Abstract
Background
Studies have indicated that graphene oxide (GO) could regulated Brassica napus L. root growth via abscisic acid (ABA) and indole-3-acetic acid (IAA). To study the mechanism and interaction between GO and IAA further, B. napus L (Zhongshuang No. 9) seedlings were treated with GO and IAA accordance with a two factor completely randomized design.
Results
GO and IAA cotreatment significantly regulated the root length, number of adventitious roots, and contents of IAA, cytokinin (CTK) and ABA. Treatment with 25 mg/L GO alone or IAA (> 0.5 mg/L) inhibited root development. IAA cotreatment enhanced the inhibitory role of GO, and the inhibition was strengthened with increased in IAA concentration. GO treatments caused oxidative stress in the plants. The ABA and CTK contents decreased; however, the IAA and gibberellin (GA) contents first increased but then decreased with increasing IAA concentration when IAA was combined with GO compared with GO alone. The 9-cis-epoxycarotenoid dioxygenase (NCED) transcript level strongly increased when the plants were treated with GO. However, the NCED transcript level and ABA concentration gradually decreased with increasing IAA concentration under GO and IAA cotreatment. GO treatments decreased the transcript abundance of steroid 5-alpha-reductase (DET2) and isochorismate synthase 1 (ICS), which are associated with brassinolide (BR) and salicylic acid (SA) biosynthesis, but increased the transcript abundance of brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1), cam-binding protein 60-like G (CBP60) and calmodulin binding protein-like protein 1, which are associated with BR and SA biosynthesis.
Last, GO treatment increased the transcript abundance of 1-aminocyclopropane-1-carboxylic acid synthase 2 (ACS2), which is associated with the ethylene (ETH) pathway.
Conclusions
Treatment with 25 mg/L GO or IAA (> 0.5 mg/L) inhibited root development. However, IAA and GO cotreatment enhanced the inhibitory role of GO, and this inhibition was strengthened with increased IAA concentration. IAA is a key factor in the response of B. napus L to GO and the responses of B. napus to GO and IAA cotreatment involved in multiple pathways, including those involving ABA, IAA, GA, CTK, BR, SA. Specifically, GO and IAA cotreatment affected the GA content in the modulation of B. napus root growth.
Keywords: Graphene oxide, Brassinolide, Gibberellin, Root growth, Transcript level
Background
Nanomaterials are defined as forms of material with at least one constituent dimension in the range of 1–100 nm. Carbon nanomaterials are types of engineered nanomaterials that are being increasingly utilized because of their excellent optical, catalytic, electrical, mechanical, and thermal properties [1]. By using carbon nanomaterials, researchers are currently resolving challenges in agriculture, such as plant disease, pesticide and stress [2]. GO is a kind of 2D nanomaterial and a functionalized form of graphene that has been increasingly applied in multiple domains since the invention of GO in 2004 [3].
Nanomaterials have been reported to improve the germination rate of rice seeds; increase the root growth of corn, tomato and cucumber; enhance the growth rate of coriander and garlic plants; protect the photosynthesis system; and aid in defense against plant disease [4, 5]. However, research has also indicated that nanomaterial treatments can result in decreased germination rates and photosynthetic efficiency, reduced root and shoot length, reduction of biomass, and reduced nutrient contents in soybean [6, 7]. The regulation of nanomaterials in plants is complex and dynamic and and depends on the type of nanoparticle, treatments (concentration, tduration and method), and phytohormone balance [8].
Although GO can regulate plant growth and development, its mechanism is not clear. Research has indicated that the response of plants to GO is closely related to the reactive oxygen species (ROS) pathway. ROS are normal products of plant cellular metabolism. However, stresses lead to excessive production of ROS, causing oxidative damage and cell death. The plant defense mechanism is activated in response to stress, and increased amounts of protective enzymes and antioxidants are synthesized, such as ascorbate peroxidase, catalase (CAT), and superoxide dismutase (SOD). Studies have shown that nanomaterials influence plant growth and development via the ROS pathway [9]. Research has shown that under stress conditions, plant growth and defense responses are regulated in a coordinated manner by the activity of several phytohormones, such as ABA, CTK, GA and IAA. In addition, studies have shown that nanomaterial treatments can alter the expression levels of genes involved in multiple pathways, including the stress responses, cell metabolism, electron transport, and ABA and IAA synthesis pathways [10].
Auxin involved in many aspects of plant growth and development in the form of IAA. This hormone is involved in regulating the growth of the main roots, lateral roots, adventitious roots, root hairs, and vascular tissue. Mostly, Low concentrations of exogenous auxin mostly promote root growth, while concentrations of exogenous auxin inhibit the expansion of the main roots and stimulate the development of lateral roots and adventitious roots. IAA is perceived by auxin receptors such as TRANSPORT INHIBITOR RESPONSE 1 (TIR1) together with Aux/IAA proteins and auxin response factors (ARFs).
Our previous experiments have proven that GO treatment regulates the root growth of Brassica napus and that this root growth was significantly correlated with the IAA content [11]. To study the mechanism by which GO regulats plant root development and crosstalk between GO and IAA further, B. napus L seedlings (Zhongshuang No. 9) were treated with GO and IAA accordance with a two factor design, and the protective enzyme activity; hormone contents; and transcript levels of key genes involved in ABA, IAA, GA, CTK, BR, and SA were measured.
Results
Phenotype and phytohormone content of B.napus subjected to GO and IAA treatments
Nanomaterials are defined as material forms with at least one constituent dimension in the range of 1–100 nm, and GO is a kind of 2D nanomaterial that has been widely applied in biology, medicine, and chemistry, as well as in environmental protection.
Seedlings growth traits, specifically, root length, root fresh weight, stem length, number of lateral roots, and endogenous phytohormone content were measured on the 10th day after GO and IAA treatments. Analysis of variance revealed indicated that GO or IAA treatment significantly affected the growth of B.napus (root length, stem length, number of adventitious roots) and the GA, IAA, CTK and ABA contents in the seedlings. GO exhibited significant crosstalk with IAA to regulate B.napus growth (Table 1). Additional IAA treatments significantly influenced the root fresh weight. GO and IAA cotreatment significantly affected the root length; number of adventitious roots; and contents of IAA, CTK and ABA. However, the cotreatment did not significantly affect the stem length or root fresh weight (P < 0.05).
Table 1.
Effects of GO and IAA treatments on the seedling growth and phytohormone content of B. napus on the 10th day after treatment
| Variation source | Root length (cm) | Stem length (cm) | NO. of Adventitious roots | Root fresh weight (g) | ABA content (ng g −1 FW) | IAA content (ng g − 1 FW) | CTK content (ng g − 1 FW) | GA content (mg g − 1 FW) |
|---|---|---|---|---|---|---|---|---|
| GO | 7.39** | 1.13** | 24.28** | 0.054 | 161.51** | 33.70** | 156.95** | 5723.51** |
| IAA | 4.95** | 1.19** | 19.62** | 0.045** | 61.26** | 67.17** | 76.61** | 8131.81** |
| GO*IAA | 4.98** | 1.09 | 14.14** | 0.044 | 60.52** | 40.43** | 47.56** | 7240.09** |
“**“Indicates a significant effect, P < 0.01
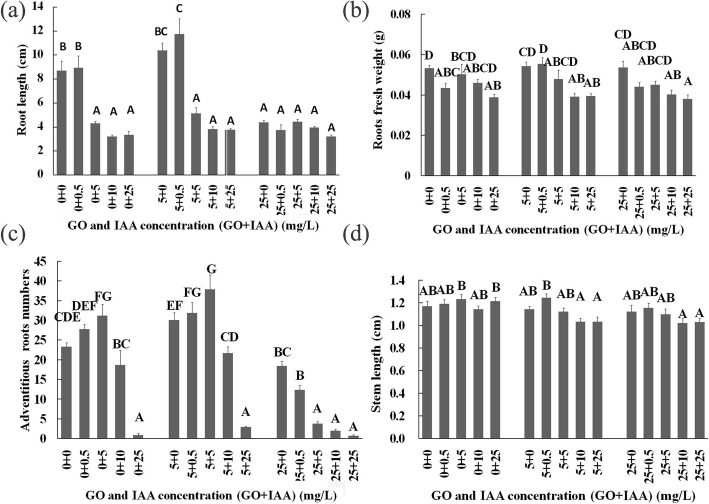
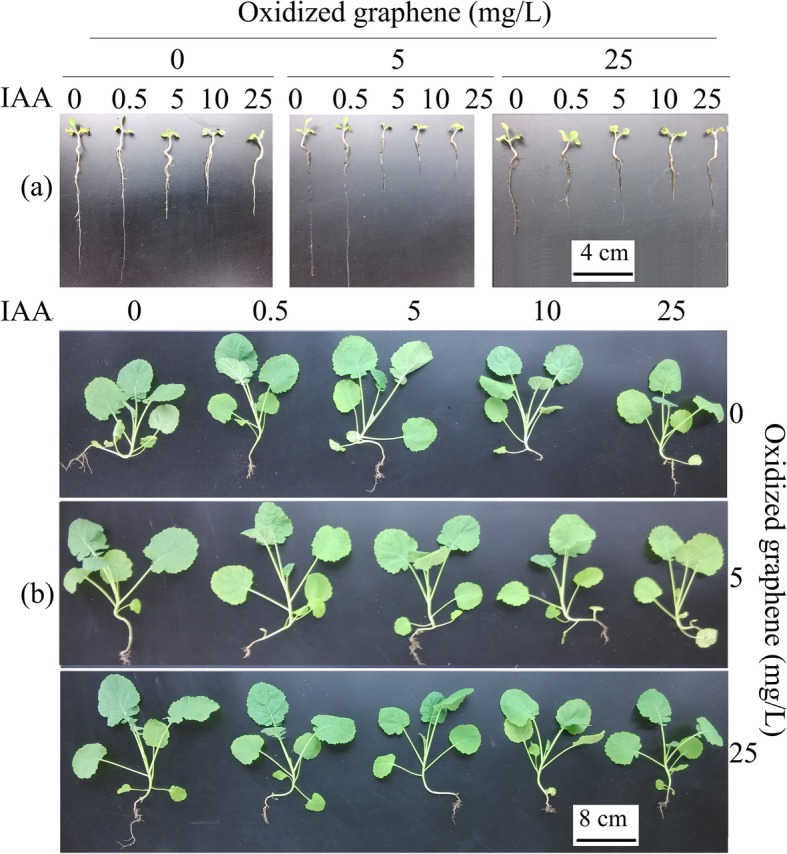
Compared with the control (CK) treatment (8.7 cm), the 5 mg/L GO treatment increased the root length (10.38 cm), but the 25 mg/L GO treatment suppressed root growth (4.39 cm) (Figs. 1 and 2a). The results showed that treatments with high concentrations of GO (> 25 mg/L) or IAA (> 0.5 mg/L) inhibited root development (Figs. 1a and 2a). Moreover, IAA cotreatment enhanced the role of the GO treatment, and the inhibition was strengthened with increasing IAA concentrations. The 0.5 mg/L IAA treatment did not significantly affect the root length, but the 5 mg/L GO cotreatment with 0.5 mg/L IAA promoted root growth, and the 0.5 mg/L IAA and 5 mg/L GO cotreatment significantly inhibited the root length, which further proved the crosstalk between GO and IAA (Fig. 2a). Parts of leaves cotreated with 25 mg/L GO and 10–25 mg/L IAA were necrotic. The results also proved that the 25 mg/L GO treatment was harmful to the seedlings and that IAA enhanced this disturbance.
Fig. 1.
Phytohormone of B.napus seedlings on the 10th (a) and 30th (b) days after GO and IAA treatments
Fig. 2.
Root length (a), root fresh weight (b), number of adventitious roots (c) and stem length (d) of B. napus seedlings on the 10th day after GO and IAA treatments. The values with different letters are significantly different; Student’s t-test, P < 0.05 (lowercase letters) or P < 0.01 (uppercaseletters)
The 5 and 25 mg/L GO treatments decreased the root fresh weight, and IAA cotreatment enhanced this effect. The 5 mg/L IAA treatment promoted adventitious root growth and increased number of adventitious root, but the 10–25 mg/L IAA treatments decreased the number of adventitious roots. Similarly, the 5 mg/L GO treatment increased the number of adventitious roots, whereas the 25 mg/L GO treatments decreased the number. The 5 mg/L GO and 0–5 mg/L IAA cotreatment increased the number of adventitious roots, but the 10–25 mg/L IAA and 5 mg/L GO cotreatment decreased the number of adventitious roots. Cotreatment with 25 mg/L GO and 0–25 mg/L IAA decreased the number of adventitious roots, and this repression was strengthened with increasing concentrations of IAA.
The IAA treatments did not affect the fresh weight or dry weight of the seedlings treated for 30 days, but the 25 mg/L GO treatment inhibited seedling growth (Fig. 2a). Cotreatment with 25 mg/L GO and 0–25 mg/L IAA inhibited the fresh weight and dry weight of seedlings treated for 30 days, and the inhibitory effect differed depending on the IAA concentration.
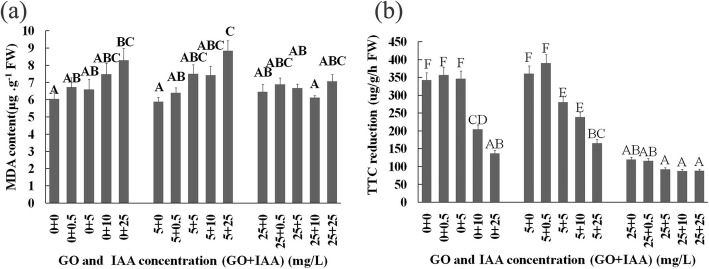
Malondialdehyde (MDA) contents and root triphenyl tetrazolium chloride (TTC) activityare affected by GO and IAA treatment
IAA and GO cotreatment resulted in a high MDA content. In addition, 10–25 mg/L IAA or 25 mg/L GO treatments decreased the root TTC activity, but low-IAA and GO treatments had no significant inhibitory effect (Fig. 3).
Fig. 3.
MDA content (a) and TTC reduction intensity (b) of seedlings on the 10th day after GO and IAA treatment. The values with different letters are significantly different; Student’s t-test, P < 0.05 (lowercase letters) or P < 0.01 (uppercaseletters)
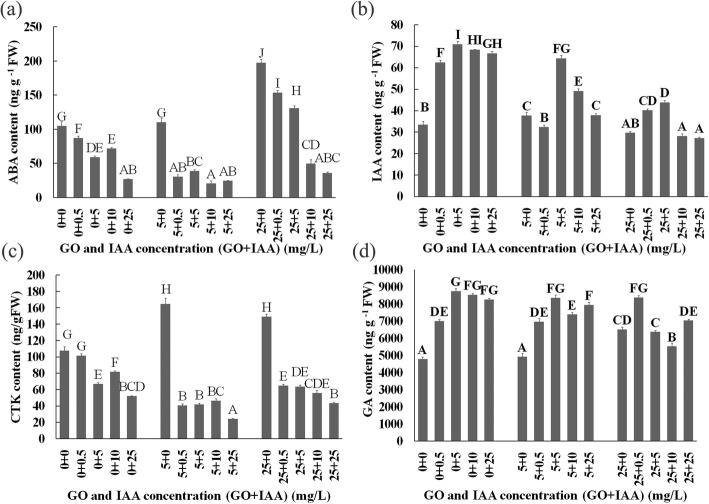
Phytohormone content s are affected by GO and IAA treatments
The results indicated that IAA treatment decreased the ABA and CTK contents but GO treatment increased the ABA and CTK contents. The ABA and CTK contents decreased with increasing IAA concentrations in response to the GO and IAA cotreatment compared with the GO treatment (Fig. 4a and c).
Fig. 4.
Contents of ABA(a), IAA(b), CTK(c) and GA (d) in B.napus seedlings on the 10th day after GO and IAA treatment. Values with different letters are significantly different (Student’s t-test, P < 0.01)
Generally, IAA contents increase with IAA increasing treatment concentrations, and our results showed a similar increase. The 5 mg/L GO treatment increased the IAA content, but the 25 mg/L GO treatment reduced the IAA content. Under the GO and IAA cotreatment, the endogenous IAA content first increased but then decreased with increasing IAA concentration from 0 to 25 mg/L (Fig. 4b).
The endogenous GA content first increased but then decreased with increasing IAA concentration. The 5 mg/L GO treatment did not alter the GA content, but the 25 mg/L GO treatment resulted in high GA content. Under GO and IAA cotreatment, the endogenous GA content also first increased but then decreased with increasing IAA concentration (Fig. 4d).
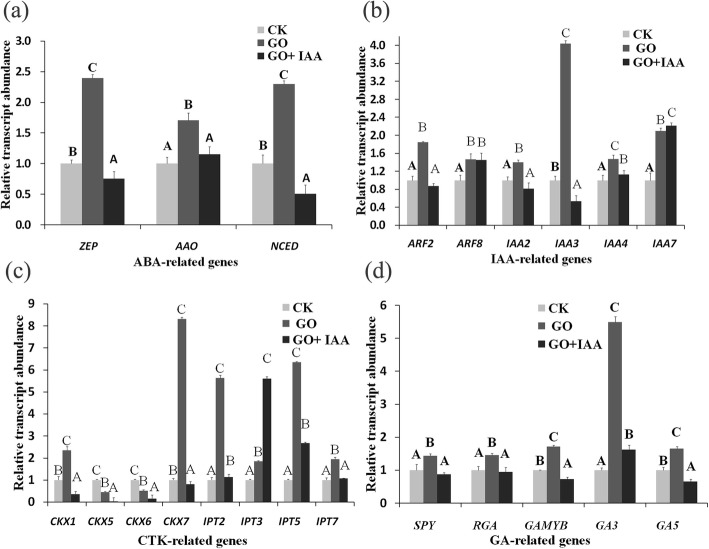
Transcript levels of key genes involved in phytohormone pathways are affected by GO and IAA treatment
Compared with the CK treatment, the 25 mg/L GO treatment increasedthe transcript levels of zeaxanthin epoxidase (ZEP), abscisic acid aldehyde oxidase (AAO) and NCED, but compared with the GO treatment, the 25 mg/L GO and 10 mg/L IAA cotreatment reduced the transcript abundance of these three genes, and the ZEP and NCED transcript levels were lower than those in the CK treatment (Fig. 5a).
Fig. 5.
Relative transcript levels of key genes involved in the ABA (a), CTK (b) and GA (c) and IAA (d) pathways in B. napus treated with 25 mg/L GO and 10 mg/L IAA on the 10th day after treatment. Values with different letters are significantly different (Student’s t-test, P < 0.01)
The transcript levels of ARF2, ARF8, IAA2, IAA3, IAA4 and IAA7 increased under the 25 mg/L GO treatment. Compared with the GO treatment, the 25 mg/L GO and 10 mg/L IAA cotreatment reduced the transcript levels of ARF2, IAA2, and IAA3 but increased the transcript level of IAA7; however, there were significant effects on the ARF8 transcript level (Fig. 5b).
The 25 mg/L GO treatment increased the transcript levels of key genes involved in CTK and GA biosynthesis, but compared with the GO treatment, the 25 mg/L GO and 10 mg/L IAA cotreatment reduced the transcript abundance, except for that of CKX5, CKX6 and IPT3 (Fig. 5c and d).
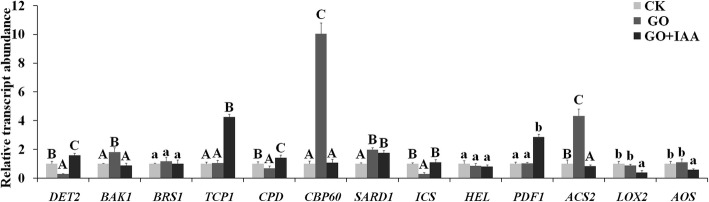
GO treatments decreased the transcript abundance of DET2 and increased the transcript abundance of BAK1; however, GO treatment did not alter the transcript abundance of serine carboxy peptidase (BRS1) and TCP1, which are involved in BR biosynthesis (Fig. 6). Compared with the CK and GO treatments, GO and IAA cotreatment improved the transcript levels of DET2 and TCP1, but compared with the GO treatment, the cotreatment inhibited the transcription of BAK1.
Fig. 6.
Relative transcript levels of key genes involved in the JA, BR, SA and ETH pathways in B. napus treated with 25 mg/L GO and 10 mg/L IAA on the 10th day after treatment. Values with different letters are significantly different (Student’s t-test, P < 0.01)
GO treatments resulted in increased transcription of ICS but decreased transcription of CBP60 and systemic acquired resistance-deficient 1 (SARD1), which are key genes involved inthe SA pathway. Compared with the GO treatment, GO and IAA cotreatment inhibited CBP60 transcription but had no significant effect on SARD1 transcription.
GO treatment did not affect the transcript abundance of LOX2 or allene oxide synthase (AOS), which are key genes involved in the jasmonic acid (JA) pathway, and had no significant effect on the transcript levels of Hevein-like protein (HEL) and PDF1, which are important genes for JA- and ETH-induced defense-related responses; however, GO treatment did increase the transcript levels of ACS2 (a key gene involved in ETH biosynthesis). Cotreatment with GO and IAA inhibited the transcription of LOX2, AOS and ACS2. By contrast, GO and IAA cotreatment improved the transcript abundance of the JA- and ETH- induced defense-related gene PDF1. Studies have shown that GO and IAA regulate plant growth via different pathways, but that crosstalk exists between GO and IAA.
Correlation analysis indicated that the root length was weakly correlated with the GA content (r = 0.26) but was not correlated with the ABA, IAA or CTK content, after GO and IAA cotreatment, which contrasted with our previous findings (GO modulation of rice root growth is dependent on the IAA content) [12]. Exogenous IAA can be applied, which could lead to a high IAA content in plants.
Discussion
Plant responses to nanomaterials depends on multiple factors
As an exogenously applied material with unique properties, GO can regulate the growth and development of plants either directly or indirectly. The accumulation of nanomaterials in plants has been shown to increase the shoot length, chlorophyll b content, number of adventitious roots, and fresh root weight of rice seedlings [13]. A 500 mg/kg CeO2 treatment was shown to increase the plant height, chlorophyll content, and biomass of barley without any toxic effects [14]. GO treatments decreased the damage caused by Cu stress by neutralizing the effects of Cu on nutrient accumulation in Lemna minor [3], and TiO2 nanoparticles have been reported to improve phosphorus uptake and improving plant growth [15]. GO treatments (25–100 mg/L) inhibited root growth and have negative effects on B. napus [8], and 2000 mg/L CeO2 inhibited the seed germination of corn, tomato and cucumber [16]. Our results indicated that GO or IAA treatment significantly affected the root length, stem length, and number of adventitious roots of B. napus seedlings. The 25 mg/L GO and 10 mg/L IAA cotreatment significantly inhibited the root growth, root fresh weight and number of adventitious roots, and inhibition was enhanced with increasing IAA concentration. The 25 mg/L GO treatment was harmful to the seedlings, which not only inhibiting root growth but also causing leaf necrosis. The effect of GO on plants depended on the concentration and treatment duration. The root length of five rice varieties treated with GO was correlated with the IAA content [11]. The research further proved that IAA had an important role in the response to GO in plants. Our results were consistent with the results in which low concentrations of GO increased plant root length, but in which high concentrations inhibited plant growth. Overall, the results indicated that the response of plants to nanomaterials depends on the plant genotype; content of endogenous phytohormone content; and the concentration, structure and localization of the nanomaterials within the plant [13, 17].
The ROS pathway clearly regulates plant growth via GO despite the complexity of the mechanism involved
Nanomaterials cause an overproduction of ROS, subsequently resulting in oxidative stress, and lipid peroxidation, causing damage to plant proteins and DNA [18]. Studies have also demonstrated that nanoparticle treatments can improve the potential to scavenge ROS and increase antioxidant enzymatic activities to regulate growth processes in plants [17].
Silver nanoparticles lead to differential expression of MSD1, CSD1 and FSD genes in rice seedlings, which is related to oxidative stress tolerance [19]. RNA-seq results indicated that hundreds of genes respond to nanoparticles, including the genes genes involved in photosynthesis-related metabolism, nitrogen metabolism, sucrose and starch metabolism and phytohormone signal transduction pathways, as well as genes involved with antioxidant enzymes [20].
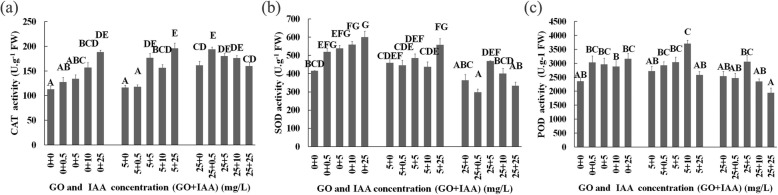
Our results showed that the high-concentration GO treatments resulted in a high MDA content and high CAT, SOD, and peroxidase (POD) activities (Fig. 7), and the 10–25 mg/L IAA and 25 mg/L GO treatments decreased the root TTC activity. The low-IAA and GO treatments had no significant inhibitory effect on root TTC activity, but the 5 mg/L IAA and 5 mg/L GO cotreatment inhibited the root TTC activity. Overall, our results proved that GO treatments regulated oxidative stress in plants, but the effect depended on the GO and IAA concentration and treatment duration, which further indicated that IAA is related to the effect of GO treatments on plant growth and development.
Fig. 7.
Activity of CAT (a), SOD(b) and POD (c) enzymes in seedlings on the 10th day after GO and IAA treatment. Values with different letters are significantly different (Student’s t-test, P < 0.01)
GO modulates plant root growth via crosstalk between multiple phytohormones
Plant hormones are considered important molecular signals that not only regulate plant growth and development but also respond to stress to improve plant tolerance. ABA is considered the primary plant stress hormone, and its content increases quickly in response to stress to improve plant stress tolerance [21]. Overexpression of the auxin response factor 5 gene increases carotenoid contents and increases tolerance to salt and drought in Arabidopsis [22]. GA is applied mainly as a growth-promoting hormone on the basis of its role in the role of increasing cell division and elongation, recently, research has shown that GA can improve plant tolerance to abiotic stress. In addition, CTK plays important roles in regulating plant growth and development, such as inhibiting lateral root initiation and leaf senescence, and regulating cell division and phloem differentiation [23]. CTK also plays an important role in controlling cell division and the maintenance of stem cells via cooperation with auxin. Additionally, cis-zeatin level increased in tissues exposed to various stresses [24], and ETH is involved in regulating seedling morphology, leaf senescence, and biotic and abiotic stress tolerance [25]. SA and JA have also been reported to play a large role in the response to biotic stress [26].
ABA biosynthesis starts with the hydroxylation and epoxidation of the C40 carotenoid β-carotene to produce the all-trans-xanthophylls zeaxanthin and violaxanthin. Violaxanthin is subsequently converted into 9-cis-epoxyxanthophylls, and further converted into xanthoxin via the protein encoded by NCED. NCED, AAO and ZEP are 3 key genes involved in ABA biosynthesis. Auxin is perceived by auxin receptors, represented by TIR1, which results in the proteolysis of Aux/IAA proteins, thereby releasing their inhibitory effect on ARFs. IAA biosynthesis occurs via two pathways: tryptophan dependent and tryptophan independent pathways. ATP/ADP adenosine phosphate isopentenyl transferases (IPTs) are responsible for the synthesis of isopentenyladenine (iP)- and trans-zeatin (tZ)-type CTKs, while CTK degradation is catalyzed by cytokinin oxidase/dehydrogenase (CKX). GA derepresses the hormone response inhibited by DELLA proteins, including the B. napus DELLA protein (RGA) ga1–3, RGL1, RGL2, and RGL3.
A series of studies have shown that GO regulates hormone content in plants. GO treatment (50 mg/L) resulted in a relatively low IAA content and a relatively high ABA content because of high transcript levels of NCED, AAO and ZEP [8]. Cu nanoparticle treatments have been reported to activate defense mechanisms against stress and to increase the content of amino acids, ABA and phenolics [27]. Moreover, silver nanoparticle treatment increased cis-zeatin in pepper, which further proved that CTK is involved in stress responses in plants [2]. However, the mechanism of how hormones interact is not clear.
Several hormones, including ABA, BR and ETH, are important for regulating lateral root growth. ABA negatively regulates lateral root growth, and CTK-deficient CKX resulted in defects in lateral root spacing [28]. In addition, a relatively low CTK contentor signaling is always accompanied by a relatively high lateral root density [29]. CTK inhibits lateral root growth by blocking the cell cycle from the G2 stage to the M stage [30]. Auxin regulates multiple stages of lateral root growth, including the establishment of pericycle cells and the emergence of lateral roots [31]. However, CTK controls lateral root formation and growth by regulating the auxin gradient [32].
Auxin regulates CTK levels in the stem by inducing the expression of CKX, suppressing the expression of IPT, and promoting the expression of strigolactone biosynthesis-related genes [33]. CTKs modulate organogenesis by down regulating PIN1 expression, and CYTOKININ RESPONSE FACTORS (CRFs) bind directly to the PIN1 promoter to control PIN1 expression in response to CTK [34].
Most importantly, GA treatment increases the number of primary roots. Studies have shown that overexpression of GA2ox1 in Populus and overexpression of RGL1 (resulting in GA-insensitive mutants) increased lateral root density and elongation [34] In addition, ETH affects lateral roots depending on the concentration: low ETH concentrations promote lateral root initiation, while higher concentrations doses inhibit lateral root initiation. The effect of BR on root elongation depends on the BR concentration; an appropriate concentration of BR promotes cell elongation, but a high concentration inhibits root growth. Moreover, compared with wild-type plants, DET2 mutants display shorter roots [35].
Researchers have shown that ABA and auxin synergistically regulate plant growth. Exogenous ABA treatments have been reported to inhibit lateral root development. However, ABA is important for primary root elongation according to studies on ABA-deficient plants [36]. IAAs inhibit auxin signaling, while ARFs positively regulate the expression levels of auxin-induced genes [37]. Generally, ABA treatment represses IAA7 expression but increases ARF2 expression [38].
The product of the NCED gene is the rate-limiting step in the ABA biosynthesis pathway. Our results proved that the NCED transcript level strongly increased when plants were treated with GO, which resulted in a high ABA content and decreased root length, further proving that ABA negatively regulates lateral root growth. A previous report also proved that GO treatment resulted in increased ABA contents and decreased IAA contents [10]. However, under increasing IAA concentrations, GO and IAA cotreatment gradually decreased NCED transcript levels and ABA concentrations. In addition, GO treatment increased the length of seminal roots of the wild-type tomato but decreased length of seminal roots of transgenic plants (overexpressing NCED) [39]. ABA may be the primary hormone that responds to GO. Our results also indicated that IAA treatments increased the IAA content, but that GO treatments decreased the IAA content.
There are 23 ARF proteins in Arabidopsis that bind specifically to the auxin-responsive element (AUXRE) TGTCTC to regulate the transcription of auxin-responsive genes. IAAs inhibit auxin signaling, while ARFs promote the transcription of auxin-induced genes [37]. Auxin signaling via ARF8 is essential for JA production [40]. ARF2 inhibits transcription of HOMEOBOX PROTEIN 33 to regulate the repressive role of ABA in primary root growth [38]. The results of that study further indicated that IAA treatments increased the ARF2 transcript level, but that IAA and GO cotreatment resulted in low ARF2 transcript levels. The results of our experiment also showed that the 10 mg/L ABA treatment increased the ARF2 transcript level but reduced the IAA content. Thirty-four dysregulated long noncoding RNAs, especially lnc37 and lnc14, were considered to be involved in the response to GO on the basis of genome-wide identification and functional analyses [41]. GO treatments significantly decreased the transcript levels of the auxin efflux carriers, PIN7 and ABCB1, and of ARR3 (a CTK response regulator) with increasing GO concentration. The low-concentration (1 mg/L) GO treatments increased the transcript levels of ARRO1 and TTG1, but the high-concentration (10 mg/L) GO treatments inhibited the transcription of these genes, which are involved in root growth [10]. It is possible that the GO treatment increased the ABA content but then decreased the IAA content under high ABA concentration.
Auxin regulates CTK levels in the stem by inducing the expression of CKX, suppressing the expression of IPT, and promoting the expression of strigolactone biosynthesis-related genes [33]. Both ABA and auxin inhibit root growth by causing excess production of ROS.
Numerous studies have shown that stress results in a low CTK content. Studies have also shown that stress causes high CTK levels because multiple factors influence stress signaling. According to transcriptome and MapMan analyses, genes that respond to CTK are involved mainly in the response to abiotic stress [42]. CTK-deficient plants have reduced levels of ABA because of low CTK levels [43]. CTK enhances cotyledon greening by promoting the proteasomal degradation of ABI5, which induces the expression of ARR5, which is involved in lateral root formation [44]. CTK can also inhibit stomatal closure via direct interaction with NO, which is an important signaling molecule that plays a role in the ABA-mediated stomatal closure pathway [45]. These results indicate that ABA and auxin can regulate the CTK content.
We assume that GO treatments increased the ABA content but then decreased the IAA content as a result of the high ABA concentration. Furthermore, the low IAA content inhibited CKX transcription and resulted in a relatively low CTK content. However, this hypothesis needs further confirmation.
BR binds to BR-insensitive 1 (BRI1) and results in a rapid association between BRI1 and its coreceptor BRI1-associated receptor kinase 1 (BAK1). BAK1 is involved in multiple signaling pathways and integrates several cell responses to regulate plant growth [46]. BRS1 is a serine carboxy peptidase that was recognized to regulate cell elongation and shape formation, both of which govern the length of hypocotyls and secondary inflorescence branches [47]. Constitutive photomorphogenesis and dwarfism (CPD) encodes an ɑ-hydroxylase that participates in a key ɑ-hydroxylationstep in BR biosynthesis [48]. TCP1 encodes a TCP transcription factor that promotes DWF4 expression for BR biosynthesis [49]. The plant lipoxygenase (LOX) enzyme catalyzes the oxidation of polyunsaturated fatty acids, after which AOS catalyzes the transformation of hydroperoxy fatty acid to SA [50]. SARD1 and CBP60 can bind to the promoter of ICS1 and positively regulate the expression of ICS1, which encodes a key enzyme involved in pathogen-induced SA synthesis [51]. Allene oxide cyclase (AOX) catalyzes the conversion of epoxyoctadecatrienoic acid (OPDA) to JA via several enzymatic reaction steps [52]. ETH levels increase under excess metal concentrations. The expression of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) and the accumulation of ETH are induced by Cd in Arabidopsis thaliana plants, mainly via the ACS pathway [47].
The transcript abundance of DET2 and ICS decreased under GO treatments; these are key genes involved in the BR and SA pathways. By contrast, GO treatment increased the transcript abundance of BAK1, which is a key gene involved in BR biosynthesis, and CBP60 and SARD1, which are important genes involved in the SA pathway. GO treatment also increased the transcript abundance of ACS2 (involved in the ETH pathway) but had no significant effect on that of LOX2 and AOS (involved in the JA pathway) or on HEL and PDF1 (involved in the JA and ETH pathways). These results indicated that the response pathways also included those of BR, SA and ETH.
Conclusions
In this study, B.napus seedlings were treated with GO and IAA, and the morphological characteristics and phytohormone contents of the treated seedlings were measured. GO and IAA significantly affected the root length; number of adventitious roots; and contents of IAA, CTK and ABA. IAA is an important phytohormone that regulates the root growth of B. napus L. under GO treatments, and the responses of B. napus to GO and IAA cotreatment involve multiple pathways, including the ABA, IAA, GA, CTK, BR, and SA pathways. Last, GO and IAA cotreatment affected the GA content in the modulation of B. napus root growth.
Methods
Plant growth and treatments
Zhongshuang No. 9 seeds were used as experimental materials and were provided Yong Chen (Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences). The seeds were germinated in the dark in a growth chamber that had a 24-h photoperiodand a temperature of 25 ± 1 °C. GO was obtained from Suzhou Carbon Science and Technology [12].
Zhongshuang No. 9 seedlings (4 days old) that displayed identical growth were selected and cotreated with GO (0, 5, and 25 mg/L) and IAA (0, 0.5, 5, 10, and 25 mg/L) in accordance with a completely randomized two-factor design reported previously [53]. More than five of the seedlings cotreated with GO and IAA for 10 days were randomly selected to measure the root length, root fresh weight, and stem height according to the previously reported methods [8].
Measurement of enzyme activities and MDA and phytohormone contents
The activity of POD, CAT and SOD enzymes was measured according to guaiacol oxidation method [54], H2O2 method [55] and nitro blue tetrazolium (NBT) method [15], respectively. The 2- thiobarbituric acid method [55] and TTC method [12] were used to measure the MDA content and root activity respectively. Phytohormones were extracted, purified and measured according to previously the reported methods [53, 56].
Determination of transcript abundance
Total RNA was extracted and reverse transcribed into cDNA for qPCR, and the relative transcript level was calculated according to previously reported method [57] in conjunction with qPCR primers [53]. DPS 7.05 software was used for analysis of variance on the basis of significance at P < 0.05 (indicated by lowercase letters in this study) or P < 0.01 (indicated by uppercase letters in this study) [58].
Acknowledgments
We are very grateful to Yong Chen (Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences) for providing the materials (Zhongshuang No. 9).
Abbreviations
- AAO
Abscisic acid aldehyde oxidase
- ABA
Abscisic acid
- ACS
Aminocyclopropane-1-carboxylate synthase
- ACS2
1-aminocyclopropane-1-carboxylic acid synthase 2
- AOS
Allene oxide synthase
- ARF
Auxin response factor
- BAK1
Brassinosteroid insensitive 1-associatedreceptor kinase 1
- BRS1
Serine carboxypeptidase
- CBP60
Cam-binding protein 60-like G
- CKX
Cytokinin oxidase/dehydrogenase
- CPD
Constitutive photomorphogenesis and dwarfism
- CTK
Cytokinin
- DET2
Steroid 5-alpha-reductase
- ETH
Ethylene
- GA
Gibberellin
- GAMYB
Transcription factor MYB65
- HEL
Hevein-like protein
- IAA
Indole-3-acetic acid
- ICS1
Isochorismate synthase 1
- IPT
Adenosine phosphate isopentenyl transferase
- JA
Jasmonate acid
- LOX
Lipoxygenase
- MDA
Malondialdehyde
- NCED
9-cis-epoxycarotenoid dioxygenase
- RGA
Brassica napus DELLA protein
- SA
Salicylic acid
- SARD1
Systemic acquired resistance-deficient 1
- SPY
UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase
- TTC
Triphenyl tetrazolium chloride
- ZEP
Zeaxanthin epoxidase
Authors’ contributions
BBX and LLX designed the research; LLX and FC conducted the research and analyzed the data; XKZ supplied the materials and analyzed the data; and LLX, BBX, FC, HWD, XGW and GXY wrote and edited the paper. All authors reviewed and approved the final version of the manuscript.
Funding
This research was supported by the National Key R&D Program of China (2017YFD0101700), The Scientific Research Foundation for Returned Overseas Chinese Scholars, and State Education Ministry, Educational Commission of Hubei Province of China (D20151303).
Availability of data and materials
The data sets supporting the results of this article are included within the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lingli Xie and Fan Chen contributed equally to this work.
References
- 1.Mukherjee A, Majumdar S, Servin AD, Pagano L, Dhankher OP, White JC. Carbon nanomaterials in agriculture: a critical review. Front Plant Sci. 2016;7:172. doi: 10.3389/fpls.2016.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinkovic T, Novak O, Strnad M, Goessler W, Jurasin DD, Paradikovic N, Vrcek IV. Cytokinin response in pepper plants (Capsicum annuum L.) exposed to silver nanoparticles. Environ Res. 2017;156:10–18. doi: 10.1016/j.envres.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Hu CW, Liu L, Li XL, Xu YD, Ge ZG, Zhao YJ. Effect of graphene oxide on copper stress in Lemna minor L.: evaluating growth, biochemical responses, and nutrient uptake. J Hazard Mater. 2018;341:168–176. doi: 10.1016/j.jhazmat.2017.07.061. [DOI] [PubMed] [Google Scholar]
- 4.Servin A, Elmer W, Mukherjee A, De la Torre-Roche R, Hamdi H, White JC, Bindraban P, Dimkpa C. A review of the use of engineered nanomaterials to suppress plant disease and enhance crop yield. J Nanopart Res. 2015;17(2):92. doi: 10.1007/s11051-015-2907-7. [DOI] [Google Scholar]
- 5.Chakravarty D, Erande MB, Late DJ. Graphene quantum dots as enhanced plant growth regulators: effects on coriander and garlic plants. J Sci Food Agr. 2015;95(13):2772–2778. doi: 10.1002/jsfa.7106. [DOI] [PubMed] [Google Scholar]
- 6.Wang XP, Yang XY, Chen SY, Li QQ, Wang W, Hou CJ, Gao X, Wang L, Wang SC. Zinc oxide nanoparticles affect biomass accumulation and photosynthesis in Arabidopsis. Front Plant Sci. 2016;6:1243. doi: 10.3389/fpls.2015.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Da Costa MVJ, Sharma PK. Effect of copper oxide nanoparticles on growth, morphology, photosynthesis, and antioxidant response in Oryza sativa. Photosynthetica. 2016;54(1):110–119. doi: 10.1007/s11099-015-0167-5. [DOI] [Google Scholar]
- 8.Cheng F, Liu YF, Lu GY, Zhang XK, Xie LL, Yuan CF, Xu BB. Graphene oxide modulates root growth of Brassica napus L. and regulates ABA and IAA concentration. J Plant Physiol. 2016;193:57–63. doi: 10.1016/j.jplph.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Siddiqi KS, Husen A. Plant response to engineered metal oxide nanoparticles. Nanoscale Res Lett. 2017;12:92. doi: 10.1186/s11671-017-1861-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li FH, Sun C, Li XH, Yu XY, Luo C, Shen YY, Qu SC. The effect of graphene oxide on adventitious root formation and growth in apple. Plant Physiol Bioch. 2018;129:122–129. doi: 10.1016/j.plaphy.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 11.Shen SS, Liu YF, Wang F, Yao GX, Xie LL, Xu BB. Graphene oxide regulates root development and influences IAA concentration in rice. J Plant Growth Regul. 2019;38(1):241–248. doi: 10.1007/s00344-018-9836-5. [DOI] [Google Scholar]
- 12.Sheng M, Tang M, Chen H, Yang BW, Zhang FF, Huang YH. Influence of arbuscular mycorrhizae on the root system of maize plants under salt stress. Can J Microbiol. 2009;55(7):879–886. doi: 10.1139/W09-031. [DOI] [PubMed] [Google Scholar]
- 13.Lin XW, Chen LY, Hu X, Feng SC, Huang L, Quan GP, Wei X, Yang ST. Toxicity of graphene oxide to white moss Leucobryum glaucum. RSC Adv. 2017;7(79):50287–50293. doi: 10.1039/C7RA10096E. [DOI] [Google Scholar]
- 14.Rico CM, Barrios AC, Tan WJ, Rubenecia R, Lee SC, Varela-Ramirez A, Peralta-Videa JR, Gardea-Torresdey JL. Physiological and biochemical response of soil-grown barley (Hordeum vulgare L.) to cerium oxide nanoparticles. Environ Sci Pollut R. 2015;22(14):10551–10558. doi: 10.1007/s11356-015-4243-y. [DOI] [PubMed] [Google Scholar]
- 15.Wang SE, Si SH. A fluorescent nanoprobe based on graphene oxide fluorescence resonance energy transfer for the rapid determination of oncoprotein Vascular Endothelial Growth Factor (VEGF) Appl Spectrosc. 2013;67(11):1270–1274. doi: 10.1366/13-07071. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Moreno ML, de la Rosa G, Hernandez-Viezcas JA, Peralta-Videa JR, Gardea-Torresdey JL. X-ray absorption spectroscopy (xas) corroboration of the uptake and storage of CeO2 nanoparticles and assessment of their differential toxicity in four edible plant species. J Agr Food Chem. 2010;58(6):3689–3693. doi: 10.1021/jf904472e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqi KS, Husen A. Engineered gold nanoparticles and plant adaptation potential. Nanoscale Res Lett. 2016;11:400. doi: 10.1186/s11671-016-1607-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arruda SCC, Silva ALD, Galazzi RM, Azevedo RA, Arruda MAZ. Nanoparticles applied to plant science: a review. Talanta. 2015;131:693–705. doi: 10.1016/j.talanta.2014.08.050. [DOI] [PubMed] [Google Scholar]
- 19.Nair PMG, Chung IM. Physiological and molecular level effects of silver nanoparticles exposure in rice (Oryza sativa L.) seedlings. Chemosphere. 2014;112:105–113. doi: 10.1016/j.chemosphere.2014.03.056. [DOI] [PubMed] [Google Scholar]
- 20.Wang YJ, Chen RY, Hao YW, Liu HC, Song SW, Sun GW. Transcriptome analysis reveals differentially expressed genes (DEGs) related to lettuce (Lactuca sativa) treated by TiO2/ZnO nanoparticles. Plant Growth Regul. 2017;83(1):13–25. doi: 10.1007/s10725-017-0280-5. [DOI] [Google Scholar]
- 21.Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32(1):40–52. doi: 10.1016/j.biotechadv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Kang C, He SZ, Zhai H, Li RJ, Zhao N, Liu QC. A sweetpotato Auxin Response Factor Gene (IbARF5) is involved in carotenoid biosynthesis and salt and drought tolerance in transgenic Arabidopsis. Front Plant Sci. 2018;9:1307. doi: 10.3389/fpls.2018.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bielach A, Podlesakova K, Marhavy P, Duclercq J, Cuesta C, Muller B, Grunewald W, Tarkowski P, Benkova E. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell. 2012;24(10):3967–3981. doi: 10.1105/tpc.112.103044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vyroubalova S, Vaclavikova K, Tureckova V, Novak O, Smehilova M, Hluska T, Ohnoutkova L, Frebort I, Galuszka P. Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol. 2009;151(1):433–447. doi: 10.1104/pp.109.142489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Q, Guo HW. Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol Plant. 2011;4(4):626–634. doi: 10.1093/mp/ssr042. [DOI] [PubMed] [Google Scholar]
- 26.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot-London. 2013;111(6):1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao LJ, Huang YX, Hu J, Zhou HJ, Adeleye AS, Keller AA. H-1 NMR and GC-MS based metabolomics reveal defense and detoxification mechanism of cucumber plant under Nano-Cu stress. Environ Sci Technol. 2016;50(4):2000–2010. doi: 10.1021/acs.est.5b05011. [DOI] [PubMed] [Google Scholar]
- 28.Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15(11):2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang L, Ramireddy E, Schmulling T. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J Exp Bot. 2013;64(16):5021–5032. doi: 10.1093/jxb/ert291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Mo X, Shou H, Wu P. Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiology. 2006;47(8):1112–1123. doi: 10.1093/pcp/pcj082. [DOI] [PubMed] [Google Scholar]
- 31.Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inze D, Beeckman T. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell. 2002;14(10):2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marhavy P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Parezova M, Petrasek J, Friml J, Kleine-Vehn J, et al. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell. 2011;21(4):796–804. doi: 10.1016/j.devcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Hayward A, Stirnberg P, Beveridge C, Leyser O. Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 2009;151(1):400–412. doi: 10.1104/pp.109.137646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elias AA, Busov VB, Kosola KR, Ma C, Etherington E, Shevchenko O, Gandhi H, Pearce DW, Rood SB, Strauss SH. Green revolution trees: semidwarfism transgenes modify gibberellins, promote root growth, enhance morphological diversity, and reduce competitiveness in hybrid poplar. Plant Physiol. 2012;160(2):1130–1144. doi: 10.1104/pp.112.200741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Xu J, Xu Z-H, Xue H-W. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell. 2005;17(10):2738–2753. doi: 10.1105/tpc.105.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PMY, Tham C, Duan L, Dinneny JR. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25(6):2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song YL, Wang L, Xiong LZ. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta. 2009;229(3):577–591. doi: 10.1007/s00425-008-0853-7. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Hua DP, He JN, Duan Y, Chen ZZ, Hong XH, Gong ZZ. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 2011;7(7):e1002172. doi: 10.1371/journal.pgen.1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiao JZ, Cheng F, Zhang XK, Xie LL, Li ZY, Yuan CF, Xu BB, Zhang LM. Preparation of graphene oxide and its mechanism in promoting tomato roots growth. J Nanosci Nanotechnol. 2016;16(4):4216–4223. doi: 10.1166/jnn.2016.12601. [DOI] [PubMed] [Google Scholar]
- 40.Pitaksaringkarn W, Ishiguro S, Asahina M, Satoh S. ARF6 and ARF8 contribute to tissue reunion in incised Arabidopsis inflorescence stems. Plant Biotechnol-Nar. 2014;31(1):49–53. doi: 10.5511/plantbiotechnology.13.1028b. [DOI] [Google Scholar]
- 41.Wu QL, Zhou XF, Han XX, Zhuo YZ, Zhu ST, Zhao YL, Wang DY. Genome-wide identification and functional analysis of long noncoding RNAs involved in the response to graphene oxide. Biomaterials. 2016;102:277–291. doi: 10.1016/j.biomaterials.2016.06.041. [DOI] [PubMed] [Google Scholar]
- 42.Brenner WG, Schmulling T. Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front Plant Sci. 2015;6:29. doi: 10.3389/fpls.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishiyama R, Le DT, Watanabe Y, Matsui A, Tanaka M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One. 2012;7(2):e32124. doi: 10.1371/journal.pone.0032124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan CM, Wang XC, Feng J, Hong SL, Liang Y, Ren B, Zuo JR. Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of abscisic acid insensitive5 protein in Arabidopsis. Plant Physiol. 2014;164(3):1515–1526. doi: 10.1104/pp.113.234740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu WZ, Kong DD, Gu XX, Gao HB, Wang JZ, Xia M, Gao Q, Tian LL, Xu ZH, Bao F, et al. Cytokinins can act as suppressors of nitric oxide in Arabidopsis. Proc Natl Acad Sci U S A. 2013;110(4):1548–1553. doi: 10.1073/pnas.1213235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Postel S, Kufner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A, Halter T, Kemmerling B, Nurnberger T. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol. 2010;89(2–3):169–174. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Deng Q, Wang X, Zhang DZ, Wang XM, Feng CZ, Xu SB. BRS1 function in facilitating lateral root emergence in Arabidopsis. Int J Mol Sci. 2017;18(7):1549. doi: 10.3390/ijms18071549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85(2):171–182. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 49.Guo ZX, Fujioka S, Blancaflor EB, Miao S, Gou XP, Li J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell. 2010;22(4):1161–1173. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyons R, Manners JM, Kazan K. Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep. 2013;32(6):815–827. doi: 10.1007/s00299-013-1400-y. [DOI] [PubMed] [Google Scholar]
- 51.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414(6863):562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 52.Mosblech A, Feussner I, Heilmann I. Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol Bioch. 2009;47(6):511–517. doi: 10.1016/j.plaphy.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Xie L-L, Chen F, Zou X-L, Shen S-S, Wang X-G, Yao G-X, Xu B-B. Graphene oxide and ABA cotreatment regulates root growth of Brassica napus L. by regulating IAA/ABA. J Plant Physiol. 2019;240:153007. doi: 10.1016/j.jplph.2019.153007. [DOI] [PubMed] [Google Scholar]
- 54.Chen GY, Chen CL, Tuan HY, Yuan PX, Li KC, Yang HJ, Hu YC. Graphene oxide triggers toll-like receptors/autophagy responses in vitro and inhibits tumor growth in vivo. Adv Healthc Mater. 2014;3(9):1486–1495. doi: 10.1002/adhm.201300591. [DOI] [PubMed] [Google Scholar]
- 55.Cakmak I, Marschner H. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 1992;98(4):1222–1227. doi: 10.1104/pp.98.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma C, Meir S, Xiao LT, Tong JH, Liu Q, Reid MS, Jiang CZ. A knotted1-like homeobox protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 2015;167(3):844–853. doi: 10.1104/pp.114.253815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and 2−ΔΔCT method. Methods (San Diego, Calif) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Tang QY, Zhang CX. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013;20(2):254–260. doi: 10.1111/j.1744-7917.2012.01519.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are included within the article.