Abstract
Background
Previous phylogenetic analyses of species within the genus Golovinomyces (Ascomycota, Erysiphales), based on ITS and 28S rDNA sequence data, revealed a co-evolutionary relationship between powdery mildew species and hosts of certain tribes of the plant family Asteraceae. Golovinomyces growing on host plants belonging to the Heliantheae formed a single lineage, comprised of a morphologically differentiated complex of species, which included G. ambrosiae, G. circumfusus, and G. spadiceus. However, the lineage also encompassed sequences retrieved from Golovinomyces specimens on other Asteraceae tribes as well as other plant families, suggesting the involvement of a plurivorous species. A multilocus phylogenetic examination of this complex, using ITS, 28S, IGS (intergenic spacer), TUB2 (beta-tubulin), and CHS1 (chitin synthase I) sequence data was carried out to clarify the discrepancies between ITS and 28S rDNA sequence data and morphological differences. Furthermore, the circumscription of species and their host ranges were emended.
Results
The phylogenetic and morphological analyses conducted in this study revealed three distinct species named, viz., (1) G. ambrosiae emend. (including G. spadiceus), a plurivorous species that occurs on a multitude of hosts including, Ambrosia spp., multiple species of the Heliantheae and plant species of other tribes of Asteraceae including the Asian species of Eupatorium; (2) G. latisporus comb. nov. (≡ Oidium latisporum), the closely related, but morphologically distinct species confined to hosts of the Heliantheae genera Helianthus, Zinnia, and most likely Rudbeckia; and (3) G. circumfusus confined to Eupatorium cannabinum in Europe.
Conclusions
The present results provide strong evidence that the combination of multi-locus phylogeny and morphological analysis is an effective way to identify species in the genus Golovinomyces.
Keywords: Erysiphaceae, Powdery mildew, Heliantheae, 28S rDNA, ITS, Golovinomyces latisporus, IGS, TUB2, CHS1
Background
Powdery mildews are obligate biotrophic ascomycetes that occur on a wide range of dicotyledonous and monocotyledonous host plants. The family Erysiphaceae has a nearly worldwide distribution, with the exception of the Antarctic region, and currently comprises around 900 species in 18 genera [1–3]. Golovinomyces was originally introduced by Braun [4] as a section of the genus Erysiphe (s. lat.) and was later raised to genus rank by Heluta [5]. Braun [6] and Braun and Takamatsu [7] accepted Golovinomyces as a distinct genus and established the new tribe Golovinomyceteae. Golovinomyces is characterized by having chasmothecia with mycelioid appendages, several, mostly 2-spored asci, an asexual morph with catenescent conidia that lack fibrosin bodies, and mostly nipple-shaped appressoria. Golovinomyces currently encompasses 57 species and 5 varieties [1, 8–13]. Erysiphe cichoracearum [14] included nearly all of the species that are now assigned to Golovinomyces. Blumer [15, 16] split E. cichoracearum sensu Salmon [14] into several species but continued to maintain the species E. cichoracearum in a very broad sense (covering collections on Asteraceae and on hosts of multiple other plant families). Braun [17] confined E. cichoracearum to powdery mildews on hosts of Asteraceae and assigned specimens on hosts belonging to other plant families to Erysiphe orontii. Phylogenetic analyses of Golovinomyces, based on ITS and 28S rDNA sequence data [18], suggested the co-evolution between Golovinomyces species and certain tribes of Asteraceae. Based on these results, Braun and Cook [1] introduced a much narrower species concept for this genus, which included two morphologically differentiated species on hosts belonging to the Heliantheae, viz., G. ambrosiae and G. spadiceus. However, in more detailed phylogenetic analyses of ITS and 28S rDNA sequences, including Golovinomyces species on Asteraceae hosts, Takamatsu et al., [19] found that powdery mildews on hosts of the Heliantheae (previously referred to as G. ambrosiae and G. spadiceus), on hosts of an Asian species of Eupatorium (G. circumfusus s. lat.) and on a multitude of other hosts, including those on other plant families, formed a single large, unresolved clade (lineage III in Takamatsu et al., [19]). The taxonomic interpretation of these results posed a serious problem since G. ambrosiae and G. spadiceus, as circumscribed in Braun and Cook [1], are two morphologically differentiated species. Hence, the resolution within phylogenetic trees based only on ITS sequences was in this case insufficient to discriminate closely allied species. Therefore, most subsequent authors followed the taxonomic treatment in Braun and Cook [1] and recognized G. ambrosiae and G. spadiceus as separate species within lineage III, based on morphological differences [20–27]. The morphological differences used to differentiate the species include above all, much broader conidia and dimorphic germ tubes belonging to the longitubus pattern within the Euoidium type of conidial germination in G. ambrosiae than in G. spadiceus [1]. Additional research has found G. spadiceus to be extremely plurivorous, occurring on hosts of the Heliantheae and other tribes of Asteraceae, e.g., Aster and Chrysanthemum [19], Chrysogonum [28], as well hosts of various other plant families, including Abelmoschus (Malvaceae) [29], Crotalaria (Fabaceae) [13], Persicaria (Polygonaceae) [11, 13, 30], Solanum (Solanaceae) [13], and Verbena (Verbenaceae) [13]. The taxonomic interpretation of the inclusion of a sequence obtained from a Japanese collection of powdery mildew on Eupatorium chinense in lineage III [19] caused an additional problem and raised the question whether the name G. circumfusus, originally described from Europe on Eupatorium cannabinum, is included in this species complex.
The purpose of the present study was to clarify and resolve the taxonomy of this Golovinomyces complex using a multilocus approach, based on ITS, 28S, IGS, TUB2 and CHS1 DNA sequences. Multi-gene analyses are currently the method of choice to analyze phylogenetically and taxonomically difficult complexes of plant pathogenic fungi, including Colletotrichum spp. [31, 32]. However, there is minimal multilocus data for the powdery mildews currently available. Most of the research involves the intraspecific genetic diversity in species such as Blumeria graminis [33, 34], Erysiphe japonica [35], E. necator [36, 37], Podosphaera xanthii [38] and Golovinomyces orontii [39]. Recently, the geographic and temporal distributions of four genotypes found in E. gracilis var. gracilis were studied based on a combination of data from the ITS, 28S rDNA and IGS regions [40]. Comprehensive applications of multilocus approaches to solve complex taxonomic-phylogenetic problems connected with the species level classification of the powdery mildews are still lacking. The present study is the first to use a multilocus approach to solve species distinction issues within the Erysiphales. An additional issue regarding the taxonomic conclusions drawn from phylogenetic results is also addressed in this study. Older taxonomic names are often available, but the application and allocation of such names are usually problematic. Because species names are based on their type collections, epitypifications, with appropriate new material, and ex-epitype sequences tend to be the main method to overcome these obstacles and to determine the application of older names. During the current study, this issue was addressed using international collaboration.
Methods
Sampling
A total of 69 specimens belonging to Golovinomyces ambrosiae, G. circumfusus, and G. spadiceus were examined, including 39 samples collected in China in recent years and 30 additional specimens from Germany, Japan, Russia, Switzerland, and the USA. Furthermore, eight specimens, consisting of three samples of G. magnicellulatus, three samples of Neoërysiphe galeopsidis, a sample of Arthrocladiella mougeotii and a sample of Erysiphe kenjiana, were used for phylogenetic analyses in this study. All of the plant materials used in this study were collected in the public gardens with Latin names or some are common ornamental plants which were identified by ourselves. Among the 69 specimens, ISC-F-0076752, ISC-F-0076753, and ISC-F-0076754 were deposited in the Herbarium of Iowa State University Fungi of Iowa, and the rest voucher specimens were deposited in the Herbarium of Mycology of Jilin Agricultural University. Names of the host plants, fungal species, locations and years of collection, voucher numbers and newly sequenced multi-gene accession numbers for the nucleotide sequence database (GenBank) in this study are given in Table 1.
Table 1.
Information of powdery mildew vouchers studied in this paper
| Species | Host | Location | Year of collection | Voucher a | GenBank accessions No. b | ||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | 28S | IGS | TUB2 | CHS1 | |||||
| Arthrocladiella mougeotii | Lycium chinense | Beijing, China | 2018 | HMJAU-PM91837 | MK452607 | MK452680 | – | – | – |
| Erysiphe kengiana | Ulmus pumila | Changchun, Jilin province, China | 2017 | HMJAU-PM91841 | MK452611 | MK452684 | – | MK452458 | – |
| Golovinomyces ambrosiae | Aster novi-belgii | Changchun, Jilin province, China | 2017 | HMJAU-PM91804 | MK452575 | MK452648 | MK452501 | MK452460 | MK452410 |
| G. ambrosiae | A. novi-belgii | Changchun, Jilin province, China | 2018 | HMJAU-PM91805 | MK452576 | MK452649 | MK452502 | MK452461 | MK452411 |
| G. ambrosiae | A. novi-belgii | Dunhua, Jilin province, China | 2018 | HMJAU-PM91806 | MK452577 | MK452650 | MK452503 | MK452462 | MK452412 |
| G. ambrosiae | A. novi-belgii | Dunhua, Jilin province, China | 2018 | HMJAU-PM91807 | MK452578 | MK452651 | MK452504 | MK452463 | MK452413 |
| G. ambrosiae | A. novi-belgii | Changchun, Jilin province, China | 2017 | HMJAU-PM91808 | MK452579 | MK452652 | MK452505 | MK452464 | MK452414 |
| G. ambrosiae | Ageratina ligustrina | Sochi city, Krasnodar region, Russia | 2018 | ERY015 | MK452643 | MK452717 | MK452570 | – | – |
| G. ambrosiae | Ambrosia artemisiifolia | Mudanjiang, Heilongjiang, China | 2017 | HMJAU-PM91809 | MK452580 | MK452653 | MK452506 | MK452465 | MK452415 |
| G. ambrosiae | A. artemisiifolia | Changchun, Jilin province, China | 2018 | HMJAU-PM91810 | MK452581 | MK452654 | MK452507 | MK452466 | MK452416 |
| G. ambrosiae | A. artemisiifolia | Tonghua, Jilin province, China | 2018 | HMJAU-PM91811 | MK452582 | MK452655 | MK452508 | MK452467 | MK452417 |
| G. ambrosiae | A. artemisiifolia | Tonghua, Jilin province, China | 2018 | HMJAU-PM91812 | MK452583 | MK452656 | MK452509 | – | MK452418 |
| G. ambrosiae | A. artemisiifolia | Guthrie County, Iowa, USA | 1987 | ISC-F-0076752 | – | – | MK452567 | – | – |
| G. ambrosiae | A. trifida | Guthrie County, Iowa, USA | 1987 | ISC-F-0076754 | – | MK452715 | – | – | – |
| G. ambrosiae | A. trifida | Guthrie County, Iowa, USA | 1997 | ISC-F-0076753 | – | – | MK452568 | – | – |
| G. ambrosiae | A. trifida | Siping, Jilin province, China | 2018 | HMJAU-PM91813 | MK452584 | MK452657 | MK452510 | MK452468 | MK452419 |
| G. ambrosiae | A. trifida | Changchun, Jilin province, China | 2018 | HMJAU-PM91814 | MK452585 | MK452658 | MK452511 | MK452469 | MK452420 |
| G. ambrosiae | A. trifida | Anshan, Liaoning, China | 2018 | HMJAU-PM91815 | MK452586 | MK452659 | MK452512 | MK452470 | MK452421 |
| G. ambrosiae | A. trifida | Shenyang, Liaoning, China | 2018 | HMJAU-PM91816 | MK452587 | MK452660 | MK452513 | – | MK452422 |
| G. ambrosiae | Dahlia pinnata | Dandong, Liaoning, China | 2012 | HMJAU-PM91817 | MK452588 | MK452661 | MK452514 | – | – |
| G. ambrosiae | D. pinnata | Changchun, Jilin province, China | 2017 | HMJAU-PM91818 | MK452589 | MK452662 | MK452515 | MK452471 | MK452423 |
| G. ambrosiae | D. pinnata | Changchun, Jilin province, China | 2017 | HMJAU-PM91819 | MK452590 | MK452663 | MK452516 | MK452472 | MK452424 |
| G. ambrosiae | D. pinnata | Changchun, Jilin province, China | 2018 | HMJAU-PM91820 | MK452591 | MK452664 | MK452517 | MK452473 | MK452425 |
| G. ambrosiae | D. pinnata | Siping, Jilin province, China | 2018 | HMJAU-PM91821 | MK452592 | MK452665 | MK452518 | MK452474 | MK452426 |
| G. ambrosiae | D. pinnata | Panzhihua, Sichuan, China | 2018 | HMJAU-PM91822 | MK452593 | MK452666 | MK452519 | MK452475 | MK452427 |
| G. ambrosiae | Dahlia sp. | Yolo Co. CA, USA | 2018 | MVAP50000445 | MK452632 | MK452705 | MK452557 | – | – |
| G. ambrosiae | Dahlia sp. | Santa Barbara Co. CA, USA | 2018 | LM0P03825217–1 | MK452637 | MK452710 | MK452562 | – | MK452457 |
| G. ambrosiae | Dahlia sp. | Seattle Washington, USA | 2018 | HMJAU-PM91854 | MK452641 | MK452714 | MK452566 | – | – |
| G. ambrosiae | Eupatorium japonicum | Aichi, Nagoya-shi, Japan | 2001 | MUMH4142 | MK452621 | MK452694 | MK452546 | – | – |
| G. ambrosiae | E. makinoi | Katashina-mura, Gunma, Japan | 2002 | MUMH4143 | MK452622 | MK452695 | MK452547 | – | – |
| G. ambrosiae | E. makinoi | Tochigi, Sano, Japan | 2002 | MUMH4424 | MK452623 | MK452696 | MK452548 | – | – |
| G. ambrosiae | E. makinoi | Okayama-shi, Okayama, Japan | 2003 | MUMH4794 | MK452625 | MK452698 | MK452550 | – | – |
| G. ambrosiae | E. makinoi | Shiga, Maibara, Japan | 2017 | MUMH7129 | MK452624 | MK452697 | MK452549 | – | – |
| G. ambrosiae | E. japonicum | Mie, Tsu-shi, Japan | 2018 | HMJAU-PM91855 | MK452626 | MK452699 | MK452551 | MK452496 | MK452453 |
| G. ambrosiae | Leucanthemum maximum | Changchun, Jilin province, China | 2016 | HMJAU-PM91836 | KX987303 | MF612182 | MK452533 | MK389490 | MK389489 |
| G. ambrosiae | Zinnia elegans | Chengdu, Sichuan, China | 2016 | HMJAU-PM91842 | MK452612 | MK452685 | MK452537 | MK452487 | MK452444 |
| G. ambrosiae | Z. elegans | Changchun, Jilin province, China | 2017 | HMJAU-PM91843 | MK452613 | MK452686 | MK452538 | MK452488 | MK452445 |
| G. ambrosiae | Z. elegans | Changchun, Jilin province, China | 2017 | HMJAU-PM91844 | MK452614 | MK452687 | MK452539 | MK452489 | MK452446 |
| G. ambrosiae | Z. elegans | Changchun, Jilin province, China | 2017 | HMJAU-PM91845 | MK452615 | MK452688 | MK452540 | MK452490 | MK452447 |
| G. ambrosiae | Z. elegans | Changchun, Jilin province, China | 2018 | HMJAU-PM91846 | MK452616 | MK452689 | MK452541 | MK452491 | MK452448 |
| G. ambrosiae | Z. elegans | Siping, Jilin province, China | 2018 | HMJAU-PM91847 | MK452617 | MK452690 | MK452542 | MK452492 | MK452449 |
| G. ambrosiae | Z. elegans | Tonghua, Jilin province, China | 2018 | HMJAU-PM91848 | MK452618 | MK452691 | MK452543 | MK452493 | MK452450 |
| G. ambrosiae | Z. elegans | Siping, Jilin province, China | 2018 | HMJAU-PM91849 | MK452619 | MK452692 | MK452544 | MK452494 | MK452451 |
| G. ambrosiae | Z. elegans | Santa Barbara Co. CA, USA | 2018 | LM0P06825217–3 | MK452633 | MK452706 | MK452558 | – | MK452456 |
| G. ambrosiae | Z. elegans | Yolo Co. CA, USA | 2018 | MVAP50000452 | MK452634 | MK452707 | MK452559 | – | – |
| G. circumfusus | Eupatorium cannabinum | Altmark, Sachsen-Anhalt, Germany | 2000 | GLM49501 | MK452630 | MK452703 | MK452553 | – | – |
| G. circumfusus | E. cannabinum | Landkreis Ostprignitz-Ruppin, Brandenburg, Germany | 2006 | GLM74796 | MK452629 | MK452702 | MK452554 | – | – |
| G. circumfusus | E. cannabinum | Spreewald, Brandenburg, Germany | 2016 | HAL 3300 F | MK452628 | MK452701 | MK452555 | MK452459 | MK452455 |
| G. latisporus | Helianthus annuus | Changchun, Jilin province, China | 2017 | HMJAU-PM91830 | MK452601 | MK452674 | MK452527 | MK452483 | MK452435 |
| G. latisporus | H. annuus | Changchun, Jilin province, China | 2017 | HMJAU-PM91828 | MK452599 | MK452672 | MK452525 | MK452481 | MK452433 |
| G. latisporus | H. annuus | Yichun, Heilongjiang, China | 2017 | HMJAU-PM91829 | MK452600 | MK452673 | MK452526 | MK452482 | MK452434 |
| G. latisporus | H. annuus | Tonghua, Jilin province, China | 2018 | HMJAU-PM91831 | MK452602 | MK452675 | MK452528 | MK452484 | MK452436 |
| G. latisporus | H. annuus | Panzhihua, Sichuan, China | 2018 | HMJAU-PM91832 | MK452603 | MK452676 | MK452529 | MK452485 | MK452437 |
| G. latisporus | H. tuberosus | Chongqing, China | 2014 | HMJAU-PM91823 | MK452594 | MK452667 | MK452520 | MK452476 | MK452428 |
| G. latisporus | H. tuberosus | Shangqiu, Henan, China | 2016 | HMJAU-PM91824 | MK452595 | MK452668 | MK452521 | MK452477 | MK452429 |
| G. latisporus | H. tuberosus | Changchun, Jilin province, China | 2017 | HMJAU-PM91825 | MK452596 | MK452669 | MK452522 | MK452478 | MK452430 |
| G. latisporus | H. tuberosus | Changchun, Jilin province, China | 2017 | HMJAU-PM91826 | MK452597 | MK452670 | MK452523 | MK452479 | MK452431 |
| G. latisporus | H. tuberosus | Changchun, Jilin province, China | 2017 | HMJAU-PM91827 | MK452598 | MK452671 | MK452524 | MK452480 | MK452432 |
| G. latisporus | H. tuberosus | Shakhty city, Rostov region, Russia | 2018 | ERY057 | MK452642 | MK452716 | MK452569 | – | – |
| G. latisporus | H. tuberosus | Shakhty city, Rostov region, Russia | 2018 | ERY061 | MK452644 | MK452718 | MK452571 | – | – |
| G. latisporus | H. tuberosus | Shakhty city, Rostov region, Russia | 2018 | ERY081 | MK452645 | MK452719 | MK452572 | – | – |
| G. latisporus | H. tuberosus | Shakhty city, Rostov region, Russia | 2018 | ERY094 | MK452646 | MK452720 | MK452573 | – | – |
| G. latisporus | H. tuberosus | Novoshakhtinsk city, Rostov region, Russia | 2018 | ERY152 | MK452647 | MK452721 | MK452574 | – | – |
| G. latisporus | H. annuus | Nyon, Vaud, Switzerland | 2018 | HAL 3299 F | MK452627 | MK452700 | MK452552 | MK452497 | MK452454 |
| G. latisporus | H. annuus | Solano Co. CA, USA | 2018 | MVAP50000419 | MK452635 | MK452708 | MK452560 | MK452498 | – |
| G. latisporus | H. annuus | Santa Barbara Co. CA, USA | 2018 | LM0P03825217–2 | MK452636 | MK452709 | MK452561 | MK452499 | – |
| G. latisporus | H. annuus | Seattle Washington, USA | 2018 | HMJAU-PM91853 | MK452640 | MK452713 | MK452565 | – | – |
| G. latisporus | H. mollis | Seattle Washington, USA | 2018 | HMJAU-PM91851 | MK452638 | MK452711 | MK452563 | – | – |
| G. latisporus | Helianthus sp. | Seattle Washington, USA | 2018 | HMJAU-PM91852 | MK452639 | MK452712 | MK452564 | MK452500 | – |
| G. latisporus | Zinnia angustifolia | Potsdam, Brandenburg, Germany | 2008 | HAL 2338 F | MK452631 | MK452704 | MK452556 | – | – |
| G. latisporus | Z. elegans | Panzhihua, Sichuan, China | 2018 | HMJAU-PM91850 | MK452620 | MK452693 | MK452545 | MK452495 | MK452452 |
| G. magnicellulatus | Physalis alkekengi | Yichun, Heilongjiang, China | 2017 | HMJAU-PM91838 | MK452608 | MK452681 | MK452535 | – | MK452441 |
| G. magnicellulatus | P. alkekengi | Changchun, Jilin province, China | 2017 | HMJAU-PM91839 | MK452609 | MK452682 | MK452536 | – | MK452442 |
| G. magnicellulatus | P. alkekengi | Changchun, Jilin province, China | 2018 | HMJAU-PM91840 | MK452610 | MK452683 | MK452534 | MK452486 | MK452443 |
| Neoërysiphe galeopsidis | Leonurus artemisia | Beijing, China | 2018 | HMJAU-PM91833 | MK452604 | MK452677 | MK452530 | – | MK452438 |
| N. galeopsidis | L. artemisia | Beijing, China | 2018 | HMJAU-PM91834 | MK452605 | MK452678 | MK452531 | – | MK452439 |
| N. galeopsidis | L. artemisia | Changchun, Jilin province, China | 2017 | HMJAU-PM91835 | MK452606 | MK452679 | MK452532 | – | MK452440 |
aHMJAU Herbarium of Mycology of Jilin Agricultural University; HAL Herbarium of Halle University; GLM Herbarium of Senckenberg Museum für Naturkunde Görlitz; MUMH Mie University Mycological Herbarium; ERY herb. Bulgakov; LM and MVAP herb. S. Rooney Latham; ISC Iowa State University. The specimens GLM74796, GLM49501 (herbarium GLM, Görlitz, Germany), HAL 2338 F, HAL 3299 F, and HAL 3300 F (herbarium HAL, Halle [Saale], Germany) were supplied by Uwe Braun. The specimens MUMH4142, MUMH4143, MUMH4424, MUMH7129, MUMH4794, and HMJAU-PM91855 (herbarium MUMH, Mie, Japan) were provided by Susumu Takamatsu. The specimens MVAP50000419, MVAP50000445, MVAP50000452, LM0P03825217–1, LM0P03825217–2, and LM0P06825217–3 were supplied by Suzanne Latham-Rooney. The specimens HMJAU-PM91851, HMJAU-PM91852, HMJAU-PM91853, HMJAU-PM91854, ISC-F-0076752, ISC-F-0076753, and ISC-F-0076754 were supplied by Michael Bradshaw; and ERY015, ERY057, ERY061, ERY057, ERY081, ERY094 and ERY152 by Timur S. Bulgakov
b“–” means failed to get sequence
Morphological examinations
For microscopic examinations, fresh samples were mounted in sterile water, and dried specimens, scraped from the leaf surface with a clean scalpel, were mounted in a drop of lactic acid on a microscope slide. Slides were examined using light microscopy with the total magnification at 200 and 400 (Zeiss Axio Scope A1, Germany). Fresh conidia were examined for the presence or absence of fibrosin bodies. A minimum of 30 measurements were made of asexual and sexual fungal structures. Germination of conidia was examined following the method of Hirata [41].
Molecular techniques and phylogenetic analyses
Whole-cell DNA was extracted from chasmothecia or conidia and mycelia by the Chelex-100 method [42, 43]. In the USA, whole-cell DNA was extracted from chasmothecia or conidia (for the herbarium specimens: ISC-F-0076752, ISC-F-0076753, and ISC-F-0076754) with the DNeasy plant mini kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. Five genomic regions (ITS, 28S rDNA, IGS, TUB2, CHS1) were selected for phylogenetic analyses. The sequences and references of primers used to amplify these regions are shown in Table 2. For the TUB2 gene, primers TubF1/TubR1 were designed based on scaffold_4647 in genome of Erysiphe necator (GenBank ID: JNVN00000000.1) [47], contig c9894 in genome of E.pisi (GenBank ID: CACM00000000) and TUB2 sequence of G. orontii (KR815663) from Pirondi et al., [39]. For the CHS1 region, primers gCS1a1/gCS1b were designed based on the CHS1 sequences of AF188934 from Blumeria graminis [48], KJ698665 from Podosphaera xanthii [38], scaffold_1559 in the genome of E. necator (GenBank ID: JNVN00000000.1), contig1307 in the genome of G. orontii from the Joint Genome Institute (JGI) (Project ID: 1055997), and contig c7151 in the genome of E. pisi (GenBank ID: CACM00000000).
Table 2.
Primer sets for multilocus sequence typing (MLST) analysis of Golovinomyces in this study
| DNA regions | Primer | Primer sequences (5′ → 3′) | Annealing temperature (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| ITS | ITS5 ITS4 | GGAAGTAAAAGTCGTAACAAGG TCCTCCGCTTATTGATATGC | 52 | 600 | [44] |
| 28S rDNA | LSU1 LSU2 | ACCCGCTGAACTTAAGCATA CCTTGGTCCGTGTTTCAAGA | 52 | 500 | [45] |
| IGS | IGS-12a NS1R | AGTCTGTGGATTAGTGGCCG GAGACAAGCATATGACTAC | 52 | 400 | [46] |
| TUB2 | TubF1 TubR1 | AGGTTCACCTCCAGACTGG CCAGCACGAACAGCATCCAT | 52 | 450 | This study |
| CHS1 | gCS1a1 gCS1b | GGTGCATTCTCGGCATATCG CGTCACCCTTGGTGCCCCAAG | 52 | 1000 | This study |
To obtain sufficient DNA for sequencing, the DNA regions of TUB2 and CHS1 were amplified by two rounds of PCR with the same primer set. All PCR reactions were conducted in 25 μL volumes. The reaction components were 2.5 μL 10 × PCR Buffer (Mg2+ plus) (TaKaRa, Japan), 2 μL dNTP Mixture (10 mM total, 2.5 mM each), 1 μL each primer (20 ng/μL), 2 μL of total genomic DNA, 0.1 μL Taq polymerase (TaKaRa, Japan) (5 U/μL) and sterile ddH2O up to a final volume of 25 μL. The PCR reactions were conducted under the following thermal cycling conditions: an initial denaturation step of 5 min at 95 °C, 35 cycles of 1 min at 94 °C, followed by 30 s at 52 °C for annealing, and 2 min at 72 °C for extension, and a final extension for 8 min at 72 °C. A negative control that lacked template DNA was included in each set of reactions. PCR products were subjected to electrophoresis in a 1.2% agarose gel in 0.5× TBE buffer. The amplified DNA products were purified using Mag-MK PCR Products Purification Kit following the protocol of the manufacturer. Amplicons were sequenced in both directions with the same PCR primers using direct sequencing in a 3730xl DNA Analyzer (Applied Biosystems) by Sangon Biotech (Shanghai, China). The sequence reactions were conducted using the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) following instructions of the manufacturer.
The reaction components for the PCR conducted at the University of Washington were 5 uL AllTaq PCR Buffer (Qiagen, Germany), 0.5 uL dNTP mixture, 0.25 μL of each primer (100 uM), 2 μL of total genomic DNA, 0.5 μL, Taq Polymerase (Qiagen, Germany) and sterile ddH2O up to a final volume of 25 μL. DNA was purified by isopropanol precipitation. These sequences [(The 28S rDNA sequence from ISC-F-0076754 and IGS sequences from ISC-F-0076752 and ISC-F-0076753] were manually trimmed using Geneious version 11.0.2 (https://www.geneious.com) and deposited in GenBank.
All other new sequences obtained in the present study were edited by DNAMAN version 6.0 and BioEdit Sequence Alignment Editor version 7.0, and then deposited in GenBank (Table 1). The ITS, 28S, IGS, TUB2 and CHS1 sequences were respectively aligned by ClastalW. Furthermore, a multilocus sequences alignment datasets file (ITS+28S + IGS + TUB2 + CHS1) including 40 strains from Table 1 was also used for phylogenetic analyses. The six alignments were further refined manually in MEGA 7.0 [49] and deposited in TreeBASE (http://www.treebase.org/) under the Accession No. of S24404 (http://purl.org/phylo/treebase/phylows/study/TB2:S24404). Phylogenetic trees were obtained from the sequence data using maximum parsimony (MP) in PAUP 4.0b [50]. The MP analyses were performed with heuristic search option using the tree bisection reconnection (TBR) algorithm with 100 random sequence additions to find the global optimum tree. All sites were treated as unordered and unweighted, with gaps treated as missing data. The strength of the internal branches of the resulting trees were tested with bootstrap (BS) analysis using 1000 replications. Tree scores, including tree length, consistency index (CI), retention index (RI), and rescaled consistency index (RC), were also calculated. Five phylogenetic trees were generated based on the ITS, 28S, IGS, TUB2 and CHS1 nucleotide sequences.
Results
Phylogenetic analyses
Parsimoniuous trees were separately constructed based on sequences of five gene regions and their combination and the numerical data including the number of taxa and characters are shown in Table 3. The information of outgroup taxon for each phylogenetic tree was also included in Table 1. The phylogenetic trees based on the ITS and 28S rDNA sequences were topologically congruent and indicated that G. ambrosiae complex on many Asteraceae plants, including Eupatorium spp. from Japan, formed a single clade with 100 and 99% bootstrap support, respectively (see Additional files 1, 2: Figure S1, S2).
Table 3.
Information of the data matrices and the respective trees based on five individual gene regions
| DNA region | ITS | 28S | IGS | TUB2 | CHS1 | ITS+28S + IGS + TUB2 + CHS1 |
|---|---|---|---|---|---|---|
| Number of sequences | 74 | 75 | 74 | 44 | 49 | 40 |
| Number of characters | 509 | 639 | 393 | 432 | 968 | 2931 |
| Number of parsimony-uninformative characters | 50 | 26 | 1 | 112 | 22 | 182 |
| Number of parsimony-informative characters | 108 | 41 | 104 | 30 | 107 | 102 |
| Tree length | 228 | 87 | 133 | 164 | 154 | 305 |
| Consistency index (CI) | 0.8684 | 0.8621 | 0.8947 | 0.9512 | 0.9156 | 0.9902 |
| Retention index (RI) | 0.9242 | 0.9250 | 0.9595 | 0.9175 | 0.9698 | 0.9855 |
| Rescaled consistency index (RC) | 0.8026 | 0.7974 | 0.8585 | 0.8728 | 0.8879 | 0.9758 |
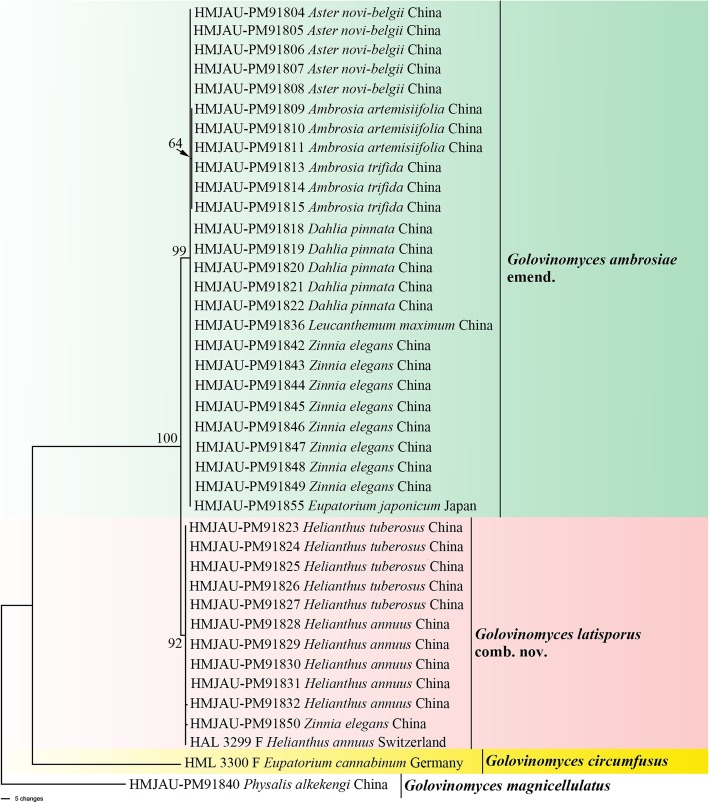
Golovinomyces circumfusus on E. cannabinum from Germany did not form a monophyletic group with G. ambrosiae complex in all phylogenies (see Additional file 1–5: Figure S1–S5 and Fig. 1). The phylogenetic tree of IGS was similar to ITS tree, with the G. ambrosiae complex formed a single clade with 100% bootstrap support based on the individual genes (see Additional file 3: Figure S3). However, the isolates from Helianthus spp. and some Zinnia spp. differed by one base from isolates on other host genera, and forming a subclade with 64% bootstrap support (see Additional file 3: Figure S3 pink clade). The G. ambrosiae complex included two groups, one identified as G. ambrosiae emend. (see Additional file 3: Figure S3 green clade) and the other as G. latisporus comb. nov. (see Additional file 3: Figure S3 pink clade), based on the phylogenetic analysis of the IGS. The G. ambrosiae complex in TUB2 and CHS1 trees was divided into two subgroups, viz. G. ambrosiae emend., including G. spadiceus with 91 and 85% bootstrap support respectively (see Additional files 4, 5: Figure S4, S5 green clade), and G. latisporus comb. nov. with 70 and 78% bootstrap support respectively (see Additional files 4, 5: Figure S4, S5 pink clade). In the G. ambrosiae emend. Clade the sequences of CHS1 from isolates on Ambrosiae artemisiifolia and A. trifida differed by one base from isolates on other hosts. Golovinomyces ambrosiae emend. is a plurivorous species that occurs on a multitude of hosts including, Ambrosia spp., multiple species from the Heliantheae and plant species of other tribes of Asteraceae including the Asian species of Eupatorium. Golovinomyces latisporus comb. nov. was confined to hosts of the Heliantheae genera Helianthus and Zinnia.
Fig. 1.
Phylogenetic analysis based on the combined sequence datasets of ITS+28S rDNA+IGS + TUB2 + CHS1 of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 40 strains from genus Golovinomyces. G. magnicellulatus (voucher: HMJAU-91840) was used as outgroup. Bootstrap values based on 1000 replications are indicated above/below the branches
Furthermore, the bootstrap values of clades G. ambrosiae emend. and G. latisporus comb. nov. (BS = 99 and 92% respectively) in combined analysis (see Fig. 1) were higher than in other trees that were constructed based on separate genes. Golovinomyces circumfusus on E. cannabinum from Europe, forming a single clade, represented a separate species based on the combined data analysis (see Fig. 1).
Taxonomy
Golovinomyces ambrosiae (Schwein.) U. Braun & R.T.A. Cook, in Cook & Braun, Mycol. Res. 113: 628 (2009). Figure 2.
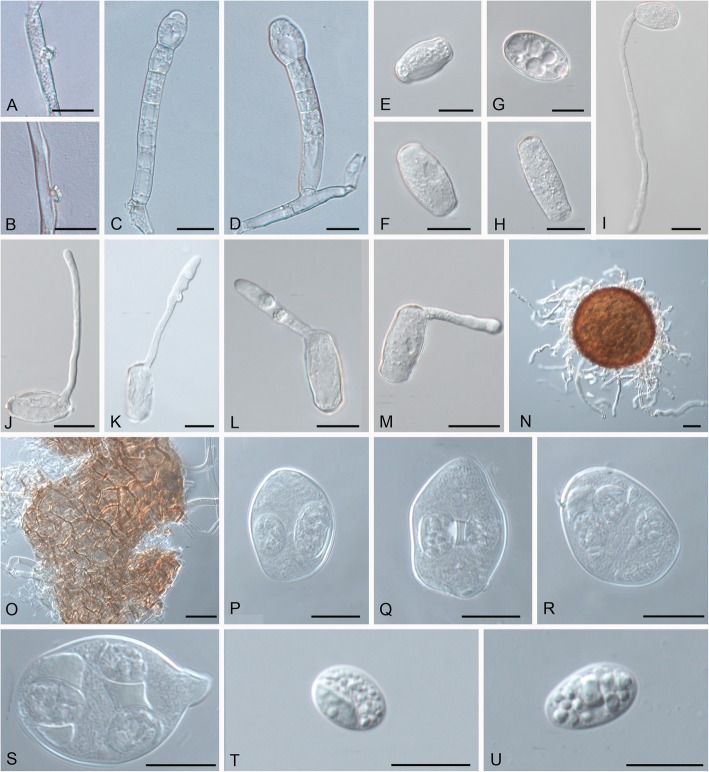
Fig. 2.
Golovinomyces ambrosiae (HMJAU-PM91814 ex Ambrosia trifida). a. Nipple-shaped hyphal appressorium. b. Slightly crenulate hyphal appressorium. c–d. Conidiophores. e–h. Conidia. i–m. Conidial germination. n. Chasmothecium. o. Peridium cells of Chasmothecium. p–q. Asci with two ascospores. r–s. Asci with three ascospores. t–u. Ascospores. Scale bars = 20 μm
≡ Erysiphe ambrosiae Schwein., Trans. Amer. Philos. Soc., N.S., 4: 270 (1834).
= Erysiphe spadicea Berk. & M.A. Curtis, Grevillea 4: 159 (1876).
≡ Golovinomyces spadiceus (Berk. & M.A. Curtis) U. Braun, in Braun & Cook, CBS Biodiversity Series 11: 329 (2012).
= Erysphe cichoracearum f. ambrosiae Jacz., Karm. Opred. Grib., Vip. 2. Muchn.-rosj. Griby (Leningrad): 186 (1927).
= Erysiphe cichoracearum f. xanthii Jacz., Karm. Opred. Grib., Vip. 2. Muchn.-rosj. Griby (Leningrad): 212 (1927).
= Oidium acanthospermi Chidd., Lloydia 18: 46 (1955).
≡ Acrosporium acanthospermi (Chidd.) Subram., Hyphomycetes (New Delhi): 835 (1971).
= Oidium lagasceae Chidd., Lloydia 18: 47 (1955).
≡ Acrosporium lagasceae (Chidd.) Subram., Hyphomycetes (New Delhi): 836 (1971).
= Oidium parthenii Satyapr. & Ushar., Curr. Sci. 50: 1081 (1981).
= E. cichoracearum var. transvaalensis G.J.M. Gorter & Eicker, S. Afr. J. Bot. 2(2): 130 (1983).
≡ Golovinomyces cichoracearum var. transvaalensis (G.J.M. Gorter & Eicker) U. Braun, Schlechtendalia 3: 51 (1999).
= Oidium blainvilleae Bappamm., Hosag. & Udaiyan, New Botanist 22: 117 (1995).
= Erysiphe cichoracearum auct. p.p.
= Golovinomyces cichoracearum auct. p.p.
Literature: Braun and Cook ([1], p. 330), Dugan [51], Takamatsu et al., [19], Khodaparast [52], Arzanlou et al., [53], Meeboon et al., ([11], p. 212), Moparthi et al., [29, 54], Trigano et al., [28], Braun et al., [13].
Illustrations: Bappammal et al., ([55], p. 110, Fig. 26, 115, Fig. 35), Nomura ([56], p. 182, Fig. 241), Braun and Cook ([1], p. 330, Fig. 359), Meeboon et al., ([11], p. 211, Figs. 9–11).
Exsiccatum: Seym. & Earle, Econ. Fungi 321.
Description: Mycelium amphigenous and caulicolous, white, in small to moderately large patches, confluent, sometimes covering entire leaves, persistent or almost so; hyphae 2–9 μm wide, thin-walled, smooth, hyaline, in old infections hyphae around ascomata sometimes turning brown; hyphal appressoria solitary, sometimes several per hyphal cell, nipple-shaped, occasionally slightly crenulate or irregular, 3–8 μm diam.; conidiophores erect, arising from the upper surface of the hyphal mother cell and usually towards one end of it; foot-cells cylindrical, straight, rarely slightly flexuous, 30–80 × 9–15 μm, followed by 1–3 shorter cells, forming catenescent conidia; conidia ellipsoid-ovoid, doliiform-subcylindrical, 25–40 × 14–20(− 24) μm, length/width ratio 1.5–2; conidial germination of the Euoidium type. Chasmothecia amphigenous, occasionally caulicolous, scattered to gregarious, 80–140 μm diam., rarely larger; peridium cells irregularly shaped, polygonal to daedaleoid, 8–30 μm diam., walls of the cells up to 2 μm wide; appendages numerous, mostly arising from the lower half, mycelioid, usually unbranched, 0.2–1.5 times as long as the chasmothecial diam., mostly shorter than the diam, (3–)4–8(− 10) μm wide, at first hyaline, later yellowish to medium brown throughout or paler towards the tips, septate, walls thin, smooth or almost so; asci numerous, mostly (5–)8–15, obovoid-saccate, 40–70 × 25–35(− 40) μm, almost sessile or short-stalked, wall thin, up to 1 μm thick, 2(− 3)-spored; ascospores broad ellipsoid-ovoid, 15–25(− 28) × 10–15(− 18) μm, colorless.
Material examined: additional collections used for molecular analyses (see Table 1); USA, Pennsylvania, Lehigh & Northampton, Bethleham, on leaves of Ambrosia sp. (Asteraceae), 1826, L. von Schweinitz, PH 62362, holotype of E. ambrosiae; USA, South Carolina, on leaves of Xanthium sp. (Asteraceae), ex herb. M.J. Berkeley, No. 2984, K(M) 164,976, holotype of E. spadiceus. JAPAN, Mie Pref., Tsu, on leaves of Xanthium strumarium (Asteraceae), 12 Nov. 1997, S. Takamatsu, TSU-MUMH 413 (reference material for Erysiphe spadicea with ex-reference material sequence – AB077644, see Braun et al. 2019). USA, Iowa, Guthrie County, Sheeder Prairie State Preserve, on leaves of Ambrosia trifida, 12 Aug. 1997, Lois H. Tiffany, ISC-F-0076753, epitype of Erysiphe ambrosiae (designated here, MycoBank MBT385758).
Host range and distribution (see [1, 13]): widespread in Asia, Australia, Europe and North America, on species of numerous host genera belonging to the families Asteraceae (Acanthospermum, Ambrosia, Aster, Blainvillea, Chrysogonum, Coreopsis, Dahlia, Eupatorium, Gerbera, Helianthus, Lagascea, Laggera, Leucanthemum, Mauranthemum [Chrysanthemum s. lat.], Melampodium, Parthenium, Telekia, Tithonia, Xanthium, Zinnia), Fabaceae (Crotalaria), Malvaceae (Abelmoschus), Polygonaceae (Persicaria), Solanaceae (Solanum), and Verbenaceae (Verbena).
Notes: Persicaria species have recently been confirmed as hosts of G. ambrosiae by molecular sequence analyses (P. alpina [30], Azerbaijan; P. decipiens [13], Australia).
Golovinomyces circumfusus (Schltdl.) U. Braun, in Braun & Cook, CBS Biodiversity Series 11: 309 (2012).
≡ Alphitomorpha circumfusa Schltdl., Verh. Ges. Naturf. Freunde Berlin 1(1): 49 (1819).
≡ Erysibe circumfusa (Schltdl.) Ehrenb., Nova Acta Phys.-Med. Acad. Caes. Leop.-Carol. Nat. Cur. 10: 169 (1821).
≡ Erysiphe circumfusa (Schltdl.) Schltdl., Fl. berol. 2: 169 (1824).
≡ Erysibe circumfusca (Schltdl.) Link, Sp. pl. 4, 6(1): 109, (1824).
≡ Erysiphe communis f. circumfusa (Schltdl.) Fr., Syst. mycol. 3: 240 (1829).
= E. communis n. corymbiferarum Fr., Syst. mycol. 3: 241 (1829), p.p.
= E. cichoracearum f. eupatorii Dearn., in Rehm, Ascomyc., Fasc. 48, No. 1950 (1911) and Ann. Mycol. 9: 290 (1911).
= E. cichoracearum auct. p.p.
= Golovinomyces cichoracearum auct. p.p.
Illustration: Braun & Cook (2012, p. 309, Fig. 331).
Literature: Jaczewski ([57], p. 197).
Exsiccatae: Barthol., Fungi Columb. 2930, 4020, 4224, 4919. Rabenh., Klotzschii Herb. Viv. Mycol. 467. Rehm, Ascomyc. 1950. Syd., Mycoth. Germ. 1530.
Description: Mycelium amphigenous, but sometimes also caulicolous, thin, white, effuse or in distinct patches, persistent on the upper leaf surface and on stems, less conspicuous and often evanescent on lower surface; hyphae branched mostly at right angles, hyaline, smooth or almost so, 3–9 μm wide; hyphal appressoria usually solitary, slightly to distinctly nipple-shaped, 3–7 μm diam.; conidiophores erect, solitary per hyphal mother cell, arising laterally or from the upper surface and usually towards one end of the mother cell, up to 160 μm long, foot-cells variable, straight to curved at the base or sinuous, 30–110 × 9–14 μm, almost cylindrical to slightly increasing in width from base to top, occasionally slightly constricted at the 7–9 μm wide basal septum that is usually at the junction with the mother cell or occasionally raised by up to 5 μm, followed by 2–3 shorter cells, forming catenescent conidia; primary conidia obovoid, secondary conidia ellipsoid-ovoid, subcylindrical, limoniform, 25–40 × 12–20 μm, length/width ratio 1.3–2.6, germ tubes terminal or almost so, short to moderately long, slightly clavate, i.e. apex with slightly swollen appressorium, Euoidium type. Chasmothecia amphigenous and caulicolous, scattered to gregarious, subglobose to somewhat depressed-globose, 85–140 μm diam., rarely larger; peridium cells irregularly polygonal, rounded to usually somewhat daedaleoid, 5–25(− 30) μm diam., walls up to 2.5 μm thick; appendages numerous, equatorial and in the lower half, mycelioid, simple, rarely branched, (0.25–)0.5–2.5(− 3.5) times as long as the chasmothecial diam., 3–8 μm wide, walls thin (up to 1 μm), smooth to faintly rough, on mature ascomata completely pale to medium dark brown throughout or somewhat paler towards the tip; asci numerous, usually 5–15, broad obovoid-saccate or almost globose, (40–)50–70(− 80) × (20–)25–35(− 40) μm, almost sessile to short-stalked, thin-walled, terminal oculus 8–15 μm diam., 2(− 3)-spored; ascospores ellipsoid-ovoid, (15–)18–25 × 10–17 μm, colourless.
Material examined: all were collected on leaves of Eupatorium cannabinum, GERMANY, ex herb. Schlechtendal, without any further data, HAL 1423 F, lectotype [designated in Dörfelt & Ali (1987)]; Brandenburg, Landkreis Ostprignitz-Ruppin, Großzerlag, 22 Sep. 2006, H. Boyle, GLM-F74796; Brandenburg, Landkreis Ostprignitz-Ruppin, north-west of Rheinsberg, 24 Sep. 2006, H. Jage, GLM-F85832; Sachsen, Zittau, Westpark, 9 Aug. 2007, H. Boyle, GLM-F80897; Sachsen-Anhalt, Salzwedel, 19 Aug. 2000, H. Jage and H. Lehmann, GLM-F49501; Sachsen-Anhalt, Halle (Saale), Osendorfer See, 12 Nov. 2000, H. Jage (GLM-F47189); Sachsen-Anhalt, Salzwedel, Hoydersburg, 11 Aug. 2004, H. Jage, GLM-F65924. Germany, Brandenburg, Spreewald, Briesensee, 8 Oct. 2016, V. Kummer, HAL 3300 F, epitype (designated here, MycoBank MBT385760).
Host range and distribution: on Eupatorium cannabinum (Asteraceae), Europe (Bulgaria, Czech Republic, Denmark, Finland, France, Germany, Hungary, Italy, Lithuania, Poland, Romania, Russia, Slovakia, Sweden, Switzerland, UK) [58–61].
Notes: Braun and Cook [1] assigned Golovinomyces on host species belonging to Eupatorium s. lat. From the northern hemisphere, including Europe, North America and northern regions of Asia, to G. circumfusus. This species seems to be confined to its type host, E. cannabinum, as collections from Asian species of Eupatorium pertain to G. ambrosiae. The affinity and identity of North American collections on Eupatorium perfoliatum, Eutrochium maculatum (≡ Eupatorium maculatum), and Eutrochium purpureum (≡ Eupatorium purpureum) remain unclear since sequence data and results of detailed morphological examinations of the asexual morphs on these hosts are not yet available. Golovinomyces on these hosts is common in North America, including several collections distributed in exsiccatae (Barthol., Fungi Columb. 2930, 4020, 4224, 4919; Rehm, Ascomyc. 1950).
Golovinomyces latisporus (U. Braun) P.-L. Qiu & S.-Y. Liu, comb. nov. Figure 3.
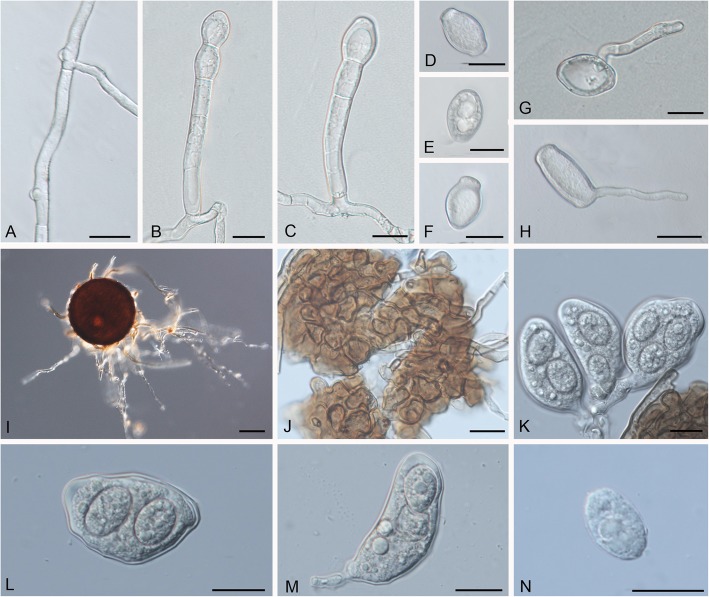
Fig. 3.
Golovinomyces latisporus comb. nov. (HMJAU-PM91828 ex Helianthus annuus). a. Nipple-shaped hyphal appressorium. b–c. Conidiophores. d–f. Conidia. g–h. Conidial germination. i. Chasmothecium. j. Peridium cells of Chasmothecium. k–m. Asci with two or three ascospores. n. Ascospores. Scale bars = 20 μm
MycoBank MB 829648.
Basionym: Oidium latisporum U. Braun, Zentralbl. Mikrobiol. 137: 315 (1982).
= Erysiphe cichoracearum f. helianthi Jacz., Karm. Opred. Grib., Vip. 2. Muchn.-rosj. Griby (Leningrad): 198 (1927).
= Erysiphe cichoracearum var. latispora U. Braun, Mycotaxon 18:117 (1983).
≡ Golovinomyces cichoracearum var. latisporus (U. Braun) U. Braun, Schlechtendalia 3: 51 (1999).
= E. cichoracearum auct. p.p.
= Golovinomyces cichoracearum auct. p.p.
Illustrations: Braun ([62], p. 316, fig. 1 [63];, p. 118, fig. 6 [17];, p. 250, pl. 66, fig. A [58];, p. 270, pl. 40, fig. A), Tanda et al., ([64], p. 254, figs. 1–2), Nomura ([56], p. 185, Fig. 249), Cook and Braun ([65], p. 627, Fig. 5), Chen et al., ([66], p. 4, fig. 1b).
Description: Mycelium amphigenous, also on stems, effuse or forming patches, thin, white, persistent or almost so; hyphae hyaline, walls thin, smooth, 3–8 μm wide; hyphal appressoria nipple-shaped, solitary or in opposite pairs, 4–8 μm diam.; conidiophores arising centrally or towards one end of hyphal mother cells and from their upper surface, erect, straight, foot-cells cylindrical, 35–80 × 9–15 μm, followed by 1–3 shorter cells, forming catenescent conidia; conidia broad ellipsoid-ovoid, doliform to somewhat limoniform, 25–45 × 15–27 μm (when fresh), length/width ratio < 2 (1.3–1.9, mostly 1.4–1.6), germ tubes dimorphic, with terminal to subterminal, occasionally lateral germination, on glass at 100% RH, long, filiform, up to 10 times as long as the width of the conidium, growing away from the glass surface (negatively hydrotropic), tips not swollen or only slightly swollen when in contact with the surface, = the longitubus pattern within the Euoidium type, a varying percentage with short, terminal to subterminal germination, subclavate with somewhat swollen apices, especially when they immediately contact a hydrophobic surface, = typical Euoidium type. Chasmothecia amphigenous, scattered to gregarious, subglobose, (65–)85–130(− 145) μm diam.; peridium cells irregularly polygonal to daedaleoid, (5–)10–25(− 35) μm diam., walls up to 2.5 μm wide; appendages numerous, arising from the lower half, mycelioid, simple, rarely branched, rarely longer than 0.5–2 times the chasmothecial diam., 3–8 μm wide, septate, walls thin, smooth or almost so, at first colourless, later completely brown or paler towards the tip; asci usually 5–15, occasionally more, clavate-saccate, 45–80 × 20–40 μm, short-stalked, wall 1–2 μm wide, 2(− 3)-spored; ascospores ellipsoid-ovoid, 18–29 × 9–20 μm, colourless.
Material examined: Additional collections used for molecular analyses (see Table 1); GERMANY, Sachsen-Anhalt, Greifenhagen, on leaves of Helianthus × laetiflorus, 20 Sep. 1981, HAL 1434 F, holotype (of O. latisporum); Sachsen-Anhalt, Greifenhagen, on leaves of Helianthus sp. (cf. maximiliani), 20 Sep. 1981, HAL 1443 F, paratype (of O. latisporum); USA, Wisconsin, Jefferson Co., Faville, Prairie Preserve, near lake Mills, on Helianthus grosseserratus, 31 Aug. 1963, H.C. Greene, DAOM 96982, holotype (of E. cichoracearum var. latispora). Switzerland, Vaud, Nyon, on leaves of Helianthus annuus, 17 Sep. 2018, HAL 3299 F, epitype (designated here, MycoBank MBT385594).
Host range and distribution: on Helianthus (angustifolius, annuus, arizonensis, atrorubens, californicus, carnosus, cusickii, debilis, debilis subsp. cucumerifolius, decapetalus, divaricatus, doronicoides, exilis, giganteus, gracilentus, grosseserratus, hirsutus, kellermanii, ×laetiflorus, laevigatus, longifolius, maximiliani, mexicanus, microcephalus, ×multiflorus, neglectus, niveus subsp. tephrodes, nuttallii, nuttallii subsp. parishii, paradoxus, parviflorus, pauciflorus, pauciflorus subsp. strumosus, subrhomboideus, petiolaris, praecox subsp. hirtus, praecox subsp. runyonii, radula, rigidus, salicifolius, scaberrimus, schweinitzii, tuberosus), Rudbeckia (amplexicaulis, bicolor, fulgida, hirta, laciniata, occidentalis, serotina, triloba), Zinnia (angustifolia, elegans) Asteraceae [Heliantheae]; Africa (South Africa, Tanzania), Asia (China, India, Israel, Japan, Korea, Nepal, Russia [Siberia, Far East], Turkey), Europe (Bulgaria, Germany, Greece, Hungary, Italy, Lithuania, Netherlands, Poland, Romania, Russia, Slovenia, Switzerland, Turkey, Ukraine, former Yugoslavia), North America (Canada, Mexico, USA), South America and West Indies (Argentina, Cuba, Bolivia, Brazil, Chile, Venezuela), Oceania (Fiji, Samoa), New Zealand (see [1, 58, 67], https://nt.ars-grin.gov/fungaldatabases/index.cfm).
Notes: Golovinomyces latisporus occurs on various Helianthus species almost worldwide. Zinnia angustifolia and Z. elegans are additional hosts proven by means of molecular methods. Golovinomyces collections found on various Rudbeckia spp. are assigned to G. latisporus with respect to the characters of the anamorph, although multilocus sequence analyses are still lacking. Taxonomy of a recently published record of “G. spadiceus” on Helianthus annuus in the United States [54] is unclear and urgently requires multilocus analyses for species identification. The identity of Golovinomyces on Iva spp. (axillaris, frutescens, xanthifolia) has not been sufficiently studied.
Discussion
The taxonomic history of the powdery mildews allied to Erysiphe cichoracearum dates back to de Candolle, in Lamarck and de Candolle [68]. He described E. cichoracearum on Scorzonera hispanica and Tragopogon porrifolius. Salmon [14] widened the concept of E. cichoracearum considerably by assigning powdery mildew on numerous hosts of various plant families to this species, including Helianthus spp. In previous circumscriptions, E. cichoracearum was characterized by having ascomata with mycelium-like appendages, several usually 2-spored asci, and conidia formed in chains without fibrosin bodies [14–17]. Braun [62] described the asexual morph of powdery mildew found on Helianthus × laetiflorus in Germany as Oidium latisporum based on the differences in conidial characters (most notably broader conidia) from collections of E. cichoracearum on various other hosts. Later, Braun [63] introduced the name E. cichoracearum var. latispora based on holomorphic North American type material, and cited E. ambrosiae as a possible synonym. Heluta [69] reallocated E. cichoracearum to Golovinomyces. Braun and Cook [1] split G. cichoracearum into several species based on molecular analyses of this complex which suggested a co-evolutionary relationship between Golovinomyces species and tribes of Asteraceae [18].
Golovinomyces on hosts of the Heliantheae was divided into two species, G. ambrosiae and G. spadiceus, distinguished by clear morphological differences in their asexual morphs [1]. Type material of E. ambrosiae was examined, and this name was used for powdery mildew on Ambrosia, Helianthus, Iva, and Rudbeckia spp. E. ambrosiae was characterized by having broad ellipsoid-ovoid, doliiform to somewhat limoniform conidia, 25–45 × 15–27 μm (when fresh) with a length/width ratio < 2 (1.3–1.9, mostly 1.4–1.6), and dimorphic germ tubes that were long and filiform (longitubus pattern with the Euoidium conidial germination type) and consisted of a varying percentage of shorter germ tubes that were often swollen at the tip (ordinary Euoidium germ tubes) [1]. Whereas, the conidial shape and size, as well as the conidial germination pattern of G. spadiceus agrees with the common Euoidium type. These morphological differences were not reflected in a comprehensive phylogenetic analyses based on ITS and 28S rDNA powdery mildews previously referred to as G. ambrosiae and G. spadiceus. In the phylogenetic analyses, G. ambrosiae and G. spadiceus formed a single undifferentiated clade (lineage III in Takamatsu et al., [19]). Furthermore, this clade also encompassed sequences obtained from Golovinomyces on Eupatorium chinense in Japan [referred to as G. circumfusus based on the circumscription of this species in Braun and Cook [1] and the assumption that all Golovinomyces collections on various Eupatorium species in Asia, Europe and North America pertain to a single species] as well as sequences from Golovinomyces on numerous Asteraceae hosts from several tribes and even other families. The extensive host range exhibited by clade 3 suggests the involvement of a plurivorous species.
Sequences from the five gene regions could not be obtained for all samples used in this study. The phylogenetic affinity of G. circumfusus could be clarified by the inclusion of sequences obtained from powdery mildew on E. cannabinum (type host) in Germany (type region). G. circumfusus on its type host does not cluster within the former “Heliantheae Clade” and is not closely allied with G. ambrosiae complex. It represents a well-supported species of its own, confined to E. cannabinum in Europe. Blumer ([16], p. 188) summarized results of previous inoculation tests carried out by himself and other authors and classified Erysiphe cichoracearum s. lat. on E. cannabinum as a biologically specialized form (f. sp. eupatorii), confined to this host. In order to stabilize the application of the old name Erysiphe circumfusa, described in the nineteenth century, an epitype has been designated. Powdery mildew on Asian Eupatorium spp. is not conspecific with G. circumfusus and pertains to a clade previously referred to as G. spadiceus [13]. This clade represents a plurivorous species on a wide range of hosts belonging to the Heliantheae and other tribes of Asteraceae as well as hosts of other plant families. However, the naming of this clade had to be corrected.
Sequences from Golovinomyces on Ambrosia spp. in Asia and North America do not cluster together with sequences obtained from Golovinomyces on Helianthus spp., but they pertain to the former plurivorous G. spadiceus. The morphological characters of the powdery mildew on Ambrosia also agree with that of G. spadiceus (the type material of Erysiphe ambrosiae contains chasmothecia, but the features of the asexual morph could not be properly examined). Hence, Braun [63] cited E. ambrosiae as a potential synonym of E. cichoracearum var. latispora. The application of the name E. ambrosiae in Braun and Cook [1], based on this questionable synonymy, must be classified as a misinterpretation. These results have nomenclatural and taxonomic consequences, viz., the older name Erysiphe ambrosiae, which has priority over G. spadiceus, is now the correct name for this plurivorous species, and G. spadiceus and its synonyms must be reduced to synonymy with G. ambrosiae. Finally, Golovinomyces on Helianthus spp., morphologically distinguished from the former G. spadiceus, turned out be genetically different as well (although undoubtedly closely allied to the latter species).
Since G. ambrosiae now represents an older name for the species previously referred to as G. spadiceus, it is necessary to rename the species on Helianthus. Hence, Oidium latisporum (= Erysiphe cichoracearum var. latispora), the oldest valid name for this taxon at the species level, is used as the basionym for the combination G. latisporus. This species is common with a near global distribution, and also occurs on Zinnia [sequences retrieved from Z. angustifolia (HAL 2338 F) refer to a German collection from a botanical garden in which the Zinnia grew close to Helianthus plants infected by G. latisporus]. Sequences retrieved from Z. elegans (HMJAU-PM91850) refer to a collection from the Sichuan province of China where no Helianthus plants grew. The powdery mildew on Rudbeckia coincides morphologically with G. latisporus. However, currently only ITS and 28S sequences are available [19]. Future examinations based on IGS, TUB2 and CHS1 are necessary to confirm the identity. In any case, the example of Zinnia shows that host plants of other genera, such as Helianthus or Iva, might also be infested by the two closely allied species, G. ambrosiae and G. latisporus. In order to answer this question, a combination of morphological examinations and phylogenetic analyses based on a multilocus approach are required in the future.
Conclusions
The phylogenetic analyses of multilocus sequence data, including ITS and 28S rDNA, IGS, TUB2, CHS1, and consideration of morphological characters enabled to resolve species delimitation in a heterogeneous complex within the genus Golovinomyces. The old names involved in this complex have been epitypified, providing ex-epitype sequence data, and three species were distinguished in the complex named G. ambrosiae emend. (including G. spadiceus), G. latisporus comb. nov. (≡ Oidium latisporum), and G. circumfusus confined to Eupatorium cannabinum in Europe. This research illustrated that such approaches are suitable and promising in cases of phylogenetically closely allied assemblages of powdery mildew species in which ITS analyses do not yield sufficient resolution.
Supplementary information
Additional file 1: Figure S1. Phylogenetic analysis of the ITS region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 73 sequences from tribe Golovinomyceteae. One sequence from Erysiphe kenjiana (accession number: MK452611) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above the branches.
Additional file 2: Figure S2. Phylogenetic analysis of the 28S rDNA region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 74 sequences from tribe Golovinomyceteae. One sequence from Erysiphe kenjiana (accession number: MK452684) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above the branches.
Additional file 3: Figure S3. Phylogenetic analysis of the IGS region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 74 sequences from tribe Golovinomyceteae. Three sequences from Neoërysiphe galeopsidis (accession numbers: MK452530, MK452531, MK452532) were used as an outgroup. Bootstrap values based on 1000 replications are indicated above the branches.
Additional file 4: Figure S4. Phylogenetic analysis of Golovinomyces ambrosiae complex and G. circumfusus based on the TUB2 region. The tree was constructed based on 43 sequences from tribe Golovinomyceteae and one sequence from Erysiphe kenjiana (accession number: MK452458) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above/below the branches.
Additional file 5: Figure S5. Phylogenetic analysis of the CHS1 region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 49 sequences from tribe Golovinomyceteae. Three sequences from Neoërysiphe galeopisidis (accession numbers: MK452438, MK452439, MK452440) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above/below the branches.
Acknowledgements
We are very grateful to Feng-Yun Zhao and Li Liu for managing the herbarium loans, and Yu-Ping Dong for collecting three specimens of Zinnia, Helianthus, and Dahlia from Sichuan Province. We also thank to Prof. Shu-Yan Liu for presenting part of our results as a poster titled “The relationships between G. ambrosiae, G. circumfusus and G. spadiceus and their hosts.” in the 2019 IS-MPMI XVIII Congress, Glasgow, Scotland.
Abbreviations
- auct
Auctorum (in the sense of other authors)
- comb. nov
Combinatio nova (new combination)
- diam
Diameter
- e.g.
For example
- emend
Emendation
- f. sp.
Forma specialis
- Figs
Figures
- No
Number
- P
Page
- p.p
Pro parte (partly)
- s
Sensu
- s. lat
Sensu lato (in the broad sense)
- spp.
Species plural
- var.
Variety
- viz.
Namely
Authors’ contributions
In this research, UB, YL, SYL and PLQ mainly provided the idea and the design the experiments; MB, SR-L, ST, TS B collected and examined the samples from abroad; PLQ, SRT and JF were in charge of the experiments. TA, DN J LLW were in charge of the collections from China. UB, SYL and PLQ mainly prepared the manuscript. All authors have read and approved the manuscript.
Funding
This research was supported by National Natural Science Foundation of China (31670022 and 31470153) which supported us to do research on taxonomy of Erysiphaceae, and by 111 Project (D17014) which support us to collaborate with foreign experts.
Availability of data and materials
The molecular data in the manuscript can be found in the GenBank database after publishing, and the materials can be found in the Herbaria shown in Table 1.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The manuscript does not infringe any personal or other copyright or property rights. The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-020-01731-9.
References
- 1.Braun U, Cook RTA. Taxonomic manual of the Erysiphales (powdery mildews). CBS biodiversity series no. 11. Utrecht: CBS, Netherlands; 2012. [Google Scholar]
- 2.Marmolejo J, Siahaan SAS, Takamatsu S, Braun U. Three new records of powdery mildews found in Mexico with one genus and one new species proposed. Mycoscience. 2018;59:1–7. [Google Scholar]
- 3.Takamatsu S. Studies on the evolution and systematics of powdery mildew fungi. J Gen Pl Pathol. 2018;84:422–426. [Google Scholar]
- 4.Braun U. Beitrag zur systematik und nomenklatur der Erysiphales. Feddes Repert. 1978;88:655–665. [Google Scholar]
- 5.Heluta VP. Filogeneticheskie vzaimosvyazi mezhdu rodami erizifal’nykh gribov i nekotorye voprosy systematika poryadki Erysiphales. Biol Zhurn Armenii. 1988;41:351–358. [Google Scholar]
- 6.Braun U. Some critical notes on the classification and generic concept of the Erysiphaceae. Schlechtendalia. 1999;3:49–55. [Google Scholar]
- 7.Braun U, Takamatsu S. Phylogeny of Erysiphe, Microsphaera, Uncinula (Erysipheae) and Cystotheca, Podosphaera, Sphaerotheca (Cystotheceae) inferred from rDNA ITS sequences – some taxonomic consequences. Schlechtendalia. 2000;4:1–33. [Google Scholar]
- 8.Liu TZ, Wen J. Golovinomyces clematidis sp. nov. from China. Mycotaxon. 2013;125:107–110. [Google Scholar]
- 9.Scholler M, Schmidt A, Siahaan SAS, Takamatsu S, Braun U. A taxonomic and phylogenetic study of the Golovinomyces biocellatus complex (Erysiphales, Ascomycota) using asexual state morphology and rDNA sequence data. Mycol Prog. 2016;15:1–13. [Google Scholar]
- 10.Bradshaw M, Braun U, Götz M, Meeboon J, Takamatsu S. Powdery mildew of Chrysanthemum × morifolium: phylogeny and taxonomy in the context of Golovinomyces species on Asteraceae hosts. Mycologia. 2017;109:508–519. doi: 10.1080/00275514.2017.1358136. [DOI] [PubMed] [Google Scholar]
- 11.Meeboon J, Kokaew J, Takamatsu S. Notes on powdery mildews (Erysiphales) in Thailand V. Golovinomyces. Trop Pl Pathol. 2018;43:202–217. [Google Scholar]
- 12.Braun U, Bradshaw M, Zhao TT, Cho SE, Shin HD. Taxonomy of the Golovinomyces cynoglossi complex (Erysiphales, Ascomycota) disentangled by phylogenetic analyses and reassessments of morphological traits. Mycobiology. 2018;46:192–204. doi: 10.1080/12298093.2018.1509512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun U, Shin HD, Takamatsu S, Meeboon J, Kiss L, Lebeda A, Kitner M, Götz M. Phylogeny and taxonomy of Golovinomyces orontii revisited. Mycol Prog. 2019;18:335–357. [Google Scholar]
- 14.Salmon E. A monograph of the Erysiphaceae. New York: Memoirs of the Torrey Botanical Club; 1900. [Google Scholar]
- 15.Blumer S. Die Erysiphaceen Mitteleuropas unter besonderer Berücksichtigung der Schweiz. Beiträge zur Kryptogamenflora der Schweiz. 1933;7:1–483. [Google Scholar]
- 16.Blumer S. Echte Mehltaupilze (Erysiphaceae) Germany: Gustav Fischer Verlag; 1967. [Google Scholar]
- 17.Braun U. A monograph of the Erysiphales (powdery mildews) Beihefte zur Nova Hedwigia. 1987;89:1–700. [Google Scholar]
- 18.Matsuda S, Takamatsu S. Evolution of host–parasite relationship of Golovinomyces (Ascomycota: Erysiphaceae) inferred from nuclear rDNA sequences. Mol Phylogenet Evol. 2003;27:314–327. doi: 10.1016/s1055-7903(02)00401-3. [DOI] [PubMed] [Google Scholar]
- 19.Takamatsu S, Matsuda S, Grigaliūnaitė B. Comprehensive phylogenetic analysis of the genus Golovinomyces (Ascomycota: Erysiphales) reveals close evolutionary relationships with its host plants. Mycologia. 2013;105:1135–1152. doi: 10.3852/13-046. [DOI] [PubMed] [Google Scholar]
- 20.Cho SE, Park JH, Park MJ, Shin HD. First report of powdery mildew caused by Golovinomyces ambrosiae on Ambrosia trifida in Korea. Pl Dis. 2011;95:1480. doi: 10.1094/PDIS-05-11-0422. [DOI] [PubMed] [Google Scholar]
- 21.Park MJ, Kim BS, Choi IY, Cho SE, Shin HD. First report of powdery mildew caused by Golovinomyces ambrosiae on sunflower in Korea. Pl Dis. 2015;99:557. [Google Scholar]
- 22.Trigiano RN, Bernard EC, Hadziabdic D, Dattito AJ, Wadl PA. First report of powdery mildew on whorled sunflower (Helianthus verticillatus) caused by Golovinomyces ambrosiae. Pl Dis. 2016;100:1017. [Google Scholar]
- 23.Huang JG, Chi MY, Sun XM, Zhao CD, Linag WX, Liang C. First report of powdery mildew caused by Golovinomyces ambrosiae on Helianthus tuberosus in China. Pl Dis. 2017;101:1317. [Google Scholar]
- 24.Choi IY, Ahn MS, Cho SE, Kim JY, Shin HD. First report of powdery mildew caused by Golovinomyces ambrosiae on English daisy (Bellis perennis) in Korea. Pl Dis. 2018;102:2032. [Google Scholar]
- 25.Han KS, Park MJ, Zhao TT, Cho SE, Shin HD. First report of powdery mildew caused by Golovinomyces ambrosiae on Brachyscome multifida in Korea. Pl Dis. 2018;102:1851. [Google Scholar]
- 26.Radisek S, Jakse J, Zhao TT, Cho SE, Shin HD. First report of powdery mildew of Helianthus tuberosus caused by Golovinomyces ambrosiae in Slovenia. J Pl Pathol. 2018;100:331. [Google Scholar]
- 27.Zhao TT, Cho SE, Kim JY, Choi IY, Shin HD. First report of powdery mildew caused by Golovinomyces ambrosiae on Helianthus salicifolius in Korea. Pl Dis. 2018;102:1453. [Google Scholar]
- 28.Trigiano RN, Boggess SL, Bernard EC. First report of powdery mildew caused by Golovinomyces spadiceus on green and gold (Chrysogonum virginianum) in the United States. Pl Dis. 2018;102:252. [Google Scholar]
- 29.Moparthi S, Bradshaw M, Frost K, Hamm PB, Buck JW. First report of powdery mildew caused by Golovinomyces spadiceus on okra in the United States. Pl Dis. 2018;102:1664. [Google Scholar]
- 30.Abasova LV, Aghayeva DN, Takamatsu S. New host records of the genus Golovinomyces (Erysiphales, Ascomycota) Proc Azerbaijan Nat Acad Sci. 2017;72:62–68. [Google Scholar]
- 31.Damm U, O'Connell RJ, Groenewald JZ, Crous PW. The Colletotrichum destructivum species complex – hemibiotrophic pathogens of forage and field crops. Stud Mycol. 2014;79:49–84. doi: 10.1016/j.simyco.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damm U, Sato T, Alizadeh A, Groenewald JZ, Crous PW. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud Mycol. 2019;92:1–46. doi: 10.1016/j.simyco.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inuma T, Khodaparast SA, Takamatsu S. Multilocus phylogenetic analyses within Blumeria graminis, a powdery mildew fungus of cereals. Molec Phylogen Evol. 2007;44:741–751. doi: 10.1016/j.ympev.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Walker AS, Bouguennec A, Confais J. Evidence of host-range expansion from new powdery mildew (Blumeria graminis) infections of triticale (×Triticosecale) in France. Pl Pathol. 2011;60:207–220. [Google Scholar]
- 35.Meeboon J, Takamatsu S. Molecular phylogeny reveals the presence of cryptic speciation within Erysiphe japonica (≡Typhulochaeta japonica), a powdery mildew on Quercus spp. Mycoscience. 2013;54:69–74. [Google Scholar]
- 36.Brewer MT, Milgroom MG. Phylogeography and population structure of the grape powdery mildew fungus, Erysiphe necator, from diverse Vitis species. BMC Evol Biol. 2010;10:268–280. doi: 10.1186/1471-2148-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira M, Cunha M. Study of the portuguese populations of the powdery mildew fungus from diverse grapevine cultivars (Vitis vinifera) J Internat Sci Vigne Vin. 2015;49:173–182. [Google Scholar]
- 38.Pirondi A, Vela-Corcía D, Dondini L, Brunelli A, Pérez-Garcia A, Collina M. Genetic diversity analysis of the cucurbit powdery mildew fungus Podosphaera xanthii suggests a clonal population structure. Fungal Biol. 2015;119:791–801. doi: 10.1016/j.funbio.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Pirondi A, Kitner M, Iotti M, Sedláková B, Lebeda A, Collina M. Genetic structure and phylogeny of Italian and Czech populations of the cucurbit powdery mildew fungus Golovinomyces orontii inferred by multilocus sequence typing. Pl Pathol. 2015;65:959–967. [Google Scholar]
- 40.Siahaan SAS, Sakamoto H, Shinoda T, Takamatsu S. Geographic and temporal distributions of four genotypes found in Erysiphe gracilis var. gracilis, a powdery mildew of evergreen oaks (Erysiphales) Mycoscience. 2018;59:110–118. [Google Scholar]
- 41.Hirata K. On the shape of the germ tubes of Erysipheae. Bull Chiba Coll Horticulture. 1942;5:34–49. [Google Scholar]
- 42.Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 43.Hirata T, Takamatsu S. Nucleotide diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience. 1996;37:283–288. [Google Scholar]
- 44.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis N, Gelfand D, Sninsky J, White T, editors. PCR Protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 45.Scholin CA, Herzog M, Sogin M, Anderson DM. Identification of group-and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J Phycol. 1994;30:999–1011. [Google Scholar]
- 46.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- 47.Jones L, Riaz S, Morales-Cruz A, Amrine KCH, McGuire B, Gubler WD, Walker MA, Cantu D. Adaptive genomic structural variation in the grape powdery mildew pathogen, Erysiphe necator. BMC Genomics. 2014;15:1081. doi: 10.1186/1471-2164-15-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang ZG, Hall A, Perfect E, Gurr SJ. Differential expression of two Blumeria graminis chitin synthase genes. Mol Plant Pathol. 2000;1:125–138. doi: 10.1046/j.1364-3703.2000.00015.x. [DOI] [PubMed] [Google Scholar]
- 49.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swofford DL. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4.0b10. Sunderland: Sinauer Associates; 2002. [Google Scholar]
- 51.Dugan FM. Golovinomyces spadiceus causing powdery mildew on Coreopsis hybrid ‘full moon’ (Heliantheae, Asteraceae) in Washington state. N Amer Fungi. 2013;8:1–3. [Google Scholar]
- 52.Khodaparast SA. Molecular identification of some anamorphic powdery mildews (Erysiphales) in Guilan province, north of Iran. Mycol Iran. 2016;3:127–133. [Google Scholar]
- 53.Arzanlou M, Torbati M, Golmohammadi H. A report of Golovinomyces sp on Zinnia in Iran. Australas Pl Dis Notes. 2018;13:47. [Google Scholar]
- 54.Moparthi S, Bradshaw MJ, Grove GG. First report of powdery mildew caused by Golovinomyces spadiceus on Helianthus annuus. Pl Dis. 2018;102:1176. [Google Scholar]
- 55.Bappammal M, Hosagoudar VB, Udaiyan K. Powdery mildews of Tamil Nadu, India. Int Quart J Pl Sci Res. 1995;22:81–175. [Google Scholar]
- 56.Nomura Y. Taxonomical study of Erysiphaceae of Japan. Tokyo: Yokendo; 1997. [Google Scholar]
- 57.Jaczewski AA. Karmanny opredelitel’ gribov. Vyp. 2. Muchnisto-rosyanye griby. Russia: Gosudarstvennogo Instituta Opytnoy Agronomii; 1927. [Google Scholar]
- 58.Braun U. The powdery mildews (Erysiphales) of Europe. Germany: Gustav Fischer Verlag; 1995. [Google Scholar]
- 59.Fakirova VI. G’bute v B’lgariya, 1 tom, razred Erysiphales (Fungi Bulgaricae, 1 tomus, ordo Erysiphales) Bulgaria: Izdatel’stvo na B’lgarskata Akademiya na Naukite; 1991. [Google Scholar]
- 60.Grigaliūnaitė B. Lietuvos grybai, III, Milteniečiai 1 (Erysiphales) Lithuania: Mokslo ir Enciklopedijų Leidybos Institutas; 1997. [Google Scholar]
- 61.Paulech C. Flóra Slovenska, X/1, Mycota (Huby), Ascomycetes (Vreckaté), Erysiphales (Múčnatkovaré) Slovakia: VEDA; 1995. [Google Scholar]
- 62.Braun U. Morphological studies in the genus Oidium (III) Zentralbl Mikrobiol. 1982;137:314–324. [Google Scholar]
- 63.Braun U. Descriptions of new species and combinations in Microsphaera and Erysiphe (IV) Mycotaxon. 1983;18:113–129. [Google Scholar]
- 64.Tanda S, Kawai T, Minamioka C. Powdery mildew fungi in Jindai Botanical Park. Tokyo J Agric Sci. 1986;30:252–260. [Google Scholar]
- 65.Cook RTA, Braun U. Conidial germination patterns in powdery mildews. Mycol Res. 2009;113:616–636. doi: 10.1016/j.mycres.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Chen RS, Chu C, Cheng CW, Chen WY, Tsay JG. Differentiation of two powdery mildews of sunflower (Helianthus annuus) by a PCR-mediated method based on ITS sequences. Eur J Pl Pathol. 2008;121:1–8. [Google Scholar]
- 67.Amano (Hirata) K. Host range and geographical distribution of the powdery mildew fungi. Tokyo Scientific Societies Press; 1986.
- 68.de Lamarck JB, de Candolle AP. Flore française. Triosieme édition, Tom second. Paris: Agasse; 1805.
- 69.Heluta VP. Novi taksonomichni kombinatsyyi v rodini Erysiphaceae. Ukrainsk Bot Zhurn. 1988;45:62–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Phylogenetic analysis of the ITS region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 73 sequences from tribe Golovinomyceteae. One sequence from Erysiphe kenjiana (accession number: MK452611) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above the branches.
Additional file 2: Figure S2. Phylogenetic analysis of the 28S rDNA region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 74 sequences from tribe Golovinomyceteae. One sequence from Erysiphe kenjiana (accession number: MK452684) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above the branches.
Additional file 3: Figure S3. Phylogenetic analysis of the IGS region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 74 sequences from tribe Golovinomyceteae. Three sequences from Neoërysiphe galeopsidis (accession numbers: MK452530, MK452531, MK452532) were used as an outgroup. Bootstrap values based on 1000 replications are indicated above the branches.
Additional file 4: Figure S4. Phylogenetic analysis of Golovinomyces ambrosiae complex and G. circumfusus based on the TUB2 region. The tree was constructed based on 43 sequences from tribe Golovinomyceteae and one sequence from Erysiphe kenjiana (accession number: MK452458) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above/below the branches.
Additional file 5: Figure S5. Phylogenetic analysis of the CHS1 region of the Golovinomyces ambrosiae complex and G. circumfusus. The tree was constructed based on 49 sequences from tribe Golovinomyceteae. Three sequences from Neoërysiphe galeopisidis (accession numbers: MK452438, MK452439, MK452440) was used as an outgroup. Bootstrap values based on 1000 replications are indicated above/below the branches.
Data Availability Statement
The molecular data in the manuscript can be found in the GenBank database after publishing, and the materials can be found in the Herbaria shown in Table 1.