Abstract
Background
Plant height, mainly decided by main stem height, is the major agronomic trait and closely correlated to crop yield. A number of studies had been conducted on model plants and crops to understand the molecular and genetic basis of plant height. However, little is known on the molecular mechanisms of peanut main stem height.
Results
In this study, a semi-dwarf peanut mutant was identified from 60Co γ-ray induced mutant population and designated as semi-dwarf mutant 2 (sdm2). The height of sdm2 was only 59.3% of its wild line Fenghua 1 (FH1) at the mature stage. The sdm2 has less internode number and short internode length to compare with FH1. Gene expression profiles of stem and leaf from both sdm2 and FH1 were analyzed using high throughput RNA sequencing. The differentially expressed genes (DEGs) were involved in hormone biosynthesis and signaling pathways, cell wall synthetic and metabolic pathways. BR, GA and IAA biosynthesis and signal transduction pathways were significantly enriched. The expression of several genes in BR biosynthesis and signaling were found to be significantly down-regulated in sdm2 as compared to FH1. Many transcription factors encoding genes were identified as DEGs.
Conclusions
A large number of genes were found differentially expressed between sdm2 and FH1. These results provide useful information for uncovering the molecular mechanism regulating peanut stem height. It could facilitate identification of causal genes for breeding peanut varieties with semi-dwarf phenotype.
Keywords: Peanut (Arachis hypogaea L.), Main stem height, RNA-seq, Cell wall related genes, Hormones, Transcription factors
Background
Plant height, a decisive factor of plant architecture, is an important agronomic trait which contributes to crop yield. Plant height is highly associated with rain or wind caused lodging and the utilization of dwarf and semi-dwarf plants could be a way to reduce the risk. Elongation of main stem is the main limiting factor of plant height. The breeding of semi-dwarf rice and wheat varieties in 1960s gave rise to the first ‘green revolution’ and increased grain yields significantly [1, 2]. Peanut (Arachis hypogaea L.) is an important oil crop in the world. In China, most of cultivated peanut varieties are erect. To prevent overgrowth and lodging, growth inhibitory substances like paclobutrazol and uniconazole are usually applied 2~3 times during the growth season. Therefore, studying the underlying molecular mechanism controlling main stem height and developing semi-dwarf peanut variety are of great significance.
Stem elongation is determined by cell growth including cell division and cell expansion. The dynamic variation of plant cell wall is critical in these processes [3]. The cellulose synthase super-family containing cellulose synthases (CesA) and cellulose synthase-like genes (CSL) is very important in cell-wall biogenesis and remodeling. CesA or CSL mutation in plants resulted in a dwarf phenotype with short internode or hypocotyl and small leaves [4–8]. Cell wall extensibility controls the rate of plant cell growth, especially cell expansion [9]. Expansins are plant cell-wall loosening proteins which promote cell enlargement by influencing cell wall extensibility and are essential in many critical developmental processes [10]. Suppression of expansin genes resulted in small, curled leaves, reduced plant height and early flowering [11–14]. Besides expansins, xyloglucanendotransglucosylase/endohydrolases (XTHs) were also suggested to regulated cell wall loosening [3, 15]. XTHs catalyze the endo cleavage of xyloglucan polymers and subsequent restructuring, which allow cellulose microfibrils moving along with the cell expansion or elongation process [16–18]. EGases (endo-1,4-β-glucanases) have been proposed to modify hemicellulose network, and were involved in processes that require cell wall weakening, including cell elongation, organ abscission, and fruit softening [19].
A number of mutants with deficiencies in phytohormone biosynthesis and signal transduction displayed dwarf phenotype [20–24]. Extensive studies showed that most of the dwarf phenotype of plants were associated with gibberellins (GAs) and brassinosteroids (BRs), and a few were caused by auxin (IAA) or strigolactone (SL) [9]. Two “Green Revolution” genes, sd1 and Rht, which encoded GA 20-oxidases and DELLA protein were cloned from rice and wheat, respectively [1, 2, 25, 26]. Later, several key genes controlling plant height have been identified. GID1 (GA-insensitive dwarf1) is the GA receptor, and its mutant gid1 in rice showed dwarf phenotype [27]. GA3-oxidases (GA3ox) catalyze the synthesis of bioactive GAs and GA2-oxidases (GA2ox) convert excess GAs to inactive forms. GA3ox deletion and GA2ox overexpression both reduced the plant height [28, 29].
Mutants of BR signaling related genes, for examples, the deetiolated (det) [30], diminuto (dim) [31], constitutive photomorphogenesis and dwarfism (cpd) [23] and dwarf4 (dwf4) [32] are all dwarf. DIM was also referred to as DWF1 or CBB1, could convert 24-methylenecholesterol to campesterol [31, 33]. DET2 was shown to encode a putative steroid 5α-reductase [34, 35]. CPD and DWF4 were members of cytochrome P450 family, encoding a putative C-3 oxidase and 22a-hydroxylase, respectively [32, 36]. BRI1 (Brassinosteroid Insensitive 1) is the membrane-localized BR receptor [37–39]. The bri1 mutant displayed severely dwarf phenotype and reduced apical dominance [40]. BAK1 (BRI1-Associated Receptor Kinase 1) is co-receptor of BRI1 and can trigger sequential transphosphorylation of BRI1/BAK1 receptor kinase complex [41–43]. The null allele of BAK1 displayed a semi-dwarf phenotype and reduced sensitivity to BRs. BIN2 plays a negative regulatory role in BR signaling and its gain-of-function mutant showed the phenotype similar to BR-deficient and BR-response mutants [44]. BES1 and BZR1 are two bHLH transcription factors, and they can directly bind to target gene promoters together with other transcription regulators to activate BR target gene expression [24, 45–48].
Auxin polar transport is critical in regulating the whole progress of plant growth and development [49]. AUX1/LAX family, PIN family and ABCB subfamily are three classes of polar auxin transporters. MDR (Multidrug Resistance) genes encode P-glycoproteins and belong to subfamily B of ATP-binding cassette (ABCB). The atmdr1 mutants were defective in basipetal auxin transport and shorter than wild-type plants [50]. SAUR (Small Auxin up RNA) genes, the largest family of auxin-responsive genes, functioned as positive effectors of cell expansion and were up-regulated in auxin-mediated cell elongation [51–53].
Many transcription factors are important regulators of plant height. SHORT INTERNODES (SHI) gene is a negative regulator of GA response and its overexpression mutant shi showed a dwarf phenotype similar to mutants defective in GA biosynthesis or response [54]. WRKY46/54/70 could be activated by BRs and were cofactors of BES1 to modulate BR target genes [55]. PACLOBUTRAZOL RESISTANT (PRE) is a positive regulator of cell elongation which can be activated by BRs, GAs and repressed by light [56, 57]. PRE together with IBH1, HBI1 and ACE constitute a triantagonistic cascade module which integrate hormonal and environmental signals to regulate cell elongation [58–60].
In peanut, genetic basis underlying plant height remains unclear. Some studies to identify QTLs for plant height have been conducted in peanut. Huang et al. (2016) constructed a high-density genetic linkage map containing 1219 mapped loci using a recombinant inbred line (RIL) population and identified 18 QTLs for plant height [61]. Li et al. (2017) detected 11 QTLs for main stem height using a RIL population [62]. Using two RIL populations, Lv et al. (2018) detected 11 QTLs for plant height. Among which, two QTLs from these two populations were co-localized in a physical interval of 3.4 Mb on A09 [63]. However, no gene closely related to plant height has been cloned in peanut until now.
In the previous research, we built a mutant library through 60Co radiation. Among which, two mutants displayed semi-dwarf phenotype. In this study, genome-wide gene expression profiling of stem and leaf from sdm2 was conducted to understand the underlying regulation network of main stem height in peanut. The expression of several genes involved in biosynthesis and signal transduction of BRs and GAs were drastically altered in sdm2. Accordingly, the downstream genes related to cell wall biogenesis and remodeling were also changed at the transcriptional level.
Results
Phenotype characteristics of peanut dwarf mutant
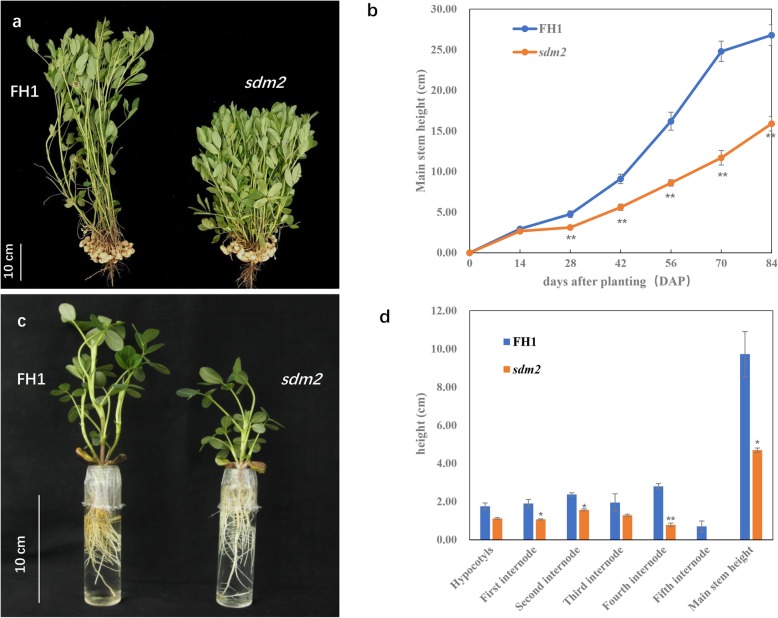
The main stem height of sdm2 is significantly shorter than that of FH1 (P < 0.01) (Fig. 1a). We measured the main stem height of mutant and FH1 grown in the experimental farm every 2 weeks during the vegetative growing stage. Results showed that during the first 2 weeks, main stem height between sdm2 and FH1 had no obviously difference. At 28 DAP (days after planting), main stem height of sdm2 was significantly shorter than that of FH1 (P < 0.01). In the following developmental stages, the height difference between sdm2 and FH1 became more and more obvious. At 84 DAP, the sdm2 was 15.9 cm in height, only about 60% of FH1 (26.8 cm) (Fig. 1b). The leaf of sdm2 was smaller, thicker and dark green in color, and showed delayed senescence compare to that of FH1. At harvest time, the number of internodes of sdm2 was 3~4 less than FH1. The number of branches (P < 0.05) and pods (P > 0.05) per plant was obviously more, while the 100-pod weight and 100-kernel weight were significantly decreased in sdm2 (P < 0.01).
Fig. 1.
Phenotypic characterization of sdm2 mutant and FH1. a Plants of sdm2 and FH1 at harvest time grown in the field; b Main stem height of sdm2 and FH1 at different growth stages grown in the field; c sdm2 and FH1 grown in Hogland solution for 14 DAP; d Main stem height measurement of sdm2 and FH1 grown in Hogland solution for 14 DAP. Three biological replicates were used for statistical analysis (t-test; *P < 0.05, **P < 0.01). Values in b, d represent means ± SE (n = 3)
To further accurately measure the change of mutant main stem height, we conducted hydroponic culture of sdm2 and FH1 in Hogland solution. The length of each internode and hypocotyl of 14 DAP seedlings were measured. We found that it was different from the results in experimental farm. For 14 DAP seedlings, the sdm2 height was already significantly shorter than that of FH1 (P < 0.05) (Fig. 1c, d). There were total five internodes in FH1 and four in sdm2 seedlings. The hypocotyl length of sdm2 was slightly shorter than FH1 and there was no significant difference (P > 0.05) for the 14 DAP seedlings. The length of most internodes of sdm2 was significantly shorter than FH1 (Fig. 1d). These results indicated that the height difference between sdm2 and FH1 was caused by both reduced total number of internode and length of each internode.
Sequencing data analysis
To clarify global gene expression changes in sdm2, 12 cDNA libraries were constructed with stem and leaf from mutant and FH1 seedlings. The libraries were sequenced using BGISEQ-500 platform. A total of 0.26 billion raw reads were generated, and the average output of each sample was 21.7 million. After removing adaptor sequences, low-quality and N-containing reads, an average of 21.58 million clean reads were obtained from each library with the clean read ratio of 99.47% (Table 1). Approximately 19.27 million and 16.00 million clean reads in each library matched the reference genome and gene set perfectly with the average mapping ratio of 89.31 and 74.18%, respectively. To investigate gene expression correlation among samples, the Pearson correlation coefficient of all gene expression levels between each two samples were calculated (Additional file 1: Fig. S1). It showed a higher correlation of similar tissue between sdm2 and FH1. The principal component analysis (PCA) could clearly divide all samples into two clusters according to two kinds of tissues (Additional file 2: Fig. S2).
Table 1.
Summary of read numbers from stem and leaf in sdm2 and FH1
| Sample name | Reads in stem | Reads in leaf | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FH1_S_1 | FH1_S_2 | FH1_S_3 | sdm2_S_1 | sdm2_S_2 | sdm2_S_3 | FH1_L_1 | FH1_L_2 | FH1_L_3 | sdm2_L_1 | sdm2_L_2 | sdm2_L_3 | |
| Total raw read(M) | 21.64 | 21.66 | 21.67 | 21.65 | 21.84 | 21.67 | 21.86 | 21.61 | 21.74 | 21.65 | 21.64 | 21.71 |
| Total clean read(M) | 21.51 | 21.54 | 21.56 | 21.53 | 21.74 | 21.52 | 21.78 | 21.50 | 21.65 | 21.47 | 21.53 | 21.60 |
| Clean read ratio(%) | 99.41 | 99.46 | 99.48 | 99.48 | 99.56 | 99.30 | 99.60 | 99.51 | 99.60 | 99.19 | 99.50 | 99.49 |
| Clean read q20(%) | 98.25 | 98.22 | 98.16 | 98.20 | 98.06 | 98.07 | 98.27 | 98.15 | 98.21 | 98.31 | 98.11 | 98.24 |
| Clean read q30(%) | 90.52 | 90.37 | 90.10 | 90.36 | 89.52 | 89.81 | 90.28 | 90.06 | 90.11 | 90.76 | 89.85 | 90.29 |
| Total mapping genome reads (M) | 20.34 | 20.53 | 20.72 | 20.56 | 20.82 | 20.15 | 17.95 | 17.05 | 19.40 | 19.03 | 16.41 | 18.30 |
| Total mapping genome ratio(%) | 94.56 | 95.33 | 96.09 | 95.49 | 95.79 | 93.63 | 82.42 | 79.30 | 89.60 | 88.65 | 76.20 | 84.70 |
| Total mapping gene reads (M) | 16.93 | 16.75 | 17.45 | 17.17 | 17.32 | 16.46 | 14.63 | 14.24 | 15.77 | 16.41 | 13.93 | 15.02 |
| Total mapping gene ratio (%) | 78.69 | 77.77 | 80.95 | 79.73 | 79.65 | 76.51 | 67.17 | 66.24 | 72.86 | 76.45 | 64.68 | 69.56 |
Note: S indicates stem, and L indicates leaf. Each sample has three replicates
DEGs between sdm2 and FH1
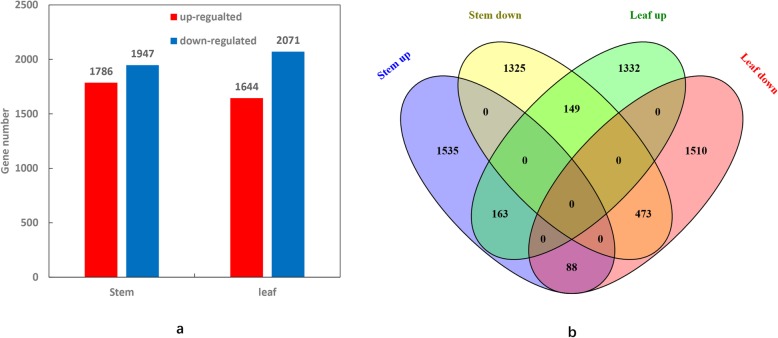
From all the samples, a total of 65,435 genes were identified. The gene expression of stem and leaf between sdm2 and FH1 was analyzed, and 3733 and 3715 differentially expressed genes (DEGs) were identified, respectively (Additional files 3 and 4: Table S1 and S2). In sdm2 stem, 1786 genes were up-regulated and 1947 genes were down-regulated to compare with FH1 (Fig. 2a and Additional file 3: Table S1). In sdm2 leaf, the up- and down-regulated genes were 1644 and 2071, respectively (Fig. 2a and Additional file 4: Table S2). The majority of DEGs had different expression pattern in stem and leaf. Among 3733 DEGs in stem, only 873 were also differentially expressed in leaf. There were 473 down-regulated and 163 up-regulated DEGs with the same change trend in stem and leaf. It is worth noting that the number of common down-regulated DEGs was obviously higher than that of up-regulated ones. Interestingly, a large number of DEGs showed opposite expression pattern in stem and leaf. For example, there were 149 genes down-regulated in mutant stem while up-regulated in leaf and 88 genes up-regulated in mutant stem but down-regulated in leaf (Fig. 2b).
Fig. 2.
Differentially expressed genes in sdm2 and FH1. a Numbers of differentially expressed genes in sdm2 and FH1; b Venn diagram showed the common and specific differentially expressed genes in stem and leaf
Functional analysis of DEGs
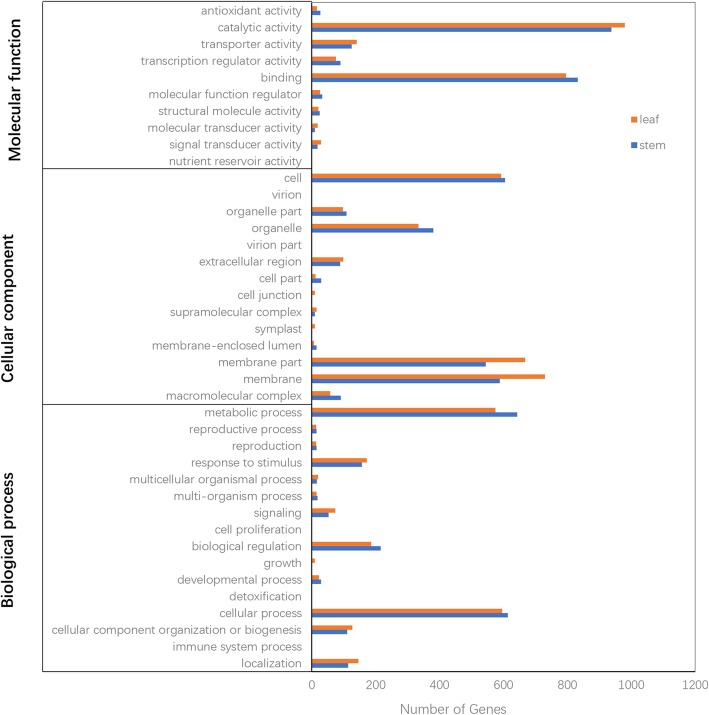
GO classification showed that 1724 DEGs from stem and 1716 DEGs from leaf were classified into three categories: biological process, cellular component, and molecular function (Fig. 3 and Additional file 5: Table S3). In molecular function category, catalytic activity and binding were the most abundant terms. For cellular component category, cell, membrane, membrane part and organelle were the main terms. The top two terms of biological process were metabolic process and cellular process. The top 20 terms in stem and leaf were shown in Additional file 6: Fig. S3 and Additional file 7: Table S4.
Fig. 3.
GO classification of DEGs in stem and leaf. The X-axis represents the number of genes annotated into the GO terms, and the Y-axis represents the functional classification
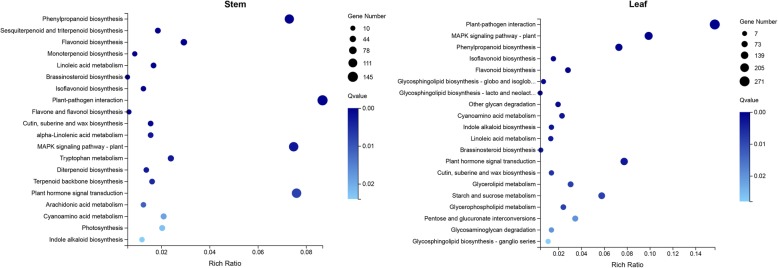
KEGG pathway analysis was also carried out. According to the enrichment results, the top 20 pathways in stem and leaf were shown in Fig. 4 and Additional file 8: Table S5. In stem, many pathways involved in hormone biosynthesis and signal transduction including terpenoid backbone biosynthesis, monoterpenoid biosynthesis, diterpenoid biosynthesis, sesquiterpenoid and triterpenoid biosynthesis, zeatin biosynthesis, and brassinosteroid biosynthesis, indole alkaloid biosynthesis as well as plant hormone signal transduction were all enriched in the mutant (Fig. 4 and Additional file 8: Table S5). In addition, MAPK signaling pathway was also enriched. These results suggested that plant hormone play major roles in regulation of the mutant phenotype. There were other pathways including photosynthesis, phenylpropanoid biosynthesis, flavonoid biosynthesis, isoflavonoid biosynthesis, and flavone and flavonol biosynthesis pathways were found to be enriched.
Fig. 4.
Bubble diagram of top 20 enriched KEGG pathways of DEGs in stem and leaf. X axis represents the Rich Ratio, which meaning the ratio of selected gene number annotated to a particular item to the total number of genes in this item in one species. The calculating formula is Rich Ratio = Term Candidate Gene Num/Term Gene Num. Y axis represents KEGG Pathway. The size of the bubbles indicates the number of genes annotated to a KEGG Pathway. And the color represents Q-value of enrichment. The deeper the color, the smaller the Q-value
In leaf, brassinosteroid biosynthesis, indole alkaloid biosynthesis, plant hormone signal transduction and MAPK signaling pathways were enriched in accordance with those in stem (Fig. 4 and Additional file 8: Table S5). Phenylpropanoid biosynthesis, flavonoid biosynthesis, isoflavonoid biosynthesis and plant-pathogen interaction were also in the top 20 enriched pathways. Different from that in stem, some glucide and glucolipid metabolic pathways were enriched in leaf.
Cell wall related genes
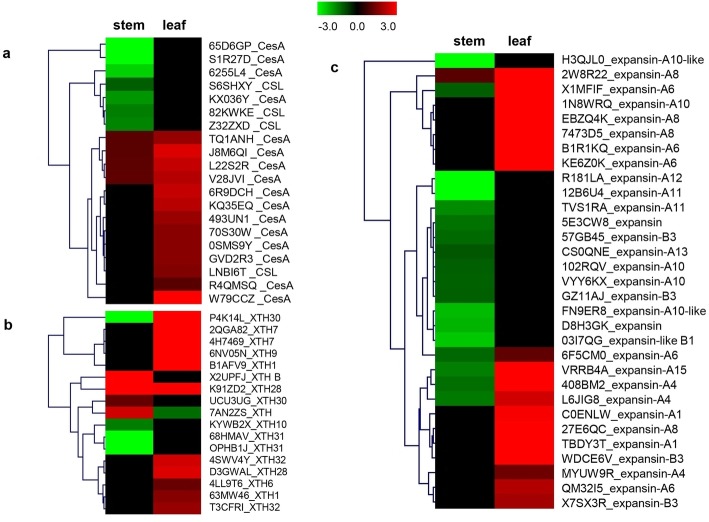
RNA-seq results showed that several CesA genes and all CSL genes were down-regulated in stem. The expression of most expansin genes was down-regulated in sdm2 stem compared with those in FH1 (Fig. 5 and Additional file 3: Table S1). Interestingly, the expression of CesA, CSL and expansin genes was up-regulated in leaf (Fig. 5 and Additional file 4: Table S2). These results were coincided with the phenotype of sdm2 leaf, dark green in color, thicker, smaller than FH1 leaf.
Fig. 5.
The expression pattern of cell wall related genes in stem and leaf of sdm2. Heatmaps represent the differential expression of CesA and CSL a, XTHs b, expansions c in stem and leaf between sdm2 and FH1. Scale bar is located at upside with log2 ratio value varying from green to red
Some XTHs were significantly up-regulated and some were down-regulated in sdm2 stem (Fig. 5 and Additional file 3: Table S1). In leaf, the expressions of all XTHs were up-regulated, and some XTHs were 16-fold higher than those in FH1 (Fig. 5 and Additional file 4: Table S2). Some genes encoding endo-1,4-β-glucanases (EGases) in stem and leaf were down-regulated.
Expression changes of hormone biosynthesis and signaling genes
DWF4 was designated as CYP90B1 and proposed to catalyze steroid 22α-hydroxylation in BR biosynthesis [64]. A gene annotated as cytochrome P450 90B1 showed 73% identity with Arabidopsis DWF4 gene, which play key roles in BR biosynthesis. In stem, the expression of this gene was detected in FH1, while not detected in sdm2. In leaf, the expression of this gene in sdm2 was also lower than that in FH1. Previous studies confirmed that cytochrome P450 monooxygenase CYP90A1 (CPD) acted as BR C-3 oxidase [36]. The expression level of CYP90A1 (CPD) in sdm2 stem decreased by 1.2-fold than that in FH1. While in leaf, its expression was slightly higher in sdm2 than in FH1. The cytochrome P450 family members, CYP90C1/D1, CYP85 (Dwarf) and CYP71A1, mediate rate-limiting reactions in BR biosynthesis [65–67]. Our results showed that the expression of CYP90C1/D1 gene decreased by 2.64-fold and 1.41-fold in stem and leaf respectively in sdm2. The Dwarf genes which encoding cytochrome P450 85A-like in stem and cytochrome P450 85A in leaf were all up-regulated. CYP71A1 which catalyze the generation of deoxocastasterone (6-deoxoCS) and castasterone (CS) was down-regulated both in stem and leaf in sdm2. A cytochrome P450 monooxygenase (CYP734A1) which degrades BRs was up-regulated in sdm2 leaf (Table 2, Additional files 3 and 4: Table S1 and S2).
Table 2.
Differential expression of hormonal biosynthesis and signaling genes in stem and leaf between sdm2 and FH1
| Gene ID | Nr Annotation | Gene name | Relative expression level (sdm2/FH1) | Expression trend (stem/leaf) | |||
|---|---|---|---|---|---|---|---|
| log2(stem) | Q-value (stem) | log2(leaf) | Q-value (leaf) | ||||
| Arahy.MA1Z2D | cytochrome P450 90B1 | DWF4 | −9.35 | 2.43E-59 | −3.99 | 1.29E-06 | down/down |
| Arahy.66GL51 | cytochrome P450 90A1 isoform X1 | CPD | −1.31 | 9.68E-13 | – | – | down/− |
| Arahy.7AH9CA | 3-epi-6-deoxocathasterone 23-monooxygenase | CYP90C1/D1 | −2.40 | 2.29E-13 | −1.59 | 2.47E-12 | down/down |
| Arahy.31R89H | cytochrome P450 85A-like | Dwarf/BR6ox | 1.99 | 1.08E-07 | – | – | up/− |
| Arahy.JZF5VE | cytochrome P450 85A-like | Dwarf/BR6ox | 2.50 | 9.71E-13 | – | – | up/− |
| Arahy.27TAQT | cytochrome P450 85A | Dwarf/BR6ox | – | – | 2.16 | 4.98E-31 | −/up |
| Arahy.2F8F1S | cytochrome P450 85A | Dwarf/BR6ox | – | – | 1.34 | 2.56E-35 | −/up |
| Arahy.TI8LFW | cytochrome P450 71A1 | CYP71A | −1.81 | 0.00E+ 00 | −2.06 | 2.09E-53 | down/down |
| Arahy.ZW245F | cytochrome P450 71A1-like | CYP71A | −1.63 | 3.04E-32 | – | – | down/− |
| Arahy.U2CXJI | cytochrome P450 734A1 | BAS1 | – | – | 1.80 | 0.00E+ 00 | −/up |
| Arahy.GJFA3Z | probable leucine-rich repeat receptor-like protein kinase At5g49770 isoform X1 | BRI1 | −2.38 | 3.16E-51 | – | – | down/− |
| Arahy.56X5WL | probable leucine-rich repeat receptor-like protein kinase At5g49770 isoform X1 | BRI1 | −1.32 | 1.91E-44 | – | – | down/− |
| Arahy.BMJ2CS | probable leucine-rich repeat receptor-like protein kinase At1g35710 | BRI1 | – | – | −2.59 | 6.71E-07 | −/down |
| Arahy.69MPHZ | probable leucine-rich repeat receptor-like protein kinase At5g49770 | BRI1 | – | – | −2.49 | 7.76E-19 | −/down |
| Arahy.DH99G0 | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 | BAK1 | −1.99 | 4.09E-07 | −2.08 | 2.39E-74 | down/down |
| Arahy.SPI7HB | BES1/BZR1 homolog protein 2 | BES1/BZR1 | −8.42 | 0.00E+ 00 | −7.30 | 0.00E+ 00 | down/down |
| Arahy.RCTC3C | BES1/BZR1 homolog protein 2 | BES1/BZR1 | −1.92 | 7.59E-08 | −1.20 | 1.08E-04 | down/down |
| Arahy.ND2ZET | BES1/BZR1 homolog protein 2 | BES1/BZR1 | −1.94 | 4.11E-05 | – | – | down/− |
| Arahy.22IVZ0 | gibberellin 20 oxidase 1-D | GA20ox | 5.77 | 3.55E-98 | – | – | up/− |
| Arahy.B5269G | gibberellin 20 oxidase 1-like | GA20ox | 1.64 | 8.35E-05 | – | – | up/− |
| Arahy.BLJ7SD | gibberellin 20 oxidase 2 | GA20ox | 5.60 | 4.62E-85 | – | – | up/− |
| Arahy.YI9B6B | probable 2-oxoglutarate-dependent dioxygenase At5g05600 isoform X1 | 2-oxoglutarate-dependent dioxygenase | 1.39 | 2.07E-06 | 1.23 | 1.02E-04 | up/up |
| Arahy.XPP88F | 2-oxoglutarate-dependent dioxygenase AOP3 | 2-oxoglutarate-dependent dioxygenase | 1.08 | 9.86E-04 | −1.90 | 4.14E-23 | up/down |
| Arahy.MT91AM | 2-oxoglutarate-dependent dioxygenase AOP3 | 2-oxoglutarate-dependent dioxygenase | 1.74 | 4.26E-07 | – | – | up/− |
| Arahy.K8MVYG | gibberellin 3-beta-dioxygenase 1 | GA3ox | −1.41 | 1.18E-49 | – | – | down/− |
| Arahy.QH4I6C | gibberellin 2-beta-dioxygenase 1 isoform X1 | GA2ox | −1.87 | 2.50E-04 | −1.82 | 1.12E-11 | down/down |
| Arahy.VR90R2 | gibberellin 2-beta-dioxygenase 8 | GA2ox | 2.74 | 1.25E-17 | – | – | up/− |
| Arahy.EPT56A | gibberellin-regulated protein 11 | GRP11 | −1.84 | 1.80E-09 | – | – | down/− |
| Arahy.F1TUFL | gibberellin-regulated protein 4 | GRP4 | −1.63 | 0.00E+ 00 | – | – | down/− |
| Arahy.FJ610L | gibberellin-regulated protein 4 | GRP4 | – | – | 4.31 | 1.92E-70 | −/up |
| Arahy.X9WA5F | PREDICTED: gibberellin-regulated protein 4 | GRP4 | −1.20 | 8.62E-115 | 3.90 | 3.18E-37 | down/up |
| Arahy.VJ6NUA | gibberellin-regulated protein 6 isoform X2 | GRP6 | −1.44 | 1.83E-145 | 3.75 | 5.91E-89 | down/up |
| Arahy.UA0Z8F | auxin transporter-like protein 1 | AUX1/LAX family | 6.46 | 2.83E-11 | 4.70 | 1.25E-15 | up/up |
| Arahy.JN7ZD3 | auxin transporter-like protein 1 | AUX1/LAX family | 3.59 | 1.64E-04 | – | – | up/− |
| Arahy.LRNE2N | auxin transporter-like protein 3 | AUX1/LAX family | – | – | 2.06 | 4.38E-72 | −/up |
| Arahy.953E9B | auxin efflux carrier component 5 isoform X1 | PIN family | – | – | −3.06 | 3.04E-06 | −/down |
| Arahy.K2GLG1 | auxin efflux carrier component 5 isoform X1 | PIN family | – | – | −1.36 | 2.22E-09 | −/down |
| Arahy.BB2RVD | ABC transporter B family member 19-like | ABCB subfamily | 3.17 | 1.24E-04 | 1.27 | 3.06E-04 | up/up |
| Arahy.RLV4PD | ABC transporter B family member 1 | ABCB subfamily | – | – | 3.48 | 2.67E-09 | −/up |
| Arahy.M63M99 | ABC transporter B family member 11 | ABCB subfamily | – | – | −1.27 | 2.65E-33 | −/down |
| Arahy.YCYQ9Q | uncharacterized protein LOC107461514 | ABCB subfamily | −5.97 | 7.24E-43 | −7.49 | 2.35E-72 | down/down |
| Arahy.M4RITN | uncharacterized protein LOC107461514 | ABCB subfamily | −5.13 | 2.19E-18 | −3.56 | 5.47E-04 | down/down |
| Arahy.8QDS1I | auxin-induced protein AUX22-like | Aux/IAA protein | −2.12 | 5.87E-54 | 1.18 | 2.98E-30 | down/up |
| Arahy.KRC5M1 | auxin-induced protein AUX22 | Aux/IAA protein | −1.52 | 3.07E-35 | 1.34 | 3.65E-35 | down/up |
| Arahy.2A4XC9 | LOW QUALITY PROTEIN: auxin-responsive protein IAA29 | Aux/IAA protein | 1.69 | 9.56E-15 | 1.79 | 1.13E-09 | up/up |
| Arahy.S4Y3WA | auxin response factor 5 isoform X2 | ARF | 2.95 | 2.80E-24 | 2.97 | 1.67E-04 | up/up |
| Arahy.1B1I67 | putative auxin response factor 23 | ARF | 2.15 | 4.14E-10 | – | – | up/− |
| Arahy.X5Q10C | auxin response factor 18-like isoform X2 | ARF | – | – | 5.44 | 5.42E-06 | −/up |
| Arahy.MJW2EP | auxin-induced protein 15A-like | SAUR family | 1.86 | 2.74E-04 | 6.59 | 5.68E-12 | up/up |
| Arahy.E8WPL7 | auxin-induced protein 6B-like | SAUR family | 1.26 | 4.39E-05 | 2.93 | 2.63E-08 | up/up |
| Arahy.W3E9BL | auxin-responsive protein SAUR50-like | SAUR family | −1.11 | 3.81E-13 | 3.98 | 1.86E-07 | down/up |
| Arahy.NWIK25 | indole-3-acetic acid-induced protein ARG7-like | SAUR family | – | – | 4.77 | 2.09E-07 | −/up |
| Arahy.G43XWA | protein SHORT INTERNODES-like | SHI | −6.32 | 1.91E-10 | −6.63 | 3.98E-12 | down/down |
| Arahy.F7N9UG | protein SHI RELATED SEQUENCE 3-like | SRS | 5.43 | 6.62E-06 | – | – | up/− |
| Arahy.R0GFR4 | protein SHI RELATED SEQUENCE 3-like | SRS | – | – | −6.63 | 3.98E-12 | −/down |
| Arahy.EXJ5K8 | probable WRKY transcription factor 30 | WRKY30 | −1.20 | 7.10E-23 | −3.09 | 1.42E-76 | down/down |
| Arahy.TC7Y0P | probable WRKY transcription factor 46 | WRKY46 | −1.39 | 5.50E-62 | −1.96 | 1.75E-96 | down/down |
| Arahy.I9PJJN | probable WRKY transcription factor 70 | WRKY70 | −2.64 | 4.37E-05 | −1.65 | 1.84E-26 | down/down |
| Arahy.2031JR | transcription factor PRE6 | PRE6 | −2.41 | 5.06E-28 | 2.91 | 6.65E-10 | down/up |
| Arahy.577H6Y | transcription factor PRE6 | PRE6 | −1.93 | 2.80E-07 | 6.24 | 4.25E-36 | down/up |
| Arahy.HR6FN8 | transcription factor PRE6 | PRE6 | −2.67 | 7.25E-71 | down/− | ||
| Arahy.YX8FPP | transcription factor PRE6-like | PRE6 | −2.33 | 5.39E-05 | down/− | ||
| Arahy.WVIE7L | transcription factor PRE6 | PRE6 | – | – | 7.87 | 2.15E-50 | −/up |
In both stem and leaf, the expression levels of BRI1 and BAK1 were down-regulated in sdm2. Noticeably, the expression levels of BES1/BZR1 gene were extremely low in stem and leaf in sdm2, 170-fold and 79-fold lower than in FH1, respectively. The expression levels of IWS1 in stem were up-regulated in sdm2, while in leaf, it was not differentially expressed. The expression of cyclin-D3–3 encoding gene was down-regulated in sdm2 stem, which was coincident with the expression trends of BR-related genes. While the expression levels of cyclin-D1 and cyclin-D5 increased in sdm2 leaf (Table 2, Additional files 3 and 4: Table S1 and S2).
Several GA biosynthesis and signal transduction genes were differentially expressed between sdm2 and FH1. In sdm2, most of GA20ox and 2-oxoglutarate-dependent dioxygenase encoding genes, which promote GA biosynthesis, were up-regulated in stem, while most 2-oxoglutarate-dependent dioxygenase decreased in leaf. While two genes encoding GA3ox were down-regulated in stem and its differential expression was not detected in leaf. GA2ox convert excess GAs to inactive forms through 2β-hydroxylation [29]. In mutant stem and leaf, genes encoding GA2ox1 and GA2ox2 were decreased in expression levels while the expression of three genes encoding GA2ox8 were up-regulated in stem (Table 2, Additional files 3 and 4: Table S1 and S2).
Two GID1 genes in stem showed opposite change trend and one GID1 gene was down-regulated in leaf. SCL3 (Scarecrow-like 3) seemed to attenuate DELLA protein and acted as a positive regulator in GA pathway [68]. In mutant stem, SCL3 gene was up-regulated while two SCL14 and one SCL21 genes were down-regulated. Interestingly, all genes encoding gibberellin-regulated protein (GRP) were reduced in sdm2 stem while up-regulated in leaf, which were consistent with the change trends of cell wall related genes including CesA, CSL, expansin and XTH genes (Table 2, Additional files 3 and 4: Table S1 and S2).
Four AUX1 genes in stem and two in leaf were all significantly up-regulated, while the expressional levels of two genes encoding auxin efflux carrier (PIN) in leaf decreased. In addition, the expression change trends of MDR/ABCB genes in stem and leaf of sdm2 were irregular, most of which were down-regulated and some were up-regulated. In sdm2 stem, two genes encoding AUX22 were down-regulated and almost all ARF members were up-regulated, while in leaf, the expressions of all Aux/IAA and ARF genes were increased. In addition, most SAUR genes in stem and almost all SAUR genes in leaf were up-regulated in sdm2 (Table 2, Additional files 3 and 4: Table S1 and S2).
Transcription factors involved in plant growth and development
Our results showed that several transcription factor genes involved in hormone signaling pathways and their downstream changed significantly. Gene encoding SHORT INTERNODES-like protein was down-regulated both in stem and leaf of sdm2. Whereas, SHI-RELATED SEQUENCE 3-like (SRS) gene increased in stem and decreased in leaf. All WRKY30/46/70 genes were down-regulated in sdm2 stem and leaf. The expression levels of four PRE6 genes decreased in stem and three PRE6 genes were up-regulated in leaf (Table 2, Additional files 3 and 4: Table S1 and S2).
Verification of DEGs using qRT-PCR
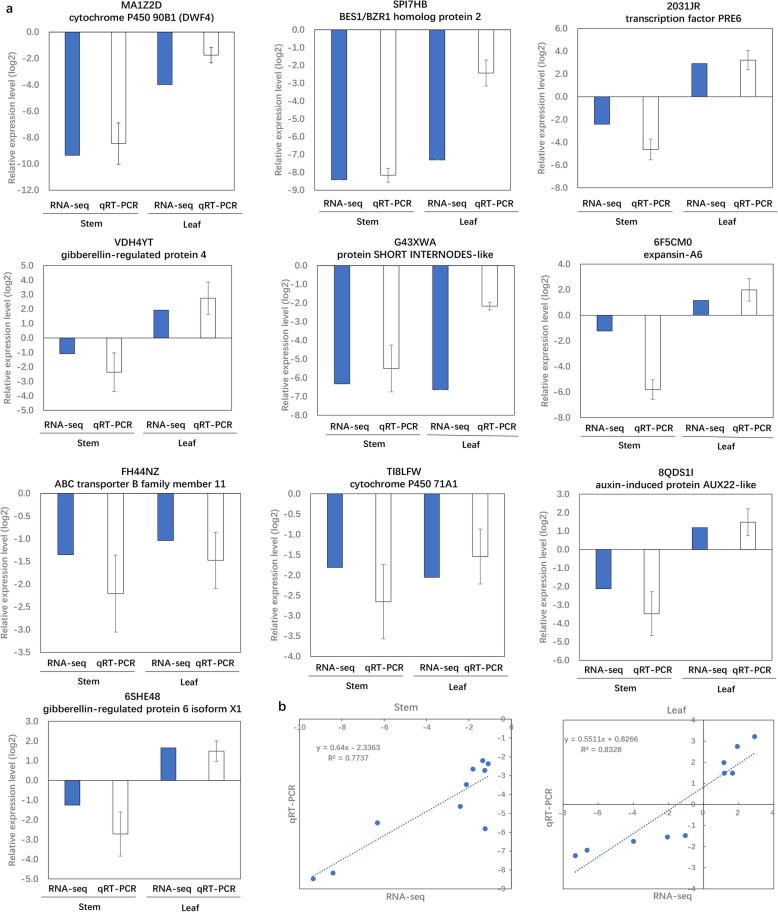
In order to validate the RNA-Seq data, the expression levels of DEGs were detected using Quantitative Real-time PCR (qRT-PCR). A total of 10 genes related to hormone signal transduction and cell wall organization were selected (Fig. 6 and Additional file 9: Table S6). The relative expression levels (log2 sdm2/FH1) of these genes estimated by qRT-PCR were generally consistent with those by RNA-seq. The overall correlation coefficient of a liner regression analysis in stem and leaf was 0.7737 and 0.8328, respectively (Fig. 6).
Fig. 6.
Verification of DEGs by qRT-PCR. a. Transcript levels of 10 genes related to hormone signal transduction and cell wall organization. Data are means of three replicates, and error bars represent±SE (n = 3). b. Pearson’s correlation of gene expression ratios between RNA-seq and qRT-PCR results. The correlation of the fold change was analyzed by RNA-seq (x-axis) with qRT-PCR (y-axis) data
Discussion
Extensive studies indicated that deficiencies in cell wall-related genes, GA, BR and IAA signaling pathways all could result in dwarfish. These deficient mutants displayed dwarf or semi-dwarf phenotype with shorter hypocotyl or internode, smaller and dark-green leaves [4–7, 20–23, 40, 50, 69]. Microscopic assays indicated that the dwarf phenotypes of BR-deficient and BR-insensitive mutants were caused by reduced cell size [23, 32]. Maize br2 mutant, deficient in the polar auxin transport, showed reduced stalk cell length and diameter [69]. In our study, sdm2 phenotype was resulted from reduction both in internode number and internode length. The difference might be due to changes in cell division and cell expansion during the stem growth process, which was supported by the identification of a number of cell wall biosynthesis and metabolism enzyme encoding genes as DEGs. Cell division and cell expansion need the biogenesis or remodeling of cell wall. The extensibility of cell wall controls the rate of cell expansion. Genes, including CesA and CSL, responsible for cell-wall biogenesis, and expansins and XTHs, involved in cell wall loosening, are all critical in plant growth and development. Our transcriptome data revealed several CesAs, almost all CSLs and expanin genes were down-regulated in sdm2 stem. Some XTHs were down-regulated in sdm2 stem. Besides, genes encoding EGases which functions in the modification of hemicellulose network were down-regulated in sdm2 stem. These results suggested that the cell wall biosynthesis and extensibility were reduced in sdm2 stem, which might result in reduced cell division and cell expansion and further cause short internode and small number of stem nodes.
Studies revealed that many cell wall organization enzymes, including CesA, XTHs, EGases, and expansins were regulated by BRs [24, 45, 48, 70–72]. The essential roles of BRs in stem elongation have been confirmed by several mutants deficient in BR biosynthesis or perception [23, 30–32, 40]. Our results showed that key BR biosynthetic genes CPD, DWF4 and CYP90C1/D1 and CYP71A1 were all down-regulated in sdm2. CYP734A1, which degrades BRs, was up-regulated in leaf of sdm2. The changing trends of above genes suggested the contents of BRs might be decreased in sdm2. BR response genes including BRI1, BAK1 and BES1/BZR1, were down-regulated, especially BES1/BZR1 in sdm2 stem. These results suggested that BRs could be a key factor in the formation of semi-dwarf phenotype of sdm2. In addition, BR signaling pathway also regulate the leaf and seed size and shape. In Arabidopsis cpd mutant, the decrease of cell size and cell number resulted in reduced leaf size [73]. The seeds of Arabidopsis det2 mutant were smaller and shorter because of decreased seed cavity, reduced endosperm volume and cell length and delayed embryo development [74]. Transgenic Arabidopsis and tobacco lines overexpressing DWF4 displayed increased plant height, longer petiole and leaf blade length, increased number of branches and siliques compared to the control [75]. In this study, sdm2 plants showed smaller leaf size, shorter petiole, and both decreased seed size and weight, which were all in accordance with phenotypic characteristics of BR-deficient mutants. We speculate that CPD, DWF4, BRI1 and BES1/BZR1 are key genes that lead to the phenotypic differences between sdm2 and FH1 line.
In this study, several GA biosynthetic genes showed irregular change trends and some seemed to promote the biosynthesis of GAs, while all GRP genes decreased in sdm2 stem which had the same expressional pattern with those of cell wall related genes. This result could be a reflection of lowered GA level in sdm2 stem, and the up-regulated expression of GA20ox and 2-oxoglutarate-dependent dioxygenases might be resulted from the feedback regulation of GA signal pathway. The polar transport of auxin was essential for plant growth and development. In sdm2, the increased transcription of auxin influx carrier and down-regulation of auxin efflux carrier together with the irregular changes of auxin polar transporters, MDR/ABCB genes, suggested the disordered auxin transport from shoot apex and young leaves to stem. It might contribute to the formation of short internode in sdm2.
BRs could affect GA biosynthesis through positively regulating GA20ox expression [76]. DELLA protein could directly interact with BZR1 and inhibit its DNA binding ability, and the promotion of GAs on cell elongation require BZR1 or BRs [57]. Application of auxin could induce the expression of GA20ox and GA3ox while reduce the GA2ox transcript [77, 78]. Auxin and BRs also could interact with each other and synergistically regulate plant development [79, 80]. Auxin could directly induce DWARF4 expression and BR biosynthesis [81]. In return, BRs influenced auxin redistribution through stimulating the expression of PIN genes [82]. There are complicated cross talk among hormones during plant growth and development. Our results suggested that the significantly down-regulated BR biosynthetic and response genes, low GA response and disordered auxin transport acted synergistically and contributed to the defects in sdm2 stem growth.
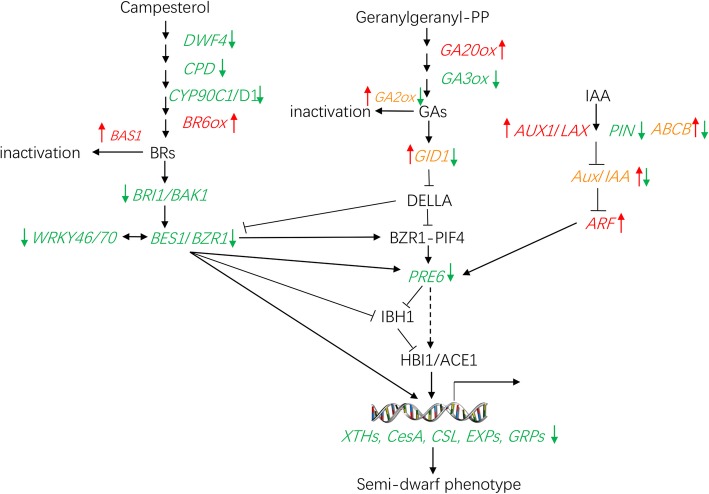
As the common target genes of the core transcription module, PRE family factors connect external and endogenous signals with downstream cell elongation components together through an antagonistic cascade reaction. The expressions of PRE6 genes in sdm2 stem and leaf were consistent with those of cell wall enzymes, which further proved the direct promotion effects of PRE on cell elongation. Tracing back to the upstream hormonal signals, the extremely low expression of BR biosynthetic and signal transduction genes and low GA signaling in sdm2 stem all contributed to the down-regulation of PRE genes, and subsequently reduced cell elongation factors which finally caused the semi-dwarf phenotype. Based on these results, we proposed a potential model regulating peanut stem development (Fig. 7). BRs, GAs and auxin signaling pathways interact cooperatively to influence PRE expression, and regulate downstream cell wall-related genes to control cell elongation during stem development. However, further research is required to verify this hypothesis after cloning the mutant gene.
Fig. 7.
The regulatory network underlying semi-dwarf phenotype in sdm2 mutant. DWF4: 22a-hydroxylase; CPD: putative C-3 oxidase; CYP90C1/D1: 3-epi-6-deoxocathasterone 23-monooxygenase; BR6ox: Brassinosteroid-6-oxidases; BAS1: Brassinosteroids C-26 hydroxylase; BRI1: Brassinosteroid insensitive 1; BAK1:BRI1-associated receptor kinase 1; BES1: BRI1-EMS suppressor 1; BZR1: Brassinazole resistant 1; GA20ox: GA 20-oxidases; GA3ox: GA 3-oxidases; GA2ox: GA 2-oxidases; GID1: GA-insensitive dwarf 1; DELLA: proteins containing conserved DELLA domains in the N terminus; PRE6: Paclobutrazol resistant 6; IBH1: ILI1 binding bHLH protein 1; HBI1: homolog of BEE2 interacting with IBH1; ACE1: activator for cell elongation 1; PHY: phytochrome; PIFs: phytochrome-interacting factors; AUX1/LAX: auxin1/like-AUX1; PIN:PIN-formed protein; ABCB: subfamily B of ATP-binding cassette; Aux/IAA: auxin resistant/indole-3-acetic acid inducible; ARF: auxin response factor; CesA: cellulose synthases; CSL: cellulose synthase-like genes; XTHs: xyloglucanendotransglucosylase/endohydrolases; EXPs: expansins; GRPs: gibberellin-regulated proteins
Conclusions
The transcriptome analysis of sdm2 identified a number of differentially expressed genes involved in hormone biosynthesis, signaling and cell wall synthetic and metabolic pathways. Especially several genes in BR pathway were significantly down-regulated in stem and leaf of sdm2 as compared to FH1. Genes in cell wall synthetic and metabolic pathway which related to cell elongation were generally down-regulated in sdm2 stem. These findings provide critical information for uncovering the molecular genetic control of peanut stem development.
Methods
Plant materials
The cultivated peanut cultivar Fenghua1 (FH1) was bred by and acquired from Yongshan Wan’s laboratory in Shandong Agricultural University, Shandong Province of China. In 2013, the seeds of FH1 was irradiated with 60Co γ-ray of 500 Gy. In M2 generation, a line with phenotypic segregation of normal and semi-dwarf plant height was screened. After self-crossing for another two generations, the stable semi-dwarf peanut line was obtained and named semi-dwarf mutant 2 (sdm2).
Planting conditions and sampling methods
In 2018, sdm2 and its wild type FH1 were sowed in experimental farm of Shandong Academy of Agricultural Sciences. The ridging mode was adapted with ridge spacing of 90 cm, ridge width of 55 cm and ridge height of 12 cm. Peanut seeds were sowed for two rows on each ridge with a single seed per hole, and the plant spacing and row spacing were 18 cm and 35 cm, respectively. All field management followed the standard agricultural practices. For each line, about 100 plants were planted. It was sowed on May 1st and harvested on September 5th with the growing period of 129 days. The days after planting (DAP) was used to calculate seedling growth time. During the vegetative period, at least 5 plants from each line were selected to investigate the main stem height through measuring the length from base of the above-ground plant to the tip of the main stem for every 2 weeks. The 1~5 internodes and the first fully developed leaf were collected from 60 DAP plants of sdm2 and FH1 for RNA sequencing. At this time, the main stem height of sdm2 and FH1 was significantly different. The samples were frozen immediately in liquid nitrogen, and stored at − 80 °C for RNA extraction. For both stem and leaf, three replicates were prepared.
The hydroponic culture experiment was conducted in illumination incubator in the laboratory. The temperature was 32 °C and the light-dark cycle was 14 h of light and 10 h of darkness. Seeds of sdm2 and FH1 were planted in glass ware filled with Hogland culture solution with the preservative film for support. About 20 plants were planted for each line. At the 14 DAP, the number of internodes and the length of each internode and hypocotyl were measured. The main stem height was the summation of each internode length. Each line had five replicates.
RNA extraction, cDNA synthesis and high throughput sequencing
Total RNA was extracted from stem and leaf using Trizol Reagent (TaKaRa, Inc., Dalian, China) according to the manufacturer’s instructions. RNA samples were treated with DNase I to remove genomic DNA contamination. RNA quality and quantity were analyzed using Agilent 2100 and NanoDrop. mRNA was enriched using magnetic beads with Oligo (dT) and cleaved into short fragments (~ 200 nt) in fragmentation buffer. The reverse transcription was conducted with random hexamer primer and then the second strand cDNA was synthesized. After end repair, the 5′ tails were phosphorylated, the 3′ tails were added with anadenine. Sequencing adaptors were ligated to the double-stranded DNA fragments. Then the fragments were amplified by PCR to construct cDNA library. The library was sequenced using BGISEQ-500 platform by Beijing Genomics Institute (BGI).
Bioinformatics analysis of RNA-Seq data
Raw reads were generated from each cDNA library. To obtain clean reads, the adaptor sequences, low-quality reads and reads containing more than 5% unknown bases were removed using SOAPunke and trimmomatic software. All clean reads were mapped to the reference genome of Arachis hypogaea cv. Tifrunner (https://www.peanutbase.org/data/public/Arachis_hypogaea/) using HISAT2 program [83]. The clean reads were aligned to reference gene by Bowtie2 (RNA-Seq by Expectation Maximization) [84]. The statistical analyses of randomness, degree of coverage, and sequencing saturation were also accomplished. The gene expression level was calculated with RSEM method [85] and normalized to FPKM (Fragments Per Kb per Million reads). The relative gene expression level between two samples was counted by log2 ratio. Pearson’s correlation coefficient between every two samples and principal component analysis (PCA) were performed. Differentially expressed genes (DEGs) were identified using DEGseq2 method and screened with the criteria of fold change≥2 and Q-value≤0.001 [86].
Gene Ontology (GO) annotation was carried out by Blast2GO program through comparing DEGs with GO terms in the GO database and GO functional classification was performed using WEGO software. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was conducted by mapping DEGs to KEGG database. The GO and KEGG functional enrichment were performed using hypergeometric test. The p-value formula in hypergeometric test can be acquired from https://en.wikipedia.org/wiki/Hypergeometric_distribution in detail. FDR (False Discovery Rate) correction of all p-values was conducted. The GO terms and KEGG pathways whose FDR ≤ 0.01 were defined as significant enriched. BLASTX (E < 0.00001) against NCBI Nr database was carried out. For transcription factor (TF) annotation, the ORF of DEGs were detected by getorf software, then these ORFs were aligned to TF protein domain (data from PlantTFDB) using hmmsearch program, and finally TFs were identified according to the TF family characteristics described in PlantTFDB (http://plntfdb.bio.uni-potsdam.de/v3.0/). The DEGs were also aligned to Plant Resistance Gene Database (PRGdb, http://prgdb.crg.eu/) by DIAMOND software to identified the resistance genes according to the query coverage and identity. The heatmap of cell wall related gene expression was charted using MeV software (https://sourceforge.net/projects/mev-tm4/) with the data of relative gene expression level between two samples counted by log2 (sdm2/FH1).
qRT-PCR validation of RNA-Seq data
We used qRT-PCR method to verify the expressional levels of 10 selected genes. RNA samples were those used for high-throughput sequencing and the reverse transcriptions were performed using PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa). The gene-specific primers were designed using PerlPrimer software and were listed in Additional file 10: Table S7. We performed qRT-PCR reaction on ABI7500 Real Time System (Applied Biosystems) using TB Green™ Premix Ex Taq™ II (TaKaRa). The parameters of thermal cycle were 94 °C for 10 min, followed by 40 cycles of 94 °C for 15 s and 60 °C for 1 min in a 20 μl volume. Each reaction was performed three biological replications with actin gene as internal reference gene. The relative expressional level of each gene between sdm2 and FH1 was calculated by 2-△△Ct method.
Supplementary information
Additional file 1 Figure S1. The correlation heatmap of each sample of sdm2 and FH1. The X and Y axes represent each sample. The color represents the Pearson correlation coefficient (the darker the color, the higher the correlation; the lighter the color, the lower the correlation).
Additional file 2 Figure S2. The principal component analysis of each sample of sdm2 and FH1. X axis represents the principal component 1 and Y axis represents the principal component 2.
Additional file 3 Table S1. Differentially expressed genes of stem between sdm2 and FH1.
Additional file 4 Table S2. Differentially expressed genes of leaf between sdm2 and FH1.
Additional file 5 Table S3. GO classification of DEGs in stem and leaf.
Additional file 6 Figure S3. Bubble diagram of top 20 enriched GO terms of DEGs in stem and leaf. X axis represents the Rich Ratio, which meaning the ratio of selected gene number annotated to a particular item to the total number of genes in this item in one species. The calculating formula is Rich Ratio = Term Candidate Gene Num/Term Gene Num. Y axis represents GO Term. The size of the bubbles indicates the number of genes annotated to a GO Term. And the color represents Q-value of enrichment. The deeper the color, the smaller the Q-value.
Additional file 7 Table S4. The top 20 enriched GO terms of DEGs in stem and leaf.
Additional file 8 Table S5. The top 20 enriched KEGG pathways of DEGs in stem and leaf.
Additional file 9 Table S6. Validation of RNA-seq results via qRT-PCR.
Additional file 10 Table S7. Primers for qRT-PCR analysis.
Abbreviations
- ABCB
Subfamily B of ATP-binding cassette
- ACE1
Activator for cell elongation 1
- ARF
Auxin response factor
- Aux/IAA
Auxin resistant/indole-3-acetic acid inducible
- AUX1/LAX
Auxin1/like-AUX1
- BAK1
BRI1-associated receptor kinase 1
- BAS1
Brassinosteroids C-26 hydroxylase
- BES1
BRI1-EMS suppressor 1
- BR6ox
Brassinosteroid-6-oxidases
- BRI1
Brassinosteroid insensitive 1
- BZR1
Brassinazole resistant 1
- CesA
Cellulose synthases
- CPD
Putative C-3 oxidase
- CSL
Cellulose synthase-like genes
- CYP90C1/D1
3-epi-6-deoxocathasterone 23-monooxygenase
- DELLA
Proteins containing conserved DELLA domains in the N terminus
- DWF4
22a-hydroxylase
- EXPs
Expansins
- GA20ox
GA 20-oxidases
- GA2ox
GA 2-oxidases
- GA3ox
GA 3-oxidases
- GID1
GA-insensitive dwarf 1
- GRPs
Gibberellin-regulated proteins
- HBI1
Homolog of BEE2 interacting with IBH1
- IBH1
ILI1 binding bHLH protein 1
- PCA
Principal component analysis
- PHY
Phytochrome
- PIFs
Phytochrome-interacting factors
- PIN
PIN-formed protein
- PRE6
Paclobutrazol resistant 6
- qRT-PCR
Quantitative Real-time PCR
- SAUR
Small auxin up RNA
- XTHs
Xyloglucanendotransglucosylase/endohydrolases
Authors’ contributions
YXZ, XJW and LH conceived and designed the study, FDG, JJM, LH, SHS, GHL, CZZ, HX, SZZ and JBS performed the experiments. FDG and JJM analyzed the data and drafted the manuscript with LH. YXZ and XJW revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by grants from the National Natural Science Foundation of China (31861143009, 31500217, 31471526), Shandong Province Natural Science Foundation (ZR2017MC005), Shandong Provincial project for crop variety improvement and application (2016LZGC025, 2017LZN032), Shandong Provincial key research and development program(2018GSF121032), Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (CXGC2018E13), Shandong Province Taishan Scholar Foundation. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The RNA-seq data in this study were available at NCBI Short Read Archive with the accession number of SRP213768 (https://www.ncbi.nlm.nih.gov/search/all/?term=SRP213768).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fengdan Guo and Junjie Ma contributed equally to this work.
Contributor Information
Xingjun Wang, Email: xingjunw@hotmail.com.
Yanxiu Zhao, Email: zhaoyx@sdnu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-6614-0.
References
- 1.Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, et al. Positional cloning of rice semidwarfing gene, sd-1: rice "green revolution gene" encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 2002;9(1):11–17. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- 2.Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, et al. 'Green revolution' genes encode mutant gibberellin response modulators. Nature. 1999;400(6741):256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 3.Van Sandt VS, Suslov D, Verbelen JP, Vissenberg K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot. 2007;100(7):1467–1473. doi: 10.1093/aob/mcm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279(5351):717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 5.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12(12):2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton RA, Gibeaut DM, Bacic A, Findlay K, Roberts K, Hamilton A, et al. Virus-induced silencing of a plant cellulose synthase gene. Plant Cell. 2000;12(5):691–706. doi: 10.1105/tpc.12.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, et al. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009;60(6):1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x. [DOI] [PubMed] [Google Scholar]
- 8.Ding Z, Lin Z, Li Q, Wu H, Xiang C, Wang J. DNL1, encodes cellulose synthase-like D4, is a major QTL for plant height and leaf width in rice (Oryza sativa L.) Biochem Biophys Res Commun. 2015;457(2):133–140. doi: 10.1016/j.bbrc.2014.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Smith SM, Li J. Genetic regulation of shoot architecture. Annu Rev Plant Biol. 2018;69:437–468. doi: 10.1146/annurev-arplant-042817-040422. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407(6802):321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- 11.Choi D, Lee Y, Cho HT, Kende H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell. 2003;15(6):1386–1398. doi: 10.1105/tpc.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho HT, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2000;97(17):9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenoni S, Reale L, Tornielli GB, Lanfaloni L, Porceddu A, Ferrarini A, et al. Downregulation of the Petunia hybrida alpha-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. Plant Cell. 2004;16(2):295–308. doi: 10.1105/tpc.018705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh HH, Sloan J, Dorca-Fornell C, Fleming A. Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiol. 2012;159(4):1759–1770. doi: 10.1104/pp.112.200881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jan A, Yang G, Nakamura H, Ichikawa H, Kitano H, Matsuoka M, et al. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol. 2004;136(3):3670–3681. doi: 10.1104/pp.104.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J. 1992;282(Pt 3):821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JE, Fry SC. Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J. 2001;26(1):23–34. doi: 10.1046/j.1365-313x.2001.01005.x. [DOI] [PubMed] [Google Scholar]
- 18.Han Y, Ban Q, Li H, Hou Y, Jin M, Han S, et al. DkXTH8, a novel xyloglucan endotransglucosylase/hydrolase in persimmon, alters cell wall structure and promotes leaf senescence and fruit postharvest softening. Sci Rep. 2016;6:39155. doi: 10.1038/srep39155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llop-Tous I, Dominguez-Puigjaner E, Palomer X, Vendrell M. Characterization of two divergent endo-beta-1,4-glucanase cDNA clones highly expressed in the nonclimacteric strawberry fruit. Plant Physiol. 1999;119(4):1415–1422. doi: 10.1104/pp.119.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King RW, Evans LT, Mander LN, Moritz T, Pharis RP, Twitchin B. Synthesis of gibberellin GA6 and its role in flowering of Lolium temulentum. Phytochemistry. 2003;62(1):77–82. doi: 10.1016/S0031-9422(02)00447-8. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134(4):1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleet CM, Sun TP. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol. 2005;8(1):77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85(2):171–182. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2(4):505–513. doi: 10.1016/S1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416(6882):701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 26.Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A. 2002;99(13):9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437(7059):693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Hou M, Liu L, Wu S, Shen Y, Ishiyama K, et al. The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014;166(4):2028–2039. doi: 10.1104/pp.114.247486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20(10):2603–2618. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58(5):991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- 31.Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, et al. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10(10):1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10(2):219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youn JH, Kim TW, Joo SH, Son SH, Roh J, Kim S, et al. Function and molecular regulation of DWARF1 as a C-24 reductase in brassinosteroid biosynthesis in Arabidopsis. J Exp Bot. 2018;69(8):1873–1886. doi: 10.1093/jxb/ery038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272(5260):398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 35.Anwar A, Liu Y, Dong R, Bai L, Yu X, Li Y. The physiological and molecular mechanism of brassinosteroid in response to stress: a review. Biol Res. 2018;51(1):46. doi: 10.1186/s40659-018-0195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohnishi T, Godza B, Watanabe B, Fujioka S, Hategan L, Ide K, et al. CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J Biol Chem. 2012;287(37):31551–31560. doi: 10.1074/jbc.M112.392720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288(5475):2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 38.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410(6826):380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 39.Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433(7022):167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 40.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111(3):671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110(2):213–222. doi: 10.1016/S0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 42.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110(2):203–212. doi: 10.1016/S0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15(2):220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295(5558):1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 45.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 46.Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120(2):249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 47.He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307(5715):1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Ye H, Guo H, Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci U S A. 2010;107(8):3918–3923. doi: 10.1073/pnas.0909198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schenck D, Christian M, Jones A, Luthen H. Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol. 2010;152(3):1183–1185. doi: 10.1104/pp.109.149591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noh B, Murphy AS, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 2001;13(11):2441–2454. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knauss S, Rohrmeier T, Lehle L. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem. 2003;278(26):23936–23943. doi: 10.1074/jbc.M212585200. [DOI] [PubMed] [Google Scholar]
- 52.Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci U S A. 2006;103(1):236–241. doi: 10.1073/pnas.0507127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, et al. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70(6):978–990. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fridborg I, Kuusk S, Moritz T, Sundberg E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell. 1999;11(6):1019–1032. doi: 10.1105/tpc.11.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in Brassinosteroid-regulated plant growth and drought responses. Plant Cell. 2017;29(6):1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14(8):802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, et al. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14(8):810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21(12):3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell. 2012;24(11):4483–4497. doi: 10.1105/tpc.112.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bai MY, Fan M, Oh E, Wang ZY. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell. 2012;24(12):4917–4929. doi: 10.1105/tpc.112.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang L, Ren X, Wu B, Li X, Chen W, Zhou X, et al. Development and deployment of a high-density linkage map identified quantitative trait loci for plant height in peanut (Arachis hypogaea L.) Sci Rep. 2016;6:39478. doi: 10.1038/srep39478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li YJ, Li LZ, Zhang XR, Zhang K, Ma DC, Liu JQ, et al. QTL mapping and marker analysis of main stem height and the first lateral branch length in peanut (Arachis hypogaeal.). Euphytica. 2017;213, 57(2).
- 63.Lv J, Liu N, Guo J, Xu Z, Li X, Li Z, et al. Stable QTLs for Plant Height on Chromosome A09 Identified From Two Mapping Populations in Peanut (Arachis hypogaea L.) Front Plant Sci. 2018;9:684. doi: 10.3389/fpls.2018.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10(2):231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, et al. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126(2):770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, et al. CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 2005;41(5):710–721. doi: 10.1111/j.1365-313X.2004.02330.x. [DOI] [PubMed] [Google Scholar]
- 67.Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell. 2001;105(5):625–636. doi: 10.1016/S0092-8674(01)00370-1. [DOI] [PubMed] [Google Scholar]
- 68.Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, et al. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108(5):2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302(5642):81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- 70.Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65(4):634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 71.Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58(2):275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19(5):765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhiponova MK, Vanhoutte I, Boudolf V, Betti C, Dhondt S, Coppens F, et al. Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 2013;197(2):490–502. doi: 10.1111/nph.12036. [DOI] [PubMed] [Google Scholar]
- 74.Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013;162(4):1965–1977. doi: 10.1104/pp.113.217703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26(6):573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- 76.Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127(2):450–458. doi: 10.1104/pp.010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ross JJ, O'Neill DP, Smith JJ, Kerckhoffs LH, Elliott RC. Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J. 2000;21(6):547–552. doi: 10.1046/j.1365-313x.2000.00702.x. [DOI] [PubMed] [Google Scholar]
- 78.Wolbang CM, Ross JJ. Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta. 2001;214(1):153–157. doi: 10.1007/s004250100663. [DOI] [PubMed] [Google Scholar]
- 79.Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004;134(4):1624–1631. doi: 10.1104/pp.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2(9):E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, et al. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 2011;66(4):564–578. doi: 10.1111/j.1365-313X.2011.04513.x. [DOI] [PubMed] [Google Scholar]
- 82.Li L, Xu J, Xu ZH, Xue HW. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell. 2005;17(10):2738–2753. doi: 10.1105/tpc.105.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Figure S1. The correlation heatmap of each sample of sdm2 and FH1. The X and Y axes represent each sample. The color represents the Pearson correlation coefficient (the darker the color, the higher the correlation; the lighter the color, the lower the correlation).
Additional file 2 Figure S2. The principal component analysis of each sample of sdm2 and FH1. X axis represents the principal component 1 and Y axis represents the principal component 2.
Additional file 3 Table S1. Differentially expressed genes of stem between sdm2 and FH1.
Additional file 4 Table S2. Differentially expressed genes of leaf between sdm2 and FH1.
Additional file 5 Table S3. GO classification of DEGs in stem and leaf.
Additional file 6 Figure S3. Bubble diagram of top 20 enriched GO terms of DEGs in stem and leaf. X axis represents the Rich Ratio, which meaning the ratio of selected gene number annotated to a particular item to the total number of genes in this item in one species. The calculating formula is Rich Ratio = Term Candidate Gene Num/Term Gene Num. Y axis represents GO Term. The size of the bubbles indicates the number of genes annotated to a GO Term. And the color represents Q-value of enrichment. The deeper the color, the smaller the Q-value.
Additional file 7 Table S4. The top 20 enriched GO terms of DEGs in stem and leaf.
Additional file 8 Table S5. The top 20 enriched KEGG pathways of DEGs in stem and leaf.
Additional file 9 Table S6. Validation of RNA-seq results via qRT-PCR.
Additional file 10 Table S7. Primers for qRT-PCR analysis.
Data Availability Statement
The RNA-seq data in this study were available at NCBI Short Read Archive with the accession number of SRP213768 (https://www.ncbi.nlm.nih.gov/search/all/?term=SRP213768).