Abstract
The tricopeptide (amino acid number in the 30s) repeats constitute some of the most common amino acid repeats in proteins of diverse organisms. The most important representatives of this class are the 34-residue and 35-residue repeats, eponymously known as tetratricopeptide repeat (TPR) and pentatricopeptide repeat (PPR), respectively. The unit motif of both consists of a pair of alpha helices. As members of the large, all-helical repeat classes, TPR and PPR share structural similarities, but also play specific roles in protein function. In this study, a comprehensive bioinformatic analysis of the PPR units and the linkers that connect them was conducted. The results suggested the existence of PPR repeats of various formats, as well as smaller, PPR-unrelated repeats. Besides their length, these repeats differed in amino acid arrangements and location of key amino acids. These findings provide a broader and unified perspective of the pentatricopeptide family while raising provocative questions about the assembly and evolution of these domains.
Keywords: TPR, PPR, tricopeptide, repeats, linker, alpha helix
Introduction
Amino acid (abbreviated here as “aa”) repeats in proteins occur in a variety of lengths,1 compositions, and structures, but a large number of them are rich in alpha helix.2 In recent years, a special class of α-helical repeats that are 34 aa and 35 aa in length, respectively, called tetratricopeptide and pentatricopeptide repeats (TPR and PPR), have received substantial attention because of their unique structure and essential roles in various cellular processes in diverse organisms.1-10 In both the Pfam11 and the SCOP (Structural Classification of Proteins) classification12 of protein domains, TPR and PPR are recognized as all-alpha domains and appear superficially similar; however, they are distinct in sequence (Figure 1) and in their interactions with specific ligands, suggesting that apparently small differences in length and sequence in the members of the tricopeptide family may have important consequences on biological roles. While the TPR domains have been recognized for their role in protein-protein interactions,3 the PPR proteins are mostly involved in various steps of RNA metabolism in mitochondria and plant chloroplasts,13-15 and have received attention relatively recently, even though they appear to be more prevalent in nature.4,5 The 3-dimensional (3D) structures of a substantial number of PPR domains have been solved, which provided a glimpse to their ability to bind to type-specific substrates and ligands.13,16,17 The PPR motifs bear several similarities to TPRs. For example, like TPR, the PPRs occur in tandem repeats, in which the properties of strategically located residues, rather than exact amino acids, are conserved, thus producing a degenerate signature string. Like TPR, the PPR unit is also bihelical, consisting of 2 antiparallel α-helices (Figure 1) that generate a helix-turn-helix motif; repetition of this motif produces a superhelix in the protein, which appears very similar in structure. Unlike TPR, however, the PPR domain in a protein often contains longer and shorter repeats that differ from PPR but appear to exhibit some degree of similarity in amino acid sequence.4,6,7 In spite of their potential importance in the structure and function of the PPR proteins, the exact patterns and relevance of these degenerate repeats and the adjoining sequences18 have not been systematically explored. In this communication, the naturally occurring PPR domains were analyzed for their primary and higher-order structural features of length, motif arrangements, and strategic amino acid location, which led to a comprehensive view of these domains.
Figure 1.

Signature residues of PPR, compared with TPR. The sequence logo plot reveals the most prevalent residues in all PPRs (Panel B), and their similarity and contrast with those of the TPR (Panel A). The 2 sequences are visually positioned to align the 2 helices (A and B, boxed in colored lines) and their signature residues (G, A, F, M). The conserved Pro32 of TPR and Pro33 of PPR are asterisked; note its positional shift from TPR to PPR, relative to the other signature residues.
PPR indicates pentatricopeptide repeat; TPR, tetratricopeptide repeat.
Methods
Retrieval of sequences and structures
TPR and PPR protein sequences were retrieved by searching with their names as key words (TPR, tetratricopeptide, PPR, pentatricopeptide) at the following sites: NCBI “Structure” and “Protein” repositories (www.ncbi.nlm.nih.gov), RCSB (http://www.rcsb.org/), and UniProt (https://www.uniprot.org/). Only the nonredundant submissions were collected and all sequences were visually checked, organized, and formatted as needed (eg, FASTA, for alignment). Synthetic peptides, incomplete and noncurated sequences, and mutated sequences were eliminated. Where mentioned, structures were predicted through Swiss-Model,19 using the homology-modeling server (https://swissmodel.expasy.org/), and downloaded as PDB files. The secondary structural elements (α-helix, β-strand, loop) were also validated by the SABLE structure prediction suite (http://sable.cchmc.org/).20 All structures were displayed by using PyMol.21
Pattern analysis
A collection of known and predicted structures of TPR and PPR proteins were used for optimal analysis.1 Most TPR and PPR protein entries in the sequence databases are predicted as such, but their structures have not been experimentally determined (eg, by crystallography or nuclear magnetic resonance (NMR)); in these cases, TPR and PPR were ascertained and their boundaries were defined by analyzing the sequences at the TPRpred website (https://toolkit.tuebingen.mpg.de/#/tools/tprpred) in the MPI Bioinformatics Toolkit suite.22 Essentially the same PPRs were also identified by using Phmmer at the European Bioinformatics Institute site (https://www.ebi.ac.uk/Tools/hmmer/search/phmmer).23 The location of the repeats in a polypeptide and the length of the linkers (amino acid number) between them were manually collected and entered in an Excel spreadsheet. Linkers of each length class were also manually collected from the same spreadsheet. Multiple sequence alignments were performed mainly using Clustal Omega (at EMBL-EBI; https://www.ebi.ac.uk/Tools/msa/).24 As the tricopeptide repeats share only weak sequence similarity, they were compared by Sequence Logo analysis (http://weblogo.berkeley.edu/logo.cgi), which detects 1 or 2 amino acid(s) dominating over the rest in a given position25 and hence considered “signature” residues.
Results
Amino acid signature and uniqueness of PPR
To start with, established algorithms were used to find the classical PPR repeats (35 aa in length) in available protein sequences in the databases, as described in “Methods” section. A sequence logo presentation of the PPRs was then created, and compared and contrasted with that of TPR, the better-known member of the tricopeptide repeat family. The logo pattern of 1137 TPR sequences (Figure 1), containing 1 to 40 repeats, matched and confirmed the previously observed general pattern in TPR sequences.2,26
The logo pattern of 22 999 PPRs (Figure 1) from 2111 proteins (Supplementary material 1) also revealed a consensus pattern, similar as well as distinct from that of the TPR in several respects.27,28 Using the established knowledge of TPR structure as guidelines,26 the boundaries of the 2 helices, designated A and B, were first demarcated by aligning the 2 repeats without regard to their residue numbers. As shown (Figure 1), both helices of PPR closely resemble their counterparts in TPR, are 12 to 13 residues in length, and are separated by short unstructured regions. The alignment reveals similarities between some of the signature residues of TPR and PPR, although most of them were shifted by 1 position. Thus, the major ones, written with TPR residue number1,26 followed by PPR, are L7/6, G15/14, A20/19, F/Y24/23, and P32/33. Two signature residues—Ala20/19, Pro32/33—previously found to be the most invariant residues in TPR, are also present in PPR, and hence act as positional landmarks (Figure 1). In a few positions, conservative replacements can be seen; for example, Ala@27-TPR (ie, Ala at position 27 of TPR) is Met@26-PPR, both being hydrophobic aliphatic amino acids. One of the notable differences is G8, which shares TPR position 8 with Ala, but is not discernible in the PPR logo. Evidently, this Gly is a near-consensus residue in TPR, but not in PPR. As mentioned earlier, the last signature residue is P@32-TPR and P@33-PPR, located only 3 residues after the TPR B-helix border, but much farther—5 residues—from the same border in PPR. Pro, and, to a smaller extent, Gly are known to be helix-breaker residues,29-31 and thus, their locations may have important implications in the helical structure of both repeats. Clearly, although PPR and TPR are both members of the tricopeptide family of repeats, PPR differs from TPR not simply by 1 residue in length, but in several aspects of primary structure. It was, therefore, reasoned that the overall architecture of a PPR domain, including the possible presence of other repeats in this domain, may also be distinct from TPR, hence deserving a thorough analysis.
Arrangement of PPR repeats in the PPR proteins
The locations of the PPR units in the PPR proteins (Supplementary material 2) were first marked, and those of the TPR units in the TPR proteins (Supplementary material 3) were also marked for comparison. Once this was done, the linker (spacer) sequences between the repeats became evident. The TPR units were found to be close to each other, with either no linker or only short linkers between them. In contrast, previous studies6 recognized that the PPR domains often contain sequence repeats of various lengths in addition to the 35-aa PPR motif with varying degrees of sequence similarity. An early study7 of Arabidopsis PPR proteins recognized 3 major length types, and designated them as follows: P (PPR, 35 aa), L (long, 35-36 aa), and S (short, ~31 aa). The L-type was farther classified into L1 and L2, which differed slightly in amino acid signature. The 3 classes (P, L1/L2, S) showed sequence similarity and shared the classical PPR signature amino acids, displayed in Figure 1, suggesting that they were all PPR-related. The PPR-containing proteins available at the time were then classified by the arrangement of these motifs, as P class (containing P motif only) or P-L-S class (containing all 3 in order). However, comprehensive analyses of the arrangement of the PPRs and their intervening linker sequences were not yet performed. In this communication, all PPRs and their connecting linkers in 2111 PPR-containing proteins were subjected to a detailed analysis, as described in “Methods” section.
To this end, all PPR sequences were located on the full-length parent proteins, and the intervening linkers were also located. Note that these are the same sequences that were used in Figure 1, and hence already validated by the signature residues characteristic of PPR. The order of their locations and the lengths were tabulated in a spreadsheet (Supplementary material 2). The collection immediately revealed that most of the PPRs are contiguously connected to one another, without any linker between them; some extreme examples are K7N504, A0A0R0LE28, A0A0R0L774, and A0A0R0G3T3, which consisted exclusively of tandem runs of 13, 15, 15, and 26 PPRs, respectively. Other proteins, however, displayed extremely diverse arrangements of the repeats and PPR-to-PPR distances, bridged by linkers that were considered worth further study.
A cursory look at the linkers (Supplementary material 2) revealed that they come in all sizes, from 1 to >100 aa in length. However, to prioritize the analysis, attention was paid to 2 most abundant size groups, the very short (1-4 aa in length) and the medium ones (26-35 aa in length) (Figure 2). For unclear reasons (see “Discussion” section), linkers of lengths in between these 2 groups were relatively scarce. The short ones were also chosen because their addition to the preceding PPR could generate potentially novel 36- to 39-aa-long repeats, all of which would be tricopeptides. The 26- to 35-aa group was also broadly in the PPR size range. Finally, for the results to be meaningful, comparison was performed only among the linkers within the same size group, and not between disparate lengths, as described below.
Figure 2.

Quantification of linkers in PPR proteins. The number of various length classes of linkers found in PPR proteins (Supplementary material 2) were counted and plotted in Excel. Note the bimodal nature of the graph, with highs near the 2 termini of the length axis (roughly 1-4 aa, and 26-35 aa), marked by light blue boxes.
PPR indicates pentatricopeptide repeat.
Short-length (1-4 aa) linkers between PPRs
These linkers were visually located (Supplementary material 3) and the total number of each size was counted, which revealed that they occur in decreasing numbers, as follows: 1 aa, 2256; 2 aa, 617; 3 aa, 355; 4 aa, 227.
To find out whether the linkers contain any distinguishing sequence feature, they were subjected to sequence logo plot, and the preceding PPR sequence was included for reference. As shown (Figure 3), the PPR sequence showed the usual signature residues, such as G14, A19, F23, M26, although their relative dominance was slightly variable in the different subsets.
Figure 3.

Signature residues of PPR, connected to short linkers (1-4 aa in length). The sequences of the linkers plus the upstream PPRs were collected and subjected to sequence logo analysis as described in “Methods” section. Number of sequences analyzed were (A) PPR with no downstream linker = 12 126, (B) PPR + 1 aa = 2256, (C) PPR + 2 aa = 617, (D) PPR + 3 aa = 355, (E) PPR + 4 aa = 227. The 35-aa-long PPR portions are boxed, and major signature residues of the PPR portion (G14, A19, F23, M26) are indicated and connected by vertical lines. The gradual downstream move of Pro33 in Panel A to Pro37 in Panel E is illustrated by the angled lines, showing the spillage of this Pro from PPR to linker. Note that the Panel A here is not the same as the Panel B of Figure 1; the logos appear similar because the directly connected PPRs (0-aa linker) dominate the global PPR population.
PPR indicates pentatricopeptide repeat.
Curiously, the signature Pro at position 33 of the PPR with no linker afterward (Panel A) appeared to shift 1 position downstream each time a linker residue was added, ie, P33 to P37, going from 0- to 4-aa linker. In addition, it gradually lost its dominance, being easily visible when only 1-aa (Panel B) and 2-aa (Panel C) linkers were added to PPR, and when it is still technically within the 35-aa-long PPR, but became very weak in the 3-aa linker (Panel D), and finally, nearly invisible in the 4-aa linker (Panel E). The apparent migration and loss of the signature Pro residue was an unexpected finding, the reason of which was unclear; however, since Pro is a helix-breaker amino acid29,30 that is generally located at the end of the B-helix of PPR, the location of the shifted Pro residues with respect to the B-helix in these particular PPRs was examined. Available experimental structures and additional predicted structures of 20 PPR-linker sequences of each length type were collected and the locations of the B-helix and the distance between the B-helix and the linker were noted for each. The data (Supplementary material 4) were schematically presented (Figure 4) for ease of visualization.
Figure 4.

B-helix to Pro distance in PPR-short linkers. The number of spacer amino acids between signature Pro residue and the base of the B-helix of the PPR (the area marked in blue) were counted (Supplementary material 4) and averaged. The lengths of the B-helix (marked in gray) in number of amino acids are also shown, and the linkers are in red. All counts are accompanied by standard errors, calculated by Excel.
PPR indicates pentatricopeptide repeat.
The results show that the B-helix was indeed slightly longer in PPRs with longer downstream linkers, increasing from a mean of 12.45 amino acids with no linker to 13.95 amino acids with a 4-aa-long linker. The distance from the end of the B-helix to the start of the linker also increased gradually, from 4.35 amino acids to 5.5 amino acids. However, these shifts were not large enough to account for the 4-aa shift of the signature Pro, from Pro33 to Pro37. The provisional conclusion is that the location of the generally accepted Pro33 signature residue of the PPR family is actually variable and appears to have evolved to move distally as the downstream linker provides more space.
Medium-length (26-35 aa) linkers between PPRs
Next, PPR linkers in the size range of 26-35 aa were analyzed similarly. They were visually identified from the same collection of PPR sequences (Supplementary materials 1 and 2) and linkers of each size class counted. As shown earlier (Figure 2), the 31-aa linker was found to be the most abundant (2015), followed by 32- (370), 27- (389), 34- (276), 33- (257), 30- (154), 28- (104), 26- (30), and 35-aa (30) linkers. Because of their low abundance, linkers <26 aa and >35 aa were not analyzed further. For the record, the number of 36-aa-, 37-aa-, 38-aa-, 39-aa-long linkers were respectively 4, 8, 2, and 1; no 40-aa-long linker was found.
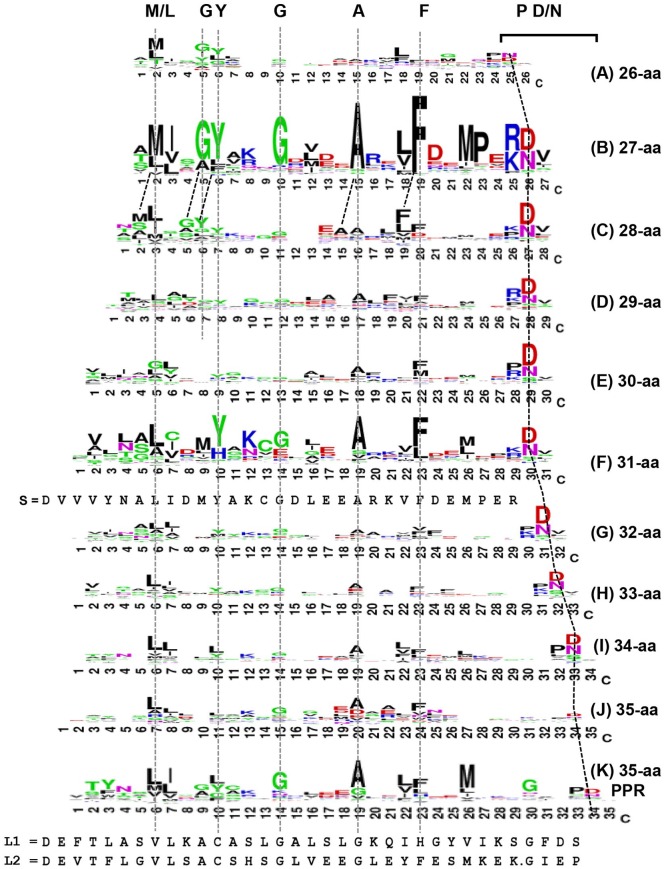
The sequences of these linkers were then analyzed by sequence logo (Figure 5), and the pattern compared with one another and with the canonical PPR (Panel K), and L1/L2 and S repeats (inserts in places of corresponding lengths).
Figure 5.
Sequence analysis of PPR-linker repeats (PLRs). These linkers, occurring between 2 PPR repeats, ranged from 26 to 35 amino acids in length (Panels A-J, respectively). The 35-aa linkers (Panel J) are not be confused with “true” PPRs (Panel K), because they do not share the PPR signature pattern (Panel B in Figure 1) and are not predicted as PPR by established algorithms. Likewise, the 34-PLRs are not TPR. Nevertheless, the various lengths display some sequence logo similarities. Residues that are relatively invariant in position among all lengths are connected by vertical broken lines, and their disappearance is indicated by the disappearance of the line. The angled lines represent the 2 visible residues (P-D/N) that appear to gradually shift position with increasing repeat length (from 24/25 to 33/34). The y-axes were deleted for space limitations, but they were similar to logos in the other figures; furthermore, all panels are presented in the same scale. Consensus S, L1, L2 sequences were previously published,7 and included here for comparison.
PPR indicates pentatricopeptide repeat; TPR, tetratricopeptide repeat.
Together, these results exposed several signature residues in each length class, many of which suggest their relatedness, but notable differences also emerged, as summarized here. (1) The strongest signatures were found in 27-aa (Panel B) and 31-aa (Panel F) linkers, which were stronger than the PPR itself (Panel K; same as Panel B in Figure 1), suggesting that these 2 classes are the most homogeneous in sequence. (2) Several signature residues appeared to be conserved in multiple size classes, as shown by the lines drawn over the most noticeable ones. (3) Each class also showed various degrees of uniqueness in signature pattern. Notably, some residues disappeared as the linker length increased, as indicated by the discontinuation of a vertical line. For example, M2, G5, and Y6 were conserved in 26-, 27-, and 28-aa classes (Panels A, B, C, respectively), but then gradually disappeared in longer linkers from Panel D onward. When properly aligned, the Y6 reappeared as Y10 in the 31-aa linker (Panel F). G10, one of the strongest residues in the 27-aa linkers (Panel B), was considerably weaker in the others. (4) The attempt to align the linkers met with difficulties, as also noted previously,7 partly due to their different lengths, and partly due to variations in the signature residues, the strength of their signals, and their locations. Nevertheless, the identity of the residues was given priority for straightforward alignment, so as to not create any gaps (Figure 5). As a result, most of the signature residues (or their conservative replacements, eg, Met/Leu) could indeed be placed on vertical straight lines. At the same time, however, a few other residues apparently shifted position, the most obvious being the Pro-Asp/Asn duo, conserved near the C-terminus in essentially all linkers; this is indicated by connecting them with slanted lines. In several other cases, an alternative set of residues could be aligned just as readily, such as the alternative M, G, Y, A, and F in the 28-aa linker (Panel C), also indicated by slanted lines. (5) It is to be reiterated that all linkers studied here (Panels A-J) were not predicted as canonical PPR by the same algorithm that correctly predicted known PPRs (see “Methods” section), and were full-length linker sequences connecting 2 PPR units (Supplementary material 2). In fact, the 35-aa linker (Panel J) has a signature pattern that is different from that of canonical PPR (Panel K). (6) When these full-length linkers were compared with the previously discussed PPR-like repeats of similar lengths, namely, the 31-aa linker (Panel F) compared with the consensus S sequence, and the 35-aa linker (Panel J) compared with consensus L1/L2 sequences,7 various degrees of divergence could be seen. Whereas the S repeat shared many residues with the 31-aa linker, including all signature residues, L1 and L2 were less similar to the 35-aa linker.
Regardless of the length of the linker, the Pro-Asp/Asn pair, mentioned above, was always the penultimate signature residues at the C-terminal end, and therefore, when the sequences were aligned by internal signature residues, this pair appeared to shift downstream with increasing linker length (Figure 5). This is reminiscent of the shift that was observed for the signature Pro in the 1- to 4-aa-long linkers (Figure 3), and therefore, the distance of PD/N from the preceding helix was interrogated as well. Since the structures of these linkers have not been experimentally solved, homology structure models were built for 2 arbitrarily chosen sequences of each length using Swiss-Model prediction, and the predicted helices were marked on the primary structure (Table 1). In addition, the corresponding PDB structure of one of each is presented for visual confirmation (Figure 6).
Table 1.
Helical regions in representative 26- to 35-aa-long linkers between PPRs.
|
26-aa:
A0A0R0E6Q8: VTFTNVLPVCARSGFLNVGKEIHAQIIRVGSSLDLFVSNALTKCGCINLAQNVLNISVREE A0A0R0KJ34: SVCNVLMSTYSKCEVPKDAKAVFECISNRNVVSWTTMISIQEEDAVSLFNAMRINGVYPND 27-aa: K7M2Y7: MVGTALIDMYAKCGRVESARLAFDQMGVRNLVSWNTMIDGYMRNGKFEDALQVFDGLPVKNA I1N4T9: YVANCLLQFYCKSSKMNYAFKVFDRMPQRDVISWNTLIFGYAGIGNMGFAQSLFDSMPERDV 28-aa: I1KSY8: ASYNAIISGLARCGRMKDAQRLFEAMPCPNVVSYTAMVDGYARVEGGIGRARALFEAMPRRNS I1KBU0: TVGNAILDAYSKCGNMEYANKMFQNLSEKRNLVTCNSLISGYVGLGSHHDANMIFSGMSETDL 29-aa: A0A0R0FZ08: SLHHNLIFALKSCETTSKIRQIHGHMVKTGLDNVPFTLSKLLAASIIDMDYAASIFSYIQTPNL I1KRU7: EHYGCMVDLLGRAGFIQEAYDIIKGMPMEPNDVILRSFLGACRNHGWVPSLDDDFLSELESELG 30-aa: K7K5P0: FLLNAFLTALVRNGRLAEAFQVFQTSPGKDIVSWNTMIGGYLQFSCGQIPEFWCCMNREGMKPDN A0A0R0G7R8: PVSNALMDMYAKCGSMEEAYLVFSQIPVKDIVSWNTMIGGYSKNSLPNEALKLFAEMQKESRPDG 31-aa: I1MT07: VTITSILSACAQLGALSFGKSVHQLIKSKNLEQNIYVSTALIDMYAKCGNISEASQLFDLTSEKNT I1MT07: STMVGLIPVSSPFGHLHLACCIQGFCVKSGTILQPSVSTALTTIYSRLNEIDLARQLFDESSEKTV 32-aa: I1MGT9: VTLASVLPACSQLERLRIGREIHCYALRNGDLIENSFVGTALVDMYCNCKQPKKGRLVFDGVVRRTV I1MGT9: DHYACLVDLLGRSGRVKEAYELINTMPSNLNKVDAWSSLLGACRIHQSVEFGEIAAKHLFVLEPNVA 33-aa: K7MRX0: YTYTGIVGACSVQEHKTCGKCLHGLVIKRGLDNSVPVSNALISMYIRFNDRCMEDALRIFFSMDLKDC I1JUQ7: YTYSSSLKACSCADAAGEGMQIHAALIRHGFPYLAQSAVAGALVDLYVKCRRMAEARKVFDRIEEKSV 34-aa: K7N0N6: AAYNQIIEILCKAQESELAESIMEDFVRSGLKPVTPSYVYLLSMYFTLELHDKLEEAFYQCLEKCRPNC K7N245: FVYNILIDGWARRGDVWEAADLMQQMRKEGLLPDIHTYTSFINACCKAGDMQATEIIQEMEASGIKPNL 35-aa: I1LZ60: VVVVAALSACARLGALELGRRIHHKYDRDSWQCGHNRGFTCAVVDMYAKCGSIEAALDVFLKTSDDMKTT A0A0R0J3I8: ESISLFLHACISRRSLEHGRKLHLHLLRSQNRVLENPTLKTKLITLYSVCGRVNEARRVFQIDDEKPPEE |
aa, amino acid; PPR, pentatricopeptide repeat.
Two examples of each linker in the range of 26 to 35 aa are presented, along with their upstream 35-aa PPR sequences (predicted by TPRpred; see “Methods” section); the full sequences of the InterPro entries (ie, the numbers shown in the left column) can be located in Supplementary material 1. Note that these–and all other linkers studied in this work–are full-length linkers with uncropped termini. In each sequence, the first 35 residues are PPR, and the rest is linker, marked in bold; all helical regions, predicted by Swiss-Model, are shaded. The predicted structures for the first members of each type (underlined) are displayed in Figure 6. The dotted lines were hand-drawn to roughly mark the end of the second helix of the linkers and the locations of the P-D/N signature. As seen, the distance between the 2 lines increases as the linkers get longer, giving the appearance of a positional shift of the P-D/N duo.
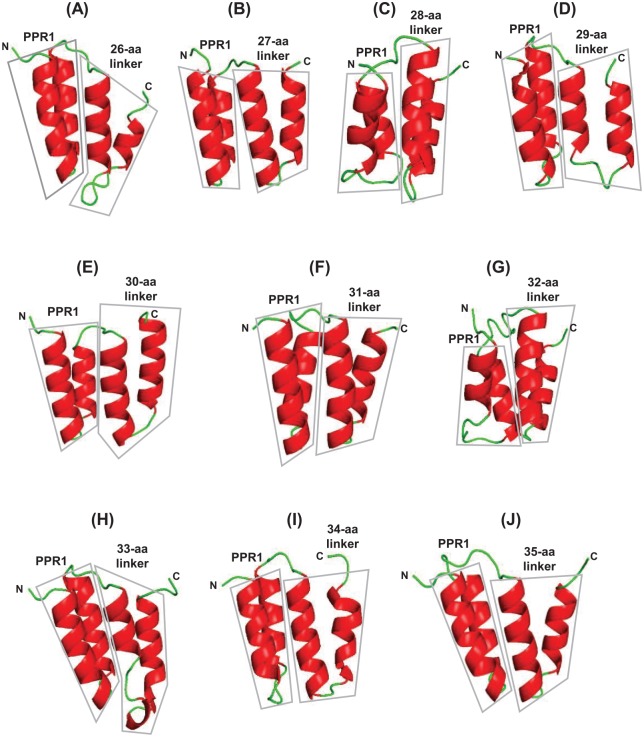
Figure 6.
Predicted bihelical structure of the midsize (26-35 aa) linkers. The structures of linkers of each length (26-35 aa, Panel A to J) and the preceding PPR (designated PPR1 in the scheme PPR1-linker-PPR2) were predicted by Swiss-Model and displayed by PyMol, as described in “Methods” section. The structures of both PPR and linker are boxed for easy recognition; the amino (N) and carboxy (C) terminal directions are also marked. Helices are in red, and the short, flexible regions connecting them are in green.
PPR indicates pentatricopeptide repeat.
Together, these structures led to the following observations. First, with few exceptions, the structures correctly predicted the 2 helices of the PPRs that preceded the linkers, which served as “positive control” and provided independent validation of PPR prediction by the TPRpred program. Second, as in the 1- to 4-aa class, the end of the B-Helix showed a trend to drift to the right (C-terminal direction) (Table 1); however, the P-D/N signature shifted much farther, as seen by the difference of slope of the 2 hand-drawn lines (Table 1). Finally, by and large all linkers in this group, regardless of the length, contained 2 helices (Table 1 and Figure 6). It can be concluded that despite their divergent signatures, these linkers are broadly PPR-like in primary and higher-order structures.
Discussion and Conclusion
The main focus of this study was the linker sequences that connect the PPRs in naturally occurring multi-PPR proteins. The study started by referring to them plainly as linkers, so as to avoid any preformed bias that they belonged to the previously designated P, L, and S types of PPR-homologous repeats. The final results show that some linkers indeed bore distant resemblance to these subtypes, but there were others with significant differences in both length and sequence. All midsize linkers showed some relatedness to PPR, especially when the sequences were allowed to slide for alignment. In fact, the diversity in these linkers is reminiscent of the difference between canonical PPR and TPR, detailed earlier (Figure 1), including positional differences of conserved signature residues. In other words, they are not much more different or similar than PPR and TPR. While detailed statistical analyses of the sequences will be needed to resolve whether the apparently unique ones represent novel subtypes, the simplest interim conclusion is that all these midsize sequences, regardless of the length, probably fall within the larger spectrum of the bihelical “tricopeptide repeat family.” Due to their bihelical nature, all repeats and linkers appear capable of being incorporated in the PPR superhelical structure (Figure 7), and hence most likely to materially participate in its function.
Figure 7.

Full superhelical PPR domains with embedded midsize linkers. The domains were predicted and presented as described in Figure 6. One example each of a 26-aa linker and a 35-aa linker, from accession numbers A0A0R0E6Q8 and I1LZ60, respectively (as in Figure 6 and Table 1), is shown. Both examples contain 11 PPRs in the full PPR domain that forms a large superhelical vortex (indicated by the spiral, white, hand-drawn curves), inside which the bihelical linkers (indicated by rectangles) fit well and maintain undisrupted continuity of the vortex. The directions of the N- and C-termini are marked. The Depth Cue (Fogging) display mode of PyMol was used to offer a sense of the front (N-terminal, bright and sharp) and rear (C-terminal, dark and foggy) of the vortex for a 3-dimensional feel.
PPR indicates pentatricopeptide repeat.
As briefly mentioned before, studies recognizing the P, L, and S subtypes also noted an organized pattern of the P-L-S triple motif in nearly half of the PPR proteins, several of which were later shown to be functional.27,28,32-34 In Arabidopsis, a more complex pattern was noted, in which 3 C-terminal motifs, named E, E +, and DYW, were associated with various combinations of P, L, and S.9,17,34,35 The current study was a global analysis of PPR proteins, conducted without regard to species, although the most PPRs were naturally from plants. In this study, attention was not paid to potential C-terminal motifs in the linkers, and the P-L-S-like combination was also not obvious. Any other combination of the length classes described in this work was also not noticeable (Supplementary material 2). However, a few arrangements of PPR and linker were occasionally observed (data not shown). For example, sometimes a long string of contiguous PPRs (ie, 0-aa linker) were tandemly connected with no linker, and 1 linker would occur near the end, before the last 1 or 2 PPRs, generating a pattern such as (PPR)n-linker-PPR; an example is G7KDN7, in which a run of 14 PPRs is connected to the last PPR with a 1-aa linker. In several polypeptides, 0-aa and 1-aa linkers repeated alternately, such that paired PPRs were connected by 1-aa linker, generating the pattern (PPR-PPR)n-1aa-(PPR-PPR). Such examples can be viewed as “super-repeats,” perhaps evolved from genetic duplication of large segments, followed by diversification of individual PPR units. In rare instances, very large linkers were encountered, such as a 100-aa linker in K7K416, and a 200-aa-long linker in G7LH09, but it was not tested whether they were composed of multiple shorter repeats. As stated, linkers larger than 39 aa were not studied because of their rarity. The PPR repeats, as a rule, are generally found clustered in a polypeptide, in which they form a functional pocket that interacts with single ligands, such as an RNA.27,36-39 A lone PPR, located far from this domain in the primary structure, may loop back in the 3-dimensional structure to serve as part of the functional PPR cluster, or it may have an independent function. In any case, the extra-long linkers and the higher-order patterns mentioned above deserve elaborate analyses, which is beyond the scope this study.
The apparent shift of the C-terminal signature in the linkers was necessary to properly align multiple other signature residues that resided in the interior of the linkers (Figure 5, Table 1); in other words, aligning the C-terminal end would have resulted in misalignment of all the other signature resides, which was energetically unacceptable. The molecular reason behind the evolution of the distance is a mystery, but it is tempting to speculate that it is rooted in the well-known role of Pro as a helix-breaker. The intricate helical architecture of the tricopeptide domains has evolved for optimal functionality, and it stands to reason that lengthening of the B-helix, or fusion of the B-helix with the A-helix of the next repeat, would severely disrupt the architecture and function of the PPR superhelix. In canonical TPRs and PPRs, the P32/33 is 2 to 4 residues away from the base of the B-helix (Figure 1). As previously noted for TPR,26,40 the P32 forces a turn to ensure the termination of the B-helix at the proper position, and the same should hold true for seamlessly connected PPR repeats, ie, those without a linker in between. The short linker sequence, devoid of secondary structure, may provide a similar cushion against fusion to the next helix, and hence obviate the need for the Pro to be near the base of the B-helix. This would allow this region greater freedom to evolve for more specialized functions.
When the number of linkers in the size range of 1 to 37 aa was counted (Figure 2), the distribution was clearly bimodal, and the 2 length classes of relatively high abundance, namely, ~1 to 4 aa and 26 to 35 aa, were studied here in detail. In contrast, linkers of the intermediate length, ie, in the range of ~10 to 25 aa occurred only infrequently. As stated earlier, the reason for this bimodal distribution with a very low trough is not clear at this time, but the effect of the linker on the structure of the adjoining PPR can be entertained as a possible reason. As the very short linkers are essentially unstructured, their only effect is to slightly increase the distance between the flanking PPRs, which may not significantly change the PPR domain (Figure 4). The 26- to 35-aa-long linkers are PPR-like in both primary and secondary structures, by virtue of possessing PPR-like residues and being bihelical (Figure 6); thus, insertion of 1 of these linkers is equivalent to adding a PPR to the series without perturbing the total tertiary structure of the PPR vortex (Figure 7). The intermediate ones (~9-25) may be at a disadvantage in both respects, ie, they are long enough to separate the flanking PPRs too far, but not long enough to be bihelical, and thus, will locally distort the PPR domain. In case a single helix is formed, and the linker is devoid of a helix-breaker residue (eg, Pro), the linker helix may fuse with the first helix of the following PPR, also causing distortion. Such structurally altered PPR domains evidently suit specific functions in select proteins in nature, but only in a few. This hypothesis remains to be tested with crystal structures, when available.
The origin of the various protein repeats remains uncertain, and the same can be said of the linkers. Regardless of their origin, and since the PPR domains have evolved to encode specific roles in their proteins of residence, one can envisage that the linkers evolved together with the overall structure of the domain. It is also worth noting that several proteins of closely related but nonidentical sequences have nearly identical number and spacing of the PPR motifs (Supplementary material 1), even though they occur in phylogenetically unrelated plants, such as A0A1U8FLB7 of Capsicum annum (hot pepper) and A0A1S3U0R6 and A0A1S3U0Q3 of Vigna radiata (mung bean), A0A2H3ZGH4 of Phoenix dactylifera (date palm), commonly known as date or date palm and D7KMD0 of Arabidopsis lyrata, all having the same pattern or minor variation thereof (linker amino acid numbers or the full range are indicated in parenthesis):
Similarly, A0A1S2Y5Q6 of Cicer arietinum (chickpea) and A0A1S4CXW1 of Nicotiana tabacum (tobacco) are virtually identical PPR proteins (69% identical, but 83% if conservative residues are included), both with C-terminal DYW domain, and thus, their PPR sequences and the spacing among them are also conserved (Supplementary material 1). These findings suggest that plants may have shared the PPR proteins extensively,41,42 either from a common ancestor or by horizontal transfer, which could also allow for joint transfer of the PPR and the adjoining linkers, and promote faster evolutionary and phylogenetic expansion of both.
Finally, experimental determination of 3D structure of the linker-PPR proteins, in both apo- and ligand (RNA?)-bound form, complemented by site-directed mutagenesis, should shed light on the exact functionality of the linkers and unravel whether they assist and/or regulate the PPR domain. Such studies will add new dimensions to the structure and function of the PPR, and may facilitate better programming of designer PPR domains for RNA-binding.27,39,43
Study Limitations
This research raised several questions that were not pursued for various reasons. First, medium-length (26-35 aa) linkers could not be studied, mainly due to their rarity, although many of them could be helix-rich, much like a PPR. Second, for the same reason, the very long linkers (eg, > 100 aa in length) were also not investigated. These linkers may have novel patterns waiting to be discovered. Finally, an occasional PPR protein appeared to contain a single PPR; examples include G7ZZ48 and K7MR95 (Supplement materials 1 and 2). These proteins may have a non-RNA-binding role, since structural studies have revealed that RNA-binding requires multiple contact points in a multi-PPR array.37 Nevertheless, future studies can now focus on these areas.
Supplemental Material
Supplemental material, Supplementary_material_1_xyz322006359eeed for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights
Supplemental material, Supplementary_material_2_xyz32200306fc177 for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights
Supplemental material, Supplementary_material_3_xyz322004a0c6789 for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights
Supplemental material, Supplementary_material_4_xyz3220001cb4de5 for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: As the sole author of this article, S.B. performed all aspects of the study, from designing and conducting the analyses, to writing and communicating the article.
Data Availability Statement: All data generated in this study are included in this published article as stated.
ORCID iD: Sailen Barik  https://orcid.org/0000-0003-1329-1786
https://orcid.org/0000-0003-1329-1786
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Kajava AV. Tandem repeats in proteins: from sequence to structure. J Struct Biol. 2012;179:279-288. doi: 10.1016/j.jsb.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 2. Sawyer N, Chen J, Regan L. All repeats are not equal: a module-based approach to guide repeat protein design. J Mol Biol. 2013;425:1826-1838. doi: 10.1016/j.jmb.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perez-Riba A, Itzhaki LS. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol. 2019;54:43-49. doi: 10.1016/j.sbi.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 4. Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415-442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 5. Manna S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie. 2015;113:93-99. doi: 10.1016/j.biochi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 6. Small ID, Peeters N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46-47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 7. Lurin C, Andres C, Aubourg S, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089-2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7:213-219. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- 9. Schallenberg-Rudinger M, Lenz H, Polsakiewicz M, Gott JM, Knoop V. A survey of PPR proteins identifies DYW domains like those of land plant RNA editing factors in diverse eukaryotes. RNA Biol. 2013;10:1549-1556. doi: 10.4161/rna.25755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sugita M, Ichinose M, Ide M, Sugita C. Architecture of the PPR gene family in the moss Physcomitrella patens. RNA Biol. 2013;10:1439-1445. doi: 10.4161/rna.24772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427-D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chandonia JM, Fox NK, Brenner SE. SCOPe: classification of large macromolecular structures in the structural classification of proteins-extended database. Nucleic Acids Res. 2019;47:D475-D481. doi: 10.1093/nar/gky1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci U S A. 2011;108:415-420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saha D, Prasad AM, Srinivasan R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol Biochem. 2007;45:521-534. doi: 10.1016/j.plaphy.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 15. Rovira AG, Smith AG. PPR proteins—orchestrators of organelle RNA metabolism. Physiol Plant. 2019;166:451-459. doi: 10.1111/ppl.12950. [DOI] [PubMed] [Google Scholar]
- 16. Zehrmann A, Verbitskiy D, Hartel B, Brennicke A, Takenaka M. PPR proteins network as site-specific RNA editing factors in plant organelles. RNA Biol. 2011;8:67-70. doi: 10.4161/rna.8.1.14298. [DOI] [PubMed] [Google Scholar]
- 17. Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663-670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 18. Richard FD, Kajava AV. In search of the boundary between repetitive and non-repetitive protein sequences. Biochem Soc Trans. 2015;43:807-811. doi: 10.1042/BST20150073. [DOI] [PubMed] [Google Scholar]
- 19. Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296-W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adamczak R, Porollo A, Meller J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins. 2004;56:753-767. [DOI] [PubMed] [Google Scholar]
- 21. DeLano WL. PyMOL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 22. Zimmermann L, Stephens A, Nam SZ, et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol. 2018;430:2237-2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 23. Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200-W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madeira F, Park YM, Lee J, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636-W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188-1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Main ER, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497-508. [DOI] [PubMed] [Google Scholar]
- 27. Yagi Y, Nakamura T, Small I. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 2014;78:772-782. [DOI] [PubMed] [Google Scholar]
- 28. Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE. 2013;8:e57286. doi: 10.1371/journal.pone.0057286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648-1652. [DOI] [PubMed] [Google Scholar]
- 30. Woolfson DN, Williams DH. The influence of proline residues on alpha-helical structure. FEBS Lett. 1990;277:185-188. doi: 10.1016/0014-5793(90)80839-b. [DOI] [PubMed] [Google Scholar]
- 31. Deville J, Rey J, Chabbert M. Comprehensive analysis of the helix-X-helix motif in soluble proteins. Proteins. 2008;72:115-135. doi: 10.1002/prot.21879. [DOI] [PubMed] [Google Scholar]
- 32. Barkan A, Rojas M, Fujii S, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hammani K, Takenaka M, Miranda R, Barkan A. A PPR protein in the PLS subfamily stabilizes the 5’-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res. 2016;44:4278-4288. doi: 10.1093/nar/gkw270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shikanai T. RNA editing in plants: machinery and flexibility of site recognition. Biochim Biophys Acta. 2015;1847:779-785. doi: 10.1016/j.bbabio.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 35. Okuda K, Chateigner-Boutin AL, Nakamura T, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21:146-156. doi: 10.1105/tpc.108.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng S, Gutmann B, Zhong X, et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016;85:532-547. doi: 10.1111/tpj.13121. [DOI] [PubMed] [Google Scholar]
- 37. Kobayashi K, Kawabata M, Hisano K, et al. Identification and characterization of the RNA binding surface of the pentatricopeptide repeat protein. Nucleic Acids Res. 2012;40:2712-2723. doi: 10.1093/nar/gkr1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci U S A. 2007;104:8178-8183. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coquille S, Filipovska A, Chia T, et al. An artificial PPR scaffold for programmable RNA recognition. Nat Commun. 2014;5:5729. doi: 10.1038/ncomms6729. [DOI] [PubMed] [Google Scholar]
- 40. Taylor P, Dornan J, Carrello A, Minchin RF, Ratajczak T, Walkinshaw MD. Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure. 2001;9:431-438. doi: 10.1016/s0969-2126(01)00603-7. [DOI] [PubMed] [Google Scholar]
- 41. O’Toole N, Hattori M, Andres C, et al. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 2008;25:1120-1128. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 42. Rivals E, Bruyere C, Toffano-Nioche C, Lecharny A. Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 2006;141:825-839. doi: 10.1104/pp.106.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McDermott JJ, Watkins KP, Williams-Carrier R, Barkan A. Ribonucleoprotein capture by in vivo expression of a designer pentatricopeptide repeat protein in Arabidopsis. Plant Cell. 2019;31:1723-1733. doi: 10.1105/tpc.19.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_material_1_xyz322006359eeed for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights
Supplemental material, Supplementary_material_2_xyz32200306fc177 for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights
Supplemental material, Supplementary_material_3_xyz322004a0c6789 for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights
Supplemental material, Supplementary_material_4_xyz3220001cb4de5 for The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them by Sailen Barik in Bioinformatics and Biology Insights