Abstract
All eukaryotic life engages in symbioses with a diverse community of bacteria that are essential for performing basic life functions. In many cases, eukaryotic organisms form additional symbioses with other macroscopic eukaryotes. The tightly linked physical interactions that characterize many macroscopic symbioses create opportunities for microbial transfer, which likely affects the diversity and function of individual microbiomes, and may ultimately lead to microbiome convergence between distantly related taxa. Here, we sequence the microbiomes of five species of clownfish-hosting sea anemones that co-occur on coral reefs in the Maldives. We test the importance of evolutionary history, clownfish symbiont association, and habitat on the taxonomic and predicted functional diversity of the microbiome, and explore signals of microbiome convergence in anemone taxa that have evolved symbioses with clownfishes independently. Our data indicate that host identity and clownfish association shapes the majority of the taxonomic diversity of the clownfish-hosting sea anemone microbiome, and predicted functional microbial diversity analyses demonstrate a convergence among host anemone microbiomes, which reflect increased functional diversity over individuals that do not host clownfishes. Further, we identify upregulated predicted microbial functions that are likely affected by clownfish presence. Taken together our study potentially reveals an even deeper metabolic coupling between clownfishes and their host anemones, and what could be a previously unknown mutualistic benefit to anemones that are symbiotic with clownfishes.
Keywords: microbiome, symbiosis, sea anemone, clownfish, coral reef
1. Introduction
The importance of symbiosis is underscored by its ubiquity—virtually all of life engages in complex multi-level symbioses that critically impact the formation and distribution of biodiversity around the globe [1–3]. Minimally, all multicellular life engages in symbioses with prokaryotic microbiota that are essential for individual health, development and nutrient acquisition, and which serve as a primary interface between individuals and their environment (e.g. [4–12]). The composition and diversity of an individual's microbial community (i.e. microbiome) has been shown to be influenced by a variety of factors, but are generally discussed as a combination of evolutionary history and ecology (e.g. [13–17]).
In addition to microbial symbioses, many multicellular eukaryotes engage in symbioses with other multicellular organisms. Central features of these interactions are the physical linkages between constituent partners which provide an opportunity for microbial transfer, adding a layer of complexity to the factors that affect the diversity and function of individual microbiomes. Many macroscopic symbioses involve diverse partners, occur across distinct habitats, and have evolved independently multiple times across the tree of life [18–20]. Consequently, macroscopic symbioses provide an important framework for exploring the processes that may lead distantly related taxa to converge on microbiomes with similar genetic and functional profiles.
Among the immense symbiotic diversity on the planet, the clownfish–sea anemone mutualism stands out as an iconic example, and holds characteristics that make it a useful system for understanding the processes that affect microbiome diversity and function within macroscopic symbioses. A classic example of mutualism, the 30 species of clownfishes (or anemonefishes) have adaptively radiated from a common ancestor to live with 10 species of sea anemones on coral reefs of the Indian and Pacific Oceans [21–23]. Within sea anemones (order Actiniaria), symbiosis with clownfishes has evolved independently at least three times [24]. The clownfish–sea anemone symbiosis is found across a range of reef habitats and involves many combinations of sea anemone–clownfish associations [21,23–27]. Clownfishes are considered obligate symbionts of sea anemones, have evolved a range of host specificities, and are never found solitarily [21,27]. Anemones in contrast, while receiving substantial benefits from hosting clownfishes [28,29], can be found solitarily [25].
The ability of clownfishes to live within the otherwise lethal tentacles of sea anemones stems from their mucus coating, which is maintained through regular physical contact with anemone hosts [30]. The long- and short-term maintenance of the symbiosis thus requires constant interaction, which should lead to regular microbial transfer. In a laboratory setting, the microbial make-up of clownfish mucus was shown to change rapidly in the presence of an anemone host [31]. However, the microbial diversity of host anemones remains uncharacterized, and the multiple origins of symbiosis with clownfishes make these animals useful for understanding if macroscopic symbioses generate microbial convergence across distantly related taxa.
Here, we use in situ field sampling to conduct the first comparative microbial study of clownfish-hosting sea anemones to test the importance of host identity, clownfish symbiont association, and habitat on the taxonomic and predicted functional diversity of the microbiome. We examine the microbiomes of five species of clownfish-hosting sea anemones that co-occur on a fine scale on coral reefs in the Maldives, but that vary in habitat and clownfish symbiont associations. Our five focal taxa come from three anemone clades that have evolved symbiosis with clownfishes independently [24] and therefore offer a unique opportunity to explore the interplay between evolutionary history, environment and macroscopic symbioses in shaping the diversity of the microbiome.
2. Material and methods
(a). Sample collection
Microbial samples were collected from five species of clownfish-hosting sea anemones in Huvadhoo Atoll, Republic of the Maldives (0°11′45.89″N, 73°11′3.53″E)—Cryptodendrum adhaesivum, Entacmaea quadricolor, Heteractis aurora, H. magnifica and Stichodactyla mertensii (table 1; electronic supplementary material, figure S1). Focal anemones come from three clades that have evolved symbiosis with clownfishes independently [24]. Samples were collected from three atoll reef habitats: (i) outer atoll fore reef (10–25 m depth), (ii) lagoonal fringing reef slope (5–25 m depth), and (iii) shallow (1 m depth) reef flat (electronic supplementary material, figure S2). Only specimens of H. magnifica were present across all three habitats (table 1). On shallow reef flats, H. magnifica exhibited a pale column phenotype and did not host fish (electronic supplementary material, figure S1F), in contrast with the common purple phenotype on deeper reefs that hosted the Maldivian endemic clownfish Amphiprion nigripes (electronic supplementary material, figure S1E). All other anemone species and individuals sampled in this study hosted Amphiprion clarkii (electronic supplementary material, figure S1A–D). Samples were collected by clipping two tentacles per anemone. A total of 94 tentacles from 47 individual anemones were sampled and preserved immediately in RNAlater.
Table 1.
Clownfish-hosting sea anemones species, habitat, sample sizes and clownfish symbiont association sampled from the Maldives and included in this study.
| species | habitat | sample size N = individuals (tissues sequenced) | fish symbiont |
|---|---|---|---|
| Cryptodendrum adhaesivum | lagoon | N = 1 (2) | Amphiprion clarkii |
| Entacmaea quadricolor | lagoon | N = 8 (16) | Amphiprion clarkii |
| Heteractis aurora | fore reef | N = 2 (4) | Amphiprion clarkii |
| lagoon | N = 5 (10) | Amphiprion clarkii | |
| Heteractis magnifica | fore reef | N = 7 (14) | Amphiprion nigripes |
| lagoon | N = 7 (14) | Amphiprion nigripes | |
| reef flat | N = 12 (24) | none | |
| Stichodactyla mertensii | fore reef | N = 3 (6) | Amphiprion clarkii |
| lagoon | N = 3 (6) | Amphiprion clarkii | |
| total | N = 48 (96) |
(b). DNA extraction and 16S amplicon sequencing
Genomic DNA was extracted from both tentacle samples collected from each individual using Qiagen DNeasy Blood and Tissue Kits. DNA was quantified and standardized to 5 ng µl−1 for each sample. Microbiome diversity was assessed via Illumina sequencing targeting a 252 base pair sequence of the hypervariable v4 region of the 16S rRNA SSU gene using primer pairs 515F and 806R. 16S amplicon libraries were prepared following the Earth Microbiome Protocol [32,33]. Sequencing of 16S amplicons was conducted on an Illumina MiSeq using a V2 500 cycle kit (250 × 250 bp) at the National Museum of Natural History's Laboratory of Analytical Biology.
(c). Microbiome data analyses
Amplicon sequence data were demultiplexed, denoised to identify amplicon sequence variants (ASVs), assembled and analysed using QIIME2 and associated plug-ins (see electronic supplementary material) [34]. Following demultiplexing and read-joining, we pooled reads from replicate samples to increase the number of sequence reads per individual anemone. Microbial taxonomy was assigned using a naive Bayes classifier trained on the SILVA 132 99% database (silva-132-99-nb-classifer). Resulting feature tables were then filtered to remove ASVs that could not be identified as bacterial, and taxonomy was visualized using QIIME2. We used BLAST to identify ASVs that remained unclassified below the Domain Bacteria after filtering. We manually removed 24 ASVs that mapped with high confidence to sea anemone mtDNA that failed to be filtered out by the SILVA database.
To test for variation in microbial taxonomic diversity across host, habitat and clownfish association, amplicon data were standardized using a rarefaction sequencing depth of 10 210 reads per individual. We tested for significant (p < 0.05) variation in microbial alpha and beta taxonomic diversity in three sample metadata categories: (i) anemone host species, (ii) clownfish symbiont association (A. clarkii, A. nigripes or none) and (iii) habitat (atoll fore reef, reef flat or lagoonal patch reef). Alpha diversity was calculated using Shannon's Diversity Index (H) and post hoc comparisons made using non-parametric Kruskal–Wallis tests. Significant variation in beta diversity between sample categories was tested for using Bray–Curtis distance measures of community dissimilarity, and post hoc comparisons made using permutational multivariate analysis of variance (perMANOVA). Ordination plots for beta diversity analyses affect using non-metric multidimensional scaling (nMDS) plots. Linear discriminant analysis effect size (LEfSe) was use to elucidate bacterial taxonomic groups that were significantly more abundant in anemones that hosted clownfishes versus those that did not (see electronic supplementary material) [35].
Finally, we predicted and compared the functional diversity of the microbial metagenomes using phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) [36]. All ASVs were normalized for 16 s copy number, and function was predicted by assigning ASVs to Kyoto Encyclopedia of Genes and Genomes Orthology categories (KEGG; see electronic supplementary material) [37]. Significant variation in predicted alpha and beta metagenomic functional diversity across sample metadata categories were tested for statistically using Shannon Diversity (H) and Bray–Curtis measures as above. We then used DESeq2 [38] and log-transformations to detect differentially abundant and highly variable functional groups across sample metadata categories.
3. Results and discussion
Our final dataset consisted of 47 individual anemones and 6257 ASVs. Final ASV counts after rarefaction ranged from 42 to 606 per individual (electronic supplementary material, figure S3). The taxonomic composition of microbial communities varied by anemone species, clownfish symbiont association and habitat (electronic supplementary material, figure S4), but were largely dominated by Gammaproteobacteria, and to a lesser extent Alphaproteobacteria (electronic supplementary material, figure S4).
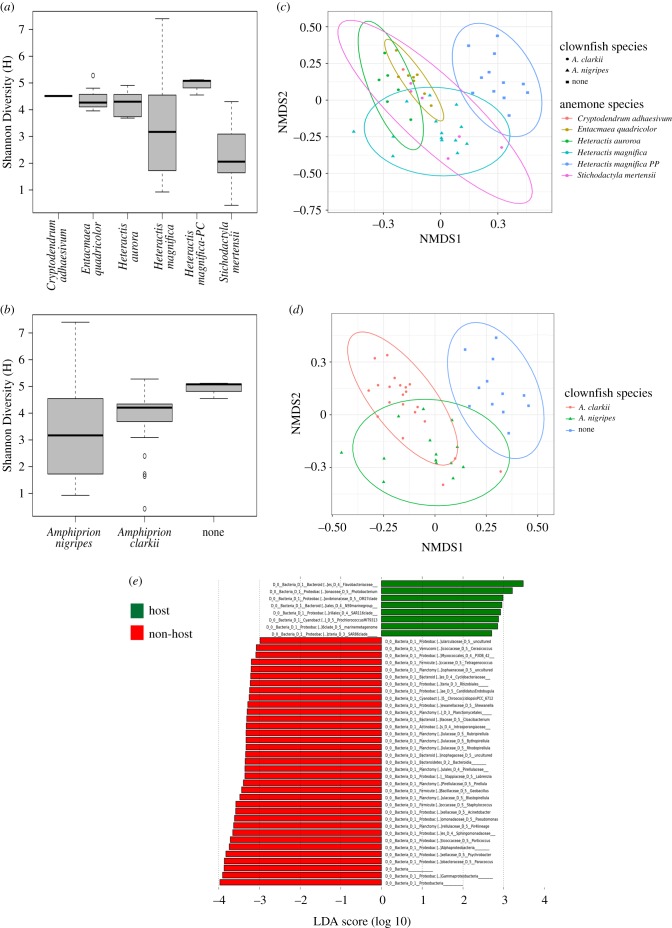
Alpha and beta diversity analyses indicate that anemone species and clownfish symbiont association, rather than habitat, drives the majority of the microbial taxonomic diversity signal in our dataset (overall alpha diversity: anemone species H = 14.22, p < 0.02; clownfish association H = 6.32, p < 0.05; habitat H = 5.70, p = 0.06; overall beta diversity: anemone species F = 5.32, p < 0.002; clownfish association F = 6.38, p < 0.002; habitat F = 1.01, p = 0.42; figure 1a–d; electronic supplementary material, tables S1–6 and figures S5 and S6). Bray–Curtis beta diversity analyses also indicate that anemones that host clownfish symbionts, regardless of host or habitat, are more similar to other anemones that host the same clownfish species than they are to anemones that host a different clownfish, or that do not host fish (perMANOVA F = 6.38, p < 0.002; figure 1d; electronic supplementary material, table S5). This implies that there is some degree of microbiome convergence across distantly related anemones that host the same clownfish species. The transfer of microbiota among macroscopic symbiotic partners is well demonstrated in plant–pollinator symbioses (e.g. [11,39]) and microbiome convergence has also been noted in some, but not all, leaf-cutter ant symbioses (e.g. [40,41]). Pratte et al. [31] recently demonstrated changes in microbiome composition between anemone-hosting and non-hosting A. clarkii clownfish in a laboratory setting, implying either direct microbial transfer from anemone to clownfish or a shift in microbial diversity in response to the interaction. Our data also suggest either direct microbial transfer from the clownfish to the host, or a shift in diversity may be occurring. Pratte et al. [31] did not sample anemone microbiomes throughout the duration of their experimental trials, and we did not sample clownfish here, so it remains to be seen how convergent the microbiomes from both symbiotic partners become when in association with each other.
Figure 1.
Host identity and symbiotic association affects the taxonomic diversity of the clownfish-hosting sea anemone microbiome. (a) Variation in microbial genetic diversity (H) grouped by host species. (b) Variation in microbial genetic diversity (H) grouped by clownfish symbiont association. (c) nMDS plot of Bray–Curtis dissimilarities coloured by anemone host species with 95% confidence ellipses. (d) nMDS plot of Bray–Curtis dissimilarities of host anemone species, coloured by clownfish symbiont association with 95% confidence ellipses. (e) Differential abundance of major microbial taxonomic groups between clownfish-hosting (green) and non-clownfish hosting (red) anemones. (Online version in colour.)
LEfSe analyses highlighted eight differentially abundant bacterial groups that are enriched in anemones that hosted clownfish versus those that did not (figure 1e). Interestingly, a number of these (e.g. Flavobacteria, OM27, SAR11, SAR86) have been implicated directly in the cycling of dissolved organic nitrogen [42], which can originate from the nitrogenous wastes of animals. It has long been known that waste by-products of clownfishes provide important sources of nitrogen to host anemones and endosymbiotic dinoflagellates [43,44]. These data potentially provide the first evidence that links this important mutualistic pathway between fish, anemone and dinoflagellates with the bacterial community structure residing on the host anemones.
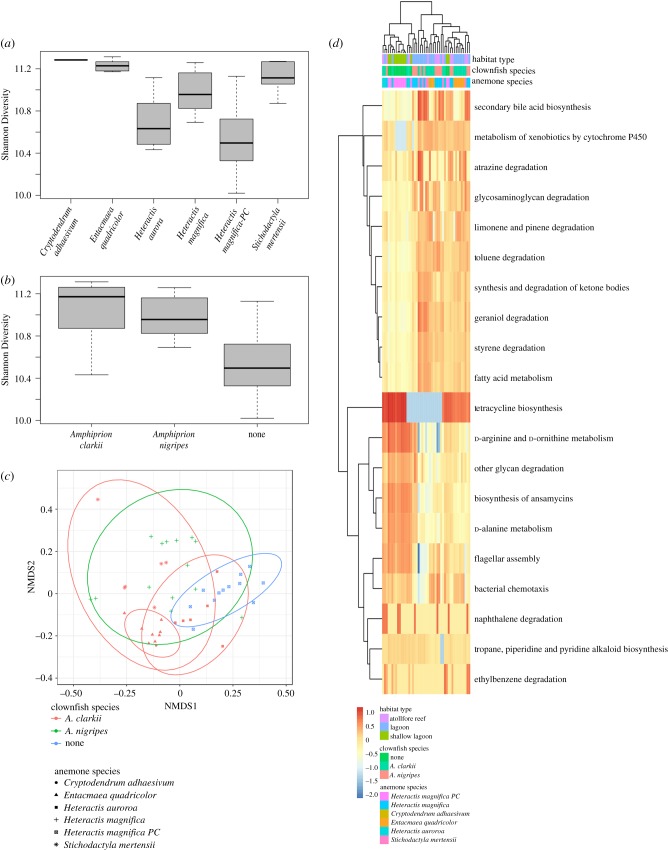
Functionally, alpha and beta diversity analyses using PICRUSt predicted metagenomes reinforce the role of anemone host identity in shaping the functional diversity of the host microbiome (Shannon Diversity Index, H = 28.38, p < 0.0001; perMANOVA F = 14.82, p < 0.002; electronic supplementary material, tables S7–8). However, interestingly, these analyses also highlight that hosting clownfish symbionts increase the functional alpha and beta microbial diversity of host anemones over those that do not (H = 15.67, p < 0.0001; F = 14.86, p < 0.002; figure 2a–c; electronic supplementary material, tables S9–10). Anemones that hosted A. clarkii and A. nigripes did not differ functionally from each other in either alpha or beta diversity measures (figure 2a–c; electronic supplementary material, table S8), and if increased functional microbial diversity is indicative of increased health, as in many other taxa, these data may represent the first evidence of a previously unknown mutualistic benefit of hosting clownfishes gained by host anemones. Metagenomic and/or metatranscriptomic data are needed to confirm these predicted metabolic functions.
Figure 2.
Host identity and symbiotic association affects the predicted functional diversity of the clownfish-hosting sea anemone microbiome. (a) Variation in predicted functional alpha diversity (H) grouped by host species. (b) Variation in predicted functional alpha diversity (H) grouped by clownfish symbiont association. (c) nMDS plot of Bray–Curtis dissimilarities of host anemone species, coloured by clownfish symbiont association, with 95% confidence ellipses. (d) Twenty most variable predicted KEGG functional categories. (Online version in colour.)
Using DESeq2 analyses, we identified predicted KEGG functions that were differentially abundant and highly variable between clownfish-hosting and non-hosting anemones (table 2 and figure 2d). In clownfish-hosting anemones, arachidonic acid (ARA) metabolic functions, part of the broader lipid metabolism pathway, were 25-fold more abundant over non-hosting anemones (table 2). No other predicted functional pathway was over threefold more abundant (table 2, electronic supplementary material, table S11). ARA is an essential polyunsaturated fatty acid (PUFA) that could be acquired by host anemones via translocated photosynthate from their Symbiodiniaceae community, heterotrophic prey capture, or via waste by-products from clownfish symbionts (e.g. [45–48]). Predicted cytochrome P450 monooxygenase pathways, which catalyse ARA and other PUFAs to biologically active, intercellular signalling molecules (eicosanoids), were also highly variable and enriched primarily in host anemones (figure 2d). Eicosanoid lipids participate in oxidative stress response and are hypothesized to play a role in the oxidative stress response in symbiotic cnidarians [48,49]. It is well documented that clownfish symbionts increase gas transfer and oxygenate their host anemones while also passing organic wastes to their anemone hosts, which functionally act as fertilizers for endosymbiotic Symbiodiniaceae [28,29]. It is possible that the increase in oxygen free radicals produced by fish movements through the host, and by the oxygen produced as waste during photosynthesis, could stimulate a metabolic response by the host anemone that mimics an oxidative stress response. Consequently, anemones that host fish could see a corresponding shift in microbiome diversity and function to compensate for increased ARA metabolites that could be harmful to the host. If so, these data could indicate a hidden cost of hosting mutualistic clownfishes for the anemones. However, ARA are used in a myriad of physiological processes, and its derivatives such as platelet activating factors, are also known to be involved in tissue growth and coral competition (e.g. [50,51]). More detailed microbial and metabolomic studies are needed to pinpoint the source of any increased levels of ARA in the host anemones in the presence of clownfish. Regardless of its origin here, the degree to which microbial communities differ in ARA functions is a striking metabolic signal that the microbial communities on host sea anemones may be responding to clownfish presence. Minimally, this finding is consistent with the literature that clownfish presence has a significant impact on the metabolism and physiology of the host anemone [28,29,43,44].
Table 2.
Top significantly enriched predicted microbial KEGG functions for clownfish hosting (grey highlighted) and non-clownfish host sea anemones. Positive log-2-fold-change values indicate microbial functions enriched in host versus non-host anemones.
| KEGG function | log-2-fold-change | p-adj | clownfish association |
|---|---|---|---|
| ARA metabolism | 25.841 | 2.57 × 10−12 | host |
| protein digestion and absorption | 2.100 | 9.71 × 10−10 | host |
| renin–angiotensin system | 1.903 | 0.040 | host |
| non-homologous end-joining | 1.730 | 0.007 | host |
| lysosome | 1.543 | 0.002 | host |
| phosphonate and phosphinate metabolism | 1.534 | 1.19 × 10−7 | host |
| linoleic acid metabolism | 1.527 | 1.96 × 10−9 | host |
| flavonoid biosynthesis | 1.516 | 0.006 | host |
| penicillin and cephalosporin biosynthesis | 1.431 | 9.71 × 10−10 | host |
| primary bile acid biosynthesis | 1.422 | 0.001 | host |
| RNA polymerase | −0.608 | 1.19 × 10−6 | non-host |
| flagellar assembly | −0.645 | 0.005 | non-host |
| d-alanine metabolism | −0.711 | 2.00 × 10−6 | non-host |
| biosynthesis of ansamycins | −0.740 | 5.29 × 10−6 | non-host |
| insulin signalling pathway | −0.803 | 2.95 × 10−6 | non-host |
| d-arginine and d-ornithine metabolism | −1.029 | 0.0008 | non-host |
| amoebiasis | −1.355 | 0.0003 | non-host |
| proteasome | −1.431 | 0.0178 | non-host |
| systemic lupus erythematosus | −1.798 | 0.0107 | non-host |
| cell adhesion molecules | −2.909 | 0.0017 | non-host |
In conclusion, we demonstrate for the first time that clownfish presence may increase the functional diversity of the anemone host microbiome, potentially revealing an even deeper metabolic coupling between clownfishes, host anemones, endosymbiotic Symbiodiniaceae, and what could be a previously unrecognized mutualistic benefit of the symbiosis at the microbial level.
Supplementary Material
Acknowledgements
We thank the Small Island Research Center for field research support. Aaron Hartmann, Melissa Ingala, Jennifer Matthews and Susan Perkins provided valuable advice on data analysis and anthozoan metabolic functions. Samples were collected under research permit 30-D/Indiv/2018/27.
Ethics
Field research and sample collection in the Maldives was conducted under permit #30-D/Indiv/2018/27.
Data accessibility
Raw sequence data, barcodes and sample metadata are publicly available through the Dryad Digital Repository: https://doi.org/10.5061/dryad.sbcc2fr28 [52].
Authors' contributions
B.M.T. and C.P.M. conceived the study. B.M.T. and C.P.M. collected the samples. H.W. conducted library preparation and sequencing. B.M.T. and R.L. analysed the data. B.M.T., R.L., E.R., H.W. and C.P.M. interpreted the data, and wrote and edited the manuscript. All authors agree to be held accountable for the content therein and approve the final version of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Gerstner Scholars Postdoctoral Fellowship and the Gerstner Family Foundation, the Lerner-Gray Fund for Marine Research, and the Richard Guilder Graduate School, American Museum of Natural History to B.M.T. Additional funding was provided by the National Museum of Natural History to C.P.M.
References
- 1.Boucher DH. 1985. The biology of mutualism: ecology and evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 2.Margulis L, Fester R. 1991. Symbiosis as a source of evolutionary innovation. Cambridge, MA: MIT Press. [PubMed] [Google Scholar]
- 3.Herre EA, Knowlton N, Mueller UG, Rehner SA. 1999. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 14, 49–53. ( 10.1016/S0169-5347(98)01529-8) [DOI] [PubMed] [Google Scholar]
- 4.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. 2015. The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. ( 10.1111/nph.13312) [DOI] [PubMed] [Google Scholar]
- 5.Sweet MJ, Bulling MT. 2017. On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 4, 9 ( 10.3389/fmars.2017.00009) [DOI] [Google Scholar]
- 6.Douglas AE. 1998. Nutritional interactions in insect–microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43, 17–37. ( 10.1146/annurev.ento.43.1.17) [DOI] [PubMed] [Google Scholar]
- 7.Christian N, Whitaker BK, Clay K. 2015. Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front. Microbiol. 6, 869 ( 10.3389/fmicb.2015.00869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. 2016. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535, 94–103. ( 10.1038/nature18850) [DOI] [PubMed] [Google Scholar]
- 9.Barea JM, Pozo MJ, Azcon R, Azcon-Aguilar C. 2005. Microbial co-operation in the rhizosphere. J. Exp. Bot. 56, 1761–1778. ( 10.1093/jxb/eri197) [DOI] [PubMed] [Google Scholar]
- 10.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid–Vibrio symbiosis. Annu. Rev. Microbiol. 68, 177–194. ( 10.1146/annurev-micro-091313-103654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ushio M, Yamasaki E, Takasu H, Nagano AJ, Fujinaga S, Honjo MN, Ikemoto M, Sakai S, Kudoh H. 2015. Microbial communities on flower surfaces act as signatures of pollinator visitation. Sci. Rep. 5, 8695 ( 10.1038/srep08695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glasl B, Herndl GJ, Frade PR. 2016. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10, 2280–2292. ( 10.1038/ismej.2016.9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blekhman R, et al. 2015. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16, 191 ( 10.1186/s13059-015-0759-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244. ( 10.1038/nature10571) [DOI] [PubMed] [Google Scholar]
- 15.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. 2016. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 ( 10.1371/journal.pbio.2000225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonseca-García C, Coleman-Derr D, Garrido E, Visel A, Tringe SG, Partida-Martínez LP. 2016. The cacti microbiome: interplay between habitat-filtering and host-specificity. Front. Microbiol. 7, 150 ( 10.3389/fmicb.2016.00150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ainsworth TD, et al. 2015. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. ( 10.1038/ismej.2015.39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bronstein JL, Alarcón R, Geber M. 2006. The evolution of plant–insect mutualisms. New Phytol. 172, 412–428. ( 10.1111/j.1469-8137.2006.01864.x) [DOI] [PubMed] [Google Scholar]
- 19.Horká I, De Grave S, Fransen CH, Petrusek A, Ďuriš Z. 2018. Multiple origins and strong phenotypic convergence in fish-cleaning palaemonid shrimp lineages. Mol. Phylogenet. Evol. 124, 71–81. ( 10.1016/j.ympev.2018.02.006) [DOI] [PubMed] [Google Scholar]
- 20.Titus BM, Daly M, Vondriska C, Hamilton I, Exton DA. 2019. Lack of strategic service provisioning by Pederson's cleaner shrimp (Ancylomenes pedersoni) highlights independent evolution of cleaning behaviors between ocean basins. Sci. Rep. 9, 629 ( 10.1038/s41598-018-37418-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litsios G, Sims CA, Wüest RO, Pearman PB, Zimmermann NE, Salamin N. 2012. Mutualism with sea anemones triggered the adaptive radiation of clownfishes. BMC Evol. Biol. 12, 212 ( 10.1186/1471-2148-12-212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litsios G, Pearman PB, Lanterbecq D, Tolou N, Salamin N. 2014. The radiation of the clownfishes has two geographical replicates. J. Biogeogr. 41, 2140–2149. ( 10.1111/jbi.12370) [DOI] [Google Scholar]
- 23.Litsios G, Salamin N. 2014. Hybridisation and diversification in the adaptive radiation of clownfishes. BMC Evol. Biol. 14, 245 ( 10.1186/s12862-014-0245-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Titus BM, et al. 2019. Phylogenetic relationships of the clownfish-hosting sea anemones. Mol. Phylogenet. Evol. 139, 106526. ( 10.1016/j.ympev.2019.106526) [DOI] [PubMed] [Google Scholar]
- 25.Fautin DC, Allen GR. 1992. Field guide to anemonefishes and their host sea anemones Perth, Australia: Western Australia Museum. [Google Scholar]
- 26.Huebner LK, Dailey B, Titus BM, Khalaf M, Chadwick NE. 2012. Host preference and habitat segregation among Red Sea anemonefish: effects of sea anemone traits and fish life stages. Mar. Ecol. Prog. Ser. 464, 1–15. ( 10.3354/meps09964) [DOI] [Google Scholar]
- 27.Camp EF, Hobbs JPA, De Brauwer M, Dumbrell AJ, Smith DJ. 2016. Cohabitation promotes high diversity of clownfishes in the Coral Triangle. Proc. R. Soc. B 283, 20160277 ( 10.1098/rspb.2016.0277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szczebak JT, Henry RP, Al-Horani FA, Chadwick NE. 2013. Anemonefish oxygenate their anemone hosts at night. J. Exp. Biol. 216, 970–976. ( 10.1242/jeb.075648) [DOI] [PubMed] [Google Scholar]
- 29.Roopin M, Henry RP, Chadwick NE. 2008. Nutrient transfer in a marine mutualism: patterns of ammonia excretion by anemonefish and uptake by giant sea anemones. Mar. Biol. 154, 547–556. ( 10.1007/s00227-008-0948-5) [DOI] [Google Scholar]
- 30.Mebs D. 2009. Chemical biology of the mutualistic relationships of sea anemones with fish and crustaceans. Toxicon 54, 1071–1074. ( 10.1016/j.toxicon.2009.02.027) [DOI] [PubMed] [Google Scholar]
- 31.Pratte ZA, Patin NV, McWhirt ME, Caughman AM, Parris DJ, Stewart FJ. 2018. Association with a sea anemone alters the skin microbiome of clownfish. Coral Reefs 37, 1119–1125. ( 10.1007/s00338-018-01750-z) [DOI] [Google Scholar]
- 32.Thompson LR, et al. 2017. A communal catalogue reveals Earth's multiscale microbial diversity. Nature 551, 457–463. ( 10.1038/nature24621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108, 4516–4522. ( 10.1073/pnas.1000080107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boleyn E, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. ( 10.1038/s41587-019-0209-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 ( 10.1186/gb-2011-12-6-r60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Beiko RG. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. ( 10.1038/nbt.2676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. ( 10.1093/nar/28.1.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McFrederick QS, Thomas JM, Neff JL, Vuong HQ, Russell KA, Hale AR, Mueller UG. 2017. Flowers and wild megachilid bees share microbes. Microb. Ecol. 73, 188–200. ( 10.1007/s00248-016-0838-1) [DOI] [PubMed] [Google Scholar]
- 40.Aylward FO, et al. 2012. Metagenomic and metaproteomic insights into bacterial communities in leaf-cutter ant fungus gardens. ISME J. 6, 1688–1701. ( 10.1038/ismej.2012.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kellner K, Ishak HD, Linksvayer TA, Mueller UG. 2015. Bacterial community composition and diversity in an ancestral ant fungus symbiosis. FEMS Microbiol. Ecol. 91, fiv073 ( 10.1093/femsec/fiv073) [DOI] [PubMed] [Google Scholar]
- 42.Orsi WD, et al. 2016. Diverse, uncultivated bacteria and archaea underlying the cycling of dissolved protein in the ocean. ISME J. 10, 2158–2173. ( 10.1038/ismej.2016.20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roopin M, Chadwick NE. 2009. Benefits to host sea anemones from ammonia contributions of resident anemonefish. J. Exp. Mar. Biol. Ecol. 370, 27–34. ( 10.1016/j.jembe.2008.11.006) [DOI] [Google Scholar]
- 44.Cleveland A, Verde EA, Lee RW. 2011. Nutritional exchange in a tropical tripartite symbiosis: direct evidence for the transfer of nutrients from anemonefish to host anemone and zooxanthellae. Mar. Biol. 158, 589–602. ( 10.1007/s00227-010-1583-5) [DOI] [Google Scholar]
- 45.Matthews JL, Oakley CA, Lutz A, Hillyer KE, Roessner U, Grossman AR, Weis VM, Davy SK. 2018. Partner switching and metabolic flux in a model cnidarian–dinoflagellate symbiosis. Proc. R. Soc. B 285, 20182336 ( 10.1098/rspb.2018.2336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Moghrabi S, Allemand D, Couret JM, Jaubert J. 1995. Fatty acids of the scleractinian coral Galaxea fascicularis: effect of light and feeding. J. Comp. Physiol. B 165, 183–192. ( 10.1007/BF00260809) [DOI] [Google Scholar]
- 47.Jiang PL, Pasaribu B, Chen CS. 2014. Nitrogen-deprivation elevates lipid levels in Symbiodinium spp. by lipid droplet accumulation: morphological and compositional analyses. PLoS ONE 9, e87416 ( 10.1371/journal.pone.0087416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, Meyer E, Grossman AR, Weis VM, Davy SK. 2017. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian–dinoflagellate symbiosis. Proc. Natl Acad. Sci. USA 114, 13 194–13 199. ( 10.1073/pnas.1710733114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lõhelaid H, Teder T, Samel N. 2015. Lipoxygenase-allene oxide synthase pathway in octocoral thermal stress response. Coral Reefs 34, 143–154. ( 10.1007/s00338-014-1238-y) [DOI] [Google Scholar]
- 50.Quinn RA, et al. 2016. Metabolomics of reef benthic interactions reveals a bioactive lipid involved in coral defence. Proc. R. Soc. B 283, 20160469 ( 10.1098/rspb.2016.0469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galtier d'Auriac I, et al. 2018. Before platelets: the production of platelet-activating factor during growth and stress in a basal marine organism. Proc. R. Soc. B 285, 20181307 ( 10.1098/rspb.2018.1307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Titus BM, Laroche R, Rodríguez E, Wirshing H, Meyer CP. 2020. Data from: Host identity and symbiotic association affects the taxonomic and functional diversity of the clownfish-hosting sea anemone microbiome Dryad Digital Repository ( 10.5061/dryad.sbcc2fr28) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Titus BM, Laroche R, Rodríguez E, Wirshing H, Meyer CP. 2020. Data from: Host identity and symbiotic association affects the taxonomic and functional diversity of the clownfish-hosting sea anemone microbiome Dryad Digital Repository ( 10.5061/dryad.sbcc2fr28) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Raw sequence data, barcodes and sample metadata are publicly available through the Dryad Digital Repository: https://doi.org/10.5061/dryad.sbcc2fr28 [52].