Abstract
L-Ascorbate (vitamin C) is ubiquitous in both our diet and the environment. Here we report that Ralstonia eutropha H16 (Cupriavidus necator ATCC 17699) uses L-ascorbate as sole carbon source via a novel catabolic pathway. RNAseq identified eight candidate catabolic genes, sequence similarity networks and genome neighborhood networks guided predictions for function of the encoded proteins, and the predictions were confirmed by in vitro assays and in vivo growth phenotypes of gene deletion mutants. L-Ascorbate, a lactone, is oxidized and ring-opened by enzymes in the cytochrome b561 and gluconolactonase families, respectively, to form 2,3-diketo-L-gulonate. A protein predicted to have a WD40-like fold catalyzes an unprecedented benzilic acid rearrangement involving migration of a carboxylate group to form 2-carboxy-L-lyxonolactone; the lactone is hydrolyzed by a member of the amidohydrolase superfamily to yield 2-carboxy-L-lyxonate. A member of the PdxA family of oxidative decarboxylases catalyzes a novel decarboxylation that uses NAD+ catalytically. The product, L-lyxonate, is catabolized to α-ketoglutarate by a previously characterized pathway. The pathway is found in hundreds of bacteria, including the pathogens Pseudomonas aeruginosa and Acinetobacter baumannii.
Graphical Abstract
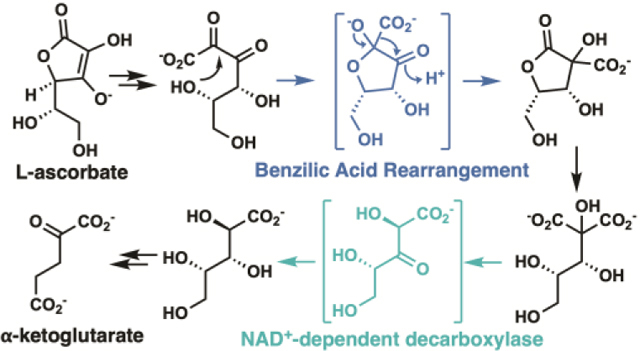
L-Ascorbate (vitamin C), ubiquitous in nature, is synthesized in plants and mammals.1,2 Two similar bacterial catabolic pathways, first characterized in Escherichia coli K-12, are encoded by two gene clusters (designated ula and sga) that encode anaerobic (ula) and aerobic (sga) pathways for the catabolism of L-ascorbate or its oxidized, hydrolyzed product, 2,3-diketo-L-gulonate (DKG).3,4
We previously characterized pathways for tetronate catabolism by Ralstonia eutropha H16 (Cupriavidus necator ATCC 17699), a soil bacterium.5 We observed that R. eutropha also can utilize L-ascorbate as sole carbon source although its genome does not encode orthologs of the Ula or Sga enzymes (Supplementary Table S1). Because plants are known to convert L-ascorbate to L-threonate,6 we hypothesized that our previously reported L-threonate catabolic pathway in R. eutropha5 could contribute to environmental L-ascorbate catabolism. However, mutants missing genes encoding the enzymes in L-threonate catabolism were able to grow with L-ascorbate (Supplementary Figure S1).
We now describe a new ubiquitous bacterial catabolic pathway for L-ascorbate. The pathway includes two novel enzymes: the first, not a member of a characterized family, likely catalyzes a benzilic acid rearrangement involving migration of a carboxylate group; the second is the first decarboxylase reported to use NAD+ as a cofactor.
RNAseq was used to quantify the abundance of whole cell transcripts in R. eutropha cells grown with L-ascorbate, succinate, or D-fructose as carbon source (Supplemental Figure S2). With L-ascorbate, eleven genes were upregulated >50-fold compared to succinate and >200-fold compared to D-fructose (Supplemental Data). Eight of the encoded proteins could be identified as enzymes in a putative catabolic pathway; these were located in three distinct gene clusters that encode sequential segments of the pathway (Figure 1).
Figure 1.
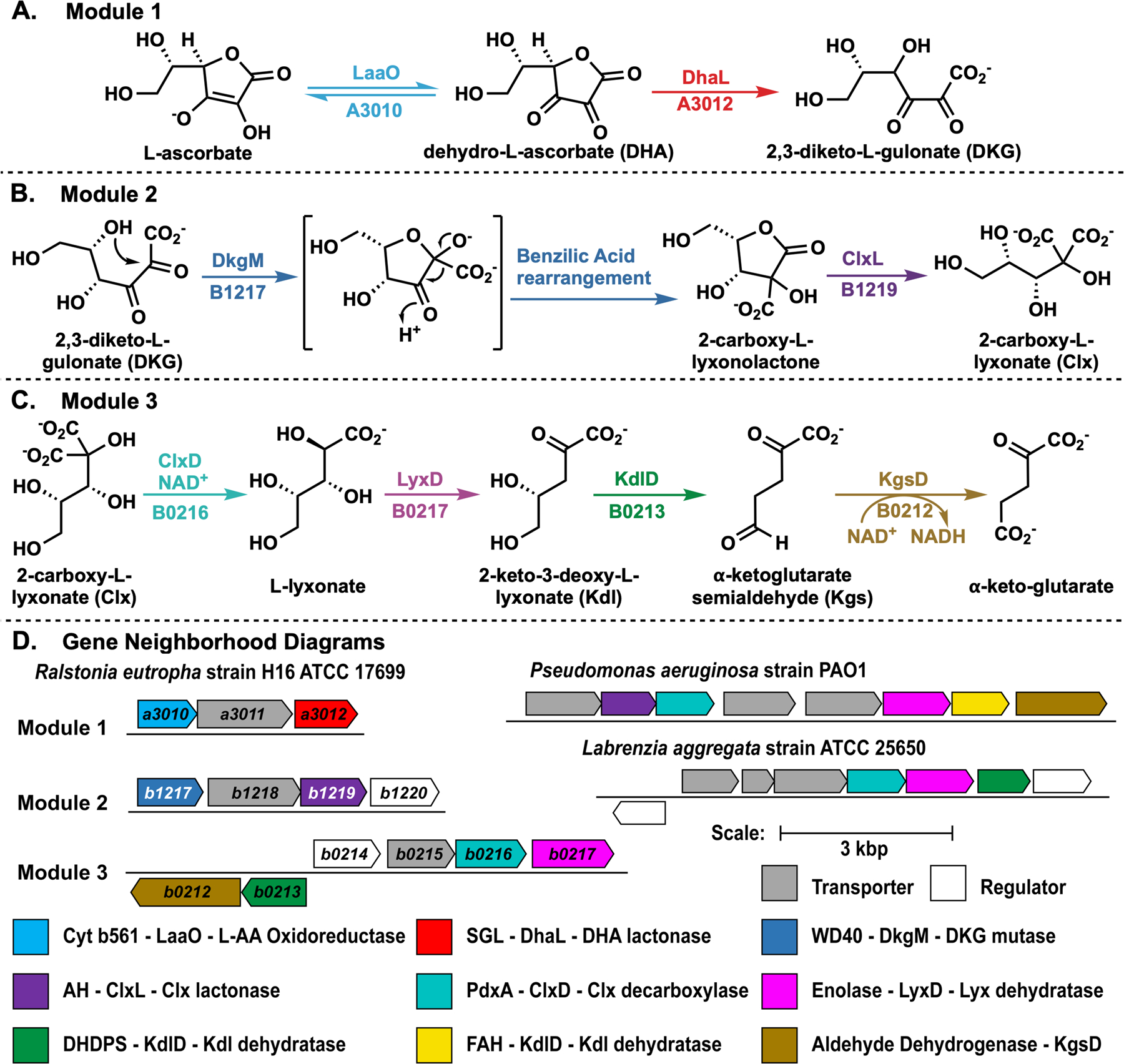
Catabolic pathway of L-ascorbate in R. eutropha occurs in three modules (A-C) that are encoded in distinct gene clusters (D). A) Module 1: L-Ascorbate is oxidized and hydrolyzed to 2,3-diketo-L-gulonate. B) Module 2: 2,3-diketo-L-gulonate is substrate for a benzilic acid rearrangement to yield 2-carboxy-L-lyxonolactone, which is ring-opened to 2-carboxy-L-lyxonate. C) Module 3: 2-carboxy-L-lyxonate is decarboxylated via oxidation and reduction of C3 to produce L-lyxonate, which is dehydrated twice and oxidized to generate α-ketoglutarate, a citric acid cycle intermediate. D) Gene neighborhood diagrams mentioned in the main text.
The EFI-EST tool (http://efi.igb.illinois.edu/efi-est/) was used to generate sequence similarity networks (SSNs) of enzymes encoded by the candidate genes (Supplementary Figures S3-5).7 SSNs were refined to generate isofunctional clusters; these SSNs were used to generate genome neighborhood networks (GNNs) using the EFI-GNT tool (http://efi.igb.illinois.edu/efi-gnt/). The proximities of the putative pathway genes with each other (Figure 1) suggested their functional relationship (Supplementary Table S2 and Figure S6).
One gene cluster (Module 3) encodes orthologs of the characterized enzymes in Pseudomonas aeruginosa PAO1 in L-lyxonate catabolism (Supplementary Table S3).8 In the P. aeruginosa pathway, an ortholog of H16_B0217 (LyxD) catalyzes the dehydration of L-lyxonate to form 2-keto-3-deoxy-L-lyxonate (Kdl). The subsequent dehydration of Kdl is catalyzed by a member (KdlD) of the fumarylacetoacetate hydrolase superfamily (FAH) to form α-ketoglutarate semialdehyde (Kgs), which is oxidized by an ortholog of H16_B0212 (KgsD) to form α-ketoglutarate, an intermediate in the citric acid cycle (Figure 1C).8 We previously reported that the KdlD from the FAH superfamily can be replaced by a member of the dihydrodipicolinate synthase (DHDPS) superfamily (e.g. UniProt ID A0NP47 from Labrenzia aggregata IAM 12614).8 These homologous enzymes from R. eutropha were heterologously expressed and display the predicted in vitro activities.
We deleted the genes encoding Module 3 in independent R. eutropha strains; strains missing lyxD or kdlD were unable to grow with L-ascorbate (Figure 2); complemented strains recovered wild type growth (Supplementary Figure S7). Strains missing kgsD had reduced growth rates with L-ascorbate (Figure 2); growth was likely enabled by the activity of two similar enzymes (H16_B0130, 61%ID/73% sim, or H16_B0322, 47% ID/58% sim, to KgsD); complemented strains recovered wild type growth (Supplementary Figure S7).
Figure 2.
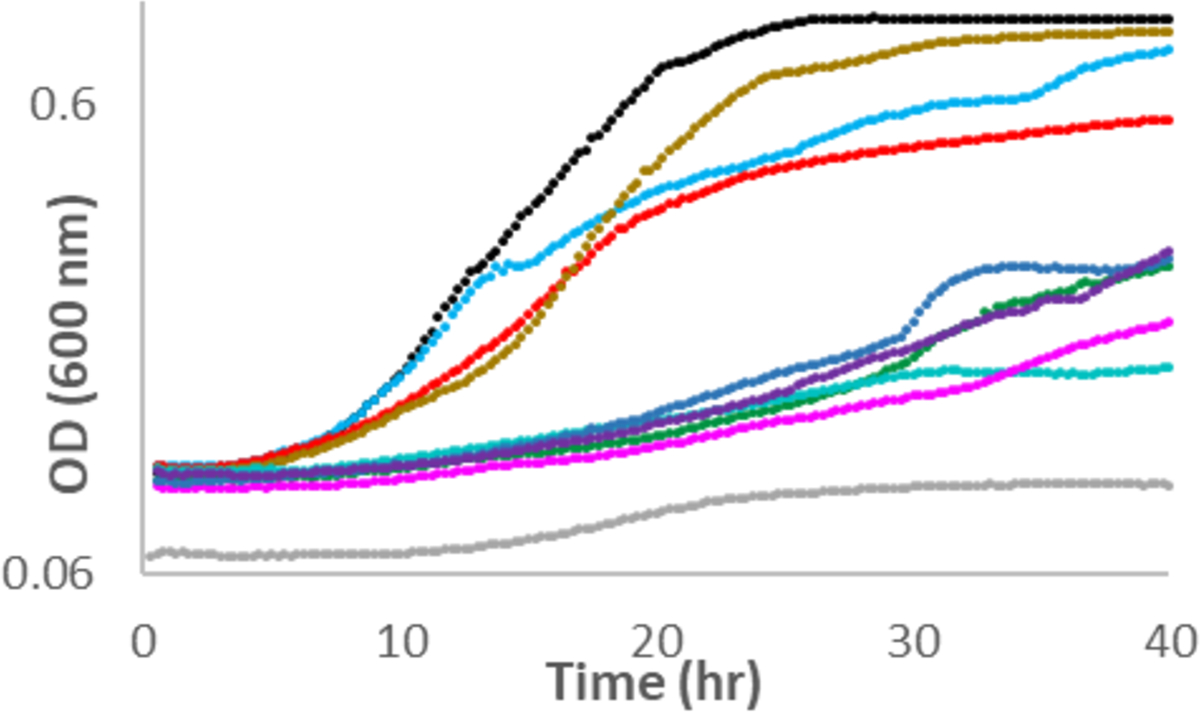
Growth of wild type (black) and mutant strains (lacking individual genes, correspondingly colored as in Figure 1) of R. eutropha with L-ascorbate as the sole carbon source.
Because R. eutropha converted L-lyxonate into α-ketoglutarate, a central metabolic intermediate, and L-lyxonate catabolic genes are essential for L-ascorbate catabolism, we hypothesized that the remaining five enzymes (encoded by Modules 1 and 2) must catalyze the upstream oxidation, lactone hydrolysis, and decarboxylation reactions to convert L-ascorbate to L-lyxonate.
Members of the transmembrane cytochrome b561 family (PF03188) that contains H16_A3010 (LaaO in Module 1) catalyze reduction of dehydro-L-ascorbate (DHA) to L-ascorbate.9 We did not characterize LaaO but predict that it catalyzes the oxidation of L-ascorbate to form DHA, the substrate for H16_A3012 (vide infra) (Figure 1A). Mutants lacking laaO grew with L-ascorbate with reduced growth rates compared to wild type (Figure 2); complemented strains grew like wild type (Supplementary Figure S7). Growth of the mutant strains with L-ascorbate was consistent with spontaneous/nonenzymatic oxidation of L-ascorbate in aerobic conditions.10
Although nonenzymatic hydrolysis of DHA is well-known,11 we observed that H16_A3012 (DhaL, PF08450 SGL gluconolactonase family in Module 1) catalyzed the hydrolysis of DHA to form DKG (kcat = 8.0 ± 0.6 s−1, Km = 490 ± 70 μM, kcat/Km = 1.6 ± 0.3 × 104 M−1 s−1, Supplementary Figures S8A and S8B). Deletion of dhaL reduced the growth rate and growth yield of R. eutropha during L-ascorbate growth (Figure 2). The complemented strains grew like wild type with each carbon source (Supplementary Figure S7).
H16_B1217 in Module 2 is predicted to have a WD40/YVTN repeat-like-containing domain (member of IPR015943). Proteins with these domains have been reported to catalyze various unrelated reactions, providing little predictive information about the function of H16_B1217.12–14 Incubation of H16_B1217 with DKG leads to the production of 2-carboxy-L-lyxonolactone (Figure 1B) (kcat = 0.6 ± 0.1 s−1, Km = 81 ± 7 μM, kcat/Km = 7.9 ± 1.8 × 103 M−1 s−1, Supplementary Figure S8C). Its identity is supported by LC-MS and 1H and 13C NMR spectroscopies as well as the observation that it is labile to hydrolysis. Labeling studies confirm the rearrangement (vide infra). The structure of 2-carboxy-L-lyxonolactone (Clx lactone) suggests that H16_B1217 (Dkg mutase or DkgM) directs addition of the C5 hydroxyl group to the C2 carbonyl.
Two mechanisms can be envisioned for the subsequent rearrangement that generates the lactone: migration of the C1 carboxylate group from C2 to C3 via a concerted benzilic acid rearrangement or stepwise decarboxylation/carboxylation (transcarboxylation) involving CO2 and a vicinal enediol as intermediate (Supplementary Figure S9). The nonenzymatic rearrangement of DKG to 2-carboxy-L-lyxonate (Clx, 2-carboxy-L-threo-pentonate) in alkaline conditions has been reported; the mechanism was assumed to be the benzilic acid rearrangement.15
To provide possible evidence to distinguish between the mechanisms, we incubated Clx with [13C]-bicarbonate at alkaline pH and also with DkgM. The nonenzymatic incubation produced no observable 13C incorporation in the product (6.1 ± 1.1% 13C-labeled Clx compared to 5.3 ± 1.4% in nonlabelled control reactions Supplementary Figure S10), providing strong support for the benzilic acid rearrangement mechanism. The incubation with DkgM also does not lead to 13C incorporation (5.5 ± 2.3% 13C-labeled Clx compared to 4.8 ± 1.0% in nonlabelled control reactions Supplementary Figure S11). However, the absence of 13C incorporation in the DkgM-catalyzed reaction is consistent with either mechanism, if the CO2 intermediate is sequestered in the active site. We prefer the benzilic acid rearrangement mechanism given the nonenzymatic counterpart.
L-Ascorbate-dependent growth of mutant strains lacking dkgM was abolished (Figure 2); complemented strains grew like wild type (Supplementary Figure S7).
We observed that H16_B1219 (PF04909 amidohydrolase family; Clx lactone lactonase, ClxL in Module 2) catalyzes the hydrolysis of Clx lactone to yield Clx, (kcat = 90 ± 10 s−1, Km = 850 ± 120 μM, kcat/Km = 1.0 ± 0.2 × 105 M−1 s−1, Supplementary Figure S8D). Clx is a geminal dicarboxylate with carboxyl groups derived from C1 and C2 of DKG.
Incubation of Clx and NAD+ with H16_B0216 (PF04166 PdxA oxidative decarboxylase family in Module 2) generates L-lyxonate (Figure 1C), even with sub-stoichiometric amounts of NAD+ (kcat = 0.33 ± 0.08 s−1, Km = 800 ± 400 μM, kcat/ Km = 4.0 ± 2.0 × 102 M−1 s−1, see Supplementary Figures S8E and S12). Characterized members of the PdxA family perform oxidative decarboxylations of β-hydroxy acids.5 We propose that H16_B0216 (Clx decarboxylase, ClxD) catalyzes the NAD+-dependent oxidation of the C3 alcohol to a ketone and NADH followed by a single decarboxylation of the β-keto diacid intermediate. ClxD then catalyzes reduction of the β-keto monoacid by the tightly bound NADH to yield L-lyxonate and regenerate the NAD+ cofactor (Scheme 1).
Scheme 1.
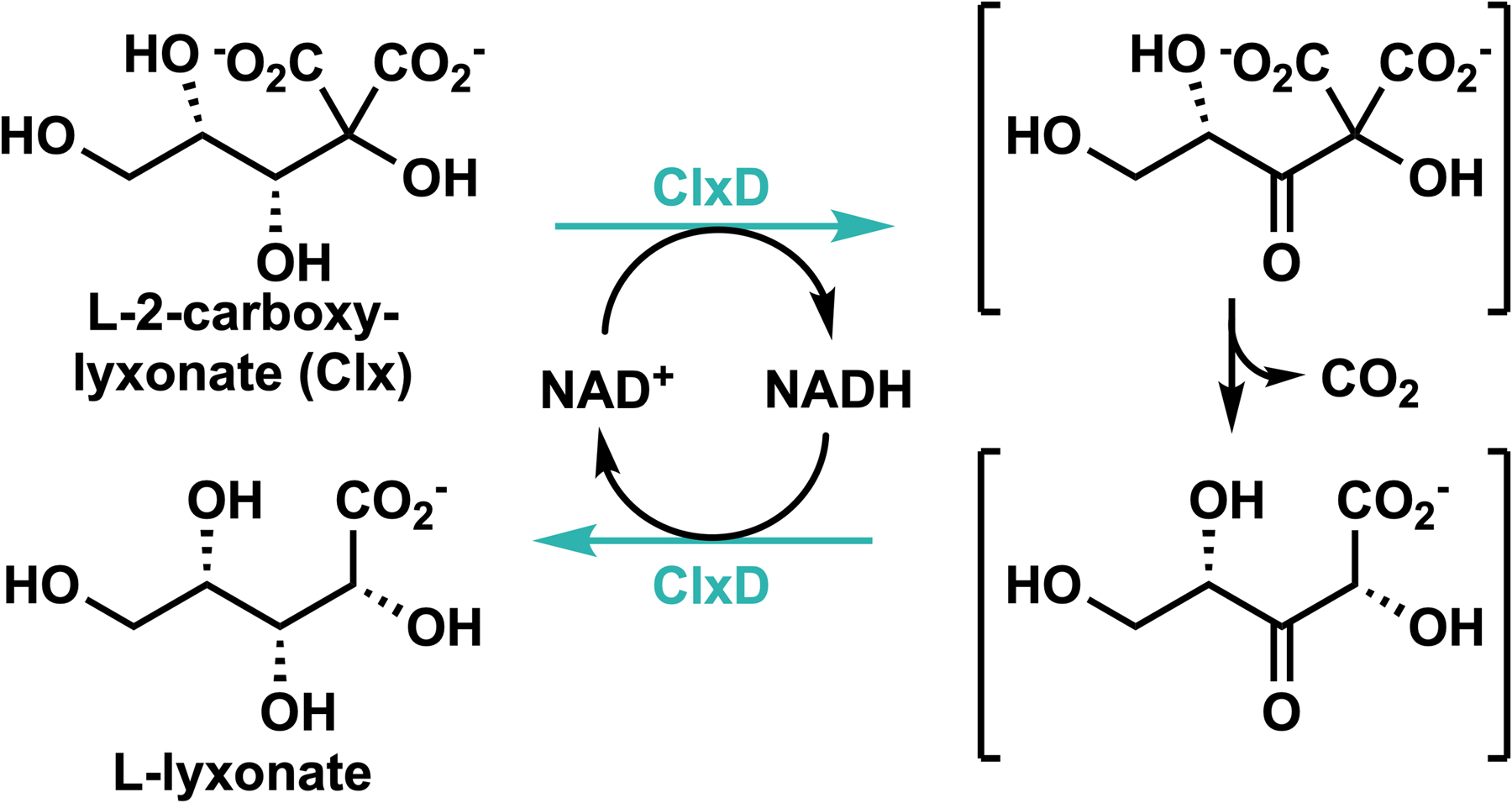
Proposed mechanism of the ClxD-catalyzed reaction
A stopped-flow experiment with equivalent amounts of enzyme and substrate revealed a small, substoichiometric transient increase in absorbance at 340 nm (Supplementary Figure S13). This result may be explained by rate-limiting oxidation of the substrate to form the β-keto diacid intermediate and NADH followed by rapid decarboxylation and reduction of the resulting β-keto monoacid intermediate.
Tightly bound NAD+ participates in other overall redox neutral enzymatic reactions,16 but ClxD represents, to our knowledge, the first decarboxylase that uses NAD+ as a cofactor.
Deletion of clxD abolished growth of R. eutropha with L-ascorbate (Figure 2); complemented strains recovered wild type growth with L-ascorbate (Supplementary Figure S7). The knockout of the orthologous gene in P. aeruginosa PAO1 (locus ID PA 2212, UniProt ID Q9I1Q5) does not affect growth with L-lyxonate.8
We used 13C-labeled L-ascorbate to confirm the reaction sequence from L-ascorbate to L-lyxonate. Clx displays two resonances in its 13C NMR spectrum at δ 177.4 and 177.7 ppm (diastereotopic). Starting with 1-[13C]-L-ascorbate, the δ 177.7 ppm resonance is enriched; starting with 2-[13C]-L-ascorbate, the δ 177.4 ppm resonance is enriched (Figure 3). Therefore, 1-[13C]-L-ascorbate produced 1-[13C]-L-lyxonate and CO2, and 2-[13C]-L-ascorbate produced L-lyxonate and 13CO2 (Supplementary Figures S14-16). This labeling requires that a bond is formed between C1 and C3 of the DKG substrate and the bond between C1 and C2 is cleaved to release CO2 in the formation of the Clx lactone product (Figure 4).
Figure 3.
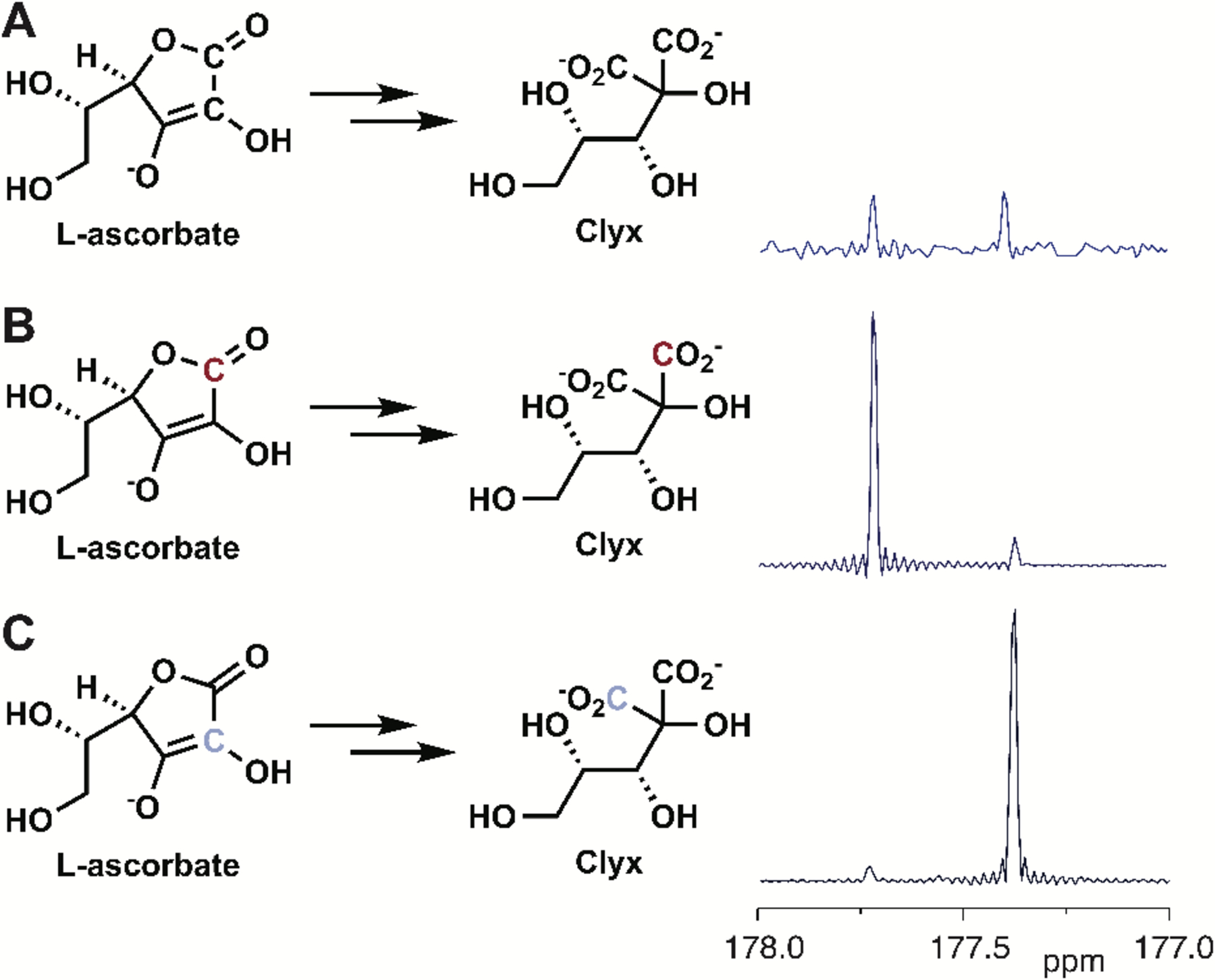
13C NMR Spectra of 2-carboxy-L-lyxonates. A) From unlabeled L-ascorbate, B) from 1-[13C]-L-ascorbate, and C) from 2-[13C]-L-ascorbate.
Figure 4.
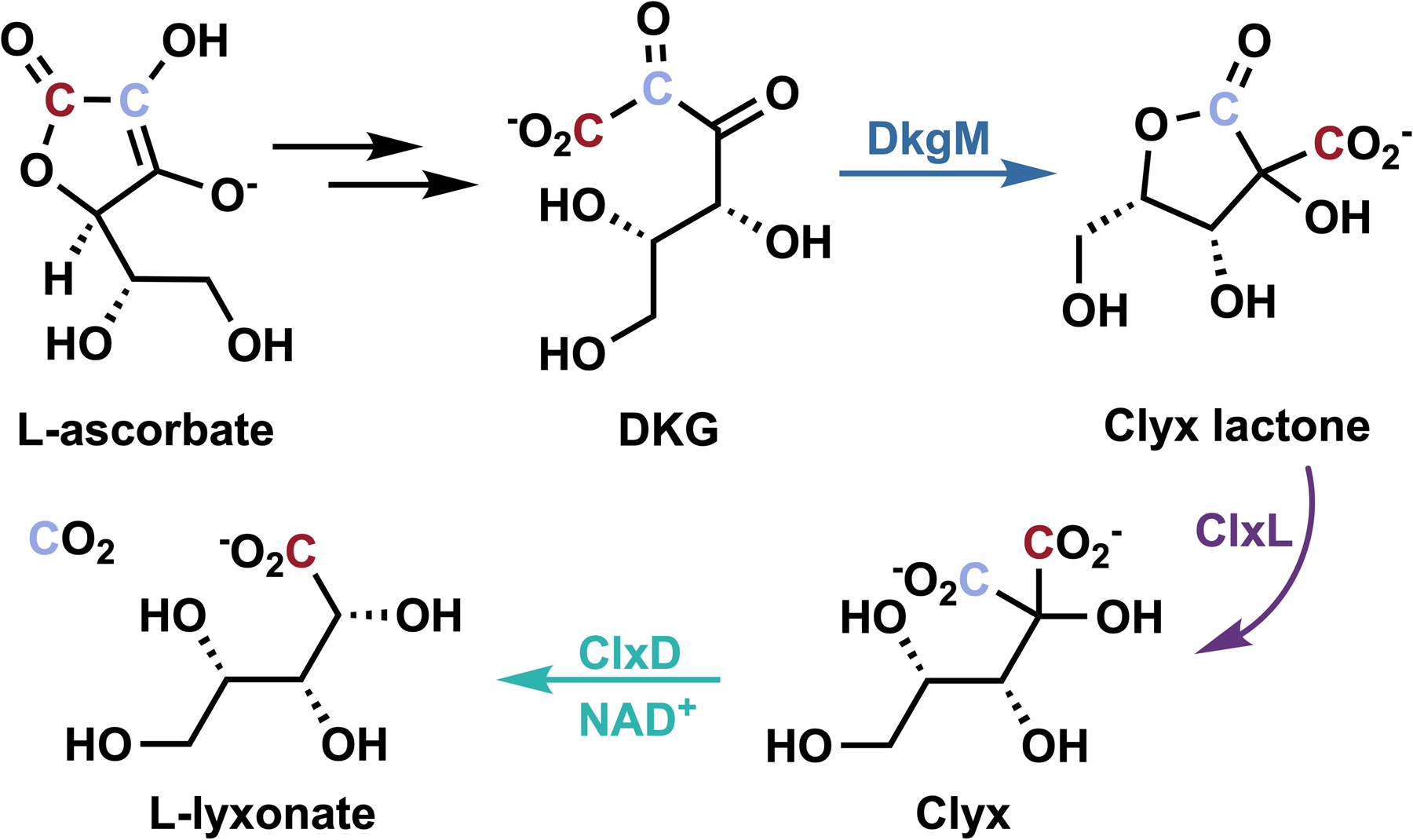
13C-labeled ascorbate was used to support our proposed pathway. The C1 of L-ascorbate becomes C1 of L-lyxonate, while C2 of L-ascorbate is removed as CO2.
As described earlier, the L-lyxonate product of the ClxD reaction is catabolized by the enzymes in module 3 (the P. aeruginosa pathway) to produce α-ketoglutarate (Figure 1C).
The complete catabolic pathway from L-ascorbate to α-ketoglutarate is encoded in the R. eutropha genome in three distinct neighborhoods, each encoding enzymes in one of the three functionally discrete modules (Figure 1). Albeit via a combination of enzymes from other families, Module 1 (from L-ascorbate to DKG) is shared with organisms that catabolize DKG using the E. coli catabolic pathways.3 In the E. coli pathways, DKG is reduced, phosphorylated and decarboxylated to form D-xylulose 5-phosphate; in the R. eutropha pathway DKG is decarboxylated to form L-lyxonate.
DKG, the substrate for the second module in the R. eutropha pathway, is a spontaneous environmental degradation product of L-ascorbate10 and is a likely catabolic starting point for the 148 organisms that have only the last two modules as determined by a combined SSN/GNN analysis (Supplemental Data).
2-Carboxy-L-lyxonate, a naturally occurring plant metabolite derived from L-ascorbate,10,17 is a probable catabolic starting point for the 415 organisms that have only the third module (Supplemental Data).
In summary, we have reported identification, characterization, and annotation of the genes and enzymes involved in a previously unknown L-ascorbate catabolic pathway (Table 1, Figure S17) found in R. eutropha, and hundreds of other organisms, including the bacterial pathogens P. aeruginosa and Acinetobacter baumanii (Supplemental Data). The pathway includes two unprecedented enzymatic reactions: the first enzyme-catalyzed benzilic acid rearrangement and the first decarboxylase to use NAD+ as a cofactor, not a cosubstrate.
Table 1.
UniProt IDs and L-Ascorbate Catabolic Pathway Enzyme Names
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the National Institutes of Health P01GM118303, Drury University, and Salisbury University. We thank Dr. Xudong Guan, NMR specialist at the Institute for Genomic Biology, University of Illinois at Urbana-Champaign for assistance with the acquisition of NMR spectra. The NMR data was collected in the IGB Core on a 600 MHz NMR funded by NIH grant number S10-RR028833. We thank Dr. Zhong Lucas Li, Director of Metabolomics at the Roy J. Carver Biotechnology Center for collecting the mass spectral data. We thank the Drury University 2016 Fall BIOL 322 Advanced Genetics course for their efforts in primer design and molecular cloning.
Funding Sources
No competing financial interests have been declared.
Footnotes
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website.
Experimental details, RNA-Seq results, sequence similarity networks, genome neighborhood networks, NMR spectra, and mass spectra values (PDF)
List of organisms encoding L-ascorbate catabolic enzymes with UniProt IDs, list of upregulated genes by RNASeq (xlsx)
REFERENCES
- (1).Hancock RD; Viola R Biosynthesis and Catabolism of L-Ascorbic Acid in Plants. Critical Reviews in Plant Sciences 2005, 24 (3), 167–188. 10.1080/07352680591002165. [DOI] [Google Scholar]
- (2).Smirnoff N Ascorbic Acid Metabolism and Functions: A Comparison of Plants and Mammals. Free Radic. Biol. Med 2018, 122, 116–129. 10.1016/j.freeradbiomed.2018.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Yew WS; Gerlt JA Utilization of L-Ascorbate by Escherichia Coli K-12: Assignments of Functions to Products of the Yjf-Sga and Yia-Sgb Operons. J. Bacteriol 2002, 184 (1), 302–306. 10.1128/JB.184.1.302-306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhang Z; Aboulwafa M; Smith MH; Milton H Saier, J. The Ascorbate Transporter of Escherichia Coli. Journal of Bacteriology 2003, 185 (7), 2243–2250. 10.1128/JB.185.7.2243-2250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhang X; Carter MS; Vetting MW; San Francisco B; Zhao S; Al-Obaidi NF; Solbiati JO; Thiaville JJ; de Crécy-Lagard V; Jacobson MP; Almo SC; Gerlt JA Assignment of Function to a Domain of Unknown Function: DUF1537 Is a New Kinase Family in Catabolic Pathways for Acid Sugars. Proc. Natl. Acad. Sci. U.S.A 2016, 113 (29), E4161–4169. 10.1073/pnas.1605546113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Truffault V; Fry SC; Stevens RG; Gautier H Ascorbate Degradation in Tomato Leads to Accumulation of Oxalate, Threonate and Oxalyl Threonate. The Plant Journal 2017, 89 (5), 996–1008. 10.1111/tpj.13439. [DOI] [PubMed] [Google Scholar]
- (7).Gerlt JA; Bouvier JT; Davidson DB; Imker HJ; Sadkhin B; Slater DR; Whalen KL Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A Web Tool for Generating Protein Sequence Similarity Networks. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 2015, 1854 (8), 1019–1037. 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ghasempur S; Eswaramoorthy S; Hillerich BS; Seidel RD; Swaminathan S; Almo SC; Gerlt JA Discovery of a Novel L-Lyxonate Degradation Pathway in Pseudomonas Aeruginosa PAO1. Biochemistry 2014, 53 (20), 3357–3366. 10.1021/bi5004298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lu P; Ma D; Yan C; Gong X; Du M; Shi Y Structure and Mechanism of a Eukaryotic Transmembrane Ascorbate-Dependent Oxidoreductase. PNAS 2014, 111 (5), 1813–1818. 10.1073/pnas.1323931111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Green MA; Fry SC Vitamin C Degradation in Plant Cells via Enzymatic Hydrolysis of 4-O-Oxalyl-L-Threonate. Nature 2005, 433 (7021), 83–87. 10.1038/nature03172. [DOI] [PubMed] [Google Scholar]
- (11).Deutsch JC Spontaneous Hydrolysis and Dehydration of Dehydroascorbic Acid in Aqueous Solution. Analytical Biochemistry 1998, 260 (2), 223–229. 10.1006/abio.1998.2700. [DOI] [PubMed] [Google Scholar]
- (12).Baker SC; Saunders NFW; Willis AC; Ferguson SJ; Hajdu J; Fülöp V Cytochrome Cd1 Structure: Unusual Haem Environments in a Nitrite Reductase and Analysis of Factors Contributing to β-Propeller Folds11Edited by K. Nagai. Journal of Molecular Biology 1997, 269 (3), 440–455. 10.1006/jmbi.1997.1070. [DOI] [PubMed] [Google Scholar]
- (13).Plata G; Fuhrer T; Hsiao T-L; Sauer U; Vitkup D Global Probabilistic Annotation of Metabolic Networks Enables Enzyme Discovery. Nature Chemical Biology 2012, 8 (10), 848–854. 10.1038/nchembio.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lin Z; Su X; Chen W; Ci B; Zhang S; Lin H Dph7 Catalyzes a Previously Unknown Demethylation Step in Diphthamide Biosynthesis. J. Am. Chem. Soc 2014, 136 (17), 6179–6182. 10.1021/ja5009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Löwendahl L; Petersson G Conversion of Dehydroascorbic Acid to a Branched Hexaric Acid in Neutral and Alkaline Aqueous Solution. Analytical Biochemistry 1976, 72 (1), 623–628. 10.1016/0003-2697(76)90575-3. [DOI] [PubMed] [Google Scholar]
- (16).Oppenheimer NJ NAD+ and NADP+ as Prosthetic Groups for Enzymes In Encyclopedia of Life Sciences (ELS); John Wiley & Sons, 2010. 10.1002/9780470015902.a0000637.pub2. [DOI] [Google Scholar]
- (17).Parsons HT; Yasmin T; Fry SC Alternative Pathways of Dehydroascorbic Acid Degradation in Vitro and in Plant Cell Cultures: Novel Insights into Vitamin C Catabolism. Biochemical Journal 2011, 440 (3), 375–385. 10.1042/BJ20110939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


