTRm-PACTk-DTA+Cas9−28 was amplified with 5′-gcatagcagagtacctgaaacagg-3′ and 5′-agcaataactactgaacccaccatc-3′ to yield a 1,067-bp fragment. The non-targeted allele from targeted clones generated with HD-23.8-CFTRm-PACTk-DTA+Cas9+0 was amplified with 5′-gcactctttgctttaggtgtgtctc-3′ and 5′-ggcatgctttgatgacgcttctg-3′ to yield a 1,912-bp fragment. These PCRs were performed using HotStarPlus (QIAGEN, Valencia, CA, USA) according to the manufacturer’s recommendations. Thermocycling conditions were as follows: 5 min at 95°C; followed by 35 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 2 min; and a final extension of 10 min at 72°C. These PCR products were sequenced with the forward primer to determine whether the non-targeted allele was ΔF508 or ΔI507 and to determine the presence of indels at the Cas9 cleavage site.
Author Contributions
This study was conceived by P.N. This study was designed and implemented by D.J.P., D.L.T., and P.N. This paper was written by P.N.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by internal Baylor College of Medicine funds.
References
- 1.Ohbayashi F., Balamotis M.A., Kishimoto A., Aizawa E., Diaz A., Hasty P., Graham F.L., Caskey C.T., Mitani K. Correction of chromosomal mutation and random integration in embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl. Acad. Sci. USA. 2005;102:13628–13633. doi: 10.1073/pnas.0506598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki K., Mitsui K., Aizawa E., Hasegawa K., Kawase E., Yamagishi T., Shimizu Y., Suemori H., Nakatsuji N., Mitani K. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl. Acad. Sci. USA. 2008;105:13781–13786. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M., Suzuki K., Qu J., Saini P., Dubova I., Yi F., Lee J., Sancho-Martinez I., Liu G.H., Izpisua Belmonte J.C. Efficient correction of hemoglobinopathy-causing mutations by homologous recombination in integration-free patient iPSCs. Cell Res. 2011;21:1740–1744. doi: 10.1038/cr.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu G.H., Suzuki K., Qu J., Sancho-Martinez I., Yi F., Li M., Kumar S., Nivet E., Kim J., Soligalla R.D. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell. 2011;8:688–694. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G.H., Qu J., Suzuki K., Nivet E., Li M., Montserrat N., Yi F., Xu X., Ruiz S., Zhang W. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aizawa E., Hirabayashi Y., Iwanaga Y., Suzuki K., Sakurai K., Shimoji M., Aiba K., Wada T., Tooi N., Kawase E. Efficient and accurate homologous recombination in hESCs and hiPSCs using helper-dependent adenoviral vectors. Mol. Ther. 2012;20:424–431. doi: 10.1038/mt.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umeda K., Suzuki K., Yamazoe T., Shiraki N., Higuchi Y., Tokieda K., Kume K., Mitani K., Kume S. Albumin gene targeting in human embryonic stem cells and induced pluripotent stem cells with helper-dependent adenoviral vector to monitor hepatic differentiation. Stem Cell Res. (Amst.) 2013;10:179–194. doi: 10.1016/j.scr.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Liu G.H., Suzuki K., Li M., Qu J., Montserrat N., Tarantino C., Gu Y., Yi F., Xu X., Zhang W. Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat. Commun. 2014;5:4330. doi: 10.1038/ncomms5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki K., Yu C., Qu J., Li M., Yao X., Yuan T., Goebl A., Tang S., Ren R., Aizawa E. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014;15:31–36. doi: 10.1016/j.stem.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida T., Ozawa Y., Suzuki K., Yuki K., Ohyama M., Akamatsu W., Matsuzaki Y., Shimmura S., Mitani K., Tsubota K., Okano H. The use of induced pluripotent stem cells to reveal pathogenic gene mutations and explore treatments for retinitis pigmentosa. Mol. Brain. 2014;7:45. doi: 10.1186/1756-6606-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W., Li J., Suzuki K., Qu J., Wang P., Zhou J., Liu X., Ren R., Xu X., Ocampo A. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto H., Ishimura M., Ochiai M., Takada H., Kusuhara K., Nakatsu Y., Tsuzuki T., Mitani K., Hara T. BTK gene targeting by homologous recombination using a helper-dependent adenovirus/adeno-associated virus hybrid vector. Gene Ther. 2016;23:205–213. doi: 10.1038/gt.2015.91. [DOI] [PubMed] [Google Scholar]
- 13.Palmer D.J., Grove N.C., Ing J., Crane A.M., Venken K., Davis B.R., Ng P. Homology requirements for efficient, footprintless gene editing at the CFTR locus in human iPSCs with helper-dependent adenoviral vectors. Mol. Ther. Nucleic Acids. 2016;5:e372. doi: 10.1038/mtna.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer D.J., Grove N.C., Turner D.L., Ng P. Gene Editing with Helper-Dependent Adenovirus Can Efficiently Introduce Multiple Changes Simultaneously over a Large Genomic Region. Mol. Ther. Nucleic Acids. 2017;8:101–110. doi: 10.1016/j.omtn.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer D.J., Turner D.L., Ng P. Bi-allelic homology-directed repair with helper-dependent adenoviruses. Mol. Ther. Methods Clin. Dev. 2019;15:285–293. doi: 10.1016/j.omtm.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crane A.M., Kramer P., Bui J.H., Chung W.J., Li X.S., Gonzalez-Garay M.L., Hawkins F., Liao W., Mora D., Choi S. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Reports. 2015;4:569–577. doi: 10.1016/j.stemcr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer D.J., Turner D.L., Ng P. Production of CRISPR/Cas9-Mediated Self-Cleaving Helper-Dependent Adenoviruses. Mol. Ther. Methods Clin. Dev. 2019;13:432–439. doi: 10.1016/j.omtm.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia E., Duan R., Shi F., Seigel K.E., Grasemann H., Hu J. Overcoming the Undesirable CRISPR-Cas9 Expression in Gene Correction. Mol. Ther. Nucleic Acids. 2018;13:699–709. doi: 10.1016/j.omtn.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]