Abstract
Organoids are powerful biomimetic tissue models. Despite their widespread adoption, methods to analyse cell-type specific post-translational modification (PTM) signalling networks in organoids are absent. Here we report multivariate single-cell analysis of cell-type specific signalling networks in organoids and organoid co-cultures. Simultaneous measurement of 28 PTMs in >1 million single small intestinal organoid cells by mass cytometry reveals cell-type and cell-state specific signalling networks in stem, Paneth, enteroendocrine, tuft, goblet cells, and enterocytes. Integrating single-cell PTM analysis with Thiol-reactive Organoid Barcoding in situ (TOBis) enables high-throughput comparison of signalling networks between organoid cultures. Multivariate cell-type specific PTM analysis of colorectal cancer tumour microenvironment organoids reveals that shApc, KrasG12D, and Trp53R172H cell-autonomously mimic signalling states normally induced by stromal fibroblasts and macrophages. These results demonstrate how standard mass cytometry workflows can be modified to perform high-throughput multivariate cell-type specific signalling analysis of healthy and cancerous organoids.
Introduction
Organoids are self-organising 3D tissue models comprising stem and differentiated cells1. Organoid monocultures typically contain one major cell class (e.g. epithelial) and can be co-cultured with heterotypic cell-types (e.g. mesenchymal2 or immune3 cells) to model cell-cell interactions in vitro. When compared with traditional 2D cell culture, organoids more accurately represent their parental tissue and are emerging as powerful models for studying multicellular diseases such as cancer4.
Post-translational modification (PTM) signalling networks underpin fundamental biological phenotypes and are frequently dysregulated in disease5. As different cell-types have different signalling networks6, 7, organoids likely contain cell-type specific PTM networks that are essential to their biology. Unfortunately, no technology currently exists to analyse cell-type specific PTM networks in organoids and organoid co-cultures.
Organoids present several technical challenges over 2D cultures for PTM analysis. Firstly, organoids are embedded in a protein-rich extracellular matrix (ECM) that confounds the application of phosphoproteomic analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS). Organoids can be removed from ECM prior to LC-MS/MS, but as dissociation of live cells alters cell signalling8, PTM measurements from dissociated live organoids do not truly represent in situ cellular states. Secondly, as organoids comprise multiple cell-types (e.g. stem and differentiated) and cell-states (e.g. proliferating, quiescent, and apoptotic), bulk phosphoproteomics cannot capture their biological heterogeneity9. Although single-cell RNA-sequencing (scRNA-seq) can describe organoid cell-types10, it cannot measure PTM signalling at the protein level. Finally, low-dimensional methods (e.g. fluorescent imaging) cannot capture the complexity of signalling networks comprising multiple PTM nodes9. Collectively, to study PTM networks in organoids, we require signalling data that is: 1) derived from cells fixed in situ, 2) cell-type specific, and 3) measures multiple PTMs simultaneously.
Mass cytometry (MC, also known as cytometry time-of-flight (CyTOF)) uses heavy metal-conjugated antibodies to measure >35 proteins in single cells11. Although MC is traditionally used for high-dimensional immunophenotyping, it can also measure PTMs in heterocellular systems (e.g. peripheral blood mononuclear cells (PBMCs)12 and tissue8). We theorised that MC workflows typically applied to immunophenotyping could be modified to study cell-type specific signalling networks in organoids.
Here we report the development of a custom multivariate-barcoded MC method to measure single-cell signalling in organoids and organoids co-cultured with stromal and immune cells. This method reveals that intestinal organoids display cell-type specific signalling networks that are intimately linked with cell-state. When applied to colorectal cancer (CRC) tumour microenvironment (TME) organoid co-cultures, we discovered that epithelial oncogenic mutations mimic signalling networks normally induced by stromal cells. These results demonstrate MC can perform multivariate single-cell analysis of cell-type specific signalling in heterocellular organoids.
Results
Single-Cell Analysis of Organoids by Mass Cytometry
We hypothesised MC could be modified to study cell-type and cell-state specific signalling in organoids. To test this, we first developed a MC workflow to measure single-cell signalling in the classical small intestinal organoid13.
In this method, we first pulse live organoids with 1275-Iodo-2′-deoxyuridine (127IdU) to identify S-phase cells14, fix organoids in Matrigel to preserve cell signalling, and stain organoids with 194/8Cisplatin to label dead epithelia15. Using a custom protocol, we then physically and enzymatically dissociate the fixed organoids into single cells prior to extra- and intracellular heavy-metal antibody staining (Fig. 1a). We performed a comprehensive screen for intestinal epithelial cell-type identification antibodies including stem (LGR5, LRIG1, OLFM4), Paneth (Lysozyme), goblet (MUC2, CLCA1), enteroendocrine (CHGA, Synaptophysin), tuft cells (DCAMKL1), and enterocytes (FABP1, Na/K-ATPase) that bind fixed antigens and are compatible with rare-earth metal conjugation for MC. Cell-type identification antibodies were validated by organoid directed differentiation16 (Supplementary Fig. 1) and integrated into a panel of 28 anti-PTM rare-earth metal antibodies spanning multiple core signalling nodes (Supplementary Table 1, 45 parameters (40 antibodies)/cell). When analysed by MC, this method enables the measurement of 28 signalling PTMs across 6 cell-types in >1 million fixed single organoid cells (Figs. 1b, c, 2a, and Supplementary Fig. 2a).
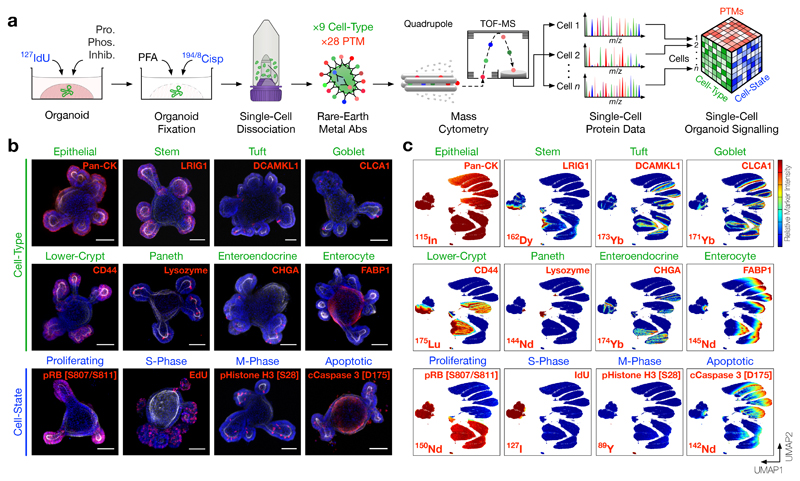
Figure 1. Cell-Type and Cell-State Identification of Single Organoid Cells by Mass Cytometry.
a) Experimental workflow. Live organoids are pulsed with 127IdU to label S-phase cells, treated with protease/phosphatase inhibitors, fixed with PFA to preserve post translational modification (PTM) signals, and stained with 194/8Cisplatin to label dead cells. Fixed organoids are then dissociated into single cells, stained with rare-earth metal-conjugated antibodies, and analysed by single-cell mass cytometry (MC). The resulting dataset contains integrated cell-type, cell-state, and PTM signalling information.
b) Confocal immunofluorescence (IF) of small intestinal organoids stained with rare-earth metal-conjugated MC antibodies highlighting individual cell-type and cell-state markers (red), F-Actin (white), and DAPI (blue), scale bars = 50 μm. Each image is representative of at least five organoids in independent IF experiments. (See Supplementary Fig. 1 for antibody validation via directed differentiation.)
c) UMAP (Uniform Manifold Approximation and Projection) distribution of 1 million single organoid cells analysed by MC resolves six major intestinal cell-types across proliferating, S-phase, M-phase, and apoptotic cell-states. Colours represent normalised local parameter intensity. (See Supplementary Fig. 2 for cell-type and cell-state classification.)
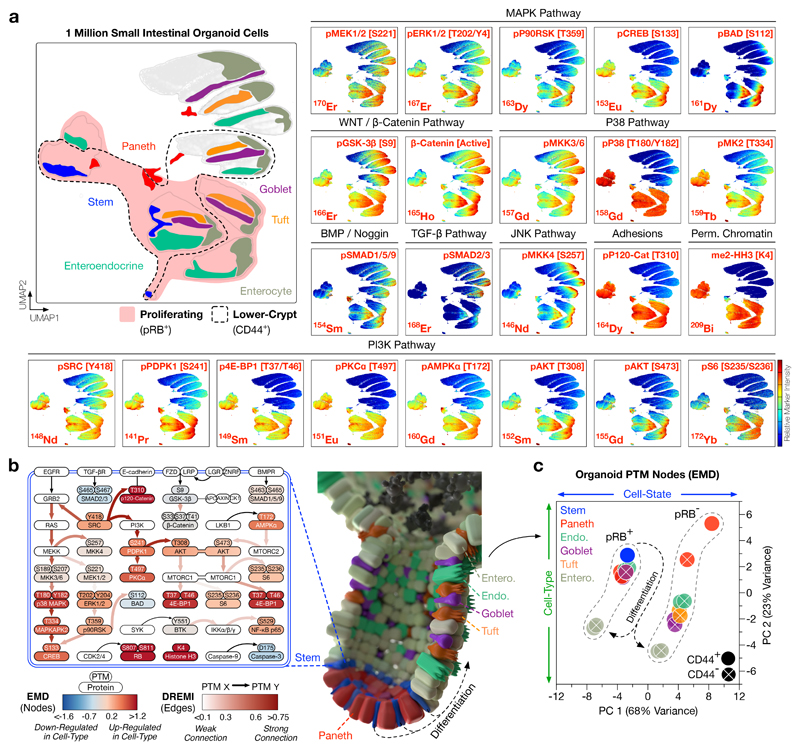
Figure 2. Cell-Type and Cell-State Specific Signalling Analysis of Intestinal Organoids.
a) UMAP distributions of PTMs across 1 million single organoid cells analysed by MC. Cell-type and cell-state UMAP guide is shown top left (see Supplementary Fig. 2 for cell-type and cell-state classification). Combining cell-type, cell-state, and PTM measurements enables cell-type specific analysis of intestinal organoid signalling. Colours represent normalised local parameter intensity.
b) Cell-type specific PTM signalling networks in small intestinal organoids, with nodes coloured by PTM-EMD (Earth Mover’s Distance) scores quantifying PTM intensity (relative to all organoid cells) and edges coloured by DREMI (Density Resampled Estimation of Mutual Information) scores quantifying PTM-PTM connectivity. Small intestinal organoids display cell-type specific signalling networks.
c) Principal Component Analysis (PCA) of PTM-EMDs for all organoid cell-types, either proliferating (pRB+)/G0 (pRB−) or in lower-crypts (CD44+)/villi (CD44−). Organoid cell signalling is dictated by cell-state (PC 1) and cell-type (PC 2). (See Supplementary Fig. 3 for complete EMD-DREMI signalling maps.)
Combining 194/8Cisplatin and 127IdU with cell-cycle markers (e.g. pRB [S807/S811], Cyclin B1, and pHistone H3 [S28]) allows clear identification of live/dead cells and classification of single organoid cells into cell-cycle stages including G0, G1, S, G2, and M-phase14,17 (Supplementary Fig. 2b). Integrated cell-type and cell-state data from small intestinal organoids confirmed that stem and Paneth cells are largely proliferative (pRB+, cCaspase3 [D175]−), whereas differentiated epithelia are often post-mitotic (pRB−) or apoptotic (cCaspase3+) (Fig. 1c). Consistent with the finding that intestinal progenitor cells have permissive chromatin in vivo18, proliferating intestinal organoid cells also present H3K4me2 whereas post-mitotic cells do not (Fig. 2a). These results confirmed that a modified MC workflow can provide cell-type and cell-state specific information from millions of single organoid cells.
Cell-Type and Cell-State Specific Signalling Networks in Intestinal Organoids
Following cell-type and cell-state identification, we next sought to construct cell-type specific PTM signalling networks in small intestinal organoids. We combined Earth Mover’s Distance (EMD)19, 20 and Density Resampled Estimation of Mutual Information (DREMI)21 to build quantitative cell-type specific signalling networks from single-cell organoid PTM data (Fig. 2b, Supplementary Fig. 3). In these networks, EMD quantifies PTM intensity (node score) for each organoid cell-type relative to the total organoid population and DREMI quantifies PTM-PTM connectivity (edge score).
EMD-DREMI analysis revealed cell-type specific PTM signalling networks in small intestinal organoids. As canonical WNT signalling is mainly driven by protein interactions, localisation, and degradation22 – not a classical PTM cascade – MC is not well suited to studying the WNT pathway. Despite this limitation, evidence of WNT flux via inhibited pGSK-3β [S9] and non-phosphorylated β-Catenin is observed in all organoid cell-types (Fig. 2a). In contrast, MAPK and PI3K pathways display unexpected cell-type specificity. For example, stem cells channel MAPK signalling through pERK1/2 [T202/Y204], pP90RSK [T359], and pCREB [S133], but fail to connect with pBAD [S112] (Fig. 2a, b). On the contrary, differentiated epithelia direct MAPK signalling away from pCREB and towards pBAD when proliferating, and lose all MAPK activity in G0 and apoptosis (Fig. 2a). Despite their strong mitogenic signalling profile, stem cells are unique among proliferating cells in their failure to phosphorylate BAD. This suggests that intestinal stem cells avoid apoptosis independent of the classical BAD-BCL-BAX/BAK axis and may compensate via high MAPK and P38 flux to CREB. Consistent with the observation that PI3K signalling is important for intestinal crypt cells23, stem and Paneth cells are enriched for pSRC [Y418] and downstream PI3K effectors such as pPDPK1 [S241], pPKCɑ [T497], pAKT [T308]/[S473], and p4E-BP1 [T37/T46] (Fig. 2a, b). Paneth cells display unexpectedly high BMP signalling (via pSMAD1/5 [S463/S465] and SMAD9 [S465/S467]), suggesting that they are either hypersensitive to BMP ligands or can cell-intrinsically activate SMAD1/5/9 (possibly via inhibition of SMAD phosphatases) (Fig. 2a, b).
Several PTM signalling events correlate with cell-state in intestinal organoids. For example, irrespective of cell-type or localisation, pP38 MAPK [T180/Y182] and pP120-Catenin [T310] are active in all proliferating cells, and pAKT [T308] and pMKK4 [S257] are hyperactivated in M-phase (Figs. 1c, 2a). In contrast, TGF-β signalling (via pSMAD2 [S465/S467] and SMAD3 [S423/S425]) is exclusively active in post-mitotic epithelia, consistent with TGF-β’s role in epithelial growth-arrest24 (Fig. 2a). To investigate the relationship between cell-type and cell-state on PTM signalling, we performed principal component analysis (PCA) of PTM-EMDs for each organoid cell-type, either proliferating or in G0 (pRB+/−), located in lower-crypts or villi (CD44+/−). PCA revealed that both cell-state (PC 1, 68% variance) and, to a lesser extent, cell-type (PC 2, 23% variance) dictate cell-signalling in small intestinal organoids (Fig. 2c, Supplementary Fig. 3). This analysis demonstrates that cell-type and cell-state are intimately linked with cell-signalling and warns against bulk PTM analysis of organoids where cell-type and cell-state resolution is lost.
Single-Cell Organoid Multiplexing using Thiol-reactive Organoid Barcoding in situ (TOBis)
We have demonstrated how a modified MC workflow can be applied to cell-type and cell-state specific signalling measurement in organoids. In order to study differential signal transduction in organoid models of healthy and diseased tissue, we must also be able to directly compare PTM networks between different organoid cultures. A major advantage of MC is its ability to perform multiplexed barcoding of experimental variables25, 26. Unfortunately, commercially available Palladium-based barcodes cannot bind organoids in situ as they react with Matrigel proteins, meaning that organoids must be removed from Matrigel and dissociated separately before barcoding (Supplementary Fig. 4a, b). We theorised that if organoids could be barcoded in situ, barcoded organoids could be pooled, dissociated, and processed as a single high-throughput MC sample. To explore this idea, we developed a new strategy to isotopically barcode organoids while still in Matrigel.
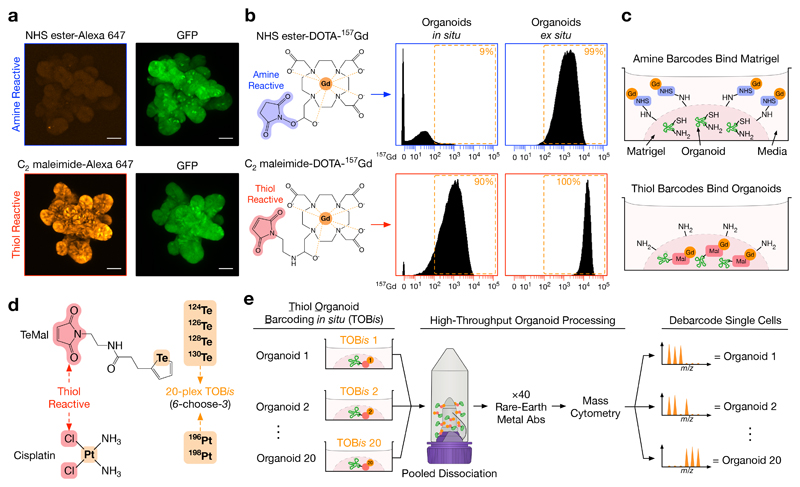
MC barcoding strategies can use amine-25 or thiol-reactive26 chemistries. We first used fluorescent probes to investigate how these chemistries react with ECM proteins and organoids. Amine-reactive probes (Alexa Fluor 647 NHS ester) bind ECM proteins and thus fail to label organoids in Matrigel. In contrast, thiol-reactive probes (Alexa Fluor 647 C2 maleimide) bypass ECM proteins and bind exclusively to organoids in situ (Fig. 3a, Supplementary Fig. 4c). We subsequently confirmed that thiol-reactive monoisotopic mass-tagged probes (C2 maleimide-DOTA-157Gd) also bind organoids in situ, whereas amine-reactive probes (NHS ester-DOTA-157Gd) only react ex situ (Fig. 3b). This data confirmed that thiol-reactive chemistries can be used to barcode organoids while still in Matrigel (Fig. 3c). Using this knowledge, we developed a custom 20-plex (6-choose-3, doublet-filtering25, 26) thiol-reactive barcoding strategy using monoisotopic tellurium maleimide (TeMal) (124Te, 126Te, 128Te, 130Te)27 and Cisplatin (196Pt, 198Pt)28 that can bypass ECM proteins and bind directly to fixed organoids in situ (Fig. 3d, Supplementary Fig. 4d). This Thiol-reactive Organoid Barcoding in situ (TOBis) approach enables high-throughput multivariate single-cell organoid signalling analysis in a single tube (Fig. 3e).
Figure 3. Thiol-reactive Organoid Barcoding in situ (TOBis) for Single-Cell Organoid Multiplexing.
a) Confocal IF of fixed GFP+ small intestinal organoids stained with either amine-reactive fluorescent probe Alexa Fluor 647 NHS ester or thiol-reactive fluorescent probe Alexa Fluor 647 C2 maleimide while still in Matrigel, scale bars = 50 μm. Each IF image is representative of three independent experiments. Amine-reactive probes bind diffusely to Matrigel with very poor binding to organoids, whereas thiol-reactive probes bypass Matrigel and bind directly to organoids (see Supplementary Fig. 4c for Matrigel background staining).
b) Small intestinal organoids stained with either amine-reactive NHS ester-DOTA-157Gd or thiol-reactive C2 maleimide-DOTA-157Gd in situ (still in Matrigel) or ex situ (removed from Matrigel) and analysed by MC. While both probes bind organoid cells ex situ, only thiol-reactive C2 maleimide-DOTA-157Gd bind organoids in situ. Data is representative of three independent experiments.
c) Model of amine- and thiol-reactive barcodes in organoid culture.
d) Thiol-reactive tellurium maleimide (TeMal) (124Te, 126Te, 128Te, 130Te) and Cisplatin (196Pt, 198Pt) isotopologs combined to form a 20-plex (6-choose-3) doublet-filtering organoid barcoding strategy.
e) Thiol-reactive Organoid Barcoding in situ (TOBis) workflow. When combined with cell-type, cell-state, and PTM probes, TOBis allows organoids to be barcoded while still in Matrigel and rapidly processed as a single sample. (See Supplementary Fig. 5 for additional details.)
It is worth noting that as Te and Pt are not typically conjugated to antibodies in MC, TOBis multiplexing does not compromise the number of antigens being measured. Moreover, as barcoding is performed on fixed organoids embedded in Matrigel, TOBis does not require the numerous centrifugation or permeabilisation steps used in traditional solution-phase barcoding. This greatly increases organoid sample-throughput (Supplementary Fig. 5a—d) and single-cell recovery (Supplementary Fig. 5e—g), thereby facilitating high-throughput organoid MC applications.
Multivariate Cell-Type Specific Signalling Analysis of Intestinal Organoid Development
Traditional mass-tag barcoding allows direct comparison of solution-phase cells between experimental conditions25. TOBis MC now enables PTM signalling networks to be directly compared between solid-phase organoid cultures in a high-throughput manner. To demonstrate this, we applied TOBis to study cell-type specific epithelial signalling during 7 days of small intestinal organoid development (Fig. 4 and Supplementary Table 1, 50 parameters (40 antibodies)/cell).
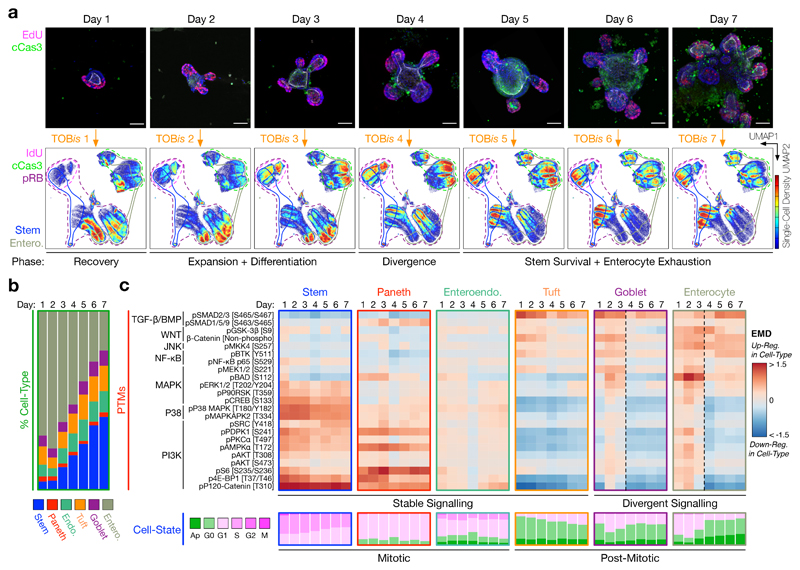
Figure 4. Cell-Type Specific Signalling During Intestinal Organoid Development.
a) Time-course confocal IF of intestinal organoid development illustrating S-phase (EdU+, magenta) and apoptotic (cCaspase 3 [D175]+, green) cells, scale bars = 50 μm. Images are representative of at least five organoids in independent time-course and IF experiments. Each time point was barcoded by TOBis, pooled into a single sample, and analysed by MC. Cell-density UMAP distributions of 2 million single organoid cells reveal changes in cell-type and cell-state during organoid development.
b) Cell-type composition of small intestinal organoids during development. Stem cells accumulate at the expense of enterocytes during organoid culture.
c) Cell-type specific PTMs and cell-states of stem, Paneth, enteroendocrine, tuft, goblet cells, and enterocytes during intestinal organoid development. Cell-state analysis shows the proportions of apoptotic, G0-, G1-, S-, G2-, and M-phase cells. Irrespective of time point, stem, Paneth, and enteroendocrine cells are stably mitotic, whereas tuft, goblet cells, and enterocytes are frequently post-mitotic. Stem, Paneth, enteroendocrine, and tuft cells display stable signalling over time, whereas goblet cell- and enterocyte-signalling diverge from Day 4.
Analysis of 28 PTMs from ~2 million single organoid cells revealed that after 1 day of culture, organoids seeded as single crypts exist in a ‘recovery’ phase where ~70% cells have entered the cell-cycle (pRB+), but <5% reach S-phase (IdU+) (compared to ~20% in developed organoids) (Fig. 4a). Days 2 and 3 mark a rapid ‘expansion and differentiation’ phase of organoid development where stem, Paneth, goblet cells, and enterocytes activate MAPK, P38, and PI3K pathways – although stem cells again fail to inhibit BAD (Fig. 4c). By Day 4, intestinal organoids reach a critical ‘divergence’ phase where crypt and villus signalling digress dramatically. While stem and Paneth cells maintain active MAPK, P38, and PI3K pathways, enterocytes lose major PI3K activity (Fig. 4c). As a result, by Days 5 to 7, enterocytes are largely post-mitotic or apoptotic with high TGF-β signalling, whereas stem cells retain mitogenic flux and cell-cycle activity (Fig. 4c). Consequently, stem cell number increases while enterocytes become exhausted at the end of intestinal organoid development (Fig. 4a, b). Notably, both stem and Paneth cells continue to display high MAPK, P38, and PI3K activity even at this late stage of organoid culture (Fig. 4c). This suggests that maintaining a stable signalling flux is a core feature of intestinal crypt cells. Such variations in organoid cell-state, cell-type, and PTM activity suggest developmental stage should be carefully considered when performing organoid experiments.
Single-Cell Signalling Analysis of CRC TME Organoids
We have demonstrated how the integration of TOBis into a MC anti-PTM workflow enables high-throughput comparison of cell-type specific signalling networks in epithelial organoids. Given that MC can theoretically resolve any cell-type, we next expanded this platform to study PTM signalling in heterocellular organoid co-culture models of colorectal cancer (CRC).
CRC develops through successive oncogenic mutations – frequently resulting in loss of APC activity, hyperactivation of KRAS, and perturbation of TP5329. In addition to oncogenic mutations, stromal fibroblasts30, 31 and macrophages32 have also emerged as major drivers of CRC33. While the underlying driver mutations of CRC have been well studied, how they dysregulate epithelial signalling relative to microenvironmental cues from stromal and immune cells is unclear.
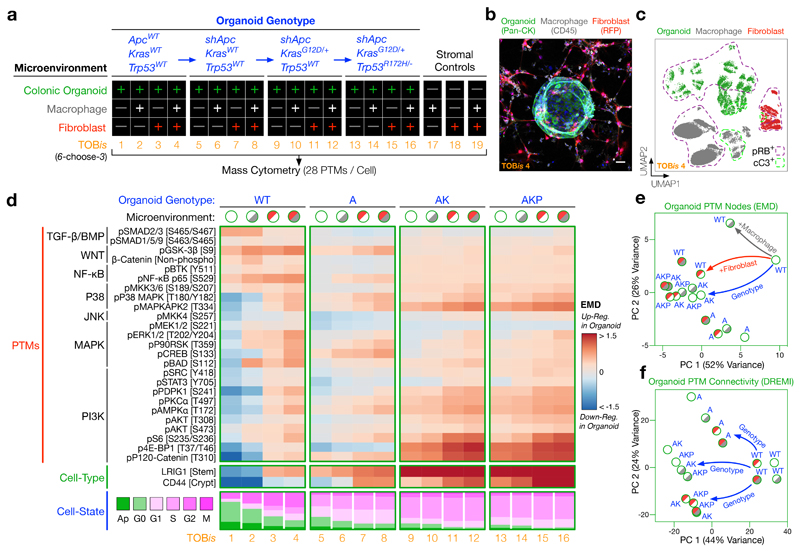
To investigate this, we cultured wild-type (WT), shApc (A), shApc and KrasG12D/+ (AK), or shApc, KrasG12D/+, and Trp53R172H/− (AKP)34, 35 colonic epithelial organoids either alone, with colonic fibroblasts, and/or macrophages (Fig. 5a, b, Supplementary Fig. 6). Each CRC genotype-microenvironment organoid culture was fixed, TOBis-barcoded, and single-cell signalling analysis was performed in one multivariate MC run (Fig. 5a and Supplementary Table 2, 50 parameters (40 antibodies)/cell). The addition of myeloid (CD68, F4/80) and mesenchymal (Podoplanin) heavy-metal antibodies enabled clear resolution of epithelial organoids, macrophages, and fibroblasts from each barcoded condition (Fig. 5c). This experimental design allowed us to directly compare mutation- and microenvironment-driven cell-type specific signalling networks in CRC organoid mono- and co-cultures.
Figure 5. Single-Cell Signalling Analysis of Colorectal Cancer (CRC) Tumour Microenvironment Organoids.
a) Experimental design. CRC organoid genotypes (wild-type (WT), shApc (A), shApc and KrasG12D/+ (AK), shApc, KrasG12D/+, and Trp53R172H/− (AKP)) were cultured in the presence or absence of colonic fibroblasts and/or macrophages (without exogenous growth factors). Each condition was TOBis-barcoded, pooled into a single sample, and analysed by MC (28 PTMs/cell).
b) Confocal IF of a WT colonic organoid (Pan-CK, green) co-cultured with colonic fibroblasts (RFP, red), and macrophages (CD45, grey) (TOBis 4), scale bar = 50 μm. Image is representative of five independent co-culture and IF experiments.
c) UMAP distribution of the colonic microenvironment model resolves single epithelial cells (green), fibroblasts (red), and macrophages (grey) (TOBis 4).
d) PTMs, progenitor cell-types, and cell-states of colonic epithelial organoids across all genotype/microenvironment combinations. The grey and red shades in the microenvironmental conditions represent macrophages and fibroblasts respectively. (See Supplementary Figs. 7 and 8 for complete EMD-DREMI signalling maps of organoids, macrophages, and colonic fibroblasts.)
e) PCA of 28 PTM-EMDs for colonic epithelial organoids across all genotype/microenvironment combinations. CRC organoids with AK/AKP mutations mimic the signalling flux driven by colonic fibroblasts. (See Supplementary Fig. 8c, d for PTM-EMD PCAs for macrophages and colonic fibroblasts.)
f) PCA of 756 PTM-DREMIs for colonic epithelial organoids across all genotype/microenvironment combinations. Epithelial signalling connectivity is regulated by genotype rather than microenvironment. (See Supplementary Fig. 8e, f for PTM-DREMI PCAs for macrophages and colonic fibroblasts.)
As expected, oncogenic mutations have a large cell-autonomous effect on epithelial signalling. Although APC mutations are known to upregulate WNT signalling22, we found that the loss of APC also activates the P38 pathway, downregulates TGF-β/BMP signalling, and activates p120-Catenin in colonic organoids (Fig. 5d). Subsequent oncogenic KrasG12D/+ and Trp53R172H/− mutations further upregulate not only the classical MAPK pathway, but also major PI3K nodes (Fig. 5d). As a result, AK and AKP organoids display increased stem/progenitor cell-type markers LRIG1 and CD44, decreased apoptosis, and increased mitogenic cell-state relative to WT and A organoids (Fig. 5d).
Both oncogenes and stromal cells can dysregulate cancer cell signalling7. However, to what extent this is driven by oncogenic mutations (cell-intrinsic) or the TME (cell-extrinsic) is less clear. To our surprise, we found that microenvironmental cues have a comparable impact on PTM regulation to oncogenic mutations (Fig. 5e). In contrast, PTM-PTM connectivity is regulated largely by genotype, not microenvironment (Fig. 5f). This observation suggests that oncogenic mutations fundamentally re-wire signalling networks, whereas stromal cells regulate acute signalling flux. Stromal cells further upregulate the PI3K pathway in CRC organoids that already contain KrasG12D and Trp53R172H mutations (Fig. 5d, Supplementary Fig. 7). Microenvironmental hyper-activation of the epithelial PI3K pathway may contribute towards the poor prognosis of CRC patients with highly stromal tumours30, 31.
In addition to mutation- and microenvironment-driven epithelial signalling, we discovered previously unreported polarity in fibroblast and macrophage cell-cell communication. For example, macrophage signalling pathways (MAPK, PI3K, and NF-κB) are heavily upregulated by fibroblasts (Supplementary Fig. 8a, c, e), whereas fibroblast signalling is scarcely altered by macrophages (Supplementary Fig. 8b, d, f). In contrast, epithelial cells upregulate MAPK and P38 signalling in fibroblasts (Supplementary Fig. 8b), which in turn, reciprocally activate MAPK and P38 signalling in epithelial cells (Supplementary Fig. 7). These results suggest that colonic fibroblasts are major regulators of intercellular signalling in the colonic microenvironment and should be further investigated as drivers of CRC.
Oncogenic Mutations Mimic Stromal Signalling Networks
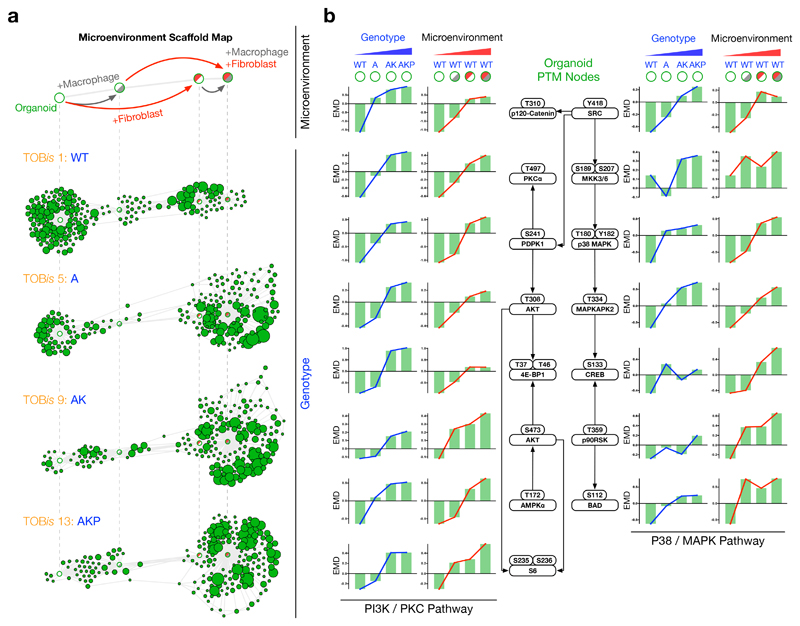
To further investigate the parity between genotypic and microenvironmental regulation of epithelial signalling, we overlaid single-cell MC data from WT, A, AK, and AKP organoids onto a fixed-node microenvironmental Scaffold map36 (Fig. 6a, Supplementary Fig. 9a). This unsupervised analysis confirmed that Apc, Kras, and Trp53 oncogenic mutations mimic the signalling profile of WT organoids in the presence of stromal cells. Inverted organoid genotype Scaffold maps also expose a striking similarity between mutation- and microenvironment-driven signalling (Supplementary Fig. 9b). Direct comparison of organoid PTMs revealed that both PI3K/PKC and P38/MAPK nodes are analogously upregulated by oncogenic mutations and microenvironmental cues (Figs. 5d, 6b).
Figure 6. Oncogenic Mutations Mimic Stromal Signalling Networks.
a) Scaffold maps constructed from WT organoids either alone or co-cultured with colonic fibroblasts and/or macrophages. Unsupervised distribution of A, AK, and AKP colonic organoids revealed that oncogenic mutations mimic signalling profiles driven by stromal fibroblasts and macrophages. (See Supplementary Fig. 9a for all genotype/microenvironment combinations and Supplementary Fig. 9b for mutation-driven Scaffold maps.)
b) PTM-EMDs for PI3K/PKC and P38/MAPK signalling nodes in colonic organoids following genotypic and microenvironmental regulation. Cell-type specific PTM analysis demonstrates oncogenic mutations and microenvironmental cues upregulate analogous signalling nodes in epithelial colonic organoids.
Taken together, TOBis-multiplexed cell-type specific PTM analysis of organoid co-cultures elucidated several fundamental processes in CRC: 1) oncogenic mutations re-structure signalling networks in cancer cells, whereas microenvironmental cues drive acute signalling flux, 2) stromal cells hyper-activate PI3K signalling in colonic epithelial cells that already carry Kras and Trp53 mutations, and 3) oncogenic mutations cell-autonomously mimic an epithelial signalling state normally induced by stromal cells.
Discussion
Organoids are heterocellular systems comprising multiple cell-types and cell-states. Cell-type specific PTM signalling networks regulate major biological processes and are frequently dysregulated in disease. As a result, understanding cell-type specific signalling networks is fundamental to the utility of organoids and organoid co-cultures. We demonstrated how a modified MC workflow that combines monoisotopic cell-type, cell-state, and PTM probes can be used to study cell-type specific signalling networks in organoids. Thiol-reactive Organoid Barcoding in situ (TOBis) enables high-throughput comparison of signalling networks across different organoid mono- and co-cultures.
While this study has focused on intestinal organoids, we expect this method to be fully compatible with organoids derived from other tissues. Cell-type identification probes for each tissue should be carefully validated, but otherwise the TOBis multiplexing and PTM analysis framework should be compatible with all organoid models (including those grown in defined hydrogels37). Moreover, our extension of MC to study colonic fibroblasts and macrophages implies that PTM signalling can be measured in any cell-type co-cultured with organoids (e.g. PBMCs co-cultured with organoids3 and air-liquid interface TME organoids38).
TOBis MC holds substantial promise for organoid screening. While drug screens of patient-derived organoid (PDO) monocultures have shown great potential39, 40, their reliance on bulk viability measurements means they cannot be used to evaluate drugs targeting stromal/immune cells or provide any mechanistic understanding of drug performance/resistance. In contrast, a TOBis-multiplexed MC screen will provide cell-type specific signalling networks, cell-cycle states, and apoptotic readouts across all cell-types in PDO co-cultures. These features make TOBis MC a powerful tool for evaluating drug/CRISPR perturbations and anti-organoid biological therapies (e.g. CAR T-cells). Future development of TOBis using additional TeMal (×7 possible) and Cisplatin (×4 possible) isotopologs will greatly expand organoid multiplexing capacity and advance this technology to high-throughput organoid screening applications.
In summary, this study demonstrates how a modified MC platform can reveal cell-type specific signalling networks in organoid monocultures and uncover novel cell-cell signalling relationships in organoid co-cultures. Given the widespread adoption of organoids as biomimetic models of healthy and diseased tissue, we propose cell-type specific PTM MC analysis as a powerful technology for multivariate organoid phenotyping.
Methods
Organoid Culture
Intestinal organoids were generated as describe by Sato et al.13. Briefly, the small intestine of 8- to 12-week-old Lgr5-EGFP-ires-CreERT2 mice was dissected, opened longitudinally, and cut into 2- to 5-mm segments. Tissue fragments were washed with ice-cold PBS and incubated with 2 mM EDTA (Sigma 03690) in PBS (Thermo 10010056) for 1 hr at 4 °C. After removal of EDTA, tissue fragments were washed vigorously in cold PBS to release the crypts. Supernatant fractions from the washes were collected and centrifuged at 1,200 rpm for 5 mins. Cells were washed with 15 mL advanced DMEM/F-12 (Thermo 12634010), passed through a 70 μm cell strainer (Fisher 11597522) to enrich for intestinal crypts, and centrifuged at 600 rpm for 2 mins. The cell pellet was resuspended in Growth Factor Reduced Matrigel (Corning 354230) and cultured at 37 °C in the presence of 5% CO2.
Small intestinal organoids were maintained in advanced DMEM/F-12 (Thermo 12634010) supplemented with 2 mM L-Glutamine (Thermo 25030081), 1 mM N-Acetyl-L-Cysteine (Sigma A9165), 10 mM HEPES (Sigma H3375), 1× B-27 Supplement (Thermo 17504044), 1× N-2 Supplement (Thermo 17502048), 50 ng/mL murine EGF (mEGF, Thermo PMG8041), 50 ng/mL murine Noggin (mNoggin, Peprotech 250-38), 500 ng/mL murine R-Spondin-1 (mR-spondin-1, Peprotech 315-32), and 1× HyClone™ Penicillin Streptomycin Solution (Fisher SV30010).
Murine colorectal cancer (CRC) organoids carrying oncogenic mutations (shApc (A), shApc and KrasG12D/+ (AK), or shApc, KrasG12D/+, and Trp53R172H/- (AKP)34, 35) were a kind gift from Prof. Lukas Dow (Cornell University) and are described in Dow & O’Rourke et al., Cell, 201534 and O’Rourke et al., Nature Biotechnology, 201735. Colonic organoids were maintained in advanced DMEM/F-12 (Thermo 12634010) supplemented with 2 mM L-Glutamine (Thermo 25030081), 1 mM N-Acetyl-L-Cysteine (Sigma A9165), 10 mM HEPES (Sigma H3375), 1× B-27 Supplement (Thermo 17504044), 1× N-2 Supplement (Thermo 17502048), 100 ng/mL murine WNT-3a (mWNT-3a, Peprotech 315-20), 50 ng/mL murine EGF (mEGF, Thermo PMG8041), 50 ng/mL murine Noggin (mNoggin, Peprotech 250-38), 500 ng/mL murine R-Spondin-1 (mR-spondin-1, Peprotech 315-32), 10 mM Nicotinamide (Sigma N0636), and 1× HyClone™ Penicillin Streptomycin Solution (Fisher SV30010).
For passaging, small intestinal organoids were retrieved from Matrigel using ice-cold PBS and broken up mechanically by passing through a 23G, 5/8” needle (Terumo AN-2316R). Colonic organoids were dissociated mechanically by pipetting or enzymatically with TrypLE™ Express Enzyme (Thermo 12604013). Organoid fragments were collected using a benchtop centrifuge, washed with ice-cold PBS, and reseeded in fresh Matrigel. The passage was performed every 4 to 7 days at a ratio of 1:2 or 1:3.
Pre-Treatment and Fixation of Organoids for Mass Cytometry
1275-Iodo-2′-deoxyuridine (127IdU) (Fluidigm 201127) was added directly to organoid culture media to a final concentration of 25 μM and incubated at 37 °C for 30 mins before fixation to identify cells in S-phase14. 5 mins before fixation, protease and phosphatase inhibitors (Sigma P8340/Sigma 4906845001) were added to organoid cultures to preserve cell signalling during fixation8. As dissociation of live tissue alters cellular states41 including PTMs8, all organoids were fixed in 4% PFA (Thermo J19943K2) for 60 mins at 37 °C to preserve cell-signalling. (Note: during method optimisation, we also trialled alternative fixatives such as Glutaraldehyde (0.2%, 1%, 2.5%), Ethanol (5%, 10%), and Glyoxal (pH 4.0, pH 5.0), but concluded that 4% PFA was optimal. 1.6% PFA can also be used but we encourage users to test a range of PFA for their specific antibody panel.) Following fixation, organoids were washed ×2 with PBS and incubated in 250 nM 194/8Cisplatin (Fluidigm 201194/8) in PBS for 10 mins on a rocker to stain dead cells15. During optimisation we found this condition yields strong 194/8Pt staining with a wide dynamic range suitable for efficient dead cell removal in silico. Organoids were then washed ×2 with PBS to remove residual Cisplatin. Fixed organoids were subsequently dissociated for single-cell analysis immediately or stored at 4 °C.
Single-Cell Dissociation of Fixed Organoids
After organoids were fixed and stained with Cisplatin, the final wash was removed and a solution of fresh 0.5 mg/mL Dispase II (Thermo 17105041), 0.2 mg/mL Collagenase IV (Thermo 17104019), and 0.2 mg/mL DNase I (Sigma DN25) in PBS was added to the organoids. (During optimisation we found Dispase II is essential for disrupting epithelial cell-cell contacts, Collagenase IV improves Matrigel degradation, and DNase I digests extracellular genomic DNA from dead organoid cells to reduce sample viscosity. We encourage users to test alternative dissociation enzymes for the specific cellular composition of their experimental system.) Organoid droplets were then scraped from the well, pooled, and the enzyme/organoid solution was transferred to a gentleMACS C-Tube (Miltenyi 130-096-334). Fixed organoids were dissociated into a single-cell suspension using the gentleMACS Octo Dissociator (with Heaters) (Miltenyi 130-096-427) at 37 °C for 50 mins using a custom program. Following dissociation, C-Tubes were centrifuged at 800 ×g for 1 min to collect cells from blades and all liquid was transferred to a fresh polypropylene FACS tube (Corning 352063). Single organoid cells were then washed ×2 in Cell Staining Buffer (CSB) (Fluidigm 201068) (5 mins, 800 ×g) to remove enzymes/cellular debris and 35 μm filtered (Fisher 10585801) (70 μm (Fisher 11597522) when the culture contains fibroblasts) to remove residual clumps.
Heavy-Metal Antibody Conjugation and Panel Design
All antibodies were custom conjugated42 with rare-earth/heavy metals using X8 polymers and monoisotopic metals from Fluidigm (Fluidigm 201300). Non-Fluidigm metals/nitrates were also used: 89Y (Sigma 217239), 113In (Trace Sciences), 115In (Trace Sciences), 157Gd (Trace Sciences), and 209Bi (Sigma 254150)43. Antibody panels (Supplementary Tables 1 and 2) were carefully designed and titrated in accordance with known monoisotopic impurities44 and antigen abundance to ensure minimal cross-channel contamination.
Mass Cytometry Analysis of Single Organoid Cells
1–5 × 106 fixed single organoid cells were blocked in CSB and stained with organoid-specific extracellular rare-earth metal antibody cocktails (Supplementary Tables 1 and 2) for 30 mins. Cells were then washed ×2 with CSB (5 mins, 800 ×g) and permeabilised in 0.1% Triton X-100 (Sigma T8787) in PBS for 30 mins. Cells were washed ×2 in CSB and further permeabilised with ice-cold 50% methanol (Fisher 10675112) for 10 mins on ice. (Note: during method optimisation we found dual 0.1% Triton X-100 and 50% Methanol incubation provides the best all-round permeabilisation for a broad range of anti-PTM antibodies.) Permeabilised cells were then washed ×2 in CSB and stained with intracellular rare-earth metal antibody cocktails (Supplementary Tables 1 and 2) for 30 mins. Stained cells were washed ×2 in CSB, fixed in fresh 1.6% formaldehyde (Thermo 28906) for 10 mins, washed in CSB, and incubated in DNA Intercalator (Fluidigm 201192A) overnight at 4 ºC. The following day, cells were washed ×2 in CSB, resuspended in Maxpar Water (Fluidigm 201069) containing 20% (v/v) EQ Beads45 (Fluidigm 201078) and 2 mM EDTA at ~0.5 × 106 cells/mL. Cells were then 35 μm filtered (Fisher 10585801) (70 μm (Fisher 11597522) when the culture contains fibroblasts) and immediately analysed using a Helios Mass Cytometer (Fluidigm) (100 – 300 events/sec). Files were normalised against EQ beads, de-barcoded25 into each experimental condition (when required), and uploaded to the Cytobank platform (http://www.cytobank.org/).
Immunofluorescence (IF) Staining of Organoids
Intestinal organoids were cultured in 8-well μ-Slides (ibidi 80826). After culture medium was removed, cells were washed with PBS and fixed with 4% PFA (Thermo J19943K2) for 30 mins at 4 °C. Cells were washed twice with PBS and permeabilised with 0.2% Triton™ X-100 (Sigma T8787) in PBS for 30 mins at room temperature. Cells were washed again, incubated with PBS containing 1% BSA (CST #9998) and 0.3% Triton™ X-100 for 30 mins, followed by incubation with primary antibodies diluted in 1% BSA/0.3% Triton™ X-100/PBS overnight at 4 °C. Cells were washed with PBS, stained with secondary antibodies and 4’,6-Diamidino-2-Phenylindole (DAPI) (Thermo D1306), with or without Alexa Fluor™ 488/568 Phalloidin (Thermo A12379/A12380) for 1 hr at room temperature, away from light. Cells were washed with PBS and mounted with Fluoromount-G™ mounting medium (Thermo 00-4958-02). Samples were imaged with a Zeiss LSM880 confocal microscope and images were analysed using FIJI46. EdU staining was performed using the Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (Thermo C10640) following the manufacture’s protocol.
Small Intestinal Organoid Directed Differentiation
Small intestinal organoids were seeded and cultured in complete organoid medium for 24 hrs to allow organoid recovery, and treated with combinations of 3 μM CHIR99021 (GSK-3β inhibitor) (Cambridge Bioscience SM13), 2 μM IWP-2 (PORCN inhibitor) (Cambridge Bioscience 13033), 1 mM Valproic Acid (HDAC inhibitor) (Cambridge Bioscience SM39), and 10 μM DAPT (γ-Secretase inhibitor) (Cambridge Bioscience SM15) for 3 days to direct organoid differentiation towards specific cell-types as described in Yin et al.16 (Supplementary Fig. 1). Directed-differentiated organoids were analysed by IF and MC as described above.
Single-Cell Dissociation of Murine Intestinal Crypts
The small intestine of 8- to 12-week-old Lgr5-EGFP-ires-CreERT2 mice was dissected and intestinal crypts were isolated as described above (see ‘Organoid Culture’). The crypts were resuspended in 5 mL of TrypLE™ Express Enzyme and incubated at 37 °C for 45 mins, mixed every 10 mins to avoid cells clumping. The cells were centrifuged at 1,200 rpm for 5 mins, resuspended in 5 mL of 4% PFA, and fixed at 37 °C for 1 hr. Fixed cells were washed once with PBS, 35 μm filtered twice to remove residual clumps, and stored at 4 °C prior to MC analysis.
Amine- versus Thiol-Reactive in situ Organoid Probes
To investigate alternative probe chemistries for in situ organoid barcoding, fixed small intestinal organoids were stained with either 50 nM Alexa Fluor™ 647 NHS ester (Thermo A20006) or 50 nM Alexa Fluor™ 647 C2 maleimide (Thermo A20347) for 1 hr while still in Matrigel. Organoid probe intensities were visualised by confocal microscopy using identical settings for each probe. To confirm IF observations using MC, fixed small intestinal organoids were stained with either 200 nM NHS-DOTA (Macrocyclics B-280) or 200 nM maleimide-DOTA (Macrocyclics B-272) coupled to 157Gd (Trace Sciences) for 1 hr in situ (in Matrigel) or ex situ (removed from Matrigel). Organoids were then dissociated into single cells and analysed by MC.
Thiol-reactive Organoid Barcoding in situ (TOBis)
Fixed organoids were washed ×2 in PBS and stained in situ with 10 nM Cisplatin (196Pt, 198Pt)28 (a kind gift from Dr. Olga Ornatsky, Fluidigm) and 1—3 μM TeMal (124Te, 126Te, 128Te, 130Te)27 barcodes for 1 hr on a rocker (barcoding matrices in Supplementary Tables 3 and 4). (Note: organoids can also be barcoded overnight at 4 °C.) The lower concentration of Cisplatin used for TOBis (10 nM) relative to dead cell stains (250 nM) is to ensure 196Pt and 198Pt signal intensities align with 124Te, 126Te, 128Te, and 130Te signals. TOBis barcoding can be performed in 6-well (3 mL barcodes), 12-well (2 mL barcodes), 24-well (1 mL barcodes), 48-well (500 μL barcodes), and 96-well (200 μL barcodes) plates. In our experience up to 10,000 cells/μL Matrigel (representing confluent intestinal organoid cultures) can be efficiently barcoded, but we suggest optimising barcode concentrations for alternative model systems. Organoids were washed ×3 in CSB containing 1 mM L-Glutathione (Sigma G6529) for 5 mins on a rocker to quench unbound barcodes (Supplementary Fig. 5h) and ×1 in PBS prior to pooled-dissociation (as described above).
To directly compare TOBis and Cell-ID™ 20-Plex Pd Barcoding Kit (Fluidigm 201060) performance for in situ barcoding, 20 wells of established small intestinal organoids in the 12-well culture format (3 × 30 μL Matrigel droplets/well) were fixed as described above and washed ×2 with PBS. TOBis Barcode 1—20 was mixed respectively with Cell-ID™ Barcode 1—20 in PBS and added to each well. The cells were incubated at room temperature for 60 mins, washed ×3 in CSB containing 1 mM L-Glutathione (Sigma G6529) for 5 mins on a rocker to quench unbound barcodes, dissociated into single cells, and analysed by MC.
Maxpar in situ versus ex situ Organoid Barcoding Comparison
Established small intestinal organoids were fixed in 4% PFA and washed ×2 with PBS. Maxpar barcodes were resuspended in Barcode Perm Buffer as per the manufacturer’s protocol (Fluidigm 201060). For in situ barcoding, cells were incubated in Barcode Perm Buffer for 10 mins followed by barcoding solution incubation at room temperature for 60 mins. The cells were washed ×2 with CSB to quench unbound barcodes and proceeded to dissociation as described above. For ex situ barcoding, cells were dissociated and barcoded according to the manufacturer’s protocol. The in situ and ex situ samples were pooled into a single tube and analysed by MC. Cells were de-barcoded and single-cell counts were analysed in GraphPad Prism 7 (two-tailed unpaired t-test).
To demonstrate the reactivity of Maxpar barcodes to Matrigel (hence its incompatibility with organoid barcoding in situ), empty Matrigel droplets were seeded in 12-well plates (3 × 30 μL droplets/well), fixed with 4% PFA, washed ×2 with PBS, and incubated with PBS, Maxpar barcode #20 (106Pd, 108Pd, and 110Pd) resuspended in PBS or CSB for 60 mins at room temperature. The Matrigel droplets were then washed ×3 with CSB, ×3 with PBS, and dissolved in ice-cold Maxpar Water (Fluidigm 201069). Matrigel concentration was measured by BCA assay (Thermo 23225) and all samples were diluted to a protein concentration of 250 μg/mL prior to analysis on solution mode using a Helios Mass Cytometer (time per reading = 1 sec, settling time = 10 msec). The dual counts of 110Pd were measured and analysed in GraphPad Prism 7 (two-tailed unpaired t-test).
TOBis versus Maxpar Cell-Recovery Comparison
During optimisation we observed that organoids barcoded using the Maxpar Cell-ID™ 20-Plex Pd Barcoding Kit (Fluidigm 201060) had much smaller cell pellets than those processed by TOBis. We hypothesised this cell-loss was due to the increased dissociation and centrifugation steps required for Maxpar barcoding when compared to TOBis. To investigate this, we directly compared both TOBis and Maxpar barcoding recoveries across a range of organoid seeding densities.
Established small intestinal organoids (4-day-culture) were removed from Matrigel and reseeded in 24-well plates (1 × 50 μL Matrigel droplet/well) across 6 serial dilutions (100—3.125%) in duplicate to generate a dynamic range of organoid cell numbers. After recovering for 12 hrs in complete organoid culture medium, organoids were fixed in 4% PFA and washed in PBS (see above). One replicate of the organoids was barcoded in situ using TOBis and the other replicate was individually dissociated and barcoded ex situ using the Maxpar kit following the manufacturer’s instructions. Cells were stained with rare-earth metal antibodies, analysed by MC, and de-barcoded. Single-cell counts were analysed in GraphPad Prism 7 (two-tailed ratio-paired t-test).
To investigate organoid cell-type recovery between Maxpar and TOBis barcoding, equal cell numbers across replicates of each barcoding strategy were identically gated in UMAP space (see below) and the percentages of stem, Paneth, enteroendocrine, tuft, goblet cells, and enterocytes were calculated. Cell-type percentages were analysed in GraphPad Prism 7 (linear regression of correlation).
Small Intestinal Organoid Time-Course
Intestinal organoids were retrieved from Matrigel using ice-cold PBS, broken up mechanically by sequentially passing through a 23G, 5/8” needle (Terumo AN-2316R) 6 times and a 26G, 1/2” needle (HSW, 4710004512) 3 times to obtain a uniform cell suspension. Organoid fragments were 70 μm filtered twice and centrifuged at 200 ×g for 5 mins. The cell pellet enriched with single crypts was washed with cold PBS, collected using a benchtop centrifuge, and resuspended in Matrigel prior to seeding. The cell ratio seeded for Days 1—7 was 30: 9: 6: 5: 4: 3: 3 to ensure comparable organoid recovery and density from each time point. At each time point, the organoids were incubated with 25 μM 127IdU, protease/phosphatase inhibitors, and fixed in 4% PFA for 60 mins at 37 °C as described above. Organoids were washed with PBS and stored at 4 °C until samples from all time points were collected. All samples were stained with 250 nM 194Cisplatin, TOBis barcoded (Supplementary Table 3), stained, and analysed in one MC experiment (Supplementary Table 1, 50 parameters (40 antibodies)/cell).
Colonic Fibroblast Isolation, Immortalisation, and Cell Culture
Colonic fibroblasts were isolated as described by Khalil et al.47. Freshly dissected murine (C57BL/6, 6- to 8-week-old) colon tissue was flushed with ice-cold PBS, cut open, washed again in PBS, and incubated in 5 mM EDTA/PBS at 250 rpm, 37 °C for 15 mins. This process was repeated for a total of ×5 EDTA/PBS washes. Washed colon tissue was transferred to a fresh tube of sterile DMEM (Thermo 41966052) supplemented with 1 mg/mL Dispase II (Thermo 17105041) and 1 mg/mL Collagenase D (Sigma 11088858001). The colon/enzyme solution was incubated at 250 rpm, 37 °C for 30—60 mins (until the tissue started to look ‘stringy’). Digested colon tissue was then centrifuged at 200 ×g, 4 °C for 5 mins. Supernatant was discarded and the pellet was resuspended in 10 mL ACK Lysing Buffer (Thermo A1049201). Cells were centrifuged at 200 ×g, 4 °C for 5 mins, and the pellet was resuspended in DMEM + 10% FBS (Thermo 10082147). Cells were 100 μm filtered (Miltenyi 130-098-463) into a T75 flask and incubated at 5% CO2, 37 °C. After 3 hrs, cells were washed ×2 with PBS to remove debris. Adhered cells were cultured with DMEM + 10% FBS + 1× Insulin-Transferrin-Selenium (ITS-G) (Thermo 41400045). After 1 week of culture, fibroblasts were observed to proliferate, while other cell-types (e.g. epithelial cells and leukocytes) senesce and/or die. Colonic fibroblasts were immortalised using pBABE-HPV-E6 retrovirus produced in Phoenix-ECO cells (a kind gift from Prof. Erik Sahai, The Francis Crick Institute, London) and stably transfected with RFP using the pCMV-DsRed-Express plasmid with Lipofectamine 3000 (Thermo L3000001) to aid co-culture visualisation. Immortalised colonic fibroblasts were cultured in DMEM + 10% FBS + 1× ITS-G at 5% CO2, 37 °C. Cells were checked for mycoplasma infection monthly using the MycoAlert™ PLUS Mycoplasma Detection Kit (Lonza LT07-701) and remained negative throughout this project. IF staining confirmed that colonic fibroblasts were positive for intestinal mesenchymal markers such as Vimentin (D21H3, CST), Podoplanin (PDPN) (8.1.1, BioLegend), PDGFRɑ (APA5, Abcam), FOXL1 (ab95286, Abcam), and GLI-1 (C-1, Santa Cruz) in both 2D and 3D cultures.
Primary Macrophage Isolation and Cell Culture
Freshly dissected murine (C57BL/6, 10- to 12-week-old females) femurs and tibias (Charles River Laboratories) were flushed ×5 with 10 mL RPMI 1640 Medium (Thermo 11875093) + 10% FBS (Thermo 10082147). Cells were centrifuged at 300 ×g for 5 mins, resuspended in RPMI + 10% FBS, 40 μm filtered (Fisher 11587522), and centrifuged at 300 ×g for 5 mins. Supernatant was discarded and the pellet was resuspended in 2 mL ACK Lysing Buffer (Thermo A1049201) for 5 mins at room temperature. Monocytes were washed in PBS, centrifuged at 300 ×g for 5 mins, resuspended in 1 mL Recovery™ Cell Culture Freezing Medium (Thermo 12648010), and stored in liquid nitrogen until use. Bone marrow-derived macrophages were expanded and activated in RPMI + 10% FBS + 25% L929-cell conditioned media (LCCM) before experiments. IF staining confirmed that the cells were positive for intestinal macrophages markers such as CD45 (30-F11, BioLegend), CD68 (FA-11, BioLegend), CD11b (M1/70, BioLegend), F4/80 (BM8, BioLegend), and CX3CR1 (SA011F11, BioLegend) in both 2D and 3D cultures.
Heterocellular CRC Tumour Microenvironment (TME) Organoid Culture
Wild-type (WT) murine colonic organoids and CRC organoids carrying oncogenic mutations A, AK, and AKP were a kind gift from Prof. Lukas Dow (Cornell University) and cultured as described above. Following expansion in complete media, organoids were cultured in the absence of exogenous growth factors (mEGF, mNoggin, mR-Spondin-1 and mWnt-3a — WENR) for 8 hrs prior to the experiment. Colonic fibroblasts were cultured in DMEM supplemented with reduced FBS (2%) and 1× ITS-G for 24 hrs before the experiment. Primary bone marrows were differentiated into macrophages using RPMI + 10% FBS + 25% LCCM for 7 days before the experiment. To establish the CRC TME culture, organoids were passaged at a ratio of ~1:2.5; colonic fibroblasts were seeded at 6,000 cells/μL, 5,000 cells/μL and 4,000 cells/μL Matrigel for monoculture, 2-way co-cultures, and 3-way co-cultures respectively; primary macrophages were seeded at 9,000 cells/μL, 8,000 cells/μL and 7,000 cells/μL Matrigel for monoculture, 2-way co-cultures, and 3-way co-cultures respectively. Organoids, fibroblasts, and macrophages were mixed in Matrigel before seeding at 3 × 30 μL droplets per well in a 12-well plate. Each microenvironment culture was maintained in WENR-free advanced DMEM/F-12 (Thermo 12634010) supplemented with 2 mM L-Glutamine (Thermo 25030081), 1 mM N-Acetyl-L-Cysteine (Sigma A9165), 10 mM HEPES (Sigma H3375), 1× B-27 Supplement (Thermo 17504044), 1× N-2 Supplement (Thermo 17502048), 1× Insulin-Transferrin-Selenium-Sodium Pyruvate (ITS-A) (Thermo 51300044), and 1× HyClone™ Penicillin Streptomycin Solution (Fisher SV30010) for 48 hrs. All cultures were incubated with 25 μM 127IdU, protease/phosphatase inhibitors, and fixed in 4% PFA for 60 mins at 37 °C (as described above). Dead cells were stained with 250 nM 194Cisplatin as described above. Organoids were barcoded using TOBis (Supplementary Table 4), pooled into a single tube, dissociated into single cells, 70 μm filtered, and stained for MC analysis (Supplementary Table 2, 50 parameters (40 antibodies)/cell). Single organoid, fibroblast, and macrophage cells were analysed by MC.
Single-Cell Signalling Data Analysis
All single cells were gated for Gaussian parameters (Event length, Centre, Residual, and Width values), DNAhigh (191Ir and 193Ir), and Cisplatinlow (194/8Pt). For small intestinal organoids and colonic organoids, intact epithelial cells were gated with EpCAM+/Pan-CK+ and CEACAM1+/Pan-CK+ respectively. Intact colonic fibroblasts were gated with RFP+/PDPN+, and primary macrophages were gated with CD68+/F4/80+. Removal of cells stained positive for mutually exclusive cell-type/cell-state markers was performed as part of data pre-processing procedures (gating strategies incorporated in the publicly deposited datasets). Cells were then clustered and visualised in UMAP (Uniform Manifold Approximation and Projection)48 space and gated for cell-type and cell-state makers before proceeding to PTM signalling analysis (Supplementary Fig. 2).
UMAP analysis was performed with the Python package umap (https://umap-learn.readthedocs.io/en/latest) using default parameters unless otherwise specified (Supplementary Table 5). UMAP was used to visualise high-dimensional MC datasets in two-dimensional space, where cell gating was performed to identify cell populations or to remove residual outliers when required. All data was arcsinh transformed with a cofactor of 5.
Earth Mover’s Distance (EMD) was computed with the Python package scprep49 (https://github.com/KrishnaswamyLab/scprep) using default parameters. EMD scores were signed by the difference of the median intensity of a given parameter between the population of interest relative to the denominator (specified below) – positive for up-regulation or negative for down-regulation. Cell populations were manually gated and exported from Cytobank, with all channels to be analysed arcsinh transformed (cofactor = 5). For single-time-point small intestinal organoids (Fig. 2b, Supplementary Fig. 3), EMD was calculated between each cell-type/state and the entire epithelial cell population. For the small intestinal organoid time-course experiment (Fig. 4c), EMD was calculated between each cell-type from each time point against the combined population of all epithelial cells across all time points. For the CRC TME model (Figs. 5d, e, 6b and Supplementary Figs. 7, 8a—d), EMD was calculated between each cell-type in each condition and the combined population of all cell-types across all conditions.
k-Nearest Neighbours Density Resampled Estimation of Mutual Information (kNN-DREMI)21 was computed with the Python package scprep49 using default parameters. Cell populations were manually gated in UMAP space and exported from Cytobank, with all the channels to be analysed arcsinh transformed (cofactor = 5). For small intestinal organoids (Fig. 2b, Supplementary Fig. 3), kNN-DREMI scores among 28 PTMs (Supplementary Table 1) were calculated, which yielded a total of 756 PTM-PTM combinations for each cell-type/cell-state. Similarly, kNN-DREMI scores for 756 PTM-PTM pairs across 28 PTMs (Supplementary Table 2) were computed for each cell-type in the CRC TME model (Fig. 5f, Supplementary Figs. 7, 8a, b, e, and f). Heatmaps were generated using the R package RColorBrewer (https://cran.r-project.org/web/packages/RColorBrewer/) based on EMD and DREMI calculations. Signalling maps were compiled in OmniGraffle Professional from the heatmaps with the nodes (PTMs) coloured by EMD scores and edges (PTM-PTM pairs) by DREMI scores.
Principal Component Analysis (PCA) was performed on z-score normalised EMD (Fig. 2c, 5e, and Supplementary Fig. 8c, d) or DREMI (Fig. 5f, Fig. 8e, f) scores using the Scikit-Learn package PCA estimator in Python (https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html) with default parameters. All measurements used for PCA are listed in Supplementary Table 5.
Force-directed Scaffold Maps36 were constructed using the R package Scaffold (https://github.com/nolanlab/scaffold) with parameters specified below. Landmark populations were manually gated and exported from Cytobank with all data arcsinh transformed (cofactor = 5). For small intestinal organoid directed differentiation (Supplementary Fig. 1e), pRB [S807/S811]+ cells were used for the Scaffold analysis. UMAP-gated stem, Paneth, enteroendocrine, goblet, tuft cells, and enterocytes from untreated organoids were used as landmark nodes, and the untreated sample was used as the reference dataset. All measurements including cell-type/state markers and PTMs were used to generate the Scaffold maps (Supplementary Table 5). For the CRC TME model (Fig. 6a, Supplementary Fig. 9), epithelial cells from each condition were clustered by the measurements of all cell-type/state markers and PTMs (Supplementary Table 5). Microenvironment-focused Scaffold maps (Fig. 6a, Supplementary Fig. 9a) were generated using epithelial cells from WT organoid monoculture and WT organoids co-cultured with macrophages and/or fibroblasts as landmark nodes, and epithelial cells from the WT organoid/macrophage/fibroblast co-culture sample as the reference dataset. For the genotype-focused Scaffold maps (Supplementary Fig. 9b), epithelial cells from monocultures of WT, A, AK, AKP organoids were used as landmark nodes, and the WT sample was used as the reference dataset. Selected cell-type and cell-state markers were used to generate the Scaffold maps (Supplementary Table 5).
Supplementary Material
Acknowledgements
We are extremely grateful to L. Dow (Cornell University) for sharing CRC organoids, X. Lu (University of Oxford) for sharing murine intestines, and O. Ornatsky (Fluidigm) for providing monoisotopic Cisplatin (195Pt and 196Pt). We thank the UCL CI Flow-Core for MC support and L. McInnes for UMAP advice. Graphical organoid renderings were designed by J. Claus (Phospho Biomedical Animation). This work was supported by Cancer Research UK (C60693/A23783, CT), UCLH Biomedical Research Centre (BRC422, CT), The Royal Society (RSG\R1\180234, CT), and Rosetrees Trust (A1989, CT).
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
All raw data, processed data, and working illustrations are available as a Community Cytobank project (https://community.cytobank.org/cytobank/experiments#project-id=1271).
Author Contributions
X.Q. Designed the study, performed organoid and MC experiments, analysed the data, and wrote the paper.
J.S. Developed TOBis, designed rare-earth metal antibody panels, performed MC analysis, and analysed data.
P.V. Isolated and characterised colonic fibroblasts, macrophages, cultured organoids, and analysed data.
P.K. Performed UMAP, EMD, DREMI, and PCA data analysis.
M.N. Developed TeMal barcodes.
S.A. Provided murine monocytes and intestines for fibroblast isolation.
V.L. Provided murine small intestines for organoid isolation.
C.T. Designed the study, analysed the data, and wrote the paper.
Competing Interests
M.N. has pending intellectual property on the use of tellurium reagents for mass cytometry applications which has been licensed to Fluidigm Corporation.
References
- 1.Clevers H. Modeling Development and Disease with Organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 2.Pastula A, et al. Three-Dimensional Gastrointestinal Organoid Culture in Combination with Nerves or Fibroblasts: A Method to Characterize the Gastrointestinal Stem Cell Niche. Stem Cells Int. 2016;2016 doi: 10.1155/2016/3710836. 3710836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dijkstra KK, et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell. 2018;174:1586–1598 e1512. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 5.Pawson T, Scott JD. Protein phosphorylation in signaling--50 years and counting. Trends Biochem Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 7.Tape CJ, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. 2016;165:910–920. doi: 10.1016/j.cell.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons AJ, et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-alpha-induced apoptosis in vivo. Mol Syst Biol. 2015;11:835. doi: 10.15252/msb.20156282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tape CJ. Systems Biology Analysis of Heterocellular Signaling. Trends Biotechnol. 2016;34:627–637. doi: 10.1016/j.tibtech.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 14.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin X, et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rapsomaniki MA, et al. CellCycleTRACER accounts for cell cycle and volume in mass cytometry data. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03005-5. 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TH, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlova DY, et al. Earth Mover's Distance (EMD): A True Metric for Comparing Biomarker Expression Levels in Cell Populations. PLoS One. 2016;11:e0151859. doi: 10.1371/journal.pone.0151859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnaswamy S, et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science. 2014;346 doi: 10.1126/science.1250689. 1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Gehart H, Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol. 2019;16:19–34. doi: 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- 24.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis LM, et al. Tellurium-based mass cytometry barcode for live and fixed cells. Cytometry A. 2018;93:685–694. doi: 10.1002/cyto.a.23495. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy RL, Mak DH, Burks JK, Barton MC. Rapid monoisotopic cisplatin based barcoding for multiplexed mass cytometry. Sci Rep. 2017;7 doi: 10.1038/s41598-017-03610-2. 3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isella C, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 31.Calon A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 32.Lan J, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2018;79:145–158. doi: 10.1158/0008-5472.CAN-18-0014. [DOI] [PubMed] [Google Scholar]
- 33.Tape CJ. The Heterocellular Emergence of Colorectal Cancer. Trends Cancer. 2017;3:79–88. doi: 10.1016/j.trecan.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dow LE, et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell. 2015;161:1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Rourke KP, et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat Biotechnol. 2017;35:577–582. doi: 10.1038/nbt.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spitzer MH, et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science. 2015;349 doi: 10.1126/science.1259425. 1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz-Acuna R, et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol. 2017;19:1326–1335. doi: 10.1038/ncb3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neal JT, et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018;175:1972–1988 e1916. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machado L, et al. In Situ Fixation Redefines Quiescence and Early Activation of Skeletal Muscle Stem Cells. Cell Rep. 2017;21:1982–1993. doi: 10.1016/j.celrep.2017.10.080. [DOI] [PubMed] [Google Scholar]
- 42.Han G, Spitzer MH, Bendall SC, Fantl WJ, Nolan GP. Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat Protoc. 2018;13:2121–2148. doi: 10.1038/s41596-018-0016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han G, et al. Atomic mass tag of bismuth-209 for increasing the immunoassay multiplexing capacity of mass cytometry. Cytometry A. 2017;91:1150–1163. doi: 10.1002/cyto.a.23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevrier S, et al. Compensation of Signal Spillover in Suspension and Imaging Mass Cytometry. Cell Syst. 2018;6:612–620 e615. doi: 10.1016/j.cels.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khalil H, Nie W, Edwards RA, Yoo J. Isolation of primary myofibroblasts from mouse and human colon tissue. J Vis Exp. 2013 doi: 10.3791/50611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McInnes L, Healy J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 2018 [Google Scholar]
- 49.van Dijk D, et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell. 2018;174:716–729 e727. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.