Abstract
Background:
Disturbed flow (d-flow)–induced senescence and activation of endothelial cells (ECs) have been suggested to have critical roles in promoting atherosclerosis. Telomeric repeat-binding factor 2 (TERF2)-interacting protein (TERF2IP), a member of the shelterin complex at the telomere, regulates the senescence-associated secretory phenotype (SASP), in which EC activation and senescence are engendered simultaneously by p90RSK-induced phosphorylation of TERF2IP S205 and subsequent nuclear export of the TERF2IP-TERF2 complex. In this study, we investigated TERF2IP-dependent gene expression and its role in regulating d-flow-induced SASP.
Methods:
A principal component analysis and hierarchical clustering were used to identify genes whose expression is regulated by TERF2IP in ECs under d-flow conditions. Senescence was determined by reduced telomere length, increased p53 and p21 expression, and increased apoptosis; EC activation was detected by NF-κB activation and the expression of adhesion molecules. The involvement of TERF2IP S205 phosphorylation in d-flow–induced SASP was assessed by depletion of TERF2IP and mutation of the phosphorylation site.
Results:
Our unbiased transcriptome analysis showed that TERF2IP caused alteration in the expression of a distinct set of genes, including rapamycin-insensitive companion of mTOR (RICTOR) and makorin-1 (MKRN1) ubiquitin E3 ligase, under d-flow conditions. In particular, both depletion of TERF2IP and overexpression of the TERF2IP S205A phosphorylation site mutant in ECs increased the d-flow and p90RSK-induced MKRN1 expression and subsequently inhibited apoptosis, telomere shortening, and NF-κB activation in ECs via suppression of p53, p21, and telomerase (TERT) induction.
Conclusions:
MKRN1 and RICTOR belong to a distinct reciprocal gene set that is both negatively and positively regulated by p90RSK. TERF2IP S205 phosphorylation, a downstream event of p90RSK activation, uniquely inhibits MKRN1 expression and contributes to EC activation and senescence, which are key events for atherogenesis.
Keywords: telomeric repeat binding factor 2-interacting protein (TERF2IP), senescence-associated secretory phenotype (SASP), p90RSK, senescence, inflammation, MKRN1
1. Introduction
Aging is one of the major risk factors for most serious chronic diseases, including cardiovascular disease, cancers, diabetes, metabolic syndromes, and kidney failure [1]. One of the most crucial causes of these prominent age-related disorders is chronic, nonmicrobial inflammation, termed sterile inflammation (indicating no detectable pathogens) or inflammaging [2]. It is well known that senescent cells produce pro-inflammatory molecules such as interleukins 1α 1β, 6, and 8 as they exhibit what is known as the senescence-associated secretory phenotype (SASP)[3]. The contributions of NF-κB, p38MAPK, and mammalian target of rapamycin (mTOR) signaling to induction of SASP factors has been reported[4–6]. p16 and p21 are known to induce senescence cell-cycle arrest but could not induce pro-inflammatory molecules, suggesting that the non-core senescence signaling pathways play a role in SASP induction[7–10]. For example, activation of DNA damage–response kinases ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR) kinases stabilize GATA4 and initiates SASP by activating NF-κB, which is independent of p53 or p16 [11].
The critical role of telomere shortening in endothelial cell (EC) senescence and subsequent development of atherosclerotic plaques, especially under disturbed (as opposed to laminar) blood flow (d-flow), has been well documented[12]. The shelterin complex (telomeric repeat-binding factors 1 and 2 [TERF1, TERF2], TERF2-interacting protein 1 [TERF2IP], POT1, TPP1, and TIN2) of the mammalian telomere[13] is known to protect telomeres, but it also regulates telomere length. We previously reported that, under d-flow, p90RSK directly associated with TERF2IP and phosphorylated the TERF2IP S205 site in ECs[14]. Furthermore, d-flow increased the EC senescence, apoptosis, and activation via p90RSK-mediated TERF2IP S205 phosphorylation. The nuclear TERF2IP-TERF2 complex—in particular, nuclear TERF2—protects telomeres [15], while cytosolic TERF2IP plays a role in NF-κB activation [13, 16, 17]. We found that depletion of TERF2IP and the TERF2IP S205A mutation in ECs inhibited nuclear export of TERF2 and protected telomeres from d-flow–induced shortening. Furthermore, TERF2IP S205 phosphorylation led to EC activation by cytosolic TERF2IP-mediated NF-κB activation. These data suggest that TERF2IP S205 phosphorylation plays a crucial role in both EC activation and senescence. However, it remains unclear how TERF2IP and TERF2IP S205 phosphorylation can transcriptionally regulate EC activation and senescence in concert, especially under d-flow conditions.
The purpose of this study was to investigate the possible role of TERF2IP in regulating gene expression in ECs under d-flow. Since we reported that laminar flow could not increase p90RSK activation, inflammation, and apoptosis[18], we focus on how d-flow can increase SASP via TERF2IP phosphorylation in this study. We used an unbiased transcriptome analysis together with functional effects of TERF2IP depletion and the mutation of the TERF2IP phosphorylation site on TERF2IP-dependent gene expression and SASP in ECs.
2. Materials and Methods
2.1. Antibodies and reagents
The following antibodies and chemicals were acquired: Anti-p16 (orb228122; Biorbyt, San Francisco, CA), anti-MKRN1 (GTX106459; Genetex, Irvine, CA), anti-p90RSK (MAB 2056; R&D Systems, Minneapolis, MN), and anti-α-tubulin (T5168; Sigma-Aldrich, St. Louis, MO) were purchased. Antibodies against TERF2IP (5433) and p21 (2947) were purchased from Cell Signaling Technology (Beverly, MA). Anti-TERT (ab32020) and anti-TERF2 (ab108997) were from Abcam (Cambridge, MA). Protease inhibitor cocktail (p8340), PMSF (36978), and NEM (E3876) were from Sigma-Aldrich.
The iQSYBR Green Supermix (1708882) was from Bio-Rad (Hercules, CA); the TL PNA kit/FITC flow cytometry kit (K5327) was from Agilent Technology (Santa Clara, CA); the FITC Annexin V apoptosis detection kit (556547) was from BD Biosciences (Franklin Lakes, NJ). We also purchased TaqMan reverse transcription reagents to generate cDNA (N808–0234; Applied Biosystems, Foster City, CA), RNeasy Plus Mini Kit (#74136; QIAGEN), a dual luciferase assay kit (E1910; Promega Life Sciences, Fitchburg, WI), and Lipofectamine 2000 transfection reagent (11668027; ThermoFisher Scientific, Waltham, MA).
2.2. siRNA
Short interfering RNA (siRNA-SMARTpool) targeting human TERF2IP (L-021219–00-0010) and MKRN1 (L-006959–00-0005) was purchased from Thermo Scientific Dharmacon. TERF2 siRNA was purchased from Santa Cruz (sc-3805). Non-specific siRNA negative control was purchased from Invitrogen (#12935–112).
2.3. Generation of plasmids and adenoviruses
Plasmids containing rat wild type (WT) p90RSK1 (WT-p90rsk) (Genebank NM031107) was generated as we previously described[19]. pLPC-human TERF2IP full length (#12542) was from Addgene. pCMV-Flag-TERF2IP-WT was obtained by subcloning TERF2IP from pLPC-human TERF2IP full length into the pCMV-Tag2B vector (Agilent Technologies, Santa Clara, CA) at sites recognized by the restriction enzymes EcoRI and XhoI. The pCMV-Flag-TERF2IP-S205A mutant was generated by site-directed mutation of pCMV-Flag-TERF2IP-WT using a QuikChange site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. FLAG-tagged adenoviral vectors containing TERF2IP-WT and - S205A (Ad-Flag-TERF2IP-WT and -S205A) were generated by cloning each corresponding insert from pCMV-Flag-TERF2IP-WT and -S205A into the pENTR1A vector (Life Technologies) at sites recognized by the restriction enzymes KpnI and XhoI, and then a recombinase reaction was performed to get a pDEST-based vector (Invitrogen Gateway LR Clonase II Enzyme mix, #11791100, ThermoFisher, Waltham, MA)) following the manufacturer’s instructions. pACT-TERF2IP (133–191 aa), -TERF2IP (1–282 aa) and -TERF2IP (full length) were obtained by subcloning TERF2IP from pENTR-Flag-TERF2IP-WT into a pACT vector at sites recognized by the restriction enzymes BamHI and XbaI. All constructs were verified by DNA sequencing by using vector specific primers. Where indicated, adenovirus containing β-galactosidase (Ad-LacZ) or green fluorescent protein (Ad-GFP) was used as a control.
2.4. Cell cultures
Human umbilical vein ECs (HUVECs) were obtained from collagenase-digested umbilical cord veins[20]. Human aortic endothelial cells (HAECs) were isolated from aortic explants of heart transplant donors of anonymous origin through the UCLA transplant program as previously described [14, 21]. No individual information, including ethnicity, history, and disease status of donors is known[22]. HUVECs and HAECs were cultured in Petri dishes or flasks coated with 0.2% gelatin type A (#901771; MP Biomedicals, Santa Ana, CA), in Endothelial Cell Medium (ECM, #1001, ScienCell, Carlsbad, CA) containing 465 mL of basal medium, 25 mL of FBS (#0025, ScienCell), 5 mL of Endothelial Cell Growth Supplement (ECGS, #1052, ScienCell) and 5 mL of penicillin/streptomycin solution (P/S, #0503, ScienCell). HUVECs were used in experiments between 3 and 7 passages as described previously[14]. HAECs were less than 15 passages. Mouse lung endothelial cells (MLECs) and mouse aortic endothelial cells (MAoECs) were isolated as described previously[19]. Cells were characterized by anti-CD31 and used for experiments within 2–3 passages as previously described[14].
2.5. Disturbed flow
We performed disturbed flow experiments using cones with radial grooves as we reported previously[14, 23]. In brief, we used cones with radial grooves that were 1-mm deep. We showed tracks of fluorescent beads suspended in culture media when grooved and nongrooved cones were rotated at the same speed. Although the non-grooved cone created straight unidirectional tracks indicating steady laminar flow, tracks made by the grooved cone were short and not oriented in the same direction, indicating non-laminar (turbulent) movement of the media in the dish. Furthermore, we confirmed the shape of ECs under grooved cone were polygonal, also supporting that this system can recapitulate the flow condition observed in the proatherogenic area of blood vessels.
2.6. Transfection and transduction
As described previously[14], ECs were transfected with siRNAs at a final concentration of 100 nM, reporter plasmids, or plasmid DNAs using Lipofectamine 2000 transfection reagent, according to the manufacturer’s instructions. After transfection, cells were allowed to recover in complete medium for 24–48 h. For adenovirus transduction, we used 20 multiplicities of infection. Prior to use, cells were cultured overnight in a low-serum (1% FBS) medium.
2.7. Gene expression profiling
Total RNA was obtained using RNeasy columns (QIAGEN) with DNA digest, according to the manufacturer’s instructions, and hybridized to GeneChip Human Transcriptome Array 2.0 (Affymetrix, Santa Clara, CA, USA). The array data were incorporated into the Transcriptome Analysis Console 3.0 software program (Affymetrix); GeneChip data were normalized using the Tukey biweight average algorithm and are represented as a biweight average on a log2 scale. Significant differences between two groups were identified using an unpaired analysis of variance (ANOVA; p<0.05) with log2-transformed normalized intensities, and transcripts with more than a 2-fold difference in expression and a p-value <0.05 were selected for each specific comparison analyzed. A principal component analysis and differential profile (heatmap) of the d-flow responses in TERF2IP-depleted cells were generated by using Qlucore Omics Explorer version 3.2. The principal component analysis was performed at a stringent p = 3.3e-5 using ANOVA on the basis of TERF2IP depletion (via siRNA) and d-flow treatment.
2.8. Mice
Mice were maintained in the vivaria of the Texas A&M Institute of Bioscience and Technology and of The University of Texas MD Anderson Cancer Center (both, Houston, Texas). All animal procedures were approved by the University Committee on Animal Resources of the Texas A&M Institute of Bioscience and Technology (2014–0231, 2017–0154) and by the Institutional Animal Care and Use Committee at MD Anderson (00001652, 00001109). All animals were housed in temperature-controlled rooms under a 12-h light/dark cycle and pathogen-free conditions.
EC-specific TERF2IP knockout mice were generated using mice expressing Cre-recombinase under the regulation of VE-cadherin promoter (C57BL/6 background), as described previously[19]. These mice were crossed with B6;129-Terf2iptm1.1Tdl/J (Stock #012346; Jackson Laboratory, Bar Harbor, ME, USA). Genotyping of pups was performed by PCR of tail clipping. DNA was extracted with a Quick-DNA miniprep plus kit (D4068; Zymo Research, Irvine, CA, USA) as per product protocol, and PCR was performed with Promega GoTaq Ggreen 2x Master Mix (M712; Promega, Fitchburg, WI, USA) as per product protocol.
2.9. Real-time PCR
At the end of experiments, ECs were washed three times with phosphate-buffered saline (PBS) and subjected to lysis in RLT Plus RNeasy lysis buffer (#74136; QIAGEN, Germantown, MD, USA). The resulting cell lysates were loaded onto a QIAshredder column (#79656; QIAGEN) and spun down to collect the eluted lysates. Total RNA was then isolated from this lysate using an RNeasy Plus Mini Kit (#74136; QIAGEN) according to the manufacturer’s instructions. cDNA was reverse-transcribed from a 50-μL reaction mixture containing 1 μg of purified RNA, 5 μL of 10X buffer, 11 μL of MgCl2, 10 μL of dNTPs, 2.5 μL of a random hexamer, 1.25 μL of oligo-dT, 1 μL of RNase inhibitor, and 0.75 μL of a reverse-transcriptase enzyme using the TaqMan Reverse Transcription Reagents (#N808–0234; made for Applied Biosystems by Roche Molecular Diagnostics, Pleasanton, CA, USA). First-strand cDNA was reverse-transcribed from total RNA by incubating reaction mixtures at 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 95°C for 5 min before soaking at 4°C in a PCR cycler. Target cDNA levels were quantified using a CFX Connect Real-Time System (Bio-Rad, Hercules, CA, USA). Each reaction mixture (10 μL) contained cDNA synthesized from 20 ng of total RNA, 5 μL of iQ SYBR Green Supermix (#1708882; Bio-Rad), and 0.5 μmol/L each of forward and reverse primers (Supplementary Table 2). RT-PCR was carried out using the MyiQTM2 Two-Color Real Time PCR System (Bio-Rad) and iQ SYBR SYBR Green (Bio-Rad) for 10 min at 95°C (thermal activation) and 40 cycles of PCR (melting for 15 s at 95°C, followed by annealing/extension for 1 min at 60°C). The ΔΔCt method was used to calculate fold-changes in expression of target RNAs: ΔCt = Ct (target gene) - Ct (housekeeping gene, GAPDH), ΔΔCt = ΔCt (treatment) - ΔCt (control), and fold-change = 2(−ΔΔCt).
2.10. Flow cytometric analyses
Following various treatments (as indicated in figures), cells were washed three times with PBS, harvested using 10 mM ethylenediaminetetraacetic acid (pH 8.0) solution at room temperature, and stained for apoptotic marker Annexin-V using Annexin V-FITC Apoptosis Detection Reagent (#ab14082; Abcam, Cambridge, MA, USA) as per the manufacturer’s instructions. Briefly, cell pellets were re-suspended in 1X Annexin V Binding Buffer and baseline measurements were taken using unstained control cells. Then, cells were stained with Annexin V-FITC (#ab14083; Abcam) at room temperature for 5 min in the dark. Measurements for all samples were carried out using an Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA). For each analysis, 10,000 cells were counted based on forward and side scatter characteristics. Results were analyzed using FlowJo software (version 10.5.0; FlowJo LLC, Ashland, OR, USA).
2.11. Telomere length assay
Telomere length was measured using the telomere PNA kit with FITC (DAKO, Glostrup, Denmark) per the manufacturer’s instructions as described previously[14].
2.12. NF-κB activity assay
NF-κB activity was measured by using a luciferase assay with a reporter gene containing five NF-κB–binding sites as an enhancer (pLuc-MCS with five repeated NF-κB–binding sites [TGGGGACTTTCCGC]; Stratagene, La Jolla, CA, USA) as we described previously[14]. Relative NF-κB activity was calculated by normalizing firefly luciferase activity to renilla luciferase activity (firefly:renilla luciferase activity ratio).
2.13. MTT assay
Cell viability was assessed using the MTT colorimetric assay (Vybrant MTT assay kit, Cat#V13154, ThermoFisher, Waltham, MA) per the manufacturer’s instructions.
2.14. Statistical analysis
Data are presented as the mean ± standard deviation (SD). Differences between two independent groups were determined by the Student t-test (two-tailed) or, when applicable, one-way ANOVA, followed by Bonferroni post hoc testing for multiple group comparisons, using Prism software (version 5.0; GraphPad Software, La Jolla, CA, USA). When groups exhibited unequal variances, the Welch ANOVA was used to perform multiple group comparisons. P-values <0.05 were considered statistically significant.
2.15. Power analysis
We performed the sample size calculation based on our previous results[24, 25]. We expected that 1) the variation (standard deviation) of annexin V (Fig. 3A, 5A) and NF-κB activity (Fig. 3C, 5C) in each group will be about 30% from the mean value, 2) 110% changes will be detected among different groups, and 3) the variation within each group will be similar. Therefore, the optimal number of samples needed to attain statistical significance of p<0.01 with a 90% probability is 6 for each group. While, based on our previous results [24, 25], we expected that 1) the variation (standard deviation) of mRNA (Fig. 1), protein expression (Fig. 2, 4, 6A and B), TL length (Fig. 3B and 5B) in each group will be about 30% from the mean value, 2) 200% changes will be detected among different groups, and 3) the variation within each group will be similar. Therefore, the optimal number of samples needed to attain statistical significance of p<0.01 with a 90% probability is 3 for each group (based on the power calculator generated by the Laboratory Animal Services Centre at The Chinese University of Hong Kong:(http://www.lasec.cuhk.edu.hk/sample-size-calculation.html).
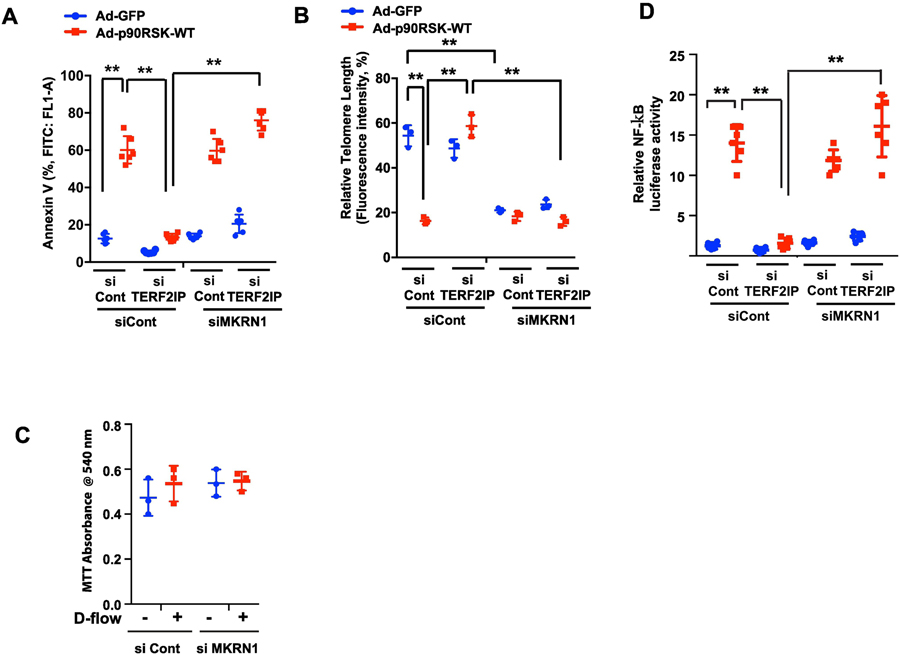
Figure 3. Reduction of apoptosis, telomere shortening, and NF-κB activation mediated by TERF2IP depletion are dependent on MKRN1 under d-flow.
(A, B) HUVECs were transfected with TERF2IP, MKRN1, or control siRNA in combinations as shown for 48 h and transduced with Ad-GFP or Ad-p90RSK-WT for 12 h. The cells were then assayed for apoptosis by quantifying FITC annexin V-positive cells (A). Telomere lengths were measured as described in the Methods (B). Data represent mean ± SD (n = 3–6). Cell viability was assessed by the MTT colorimetric assay as described in the Methods (C). Data present mean ± SD (n = 3). (D) HUVECs were transfected with TERF2IP, MKRN1, or control siRNA in combinations as indicated together with NF-κB reporter gene for 48 h and transduced with Ad-GFP or Ad-p90RSK-WT for 12 h. NF-κB activity was assayed by the dual-luciferase reporter assay. Data represent mean ± SD (n = 6). **p <0.01. All data were analyzed by one-way ANOVA followed by the Bonferroni post hoc test.
Figure 5. Reduction of apoptosis, telomere shortening, and NF-κB activation mediated by TERF2IP S205A mutant overexpression are dependent on MKRN1 under d-flow.
(A, B) HUVECs were transfected with MKRN1 or control siRNA for 48 h and transduced with Ad-GFP or Ad-TERF2IP S205A for 12 h. The cells were then assayed for apoptosis by quantifying FITC annexin V-positive cells (A). Telomere lengths were measured as described in the Methods (B). Data represent mean ± SD (n = 3–6). (C) HUVECs were transfected with MKRN1 or control siRNA and also with NF-κB reporter gene for 48 h and transduced with Ad-GFP or Ad-TERF2IP S205A for 12 h. NF-κB activity was assayed by the dual-luciferase reporter assay. Data represent mean ± SD (n = 6). **p <0.01. All data were analyzed by one-way ANOVA followed by the Bonferroni post hoc test.
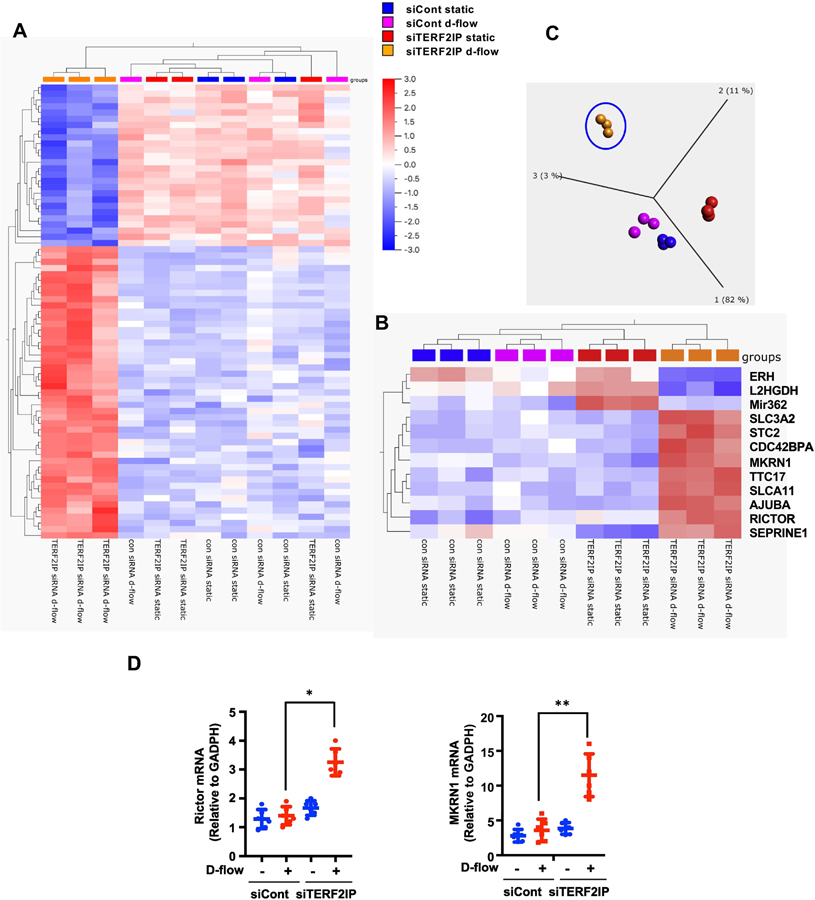
Figure 1. TERF2IP-mediated changes in gene and miRNA expression in human aortic endothelial cells (HAECs) under d-flow conditions.
(A, B) Gene and miRNA expression profiles regulated by TERF2IP and d-flow were determined by hierarchical clustering with p = 6.6e-5 (A) or p = 3.3e-5 (B). The gene abbreviations are defined in Supplementary Table 1. (C) The principal component analysis, based on the conditions of TERF2IP depletion by siRNA and d-flow, was performed at a stringent p = 3.3e-5. (D) HAECs were transfected by incubation with control siRNA or TERF2IP siRNA for 48 h and subjected to d-flow for 16 h. Expression of RICTOR and MKRN1 mRNAs was determined by quantitative real-time PCR. Data represent mean ± SD (n = 4; **p<0.01, *p<0.05. All data were analyzed by one-way ANOVA followed by the Bonferroni post hoc test.
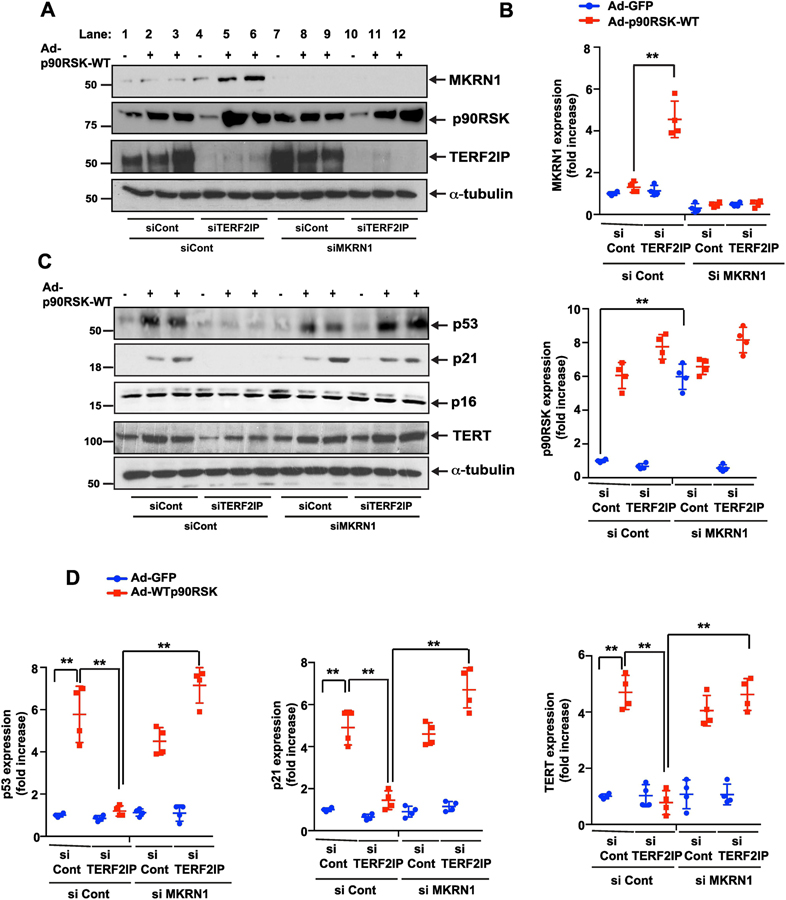
Figure 2. Depletion of TERF2IP increases MKRN1 expression and inhibits the expression of p53, p16, and TERT.
Since we found no difference of d-flow-induced SASP induction between HAECs and HUVECs, we used HUVECs to study the mechanisms of d-flow-induced SASP in Fig.2–5. HUVECs were transfected with control, TERF2IP, or MKRN1 siRNA in combinations as shown for 48 h and transduced with Ad-GFP or Ad-p90RSK-WT for 12 h. Western blotting was performed with specific antibodies as indicated (A, C). The graphs represent densitometry data from immunoblots. Mean ± SD (n = 3) (B, D). **p<0.01.
Figure 4. Overexpression of TERF2IP S205A mutant increases MKRN1 expression and inhibits the expression of p53, p16, and TERT.
HUVECs were transfected with control siRNA or MKRN1 siRNA for 48 h and transduced with Ad-GFP or Ad-TERF2IP S205A for 12 h. Expression of indicated proteins was detected by Western blotting performed using specific antibodies (A, C). Graphs represent densitometry data from immunoblots. Data represent mean ± SD (n = 3) (B, D). **p <0.01.
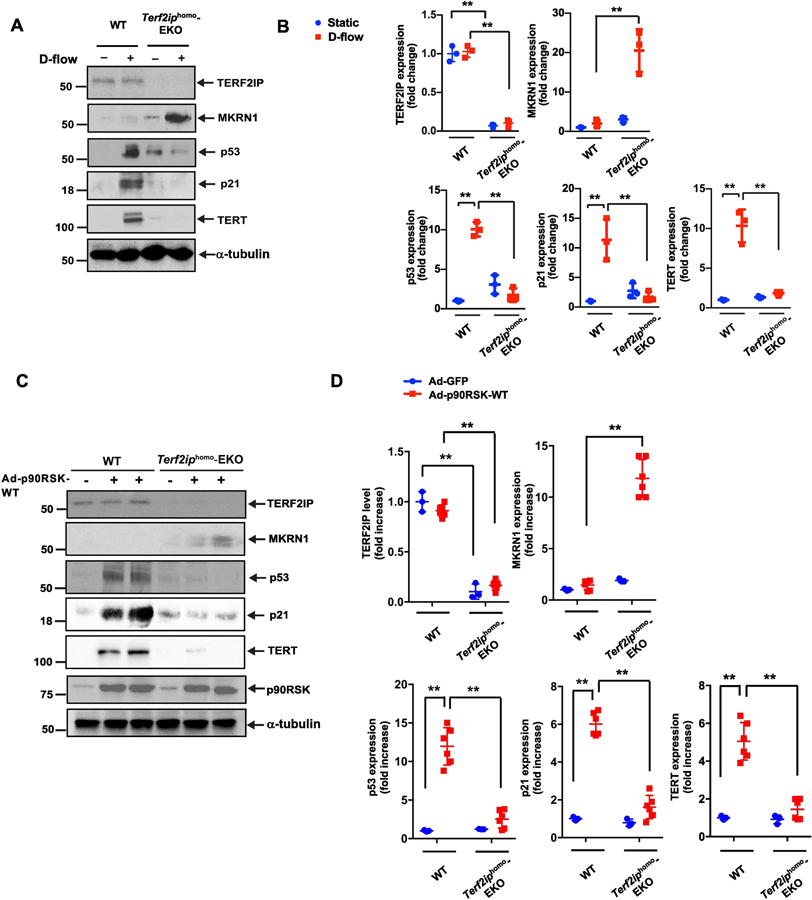
Figure 6. Depletion of TERF2IP inhibits both senescence and activation induced by d-flow and overexpression of wild-type p90RSK.
(A, B) Mouse lung endothelial cells (MLECs) were isolated from wild-type (WT) and Terf2iphomo-EC-specific knockout (EKO) mice. After cells were treated with d-flow (+) or no flow (i.e. static culture) (−) conditions for 12 h, protein expression was analyzed by Western blotting using specific antibodies as indicated (A) and the levels of expression of each protein under different conditions were quantified (B). Data represent mean ± SD (n = 3). (C, D) MLECs isolated from WT and Terf2iphomo-EKO mice were transduced with Ad -p90RSK-WT (+) or Ad-GFP (−) as a control. After 12 h, Western blotting was performed using specific antibodies as indicated (C). Graphs represent densitometry data of immunoblots from three independent experiments (D). Data represent mean ± SD (n = 3–6). **p <0.01. All data were analyzed by one-way ANOVA followed by the Bonferroni post hoc test.
2.16. Data availability
The microarray data are available in a public repository from the NCBI website (GEO accession number GSE119407). The authors declare that all of the other data supporting the findings of this study are available in the article, in the Supplementary Information files, or from the corresponding author upon request.
3. Results
3.1. D-flow activates a distinct gene and miRNA expression program in TERF2IP-depleted ECs
The shelterin complex not only protects chromosome ends from degradation and DNA damage but also can function outside the telomere region and regulate transcription of various genes[17, 26]. In particular, TERF2IP regulates metabolism- and immunity-related gene networks[17, 27]. Therefore, to investigate gene expression profiles regulated by TERF2IP in ECs under d-flow conditions, we used the principal component analysis and hierarchical clustering in four groups of human aortic endothelial cells (HAECs) treated in static or d-flow conditions and with or without TERF2IP knockout by siRNA (Fig. 1). We found a greater degree of gene clustering in TERF2IP-depleted ECs under d-flow conditions than in any other treatment group (p = 6.6e-5 and q = 0.029, variance = 0.0034). Forty-seven genes and miRNAs were differentially expressed, of which 26 were downregulated (Fig. 1A). At a more stringent filtering condition (p = 3.3e-5 and q = 0.088, variance = 0.0188), the principal component analysis plot shows that the TERF2IP depleted cells under d-flow conditions revealed changes in distinct sets of genes and miRNAs (Fig. 1B): seven genes were upregulated and one miRNA (miR-362) was downregulated compared to the expression levels in the other three groups (Fig. 1C). It has been reported that MKRN1 and RICTOR can regulate EC proliferation and apoptosis (40–43). Our results showing that the mRNA levels of MKRN1 and RICTOR were significantly increased only in TERF2IP deleted HAECs under d-flow can be consistent with those of the previous studies (Fig. 1D), which revealed the critical role of d-flow-induced TERF2IP S205 phosphortylation in regulating EC proliferation and apoptosis[14].
3.2. Phosphorylated TERF2IP augments p53, p21, and TERT expression by inhibiting p90RSK-induced MKRN1 expression
We investigated the functional role of MKRN1, a member of the E3 ubiquitin-protein ligase family, in p90RSK-mediated TERF2IP signaling. Only the combination of p90RSK overexpression and TERF2IP depletion, and not either alone, increased MKRN1 expression (Fig. 2A, lanes 1–6, and 2B). Since MKRN1 has been reported to be an E3 ubiquitin ligase for p53, p21, and telomerase reverse transcriptase (TERT) [28–30], we first investigated the role of TERF2IP in regulating expression of p53, p21, and p16, another cellular senescence gene, and TERT. Expression of p53, p21, and TERT, but not p16, was significantly increased in ECs in which p90RSK was overexpressed (Fig. 2A, C, lane 2–3), and this increased expression was abolished by the depletion of TERF2IP (Fig. 2C, lanes 5–6, and 2D).
Since TERF2IP depletion and p90RSK overexpression increased MKRN1 expression (Fig. 2A, lanes 5 and 6) and since MKRN1 can cause p53, p21, and TERT degradation[28–30], we investigated the role of MKRN1 in p90RSK-induced p21, p53, and TERT expression. Since p90RSK overexpression alone did not increase MKRN1 expression (Fig. 2A, lanes 1–3), MKRN1 depletion had no effect on p21, p53, and TERT expression (Fig. 2A, 2C, lanes 7–9). In contrast, MKRN1 depletion in the setting of p90RSK overexpression and TERF2IP depletion completely reversed the depletion of the TERF2IP-mediated inhibitory effect in p90RSK-induced p21, p53, and TERT expression (Fig. 2C, lanes 4–6 and 10–12, 2D). The roles of p21 and p53 in apoptosis and senescence have been well established, and TERF2IP depletion abolished p90RSK-induced EC apoptosis and telomere shortening (Fig. 3A, 3B). Of note, under these conditions, we could not find any changes in terms of cell viability under the condition of d-flow and MKRN1 depletion, suggesting that d-flow induces apoptosis and TL shortening, but not necrosis, under this condition (Fig. 3C). MKRN1 depletion played no role in p90RSK-induced apoptosis and telomere shortening but completely diminished the protective effect of TERF2IP depletion by siRNA. Interestingly, depletion of MKRN1 by siRNA induced telomere shortening but did not increase apoptosis, suggesting that telomere shortening alone is not sufficient to induce EC apoptosis.
As shown in Figs. 2 and 3, p90RSK activation induced both TERT expression and telomere shortening at the same time. TERT can prolong telomere length[31], but we found telomere shortening rather than elongation under p90RSK activation. In p90RSK signaling, therefore, TERT may not be the major factor for inducing telomere shortening. The critical role of TERT in regulating NF-κB activation has been reported[32]. In addition, we showed the key role of both TERF2IP and TERF2IP S205 phosphorylation in p90RSK-induced EC activation[14]. Therefore, we investigated NF-κB activation and found that TERF2IP depletion inhibited p90RSK-induced NF-κB activation, which was completely diminished by the reduction of MKRN1 expression by MKRN1 siRNA transfection (Fig. 3D).
Next, we examined the role of TERF2IP S205 phosphorylation in p90RSK-mediated MKRN1 expression and also expression of MKRN1 substrates, including p53, p21, and TERT. MKRN1 expression was significantly increased in HUVECs co-transduced with Ad-p90RSK and Ad-TERF2IP S205A (Fig. 4A, top, lanes 4–6). We confirmed that TERF2IP S205 phosphorylation was significantly inhibited in cells transduced with Ad-TERF2IP S205A (Fig. 4A, lanes 4–6), supporting the critical role of TERF2IP S205 phosphorylation in p90RSK-mediated inhibition of MKRN1 expression. We also determined the contribution of MKRN1 in TERF2IP S205A mutant-mediated inhibition of p53, p21, and TERT expression (Fig. 4A, 4C, lanes 4–6 and 10–12, Fig. 4B, and Fig. 4D) and found that MKRN1 had a crucial role in TERF2IP S205 phosphorylation-induced senescence and activation (Fig. 5A–C). Taken together, these data suggest that MKRN1 has a key athero-protective role by simultaneously inhibiting both senescence and activation of ECs and that this protection is mediated by the abatement of TERF2IP and TERF2IP S205 phosphorylation. Furthermore, MKRN1 and RICTOR belong to a distinct reciprocal gene set that is regulated by p90RSK. Although p90RSK regulates this yin-yang gene set both negatively and positively, TERF2IP S205 phosphorylation, which is a downstream event of p90RSK activation, uniquely inhibits MKRN1 expression.
3.3. TERF2IP simultaneously regulates EC activation and senescence
To specifically study the role of TERF2IP in ECs in vivo, we generated EC-specific heterozygous and homozygous Terf2ip-knockout mice. These mice were developed from mice that expressed Cre-recombinase under the regulation of the vascular endothelial-cadherin (VE-Cad) promoter[33] (VE-Cad-Cre Tg [Terf2ipfl/+ or Terf2ipfl/fl], herein referred to as Terf2iphet-EC-specific knockout [EKO] or Terf2iphomo-EKO, respectively). As controls, we used non-transgenic littermates: VE-Cad-Cre+/–Tg [Terf2ip+/+] or VE-Cad-Cre–/–Tg[Terf2ipfl/+ or Terf2ipfl/fl]). First, we established primary cultures of mouse lung endothelial cells (MLECs) and mouse aortic endothelial cells (MAoECs) from Terf2iphomo-EKO mice and exposed them to d-flow. EC activation (NF-κB activation, and TERT) and EC senescence (telomere shortening, apoptosis, and expression of p53 and p21) induced by d-flow (Fig. 6A, 6B, MLECs) or by overexpression of p90RSK (Fig. 6C and 6D, MLECs; Fig. 7A, 7B, MAoECs) were all attenuated in ECs isolated from Terf2iphomo-EKO mice. These attenuations were accompanied by MKRN1 induction (Figs. 6 and 7).
Figure 7. Both senescence and expression of inflammation-related molecules induced by overexpression of wild-type p90RSK are inhibited by depletion of TERF2IP.
(A, B) Mouse aortic endothelial cells (MAoECs) were isolated from WT and Terf2iphomo-EC-specific knockout (EKO) mice. After transduction with Ad-p90RSK-WT (+) or Ad-GFP (−) as a control for 12 h, expression of indicated proteins was analyzed by Western blotting using specific antibodies (A). Graphs represent densitometry data from immunoblots for selected proteins (B). Data represent mean ± SD (n = 3). **p <0.01. All data were analyzed by one-way ANOVA followed by the Bonferroni post hoc test. (C) D-flow–induced p90RSK activation simultaneously induces EC senescence and activation via TERF2IP S205 phosphorylation. TERF2IP S205 phosphorylation induces nuclear export of the TERF2IP-TRF2 complex, leading to senescence induced by telomere shortening/dysfunction and EC activation (i.e., NF-κB activation) induced by TERF2IP-IkB kinase binding in the cytosol. Telomere dysfunction or dislocation of the TERF2IP-TRF2 complex from telomeres precipitates trans-repression of the distinct reciprocal gene set, including MKRN1, enhancing EC senescence and activation under d-flow conditions.
4. Discussion
We previously reported that TERF2IP has a crucial role in concurrently regulating EC activation/inflammation including TNF-α expression, and senescence, which are induced by phosphorylation of TERF2IP at S205, instigating atherosclerotic plaque formation[14]. In this study, we found that TERF2IP S205 phosphorylation is also involved in regulation of a unique set of genes, including MKRN1 and RICTOR. In particular, downregulation of MKRN1 expression by TERF2IP S205 phosphorylation promotes both senescence and inflammation in ECs by upregulating the expression of p21, p53, and TERT. These results suggest the crucial role of TERF2IP S205 phosphorylation in d-flow–induced SASP, which leads to plaque formation.
When ECs with depleted TERF2IP were exposed to d-flow, the expression of a certain gene set and miRNAs was affected compared with ECs treated under three different control conditions (Fig. 1). With the stringent filtered condition at p = 3.3e-5, seven genes (AJUBA, SLC7A11, SLC3A2, STC2, CDC42BPA, MKRN1, and RICTOR) were upregulated and one miRNA (mir362) was downregulated compared with their expression levels in the control groups. There are several possible mechanisms for this distinct transcriptional pattern. First, TERF2IP and TRF2 that are not associated with telomeres may bind non-telomeric sites of genomic DNA, such as the satellite III repeat sequence-enriched region, and regulate the transcription of the genes to which they are bound as transcription factors[34, 35]. It has been suggested that telomere dysfunction itself can be coupled to transcriptional regulation. For example, telomere shortening induced by extra-nuclear translocation of the TERF2IP-TRF2 complex can regulate sub-telomere heterochromatin[34] or disturb the telomere terminal loop and change the expression of sub-telomere or distantly located genes, respectively[26]. In our study, we found trans-repression of seven genes, suggesting that the TERF2IP-TRF2 complex plays a role not only as a transcriptional activator but also as a repressor. Further investigation is necessary to determine the exact mechanism of this trans-repression, but considering the results of our current study and previous reports[26] together, it is clear that the shelterin complex and telomere shortening itself can regulate a distinct transcriptional program and simultaneously control senescence and other physiological events, such as inflammation, by regulating the intra-cellular localization of the shelterin complex.
MKRN1is a transcriptional co-regulator and a ubiquitin E3 ligase. It can destabilize p53, p21, and TERT through ubiquitination and eventually reduce their expression by proteasome-dependent degradation. In addition, MKRN1 can act as an RNA-binding protein to control apoptosis, transcript transport, and translation. Ghosh et al. reported that TERT directly regulates NF-κB transcriptional activity by binding to the NF-κB p65 subunit[36, 37]. Since we observed that depletion of MKRN1 in ECs significantly increased TERT expression, we investigated NF-κB activation and found that TERF2IP siRNA inhibited p90RSK-induced NF-κB activation and that this inhibition was completely reversed by the depletion of MKRN1. Together, these results suggestthat TERF2IP-MKRN1-TERT signaling has a crucial role in regulating p90RSK-mediated NF-κB activation in ECs. Notably, we found that TERT expression was decreased by the depletion of TERF2IP in ECs exposed to d-flow, but the telomere length was restored by TERF2IP siRNA treatment (Fig. 3B). In addition, the TERF2IP S205A mutation inhibited the d-flow–induced NF-κB activation and TERT expression by increasing MKRN1 expression (Fig. 5). These results suggest that TERF2IP-MKRN1 signaling has a key role in regulating NF-κB via inhibition of TERT expression and may have a more significant effect on controlling the telomere length than TERT in fully differentiated ECs, as previously suggested[38].
Telomere shortening can induce p53, p21, and p16 expression and cause replicative senescence [39]. In our study, overexpression of p90RSK induced p53 and p21, but not p16, expression. This may be due to the fact that both p53 and p21, but p16 was not reported as a substrate of MKRN1[29]. Taken together, these findings suggest that phosphorylated TERF2IP–mediated reduction of MKRN1 expression has a crucial role in simultaneously upregulating both activation and senescence in ECs exposed to d-flow. As shown in Fig. 7, p90RSK activation can positively and negatively regulate a distinct reciprocal gene set, including Mkrn1 and Rictor. Although phosphorylation of TERF2IP S205 acts as a negative regulator of p90RSK, it remains unclear how p90RSK activation can positively regulate this yin-yang–regulated gene set (Fig. 7C).
In summary, we have provided evidence that TERF2IP S205 phosphorylation regulates a distinct set of genes, including MKRN1. We showed that the reduction of MKRN1 expression induced by TERF2IP S205 phosphorylation is critical for d-flow–induced SASP. We suggest that the balance between TERF2IP-dependent inhibition and TERF2IP-independent increase of MKRN1 expression regulated by d-flow determines how d-flow induces EC activation and senescence in concert (Fig. 7C). Future studies are necessary to determine how nuclear export of TERF2IP-TRF2 or TERF2IP S205 phosphorylation contributes to the regulation of the distinct gene set regulating SASP.
Supplementary Material
Highlights.
Disturbed flow induces senescence-associated secretory phenotype via TERF2IP.
TERF2IP S205 phosphorylation is crucial for regulating EC senescence and activation.
TERF2IP provokes trans-repression of the distinct reciprocal gene set.
MKRN1 is a key molecule for the inhibition of TERF2IP-dependent SASP.
Acknowledgments
We thank the Department of Scientific Publications at MD Anderson Cancer Center for editing the article. This work was supported by the NIH/NCI through the MD Anderson Cancer Center Support Grant (P30CA016672), which supports the Sequencing and Microarray Facility, and by funding from the NIH to J.A. (HL-130193, HL-123346, and HL-118462) and N.-T.L. (HL-134740).
Abbreviations:
- ANOVA
analysis of variance
- d-flow
disturbed flow
- EC
endothelial cell
- EKO
EC-specific knockout
- FITC
fluorescein isothiocyanate
- HAECs
human aortic ECs
- HUVEC
human umbilical vein EC
- MKRN1
makorin-1 E3 ubiquitin-protein ligase
- miRNA
microRNA
- p90RSK
p90 ribosomal S6 kinase
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RICTOR
rapamycin-insensitive companion of mTOR
- SASP
senescence-associated secretory phenotype
- TERF2
telomeric repeat-binding factor 2
- TERF2IP
TERF2-interacting protein 1
- TERT
telomerase reverse transcriptase
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests
None.
References
- [1].Aging Campisi J., cellular senescence, and cancer. Annu Rev Physiol 2013;75:685–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yu HT, Park S, Shin EC, Lee WW. T cell senescence and cardiovascular diseases. Clin Exp Med 2016;16:257–63. [DOI] [PubMed] [Google Scholar]
- [3].Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med 2015;21:1424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Salminen A, Kauppinen A, Kaarniranta K. Emerging role of NF-kappaB signaling in the induction of senescence-associated secretory phenotype (SASP). Cell Signal 2012;24:835–45. [DOI] [PubMed] [Google Scholar]
- [5].Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J 2011;30:1536–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Newgard CB, Sharpless NE. Coming of age: molecular drivers of aging and therapeutic opportunities. J Clin Invest 2013;123:946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 2008;6:2853–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 2009;11:973–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Coppe JP, Rodier F, Patil CK, Freund A, Desprez PY, Campisi J. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J Biol Chem 2011;286:36396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Watanabe S, Kawamoto S, Ohtani N, Hara E. Impact of senescence-associated secretory phenotype and its potential as a therapeutic target for senescence-associated diseases. Cancer Sci 2017;108:563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Heo KS, Chang E, Le NT, Cushman H, Yeh ET, Fujiwara K, et al. De-SUMOylation enzyme of sentrin/SUMO-specific protease 2 regulates disturbed flow-induced SUMOylation of ERK5 and p53 that leads to endothelial dysfunction and atherosclerosis. Circ Res 2013;112:911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Martinez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nature reviews Cancer. 2011;11:161–76. [DOI] [PubMed] [Google Scholar]
- [14].Kotla S, Vu HT, Ko KA, Wang Y, Imanishi M, Heo KS, et al. Endothelial senescence is induced by phosphorylation and nuclear export of telomeric repeat binding factor 2-interacting protein (TERF2IP) JCI Insight. 2019;in press. [DOI] [PMC free article] [PubMed]
- [15].Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 2011;18:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kabir S, Sfeir A, de Lange T. Taking apart Rap1: an adaptor protein with telomeric and non-telomeric functions. Cell Cycle. 2010;9:4061–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nature cell biology. 2010;12:758–67. [DOI] [PubMed] [Google Scholar]
- [18].Heo KS, Le NT, Cushman HJ, Giancursio CJ, Chang E, Woo CH, et al. Disturbed flow-activated p90RSK kinase accelerates atherosclerosis by inhibiting SENP2 function. J Clin Invest 2015;125:1299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Le NT, Heo KS, Takei Y, Lee H, Woo CH, Chang E, et al. A Crucial Role for p90RSK-Mediated Reduction of ERK5 Transcriptional Activity in Endothelial Dysfunction and Atherosclerosis. Circulation. 2013;127:486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Takahashi M, Berk BC. Mitogen-activated protein kinase (ERK1/2) activation by shear stress and adhesion in endothelial cells. Essential role for a herbimycin-sensitive kinase. J Clin Invest 1996;98:2623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Navab M, Hough GP, Stevenson LW, Drinkwater DC, Laks H, Fogelman AM. Monocyte migration into the subendothelial space of a coculture of adult human aortic endothelial and smooth muscle cells. J Clin Invest 1988;82:1853–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Romanoski CE, Lee S, Kim MJ, Ingram-Drake L, Plaisier CL, Yordanova R, et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet 2010;86:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Heo KS, Lee H, Nigro P, Thomas T, Le NT, Chang E, et al. PKCzeta mediates disturbed flow-induced endothelial apoptosis via p53 SUMOylation. J Cell Biol 2011;193:867–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh MV, Kotla S, Le NT, Ae Ko K, Heo KS, Wang Y, et al. Senescent Phenotype Induced by p90RSK-NRF2 Signaling Sensitizes Monocytes and Macrophages to Oxidative Stress in HIV-Positive Individuals. Circulation. 2019;139:1199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kotla S, Vu HT, Ko KA, Wang Y, Imanishi M, Heo KS, et al. Endothelial senescence is induced by phosphorylation and nuclear export of telomeric repeat binding factor 2-interacting protein. JCI Insight 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ye J, Renault VM, Jamet K, Gilson E. Transcriptional outcome of telomere signalling. Nat Rev Genet 2014;15:491–503. [DOI] [PubMed] [Google Scholar]
- [27].Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, et al. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nature cell biology. 2010;12:768–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ko A, Shin JY, Seo J, Lee KD, Lee EW, Lee MS, et al. Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J Natl Cancer Inst 2012;104:1660–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, et al. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J 2009;28:2100–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Salvatico J, Kim JH, Chung IK, Muller MT. Differentiation linked regulation of telomerase activity by Makorin-1. Mol Cell Biochem 2010;342:241–50. [DOI] [PubMed] [Google Scholar]
- [31].Donate LE, Blasco MA. Telomeres in cancer and ageing. Philos Trans R Soc Lond B Biol Sci 2011;366:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wu XQ, Yang Y, Li WX, Cheng YH, Li XF, Huang C, et al. Telomerase reverse transcriptase acts in a feedback loop with NF-kappaB pathway to regulate macrophage polarization in alcoholic liver disease. Sci Rep 2016;6:18685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Heo K, Le N, Cushman HJ, Giancursio CJ, Chang E, Woo CH, et al. Disturbed flow-activated p90RSK-SENP2 module accelerates atherosclerosis. J Clin Invest 2015;in press. [DOI] [PMC free article] [PubMed]
- [34].Simonet T, Zaragosi LE, Philippe C, Lebrigand K, Schouteden C, Augereau A, et al. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res 2011;21:1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Biroccio A, Cherfils-Vicini J, Augereau A, Pinte S, Bauwens S, Ye J, et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat Cell Biol 2013;15:818–28. [DOI] [PubMed] [Google Scholar]
- [36].Akiyama BM, Loper J, Najarro K, Stone MD. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA 2012;18:653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, et al. Telomerase directly regulates NF-kappaB-dependent transcription. Nat Cell Biol 2012;14:1270–81. [DOI] [PubMed] [Google Scholar]
- [38].Mattson MP, Fu W, Zhang P. Emerging roles for telomerase in regulating cell differentiation and survival: a neuroscientist’s perspective. Mech Ageing Dev 2001;122:659–71. [DOI] [PubMed] [Google Scholar]
- [39].Montes M, Lund AH. Emerging roles of lncRNAs in senescence. FEBS J 2016;283:2414–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data are available in a public repository from the NCBI website (GEO accession number GSE119407). The authors declare that all of the other data supporting the findings of this study are available in the article, in the Supplementary Information files, or from the corresponding author upon request.