Abstract
Background:
Environmental pollution contributes to fatty liver disease pathogenesis. Polychlorinated biphenyl (PCB) exposures have been associated with liver enzyme elevation and suspected steatohepatitis in cohort studies. Male mice treated with the commercial PCB mixture, Aroclor 1260 (20 mg/kg), and fed high fat diet (HFD) for 12 weeks developed steatohepatitis. Receptor-based modes of action including inhibition of the epidermal growth factor (EGF) receptor were previously proposed, but other mechanisms likely exist.
Objectives:
To identify and validate the pathways, transcription factors, and mechanisms responsible for the steatohepatitis associated with PCB and HFD co-exposures.
Methods:
Comparative proteomics analysis was performed in archived mouse liver samples from the aforementioned chronic exposure study. Pathway and transcription factor analysis (TFA) was performed, and selected results were validated.
Results:
Liver proteomics detected 1103 unique proteins. Aroclor 1260 upregulated 154 and downregulated 93 of these. Aroclor 1260+HFD co-exposures affected 55 pathways including glutathione metabolism, intermediary metabolism, and cytoskeletal remodeling. TFA of Aroclor 1260 treatment demonstrated alterations in the function of 42 transcription factors including downregulation of NRF2 and key nuclear receptors previously demonstrated to protect against steatohepatitis (e.g., HNF4α, FXR, PPARα/δ/γ, etc.). Validation studies demonstrated that Aroclor 1260 significantly reduced HNF4α protein levels; while Aroclor 1260+HFD reduced expression of the HNF4α target gene, albumin, in vivo. Aroclor 1260 attenuated EGF-dependent HNF4α phosphorylation and target gene activation in vitro. Aroclor 1260 reduced levels of NRF2, its target genes, and glutathione in vivo. Aroclor 1260 attenuated EGF-dependent NRF2 upregulation, in vitro. Aroclor 1260 indirectly activated hepatic stellate cells in vitro via induction of hepatocyte-derived TGFβ.
Conclusion:
PCB exposures adversely impacted transcription factors regulating liver protection, function, and fibrosis. PCBs, thus, compromised the liver by reducing its protective responses against nutritional stress to promote diet-induced steatohepatitis. The identified mechanisms by which environmental pollutants influence fatty liver disease pathogenesis require confirmation in humans.
Graphical Abstract

INTRODUCTION:
Non-alcoholic fatty liver disease (NAFLD) has an estimated 25% prevalence worldwide (1). While NAFLD is associated with obesity, obesity does not explain the genesis of NAFLD in all cases. Moreover, only some subjects with NAFLD develop progressive liver disease characterized by steatohepatitis and fibrosis. More recently, environmental pollution exposures have been shown to impact the genesis and progression of NAFLD [reviewed (2–6)]. The term, toxicant associated steatohepatitis (TASH), has been coined to denote the steatohepatitis associated with chemical pollutants (7). However, environmental hepatology remains an understudied field. The mechanisms by which pollution impacts NAFLD pathogenesis are largely unknown.
Polychlorinated biphenyls (PCBs) are hepatotoxic, endocrine and metabolism disrupting chemicals which contaminate the food supply and accumulate in human adipose and liver (2, 4). All evaluated adult subjects enrolled in the National Health and Nutrition Examination Survey (NHANES) 2003–2004 had detectable PCB levels (8). Multiple cohort studies demonstrate associations between PCB exposures and liver enzymes and/or suspected NAFLD (8–11). Animal studies suggest a causal role for PCBs in NAFLD (12–14). We recently demonstrated that exposure to Aroclor 1260 (A1260), a commercially produced PCB mixture enriched in non-dioxin-like PCBs, in combination with a high fat diet (HFD) caused steatohepatitis in mice (12, 14). The PCB congeners in Aroclor 1260 mixture are representative of the PCBs composition found in human fat. Receptor-based modes of action have been proposed for endocrine disrupters including PCBs (2). PCBs have been shown to activate human and murine aryl hydrocarbon receptor (AhR) and some nuclear receptors (NRs) [e.g., the pregnane X receptor (PXR) and the constitutive androstane receptor (CAR)]; while inhibiting epidermal growth factor receptor (EGFR) signaling (12–18). Knocking out PXR or CAR modulated, but did not prevent the steatohepatitis associated with Aroclor 1260 and high fat diet co-exposures, implicating additional mechanisms (14).
Proteomics is a well-established discovery tool in both hepatology and environmental health research. We hypothesized that liver proteomics would discover new mechanisms for PCBs in liver disease progression using our established mouse model (12). Selected mechanisms were validated, and the potential influence of epidermal growth factor signaling was investigated in some cases.
METHODS:
Animal Exposures
The mouse liver samples used for proteomics analysis in this study were obtained from archived (−80°C) tissues from a prior study (12). Briefly, 8-week old C57BL/6 male mice were treated with Aroclor 1260 (20 mg/kg) (Accustandard, CT), or vehicle control (corn oil) by one-time gavage and fed a control or high fat diet (42% milk fat) for 12 weeks (n=10 per group). Male mice were selected as PCB-exposed men in the Anniston Community Health Survey (ACHS) had a higher prevalence of TASH (9). Aroclor 1260 was selected because its composition is similar to the PCBs previously shown to bio-accumulate in human adipose (18), and the dose was selected to model the high end of human exposures in ACHS. Mice fed HFD (the HFD group) developed steatosis, while HFD-fed mice co-exposed to Aroclor 1260 (the HFD+ group) developed steatohepatitis (12). Control diet-fed mice did not develop fatty liver disease whether they were exposed to PCB vehicle control (the CD group) or Aroclor 1260 (the CD+ group). The workflow of this study is illustrated in Figure 1A. A n=3 of each of the 4 groups were used for the hepatic proteomic analysis. The hepatic concentrations of 90 PCB congeners were measured at the University of Kentucky Superfund Research Center, and the sum of these means was reported (See Supporting Information Supplemental Methods). These PCB levels were measured in nine additional wild type C57BL/6 mice identically exposed to Aroclor 1260 (20 mg/kg) and fed HFD for 12 weeks. Archived liver samples from these mice were provided from another previously published study (14). All animal protocols were approved by the University of Louisville’s Institutional Animal Care and Use Committee (IACUC), and all animals were treated humanely as outlined in the “Guide for the Care and Use of Laboratory Animals” (19).
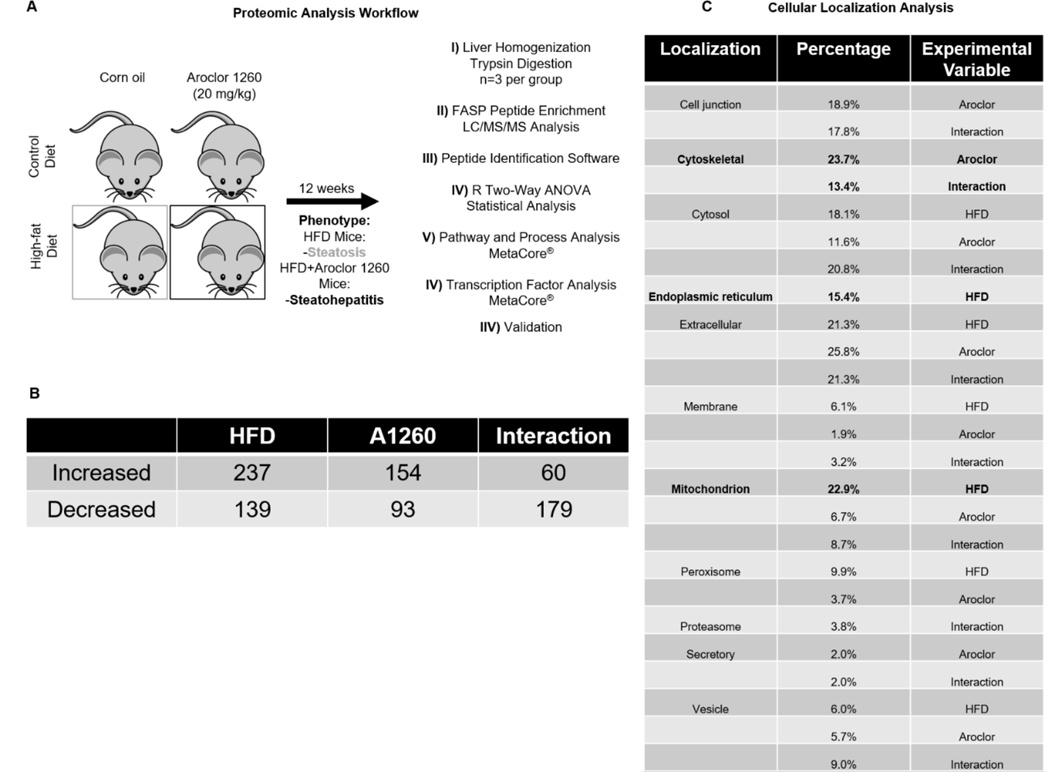
Figure 1: Liver proteomic analysis identifies protein alterations due to HFD, Aroclor 1260, and their interaction in a mouse model of fatty liver disease.
A. Proteomic analysis workflow of mouse liver samples. B. Significant (p=0.05) differential abundances of hepatic proteins due to a HFD, Aroclor 1260 exposure, and their interaction. A complete list of significant results is provided in Supporting Information Tables S2A–C. Cellular localization of the significant proteins for each variable. Three biological replicates per group was used for the proteomic analysis (12 in total). Peptide abundances were compared by two-way ANOVA, and only peptides that were significant (p<0.05) were investigated further.
Peptide Extraction, LC/MS/MS Analysis, and Data Sharing
Three samples per group were randomly selected for proteomic analysis. Mouse livers were homogenized in 1% SDS RIPA buffer. Proteins were reduced with DTT followed by denaturation with 8M Urea, alkylation with iodoacetamide, these reagents were purchased from Sigma-Aldrich (St. Louis, MO). Peptides were digested with Trypsin purchased from Promega (Madison, WI) followed by enrichment and clean up with C18 filters purchased from The Nest Group (Southborough, MA). Protein lysates (100 μg per sample) were trypsinized using filter aided sample preparation (20). Peptide concentrations were measured by a bicinchoninic acid assay, and equal amounts of peptides were injected into the high-performance liquid chromatography machine in a 0.1% formic acid and 80% acetonitrile gradient. A Finnigan LTQ mass spectrometer (ThermoFisher) was used to collect mass spectral data from the LC eluate. The generated msf files from Proteome Discoverer were loaded into Scaffold Q+S v4.4.5. Scaffold was used to calculate the false discovery rate (FDR) using the Peptide and Protein Prophet algorithms. Data files for acquired LCMS data (.RAW), search engine files (.mgf), peak list files (.mzML) files, and search results aggregated into a Scaffold3 (.sf3, ProteomeSoftware.com) were deposited with MassIVE (http://massive.ucsd.edu/) data repository with the Center for Computational Mass Spectrometry at the University of California, San Diego and shared with the ProteomeXchange (www.proteomexchange.org).
Protein Mapping and MetaCore Analysis
The data were analyzed with Proteome Discoverer v1.4.1.14. The database used in the Mascot v2.5.1 and SequestHT algorithms was the 3/7/2016 version of the UniprotKB Mus musculus reference proteome canonical and isoform sequences with the nonmurine sequences from the 1/1/2012 version of the gpm.org cRAP database appended to it. Cellular Localization Analysis, Pathways and Processes Analysis, as well as Transcription Factor Analysis (TFA) were performed using Metacore Software (Clarivate Analytics, Philadelphia, PA). Hepatic proteins that had significantly differential abundance (p-value < 0.05, by two-way ANOVA examining the diet and exposure factors) were analyzed by MetaCore software. Results that met the FDR threshold (< 0.05) were accepted. For the MetaCore TFA, when making direct comparisons between proteins of the HFD group and the HFD+, a criterion of at least a 3-fold increase or 2-fold decrease was used to select the proteins analyzed. Transcription factors are listed by highest activity (largest z-score/target gene differential) to the lowest activity.
Cell Culture
Cells were seeded at 1×104 per well in 12-well plates and grown to confluence for 24 hours. HepG2 (10% FBS), A-431 (10% FBS) and LX-2 cells (2% FBS) were grown in Dulbecco’s modified Eagle’s medium (DMEM) (VWR, Radnor, PA) supplemented with fetal bovine serum and 1% antimycotic/antibiotic solution (Mediatech, Manassas, VA). AML-12 cells were grown in 1:1 DMEM/F12 media (VWR Radnor, PA) supplemented with ITS Universal Culture Supplement Premix (VWR Radnor, PA), dexamethasone (40 ng/ml), 1% antimycotic/antibiotic solution (Mediatech, Manassas, VA) and 10% fetal bovine serum (VWR Radnor, PA). The cells were incubated in a 5% carbon dioxide atmosphere and 95% humidity at 37° and sub-cultured every 2 days.
HepG2 cells are a male human hepatoblastoma cell line. AML-12 cells are mouse hepatocytes derived from a male transgenic mouse that expresses human TGFα. The A-431 cell line is a human epidermoid carcinoma cell line derived from a female patient. LX-2 cells are a human hepatic stellate cell (HSC) line used for studying liver fibrosis and HSC activation.
Hepatocyte nuclear factor 4 alpha (HNF4α) phosphorylation assay
AML-12 (CRL-2254) cells obtained from ATCC (Manassas, VA) were exposed to DMEM/F-12 media containing either EGF (1.2 nM) (EMD Millipore, Burlingtion, MA), Aroclor 1260 (10 μg/mL) and EGF (1.2 nM), or 0.1% DMSO for 30 minutes then protein was extracted to measure phosphorylated HNF4α, total HNF4α, phosphorylated EGFR and total EGFR.
Nuclear factor (erythroid-derived 2)-like 2 (NRF2) protein stability assay
A-431 (CRL-1555) cells obtained from ATCC (Manassas, VA) were exposed to media containing either EGF (1.2 nM), Aroclor 1260 (10 μg/mL) and EGF (1.2 nM), or 0.1% DMSO for 2 hours then protein was extracted to measure total NRF2 and β-actin. The A-431 cell line was used as it has adequate EGFR and NRF2 expression and the protocol had previously been validated by others (21).
Hepatic Stellate Cell activation assay
LX-2 cells were obtained from Sigma-Aldrich (St. Louis, MO) while HepG2 cells (HB-8065) were obtained from ATCC (Manassas, VA). Initially LX-2 and HepG2 cells were cultured in DMEM media containing either 10 μg/mL of Aroclor 1260, or 0.1% DMSO. RNA was extracted at 24 hours, and 48 hours. Subsequently, some LX-2 cells were treated with conditioned media from the HepG2 cells and RNA was again prepared.
Western Blot Analysis
Liver tissue and cells were lysed in RIPA Buffer (100 mg tissue/0.5 mL RIPA supplemented with protease, and phosphatase inhibitors (10 μL/mL, Sigma-Aldrich, St. Louis, MO). Protein concentration was determined by the bicinchoninic acid protein assay (Sigma-Aldrich, St. Louis, MO). Protein (15 μg) was separated on 7.5% SDS Gel (BioRad, Hercules, CA), transferred to polyvinylidene difluoride membranes. Membranes were incubated in phosphate buffered saline (PBS), pH 7.5 containing 1% Tween 20 (PBS-T) and 5% fat free Milk for 1 hour at room temperature. Membranes were incubated at 4° C overnight with a primary antibody then washed followed by incubation with secondary antibody (Cell Signaling Technology, Danvers, MA). Membranes were incubated with ECL (Thermo Scientific, Waltham, MA) and luminescent signals were captured with BioRad Chemidoc Imaging System (Hercules, CA). For detection of phosphoproteins, the phospho-antibody blots were stripped for 15 minutes in Restore PLUS Western Blotting Stripping Buffer (Thermo Scientific, Waltham, MA) then re-probed using primary antibody against total protein overnight in 5% FF Milk in PBS-T. The following antibodies were used and purchased from Abcam (Cambridge, MA): Hnf4α, Nrf2, Y1173 EGFR; Thermo-scientific (Waltham, MA): S313 Hnf4α; Cell Signaling Technology (Danvers, MA): β-actin; and Santa Cruz (Dallas, TX) EGFR.
Real-time PCR Analysis
Mouse livers or cells were homogenized, and total RNA was extracted using the RNA-STAT 60 protocol (Tel-Test, Austin, Texas). RNA integrity and quantity were determined with the Nanodrop (ND-1000, Thermo Scientific, Wilmington Delaware) using the ND-1000 V3.8.1 software package. cDNA was synthesized from total RNA using the QuantiTect Reverse Transcription Kit (Qiagen, Valenci, California). PCR was performed on the BioRad CFX384 Real-Time System using the BioRad iTaq Universal Probes Supermix. Taqman Gene Expression Assays (Thermo-Fisher). Gene expression levels were calculated according to the 2-ΔΔCt method, and Gapdh was used as the endogenous control. The PCR primers utilized are provided in Supporting Information Table S1.
Total Glutathione Assay
Total glutathione concentration was determined by the method of Griffith (22, 23). Liver tissue was homogenized in PBS and centrifuged allowing determination of total thiol (predominately glutathione) through a colorimetric recycling assay at 412 nm. Total glutathione was normalized to liver protein.
Statistical Analysis
Western blot densitometry values, RNA expression values, and glutathione measurements were statistically analyzed using GraphPad Prism Version 7 for Macintosh (San Diego, CA). The data are expressed as box and whisker plots. Data in figure 3C were compared using one-way ANOVA. Data in figure 5A–C were statistically compared using an unpaired t-test. All other in vitro and in vivo datasets were compared using two-way ANOVA. P<0.05 was considered statistically significant. Quantified peptides from MS analysis were statistically compared by two-way ANOVA with a looping two-way ANOVA script in the R software package.
Figure 3: Aroclor 1260 reduced HNF4α protein expression and activity, and HNF4α is a downstream phosphorylation target of the EGFR inhibited by PCBs.
A. (i) Immunoblot analysis, and (ii) RT qPCR analysis of hepatic HNF4α protein and mRNA from mice fed a control diet (CD), control diet and exposed to Aroclor 1260 (20 mg/kg) (CD+), fed a HFD (HFD), and fed a HFD and exposed to Aroclor 1260 (20 mg/kg) (HFD+). B. RT qPCR Analysis of HNF4α target genes (i) Albumin, (ii) Cyp2c29, and (iii) Ttpa from murine liver. C. Western blot analysis of (i) Y1173 EGFR and (ii) HNF4α S313 phosphorylation in AML-12 cell lysates exposed to DMSO (0.1%), EGF (1.2 nM), and EGF+A1260 (1.2 nM EGF, 10 μg/mL). D. RT qPCR analysis of HNF4α target gene Pklr in AML-12s after 6-hour incubation with either DMSO (0.1%), EGF (1.2 nM), EGF+A1260 (1.2nM EGF, 10 μg/mL), or A1260 (10 μg/mL). All data are represented as box and whisker plots. An n=5 was used for the HNF4α protein levels analysis in vivo, an n=10 for Hnf4α mRNA and HNF4α target gene mRNA in vivo, an n=4 for HNF4α S313 phosphorylation, and an n=4 for Pklr mRNA. A P<0.05 is denoted with *. In the in vivo datasets an a denotes significance due to Aroclor, b HFD, and c interaction. Two-way ANOVA was used to statistically compare the in vivo data. One-way ANOVA was used for the statistical analysis for Fig 3Ci-ii. A two-way ANOVA was used for the statistical analysis in Fig 3D; an a denotes significance due to EGF, b due to Aroclor 1260.
Figure 5: Aroclor exposure activates HSCs through hepatocyte-derived TGF-β.
A. RT qPCR analysis of (i) ACTA2 and (ii) PLIN2 in LX-2 cells directly exposed to either DMSO (0.1%) or A1260 (10 μg/mL) for 24 or 48 hours. B. RT qPCR analysis of (i) ACTA2 and (ii) PLIN2 in LX-2 cells exposed to HepG2 media from HepG2 cells exposed to either DMSO (0.1%) or A1260 (10 μg/mL) for 24 hours. C. Gene expression analysis of (i) PDGFα and (ii) TGF-β in HepG2 cells directly exposed to either DMSO (0.1%) or A1260 (10 μg/mL) for 24 hours. D. RT qPCR analysis of MEF2C in murine liver of mice fed a control or HFD with either exposure to Aroclor 1260 (20 mg/kg) or vehicle control, denoted as CD, CD+, HFD, HFD+. For in vitro studies, p<0.05 is denoted with *, p<0.01 **. For in vivo studies, a p<0.05 is denoted by a for Aroclor, b for HFD or c for interaction. A n=4 was used for figure 5A-C, n=10 for figure 5D and the data are presented as box and whisker plots. A two-tailed t-test was used to statistically compare datasets Fig 5A-C and a two-way ANOVA for 5D.
RESULTS:
Liver proteomic analysis identifies protein alterations due to HFD, Aroclor 1260, and their interaction in a steatohepatitis mouse model
Neither the CD nor the CD+ groups developed fatty liver disease. The HFD group developed steatosis, while the HFD+ group developed steatohepatitis (12). The concentrations of ninety PCB congeners were measured in the livers of Aroclor 1260-exposed mice fed HFD. The average ΣPCB concentration was 22,751±23,350 ng/g. Liver proteomics was performed and 1103 unique proteins were detected. Quantitative values for each measured protein were similar between biological replicates (Figure S1). Significant alterations in peptide abundance occurred with Aroclor 1260 exposure (154 increased, 93 decreased); HFD (237 increased, 139 decreased); and their interaction (60 increased, 179 decreased) (Figure 1B & Supporting Information Tables S2A–C). The cellular localization of affected proteins is shown in Figure 1C. The proteins significantly altered by a HFD were localized primarily to the mitochondria (p=3e−25) and the endoplasmic reticulum (ER, p=3.5e−19). Proteins significantly altered by Aroclor 1260 exposure were localized primarily to the cytoskeleton (p=8.4e−10). Proteins localized to cytoskeletal regions were also significantly affected by the interaction between HFD and Aroclor exposure (p=8.6e−7).
Analysis of pathways and processes impacted by differentially expressed proteins due to HFD, Aroclor 1260, or the interaction between HFD and Aroclor 1260
A number of pathways and processes were significantly affected by HFD (n=78), Aroclor 1260 (n=30), or the interaction between the two (n=55). The complete results of these analyses are provided in Supporting Information Tables S3A–F, and a subset of these are summarized in Figure 2. Pathways altered by HFD included enhanced ER stress (p=3.02e−13) and diminished oxidative phosphorylation (p=8e−5). These findings are consistent with previously published studies (24, 25). Pathways and processes altered by Aroclor exposure included cytoskeletal processes and muscle contraction (p=5.8e−13), possibly consistent with hepatic stellate cell activation (26); and reduced FXR regulation of glucose and lipid metabolism (p=0.002). Aroclor 1260 and HFD interacted to enrich similar cytoskeletal pathways (p=0.0001); while diminishing glutathione metabolism (p=4e−5); and altering intermediary metabolism.
Figure 2: Pathway analysis of proteins altered by either a HFD, Aroclor 1260 exposure or their interaction.
A, –Log(p-value) plots and pathway analysis of proteins significantly altered by HFD; B, Aroclor exposure; or C, their interaction. The complete list of altered pathways and processes are provided in Supporting Information Tables S3A–F. All pathways and processes identified through MetaCore analysis had to meet a FDR threshold <0.05.
Transcription factor analysis results
TFA predicted significant alterations in transcription factor activity with HFD (n=51), Aroclor 1260 (n=42), or their interaction (n=19) (Tables 1A–C) as determined by two-way ANOVA. HFD increased activity of transcription factors associated with cellular stress [e.g., transcription factor AP-1 (cJun, p=5.5e−5) and nuclear factor NF-kappa-B p65 subunit (NFkB, p=0.004)]; and ER stress [e.g., activating transcription factor-6 (ATF-6, p=1.3e−7), activating transcription factor-4 (ATF-4, p=0.0006), and X-box binding protein-1 (XBP1, p=0.0008)].
Tables 1A-C: Transcription factor analysis results.
TFA of proteins significantly diminished or enriched by experimental variables. Table 1A: HFD Transcription Factor Analysis, Table 1B: Aroclor 1260 Transcription Factor Analysis, Table 1C: HFD and Aroclor 1260 Interaction Transcription Factor Analysis
| Transcription Factor | Target Gene Differential | Z-Score Differential | p-value |
|---|---|---|---|
| ATF-6 alpha (50kDa) | 6 | 11.69 | 8.5e-08 |
| TAFII70 | 3 | 7.937 | 0.0003 |
| Androgen receptor | 3 | 7.937 | 0.0008 |
| TAF12 | 2 | 7.567 | 0.0007 |
| C/EBPalpha | 2 | 7.567 | 0.0003 |
| PPAR-alpha | 12 | 7.241 | 3.12e-06 |
| TFIID 30 kDa subunit | 2 | 6.609 | 0.0009 |
| TFIID 31 kD subunit | 2 | 6.609 | 0.003 |
| HNF3-beta | 2 | 6.609 | 0.001 |
| ATF-6 alpha (90kDa) | 3 | 6.51 | 0.001 |
| TR-beta | 8 | 5.803 | 1.8e-09 |
| GATA-4 | 9 | 5.775 | 2.9e-05 |
| TAF4 (TAFII130) | 2 | 5.64 | 0.005 |
| TAF5 | 2 | 5.64 | 0.006 |
| ESR1 (nuclear) | 30 | 5.502 | 2.7e-06 |
| c-Jun | 21 | 4.817 | 2.4e-05 |
| ATF-4 | 7 | 4.691 | 0.0005 |
| XBP1 | 7 | 4.645 | 0.0006 |
| PPAR-gamma | 13 | 4.449 | 6.2e-05 |
| FXR | 7 | 4.447 | 0.0008 |
| RelA (p65 NF-kB subunit) | 21 | 4.318 | 0.0008 |
| p53 | 25 | 4.046 | 0.0003 |
| HSF1 | 9 | 3.898 | 0.001 |
| RelB (NF-kB subunit) | 7 | 3.861 | 0.0004 |
| NRF2 | −3 | 3.78 | 0.0008 |
| LHX2 | 11 | 3.512 | 0.002 |
| HNF3-alpha | 7 | 3.427 | 0.004 |
| FOXO3A | 7 | 3.357 | 0.0003 |
| NF-kB1 (p50) | 11 | 3.276 | 0.003 |
| FKHR | 8 | 3.212 | 0.007 |
| c-Fos | 9 | 3.133 | 0.006 |
| HNF4-alpha | 10 | 3.073 | 9.6e-12 |
| GCR | 2 | −0.025 | 6.5e-06 |
| SP1 | −4 | −1.64 | 6.6e-06 |
| C/EBPbeta | 8 | −2.245 | 5.8e-06 |
| c-Myc | 0 | −3.767 | 1.56e-16 |
| YY1 | −18 | −3.85 | 0.0003 |
| ERR1 | −6 | −3.865 | 0.002 |
| LXR-alpha | −4 | −4.371 | 6.9e-05 |
| NURR1 | −3 | −5.265 | 0.002 |
| SP4 | −4 | −5.305 | 0.0009 |
| ROR-gamma | −3 | −5.521 | 0.002 |
| BACH1 | −3 | −5.709 | 0.001 |
| SREBP1 precursor | −4 | −5.753 | 0.0008 |
| NRF1 | −7 | −6.094 | 0.0002 |
| CREB-H | −3 | −6.659 | 0.0007 |
| ERR3 | −7 | −7.256 | 0.002 |
| MLX | −2 | −7.35 | 0.002 |
| SREBP1 (nuclear) | −9 | −8.192 | 6.6e-05 |
| HNF1-alpha | −14 | −9.272 | 3.82e-10 |
| SREBP2 (nuclear) | −7 | −9.464 | 3.1e-07 |
| Transcription Factor | Target Gene Differential | Z-Score Differential | p-value |
|---|---|---|---|
| BARX2 | 4 | 8.939 | 2.4e-06 |
| TR-beta | 9 | 8.623 | 1.9e-07 |
| TAF3 | 2 | 7.383 | 0.002 |
| C/EBPdelta | 6 | 5.317 | 0.0002 |
| GATA-4 | 6 | 5.021 | 0.0004 |
| C/EBPbeta | 13 | 4.922 | 1.1e-05 |
| Androgen receptor | 17 | 4.892 | 8.1e-06 |
| TBP | 5 | 4.855 | 0.0008 |
| MYOD | 6 | 4.847 | 0.0006 |
| p53 | 19 | 4.616 | 3.8e-05 |
| XBP1 | 5 | 4.408 | 0.002 |
| C/EBPalpha | 8 | 3.873 | 0.002 |
| LHX2 | 8 | 3.576 | 0.002 |
| CREB-H | 1 | 0.768 | 0.0004 |
| ESR1 (nuclear) | 7 | 0.718 | 0.0001 |
| c-Myc | 12 | 0.698 | 4e-06 |
| LXR-beta | 1 | 0.557 | 0.001 |
| SP1 | 8 | 0.454 | 0.0002 |
| HNF4-alpha | 0 | −2.204 | 2.5e-05 |
| HNF1-alpha | −4 | −3.536 | 0.007 |
| SREBP1 (nuclear) | −3 | −3.816 | 0.008 |
| PPAR-gamma | −5 | −3.866 | 0.004 |
| HNF3-alpha | −4 | −3.913 | 0.005 |
| USF1 | −4 | −3.99 | 0.004 |
| NRF2 | −4 | −4.003 | 0.004 |
| PPAR-alpha | −4 | −4.097 | 0.004 |
| PPAR-beta(delta) | −3 | −4.136 | 0.006 |
| NFYA | −4 | −4.52 | 0.002 |
| PBX1 | −3 | −4.769 | 0.004 |
| Oct-3/4 | −14 | −5.119 | 1.6e-05 |
| HIF1A | −10 | −5.268 | 0.0003 |
| BACH1 | −2 | −5.729 | 0.005 |
| TR-alpha | −4 | −5.782 | 0.0005 |
| HOXD13 | −2 | −5.925 | 0.004 |
| RXRA | −6 | −6.718 | 3.4e-05 |
| Securin | −2 | −6.933 | 0.002 |
| FXR | −6 | −7.6 | 1.1e-5 |
| ZNF202 | −2 | −7.856 | 0.001 |
| SREBP1 precursor | −4 | −8.937 | 3.8e-05 |
| MLX | −2 | −10.99 | 0.0004 |
| ZNF395 | −1 | −12.4 | 0.006 |
| LXR-alpha | −8 | −13.17 | 9.6e-10 |
| Transcription Factor | Target Gene Differential | Z-Score Differential | p-value |
|---|---|---|---|
| hnRNP D-like | 1 | 20.68 | 0.002 |
| ZGPAT | 1 | 20.68 | 0.002 |
| GABP beta1 | 2 | 9.571 | 0.0009 |
| ATF-6 alpha (50kDa) | 2 | 7.889 | 0.001 |
| HNF3-alpha | 5 | 6.269 | 0.0001 |
| TR-beta | 4 | 6.057 | 0.0004 |
| p53 | 9 | 3.713 | 0.001 |
| ESR1 (nuclear) | −19 | −3.557 | 0.0002 |
| NFYA | −6 | −4.235 | 0.002 |
| HNF4-alpha | −12 | −4.418 | 0.0002 |
| C/EBPbeta | −14 | −4.521 | 9.7e-05 |
| NRF2 | −8 | −5.316 | 0.0001 |
| KLF15 | −3 | −5.765 | 0.001 |
| NFYC | −3 | −6.092 | 0.001 |
| ChREBP | −5 | −6.342 | 0.0001 |
| LXR-alpha | −7 | −7.281 | 6.1e-06 |
| HNF4-gamma | −2 | −7.298 | 0.002 |
| MLX | −2 | −7.298 | 0.002 |
| c-Myc | −20 | −7.298 | 1.6e-05 |
Aroclor 1260 exposure diminished the activity of nuclear factor (erythroid-derived 2)-like 2 (NRF2, p=0.004) and several nuclear receptors implicated in hepatic lipid metabolism and NASH [e.g., hepatic nuclear factor 4 alpha (HNF4α, p=5.5e−5), hepatic nuclear factor alpha (HNF1α, p=0.007), farnesoid X receptor (FXR, p=1.1e−5), peroxisome proliferator-activated receptor alpha/delta/gamma (PPARα/δ/γ, p=0.004/p=0.006/p=0.004), retinoic X receptor alpha (RXRα, p=3.4e-5), liver x receptor α (LXRα, p=9.3e-10), and others.
Aroclor 1260 exposure increased transcriptional activity of myofibroblast specific transcription factors including myogenic differentiation factor 1 (MYOD, p=0.0007) and BarH-like homeobox gene 2 (BARX2, p=3.3e−5). Some PCB congeners are known to impact thyroid and sex steroid signaling. Aroclor 1260 exposure was associated with increased activities of thyroid receptor β (TRβ, p=1.9e−7), androgen receptor (AR, p=8.1e−6), and estrogen receptor 1 (ESR1, p=0.0001), but decreased activity of the thyroid receptor α (TRα, p=0.0005). Aroclor 1260 exposure was also associated with increased transcriptional activity of multiple CCAAT enhancer binding proteins (C/EBPs) including C/EBPα (p=0.02), C/EBPβ (p=1.1e−5), and C/EBPδ (p=0.0002). The interaction between HFD and Aroclor decreased the transcriptional activities of NRF2 (p=0.0002), C/EBPβ (p=9.7e−5), and three NRs [e.g., HNF4α (p=0.0004), hepatocyte nuclear factor gamma (HNF4γ) (p=0.002), estrogen receptor 1 (ESR1) (p=0.0002)].
TFA was also performed through comparison of the HFD+ vs. HFD treated groups of mice by fold-change thresholds (Supporting Information Table S4). Although some differences exist in the TFA results generated by the different methods, NRF2, HNF4α, and ESR1 remained negatively regulated in both. Myocyte enhancer factor 2c (MEF2C), a positive regulator of HSC activation was increased (p=1.6e−6) in HFD+ vs. HFD. CAMP responsive element binding protein 1 (CREB1) activity was decreased in HFD+ vs. HFD (p=1.8e−6). By and large, the TFA results support the pathway and process results. Based on consistency, intermediary metabolism (HNF4α), glutathione metabolism (NRF2), and cytoskeletal remodeling/fibrosis (MEF2C) were selected for validation.
Aroclor 1260 reduced HNF4α protein expression and activity, and HNF4α is a downstream phosphorylation target of the EGFR inhibited by PCBs
The TFA demonstrated that HNF4α activity was downregulated by Aroclor 1260 and the Aroclor 1260-HFD interaction. Thus, HNF4α was selected for validation and further investigation. Aroclor 1260 exposure significantly reduced HNF4α protein levels in vivo with either control diet (2.2-fold) or HFD (1.9-fold) feeding (p=0.02); and HFD significantly reduced HNF4α mRNA levels 3.1-fold (p=0.006) (Figure 3Ai–ii). Proteomics analysis demonstrated that several HNF4α target genes were reduced by Aroclor 1260, so the mRNA expression for some of these genes was determined. Albumin mRNA was reduced by Aroclor 1260 exposure (p=0.0001) and was markedly suppressed (4.6-fold) by the Aroclor 1260-HFD interaction (p=0.0003, Figure 3Bi). Likewise, cytochrome P450 family 2 subfamily c (Cyp2c29) mRNA was decreased 1.1-fold by the Aroclor 1260-HFD interaction (p=0.03, Figure 4Bii). Alpha tocopherol transfer protein (Ttpa) was diminished 1.2-fold due to the pollutant-diet interaction (p=0.01, Figure 3Biii).
Figure 4: Aroclor 1260 reduced NRF2 protein/mRNA expression and glutathione levels, and NRF2 is an EGF-sensitive target negatively impacted by PCBs.
A. (i) Immunoblot analysis, (ii) RT qPCR of hepatic NRF2 protein and mRNA, respectively, and total glutathione measurements from mice fed a control diet (CD), control diet and exposed to Aroclor 1260 (20 mg/kg) (CD+), a HFD (HFD), and HFD and exposed to Aroclor 1260 (20 mg/kg) (HFD+). B. RT qPCR analysis of hepatic NRF2 target genes (i) Nqo1, (ii), Gstm1, (iii) Me1, and (iv) Gclc in murine liver. C. (i) Immunoblot analysis of NRF2 protein stability in A-431 cells exposed to either DMSO (0.1%), EGF (1.2 nM), EGF+A1260 (1.2nM EGF, 10 μg/mL), or A1260 (10 μg/mL) for 2 hours. A p<0.05 is denoted by a for Aroclor, b for HFD/EGF or c for interaction in the in vivo datasets. All data are represented as box and whisker plots. A sample size of 5 was used for the analysis of the NRF2 protein levels in vivo, an n=10 for NRF2 mRNA and NRF2 target gene mRNA in vivo. An n=4 was used for the NRF2 in vitro assay and an a denotes significance due to EGF and c for interaction. Statistical significance was determined by two-way ANOVA.
Since PCBs disrupt EGFR signaling and HNF4α is regulated by phosphorylation, interactions between EGF and Aroclor 1260 on HNF4α were evaluated in vitro (Figure 3C). Compared to vehicle control, EGF treatment significantly increased the relative phosphorylation of both EGFR (p=0.02) and HNF4α (p=0.02) in AML-12 cells (Figure 3Ci–ii). These changes were significantly attenuated by Aroclor 1260 exposure as seen by a 1.5-fold decrease in phosphorylated EGFR; and a 2.8-fold decrease in phosphorylated HNF4α; Figure 3Ci–ii). Expression of the HNF4α target gene, pyruvate kinase, liver and RBC (Pklr) was increased by EGF (p=0.0005) at the mRNA level, and this was attenuated 2-fold by Aroclor 1260 (p=0.002, Figure 3D). The full list of mean values and standard deviations are listed in Supporting Information Table S5. In total, these data demonstrate that HNF4α protein, but not mRNA expression, is downregulated by Aroclor 1260 exposure. HNF4α function was reduced by Aroclor 1260 and/or the interaction between Aroclor 1260 and HFD. The latter interaction may potentially contribute to the observed transition from steatosis to steatohepatitis. Reduction in EGF-dependent HNF4α phosphorylation by Aroclor 1260 partially explains the diminished HNF4α activity. These data validate the observed alterations in some of the intermediary metabolism pathways/processes and TFA of the proteomic data.
Aroclor 1260 reduced NRF2 protein/mRNA expression and glutathione levels, and NRF2 is an EGF-sensitive target negatively impacted by PCBs
The TFA and pathway analysis of the proteomic data suggested that NRF2 and glutathione metabolism were downregulated by Aroclor 1260 and/or its interaction with HFD. Thus, NRF2 was selected for validation. Hepatic NRF2 protein, NRF2 mRNA, and total glutathione were diminished by Aroclor 1260 exposure in vivo (p=0.04, p=0.02, and p=0.03, respectively; Figure 4Ai–iii). In control diet-fed mice, Aroclor 1260 reduced NRF2 protein 3.3-fold, NRF2 mRNA by 1.4-fold, and glutathione by 3.1-fold. In HFD-fed mice, Aroclor 1260 reduced hepatic NRF2 mRNA by 1.3-fold and glutathione by 2-fold. Expression (mRNA) of NRF2 target genes was then evaluated in vivo (Figure 4Bi–iv). Aroclor 1260 reduced expression of NAD(P)H quinone dehydrogenase 1 (Nqo1, p=0.014), glutathione S-transferase mu 1 (Gstm1, p=0.015), and malic enzyme 1 (Me1, p=0.004). Expression of glutamate-cysteine ligase catalytic subunit (Gclc) was impacted by a significant interaction between Aroclor 1260 and HFD. Gclc expression was decreased 1.7-fold in HFD+ vs. HFD (p=0.045).
Since NRF2 has been shown to be regulated by EGFR signaling in C. Elegans (9) and PCBs inhibit EGFR signaling, the potential interactive effects of EGF and PCBs were determined on NRF2 protein stability in vitro (Figure 4C). A-431 cells were used as they express EGFR and NRF2 at detectable levels (21). EGF-treated A-431 cells had significantly higher NRF2 protein levels after two hours, suggesting that NRF2 is stabilized by EGF treatment alone as seen by a 3.6-fold increase. Aroclor 1260 greatly attenuated this effect (p=0.01) as demonstrated by a 2-fold decrease with EGF and Aroclor 1260. The full list of mean values and standard deviations are listed in Supporting Information Table S5.
These data by and large confirm the proteomic results regarding glutathione metabolism and NRF2 activity with some caveats. While the TFA demonstrated that both Aroclor 1260 and the pollutant-diet interaction impacted NRF2 activity; the glutathione metabolism pathway/process was only affected by the interaction. However, the protein and gene expression validation studies demonstrate that the reduction in glutathione metabolism and NRF2 expression/activity are predominantly Aroclor 1260 effects, rather than pollutant-diet interaction effects. PCBs may inhibit EGF-dependent NRF2 stabilization by disrupting EGFR signaling.
Aroclor exposure activates HSCs through hepatocyte-derived TGF-β
Hepatic stellate cells (HSCs) normally store Vitamin A and can transdifferentiate into proliferative myofibroblasts or “activated” HSCs. Since pathway/process analysis and TFA demonstrate that cytoskeletal processes were increased by both Aroclor exposure and the pollutant-HFD interaction, we hypothesized that Aroclor 1260 could activate HSCs. This was tested in vitro with LX-2 cells, an immortalized human hepatic stellate cell line commonly used for assessing mechanisms of liver fibrosis. In cultured LX-2 cells, Aroclor 1260 did not directly increase alpha-actin-2 (ACTA2, an activated HSC marker) or decrease perilipin 2 (PLIN2 a quiescent HSC marker) mRNA expression (Figure 5Ai–ii). In contrast, conditioned media from Aroclor 1260-exposed HepG2 cells increased ACTA2 gene expression in LX-2 at 48 hours by 3.3-fold (P=0.006, Figure 5Bi–ii). PLIN2 was unaffected. Aroclor 1260 exposure increased expression of the key fibrogenic cytokine, transforming growth factor-beta (TGF-β), in HepG2 cells by 2-fold (p=0.03, Figure 5Cii); but it did not change platelet derived growth factor alpha (PDGFα) expression (Figure 5Ci). Hepatic mRNA expression of Mef2c, a myofibroblast identity factor, was significantly increased 1.3-fold (HFD+ vs. HFD) by Aroclor 1260 exposure in vivo, confirming the TFA results. The full list of mean values and standard deviations are listed in Supporting Information Table S5. These data confirm the proteomics results and analyses and suggest that PCBs may be pro-fibrotic. PCB-induced stellate cell activation may be indirectly mediated by hepatocyte-derived TGF-β.
DISCUSSION:
Cohort studies, including the ACHS, reproducibly demonstrate associations between PCB exposures and liver injury. The ACHS cohort is comprised of PCB-exposed subjects with a high prevalence of obesity (9). In ACHS, PCB exposures were associated with suspected TASH characterized by increased circulating biomarkers of hepatocellular necrosis, insulin resistance, and inflammation (9). The animal exposure protocol utilized in the present study was designed to model the increased PCB exposures and body mass indices encountered in ACHS in order to examine potential liver disease mechanisms. Previous analysis of this model demonstrated that Aroclor 1260 did not cause fatty liver pathology in mice fed a healthy control diet, but worsened HFD-induced fatty liver disease (12). More specifically, while the HFD group developed steatosis, the HFD+ group developed steatohepatitis. The present proteomics analysis elucidated several novel mechanisms by which PCBs compromised the liver to promote diet-induced steatohepatitis.
The most important novel mechanistic concept revealed by this proteomics study is that PCBs attenuated the function of key hepatic transcription factors which protect against over-nutrition and steatohepatitis (e.g, NRF2, FXR, PPARα/δ/γ, CREB-1, HNF4α, etc.) while indirectly activating HSCs. The protective roles of these transcription factors include the regulation of liver antioxidant response, metabolism, function, and inflammation. While PCB research has historically focused on the positive PCB responses (e.g., activation of AhR, PXR, CAR, etc.), a key finding of this study is that the negative responses to Aroclor 1260 generally outnumbered the positive ones (e.g., pathway/receptor inhibition rather than activation). While the mechanisms of negative regulation of transcription factors by PCBs is largely unknown, this work demonstrates that their regulation may be indirect and related to reduced transcription factor expression and EGF-dependent phosphorylation (e.g., NRF2, HNF4α). Recently, PCBs were identified to be ‘signaling disrupting chemicals’ which reduced the hepatic phosphoproteome by 25% in TASH (15, 17). While direct ligand activation of NRs is well established, indirect phosphoregulation has been demonstrated to be just as critical for NR function (27, 28).
The observed Aroclor 1260-induced reduction in the function of the aforementioned transcription factors was not associated with significant histopathologic changes in mice fed CD. However, it likely played a causal role in promoting HFD-induced steatohepatitis. The rationale supporting this hypothesis stems from the observation that medications activating these transcription factors or related pathways are already being used clinically or are in late stage clinical trials for NASH (Table 2). If antioxidants and transcription factor agonists have therapeutic roles in NASH, it is plausible that their reduction by PCBs may be pathogenic. A two ‘hit’ model of fatty liver disease has previously been proposed. We previously hypothesized that Aroclor 1260 exposure is a second ‘hit’ mediating the progression from diet-induced steatosis to the more advanced steatohepatitis (12). However, based on the data generated by this proteomics study, it now appears that PCB exposures are the first ‘hit’ which compromise the liver to promote HFD (the second ‘hit’)-induced steatohepatitis (see TOC Graphic).
Table 2:
Pathways/Transcription Factors Inhibited by PCBs and Activated by NASH Therapies
Human confirmation of these results is required. However, if confirmed, the results could have meaningful clinical implications with regards to NASH pathogenesis and treatment. Moreover, it would solve an important paradox facing endocrine disrupter research. While metabolic diseases like NAFLD have increased in prevalence, some environmental pollutants, including PCBs, have declined in the US. Perhaps only low-dose exposures to environmental pollutants are required to promote the development of obesity-associated diseases like steatohepatitis. Regarding steatohepatitis therapy in PCB-exposed populations, a three-pronged approach targeting overweight/obesity, PCB levels, and PCB effects requires investigation. However, should the transcription factor data be confirmed in humans, they would inform strategies to pharmacologically target PCB effects. For example, TRβ agonists (currently in clinical trials for NASH) might not be effective for patients with increased PCB levels, since TRβ activity was up-regulated by Aroclor 1260. Drugs targeting down-regulated transcription factors (e.g., obeticholic acid, elafibranor, pentoxifylline, etc.) and antioxidants would theoretically be more effective. Alternatively, medications targeting PCB-induced phosphoprotein signaling disruption could be examined. While more data are clearly needed, environmental exposure assessment could play a role in personalized medicine for NASH in the future.
PCB-induced alterations in HNF4α, NRF2, and MEF2C were validated, and these results warrant discussion. HNF4α protein expression, EGF-dependent S313 phosphorylation, and transcriptional activity were reduced by Aroclor 1260 exposure. HNF4α is a critical NR for hepatocyte identity and function, and it transcriptionally regulates 60% of liver-specific genes (29). HNF4α-null mice spontaneously develop NAFLD (29). HNF4α has inconsistently been shown to be down-regulated in human NASH (30, 31). Altered liver HNF4α has been demonstrated following exposures to other metabolism disrupting chemicals (e.g., perfluoroalkyl substances (32)), but it has not previously been shown for PCBs. Aroclor 1260 impaired liver function, particularly in combination with HFD, by decreasing gene expression of the HNF4α regulated genes, albumin and Cyp2c29. HNF4α is also important for beta cell identity and function, and HNF4α polymorphisms have been associated with maturity onset diabetes of the young. Perhaps PCB-induced HNF4α inhibition in pancreas could account for the decreased insulin levels associated with PCB exposures in ACHS (9), but this concept requires confirmation.
NRF2 is a critical transcription factor that regulates the liver’s antioxidant response to oxidative stress. Steatohepatitis (33) and PCB exposures (34) have previously been associated with increased oxidative stress. Pathway and transcription factor analyses identified both glutathione metabolism and NRF2 as being downregulated in the steatohepatitis due to Aroclor 1260 and HFD co-exposures. This confirms the results of our previous metabolomics study demonstrating hepatic glutathione depletion in a PCB 153-treated HFD fed mouse model (35). Aroclor 1260 decreased hepatic NRF2 mRNA/protein levels, target gene expression, and total glutathione levels. Aroclor 1260 adversely impacted the EGFR-NRF2 signaling pathway. While this pathway is well-characterized in C. elegans and cancer; here it was demonstrated, perhaps for the first time, in mammalian liver. The increased oxidative stress associated with PCB exposures is due, at least in part, to decreased hepatic glutathione production. A limitation of this research is that the GSH:GSSG ratios and pro-oxidant levels were not measured. However, we postulate that the livers of HFD+ mice would have increased pro-oxidants and a decreased GSH:GSSG ratio. This would mean that the transition from steatosis (HFD) to steatohepatitis (HFD+) could be associated with increased oxidative stress in the absence of a compensatory antioxidant response to that stress. Intracellular glutathione levels modulate cell death mechanisms, and glutathione depletion is associated with necrosis rather than apoptosis (36). Perhaps that is one reason why PCB exposures in ACHS were associated with liver necrosis and TASH (9).
Pathway analysis demonstrated up-regulated cytoskeletal remodeling in response to PCBs and the PCB-HFD interaction. Because fibrosis a component of cytoskeletal remodeling in MetaCore and it determines survival in steatohepatitis, fibrosis was further examined. MEF2C is an HSC transcription factor that regulates fibrotic gene expression upon HSC activation (37); and TFA demonstrated its activation in the HFD+ (vs. HFD) group. Increased MEF2C expression was validated at the RNA level in vivo. In cell culture models, PCBs did not directly activate HSCs. However, PCBs stimulated increased expression of the potent HSC activating cytokine, TGF-β, in HepG2 cells. Conditioned media from Aroclor 1260-treated HepG2 cells activated LX-2 cells, and this activation was probably due to the TGF-β elaborated by the HepG2 cells. These data suggest that PCB exposures can promote a fibrotic response in liver through a mechanism involving TGF-β production from hepatocytes and MEF2C upregulation. A limitation of this research is that histological fibrosis was not increased in the 12-week mouse exposure model. However, activation of pro-fibrotic pathways at the molecular level is known to precede the histological development of fibrosis. With longer exposures (e.g., 16–24 weeks), the development of histological fibrosis would be anticipated with Aroclor 1260-HFD co-exposures in mice. In addition to fibrosis, cytoskeletal remodeling may have also been increased in hepatocytes. However, this possibility was not further examined in vivo. PCB exposures were, however, associated with increased circulating levels of the liver cytoskeletal protein, cytokeratin 18, in ACHS (9).
Other findings worthy of discussion include the impact of PCBs on sex hormone signaling and C/EBPs. Both NAFLD and PCBs chemicals have previously been associated with sex differences (2). In this study investigating male mice, estrogen and androgen receptor signaling were simultaneously upregulated by Aroclor 1260 exposure. However, TFA revealed a PCB-diet interaction which reduced ESR1 function. The potential role of PCB-mediated endocrine disruption in steatohepatitis requires further investigation. C/EBPs are a family of transcription factors which regulate diverse hepatic processes including metabolism, inflammation, and regeneration. In this study, the functions of C/EBPα, C/EBPβ, and C/EBPδ were up-regulated by Aroclor 1260. However, a PCB-diet interaction downregulated C/EBPβ. The role of C/EBPs and their modulation by PCBs in steatohepatitis warrants further investigation. However, in PCB-exposed steatotic livers, simultaneous inhibition of the EGFR and C/EBPβ function should impair liver regeneration.
In contrast to PCB effects, the hepatic pathways and processes impacted by HFD are well characterized. Our HFD results (e.g., mitochondrial dysfunction promoting oxidative stress as well as the upregulation of ER stress and inflammatory pathways) are consistent with previous studies (24, 25, 38, 39). These consistencies serve to further validate the results of this study. PCBs disrupted key hepatic functions and protective responses promoting HFD-induced cellular damage and steatohepatitis histopathology.
The Aroclor 1260 dose utilized in this study was selected to model the high end of human serum PCB levels in the ACHS cohort. This dose was based on a modification of a previously published chronic PCB exposure protocol in female rats (40). In that study, rats were fed a chow diet and exposure assessment was performed in liver, blood, and adipose. Although similar cumulative PCB doses were administered, liver levels were approximately 6-fold higher in the present study. Because PCBs are lipophilic, the observed difference was probably related to HFD-induced hepatic steatosis. If this is confirmed, then it would help to explain why PCB hepatotoxicity is enhanced by high fat feeding. However, sex, species, and PCB congener-specific differences may have also contributed. In the future, PCB levels should be simultaneously assessed in blood, liver, and adipose compartments in both animal models and human subjects with and without diet-induced steatosis. Because mean PCB levels were increased in the ACHS vs. NHANES cohorts (41), lower Aroclor 1260 doses could be used in the future to model the effects of PCB exposures in the general population.
In summary, this murine proteomics study elucidated novel modes of PCB action in steatohepatitis. PCB exposures adversely impacted nuclear receptors and other transcription factors regulating liver protection, function, and fibrosis. PCBs compromised the liver by reducing its protective responses against nutritional stress to promote diet-induced steatohepatitis. An indirect mode of PCB action in reducing NRF2 and HNF4α function was demonstrated, involving altered EGF-dependent transcription factor phosphorylation. PCBs induced cytoskeletal remodeling and indirectly increased HSC activation via hepatocyte-derived TGFβ. These results require confirmation in human subjects.
Supplementary Material
Tables S2A-C: Differentially Expressed Proteins
Table S1: PCR Primers
Figure S1: Linear Relationship for Peptides Between Biological Replicates in LC/MS/MS
Tables S3A-F: Significantly Altered Pathways and Processes
Table S4: Transcription factor analysis of HFD+ (steatohepatitis) vs. HFD (steatosis) groups
Table S5: List of Mean ± SD for data presented in Figures 3,4,5
Acknowledgments
Financial Support & Acknowledgements:
This work was supported in part by the National Institute of Environmental Health Sciences [F31ES028982, R35ES028373, R01ES021375, P42ES023716, and P42ES007380], the National Institute of General Medical Sciences [P20GM113226], and the National Institute on Alcohol Abuse and Alcoholism [P50AA024337]. The authors thank Dr. Heather Clair for her consultation on this study.
List of Abbreviations:
- ACTA2
alpha-actin-2
- ATF-4
activating transcription factor-4
- ATF-6
activating transcription factor-6
- AhR
aryl hydrocarbon receptor
- A1260
Aroclor 1260
- BARX2
BarH-like homeobox gene 2
- cJun
transcription factor AP-1
- CAR
Constitutive androstane receptor
- CD
control diet
- CD+
control diet plus Aroclor 1260 exposure
- C/EBP
CCAAT enhancer binding protein
- CREB1
cAMP responsive element binding protein 1
- Cyp2c29
cytochrome P450, family 2, subfamily c, polypeptide 29
- EGFR
epidermal growth factor receptor
- ER
endoplasmic reticulum
- ESR1
estrogen receptor 1
- FDR
false discovery rate
- FXR
farnesoid X receptor
- GCLC
glutamate-cysteine ligase catalytic subunit
- GSTM1
glutathione s-transferase mu 1
- HFD
high fat diet
- HFD+
high fat diet plus Aroclor 1260 exposure
- HNF1α
hepatic nuclear factor 1 alpha
- HNF4α
hepatocyte nuclear factor 4 alpha
- HNF4γ
hepatocyte nuclear factor gamma
- HSCs
Hepatic stellate cells
- LXRα
liver x receptor α
- ME1
Malic enzyme 1
- MEF2C
myocyte enhancer factor 2c
- MYOD
myocyte differentiation factor
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- NHANES
National Health and Nutrition Examination Survey
- NQO1
NAD(P)H quinone dehydrogenase 1
- NFkB
nuclear factor NF-kappa-B p65 subunit
- NRF2
nuclear factor, erythroid 2 like 2
- NRs
nuclear receptors
- PCBs
polychlorinated biphenyls
- PDGFα
platelet derived growth factor alpha
- PKLR
pyruvate kinase, liver and RBC
- PLIN2
perilipin 2
- PPAR α/δ/γ
peroxisome proliferator-activated receptors alpha/delta/gamma
- PXR
pregnane X receptor
- RIPA
radio immunoprecipitation assay
- TASH
toxicant associated steatohepatitis
- TFA
transcription factor analysis
- TGF-β
transforming growth factor-beta
- TRα
thyroid receptor alpha
- TRβ
thyroid receptor β
- TTPA
alpha-tocopherol transfer protein
- XBP1
X-box binding protein-1
Footnotes
Declaration of competing financial interests: The authors declare they have no actual or potential competing financial interests regarding this work.
Supporting Information:
Supplemental Methods: Exposure Assessment
REFERENCES:
- 1.Younossi ZM; Koenig AB; Abdelatif D; Fazel Y; Henry L; Wymer M, Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, (1), 73–84. [DOI] [PubMed] [Google Scholar]
- 2.Heindel JJ.; Blumberg B.; Cave M.; Machtinger R.; Mantovani A.; Mendez MA.; Nadal A.; Palanza P.; Panzica G.; Sargis R.; Vandenberg LN.; Vom Saal F., Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol 2017, 68, 3–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang AL; Beier JI, Interaction of volatile organic compounds and underlying liver disease: a new paradigm for risk. Biol Chem 2018, 399, (11), 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahlang B; Beier JI; Clair HB; Bellis-Jones HJ; Falkner KC; McClain CJ; Cave MC, Toxicant-associated steatohepatitis. Toxicol Pathol 2013, 41, (2), 343–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foulds CE; Trevino LS; York B; Walker CL, Endocrine-disrupting chemicals and fatty liver disease. Nat Rev Endocrinol 2017, 13, (8), 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang AL; Beier JI, Interaction of volatile organic compounds and underlying liver disease: a new paradigm for risk. Biol Chem 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cave M; Falkner KC; Ray M; Joshi-Barve S; Brock G; Khan R; Bon Homme M; McClain CJ, Toxicant-associated steatohepatitis in vinyl chloride workers. Hepatology 2010, 51, (2), 474–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cave M; Appana S; Patel M; Falkner KC; McClain CJ; Brock G, Polychlorinated biphenyls, lead, and mercury are associated with liver disease in American adults: NHANES 2003–2004. Environ Health Perspect 2010, 118, (12), 1735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clair HB; Pinkston CM; Rai SN; Pavuk M; Dutton ND; Brock GN; Prough RA; Falkner KC; McClain CJ; Cave MC, Liver Disease in a Residential Cohort With Elevated Polychlorinated Biphenyl Exposures. Toxicol Sci 2018, 164, (1), 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rantakokko P; Mannisto V; Airaksinen R; Koponen J; Viluksela M; Kiviranta H; Pihlajamaki J, Persistent organic pollutants and non-alcoholic fatty liver disease in morbidly obese patients: a cohort study. Environ Health 2015, 14, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serdar B; LeBlanc WG; Norris JM; Dickinson LM, Potential effects of polychlorinated biphenyls (PCBs) and selected organochlorine pesticides (OCPs) on immune cells and blood biochemistry measures: a cross-sectional assessment of the NHANES 2003–2004 data. Environ Health 2014, 13, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahlang B; Song M; Beier JI; Cameron Falkner K; Al-Eryani L; Clair HB; Prough RA; Osborne TS; Malarkey DE; Christopher States J; Cave MC, Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and non-alcoholic fatty liver disease. Toxicol Appl Pharmacol 2014, 279, (3), 380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahlang B; Perkins JT; Petriello MC; Hoffman JB; Stromberg AJ; Hennig B, A compromised liver alters polychlorinated biphenyl-mediated toxicity. Toxicology 2017, 380, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahlang B; Prough RA; Falkner KC; Hardesty JE; Song M; Clair HB; Clark BJ; States JC; Arteel GE; Cave MC, Polychlorinated Biphenyl-Xenobiotic Nuclear Receptor Interactions Regulate Energy Metabolism, Behavior, and Inflammation in Non-alcoholic-Steatohepatitis. Toxicol Sci 2016, 149, (2), 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardesty JE; Al-Eryani L; Wahlang B; Falkner KC; Shi H; Jin J; Vivace BJ; Ceresa BP; Prough RA; Cave MC, Epidermal Growth Factor Receptor Signaling Disruption by Endocrine and Metabolic Disrupting Chemicals. Toxicol Sci 2018, 162, (2), 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardesty JE; Wahlang B; Falkner KC; Clair HB; Clark BJ; Ceresa BP; Prough RA; Cave MC, Polychlorinated biphenyls disrupt hepatic epidermal growth factor receptor signaling. Xenobiotica 2017, 47, (9), 807–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardesty JE.; Wahlang B.; Falkner KC.; Shi H.; Jin J.; Wilkey D.; Merchant M.; Watson C.; Prough RA.; Cave MC., Hepatic signalling disruption by pollutant Polychlorinated biphenyls in steatohepatitis. Cell Signal 2019, 53, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlang B; Falkner KC; Clair HB; Al-Eryani L; Prough RA; States JC; Coslo DM; Omiecinski CJ; Cave MC, Human receptor activation by aroclor 1260, a polychlorinated biphenyl mixture. Toxicol Sci 2014, 140, (2), 283–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Council NR, Guide for the Care and Use of Laboratory Animals: Eighth Edition The National Academies Press: Washington, DC, 2011; p 246. [Google Scholar]
- 20.Wisniewski JR; Zougman A; Nagaraj N; Mann M, Universal sample preparation method for proteome analysis. Nat Methods 2009, 6, (5), 359–62. [DOI] [PubMed] [Google Scholar]
- 21.Huo L; Li CW; Huang TH; Lam YC; Xia W; Tu C; Chang WC; Hsu JL; Lee DF; Nie L; Yamaguchi H; Wang Y; Lang J; Li LY; Chen CH; Mishra L; Hung MC, Activation of Keap1/Nrf2 signaling pathway by nuclear epidermal growth factor receptor in cancer cells. Am J Transl Res 2014, 6, (6), 649–63. [PMC free article] [PubMed] [Google Scholar]
- 22.Tamasi V; Jeffries JM; Arteel GE; Falkner KC, Ebselen augments its peroxidase activity by inducing nrf-2-dependent transcription. Arch Biochem Biophys 2004, 431, (2), 161–8. [DOI] [PubMed] [Google Scholar]
- 23.Griffith OW, Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem 1980, 106, (1), 207–12. [DOI] [PubMed] [Google Scholar]
- 24.Dara L; Ji C; Kaplowitz N, The contribution of endoplasmic reticulum stress to liver diseases. Hepatology 2011, 53, (5), 1752–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malhi H; Kaufman RJ, Endoplasmic reticulum stress in liver disease. J Hepatol 2011, 54, (4), 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchida T; Friedman SL, Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol 2017, 14, (7), 397–411. [DOI] [PubMed] [Google Scholar]
- 27.Lalevee S; Ferry C; Rochette-Egly C, Phosphorylation control of nuclear receptors. Methods Mol Biol 2010, 647, 251–66. [DOI] [PubMed] [Google Scholar]
- 28.Negishi M, Phenobarbital Meets Phosphorylation of Nuclear Receptors. Drug Metab Dispos 2017, 45, (5), 532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunewardena S; Walesky C; Apte U, Global Gene Expression Changes in Liver Following Hepatocyte Nuclear Factor 4 alpha deletion in Adult Mice. Genom Data 2015, 5, 126–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lake AD; Chaput AL; Novak P; Cherrington NJ; Smith CL, Transcription factor binding site enrichment analysis predicts drivers of altered gene expression in nonalcoholic steatohepatitis. Biochem Pharmacol 2016, 122, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baciu C; Pasini E; Angeli M; Schwenger K; Afrin J; Humar A; Fischer S; Patel K; Allard J; Bhat M, Systematic integrative analysis of gene expression identifies HNF4A as the central gene in pathogenesis of non-alcoholic steatohepatitis. PLoS One 2017, 12, (12), e0189223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beggs KM; McGreal SR; McCarthy A; Gunewardena S; Lampe JN; Lau C; Apte U, The role of hepatocyte nuclear factor 4-alpha in perfluorooctanoic acid- and perfluorooctanesulfonic acid-induced hepatocellular dysfunction. Toxicol Appl Pharmacol 2016, 304, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanyal AJ; Chalasani N; Kowdley KV; McCullough A; Diehl AM; Bass NM; Neuschwander-Tetri BA; Lavine JE; Tonascia J; Unalp A; Van Natta M; Clark J; Brunt EM.; Kleiner DE.; Hoofnagle JH.; Robuck PR.; Nash CRN., Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med 2010, 362, (18), 1675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buha A; Antonijevic B; Milovanovic V; Jankovic S; Bulat Z; Matovic V, Polychlorinated biphenyls as oxidative stress inducers in liver of subacutely exposed rats: implication for dose-dependence toxicity and benchmark dose concept. Environ Res 2015, 136, 309–17. [DOI] [PubMed] [Google Scholar]
- 35.Shi X; Wahlang B; Wei XL; Yin XM; Falkner KC; Prough RA; Kim SH; Mueller EG; McClain CJ; Cave M; Zhang X, Metabolomic Analysis of the Effects of Polychlorinated Biphenyls in Nonalcoholic Fatty Liver Disease. Journal of Proteome Research 2012, 11, (7), 3805–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandes RS; Cotter TG, Apoptosis or necrosis: intracellular levels of glutathione influence mode of cell death. Biochem Pharmacol 1994, 48, (4), 675–81. [DOI] [PubMed] [Google Scholar]
- 37.Wang X; Tang X; Gong X; Albanis E; Friedman SL; Mao Z, Regulation of hepatic stellate cell activation and growth by transcription factor myocyte enhancer factor 2. Gastroenterology 2004, 127, (4), 1174–88. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Ruiz I; Solis-Munoz P; Fernandez-Moreira D; Grau M; Colina F; Munoz-Yague T; Solis-Herruzo JA, High-fat diet decreases activity of the oxidative phosphorylation complexes and causes nonalcoholic steatohepatitis in mice. Dis Model Mech 2014, 7, (11), 1287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lockman KA; Htun V; Sinha R; Treskes P; Nelson LJ; Martin SF; Rogers SM; Le Bihan T; Hayes PC; Plevris JN, Proteomic profiling of cellular steatosis with concomitant oxidative stress in vitro. Lipids Health Dis 2016, 15, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.NTP, Toxicology and carcinogenesis studies of a binary mixture of 3,3’,4,4’,5-pentachlorobiphenyl (PCB 126) (Cas No. 57465–28-8) and 2,2’,4,4’,5,5’-hexachlorobiphenyl (PCB 153) (CAS No. 35065–27-1) in female Harlan Sprague-Dawley rats (gavage studies). Natl Toxicol Program Tech Rep Ser 2006, (530), 1–258. [PubMed] [Google Scholar]
- 41.Pavuk M; Olson JR; Sjodin A; Wolff P; Turner WE; Shelton C; Dutton ND; Bartell S, Serum concentrations of polychlorinated biphenyls (PCBs) in participants of the Anniston Community Health Survey. Sci Total Environ 2014, 473–474, 286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neuschwander-Tetri BA; Loomba R; Sanyal AJ; Lavine JE; Van Natta ML; Abdelmalek MF; Chalasani N; Dasarathy S; Diehl AM; Hameed B; Kowdley KV; McCullough A; Terrault N; Clark JM; Tonascia J; Brunt EM; Kleiner DE; Doo E; Network NCR, Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 2015, 385, (9972), 956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ratziu V; Harrison SA; Francque S; Bedossa P; Lehert P; Serfaty L; Romero-Gomez M; Boursier J; Abdelmalek M; Caldwell S; Drenth J; Anstee QM; Hum D; Hanf R; Roudot A; Megnien S; Staels B; Sanyal A; Group G-IS, Elafibranor, an Agonist of the Peroxisome Proliferator-Activated Receptor-alpha and -delta, Induces Resolution of Nonalcoholic Steatohepatitis Without Fibrosis Worsening. Gastroenterology 2016, 150, (5), 1147–1159 e5. [DOI] [PubMed] [Google Scholar]
- 44.Singh S; Khera R; Allen AM; Murad MH; Loomba R, Comparative effectiveness of pharmacological interventions for nonalcoholic steatohepatitis: A systematic review and network meta-analysis. Hepatology 2015, 62, (5), 1417–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S2A-C: Differentially Expressed Proteins
Table S1: PCR Primers
Figure S1: Linear Relationship for Peptides Between Biological Replicates in LC/MS/MS
Tables S3A-F: Significantly Altered Pathways and Processes
Table S4: Transcription factor analysis of HFD+ (steatohepatitis) vs. HFD (steatosis) groups
Table S5: List of Mean ± SD for data presented in Figures 3,4,5