Abstract
Spinocerebellar ataxia type-1 (SCA1) is caused by an abnormally expanded polyglutamine (polyQ) tract in ataxin-1. These expansions are responsible for protein misfolding and self-assembly into intranuclear inclusion bodies (IIBs) that are somehow linked to neuronal death. However, owing to lack of a suitable cellular model, the downstream consequences of IIB formation are yet to be resolved. Here, we describe a nuclear protein aggregation model of pathogenic human ataxin-1 and characterize IIB effects. Using an inducible Sleeping Beauty transposon system, we overexpressed the ATXN1(Q82) gene in human mesenchymal stem cells that are resistant to the early cytotoxic effects caused by the expression of the mutant protein. We characterized the structure and the protein composition of insoluble polyQ IIBs which gradually occupy the nuclei and are responsible for the generation of reactive oxygen species. In response to their formation, our transcriptome analysis reveals a cerebellum-specific perturbed protein interaction network, primarily affecting protein synthesis. We propose that insoluble polyQ IIBs cause oxidative and nucleolar stress and affect the assembly of the ribosome by capturing or down-regulating essential components. The inducible cell system can be utilized to decipher the cellular consequences of polyQ protein aggregation. Our strategy provides a broadly applicable methodology for studying polyQ diseases.
Keywords: Ataxin-1, Polyglutamine, Sleeping beauty transposon, Oxidative stress, Protein network, Ribosome
Abbreviations: SCA1, Spinocerebellar ataxia type-1; polyQ, polyglutamine; IIBs, intranuclear inclusion bodies; SB, Sleeping Beauty; MSC, mesenchymal stem cell; FTIR, Fourier-transform infrared spectroscopy; AFM, atomic force microscopy; MS, mass spectrometry; PPI, protein-protein interaction; iPSC, induced pluripotent stem cell; NPC, neural progenitor cell; ROS, reactive oxygen species; DE, differentially expressed genes; GSEA, Gene Set Enrichment Analysis
1. Introduction
Polyglutamine (polyQ) diseases are neurodegenerative disorders caused by trinucleotide (CAG) repeat expansions in the coding region of human genes, which result in abnormally large polyQ tracts in the produced proteins. In spinocerebellar ataxia type-1 (SCA1), the disease is caused by CAG expansions in the ATXN1 gene [1]. The polyQ-expanded ataxin-1 (ATXN1) protein forms small oligomers and slowly aggregates into larger insoluble nuclear inclusions in the affected neurons [2]. These are specifically detectable in the Purkinje cells of the cerebellum in SCA1 patients [1]. Several lines of evidence suggest that the deposition of large inclusions may be protective due to the sequestration of smaller cytotoxic oligomers [3]. However, recent findings indicate that insoluble inclusions might be also toxic, as they cause quiescence and activate necrotic mechanisms in cells [4]. These events are thought to be induced by oxidative stress and the activation of checkpoint kinases which regulate the cell cycle [5].
PolyQ aggregates have been extensively investigated in cell-free assays, using recombinant proteins produced in bacteria [[6], [7], [8]]. Studies in yeast, murine neurons and HeLa cells overexpressing fragments of polyQ-expanded genes provided valuable information on the structure and interactions of pathogenic inclusions in cellular systems [9,10]. However, a reliable aggregation model is still missing. While formation of insoluble inclusions was also observed in human induced pluripotent stem cell (iPSC)-derived neurons [11], this phenotype cannot be easily reproduced [12]. Therefore, novel protein aggregation models are needed that could be readily used to characterize polyQ inclusions generated in primary human cells.
Human mesenchymal stem cells (MSCs) have been previously used for modeling of polyQ diseases [13]. These cells have a partial ability to fuse with Purkinje neurons [14], suggesting that they might contribute to the restoration of their physiological neuronal function. MSCs can be easily modified genetically using the Sleeping Beauty (SB) transposon system [15], a safer alternative to viral approaches [16]. Engineered cells proliferate and stably express human transgenes for several passages [[17], [18], [19]]. MSCs can also model replicative senescence, a cellular event that resembles aging and is associated with protein aggregation [20]. Expression of aberrant, polyQ-expanded genes in human MSCs is expected to generate suitable cellular models supporting robust, measurable and reproducible pathogenic phenotypes.
In this study, we report an inducible version of a previously described SB system [21] that can be utilized for the controlled expression of pathogenic genes in cultured primary cells. We used this system to overexpress polyQ-expanded ATXN1 in human MSCs and observed that cells reproducibly accumulate intranuclear inclusion bodies (IIBs). First, the IIBs generate cellular oxidative stress and following prolonged induction become insoluble. Using a combination of biophysical methods [Fourier-transform infrared spectroscopy (FTIR), atomic force microscopy (AFM) and mass spectrometry (MS)], we characterized their structure and protein composition. In addition, we analyzed transcriptional changes and mechanisms of dysfunction caused by the overexpression of mutant ATXN1 and the gradual accumulation of its protein product. We assessed the similarity of this model with human SCA1 cerebellum at the end-stage of the disease in terms of mechanisms leading to disease. In particular, our data indicate that aggregation of pathogenic ATXN1 perturbs the cellular protein synthesis machinery in a cerebellar protein-protein interaction (PPI) network. The potential implications of these results for the homeostasis of cerebellar neurons are discussed.
2. Materials and methods
2.1. Construction of Tet-On Sleeping Beauty transposon plasmids
For the generation of a pT2-CMV/TetO2-EYFP-GW transposon plasmid, the CMV promoter was excised by HindIII-NheI digestion from a pT2-CMV-EYFP-GW plasmid [21] and replaced by a CMV/TetO2 promoter. ATXN1(Q30) (wild-type ATXN1; GeneID: 6310) or (Q82) cDNA was shuttled into pT2-CMV/TetO2-EYFP-GW plasmid by LR recombination, as previously described [22]. Correct recombination was verified by BsrGI restriction digestion.
An expression cassette encoding the SV40 promoter, β-globin intron, Tet-repressor (TetR) and bGH polyA signal flanked by SpeI restriction sites was synthesized and cloned into a pU57C plasmid. The cassette was subcloned by SpeI digestion into the MCS of a pT2-HB plasmid resulting in the generation of a pT2-TetR plasmid. Then, an expression cassette encoding the SV40 promoter, neomycin resistance gene (neoR) and SV40 polyA site was excised by HindIII from a pT2-neoR plasmid and inserted into the pT2-TetR plasmid, resulting in the generation of a bicistronic pT2-TetR-neoR transposon. The orientation of TetR and neoR expression cassettes was verified by EcoRV digestion. Plasmid isolation was performed using a DNA plasmid purification kit (Macherey-Nagel).
2.2. MSC and neural precursor cell (NPC) culture
Isolation and characterization of human MSCs have been previously described [17,18]. Cells were cultured and expanded ex-vivo using Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 IU/mL) and streptomycin (100 μg/mL). NPCs were obtained from healthy human iPSCs as previously described [23]. Cells were seeded on Matrigel-coated plates and cultured in DMEM-F12/NeuroCult 1:1 supplemented with 1:200 N2 supplement, 1:100 B27 supplement lacking vitamin A, 3 μM CHIR 99021, 0.5 μM purmorphamine, 150 μM ascorbic acid (Sigma-Aldrich) and 1% penicillin/streptomycin/glutamine [24].
2.3. Generation of stable cell lines
MSCs at passage 5 or NPCs at passage 9 were transfected with a mixture of SB100X/pT2 plasmids using Xfect reagent (Clontech), according to manufacturer's instructions. Briefly, 2 × 105 cells were seeded in a 6-well tissue culture plate. The next day, cells were transfected with 5 μg of total plasmid DNA encoding SB100X transposase and pT2 plasmids (1:9 ratio). The ratio of pT2-TetO2-YFP-ATXN1(Q30) or (Q82) and pT2-TetR-neoR transposon plasmids was 5:1. For the generation of MSCs stably producing Venus, a pT2-GAGGS-Venus-neoR transposon plasmid was used. Transfected cells were selected at day 7 post-transfection with 100 μg/mL G418 and expanded in the presence of the antibiotic. For transgene induction, fresh doxycycline (Dox) (2 μg/mL) was added every 48hrs in the culture medium.
2.4. Immunofluorescence
Tet-On YFP-ATXN1(Q82) MSCs or NPCs were cultured in the presence or absence of doxycycline in CELLview glass bottom plates (Greiner Bio One). Then, cells were fixed with 4% paraformaldehyde (PFA) in PBS and permeabilized with 0.1% Triton-X 100 PBS before mounting with DAPI ProLong Gold antifade reagent (ThermoFisher). Cells were visualized in an Axiovert microscope equipped with an HBO 50 mercury lamp and reflectors with fluorescence filter sets. Image acquisition was performed with the Fluorescence Lite software module of AxioVision LE (Carl Zeiss).
2.5. Flow cytometry
For quantification of transgene expression, Tet-On YFP-ATXN1(Q82) MSCs were measured in a Cytomics FC500 flow cytometer (Beckman Coulter) using the CXP2.2 software. For phenotypic characterization, 1 × 106 cells were stained with antibodies against hemopoietic (CD34, 45) or mesenchymal (CD29, 73, 90 and CD105) stem cell markers. Unstained cells or cells stained with IgG isotype antibodies were used as negative controls.
2.6. Immunoblotting
MSCs (5 × 105) were lysed in 1% SDS PBS containing protease inhibitors (Calbiochem) and benzonase (Novagen). Cell extracts were analyzed in an 8% SDS-PAGE. Ectopic expression of YFP-ATXN1(Q30) or (Q82) was validated by Western blotting with the polyclonal anti-ATXN1 SA4645 antibody [22] or anti-GFP antibody (MAB2510, Millipore). β-actin was detected with mouse monoclonal anti-β-actin antibody (sc-47778, Santa Cruz). After incubation with an appropriate alkaline-phosphatase conjugated secondary antibody, proteins were detected with NBT-BCIP (Applichem).
2.7. Filter retardation assay
Insoluble protein inclusions were quantified using a filter retardation assay [25]. Cell extracts of Tet-On YFP-ATXN1(Q30) or (Q82) MSCs (150 μg total protein) were mixed with equal volumes of 4% SDS and 100 mM DTT and heated at 95 °C for 5 min. Samples were diluted with 100 μl 0.2% SDS and filtered through a 0.2 μm cellulose acetate membrane. SDS-resistant inclusions retained on the membrane were detected using the polyclonal anti-ATXN1 SA4645 antibody (1:1000) [22].
2.8. FTIR spectroscopy
Tet-On YFP-ATXN1(Q82) MSCs were cultured on MirrIR low-e-glass slides (Kevley Technologies, USA) washed with PBS and air dried. The spectra were taken using a Renishaw-Smiths micro Raman-FTIR microscope (Smiths-IlluminatIR, Smiths Detection, Hertfordshire, UK) and an ARO microscope lens (smithsARO 15x/N.A.88) at a range of 600–4000 cm−1 wavenumbers with 100 measurements for the background spectrum and 300 for the sample spectrum with a 4 cm−1 resolution. The low-e glass slide was placed on an automated holder and exposed to the instrument hood-controlled temperature and humidity environment of 22 °C and 37%, respectively. Background and sample absorption spectra were taken at a 100 μm diameter circular region. The sample absorption spectrum was calculated after background subtraction using the SynchronizIR software (Soft Imaging System GmbH, Germany). The second derivative spectra were obtained by a 5-point Savitzky-Golay algorithm of the smoothed spectrum (11-point binomial) using the GRAMS/32 software (Galactic Industries Corporation, Salem, NH, USA), according to relative processing [26].
2.9. Cytotoxicity assay
Tet-On YFP-ATXN1(Q82) MSCs were cultured in 96-well tissue culture plates (2 × 104 per well). Equal volume of Caspase 3/7 Glo substrate (Promega) was added in each well and luminescence was measured in a Victor3 multilabel plate reader (PerkinElmer). Each experiment was performed in triplicate.
2.10. Detection of reactive oxygen species (ROS)
2′7′-Dichlorodihydrofluorescein diacetate (H2DCFDA) (ThermoFischer) was used for detection of ROS. Tet-On YFP-ATXN1(Q82) were resuspended in PBS with or without the dye at a final concentration of 10 μM (loading buffer) and incubated at 37 °C for 30 min. The loading buffer was removed, cells were resuspended in prewarmed complete medium and further incubated at 37 °C for 5 min. The absorption and emission of the oxidation product were measured at 493 and 520 nm, respectively. Each experiment was performed in triplicate.
2.11. Proteasome peptidase assay
Chymotrypsin-like (CT-L) proteasome activity was assayed in protein extracts with the hydrolysis of a specific fluorogenic peptide, namely LLVY-AMC, at 37 °C for 30 min, as previously described [27]. Proteasome activity was determined as the difference between the total activity in protein extracts and the remaining activity in the presence of 20 μM MG132. Fluorescence was measured using a VersaFluor fluorescence spectrophotometer (Bio-Rad). Protein concentrations were determined using the Bradford method with bovine serum albumin as standard.
2.12. Preparation of samples for AFM
Cells were either kept in culture medium or fixed with 4% PFA PBS before observation. For imaging of nuclei, Tet-On YFP-ATXN1(Q82) MSCs were treated as previously described [28]. In brief, AFM images of nuclei were obtained by cell lysis in 0.25% NP-40 hypotonic Tris-HCl pH 7.4 buffer followed by fixation. For observation of polyQ inclusions, cells were lysed in 0.1% SDS PBS and purified inclusions attached to the plastic surface were immersed in PBS.
2.13. AFM imaging and mechanical characterization of nuclear content
Hybrid mode imaging, contact mode and Young's modulus mapping were performed using either Bruker Dimension FastScan (Bruker Nano Surfaces) AFM or JPK NanoWizard 3 (JPK). Bruker SCANASYST-FLUID+ and SNL-10 A and B silicon nitride cantilevers equipped with silicon tip (both from Bruker Nano Surfaces) were used for imaging. Sensitivity and cantilever spring constant were calibrated by the routine procedure recommended by the instrument producer.
Hybrid mode imaging (Bruker QNM mode) with SCANASYST-FLUID + probe was employed to record semi-quantitative images of isolated nucleus in PBS buffer. Images covering an area of 15 × 15 μm2 and 2.5 × 2.5 μm2 area (1500 × 1500 px2 resolution) were recorded with a set point of 0.75 nN and lifting height of 150 nm, (Z-piezo range 0.75 μm and 0.35 μm, respectively). Images covering the whole nucleus (20 × 20 μm2, 1024 × 1024 px2 resolution) as well as the detailed view of 5 × 5 and 2 × 2 μm2 were recorded by JPK Quantitative Imaging mode (hybrid mode) with a set point of 0.35 nN and lifting height of 90 nm (Z-piezo range was shortened to 3.0 μm).
Cells fixed on a standard microscopic glass slide were visualized by constant force contact mode (JPK NanoWizard 3), when the Bruker SNL-10A cantilever was used. Set-point equal to 5.0 nN, feedback setting iGain = 50.0 Hz and PGain = 0.001 were the operating parameters for capturing images of 50 × 50, 25 × 25 and 10 × 10 μm2 (2048 × 2048 px2).
For mapping of Young's modulus of living cells a previously published procedure [29,30] was adopted. Briefly, Bruker SNL-10B probe was used to record 64 × 64 maps of force-distance curves (set-point 1 nN, Z-length 15 μm, time per curve 0.5 s). The recorded force-distance (FD) curves were fitted with the Bilodeau modification of the Hertzian model. Final post-processing and editing of the images and stiffness maps was performed using the Gwyddion software ver. 2.44 [31].
2.14. Force-distance curves post-processing
Force mapping provides a network of force-distance curves, measuring the dependence of the tip-sample interaction force on the tip height above the surface. The absolute value of Young's modulus can be determined by fitting the FD curve to the modified Hertzian-Sneddon equation:
where F is the measured force, E is the Young's modulus, ν is the Poisson's ratio (0.5 for incompressible materials), δ is the tip-sample separation (obtained by correction of the cantilever height to its bending) and α is the half-angle to face of pyramidal tip (reflects the tip geometry). The data processing module of the JPK software was used to process the maps of FD curves. The resulting Young's modulus maps were exported in order to be post-processed using the Gwyddion software. Data masking and final export of stiffness maps were performed also using the Gwyddion software.
2.15. Mass spectrometry
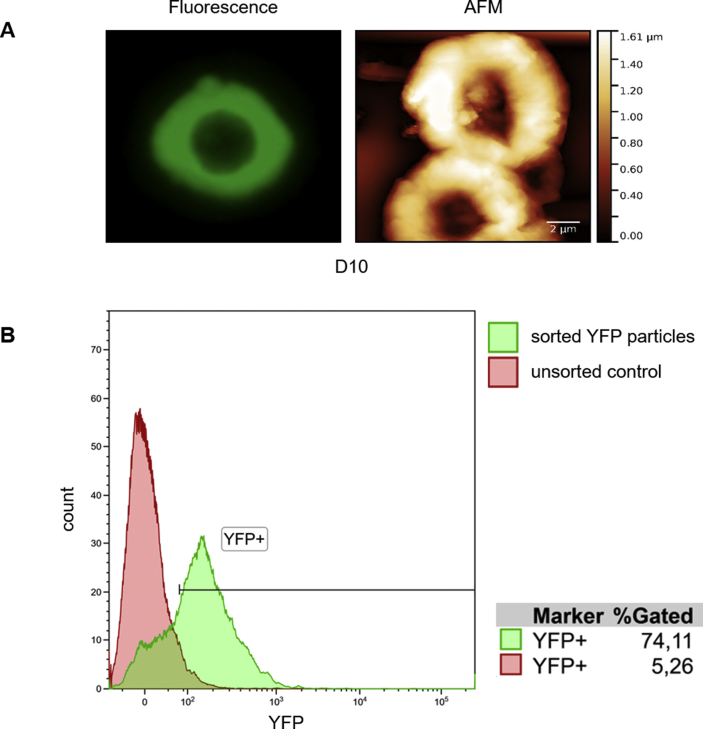
For proteomic analysis of insoluble polyQ inclusions, Tet-On YFP-ATXN1(Q82) MSCs at D10 were lysed in 0.1% SDS PBS. Intact fluorescent polyQ inclusions were sorted from cell extracts in a BD FACSAria IIu instrument (BD Biosciences) after doublet exclusion and using an unstained control sample to exclude possible autofluorescence. Protein samples were processed by the filter-aided sample preparation (FASP) method [32]. Proteins were alkylated, digested by trypsin on filter unit membrane and resulting peptides were eluted by ammonium bicarbonate buffer. The peptide mixture was dried under vacuum and transferred using peptide extraction procedure to TPX vial. LC-MS/MS analysis was done using the RSLCnano system (Thermo Fisher Scientific) on-line connected to Impact II Ultra-High Resolution Qq-Time-Of-Flight mass spectrometer (Bruker). The analytical column outlet was directly linked to the CaptiveSpray nanoBooster ion source (Bruker ). MS and MS/MS spectra were acquired in a data-dependent strategy with 3 s long cycle time.
For quantitative mass spectrometry, technical triplicates of Tet-On YFP-ATXN1(Q82) MSCs at D10 or D0 were lysed in SDT buffer. LC-MS/MS analysis of peptide mixture was done using RSLCnano system (Thermo Fisher Scientific) on-line connected to Orbitrap Q-Exactive HF X system (Thermo Fisher Scientific). The analytical column outlet was directly linked to the Digital PicoView 550 (New Objective) ion source. ABIRD (Active Background Ion Reduction Device, ESI Source Solutions) was installed. MS data were acquired in a data-dependent strategy selecting up to top 20 precursors.
The analysis of the mass spectrometric data was carried out using Maxquant (version 1.6.3.3) with Andromeda search engine utilization. Search was done against the UniprotKB Human (version 20180912) and Maxquant's contamination databases (downloaded with given version). Mass tolerances for peptides and MS/MS fragments were 4.5–10 ppm and 0.05 Da, respectively. Oxidation of methionine, deamidation (N, Q) and protein N-terminal acetylation as optional modifications, carbamidomethylation (C) as fixed modification and two enzyme miss cleavages were set for search. Peptides and proteins with false discovery rate (FDR; q-value) < 1% were considered. Maxquant label free quantification algorithm (MaxLFQ) was applied for global data normalization [33] (minimal ratio count 1). MQ protein group list was processed via KNIME Analytics Platform (v. 3.7.1). Results have been deposited in the PRIDE Archive (https://www.ebi.ac.uk/pride/archive) under accession number PXD012709.
2.16. RNA-sequencing
Total RNA was extracted from Tet-On YFP-ATXN1(Q82) MSCs using the AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN), according to manufacturer's instructions. For library preparation, mRNA was enriched from 250 ng total RNA using the NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs), according to manufacturer's protocol. Sequencing libraries were prepared from mRNA using SENSE Total RNA-seq Library Prep Kit (Lexogen), according to manufacturer's protocol. Single-end 75 bp sequencing was performed in a NextSeq 500 instrument (Illumina). In average, more than 30 million reads per time-point were obtained.
Post-mortem human cerebellum from a 74-year-old female SCA1 patient and an age-/sex-matched healthy individual (Sample ID: A136/02 and A158/14, respectively) were obtained from the MRC London Neurodegenerative Diseases Brain Bank. Total RNA was extracted from frozen samples using the Direct-zol RNA Kit (Zymo Research). An aliquot of total RNA was loaded on Total RNA Pico Chip and measured for its integrity using Bionalyzer 2100 (Agilent Technologies). Sequencing libraries were prepared using KAPA RNA HyperPrep with Ryboerase kit, according to manufacturer's instructions (Roche). High throughput single-indexed, paired-end 75 bp sequencing was performed in a HiSeq 4000 instrument (Illumina). RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-7713 and E-MTAB-7714.
2.17. Data analysis and visualization of RNA-seq data
The quality control was performed based on the FastQC software (Babraham Institute, Cambridge, UK). Samples with a low number of raw reads (less than 10 million per sample) were excluded from downstream analysis. Reads were trimmed and filtered using Trim Galore! and consequently aligned against the UCSC hg19 reference genome using the HΙSAT2 protocol [34]. Overall alignment rate for all samples ranged between 89% and 93%. Transcriptome assembly was performed using StringTie and differential expression calculation was performed using Ballgown. Gene expression levels were normalized to Fragments Per Kilobase of transcript per Million (FPKM).
Subsequent data analysis was carried out in the R environment. T-test was applied to compare FPKM levels between the two different groups. Hierarchical clustering analysis was performed applying the R package “gplots” and FPKMs were normalized to z-score. The agglomerative hierarchical clustering was performed using the Euclidean distance metric and the complete agglomeration method. To compute the principal components (PCs), the prcomp function was applied. Gene set enrichment analysis (GSEA) was calculated using the clusterProfiler R package [35]. Gene Ontology (GO) and Pathway enrichment analyses were performed using the online tool Enrichr where KEGG and PANTHER pathways were examined [36]. Venny 2.1 (BioinfoGP, CNB–CSIC, http://bioinfogp.cnb.csic.es/tools/venny/index.html) was utilized to overlap the differentially expressed genes from different comparisons.
2.18. Construction of a PPI network
The PPI network in which the products of differentially expressed (DE) genes participate was constructed using release 2 of the Human Integrated Protein-Protein Interaction rEference (HIPPIE) [37]. Only cerebellum-specific interactions with HIPPIE confidence scores of at least 0.63 (median of all scores in the database) were considered, which resulted in a network of 516 proteins.
2.19. Statistical analysis
Statistical analysis was performed using the GraphPad Prism software (San Diego, USA). All cell-based assays were performed in triplicates and results are shown as mean ± SEM calculated by a T-test. For AFM measurement of cell stiffness, a one-way ANOVA with Bonferroni post-hoc test was used.
To determine if the size of the largest connected component (LCC) of the constructed PPI network (328 proteins) was greater than expected by chance, it was compared with a distribution of LCC sizes using a z-test. This distribution resulted from randomly sampling 516 proteins 1,000 times from the complete cerebellum-specific PPI network. A z-test was also employed to determine if the number of DE genes, common to cells and the SCA1 patient and also present in the PPI network, was significantly larger than expected by chance. Specifically, we randomly sampled 185 genes from the 3,923 SCA1-differentially-expressed pool 1,000 times and intersected the sample with the 328 members of the PPI LCC. Then, we checked whether 109 (the actual size of the intersection between the 185 SCA1-DE genes and the LCC) was significantly greater than the constructed random distribution. Differences in protein levels of the network were investigated by LIMMA test. P-values were adjusted with the Benjamini-Hochberg method.
3. Results
3.1. Inducible Sleeping Beauty transposon system to express ATXN1 variants
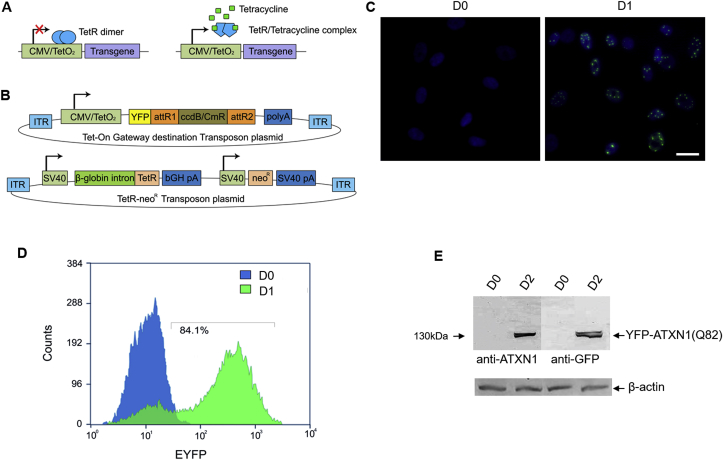
In order to develop a stable ATXN1 expression model, an inducible Tet-On SB transposon system was generated. The system consists of two pT2-based expression plasmids, a Tet-On YFP-tagged Gateway destination plasmid, in which ATXN1(Q82) was shuttled [resulting in YFP-ATXN1(Q82)], and a TetR-neoR bicistronic transposon plasmid encoding for the TetR protein and the neomycin resistance gene (Fig. 1A–B). Using the hyperactive SB100X transposase [15], a stable expression cell line was generated in human MSCs.
Fig. 1.
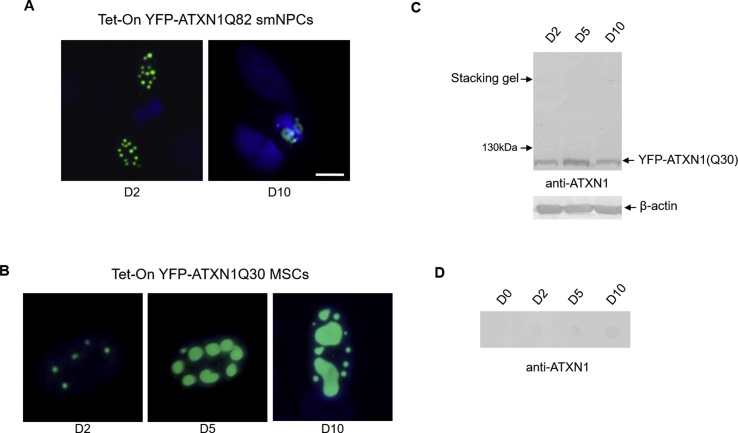
Generation of Tet-On YFP-ATXN1(Q82) MSCs using a novel inducible SB transposon system. (A) Schematic overview of the Tet-On principle. In the absence of tetracycline, TetR protein dimers bind to the CMV/TetO2 promoter and suppress transgene expression. In the presence of tetracycline, TetR adopts a different conformation and loses its affinity for the CMV/TetO2 promoter allowing transgene expression. (B) Components of the Tet-On SB transposon system. The system is comprised of two pT2-based plasmids, a Tet-On Gateway compatible and a bicistronic TetR-neoR transposon. (C) Fluorescence microscopy of Tet-On YFP-ATXN1(Q82) MSCs in the absence (D0, left) or presence (D1, right) of Dox (scale bar = 25 μm). (D) Flow cytometry (histogram plot) of uninduced (blue color) and induced (green color) cells. (E) Immunoblot detection of transgene expression in extracts of uninduced and induced cells using anti-ATXN1 or anti-GFP antibodies. β-actin was used as a loading control. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Tet-On YFP-ATXN1(Q82) MSCs displayed no fluorescent signal in the absence of the inducer Dox (D0), whereas inducing the cells for 24 h (D1) resulted in readily detectable fluorescent YFP-ATXN1(Q82) signal (Fig. 1C). Following quantification by flow cytometry, cells at D1 were 84.1% positive for YFP fluorescence signal (Figs. 1D) and 95% after 48 h (D2) of induction. The induced cells produced high levels of YFP-ATXN1(Q82) after 48hrs (D2). An identical molecular weight band (~130 kDa) was also detectable with an anti-GFP antibody (Fig. 1E), corresponding to the exogenous YFP-tagged polyQ-expanded protein, and not the endogenous ATXN1.
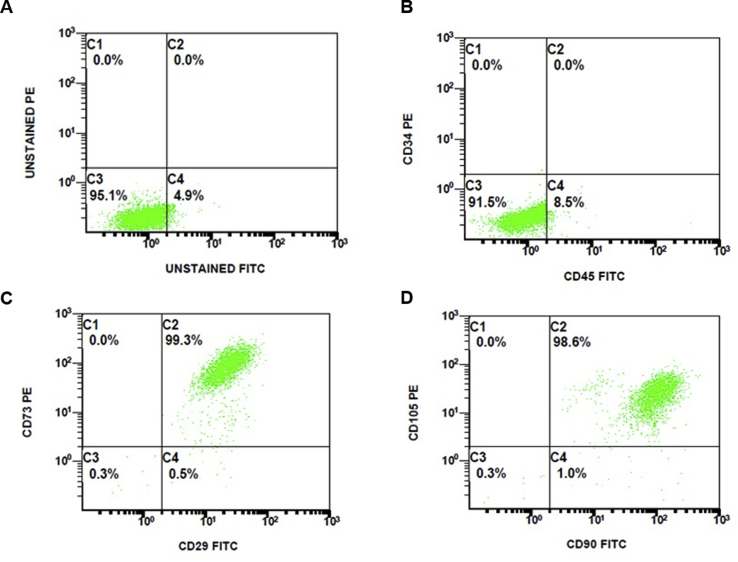
We also investigated whether Tet-On YFP-ATXN1(Q82) MSCs retain their mesenchymal phenotype. As indicated by flow cytometry, uninduced/D0 cells were negative for Cluster of Differentiation (CD) markers of the hematopoietic lineage (CD34 and CD45). In contrast, they were >95% double positive for CD markers of the mesenchymal lineage (CD29, CD73, CD90 and CD105) (Supplementary Fig. 1). This indicates that SB-mediated genetic modification did not affect the mesenchymal properties of the stable lines.
3.2. The pathogenic ATXN1 forms specific annular IIBs in MSCs
Next, we studied the morphology of YFP-ATXN1(Q82) IIBs at different time-points post induction. Cells accumulated small inclusions at D2 that increased in size and partially fused by day 5 (D5). At this point, inclusions consisted of distinct subdomains and seemed to expand around a brighter central core. At day 10 (D10), IIBs adopted an annular structure and dramatically increased in size, occupying most of the nucleus (Fig. 2A). Annular IIBs were also formed in neural progenitor cells (NPCs) derived from healthy iPSCs (Supplementary Fig. 2A). However, only a few of them contained fluorescent inclusions at D10 post induction, suggesting that iPSC-derived NPCs, by contrast to MSCs, may efficiently remove small inclusions or suppress long-term production of the pathogenic ATXN1. Therefore, further experiments were performed using genetically-modified MSCs.
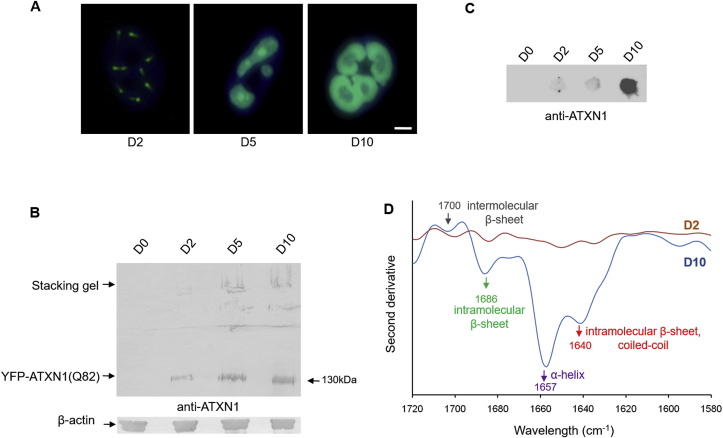
Fig. 2.
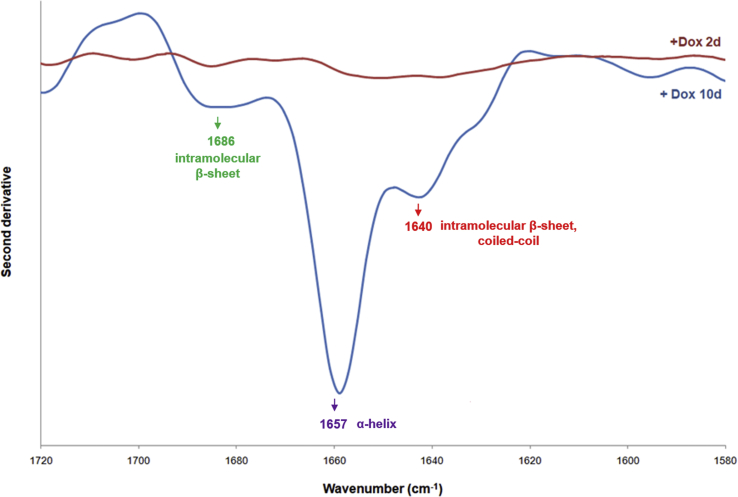
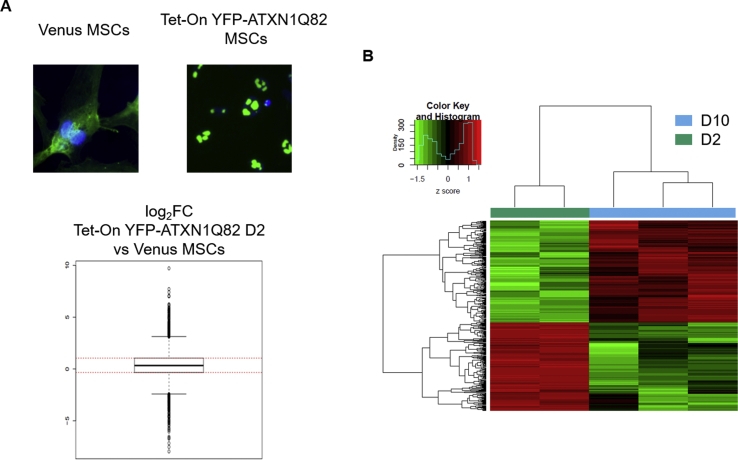
MSCs expressing YFP-ATXN1(Q82) accumulate insoluble IIBs. (A) Fluorescence microscopy of Tet-On YFP-ATXN1(Q82) MSCs at D2-D10. Nuclei are stained with DAPI (blue color) (scale bar = 10 μm). (B–C) Immunoblots for ATXN1 in B. SDS-PAGE and C. Filter retardation assays using extracts of uninduced or induced cells. β-actin was used as loading control. (D) Second derivative of the FTIR absorption spectrum of Tet-On YFP-ATXN1(Q82) MSCs at D2 or D10. Spectra were corrected after subtraction of the Venus MSCs spectrum. The D10 spectrum shows a peak at 1657 cm−1 indicative of α-helix conformation and three peaks at 1640, 1686 and 1700 cm−1, corresponding to coiled-coils, intra- and inter-molecular β-sheets in ATXN1(Q82). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In order to investigate whether polyQ-expansions in ATXN1 are responsible for the annular structure of IIBs, we also generated an inducible control MSC line, producing the wild-type protein YFP-ATXN1(Q30). YFP-ATXN1(Q30) formed spherical IIBs which also increased in size upon prolonged induction with Dox (Supplementary Fig. 2B). However, in contrast to mutant ATXN1, these IIBs did not adopt an annular shape at D10. Therefore, ATXN1 with an expanded polyQ tract specifically forms annular IIBs in MSCs.
To analyze the solubility of ATXN1 proteins, equal amounts of Tet-On YFP-ATXN1(Q30) as well as YFP-ATXN1(Q82) MSC extracts, were analyzed by SDS-PAGE. While, soluble YFP-ATXN1(Q30) and YFP-ATXN1(Q82) were detected at the expected molecular weight at D2, D5 and D10 (Supplementary Fig. 2C and 2B, lower band), a fraction of YFP-ATXN1(Q82) appeared in a higher molecular weight region of the gel in extracts from D5 and D10 cells. These protein species failed to enter the resolving gel or accumulated at the bottom of the well of the loading gel (Fig. 2B). Finally, we examined whether YFP-ATXN1(Q82) IIBs are detectable in a filter retardation assay. This method allows quantification of large, insoluble SDS-stable protein inclusions [25]. Pathogenic ATXN1(Q82) readily formed SDS-stable inclusions at D10 (Fig. 2C), whereas no such inclusions were immunodetected in MSCs inducibly producing YFP-ATXN1(Q30) (Supplementary Fig. 2D). Taken together, these data indicate that mutant ATXN1 forms insoluble IIBs in MSCs with a potential pathogenic conformation after prolonged induction.
3.3. ATXN1(Q82) IIBs are enriched in coiled-coils and β-sheets
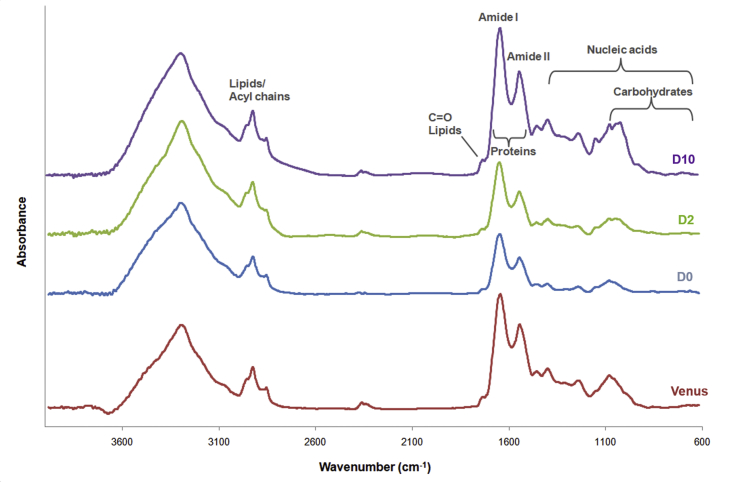
We then sought to investigate whether pathogenic ATXN1 adopts a β-sheet conformation in IIBs. To do so, we determined the nominal FTIR absorption spectrum of Tet-On YFP-ATXN1(Q82) MSCs at D10 and observed typical peaks for the main biological macromolecules (Supplementary Fig. 3). Proteins were detected by two different absorption peaks corresponding to amide I and II signals (1400-1800 cm−1) that are related to the C O stretching vibration of the peptide bond [38]. Similar absorption spectra were also obtained from Tet-On YFP-ATXN1(Q82) MSCs at D0 or D2 (Supplementary Fig. 3).
The measurement from D0 cells was taken as background and subtracted from the absorption spectrum of Tet-On YFP-ATXN1(Q82) MSCs at D2 or D10. The second derivative of the corrected spectra showed no distinct peaks in the range of amide I at D2 in YFP-ATXN1(Q82) expressing MSCs. This indicates that these cells contain proteins with a similar conformation to D0 MSCs. In contrast, Tet-On YFP-ATXN1(Q82) MSCs at D10 exhibited strong peaks at 1657, 1640 and 1686 cm−1 (Supplementary Fig. 4). The distinct high-intensity peak at 1657 cm−1 may be attributed to α-helical protein structures [38] of YFP-ATXN1(Q82). However, the peak at 1640 cm−1 combined with a low-intensity peak at 1685 cm−1 suggests an enrichment for coiled-coils [39] and β-sheets [40,41] in the aggregated polyQ-expanded protein.
To exclude the possibility that these peaks result from the fluorescent protein tag, a Venus-tagged MSC line, stably producing a fluorescent variant of YFP [42], was generated as a control. The absorption spectrum of Venus MSCs (Supplementary Fig. 3) was subtracted from the relevant spectra of Tet-On YFP-ATXN1(Q82) MSCs at D2 or D10 and their second derivatives are illustrated in Fig. 2D. The spectrum of D10 Tet-On YFP-ATXN1(Q82) MSCs showed strong peaks at 1657, 1640 and 1686 cm−1 along with an additional low peak at 1700 cm−1. No such peaks were observed in D2 cells. These results suggest that the detected peaks can be attributed merely to polyQ-expanded ATXN1. Furthermore, the peaks at 1640 and 1686 cm−1 indicate that aggregated YFP-ATXN1(Q82) is enriched in coiled-coils and intra-molecular β-sheets, while the 1700 cm−1 peak suggests that the pathogenic protein also forms inter-molecular β-sheets in IIBs [40,43]. This finding suggests that the insolubility of YFP-ATXN1(Q82) IIBs (Fig. 2C) is due to the enrichment of the number of coiled-coils, intra- and inter-molecular β-sheets in the pathogenic protein.
3.4. Intranuclear accumulation of ATXN1(Q82) IIBs affects the mechanical properties of MSCs
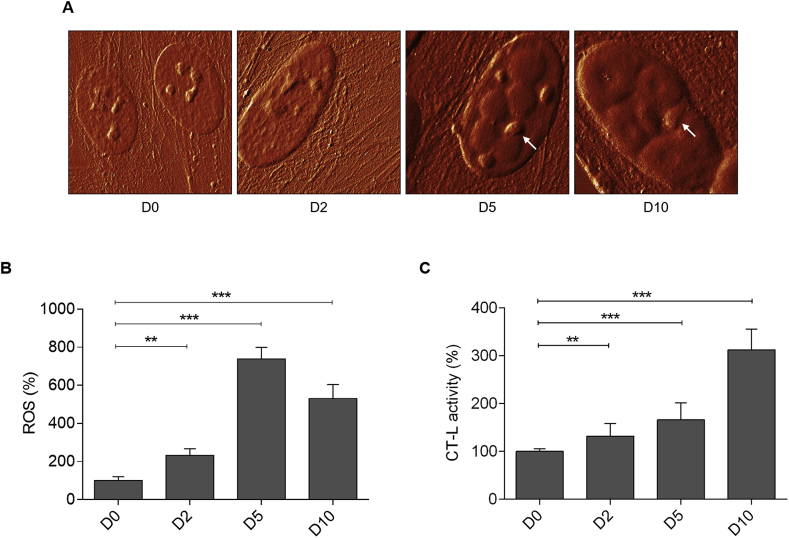
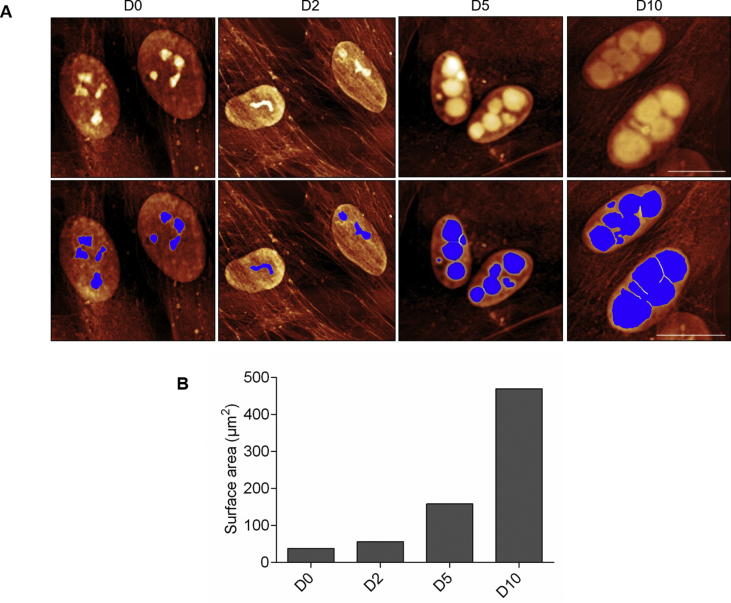
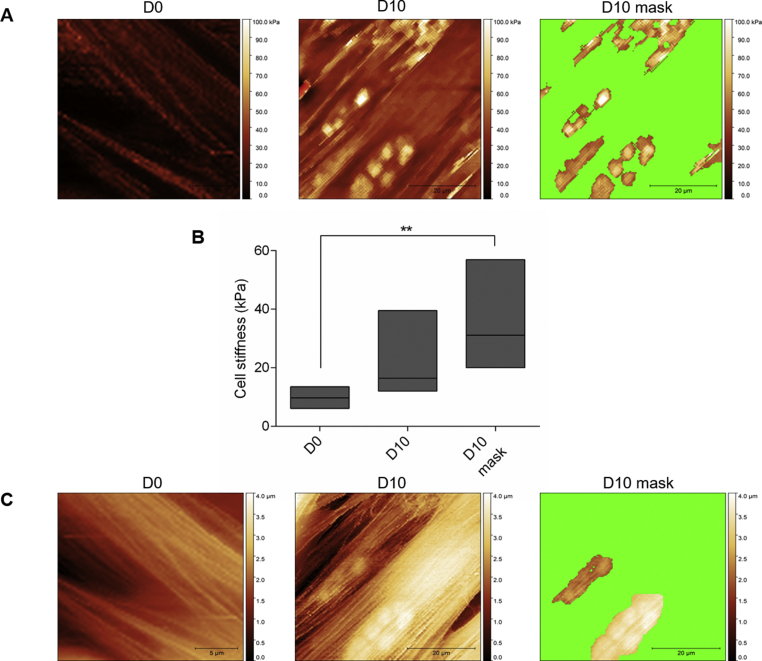
Next, we determined the biophysical properties of YFP-ATXN1(Q82) expressing MSCs using AFM. This method provides structural, mechanical and functional information of non-stained cells or molecules at high resolution under physiological conditions [44,45]. The occupied nuclear area was similar in D0 and D2 MSCs indicating that polyQ inclusions were not detectable by AFM at these time-points. In contrast, a significant increase of the nuclear content was observed in D5 and D10 MSCs (Supplementary Fig. 5A). In the contact mode images, the nuclear content appeared above the threshold (blue masked area). Quantification of the masked area revealed an approximately 12-fold increase in occupied nuclear surface area of D10 MSCs compared to D0 cells (Supplementary Fig. 5B). These data illustrate that the growth of YFP-ATXN1(Q82) IIBs can be efficiently quantified by AFM. Hybrid mode AFM images provided a detailed visualization of the nuclear content. Processing of force-distance curves by the surface distribution of the slope of indentation curve, allowed us to distinguish between natural nuclear structures (e.g. nucleoli - marked with the white arrow) and polyQ inclusions (Fig. 3A). Overlay of AFM and fluorescence images showed that the marked structures were non-fluorescent (Supplementary Fig. 6) excluding the possibility that they represent smaller polyQ protein inclusions. These images indicate that annular IIBs compress the nucleoli (Fig. 3A), suggesting that the accumulation of YFP-ATXN1(Q82) may interfere with their physiological function.
Fig. 3.
Cellular effects of YFP-ATXN1(Q82) IIBs. (A) Detailed view of the cell nucleus. Cells at D0-D10 were fixed and visualized by AFM in hybrid mode (Quantitative Imaging). White arrows indicate natural nuclear content (nucleoli). The white scale bar in the right image is equal to 20 μm. Z-scaling of all images ranges from 85 to 190 mN/m (B–C) Percentages of (B) ROS and (C) CT-L proteasome activity in uninduced and induced Tet-On YFP-ATXN1(Q82) MSCs at D2, D5 and D10. ROS levels and CT-L activity of control cells (D0) were arbitrarily set to 100%. Error bars denote ±SEM (** p-value < 0.01, *** p-value < 0.001).
Mechanical properties of Tet-On YFP-ATXN1(Q82) MSCs were studied using the force-volume/nanoindentation method [30,46]. The Young's modulus (stiffness) of D0 and D10 MSCs was compared. There was no significant difference when Young's modulus of whole cells was compared (10.2 ± 2.8 kPa and 23 ± 15 kPa, for D0 and D10 cells, respectively), possibly due to the small occupied area of protein inclusions. However, a significant difference was observed when only the area containing IIBs was compared with the stiffness of an uninduced cell (10.2 ± 2.8 kPa and 36 ± 19 kPa for D0 and D10 cells, respectively, p-value < 0.01) (Supplementary Figs. 7A–B). Similarly, the topography of these cells was compared. Using the set point height technique, YFP-ATXN1(Q82) IIBs were efficiently detected only in D10 MSCs (Supplementary Fig. 7C).
3.5. Accumulation of large ATXN1(Q82) IIBs is an irreversible process
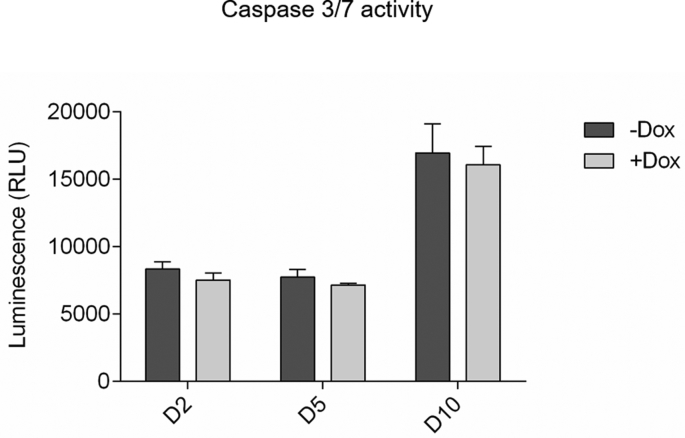
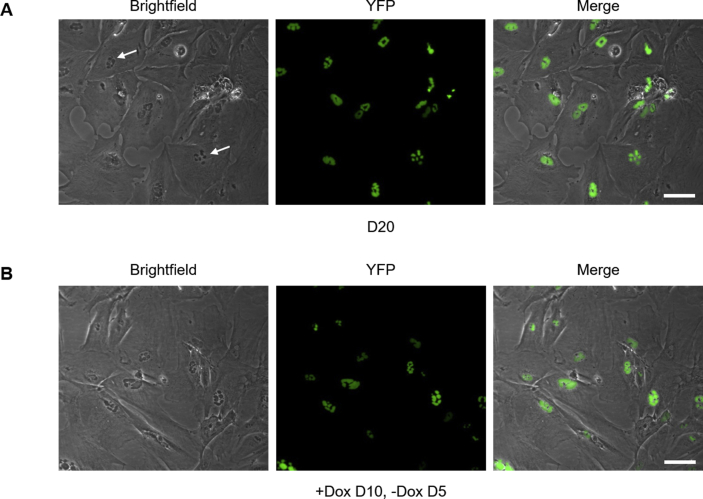
We then investigated whether polyQ inclusions are cytotoxic. Activated caspase 3/7 was measured in Tet-On YFP-ATXN1(Q82) MSCs in the presence or absence of Dox. Induced cells showed similar levels of caspase activity compared to uninduced cells at all time-points (Supplementary Fig. 8). Deposition of YFP-ATXN1(Q82) IIBs does not induce apoptosis in these cells. Nevertheless, after prolonged (10 days) induction, Tet-On YFP-ATXN1(Q82) MSCs acquired an enlarged and flattened phenotype typical of senescent cells. By D20, the majority of cells lost their proliferative capacity and transited towards a growth-arrested state. IIBs were readily visible by optical microscopy (Supplementary Fig. 9A, white arrows); however, they did not significantly increase in size compared to D10. Cells did not die; they rather maintained this state for up to 30 days post induction. These findings suggest that the intranuclear accumulation of mutant ATXN1 does not result in the activation of apoptosis but rather affects the normal cell cycle of MSCs.
Then, we tested whether aggregation of YFP-ATXN1(Q82) induces oxidative stress in MSCs. As shown in Fig. 3B, production of the pathogenic polyQ protein for 2 days resulted in a significant increase in the levels of ROS. This effect was significantly enhanced in both D5 and D10 cells (~7- or 5-fold higher ROS levels, respectively compared to D0 MSCs). Since oxidative stress induces alterations in the protein degradation machinery, chymotrypsin-like (CT-L) activity of the proteasome was measured. CT-L activity was enhanced in D2 cells and further elevated at later time-points. D10 cells showed approximately 3-fold higher proteasome activity compared to D0 MSCs (Fig. 3C). To check if the aggregation was reversible, we asked whether removal of Dox from induced cells would result in the clearance of polyQ inclusions. Tet-On YFP-ATXN1Q82MSCs were induced for 10 days; they were then cultured for 5 additional days in medium without Dox. Cells containing large YFP-ATXN1(Q82) IIBs failed to dissolve them (Supplementary Fig. 9B), indicating that aggregated polyQ proteins are resistant to degradation. This observation supports our finding that Tet-On YFP-ATXN1(Q82) MSCs at D10 contain insoluble IIBs (Fig. 2C–D).
3.6. Insoluble ATXN1(Q82) IIBs recruit proteins associated with neurodegenerative processes
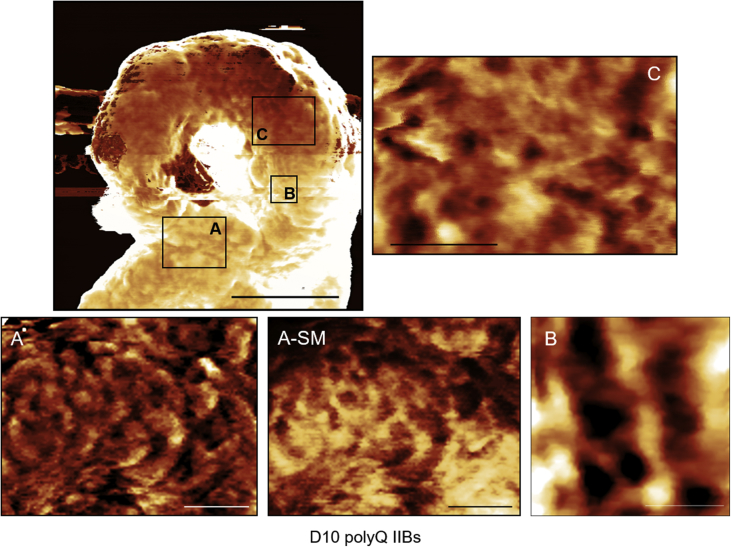
Based on our finding that polyQ inclusions are insoluble in SDS (Fig. 2C), we sought to isolate and characterize them. Intact annular inclusions of YFP-ATXN1(Q82) at D10 were observed by fluorescence microscopy and AFM (Supplementary Fig. 10A). A detailed view of the inclusion surface using AFM hybrid mode indicated the presence of a lamellar structure (Fig. 4A and C). A higher magnification also showed the presence of pores between the layers of the inclusion (Fig. 4B). The average height of an inclusion was 14.8 ± 5.2 nm. Stiffness measurement on a 6 μm2 area of the polyQ inclusion showed the distribution of Young's modulus with an average value of 33.6 MPa (Fig. 4A-SM).
Fig. 4.
Detailed view of polyQ ATXN1 protein inclusions visualized in-situ. IIBs from Tet-On YFP-ATXN1(Q82) at D10 were immobilized on a plastic surface after lysis of cells in 0.1% SDS PBS. Measurements were performed in isotonic PBS by AFM hybrid mode imaging. Closer view of different parts of an IIB is shown below the main image in A–C. A-SM shows the distribution of stiffness over the area of image A. Inserted scale bars and Z-scale ranges are as follows: main image 3 μm/100 nm, (A) 1 μm/15 nm, (A-SM) 1 μm/3–130 MPa, (B) 300 nm/30 nm and (C) 750 nm/30 nm.
Annular polyQ inclusions were then purified from extracts by particle sorting. Efficient isolation was validated post-sorting by flow cytometry (Supplementary Fig. 10B). Proteins recruited into polyQ inclusions were identified by mass spectrometry (MS). In total, 303 different proteins were detected (Supplementary Table 1A) including proteasome and ribosome subunits, ubiquitin-related enzymes and heat-shock proteins. We identified proteins with nuclear localization such as histones, heterogeneous nuclear ribonucleoproteins and nucleophosmin, indicating the nuclear origin of the isolated inclusions. GO enrichment analysis showed that the majority of the identified proteins fall into two distinct clusters, neutrophil degranulation/exocytosis and translation (Supplementary Table 1B) and are functionally similar with protein components of polyQ-expanded Httex1 inclusions (Supplementary Table 1C) [4,10,47,48]. Notably, pathway analysis using the PANTHER database showed that the fraction of proteins recruited into insoluble polyQ IIBs is significantly enriched for the terms Parkinson's and Huntington's disease (Table 1), supporting their relevance for neurodegenerative disease modeling.
Table 1.
Pathway enrichment analysis of proteins detected in insoluble polyQ IIBs.
| Enrichment term | Overlap | p-value | Genes/Proteins |
|---|---|---|---|
| Cytoskeletal regulation by Rho GTPase (P00016) | 12/70 | <0.0001 | TUBB2B; TUBB6; ACTC1; TUBB3; TUBB; CFL1; ARPC4-TTLL3; MYH9; PFN1; TUBB4B; MYH10; ACTG1 |
| Integrin signaling pathway (P00034) | 15/156 | <0.0001 | ITGB1; ARF1; ACTN1; COL12A1; FN1; ACTN4; ACTG1; COL1A1; COL3A1; COLA2; COL6A1; FLNA; FLNB; TLN1; VCL |
| Parkinson's disease (P00049) | 11/81 | <0.0001 | HSPA9; YWHAE; PSMA5; HSPA8; UCHL; HSPA5; YWHAQ; YWHAB; YWHAZ; YWHAG; PSMA7 |
| Huntington's disease (P00029) | 10/124 | <0.0001 | ARF4; TUBB6; TUBB2B; ARF1; ACTC1; TUBB3; TUBB; TUBB4B; GAPDH; ACTG1 |
| Glycolysis (P00024) | 6/17 | <0.0001 | TPI1; PKM; PGK1; ENO1; ALDOA; GAPDH |
Top five statistically significant enriched pathway terms and overlap of identified genes/proteins with the components of the pathway. The analysis was performed using the PANTHER database.
3.7. Partially soluble and insoluble ATXN1(Q82) IIBs generate different transcriptional responses
ATXN1 is an RNA-binding protein participating in transcriptional regulation [49,50]. Therefore, we investigated transcriptional changes induced by its overexpression in MSCs. Transcriptome profiles of D0-D10 cells were generated and compared to those of Venus MSCs. Genes with a similar expression profile in both overexpression models (Supplementary Fig. 11A) were excluded from downstream analysis. This experimental setup identified 3,984 differentially expressed (DE) genes (hereafter, ATXN1-DE) due to YFP-ATXN1(Q82) overexpression (Supplementary Table 2). Gene Set Enrichment Analysis (GSEA) of ATXN1-DE genes dysregulated in D2 compared to Venus MSCs (|log2fold change| ≥ 1, p-value < 0.05) indicated that expression of the mutant ATXN1 significantly affected genes encoding proteins with nuclear localization (Supplementary Table 3).
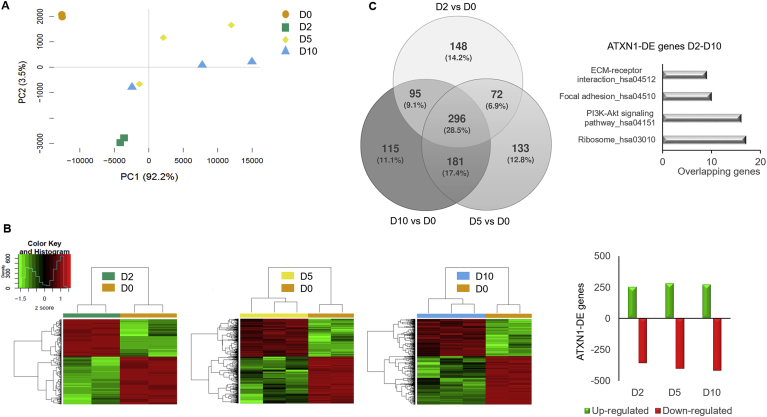
Principal component analysis (PCA) indicated that D2, D5 and D10 cells had a distinct expression profile compared to D0. D2 cells were also distinct from D5 and D10 MSCs, which grouped closely and shared a more common expression profile (Fig. 5A). ATXN1-DE clusters included 611, 682 and 687 genes for D2, D5 and D10, respectively and 1040 unique genes in total. The majority of genes were down-regulated (Fig. 5B). A total of 296 genes (28.5%) were found to be dysregulated at all time-points compared to D0 cells. Pathway term enrichment analysis using the KEGG database showed that these genes were enriched in components of the ribosome or the PI3K-Akt signaling pathway (Fig. 5C and Supplementary Table 4A).
Fig. 5.
Transcriptional changes in Tet-On YFP-AXN1(Q82) MSCs containing IIBs. (A) Principal Component Analysis (PCA) of the expression vectors of the 3,984 genes specifically dysregulated in Tet-On YFP-AXN1(Q82) MSCs in four time-points according to their PC1 and 2 coordinates. Percentages (%) indicate the proportion of variance of analyzed data per axis. (B) Hierarchical clustering analysis and heatmap of ATXN1-DE genes between cells at D2, D5 or D10 vs D0 (|log2fold change| ≥ 1 and p-value < 0.05). Each row represents one gene and each column one sample. Figure also shows the absolute numbers of up- or down-regulated ATXN1-DE genes in each time point. (C) Venn diagram showing the overlap of ATXN1-DE genes per comparison (described in B). KEGG pathway enrichment analysis of ATXN1-DE genes at all time points (D2-D10) is also shown.
The identity of 148 genes (14.2% of 1,040 genes) specifically dysregulated in D2 cells was investigated. GO Biological Process (BP) indicated several genes involved in positive regulation of neuron apoptotic process (Supplementary Table 4B). Interestingly, analysis of genes dysregulated specifically in D5 and D10 cells (181 genes, 17.4% of the 1,040 genes) revealed a set involved in the opposite process, negative regulation of neuron apoptotic process (Supplementary Table 4C). This indicates that soluble YFP-ATXN1(Q82) produced in MSCs at the early time-point (D2) has a different effect in cells than the partially insoluble protein which forms IIBs at later time-points (D5 and D10). We also investigated the identity of 115 genes (11.1% of the 1040 genes) exclusively dysregulated in D10 cells containing insoluble IIBs. GO Cellular Component (CC) analysis revealed enrichment terms for PML body and other subnuclear structures (Supplementary Table 4D) that are associated with response to oxidative stress [51]. This is in agreement with our observation that production of mutant ATXN1 induces oxidative stress in MSCs (Fig. 3B) and the related cell response pathways. These data illustrate the complexity of the dynamic processes that occur in cells expressing pathogenic ATXN1 in consecutive time-points.
Enrichment analysis using the KEGG database on the significantly dysregulated ATXN1-DE genes at D2 indicated that up-regulated genes participate in ECM-receptor interaction or focal adhesion, while down-regulated genes were involved in the ribosome, DNA replication, cell cycle and oxidative phosphorylation. Importantly, a significant number of down-regulated genes participate in pathways related to neurodegenerative diseases, validating our polyQ protein aggregation model (Table 2A).
Table 2.
Pathway enrichment analysis of up- or down-regulated genes in Tet-On YFP-ATXN1(Q82) MSCs containing polyQ IIBs.
|
A. D2 versus D0 | |||||
|---|---|---|---|---|---|
|
Up-regulated |
Down-regulated |
||||
| Enrichment term | Overlap | p-value | Enrichment term | Overlap | p-value |
| ECM-receptor interaction_hsa04512 | 12/82 | >0.0001 | Ribosome_hsa03010 | 30/137 | >0.0001 |
| Focal adhesion_hsa04510 | 16/202 | >0.0001 | DNA replication_hsa03030 | 6/36 | >0.0001 |
| PI3K-Akt signaling pathway_hsa04151 | 18/341 | >0.0001 | Alzheimer's disease_hsa05010 | 10/168 | 0.0010 |
| Proteoglycans in cancer_hsa05205 | 13/203 | >0.0001 | Cell cycle_hsa04110 | 8/124 | 0.0018 |
| Rap1 signaling pathway_hsa04015 | 13/211 | >0.0001 | Oxidative phosphorylation_hsa00190 | 8/133 | 0.0028 |
| Regulation of actin cytoskeleton_hsa04810 |
12/214 |
>0.0001 |
Huntington's disease_hsa05016 |
9/193 |
0.0084 |
| B. D10 versus D2 | |||||
| Up-regulated | Down-regulated | ||||
|
Enrichment term |
Overlap |
p-value |
Enrichment term |
Overlap |
p-value |
| Protein digestion and absorption_hsa04974 | 7/90 | 0.0001 | DNA replication_hsa03030 | 4/36 | 0.0003 |
| AGE-RAGE signaling pathway in diabetic complications_hsa04933 | 6/101 | 0.0008 | Cell cycle_hsa04110 | 7/124 | 0.0002 |
| PI3K-Akt signaling pathway_hsa04151 | 9/341 | 0.0114 | p53 signaling pathway_hsa04115 | 5/69 | 0.0004 |
| p53 signaling pathway_hsa04115 | 4/69 | 0.0064 | Axon guidance_hsa04360 | 6/127 | 0.0012 |
| Lysosome_hsa04142 | 5/123 | 0.0104 | Wnt signaling pathway_hsa04310 | 6/142 | 0.0020 |
| Transcriptional misregulation in cancer_hsa05202 | 6/180 | 0.0131 | MAPK signaling pathway_hsa04010 | 7/255 | 0.0095 |
Statistically significant enriched pathway terms for up- or down-regulated genes in the relevant comparisons (A. D2 versus D0 and B. D10 versus D2) and the overlap with the components of the pathway. The analysis was performed using the KEGG database.
Regarding cellular pathways that were affected by the accumulation of insoluble IIBs in MSCs, we compared D10 and D2 MSCs (S11B Fig). Up-regulated genes were enriched in functions related to protein digestion and absorption and AGE-RAGE, PI3K-Akt, p53 signaling pathways. In contrast, down-regulated genes were involved in DNA replication and cell cycle (Table 2B), consistent with our observation that MSCs containing insoluble IIBs enter a growth-arrest phase (Supplementary Fig. 9A).
3.8. Tet-On YFP-ATXN1(Q82) MSCs share transcriptional similarities with human SCA1 cerebellum
Does our inducible YFP-ATXN1(Q82) MSC system share transcriptional changes with a pathogenic SCA1 human cerebellum? To answer this question, we performed RNA-seq in post-mortem cerebellar tissue both from a late stage SCA1 patient diagnosed with dementia and spinal cerebellar atrophy with polyglutamine inclusions, and from an age/sex-matched healthy individual. This comparative approach identified 3,923 dysregulated genes in the SCA1 patient (|log2fold change| ≥ 1), hereafter named as SCA1-DE genes (Supplementary Table 5). GSEA showed that this dataset was enriched for genes encoding nuclear proteins (Supplementary Table 6), similar to our findings in MSCs overexpressing YFP-ATXN1(Q82) (Supplementary Table 3).
PolyQ protein inclusions are mainly detected in the cerebellum of SCA1 patients [1]. We therefore compared the patient data with the D10 ATXN1-DE genes in cells containing insoluble IIBs. The down-regulated SCA1-DE and ATXN1-DE genes participate in several common pathways, including cell cycle, oxidative phosphorylation or pathways involved in RNA metabolism (e.g. ribosome, spliceosome and RNA transport) (Table 3A). Similarly, common pathways were identified between the up-regulated genes of the two sets. The up-regulated SCA1-DE genes participate mainly in focal adhesion, as also observed for up-regulated ATXN1-DE genes (Table 3B).
Table 3.
Pathway enrichment analysis of down- and up-regulated genes in human SCA1 cerebellum and D10 Tet-On YFP-ATXN1(Q82) MSCs.
|
A. Down-regulated | |||||
|---|---|---|---|---|---|
|
SCA1 patient versus control |
D10 versus D0 cells |
||||
| Enrichment term | Overlap | p-value | Enrichment term | Overlap | p-value |
| Protein processing in endoplasmic reticulum_hsa04141 | 66/169 | >0.0001 | Ribosome_hsa03010 | 32/137 | >0.0001 |
| Spliceosome_hsa03040 | 52/134 | >0.0001 | Huntington's disease_hsa05016 | 12/193 | 0.0008 |
| RNA degradation_hsa03018 | 31/77 | >0.0001 | DNA replication_hsa03030 | 5/36 | 0.0009 |
| Cell cycle_hsa04110 | 41/124 | >0.0001 | Cell cycle_hsa04110 | 9/124 | 0.0012 |
| RNA transport_hsa03013 | 52/172 | >0.0001 | Alzheimer's disease_hsa05010 | 10/168 | 0.0029 |
| Proteasome_hsa03050 | 20/44 | >0.0001 | Parkinson's disease_hsa05012 | 9/142 | 0.0030 |
| Protein export_hsa03060 | 13/23 | >0.0001 | Non-alcoholic fatty liver disease (NAFLD)_hsa04932 | 9/151 | 0.0045 |
| Ubiquitin mediated proteolysis_hsa04120 | 42/137 | >0.0001 | Oxidative phosphorylation_hsa00190 | 8/133 | 0.0069 |
| FoxO signaling pathway_hsa04068 | 40/133 | >0.0001 | Nucleotide excision repair_hsa03420 | 4/47 | 0.0165 |
| DNA replication_hsa03030 | 15/36 | 0.0002 | Spliceosome_hsa03040 | 7/134 | 0.0227 |
| Non-homologous end-joining_hsa03450 | 8/13 | 0.0003 | Pyrimidine metabolism_hsa00240 | 6/105 | 0.0229 |
| mRNA surveillance pathway_hsa03015 | 28/91 | 0.0003 | Ubiquitin mediated proteolysis_hsa04120 | 7/137 | 0.0252 |
| Nucleotide excision repair_hsa03420 | 16/47 | 0.0018 | RNA transport_hsa03013 | 8/172 | 0.0285 |
| Oxidative phosphorylation_hsa00190 | 34/133 | 0.0029 | Fatty acid biosynthesis_hsa00061 | 2/13 | 0.029 |
| Ribosome_hsa03010 |
34/137 |
0.0049 |
Base excision repair_hsa03410 |
3/33 |
0.0311 |
| B. Up-regulated | |||||
| SCA1 patient versus control | D10 versus D0 cells | ||||
|
Enrichment term |
Overlap |
p-value |
Enrichment term |
Overlap |
p-value |
| Focal adhesion_hsa04510 | 20/202 | >0.0001 | Focal adhesion_hsa04510 | 17/202 | >0.0001 |
| Chronic myeloid leukemia_hsa05220 | 11/73 | 0.0001 | ECM-receptor interaction_hsa04512 | 11/82 | >0.0001 |
| Hippo signaling pathway_hsa04390 | 16/153 | 0.0001 | PI3K-Akt signaling pathway_hsa04151 | 17/341 | >0.0001 |
| Cell adhesion molecules (CAMs)_hsa04514 | 15/142 | 0.0002 | Proteoglycans in cancer_hsa05205 | 11/203 | >0.0001 |
| Notch signaling pathway_hsa04330 | 8/48 | 0.0003 | Protein digestion and absorption_hsa04974 | 8/90 | >0.0001 |
| AGE-RAGE signaling pathway in diabetic complications_hsa04933 | 11/101 | 0.0011 | Regulation of actin cytoskeleton_hsa04810 | 11/214 | 0.0002 |
| Proteoglycans in cancer_hsa05205 | 17/203 | 0.0012 | Pathways in cancer_hsa05200 | 15/397 | 0.0003 |
| Regulation of actin cytoskeleton_hsa04810 | 17/214 | 0.0021 | Rap1 signaling pathway_hsa04015 | 10/211 | 0.0006 |
| Long-term potentiation_hsa04720 | 8/66 | 0.0025 | Phagosome_hsa04145 | 7/154 | 0.0049 |
| ECM-receptor interaction_hsa04512 | 9/82 | 0.0028 | Leukocyte transendothelial migration_hsa04670 | 6/118 | 0.0053 |
| Inositol phosphate metabolism_hsa00562 | 8/71 | 0.0040 | Glioma_hsa05214 | 4/65 | 0.0114 |
| ErbB signaling pathway_hsa04012 | 9/87 | 0.0042 | AGE-RAGE signaling pathway in diabetic complications_hsa04933 | 5/101 | 0.0118 |
The table shows statistically significant enriched pathway terms for A. down- or B. up-regulated genes in the relevant comparisons and the overlap with the components of the pathway. The analysis was performed using the KEGG database.
We also found that 27% (185/687) of the dysregulated D10 ATXN1-DE genes were shared with the SCA1-DE gene set. As indicated by KEGG enrichment analysis, the commonly dysregulated genes included genes involved in the ribosomal function (Supplementary Tables 7A–B). Collectively, these results indicate that the transcriptional changes at end-stage human SCA1 cerebellum resemble the cellular response observed in MSCs containing insoluble IIBs of the mutant ATXN1 at D10.
3.9. ATXN1(Q82) IIBs perturb the ribosomal protein interaction network
The protein inclusions in neurodegenerative diseases produce pathogenic effects due to a trapping effect [52]. According to this hypothesis, protein inclusions act as molecular “black-holes”, trapping proteins along with their downstream interactors. We sought to model this scenario by investigating the PPI network of the ATXN1-DE proteins.
To gain insights into the perturbation of PPIs caused by insoluble IIBs, we used the dataset of dysregulated D10 ATXN1-DE genes and compared it to a cerebellum-specific human PPI network, using HIPPIE v2.0 [37]. This database collects interactions between human proteins scored according to the reliability of the associated experimental evidence. High-confidence interactions (HIPPIE score ≥ 0.63) were considered between the products of the D10 versus D0 ATXN1-DE genes that were expressed in the cerebellum [37], the most affected tissue in SCA1 disorder [53]. The resulting network was formed by 516 gene products.
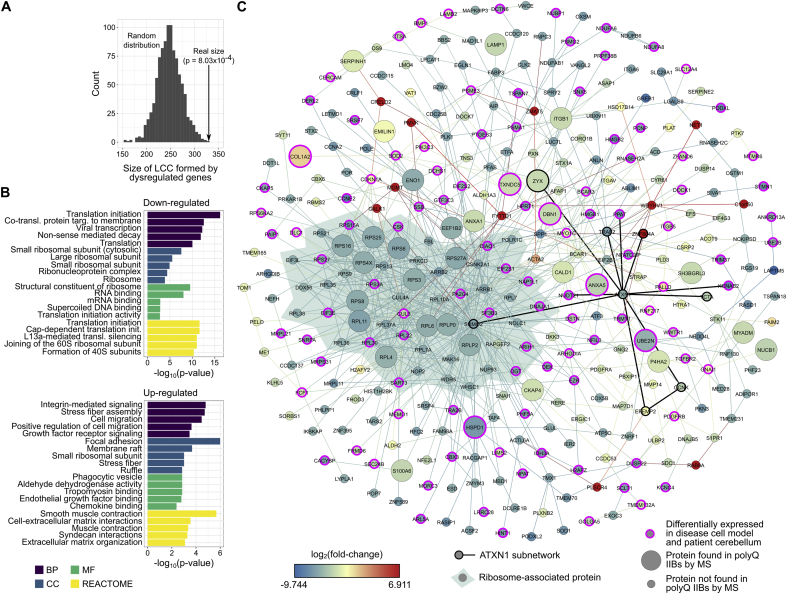
Importantly, the subnetwork containing the largest connected component (LCC) of the constructed PPI network had a statistically significant LCC size (p-value = 8.03 × 10−4, 328 proteins, Supplementary Table 8) when compared to a randomly sampled control network (Fig. 6A). This strategy identified a highly interconnected PPI module that was perturbed by the dysregulation of gene expression and induced by the accumulation of insoluble polyQ IIBs in cells. Most of the proteins in the LCC are down-regulated and their functional enrichment analysis points to the perturbation of key proteins required for proper ribosome assembly and translation (Fig. 6B–C). The most densely connected region of the LCC is formed, almost entirely, by ribosomal subunits whose protein levels are significantly reduced in D10 MSCs containing insoluble IIBs (Table 4 and Supplementary Table 9). Also, a significant fraction of the proteins identified in polyQ inclusions (Supplementary Table 1) is also part of the LCC (n = 33, p-value < 2 × 10−16), further supporting the role of IIBs in the disruption of nucleolus-related functions. Intriguingly, this network also contains 109 human SCA1-DE components (p-value < 2 × 10−16, Fig. 6C and Supplementary Table 8), indicating their potential relevance for SCA1 pathogenesis. Thus, these data support the hypothesis that the pathogenic effect of the mutant ATXN1 in our cell model is associated with the trapping of key protein synthesis factors and their interactors by the polyQ IIBs.
Fig. 6.
ATXN1 IIBs dysregulate ribosome assembly and translation-associated protein interactions. (A) ATXN1-DE genes significantly dysregulated between D10 and D0 cells form a highly interconnected PPI network module (largest connected component or LCC) whose size (black arrow) is significantly greater than expected by chance. (B) The down-regulated members of this LCC play important roles in ribosome assembly and translation. By contrast, the up-regulated members of the LCC are involved in cellular organization functions and compartments. (C) The LCC of the constructed cerebellum-specific PPI network has a densely connected region formed almost entirely by ribosomal subunits and contains a significant number of proteins detected by MS in the insoluble IIBs at D10 (n = 33, p-value < 0.001). This depiction also highlights the interaction partners of ATXN1 in the LCC and human SCA1-DE genes (n = 109, p-value < 0.001, magenta color). Expression levels of each gene in the disease cell model are also shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 4.
Protein levels of LCC ribosome components in cells containing insoluble polyQ IIBs.
| Protein Name | Accession # |
log2fold change D10 versus D0 |
p-value | adj. p-value |
|---|---|---|---|---|
| Ataxin-1 | P54253 | 9.05 | >0.001 | >0.001 |
| Eukaryotic translation initiation factor 2 subunit 1 | P05198 | −0.51 | >0.001 | 0.001 |
| Eukaryotic translation initiation factor 3 subunit E | P60228 | −0.53 | >0.001 | 0.001 |
| Eukaryotic translation initiation factor 3 subunit L | Q9Y262 | −0.60 | >0.001 | 0.001 |
| rRNA 2′-O-methyltransferase fibrillarin | P22087 | −0.17 | 0.036 | 0.065 |
| 60S ribosomal protein L4 | P36578 | −0.50 | >0.001 | 0.001 |
| 60S ribosomal protein L6 | Q02878 | −0.56 | >0.001 | 0.001 |
| 60S ribosomal protein L7 | P18124 | −0.63 | >0.001 | 0.001 |
| 60S ribosomal protein L7a | P62424 | −0.32 | 0.014 | 0.030 |
| 60S ribosomal protein L10a | P62906 | −0.60 | >0.001 | 0.001 |
| 60S ribosomal protein L11 | P62913 | −0.70 | >0.001 | 0.001 |
| 60S ribosomal protein L22 | P35268 | −0.63 | >0.001 | 0.001 |
| 60S ribosomal protein L30 | P62888 | −0.66 | >0.001 | 0.002 |
| 60S ribosomal protein L35 | P42766 | −0.65 | >0.001 | 0.001 |
| 60S ribosomal protein L36 | Q9Y3U8 | −0.54 | >0.001 | 0.002 |
| 60S ribosomal protein L37a | P61513 | −0.50 | >0.001 | 0.002 |
| 60S ribosomal protein L38 | P63173 | −0.25 | 0.009 | 0.021 |
| 60S acidic ribosomal protein P0 | P05388 | −0.59 | >0.001 | >0.001 |
| 60S acidic ribosomal protein P2 | P05387 | −0.62 | >0.001 | 0.001 |
| 40S ribosomal protein S3 | P23396 | −0.73 | >0.001 | 0.001 |
| 40S ribosomal protein S3a | P61247 | −0.64 | >0.001 | 0.001 |
| 40S ribosomal protein S4, X isoform | P62701 | −0.61 | >0.001 | 0.001 |
| 40S ribosomal protein S6 | P62753 | −0.67 | >0.001 | 0.000 |
| 40S ribosomal protein S8 | P62241 | −0.63 | >0.001 | >0.001 |
| 40S ribosomal protein S9 | P46781 | −0.61 | >0.001 | 0.001 |
| 40S ribosomal protein S13 | P62277 | −0.62 | >0.001 | 0.001 |
| 40S ribosomal protein S15a | P62244 | −0.62 | >0.001 | 0.001 |
| 40S ribosomal protein S16 | P62249 | −0.45 | 0.002 | 0.006 |
| 40S ribosomal protein S21 | P63220 | −0.88 | >0.001 | 0.001 |
| 40S ribosomal protein S25 | P62851 | −0.64 | >0.001 | >0.001 |
| 40S ribosomal protein S27 | P42677 | −1.08 | >0.001 | >0.001 |
| Ubiquitin-40S ribosomal protein S27a | P62979 | 0.52 | >0.001 | 0.001 |
The table shows the log2fold change in the protein levels of LCC components which are related to protein synthesis.
4. Discussion
4.1. Inducible, Sleeping Beauty transposon-based expression of ATXN1(Q82) in human MSCs
Cellular protein aggregation models are invaluable to elucidate the pathogenic effects of polyQ IIBs in neurodegenerative diseases, including SCA1. These models are also important for screening assays and for drug development [13]. SCA1 patient-derived iPSCs [54], differentiated into cerebellar neurons [55] provide an opportunity for studying disease progression in the exact match of the affected cell type. Nevertheless, the developmental reset occurring upon iPSC derivation erases aging-associated signatures [56], which may interfere with the aggregation process occurring in adult neurons. In fact, in the context of Huntington's disease, protein aggregation could be detected in neurons directly derived from fibroblasts but not in neurons differentiated from iPSCs [57]. In line with these findings, we observed that iPSC-derived NPCs efficiently removed small inclusions and failed to support long-term production of the pathogenic ATXN1.
As an alternative approach, our inducible polyQ-expanded ATXN1-based model generates reproducibly large nuclear inclusions in human MSCs. This model overexpresses the full-length ATXN1(Q82), harboring all domains that affect protein dimerization and aggregation [58,59] in an inducible manner, using the Tet-On SB transposon system. The SB system is suitable for the modification of human stem cells with high reproducibility [[60], [61], [62]]. In combination with the hyperactive transposase SB100X [15], our Gateway-compatible system [63] can be used for rapid and highly efficient generation of expression vectors. The inducible expression system is not leaky and provides a tight control. It is also suitable for a stable, forced expression of pathogenic transgenes and a reproducible formation of the nuclear IIBs in cultured primary human cells.
Even though MSCs are adult stem cells of mesodermal origin [64] with a limited neurogenic differentiation potential [65], they have several advantages compared to immortalized laboratory lines previously utilized to study polyQ protein aggregation [9,66]. These cells can be modified using the SB system and cultured in cost-effective conditions [17,18]. More importantly, MSCs are resistant to the proteotoxic insults of ATXN1(Q82), which allowed a detailed investigation of the gradual accumulation of the toxic protein. Also, MSCs are subjected to replicative senescence, a phenomenon that resembles aging [67] and promotes polyQ protein aggregation. Thus, MSCs proved to be a proper choice in our studies aiming at characterizing the IIB formation of the pathogenic ATXN1.
4.2. ATXN1(Q82) gradually forms insoluble IIBs in MSCs
Prolonged induction of MSCs resulted in the formation of partially insoluble IIBs. Using AFM, we established a set of methods to efficiently measure nuclear coverage, determine topology of IIBs and quantify cell stiffness in non-stained cells at different stages of their formation. In our cell model, purified annular IIBs exhibit a significantly lower stiffness compared to polyQ aggregates prepared in vitro [68,69], suggesting that they contain a decreased amount of amyloid fibrils. However, the annular structure might be a common feature of insoluble IIBs. Such inclusions have been observed in cell-free systems and transgenic mouse models of polyQ diseases [70,71], indicating that they are polyQ length-dependent and are specifically induced by the pathogenic protein.
Accumulation of the pathogenic ATXN1 was accompanied by discrete changes in the expression profiles of nuclear proteins. This observation is consistent with the topology of IIBs. Importantly, our inducible ATXN1(Q82) MSCs are suitable to model both the early and late stage effects of protein aggregation (D2 vs D10). For example, components of the focal adhesion complex were up-regulated at the early stage [e.g. D2 ATXN1(Q82) MSCs], genes of the PI3K-Akt pathway were constantly up in D2-D10, while members of the p53 signaling pathway specifically featured in the late stage.
The downregulation of genes participating in oxidative phosphorylation may cause defects in the respiratory chain of both the MSCs and the cerebellar cells of the SCA1 patient. These defects may be responsible for the cellular oxidative stress while the activation of proteasome can be considered as a secondary line of defense against it. Our findings are consistent with the stimulation of ROS production by aggregating polyQ-expanded proteins [5,72] and connect to early hypotheses implicating oxidative stress in the pathology of neurodegenerative diseases [73].
The accumulation of IIBs also interferes with the nucleolus, suggesting that MSCs undergo nucleolar stress [74] and eventually enter a growth-arrest phase. This hypothesis is consistent with the down-regulation of genes involved in DNA replication and cell cycle, generally observed here and in models of polyQ diseases [75]. In fact, proteins oligomers were shown to dysregulate the cell cycle through the PI3K-Akt pathway [76] which promotes necrotic cell death [77].
4.3. PolyQ-induced stress perturbs the protein synthesis machinery
PolyQ tracts are normally present in a significant percentage of human proteins. Experimental evidence suggests that these domains mediate PPIs during the assembly of protein complexes [78]. Pathogenic polyQ proteins are enriched in coiled-coils [22,79] and spontaneously self-assemble into insoluble β-sheet rich inclusions [59,80] which are cytotoxic when they are transported into the nucleus [81]. The enrichment of aggregated ATXN1(Q82) in coiled-coils and β-sheets, suggests that insoluble IIBs might act as scaffolds for the sequestration of various protein molecules, perturbing protein interaction networks. The recruitment of ribosomal proteins and nucleophosmin, a nucleolar-stress marker which is critical for ribosome biogenesis [82] in insoluble IIBs indicates that these proteins are not able to perform their normal functions. In parallel, the down-regulation of ribosomal gene expression depletes the available ribosomal proteins in cells containing insoluble IIBs. Our results indicate a perturbed cerebellum-specific PPI network, which contains a densely connected subnetwork formed by components involved in RNA metabolism and protein synthesis.
Even though polyQ diseases may not share the same pathology at early stages, perturbation of the ribosomal complex might be a common feature at a later stage. PolyQ oligomers recruit ribosomal proteins [4,47] and promote the cytosolic mislocalization of fibrillarin (Fbl), a nucleolar protein involved in rRNA processing [47]. The combination of these events results in the retention of ribosomal proteins in the nucleus and prevents proper ribosome biogenesis. Furthermore, nucleolar stress induced by pathogenic misfolded proteins results in the extensive oxidation of ribosomal RNA, the reduction of ribosomes and the decrease in protein synthesis [reviewed in Ref. [74]]. Eventually, insoluble polyQ inclusions may cause similar mechanical and molecular defects in cells containing them.
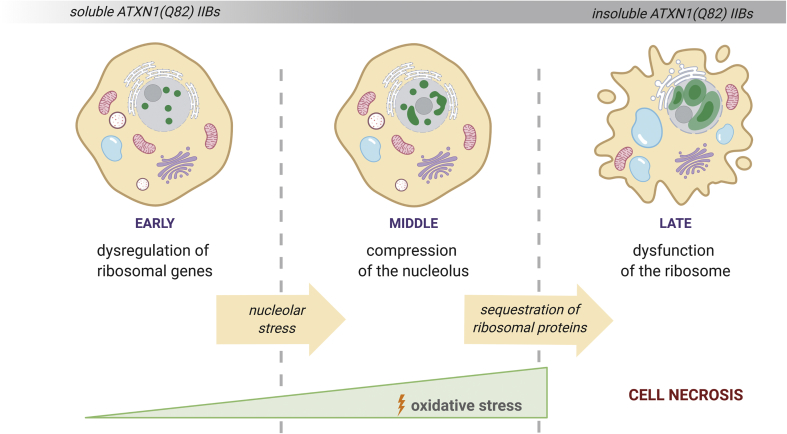
We propose a series of events induced by the pathogenic ATXN1, including (a) down-regulation of ribosomal genes; (b) gradual induction of oxidative and nucleolar stress by expanding IIBs; and (c) sequestration of ribosomal proteins into insoluble IIBs. Altogether, these events cause defects in RNA metabolism, ribosome assembly and protein synthesis leading to cell necrosis (Fig. 7). Notably, our findings correlate with the effects of the glutamine-rich PQN-41C protein in C. elegans. PQN-41C forms non-toxic cytoplasmic aggregates and controls non-apoptotic developmental cell death [83], supporting the hypothesis that similar mechanisms might be activated by polyQ-expanded proteins in vertebrates. Furthermore, the disturbed PPI network presented here, contains a significant number of genes that are dysregulated in human SCA1 cerebellum. Experimental data indicate that disrupted homeostasis of ribosomes triggers dendritic degeneration and disconnection of neuronal circuitries [84]. In the long-term, perturbation of the protein synthesis machinery may cause dysfunction of cerebellar neurons driving them towards a necrotic state in SCA1.
Fig. 7.
Cellular events caused by the gradual formation of insoluble ATXN1(Q82) IIBs. Production of ATXN1(Q82) dysregulates ribosomal genes and causes oxidative and nucleolar stress at early stages of protein aggregation. Expanding IIBs (shown with green color) compress the nucleolus and sequester ribosomal proteins. Eventually, insoluble ATXN1(Q82) IIBs disrupt the biogenesis of ribosomes and cause cell necrosis. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
5. Conclusions
Our study establishes a nuclear aggregation model of pathogenic ATXN1, the causative mutation of the SCA1 disease. Using an inducible SB-based expression system in MSCs, we generated primary human cells gradually accumulating insoluble IIBs. We show that IIBs generate cellular oxidative stress and characterized their structure and morphology. Furthermore, we identified the transcriptional changes induced by the presence of polyQ-expanded ATXN1, which are partially observed in end-stage SCA1 cerebellum, as well. The transcriptional changes in senescent cells containing insoluble IIBs indicate a cerebellum-specific perturbed protein interaction network involved in protein synthesis. Our model can be utilized for detailed investigations of the biochemical pathways affecting polyQ protein aggregation and the development of protective approaches against it.
Authors' contributions
Conceived and designed the experiments: SL GAL JP TR NC SPos ZIz PS MAA SP. Performed the experiments: SL JP TR BT KM GK ML. Analyzed the data: SL GAL JP TR KM MT JO GK MS FP. Contributed material: AZ AP ZIv ZIz. Wrote the paper: SP GAL JP GK. Edited the manuscript: SL TR MT JO NC AP ZIv SPos ZIz PS MAA. Designed the study: SP. All authors read and approved the final manuscript.
Funding
This work was supported by the European Union’s Horizon 2020 research and innovation programme [grant agreement No. 692298], the Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Technology (GSRT) [grant agreement No. 122 to SP]. Proteomics and Nanobiotechnology Core Facilities were supported by the CIISB research infrastructure project LM2018127 funded by the Ministry of Education, Youth and Sports of the Czech Republic. Genomics Core Facility was supported by the NCLG research infrastructure project LM2018132. Computational resources were supplied by the project "e-Infrastruktura CZ" (e-INFRA LM2018140) provided within the programme “Projects of Large Research, Development and Innovations Infrastructures”. The work was supported by the European Regional Development Fund-Project “CIISB4HEALTH” (No. CZ.02.1.01/0.0/0.0/16_013/0001776). AP was supported by the German Federal Ministry of Education and Research-BMBF (grant AZ.031A318). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
The authors declare no commercial or financial conflict of interest.
Acknowledgements
The authors wish to thank Stepan Solny, Anastasia Apostolidou, Evangelia Siska and Anna Dalda for technical assistance and Erich Wanker for critical suggestions. Bioinformatics Core Facility of CEITEC Masaryk University is gratefully acknowledged for obtaining the scientific data presented in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101458.
Contributor Information
Zsuzsanna Izsvák, Email: zizsvak@mdc-berlin.de.
Spyros Petrakis, Email: spetrak@certh.gr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
List of proteins identified by MS in polyQ IIBs and functional enrichment analysis. The table shows the ID of identified protein groups and relevant gene name(s), fasta header(s) of protein(s) contained within the group, the total number of identified peptide sequences associated with the protein group and number of unique peptides associated with the protein group. It also shows protein sequence coverage of the identified protein and its molecular weight. For each protein, an intensity-based absolute quantification (iBAQ) value is shown. A protein's total intensity was divided by the number of theoretically observable tryptic peptides (between 6 and 30 amino acids in length). UPS standards were not spiked into the sample. All contaminants (trypsin, keratins from human skin) were filtered out from the protein list. Each remaining protein's iBAQ value was then divided by the sum of all non-contaminant iBAQ values and multiplied by 1e6 (ppm scale), generating a riBAQ value for each protein. Supplementary Table 1B shows enrichment analysis for the identified protein components of the polyQ IIBs. Supplementary Table 1C presents enrichment analysis and comparisons with similar studies reporting protein components of polyQ-expanded Httex1 inclusions.
Expression changes in 3,984 genes dysregulated by mutant ATXN1 (ATXN1-DE genes). The table shows fold change, log2fold change and p-value for each gene per comparison.
GSEA for significantly dysregulated ATXN1-DE genes in Tet-On YFP-ATXN1(Q82) at D2 compared to Venus MSCs. The GSEA table indicates GO/pathway ID and description, SetSize, enrichment Score, NES/normalized enrichment score, p-value, adjusted p-value and q-value for the test. Rank is the position in the ranked list at which the maximum enrichment score occurred and a list of GeneIDs in the category.
Enrichment analysis for different sets of genes shown in the Venn diagram from Fig. 5C.
Expression changes in 3,923 genes dysregulated in the cerebellum of a SCA1 patient (human SCA1-DE genes). The table shows FPKM expression values for each gene in the cerebellum of a SCA1 patient and a healthy individual, fold change and log2fold change for each gene in the comparison between SCA1 patient versus control.
GSEA for human SCA1-DE genes. The GSEA table indicates GO/pathway ID and description, SetSize, enrichment Score, NES/normalized enrichment score, p-value, adjusted p-value and q-value for the test. Rank is the position in the ranked list at which the maximum enrichment score occurred and a list of Gene Symbols in the category.
Dysregulated genes in both D10 MSCs and human SCA1 cerebellum. The table shows log2fold change for common DE genes in cells and disease tissue (n = 185) and enrichment analysis.
Components of the LCC subnetwork formed by 328 proteins. For each gene (node), the table indicates log2fold change in D10 vs D0 Tet-On YFP-ATXN1(Q82) MSCs/SCA1 patient vs control human cerebellum and whether it was identified by MS in insoluble polyQ IIBs. It also indicates edges between nodes of the LCC.
Quantitative proteomics analysis (D10 vs D0) of ribosome components and associated proteins in LCC subnetwork. The table shows the accession number of each identified protein and the relevant gene name. For each protein identified per sample, it shows the number of unique peptides and the sequence coverage. It also includes absolute/relative iBAQ values and LQF values used for relative protein quantification. Protein abundance among D10 vs D0 sample groups is shown as a log2fold change.
Supplementary Fig. 1.
Flow cytometry of Tet-On YFP-ATXN1(Q82) MSCs. Measurements of (A) unstained cells and cells stained for (B) CD34/CD45 hemopoietic, (C) CD29/CD73 and (D) CD90/CD105 mesenchymal stem cell markers.
Supplementary Fig. 2.
IIBs in Tet-On YFP-ATXN1(Q82) NPCs and Tet-On YFP-ATXN1(Q30) MSCs. (A) Fluorescence microscopy of Tet-On YFP-ATXN1(Q82) NPCs at D2 and D10. Nuclei are stained with DAPI (blue color). (B) Fluorescence microscopy of Tet-On YFP-ATXN1(Q30) MSCs at D2, D5 and D10. Nuclei are stained with DAPI (blue color) (scale bar = 10 μm). (C–D) Immunoblots for ATXN1 in C. SDS-PAGE and D. Filter retardation assays using extracts of uninduced and induced Tet-On YFP-ATXN1(Q30) MSCs. β-actin was used as a loading control.
Supplementary Fig. 3.
FTIR absorption spectrum of cell models used in this study. Spectra of Venus MSCs (background) and Tet-On YFP-ATXN1(Q82) MSCS at D0-D10. The absorption regions of the main biological macromolecules are indicated.
Supplementary Fig. 4.
The second derivative of the (D0 background corrected) FTIR absorption spectrum of Tet-On YFP-ATXN1(Q82) MSCs at D2 or D10. Spectra were corrected after subtraction of the spectrum from uninduced MSCs (D0 in Fig. S3). The spectrum for D10 shows a peak at 1657 cm−1 indicative of α-helix and two peaks at 1640 and 1686 cm−1, corresponding to coiled-coils and intra-molecular β-sheets in YFP-ATXN1(Q82).
Supplementary Fig. 5.
The occupied nuclear surface area in Tet-On YFP-ATXN1(Q82) MSCs. (A) Cells fixed on a standard microscope glass slide were observed by contact mode AFM imaging. When pointing at two surrounding nuclei, the occupied nuclear surface area was shown by a blue mask. Scale bar of the images is 15 μm and Z-scale range 1 μm. (B) Quantification of occupied nuclear surface area in D0-D10 cells.
Supplementary Fig. 6.
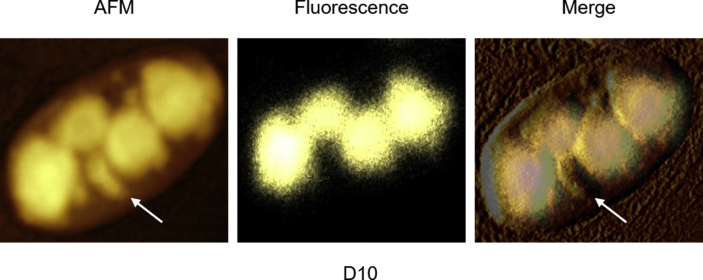
Overlay imaging of polyQ inclusions. AFM, fluorescence and merged images of Tet-On YFP-ATXN1(Q82) MSC nuclei induced for 10 days. The white arrow indicates natural nuclear content, i.e. nucleoli.
Supplementary Fig. 7.
Mechanical properties and topography of Tet-On YFP-ATXN1(Q82) MSCs characterized by nanoindentation. (A) Left: elastic properties of uninduced and control cells. Middle: properties of cells at D10. Right: selective representation of the stiffness of polyQ IIBs bounded by a green mask. (B) Comparison of average stiffness values for cells show in A (** p-value < 0.01). (C) The left image shows the topography of uninduced and control cells. The surface of cells at D10 is shown in the middle. The right image indicates polyQ IIBs bounded by a green mask.
Supplementary Fig. 8.
Cytotoxicity in Tet-On YFP-ATXN1(Q82) MSCs cultured in the presence or absence of Dox. Measurement of caspase 3/7 activity in Tet-On YFP-ATXN1(Q82) MSCs. Error bars denote ±SEM.
Supplementary Fig. 9.
Tet-On YFP-ATXN1(Q82) MSCs at various time-points post induction. (A) Tet-On YFP-ATXN1(Q82) MSCs at D20 shown by optical or fluorescence microscopy. White arrows indicate polyQ IIBs. (B) Tet-On YFP-ATXN1(Q82) MSCs at D10 and subsequent removal of the inducer shown by optical or fluorescence microscopy. YFP-ATXN1(Q82) IIBs are shown in green (scale bar = 30 μm).
Supplementary Fig. 10.
Detection methods of YFP-ATXN1(Q82) IIBs. (A) Purified IIBs were observed in fluorescence mode or AFM hybrid mode under physiological pH and ionic strange (scale bar = 2 μm). (B) Flow cytometry of YFP particles post sorting. Overlay histogram plot of unsorted control (red color) and YFP + sorted particles (green color).
Supplementary Fig. 11.
Transcriptional changes in Tet-On YFP-ATXN1(Q82) MSCs. (A) Distribution of log2fold expression gene changes between Tet-On YFP-ATXN1(Q82) MSCs at D2 and Venus MSCs. Genes with a similar expression profile in both overexpression models (2nd and 3rd quartile) were excluded from downstream analysis. The figure also shows fluorescence images of the cells. Produced proteins are shown in green color. Nuclei are stained with DAPI (blue color). (B) Hierarchical clustering analysis and heatmap of DE genes between Tet-On YFP-ATXN1(Q82) MSCs at D10 and D2 (log2fold change ≥ |1| and p-value < 0.05). Every row represents one gene and every column one sample.
References
- 1.Orr H.T., Zoghbi H.Y. SCA1 molecular genetics: a history of a 13 year collaboration against glutamines. Hum. Mol. Genet. 2001;10(20):2307–2311. doi: 10.1093/hmg/10.20.2307. [DOI] [PubMed] [Google Scholar]
- 2.Skinner P.J., Koshy B.T., Cummings C.J., Klement I.A., Helin K., Servadio A., et al. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389(6654):971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi T., Kikuchi S., Katada S., Nagai Y., Nishizawa M., Onodera O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum. Mol. Genet. 2008;17(3):345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 4.Ramdzan Y.M., Trubetskov M.M., Ormsby A.R., Newcombe E.A., Sui X., Tobin M.J., et al. Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep. 2017;19(5):919–927. doi: 10.1016/j.celrep.2017.04.029. [DOI] [PubMed] [Google Scholar]
- 5.Bertoni A., Giuliano P., Galgani M., Rotoli D., Ulianich L., Adornetto A., et al. Early and late events induced by polyQ-expanded proteins: identification of a common pathogenic property of polYQ-expanded proteins. J. Biol. Chem. 2011;286(6):4727–4741. doi: 10.1074/jbc.M110.156521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke K.A., Legleiter J. Atomic force microscopy assays for evaluating polyglutamine aggregation in solution and on surfaces. Methods Mol. Biol. 2013;1017:21–40. doi: 10.1007/978-1-62703-438-8_2. [DOI] [PubMed] [Google Scholar]
- 7.Dahlgren P.R., Karymov M.A., Bankston J., Holden T., Thumfort P., Ingram V.M., et al. Atomic force microscopy analysis of the Huntington protein nanofibril formation. Nanomed. Nanotechnol. Biol. Med. 2005;1(1):52–57. doi: 10.1016/j.nano.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Duim W.C., Chen B., Frydman J., Moerner W.E. Sub-diffraction imaging of huntingtin protein aggregates by fluorescence blink-microscopy and atomic force microscopy. ChemPhysChem : a European journal of chemical physics and physical chemistry. 2011;12(13):2387–2390. doi: 10.1002/cphc.201100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauerlein F.J.B., Saha I., Mishra A., Kalemanov M., Martinez-Sanchez A., Klein R., et al. In situ architecture and cellular interactions of PolyQ inclusions. Cell. 2017;171(1):179–187 e10. doi: 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Gruber A., Hornburg D., Antonin M., Krahmer N., Collado J., Schaffer M., et al. Molecular and structural architecture of polyQ aggregates in yeast. Proc. Natl. Acad. Sci. U. S. A. 2018;115(15):E3446–E3453. doi: 10.1073/pnas.1717978115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch P., Breuer P., Peitz M., Jungverdorben J., Kesavan J., Poppe D., et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480(7378):543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 12.Hansen S.K., Stummann T.C., Borland H., Hasholt L.F., Tumer Z., Nielsen J.E., et al. Induced pluripotent stem cell - derived neurons for the study of spinocerebellar ataxia type 3. Stem Cell Res. 2016;17(2):306–317. doi: 10.1016/j.scr.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Siska E.K., Koliakos G., Petrakis S. Stem cell models of polyglutamine diseases and their use in cell-based therapies. Front. Neurosci. 2015;9:247. doi: 10.3389/fnins.2015.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huda F., Fan Y., Suzuki M., Konno A., Matsuzaki Y., Takahashi N., et al. Fusion of human fetal mesenchymal stem cells with "degenerating" cerebellar neurons in spinocerebellar ataxia type 1 model mice. PloS One. 2016;11(11) doi: 10.1371/journal.pone.0164202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mates L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41(6):753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 16.Hudecek M., Izsvak Z., Johnen S., Renner M., Thumann G., Ivics Z. Going non-viral: the Sleeping Beauty transposon system breaks on through to the clinical side. Crit. Rev. Biochem. Mol. Biol. 2017;52(4):355–380. doi: 10.1080/10409238.2017.1304354. [DOI] [PubMed] [Google Scholar]
- 17.Kapetanou M., Chondrogianni N., Petrakis S., Koliakos G., Gonos E.S. Proteasome activation enhances stemness and lifespan of human mesenchymal stem cells. Free Radic. Biol. Med. 2016;103:226–235. doi: 10.1016/j.freeradbiomed.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Siska E.K., Weisman I., Romano J., Ivics Z., Izsvak Z., Barkai U., et al. Generation of an immortalized mesenchymal stem cell line producing a secreted biosensor protein for glucose monitoring. PloS One. 2017;12(9) doi: 10.1371/journal.pone.0185498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trachana V., Petrakis S., Fotiadis Z., Siska E.K., Balis V., Gonos E.S., et al. Human mesenchymal stem cells with enhanced telomerase activity acquire resistance against oxidative stress-induced genomic damage. Cytotherapy. 2017;19(7):808–820. doi: 10.1016/j.jcyt.2017.03.078. [DOI] [PubMed] [Google Scholar]
- 20.Saarikangas J., Barral Y. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife. 2015;4 doi: 10.7554/eLife.06197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrakis S., Rasko T., Mates L., Ivics Z., Izsvak Z., Kouzi-Koliakou K., et al. Gateway-compatible transposon vector to genetically modify human embryonic kidney and adipose-derived stromal cells. Biotechnol. J. 2012;7(7):891–897. doi: 10.1002/biot.201100471. [DOI] [PubMed] [Google Scholar]
- 22.Petrakis S., Rasko T., Russ J., Friedrich R.P., Stroedicke M., Riechers S.P., et al. Identification of human proteins that modify misfolding and proteotoxicity of pathogenic ataxin-1. PLoS Genet. 2012;8(8) doi: 10.1371/journal.pgen.1002897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., et al. Human iPSC-derived neural progenitors are an effective drug discovery model for neurological mtDNA disorders. Cell stem cell. 2017;20(5):659–674 e9. doi: 10.1016/j.stem.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C.S., Hoing S., et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PloS One. 2013;8(3) doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wanker E.E., Scherzinger E., Heiser V., Sittler A., Eickhoff H., Lehrach H. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 26.Ami D., Natalello A., Doglia S.M. Fourier transform infrared microspectroscopy of complex biological systems: from intact cells to whole organisms. Methods Mol. Biol. 2012;895:85–100. doi: 10.1007/978-1-61779-927-3_7. [DOI] [PubMed] [Google Scholar]
- 27.Chondrogianni N., Petropoulos I., Franceschi C., Friguet B., Gonos E.S. Fibroblast cultures from healthy centenarians have an active proteasome. Exp. Gerontol. 2000;35(6–7):721–728. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore J.L., Yoshida A., Takahashi H., Deguchi K., Kobori T., Louvet E., et al. Analyses of nuclear proteins and nucleic acid structures using atomic force microscopy. Methods Mol. Biol. 2015;1262:119–153. doi: 10.1007/978-1-4939-2253-6_8. [DOI] [PubMed] [Google Scholar]
- 29.Golan M., Jelinkova S., Kratochvilova I., Skladal P., Pesl M., Rotrekl V., et al. AFM monitoring the influence of selected cryoprotectants on regeneration of cryopreserved cells mechanical properties. Front. Physiol. 2018;9:804. doi: 10.3389/fphys.2018.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nardone G., Oliver-De La Cruz J., Vrbsky J., Martini C., Pribyl J., Skladal P., et al. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat. Commun. 2017;8:15321. doi: 10.1038/ncomms15321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Necas D., Klapetek P. Gwyddion: an open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012;10(1):181–188. [Google Scholar]
- 32.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 33.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & cellular proteomics. MCP. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alanis-Lobato G., Andrade-Navarro M.A., Schaefer M.H. HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 2017;45(D1):D408–D414. doi: 10.1093/nar/gkw985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arrondo J.L., Goni F.M. Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog. Biophys. Mol. Biol. 1999;72(4):367–405. doi: 10.1016/s0079-6107(99)00007-3. [DOI] [PubMed] [Google Scholar]
- 39.Heimburg T., Schuenemann J., Weber K., Geisler N. Specific recognition of coiled coils by infrared spectroscopy: analysis of the three structural domains of type III intermediate filament proteins. Biochemistry. 1996;35(5):1375–1382. doi: 10.1021/bi9515883. [DOI] [PubMed] [Google Scholar]
- 40.Natalello A., Frana A.M., Relini A., Apicella A., Invernizzi G., Casari C., et al. A major role for side-chain polyglutamine hydrogen bonding in irreversible ataxin-3 aggregation. PloS One. 2011;6(4) doi: 10.1371/journal.pone.0018789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka M., Morishima I., Akagi T., Hashikawa T., Nukina N. Intra- and intermolecular beta-pleated sheet formation in glutamine-repeat inserted myoglobin as a model for polyglutamine diseases. J. Biol. Chem. 2001;276(48):45470–45475. doi: 10.1074/jbc.M107502200. [DOI] [PubMed] [Google Scholar]
- 42.Rekas A., Alattia J.R., Nagai T., Miyawaki A., Ikura M. Crystal structure of venus, a yellow fluorescent protein with improved maturation and reduced environmental sensitivity. J. Biol. Chem. 2002;277(52):50573–50578. doi: 10.1074/jbc.M209524200. [DOI] [PubMed] [Google Scholar]
- 43.Zandomeneghi G., Krebs M.R., McCammon M.G., Fandrich M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 2004;13(12):3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trache A., Meininger G.A. Atomic force microscopy (AFM) Curr Protoc Microbiol. 2008 doi: 10.1002/9780471729259.mc02c02s8. 2C.2.1-2C.2.17. [DOI] [PubMed] [Google Scholar]
- 45.Vahabi S., Nazemi Salman B., Javanmard A. Atomic force microscopy application in biological research: a review study. Iran. J. Med. Sci. 2013;38(2):76–83. [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas G., Burnham N.A., Camesano T.A., Wen Q. Measuring the mechanical properties of living cells using atomic force microscopy. JoVE. 2013;76 doi: 10.3791/50497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y.E., Hosp F., Frottin F., Ge H., Mann M., Hayer-Hartl M., et al. Soluble oligomers of PolyQ-expanded huntingtin target a multiplicity of key cellular factors. Mol. Cell. 2016;63(6):951–964. doi: 10.1016/j.molcel.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 48.Kim J., Waldvogel H.J., Faull R.L., Curtis M.A., Nicholson L.F. The RAGE receptor and its ligands are highly expressed in astrocytes in a grade-dependant manner in the striatum and subependymal layer in Huntington's disease. J. Neurochem. 2015;134(5):927–942. doi: 10.1111/jnc.13178. [DOI] [PubMed] [Google Scholar]
- 49.Tong X., Gui H., Jin F., Heck B.W., Lin P., Ma J., et al. Ataxin-1 and Brother of ataxin-1 are components of the Notch signalling pathway. EMBO Rep. 2011;12(5):428–435. doi: 10.1038/embor.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue S., Serra H.G., Zoghbi H.Y., Orr H.T. The spinocerebellar ataxia type 1 protein, ataxin-1, has RNA-binding activity that is inversely affected by the length of its polyglutamine tract. Hum. Mol. Genet. 2001;10(1):25–30. doi: 10.1093/hmg/10.1.25. [DOI] [PubMed] [Google Scholar]
- 51.Sahin U., Ferhi O., Jeanne M., Benhenda S., Berthier C., Jollivet F., et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 2014;204(6):931–945. doi: 10.1083/jcb.201305148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hosp F., Vossfeldt H., Heinig M., Vasiljevic D., Arumughan A., Wyler E., et al. Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. 2015;11(7):1134–1146. doi: 10.1016/j.celrep.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koeppen A.H. The pathogenesis of spinocerebellar ataxia. Cerebellum. 2005;4(1):62–73. doi: 10.1080/14734220510007950. [DOI] [PubMed] [Google Scholar]
- 54.Buijsen R.A.M., Gardiner S.L., Bouma M.J., van der Graaf L.M., Boogaard M.W., Pepers B.A., et al. Generation of 3 spinocerebellar ataxia type 1 (SCA1) patient-derived induced pluripotent stem cell lines LUMCi002-A, B, and C and 2 unaffected sibling control induced pluripotent stem cell lines LUMCi003-A and B. Stem Cell Res. 2018;29:125–128. doi: 10.1016/j.scr.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 55.Ishida Y., Kawakami H., Kitajima H., Nishiyama A., Sasai Y., Inoue H., et al. Vulnerability of Purkinje cells generated from spinocerebellar ataxia type 6 patient-derived iPSCs. Cell Rep. 2016;17(6):1482–1490. doi: 10.1016/j.celrep.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 56.Mertens J., Reid D., Lau S., Kim Y., Gage F.H. Aging in a dish: iPSC-derived and directly induced neurons for studying Brain aging and age-related neurodegenerative diseases. Annu. Rev. Genet. 2018;52:271–293. doi: 10.1146/annurev-genet-120417-031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Victor M.B., Richner M., Olsen H.E., Lee S.W., Monteys A.M., Ma C., et al. Striatal neurons directly converted from Huntington's disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat. Neurosci. 2018;21(3):341–352. doi: 10.1038/s41593-018-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Chiara C., Menon R.P., Dal Piaz F., Calder L., Pastore A. Polyglutamine is not all: the functional role of the AXH domain in the ataxin-1 protein. J. Mol. Biol. 2005;354(4):883–893. doi: 10.1016/j.jmb.2005.09.083. [DOI] [PubMed] [Google Scholar]
- 59.Petrakis S., Schaefer M.H., Wanker E.E., Andrade-Navarro M.A. Aggregation of polyQ-extended proteins is promoted by interaction with their natural coiled-coil partners. Bioessays. 2013;35(6):503–507. doi: 10.1002/bies.201300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belay E., Matrai J., Acosta-Sanchez A., Ma L., Quattrocelli M., Mates L., et al. Novel hyperactive transposons for genetic modification of induced pluripotent and adult stem cells: a nonviral paradigm for coaxed differentiation. Stem Cell. 2010;28(10):1760–1771. doi: 10.1002/stem.501. [DOI] [PubMed] [Google Scholar]
- 61.Jackel C., Nogueira M.S., Ehni N., Kraus C., Ranke J., Dohmann M., et al. A vector platform for the rapid and efficient engineering of stable complex transgenes. Sci. Rep. 2016;6:34365. doi: 10.1038/srep34365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kowarz E., Loscher D., Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10(4):647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- 63.Hartley J.L., Temple G.F., Brasch M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10(11):1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vodyanik M.A., Yu J., Zhang X., Tian S., Stewart R., Thomson J.A., et al. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell stem cell. 2010;7(6):718–729. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scuteri A., Miloso M., Foudah D., Orciani M., Cavaletti G., Tredici G. Mesenchymal stem cells neuronal differentiation ability: a real perspective for nervous system repair? Curr. Stem Cell Res. Ther. 2011;6(2):82–92. doi: 10.2174/157488811795495486. [DOI] [PubMed] [Google Scholar]
- 66.Guo L., Giasson B.I., Glavis-Bloom A., Brewer M.D., Shorter J., Gitler A.D., et al. A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol. Cell. 2014;55(1):15–30. doi: 10.1016/j.molcel.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wagner W., Bork S., Horn P., Krunic D., Walenda T., Diehlmann A., et al. Aging and replicative senescence have related effects on human stem and progenitor cells. PloS One. 2009;4(6) doi: 10.1371/journal.pone.0005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruggeri F.S., Longo G., Faggiano S., Lipiec E., Pastore A., Dietler G. Infrared nanospectroscopy characterization of oligomeric and fibrillar aggregates during amyloid formation. Nat. Commun. 2015;6:7831. doi: 10.1038/ncomms8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruggeri F.S., Vieweg S., Cendrowska U., Longo G., Chiki A., Lashuel H.A., et al. Nanoscale studies link amyloid maturity with polyglutamine diseases onset. Sci. Rep. 2016;6:31155. doi: 10.1038/srep31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sathasivam K., Lane A., Legleiter J., Warley A., Woodman B., Finkbeiner S., et al. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington's disease. Hum. Mol. Genet. 2010;19(1):65–78. doi: 10.1093/hmg/ddp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wacker J.L., Zareie M.H., Fong H., Sarikaya M., Muchowski P.J. Hsp 70 and Hsp 40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat. Struct. Mol. Biol. 2004;11(12):1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- 72.Hands S., Sajjad M.U., Newton M.J., Wyttenbach A. In vitro and in vivo aggregation of a fragment of huntingtin protein directly causes free radical production. J. Biol. Chem. 2011;286(52):44512–44520. doi: 10.1074/jbc.M111.307587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beal M.F. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann. Neurol. 1995;38(3):357–366. doi: 10.1002/ana.410380304. [DOI] [PubMed] [Google Scholar]
- 74.Hetman M., Pietrzak M. Emerging roles of the neuronal nucleolus. Trends Neurosci. 2012;35(5):305–314. doi: 10.1016/j.tins.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu M., Boschetti C., Tunnacliffe A. Long term aggresome accumulation leads to DNA damage, p53-dependent cell cycle arrest, and steric interference in mitosis. J. Biol. Chem. 2015;290(46):27986–28000. doi: 10.1074/jbc.M115.676437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhaskar K., Miller M., Chludzinski A., Herrup K., Zagorski M., Lamb B.T. The PI3K-Akt-mTOR pathway regulates Abeta oligomer induced neuronal cell cycle events. Mol. Neurodegener. 2009;4:14. doi: 10.1186/1750-1326-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Y.T., Tan H.L., Huang Q., Ong C.N., Shen H.M. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy. Autophagy. 2009;5(6):824–834. doi: 10.4161/auto.9099. [DOI] [PubMed] [Google Scholar]
- 78.Schaefer M.H., Wanker E.E., Andrade-Navarro M.A. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012;40(10):4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pelassa I., Cora D., Cesano F., Monje F.J., Montarolo P.G., Fiumara F. Association of polyalanine and polyglutamine coiled coils mediates expansion disease-related protein aggregation and dysfunction. Hum. Mol. Genet. 2014;23(13):3402–3420. doi: 10.1093/hmg/ddu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner A.S., Politi A.Z., Ast A., Bravo-Rodriguez K., Baum K., Buntru A., et al. Self-assembly of mutant huntingtin exon-1 fragments into large complex fibrillar structures involves nucleated branching. J. Mol. Biol. 2018;430(12):1725–1744. doi: 10.1016/j.jmb.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 81.Yang W., Dunlap J.R., Andrews R.B., Wetzel R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum. Mol. Genet. 2002;11(23):2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- 82.Itahana K., Bhat K.P., Jin A., Itahana Y., Hawke D., Kobayashi R., et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12(5):1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 83.Blum E.S., Abraham M.C., Yoshimura S., Lu Y., Shaham S. Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science. 2012;335(6071):970–973. doi: 10.1126/science.1215156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Slomnicki L.P., Pietrzak M., Vashishta A., Jones J., Lynch N., Elliot S., et al. Requirement of neuronal ribosome synthesis for growth and maintenance of the dendritic tree. J. Biol. Chem. 2016;291(11):5721–5739. doi: 10.1074/jbc.M115.682161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of proteins identified by MS in polyQ IIBs and functional enrichment analysis. The table shows the ID of identified protein groups and relevant gene name(s), fasta header(s) of protein(s) contained within the group, the total number of identified peptide sequences associated with the protein group and number of unique peptides associated with the protein group. It also shows protein sequence coverage of the identified protein and its molecular weight. For each protein, an intensity-based absolute quantification (iBAQ) value is shown. A protein's total intensity was divided by the number of theoretically observable tryptic peptides (between 6 and 30 amino acids in length). UPS standards were not spiked into the sample. All contaminants (trypsin, keratins from human skin) were filtered out from the protein list. Each remaining protein's iBAQ value was then divided by the sum of all non-contaminant iBAQ values and multiplied by 1e6 (ppm scale), generating a riBAQ value for each protein. Supplementary Table 1B shows enrichment analysis for the identified protein components of the polyQ IIBs. Supplementary Table 1C presents enrichment analysis and comparisons with similar studies reporting protein components of polyQ-expanded Httex1 inclusions.
Expression changes in 3,984 genes dysregulated by mutant ATXN1 (ATXN1-DE genes). The table shows fold change, log2fold change and p-value for each gene per comparison.
GSEA for significantly dysregulated ATXN1-DE genes in Tet-On YFP-ATXN1(Q82) at D2 compared to Venus MSCs. The GSEA table indicates GO/pathway ID and description, SetSize, enrichment Score, NES/normalized enrichment score, p-value, adjusted p-value and q-value for the test. Rank is the position in the ranked list at which the maximum enrichment score occurred and a list of GeneIDs in the category.
Enrichment analysis for different sets of genes shown in the Venn diagram from Fig. 5C.
Expression changes in 3,923 genes dysregulated in the cerebellum of a SCA1 patient (human SCA1-DE genes). The table shows FPKM expression values for each gene in the cerebellum of a SCA1 patient and a healthy individual, fold change and log2fold change for each gene in the comparison between SCA1 patient versus control.
GSEA for human SCA1-DE genes. The GSEA table indicates GO/pathway ID and description, SetSize, enrichment Score, NES/normalized enrichment score, p-value, adjusted p-value and q-value for the test. Rank is the position in the ranked list at which the maximum enrichment score occurred and a list of Gene Symbols in the category.
Dysregulated genes in both D10 MSCs and human SCA1 cerebellum. The table shows log2fold change for common DE genes in cells and disease tissue (n = 185) and enrichment analysis.
Components of the LCC subnetwork formed by 328 proteins. For each gene (node), the table indicates log2fold change in D10 vs D0 Tet-On YFP-ATXN1(Q82) MSCs/SCA1 patient vs control human cerebellum and whether it was identified by MS in insoluble polyQ IIBs. It also indicates edges between nodes of the LCC.
Quantitative proteomics analysis (D10 vs D0) of ribosome components and associated proteins in LCC subnetwork. The table shows the accession number of each identified protein and the relevant gene name. For each protein identified per sample, it shows the number of unique peptides and the sequence coverage. It also includes absolute/relative iBAQ values and LQF values used for relative protein quantification. Protein abundance among D10 vs D0 sample groups is shown as a log2fold change.