The collective response of mixed bacterial ecosystems to antibiotics can be opposite to that of single-species cultures.
Abstract
Much is known about the effects of antibiotics on isolated bacterial species, but their influence on polybacterial communities is less understood. Here, we study the joint response of a mixed community of nonresistant Bacillus subtilis and Escherichia coli bacteria to moderate concentrations of the β-lactam antibiotic ampicillin. We show that when the two organisms coexist, their population response to the antibiotic is opposite to that in isolation: Whereas in monoculture B. subtilis is tolerant and E. coli is sensitive to ampicillin, in coculture it is E. coli who can proliferate in the presence of the antibiotic, while B. subtilis cannot. This antithetic behavior is predicted by a mathematical model constrained only by the responses of the two species in isolation. Our results thus show that the collective response of mixed bacterial ecosystems to antibiotics can run counter to what single-species potency studies tell us about their efficacy.
INTRODUCTION
Bacteria inhabit virtually every ecological habitat in our planet, from the interior of our bodies to the oceans, usually living in mixed communities that contain a variety of distinct species (1–3). Within these communities, individuals from different species signal each other (4, 5), compete for nutrients (6–8), and produce substances that benefit other species in the community (9, 10). A particularly relevant situation in which interaction among bacterial strains has been seen to be important is in their response to antibiotics (11–14). In this condition, mixed-species communities can exhibit, for instance, spreading of antibiotic resistance (15, 16) and cross-protection against multiple antibacterial agents (17–19). Most of these studies involve genetic antibiotic resistance (20, 21), however, although antibiotic survival is also commonly driven by nongenetic mechanisms that underlie responses such as persistence (22, 23).
Mounting evidence shows that the nongenetic response of multispecies bacterial communities to antibiotics goes beyond their behavior in isolation (24, 25). However, the mechanisms underlying the modulation of these responses are still poorly understood. To address this question, here, we studied how a mixed community of nonresistant bacteria responds to moderate concentrations of a β-lactam antibiotic, which attacks cells by interfering with membrane formation. The situation is represented schematically in the left side of Fig. 1, which shows the response of two isolated bacterial species, one tolerant and one sensitive, to the antibiotic. The tolerant response is characterized by exponential growth after a lag time that increases gradually with antibiotic concentration (26, 27). The sensitive species, in contrast, exhibits a sudden loss in its ability to proliferate as the antibiotic levels increase.
Fig. 1. Scheme illustrating the bacterial response of single-species cultures (left) and mixed-species cultures (right) to a specific antibiotic.
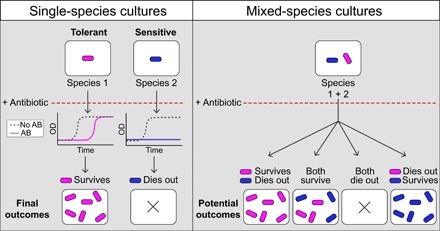
Species 1 (magenta) has a tolerant response to an antibiotic (AB), while species 2 (blue) presents a sensitive response. Both sensitivities can be captured by measuring bacterial growth [optical density (OD)] over time and observing the resulting final outcomes. The mixed-species culture can have one of four different potential outcomes, described in the text.
When the two species are cultured together (right side of Fig. 1), we can expect one of four potential outcomes: (i) The response of the two species could be the same as in isolation (first panel from the left), i.e., cross-species interactions would not alter the behavior of the individual species. (ii) The tolerant species could affect positively the sensitive one (second panel from the left) so that the mixed-species community would become resilient as a whole to the antibiotic because of interspecies coupling. (iii) Alternatively, the tolerant species could become negatively affected by the sensitive one (third panel from the left), rendering the whole bacterial community unviable. Last, and somewhat counterintuitively, (iv) the response of the two bacterial species could swap such that the species that survived the antibiotic in isolation dies out in the presence of the other species and vice versa (panel on the right).
Our results show that, at least for the bacterial species and antibiotic chosen here, interspecies coupling in the presence of antibiotic leads to the fourth scenario described above, i.e., the response of the two species when growing together is opposite to that in isolation. To understand this counterintuitive behavior, we first developed a common theoretical framework to account for the two responses in isolation, by means of a mathematical model of the binding between the antibiotic and its target molecule. The model reproduces the experimentally observed responses of the two bacterial species in monoculture and anticipates that, in a mixed community, the two bacterial species exchange their response patterns due to their sharing of a common pool of antibiotic, an effect that we confirm experimentally. It is worth emphasizing that what changes here is not the molecular sensitivity of the bacteria to the antibiotic (as established typically by single-species efficacy studies) but rather their collective response to the drug, as a result of the change in drug availability for each bacterial type in the presence of the other. Together, our results suggest that the reaction of mixed-species communities to antibiotics cannot be extrapolated from dose-response studies in isolated species, arguing instead for the need of a community-specific analysis in each case.
RESULTS
Single-species response to ampicillin
We began by examining systematically the behavior of liquid monocultures of Bacillus subtilis and Escherichia coli cells (gram positive and gram negative, respectively) upon addition of increasing concentrations of antibiotic. Specifically, we used the β-lactam antibiotic ampicillin, at a maximal concentration of 50 μg/ml, a value higher than the minimal inhibitory concentration for E. coli (28, 29). We then considered nine serial halving dilutions of the antibiotic, together with the baseline antibiotic-free case as a reference. The typical responses of the two bacterial species to increasing antibiotic concentrations are shown in Fig. 2 (A and B), which plots the cell density of the liquid cultures, as monitored by their light absorbance [optical density (OD)] at 600 nm (fig. S1, A and B, shows the mean and SE curves of all the experiments performed, four biological replicates with two technical replicates each, for a total of eight replicates).
Fig. 2. Single-species responses to ampicillin.
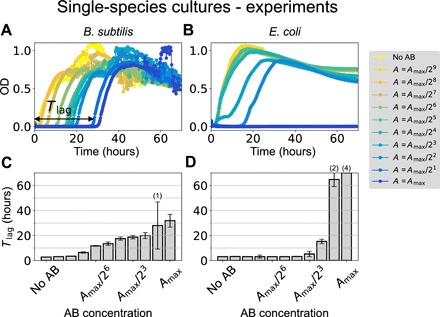
(A and B) Growth curves of B. subtilis (A) and E. coli (B) under increasing concentrations of ampicillin. The maximum antibiotic concentration is Amax = 50 μg/ml in both cases. The horizontal double-headed arrow in (A) shows schematically the meaning of the time lag (Tlag) for the highest antibiotic concentration. (C and D) The corresponding median time lags for (A) and (B), respectively, as a function of the initial antibiotic concentration. Error bars represent SD over four replicates (see also fig. S1). The numbers in parentheses on top of some bars indicate the number of replicates in which growth was not observed within the time window of our experiment (70 hours). For the bar on the far right reaching the maximum of the y axis, none of the four replicates exhibited growth in 70 hours.
As shown in Fig. 2A, B. subtilis grows after a lag time that increases gradually with antibiotic concentration. Once the population starts to grow, its behavior is basically independent of the amount of antibiotic. The cell density fluctuations observed in the stationary phase arise from the fact that B. subtilis cells tend to aggregate in liquid culture, exhibiting vortex-like collective motions that can affect absorbance measurements (30). These fluctuations do not affect the phenomena that we are reporting here, which focus on the initial growth of the cultures, when they are very far from the stationary phase and thus free from aggregates.
To study whether the tolerant response of B. subtilis to ampicillin could be due to gain of resistance, we cultured this species in the presence of antibiotic and extracted the cells after they had started growing (fig. S2). We then put those cells in new media under various concentrations of antibiotic, using again the same protocol used in Fig. 2. Measuring population growth over time shows that these surviving cells exhibit the same gradual lag response to the antibiotic as the naïve cells. Thus, the delayed growth is not the result of a gain of resistance.
The same response can be observed in the presence of carbenicillin (fig. S3A), another β-lactam antibiotic similar to ampicillin, but not with other types of antibiotics from different families (fig. S4, A to C). The behavior also arises in a different strain (fig. S4, D to G). The reaction of B. subtilis to ampicillin contrasts markedly with that of E. coli, which is abrupt and devoid of any gradual delayed growth (Fig. 2B).
The lag response reported for B. subtilis is consistent with bacterial tolerance (22, 31) and provides us with a marker to study the influence of cross-species interaction to antibiotics stress. To that end, we used the growth curves to calculate the lag time (Tlag), defined as the time the population takes to start growing after adding antibiotic to the medium (see double-headed arrow in Fig. 2A). We calculate this quantity by determining the times at which the growth curves cross an arbitrary threshold, fixed at 0.1 OD. The corresponding lag times for B. subtilis at increasing antibiotic concentrations are plotted in Fig. 2C, showing the progressive and tolerant response to ampicillin of this bacterial species. This again contrasts with the abrupt response of E. coli (Fig. 2D), with no growth observed within our 70-hour observation window for high concentrations of the antibiotic.
Chemical kinetics description of the response to ampicillin
The β-lactam antibiotic ampicillin interferes with bacterial growth by binding and inactivating membrane precursor proteins known as penicillin-binding proteins (PBPs), which are involved in the final stages of cell wall production. In that way, the drug causes an imbalance in the metabolism of the cell wall, which impairs membrane formation. The chemical processes underlying PBP inhibition can be described by the following reaction scheme (32, 33)
| {1} |
where P stands for free PBP, A stands for free ampicillin, and I represents the inactivated form of the antibiotic, which results from the acylation and subsequent hydrolyzation of the complex P ⋅ A formed by the antibiotic and PBP. Because the hydrolyzation step is much faster than the time scale of cell doubling (32), we combine those two steps into one, whose rate we represent by γ. We consider that the decrease of the active form of the antibiotic is only produced by the inactivation of the antibiotic by the complex (not by a passive degradation of the antibiotic in the media; fig. S5) (32). Furthermore, the binding between P and A is usually assumed to be much stronger than the unbinding (34). The kinetics of reaction scheme {1} can be described by the following pair of ordinary differential equations
| (1) |
| (2) |
where P denotes the concentration of free PBP in a cell and A denotes the corresponding intracellular concentration of free antibiotic (AB). The total PBP concentration Pt = P + [P · A] is assumed to be conserved. We also assume fast equilibration between the intracellular and extracellular concentrations of antibiotic [on the order of minutes (35, 36)], in comparison with the time scales involved in the growth phenomena that we are describing here (on the order of hours).
The dynamics of this system is determined by the nullclines of P and A (green and dashed magenta lines in Fig. 3A, respectively)
| (3) |
| (4) |
Fig. 3. Monoculture growth dynamics of the molecular titration model.
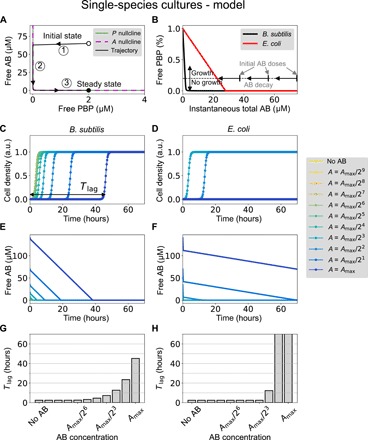
(A) Sample trajectory (black line) in the phase space defined by the two variables P and A. The two nullclines of the system are shown in solid green and dashed magenta lines. Labels 1 to 3 correspond to the three portions of the trajectories indicated in fig. S6. The parameters are k1 = 0.15 (μM · s)−1, k2 = 0.0015 s−1, Pt = 2 μM, At = 65 μM, and γ = 5 × 10−4 s−1. (B) Free PBP as a function of instantaneous total antibiotic (AB) concentration, under the assumption that AB varies slowly [which happens in portions 2 and 3 of the trajectory shown in (A)]. The parameters for B. subtilis (black line) are the ones given above, while E. coli (red line) is described by two parameter changes: Pt = 28 μM and γ = 6 × 10−6 s−1. (C and D) Dynamics of the cell density resulting from the logistic equation (Eq. 8) for both species and different antibiotic concentrations. Parameters are those of the previous panels, plus kp = 2 μM, n = 2, μ = 6 × 10−4 s−1, and μ0 = − μ/10. The maximum antibiotic concentration is Amax = 140 μM, which approximately corresponds to the antibiotic concentration used in the experiments shown in Fig. 2 above. a.u., arbitrary units. (E and F) Corresponding temporal behavior of the free antibiotic. (G and H) Growth time lag (Tlag) as a function of the total antibiotic concentration.
Figure 3A shows a typical trajectory of the system (black line) in the phase space defined by P and A, for an initial condition where all the antibiotic and all the PBP molecules are free (empty circle). Starting from that initial state, the AB quickly eliminates nearly all free PBP (portion of the trajectory labeled as 1 in Fig. 3A), until the trajectory hits the P nullcline. At that point, A starts decreasing, but it does so very slowly, as can be seen by substituting Eq. 3 into Eq. 2. This leads to
| (5) |
given that P is much smaller than Pt at this point. This corresponds to a linear decrease of A in time, as shown in fig. S6A (portion 2 of the time trace).
Threshold-linear response of free PBP to the antibiotic
The nonexponential decay of A given by Eq. 5 allows us to assume that the total amount of free antibiotic is approximately constant in the time scale of P (whose dynamics still contains an exponential decay term), as shown in fig. S6B (portion 2 of the time trace)
| (6) |
Substituting Eq. 6 into Eq. 1 and calculating the steady state of that equation leads to the stationary value of the free PBP as a function of the total AB concentration At
| (7) |
In the limit of quasi-irreversible binding between A and P, given by k2 ≪ k1Pt, the amount of free PBP depends on total antibiotic concentration in a threshold-linear manner, as shown in Fig. 3B. This behavior results from the fact that due to the quasi-irreversible binding between AB and PBP, the antibiotic acts as a titrating molecule that leaves no free PBP as long as the AB concentration is higher than the total concentration of PBP (which sets the threshold, see longer gray vertical arrow in Fig. 3B). On the other hand, when AB decreases below total PBP, the free amount of PBP builds up linearly. Around the threshold, when the AB level is close to the total amount of PBP, this molecular titration leads to an ultrasensitive response (37).
Molecular titration of the antibiotic underpins delayed growth
The threshold-linear response of free PBP with respect to the instantaneous total AB concentration can explain the delayed growth behavior observed experimentally. First, we consider that growth occurs when free PBP is larger than a certain threshold (shown by the horizontal dashed line in Fig. 3B). Next, we take into account that the total concentration of available antibiotic decays slowly (linearly) in part 2 of the system trajectory (as shown in fig. S6A). Under these two conditions, we can expect that starting from a certain antibiotic level larger than Pt, the population does not grow initially (PBP is below threshold), but as AB decreases (horizontal leftward arrows in Fig. 3B) according to Eq. 5, eventually (and suddenly) free PBP will go above the growth threshold, and the population will start proliferating. The time needed for the threshold to be crossed naturally depends on the initial AB concentration. Furthermore, the difference between the two bacterial species can be accounted for by considering different amounts of total PBP, Pt (compare the black and red lines in Fig. 3B). In addition, we can account for the higher sensitivity of E. coli by assuming that the inactivation rate of the antibiotic, γ, is smaller in that species than in B. subtilis.
Our expectations regarding the behavior of dynamical systems 1 and 2 are confirmed in Fig. 3 (C to H). The figure compares the growth dynamics of the two bacterial species, resulting from assuming a simple logistic growth in the two cases, with a growth rate that depends on the amount of free PBP
| (8) |
Here, the cell density ρ is normalized by the carrying capacity of the medium, and the growth rate depends on the average free PBP per cell, which in our model (that neglects population heterogeneity) is represented by P. This dependence is, in principle, assumed to be switch-like, with threshold kp (corresponding to the horizontal dashed line in Fig. 3B) and effective cooperativity n. The coefficient μ0 represents the basal growth rate in the total absence of PBP, which is considered negative (net death), because PBP is necessary for cellular proliferation.
Figure 3 (C and D) shows the dynamics of the cell density ρ(t) for both bacterial species, as resulting from the parameter choices made in Fig. 3 (A and B). The corresponding dynamics of free antibiotic are plotted in Fig. 3 (E and F), which shows that AB decays much faster in B. subtilis than in E. coli. Last, we compute the time lag (Tlag) by following a method similar to that used in the experiments, applying it directly to ρ. The results are shown in Fig. 3 (G and H) for the two bacterial species and confirm the gradual tolerant response of B. subtilis, and the abrupt increase in sensitivity of E. coli, for increasing antibiotic levels.
Experimental evidence of differential ampicillin inactivation in B. subtilis and E. coli
As mentioned above, the model assumes that the inactivation rate of ampicillin is higher in B. subtilis than in E. coli. This leads to a faster decrease of free antibiotic in the media in the monoculture of B. subtilis compared to E. coli (Fig. 3, E and F). To test this assumption, we grew monocultures of B. subtilis and E. coli, and a mixed culture of both species, in the presence of the maximum ampicillin concentration used in this study (Fig. 4A). After 20 hours of culture, no growth was observed in any of the conditions, and we used the supernatants of the different cultures as a medium for three new separated monocultures of B. subtilis. Because this species adapts its lag time in relation to the concentration of the antibiotic in the medium (Fig. 2A), we can use this lag time as a reporter of the amount of the remaining free antibiotic in the different supernatants, allowing us to compare its inactivation rate for the two species. Figure 4B shows the different growth curves for the three AB reporter cultures of B. subtilis, grown on the different supernatants. The quantification of the lag times for the different conditions shows that the time lag for the supernatant from the B. subtilis monoculture is clearly shorter (∼5 hours) than the one arising from the supernatant of the E. coli monoculture (∼35 hours) (Fig. 4C). These results support the model assumption that B. subtilis inactivates ampicillin faster than E. coli.
Fig. 4. Test to compare the ampicillin inactivation rates of B. subtilis and E. coli.
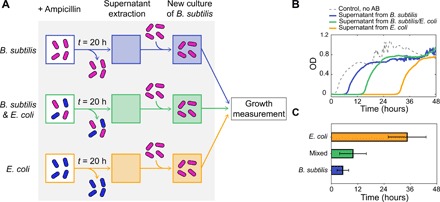
(A) Scheme of the experimental strategy. Cultures of B. subtilis, B. subtilis/E. coli, and E. coli are grown in the presence of ampicillin. After 20 hours, cells are removed from the media and the supernatant is used as media for independent new cultures of B. subtilis. Last, growth measurements are performed for the different cultures. (B) Bacterial growth (OD) as a function of time for B. subtilis in the supernatant from the B. subtilis monoculture (blue line), the mixed culture (green line), and the E. coli monoculture (orange line). The dashed line corresponds to a control, which contains B. subtilis in the absence of antibiotic. (C) Bar plot showing the quantification of the lag times of the B. subtilis monocultures in the presence of the three different supernatants. Error bars represent SE for three different replicas.
The time lag in response to a supernatant from the mixed culture lies in between the monoculture ones (green data in Fig. 4, B and C, ∼10 hours), which suggests that the mixed-species culture inactivates the antibiotic at an intermediate rate. We asked whether this behavior can be recapitulated by our mathematical model and what are its consequences. We thus turn next to modeling the response of the mixed-species community to the antibiotic.
Modeling the response of a mixed-species community to increasing antibiotic dosage
In the simplest scenario, the way in which B. subtilis and E. coli would interact when cultured together is by participating in a common pool of antibiotic. Using the principle of parsimony, we assume that the antibiotic diffuses freely between the bacteria and the extracellular medium, at equal rates for the two species, and that the extracellular antibiotic concentration is in quasi-steady state (in the time scale in which PBP and AB vary when the cells start growing; see the Supplementary Materials). These assumptions lead to the following mathematical model of the mixed system
| (9) |
| (10) |
where the subindices s and c denote the B. subtilis and E. coli populations, respectively, and ka corresponds to the effective diffusion between the bacteria and the medium.
Adding to these equations the corresponding ones for the concentrations of PBP and the densities of the two bacterial species leads to a model of the mixed-species community, whose dynamics is compared in Fig. 5 (A and B) with that of the single-species culture, for a moderate antibiotic concentration. The results show that, due to the coupling between them, the two species change their behavior oppositely with respect to the one exhibited in isolation: B. subtilis delays its growth (Fig. 5A) and E. coli advances it (Fig. 5B). The effect can be ascribed to the fact that, in the presence of the two species (which inactivate ampicillin at very different rates), the free antibiotic decays at an intermediate rate: slower for B. subtilis (Fig. 5C) and faster for E. coli (Fig. 5D). This leads to an effective growth induction of E. coli by B. subtilis, and growth inhibition of B. subtilis by E. coli. The effect is examined systematically in Fig. 5 (E and F), which shows the difference in time lags, ΔTlag = Tlag(single) − Tlag(mixed), as a function of the antibiotic level. ΔTlag is defined such that a positive value indicates an advance in population growth in the presence of the other species, and a negative value corresponds to a delay in the timing of growth (see the wide vertical arrows between Fig. 5, E and F). Thus, the model predicts that the collective response to the antibiotic when the two bacterial species are cultured in the presence of each other is opposite to that in isolation. This result contrasts with previous studies that report mutual cooperation between different species (18) in the presence of antibiotics or mutual competition (8). Here, the effect of cross-species interaction is asymmetric for the two species, a situation that is reminiscent of cooperator-cheater dynamics (12, 20), but which involves here no intrinsically resistant bacterial strain. This behavior is robust to parameter changes (fig. S6C). In particular, the binding affinities between PBP and the antibiotic (k1 and k2) need not be equal between the two species for the phenomenon to arise. Cooperativity in the growth response to PBP (Eq. 8) is not necessary either, because the behavior persists for n = 1.
Fig. 5. Growth dynamics of the molecular titration model for B. subtilis (left column) and E. coli (right column) in the case of a mixed-species community.
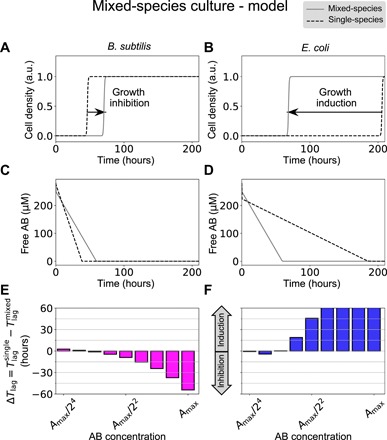
(A and B) Dynamics of the cell density for the two species in mixed cultures (gray solid lines) in comparison with their behavior in isolation (dashed lines) for a total antibiotic concentration of 280 μM. (C and D) Corresponding temporal behavior of the free antibiotic (the single-culture case has been scaled up twofold for comparison). (E and F) Predicted time lag difference between the monoculture and the coculture conditions for the two bacterial species. Parameters are those of Fig. 3, plus ka = 0.2 s−1. The maximum antibiotic concentration is Amax = 280 μM.
Experimental response of mixed cultures
We now test the model’s expectation that the interaction between B. subtilis and E. coli in a mixed-species community swaps the response of the two organisms to increasing amounts of the antibiotic. To that end, first we tagged the two bacterial species with different fluorescent proteins, with B. subtilis expressing yellow fluorescent protein (YFP) and E. coli expressing cyan fluorescent protein (CFP), to distinguish their growth when cultured together (fig. S1, C and D). We then cocultured the two species and measured their growth by monitoring the emission from their respective fluorescence markers. As shown in Fig. 6A (see also fig. S7), in the presence of E. coli, B. subtilis cells are able to grow only for low levels of ampicillin. The time lag changes abruptly with antibiotic concentration (Fig. 6C), oppositely to the delayed growth response displayed in isolation (c.f. Fig. 2A).
Fig. 6. Experimental observations of mixed-species cultures of B. subtilis and E. coli.
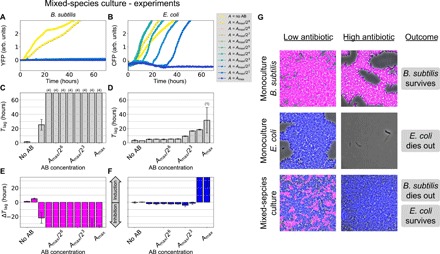
(A and B) Fluorescence marker signal reporting on the growth of the two species: YFP for B. subtilis and CFP for E. coli, respectively. (C and D) Median time lags for (A) and (B), respectively, as a function of the initial antibiotic concentration. Error bars represent SD over four replicates. The numbers in parentheses on top of some bars indicate the number of replicates in which growth was not observed within the time window of our experiment (70 hours). (E and F) Time lag difference between the mixed-species and single-species conditions as a function of the total antibiotic concentration (Amax = 100 μg/ml). (G) Composite phase and fluorescence microscopy images after 48 hours in coculture. B. subtilis cells express YFP (represented here in magenta), and E. coli cells express CFP (represented in blue). The low antibiotic concentration condition (left column) contains media with ampicillin (0.39 μg/ml), and the high antibiotic condition (middle column) contains 25 μg/ml.
In contrast, E. coli cells are now able to grow in the presence of B. subtilis for antibiotic levels that prevented growth in isolation (Fig. 6B). Concurrently, E. coli displays a gradual increase in time lag as the antibiotic concentration increases (Fig. 6D). This behavior differs from the abrupt response of E. coli to ampicillin in isolation (c.f. Fig. 2B) and is similar to the response of B. subtilis monocultures. We quantify this antithetic response systematically in Fig. 6 (E and F), which plots the lag-time differences (ΔTlag) between the monocultures and the cocultures for the two species. The results show that E. coli inhibits the growth of B. subtilis (magenta bars in Fig. 6E), while B. subtilis induces the growth of E. coli (blue bars in Fig. 6F). The population measurements discussed above were confirmed by microscopy, as shown in Fig. 6G. As can be seen in the figure, B. subtilis in isolation survives under high ampicillin concentrations after 48 hours of culture (top row), while E. coli dies out in the presence of the same level of antibiotic (bottom row). Conversely, in the mixed culture, it is E. coli, and not B. subtilis, the bacterial species that survives antibiotic treatment (middle row).
DISCUSSION
The results discussed above show that the joint response of cocultures of nonresistant B. subtilis and E. coli cells to ampicillin is reversed with respect to their behavior in isolation. This effect can be understood in terms of a minimal mathematical model of the binding of this β-lactam antibiotic to PBP and the posterior inactivation of the antibiotic. The model is generic and reproduces qualitatively the behaviors of the two monocultures: (i) the gradual increase in time lag of B. subtilis with respect to the antibiotic level, characteristic of a tolerant response, and (ii) the sudden jump in sensitivity exhibited by E. coli when the antibiotic concentration surpasses a threshold value. This threshold behavior results from an ultrasensitive response caused by the quasi-irreversible binding of the antibiotic to PBP, which occurs in E. coli but not appreciably in B. subtilis, where the critical antibiotic concentration is small (Fig. 3B). Our model shows that this difference in thresholds can be ascribed to a difference in the total amount of PBPs between the two species, which is expected to be higher in E. coli than in B. subtilis. This might seem a counterintuitive assumption, given that the membrane of gram-negative bacteria such as E. coli has a thinner peptidoglycan layer than gram-positive bacteria such as B. subtilis. Thus, it could be expected that E. coli would have less PBP. However, experimental evidence shows that E. coli has more PBPs than gram-positive bacteria such as Staphylococcus aureus (38). In addition, our experimental data show that low antibiotic concentrations, which delay growth in B. subtilis, have no effect whatsoever on E. coli (Fig. 2, A and B). These observations are consistent with our assumption that high PBP levels in E. coli titrate out the antibiotic, while this is not the case in B. subtilis due to lower PBP levels.
The gradual dependence of the time lag on the antibiotic levels in B. subtilis, as well as the sensitive response of E. coli to large amounts of antibiotic, is reproduced by the model if we consider a differential inactivation of the antibiotic. Specifically, the antibiotic is assumed to be inactivated at a smaller rate in E. coli than in B. subtilis, hypothesis that we verified experimentally (c.f. Fig. 4). The fact that the β-lactam antibiotic carbenicillin also elicits a lagged growth response in B. subtilis (fig. S3A), but not in E. coli (fig. S3B), suggests that differential antibiotic inactivation is a generic feature of β-lactam antibiotics. We note that the model indicates that the decay of the antibiotic results solely from its inactivation (γ), not from the affinity of the binding to its molecular target (k1, k2), as shown by Eq. 5.
Our minimal model makes the unexpected observation that the response patterns described above swap in coculture due to the sharing of a common pool of antibiotic among the two species. In coculture, B. subtilis reduces the amount of free ampicillin in the medium, which promotes the growth of E. coli for relatively high concentrations of the antibiotic. Conversely, E. coli acts as a buffer that delays the suppression of the antibiotic in the medium and thus inhibits the growth of B. subtilis at time lags for which the antibiotic would have already been lost if B. subtilis were alone.
Experiments confirm the predictions of the model, showing, in fact, a more marked antithetic response than the one anticipated. This can be ascribed to the simplicity of our assumptions, which include a symmetric response to the antibiotic by the two bacterial species in terms, for instance, of PBP binding (where we assume equal rates k1 and k2 for the two species). We also assume simple diffusion between the intracellular and extracellular media; an additional source of asymmetry would arise if the antibiotic were transported actively. Changing some of our assumptions should provide a better quantitative fit to the model, but this would require extra hypotheses regarding the molecular mechanisms through which ampicillin interacts with the two bacterial species considered here, hypotheses that, in most cases, cannot be corroborated with our current knowledge. For those reasons, we decided to keep our model as simple and parsimonious as possible.
The behavior reported here can be interpreted in terms of cooperator-cheater dynamics (39). In our system, the cooperator (B. subtilis) experiences a decrease in fitness by becoming unable to grow in the presence of the antibiotic, which is retained by the cheater (E. coli) through slower inactivation. The cheater, in turn, increases its fitness and is able to grow as a result of the inactivation of the antibiotic by the cooperator. Our work is, however, fundamentally different from traditional game theoretical studies, both in the way in which the cross-species interaction occurs (via nongenetic inactivation mechanisms) and in its outcome (that resilience to antibiotics depends on the ecological context in which bacteria reside). Notably, the results shown here indicate that the knowledge gained by studying the antibiotic response of nonresistant bacterial species in isolation may not be directly transferred to situations in which these species coexist with others in mixed communities. Given the ubiquity of multispecies bacterial consortia, it might be advisable to revise our approaches to the characterization of how antibiotics affect bacterial physiology, and how bacterial cells evade those effects.
MATERIALS AND METHODS
Bacterial strains and cultures
The bacterial strains used in this study are listed in table S1. Plasmid pSB1C3 containing the promoter BBa_K880005 controlling the expression of CFP was transformed into E. coli MG1655 strain. Overnight liquid cultures were grown at 37°C in LB (Miller’s modification) in an incubator with shaking. Appropriate antibiotics for selection were added to the following final concentrations: spectinomycin (10 μg/ml) and chloramphenicol (10 μg/ml). The next day, the cultures were diluted to a final OD600 of 0.1 with the corresponding antibiotics, and cells were grown at 37°C with shaking. After 2 hours, temperature was switched to 30°C, and cells continue growing for 2 hours more. The saturated culture was diluted to OD600 of 0.1, and if required, isopropyl-β-d-thiogalactopyranoside (IPTG) was added with a final concentration of 1 mM. Cells were grown for 2 hours at 30°C. For monocultures, cells were diluted in LB with 1 mM IPTG to a final OD600 of 0.02. For mixed cultures, cells were diluted to a final OD600 of 0.04, containing an OD600 of 0.02 for each bacterial type.
Plate reader measurements
For the plate reader measurements, we used 96-well black microwell plates with clear lid (Nunc, Denmark). A serial twofold dilution of the selected antibiotic was prepared, starting by the corresponding maximum concentration (50 μg/ml ampicillin for monocultures and 100 μg/ml for mixed cultures), in a final volume of 150 μl per well. Other antibiotics with their maximum concentrations tested in this study were as follows: carbenicillin (25 μg/ml), kanamycin (50 μg/ml), and spectinomycin (50 μg/ml). One well per condition was used as a control without antibiotics, and another well was used as a blank. IPTG was added to a final concentration of 1 mM, if needed. Last, an inoculum of bacteria was added to a final OD600 of 0.01 for monocultures and 0.02 for mixed cultures, containing an OD600 of 0.01 for each bacterial type.
The measurements were acquired using a microtiter plate reader (Tecan Infinite M200 Pro, Tecan Group Ltd., Switzerland). The temperature was set at 30°C with linear shaking. The absorbance wavelength for OD was set at 600 nm. Measurements were made every 10 min. Each condition had two biological replicates (taken in four experiments in different days).
Relative determination of ampicillin inactivation rates
To establish whether B. subtilis has an antibiotic inhibition rate higher than E. coli, inocula from the monocultures of B. subtilis and E. coli were placed in independent Falcon tubes to a final OD600 of 0.01, containing 10 ml of LB conditioned with ampicillin (50 μg/ml). For the mixed culture, cells were diluted to a final OD600 of 0.02, containing an OD600 of 0.01 for each bacteria type in 10 ml of LB conditioned with ampicillin (100 μg/ml). Cells were grown at 30°C with shaking for 20 hours. To extract supernatants, the different cultures were centrifuged (at 4000g for 4 min) and filtered using 0.2-μm pore size filters. The supernatants from the different conditions were then used to set up the experiment for the plate reader measurements.
Mathematical modeling and data analysis
The mathematical model described in the main text was simulated with custom-made Python code. In particular, the ordinary differential equations were integrated with the function odeint from SciPy. The experimental data were also analyzed with custom-made code in Python.
Supplementary Material
Acknowledgments
We acknowledge D.-y. D. Lee and L. Espinar for helpful discussions. We also acknowledge C. Toscano-Ochoa for providing us with the E. coli plasmid pSB1C3-BBa-K880005-cfp. Funding: This work was supported by the Spanish Ministry of Economy and Competitiveness and FEDER (project PGC2018-101251-B-I00) and by the Generalitat de Catalunya (project 2017SGR1054). J.G.-O. acknowledges support from the ICREA Academia programme and from the “María de Maeztu” Programme for Units of Excellence in R&D (Spanish Ministry of Economy and Competitiveness, MDM-2014-0370). Author contributions: L.G.-L. and J.G.-O. designed the research. L.G.-L. performed the experiments. L.G.-L. and J.G.-O. performed the data analysis. J.G.-O. performed the mathematical modeling. L.G.-L. and J.G.-O. discussed and wrote the manuscript. J.G.-O. supervised and acquired the financial support for the project. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/10/eaaz5108/DC1
Supplementary Text
Fig. S1. Growth curves of monocultures of B. subtilis and E. coli in the presence of increasing concentrations of ampicillin.
Fig. S2. B. subtilis tolerance to ampicillin is not due to gain of resistance.
Fig. S3. Growth curves of monocultures of B. subtilis and E. coli in the presence of low concentrations of another β-lactam antibiotic.
Fig. S4. Growth curves of monocultures of B. subtilis PY79 and NCBI 3610 in the presence of increasing concentrations of different antibiotics.
Fig. S5. Ampicillin is not degraded passively in the medium.
Fig. S6. Model dynamics and sensitivity analysis.
Fig. S7. Growth curves and fluorescent signal of mixed culture in the presence of increasing concentrations of ampicillin.
Table S1. Strains used in this study.
Notebook S1. Code to reproduce the modeling results of Fig. 3.
Notebook S2. Code to reproduce the modeling results of Fig. 5.
Notebook S3. Code to reproduce the modeling results of fig. S6 (A and B).
Notebook S4. Code to reproduce the modeling results of fig. S6C.
REFERENCES AND NOTES
- 1.Torsvik V., Øvreås L., Thingstad T. F., Prokaryotic diversity—Magnitude, dynamics, and controlling factors. Science 296, 1064–1066 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium , Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunagawa S., Coelho L. P., Chaffron S., Kultima J. R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D. R., Alberti A., Cornejo-Castillo F. M., Costea P. I., Cruaud C., d’Ovidio F., Engelen S., Ferrera I., Gasol J. M., Guidi L., Hildebrand F., Kokoszka F., Lepoivre C., Lima-Mendez G., Poulain J., Poulos B. T., Royo-Llonch M., Sarmento H., Vieira-Silva S., Dimier C., Picheral M., Searson S., Kandels-Lewis S.; Tara Oceans coordinators, Bowler C., de Vargas C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Jaillon O., Not F., Ogata H., Pesant S., Speich S., Stemmann L., Sullivan M. B., Weissenbach J., Wincker P., Karsenti E., Raes J., Acinas S. G., Bork P., Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Bassler B. L., Small talk: Cell-to-cell communication in bacteria. Cell 109, 421–424 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Keller L., Surette M. G., Communication in bacteria: An ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Gjødsbøl K., Christensen J. J., Karlsmark T., Jørgensen B., Klein B. M., Krogfelt K. A., Multiple bacterial species reside in chronic wounds: A longitudinal study. Int. Wound J. 3, 225–231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hibbing M. E., Fuqua C., Parsek M. R., Peterson S. B., Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghoul M., Mitri S., The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845 (2016). [DOI] [PubMed] [Google Scholar]
- 9.West S. A., Diggle S. P., Buckling A., Gardner A., Griffin A. S., The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38, 53–77 (2007). [Google Scholar]
- 10.Celiker H., Gore J., Cellular cooperation: Insights from microbes. Trends Cell Biol. 23, 9–15 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Cordero O. X., Wildschutte H., Kirkup B., Proehl S., Ngo L., Hussain F., Roux F. L., Mincer T., Polz M. F., Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337, 1228–1231 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Vega N. M., Gore J., Collective antibiotic resistance: Mechanisms and implications. Curr. Opin. Microbiol. 21, 28–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong E. F., Tsui C., Kucharíková S., Andes D., Van Dijck P., Jabra-Rizk M. A., Commensal protection of staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. MBio 7, e01365-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost I., Smith W. P. J., Mitri S., Millan A. S., Davit Y., Osborne J. M., Pitt-Francis J. M., MacLean R. C., Foster K. R., Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J. 12, 1582–1593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alekshun M. N., Levy S. B., Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Gerardin Y., Springer M., Kishony R., A competitive trade-off limits the selective advantage of increased antibiotic production. Nat. Microbiol. 1, 16175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brook I., The role of β-lactamase-producing-bacteria in mixed infections. BMC Infect. Dis. 9, 202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yurtsev E. A., Conwill A., Gore J., Oscillatory dynamics in a bacterial cross-protection mutualism. Proc. Natl. Acad. Sci. U.S.A. 113, 6236–6241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorg R. A., Lin L., van Doorn G. S., Sorg M., Olson J., Nizet V., Veening J.-W., Collective resistance in microbial communities by intracellular antibiotic deactivation. PLOS Biol. 14, e2000631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yurtsev E. A., Chao H. X., Datta M. S., Artemova T., Gore J., Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol. Syst. Biol. 9, 683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beardmore R. E., Cook E., Nilsson S., Smith A. R., Tillmann A., Esquivel B. D., Haynes K., Gow N. A. R., Brown A. J. P., White T. C., Gudelj I., Drug-mediated metabolic tipping between antibiotic resistant states in a mixed-species community. Nat. Ecol. Evol. 2, 1312–1320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kester J. C., Fortune S. M., Persisters and beyond: Mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit. Rev. Biochem. Mol. Biol. 49, 91–101 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Brauner A., Fridman O., Gefen O., Balaban N. Q., Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 24.de Vos M. G. J., Zagorski M., McNally A., Bollenbach T., Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc. Natl. Acad. Sci. U.S.A. 114, 10666–10671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radlinski L., Rowe S. E., Kartchner L. B., Maile R., Cairns B. A., Vitko N. P., Gode C. J., Lachiewicz A. M., Wolfgang M. C., Conlon B. P., Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against staphylococcus aureus. PLOS Biol. 15, e2003981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fridman O., Goldberg A., Ronin I., Shoresh N., Balaban N. Q., Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Srimani J. K., Huang S., Lopatkin A. J., You L., Drug detoxification dynamics explain the postantibiotic effect. Mol. Syst. Biol. 13, 948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.May T., Ito A., Okabe S., Induction of multidrug resistance mechanism in Escherichia coli biofilms by interplay between tetracycline and ampicillin resistance genes. Antimicrob. Agents Chemother. 53, 4628–4639 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohanski M. A., DePristo M. A., Collins J. J., Sub-lethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37, 311–320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinberg N., Rosenberg G., Keren-Paz A., Kolodkin-Gal I., Collective vortex-like movement of bacillus subtilis facilitates the generation of floating biofilms. Front. Microbiol. 9, 590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin-Reisman I., Ronin I., Gefen O., Braniss I., Shoresh N., Balaban N. Q., Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Zapun A., Contreras-Martel C., Vernet T., Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 32, 361–385 (2008). [DOI] [PubMed] [Google Scholar]
- 33.zur Wiesch P. A., Abel S., Gkotzis S., Ocampo P., Engelstädter J., Hinkley T., Magnus C., Waldor M. K., Udekwu K., Cohen T., Classic reaction kinetics can explain complex patterns of antibiotic action. Sci. Transl. Med. 7, 287ra73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu W.-P., Kincaid E., Sun Y., Bauer M. D., Kinetics of β-lactam interactions with penicillin-susceptible and -resistant penicillin-binding protein 2x proteins from Streptococcus pneumoniae. J. Biol. Chem. 276, 31494–31501 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Krishnamoorthy G., Leus I. V., Weeks J. W., Wolloscheck D., Rybenkov V. V., Zgurskaya H. I., Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into gram-negative bacteria. MBio 8, e01172-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westfall D. A., Krishnamoorthy G., Wolloscheck D., Sarkar R., Zgurskaya H. I., Rybenkov V. V., Bifurcation kinetics of drug uptake by gram-negative bacteria. PLOS ONE 12, e0184671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchler N. E., Louis M., Molecular titration and ultrasensitivity in regulatory networks. J. Mol. Biol. 384, 1106–1119 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Pucci M. J., Dougherty T. J., Direct quantitation of the numbers of individual penicillin-binding proteins per cell in Staphylococcus aureus. J. Bacteriol. 184, 588–591 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez A., Gore J., Feedback between population and evolutionary dynamics determines the fate of social microbial populations. PLOS Biol. 11, e1001547 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Ojalvo J., Elowitz M. B., Strogatz S. H., Modeling a synthetic multicellular clock: Repressilators coupled by quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 101, 10955–10960 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milo R., Jorgensen P., Moran U., Weber G., Springer M., BioNumbers–The database of key numbers in molecular and cell biology. Nucleic Acids Res. 38, D750–D753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Süel G. M., Kulkarni R. P., Dworkin J., Garcia-Ojalvo J., Elowitz M. B., Tunability and noise dependence in differentiation dynamics. Science 315, 1716–1719 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/10/eaaz5108/DC1
Supplementary Text
Fig. S1. Growth curves of monocultures of B. subtilis and E. coli in the presence of increasing concentrations of ampicillin.
Fig. S2. B. subtilis tolerance to ampicillin is not due to gain of resistance.
Fig. S3. Growth curves of monocultures of B. subtilis and E. coli in the presence of low concentrations of another β-lactam antibiotic.
Fig. S4. Growth curves of monocultures of B. subtilis PY79 and NCBI 3610 in the presence of increasing concentrations of different antibiotics.
Fig. S5. Ampicillin is not degraded passively in the medium.
Fig. S6. Model dynamics and sensitivity analysis.
Fig. S7. Growth curves and fluorescent signal of mixed culture in the presence of increasing concentrations of ampicillin.
Table S1. Strains used in this study.
Notebook S1. Code to reproduce the modeling results of Fig. 3.
Notebook S2. Code to reproduce the modeling results of Fig. 5.
Notebook S3. Code to reproduce the modeling results of fig. S6 (A and B).
Notebook S4. Code to reproduce the modeling results of fig. S6C.


