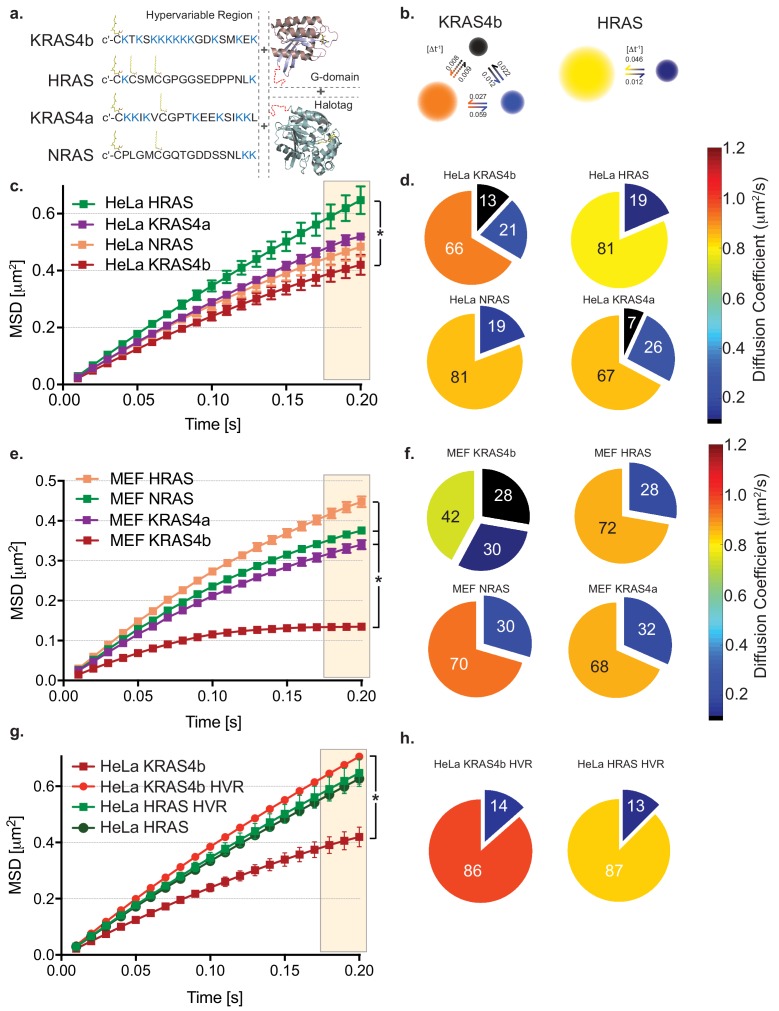
Figure 1. HMM and MSD analysis results of KRAS4b and isoforms in HeLa and MEF cells.
(a) Illustration of combinations of fusion proteins between hypervariable region (HVR) and G-domain of RAS isoforms with HaloTag that were expressed in HeLa and RAS-less MEF cells. (b) Hidden Markov modeling (HMM) using vbSPT of single molecule tracking (SMT) measurements showed different mobile patterns and was described with three and two diffusive states for KRAS4b and HRAS, respectively. Each diffusion (state) coefficient is pseudo-color coded, as marked by the rainbow scale bar and probability of transitions between states per frame rate (Δt = 10 ms). (c) The Mean Squared Displacement (MSD) vs. time plot showed highest confinement of KRAS4b whereas the least for HRAS (* indicates three displacement values under shaded area are significantly different, p<0.05). (d) Diffusion coefficients and occupancy (HMM analysis) from various isoforms of RAS molecules on HeLa cell membrane are summarized in pie charts. (e) MSD analysis on SMT measurements on RAS-less MEF cells for various RAS isoforms (* indicates three displacement values under shaded area are significantly different, p<0.05). (f) The summary of diffusive states from HMM analysis on MEF cells are presented in pie charts for KRAS4b, HRAS, KRAS4a and NRAS. (g) MSD plot showing a significant difference (p<0.05) in confinement (bending of curve) of diffusion between KRAS4b full-length and its truncated HVR, whereas no significant difference was found in case of HRAS. (h) Pie charts show both KRAS4b and HRAS HVRs diffusion were best described by a two-state model using HMM analysis.