Abstract
The mu opioid receptor gene, OPRM1, undergoes extensive alternative splicing, creating an array of splice variants that are conserved from rodent to human. Both mouse and human OPRM1 have five exon 5-associated 7 transmembrane full-length carboxyl terminal variants, MOR-1B1, MOR-1B2, MOR-1B3, MOR-1B4 and MOR-1B5, all of which are derived from alternative 3’ splicing from exon 3 to alternative sites within exon 5. The functional relevance of these exon 5-associated MOR-1Bs has been demonstrated in mu agonist-induced G protein coupling, adenylyl cyclase activity, receptor internalization and desensitization, and post-endocytic sorting, as well as region-specific expression at the mRNA level. In the present study we mapped a polyadenylation site for both mouse and human MOR-1Bs that defines the 3’-untranslated regions (3’-UTR) of MOR-1Bs and stabilizes mMOR-1Bs mRNAs. We identified a conserved miR378a-3p sequence in the 3’-UTR of both mouse and human MOR-1BS transcripts through which miR-378a-3p can regulate expression of MOR-1Bs at the mRNA level. Chronic morphine treatment significantly increased miR-378–3p level in Be(2)C cells and the brainstem of the morphine tolerant mice, contributing to the decreased expression of the mouse and human MOR-1B3 and MOR-1B4. Our study provides new insights into the role of miRNAs and Oprm1 splice variants in morphine tolerance.
Keywords: Morphine, OPRM1, splicing, opioid, microRNA
Introduction
The mu opioid receptor primarily mediates the actions of most clinically used opioids, including morphine and fentanyl, as well as heroin. A single copy of the mu opioid receptor gene, OPRM1, undergoes extensive alternative pre-mRNA splicing, generating an array of splice variants that are conserved from rodent to human (1, 2). These splice variants can be categorized into three structurally distinct classes: (1) full-length 7-transmembrane (7TM) carboxy- (C-) terminal variants; (2) truncated 6TM variants that lack the first TM; and (3) truncated single TM variants containing the first TM (1, 2). Increasing evidence suggest that these splice variants are functionally relevant in opioid pharmacology (1, 2).
All the full-length 7TM C-terminal variants have an identical receptor sequence encoded by exons 1/2/3, but differ from each other at the tip of the intracellular C-terminus encoded by alternative exons downstream of exon 3. Both mouse and human OPRM1 have five exon 5-associated 7TM C-terminal variants, MOR-1B1, MOR-1B2, MOR-1B3, MOR-1B4 and MOR-1B5 (2)(Fig. 1). All these exon 5-associated variants were generated through alternative 3’ splicing from exon 3 donor site to five alternative acceptor sites within exon 5 to create unique intracellular C-terminal tails encoded by exons 5a (MOR-1B1), 5b (MOR-1B2), 5c (MOR-1B3), 5d (MOR-1B4) and 5e (MOR-1B5). The mouse exon 5a in mMOR-1B1 encodes five amino acids that are identical to the first five amino acids from the human exon 5a except that the human exon 5a predicts additional 13 amino acids beyond five amino acids (3, 4). The predicted amino acids from exons 5b – 5e in MOR-1B2 - MOR-1B5 between mouse and human are quite different. However, the genomic location of exon 5 and splicing pattern are conserved from mouse to human. The 3’-untranslated regions (3’-UTR) and polyadenylation (poly(A)) sites of the mouse and human MOR-1Bs have not been reported. The functional relevance of these exon 5-associated MOR-1Bs has been demonstrated in mu agonist-induced G protein coupling (3, 4), adenylyl cyclase activity (4), receptor desensitization (5), internalization (5) and post-endocytic sorting (6), as well as region-specific or strain-specific expression at the mRNA level (3, 7, 8).
Fig. 1.
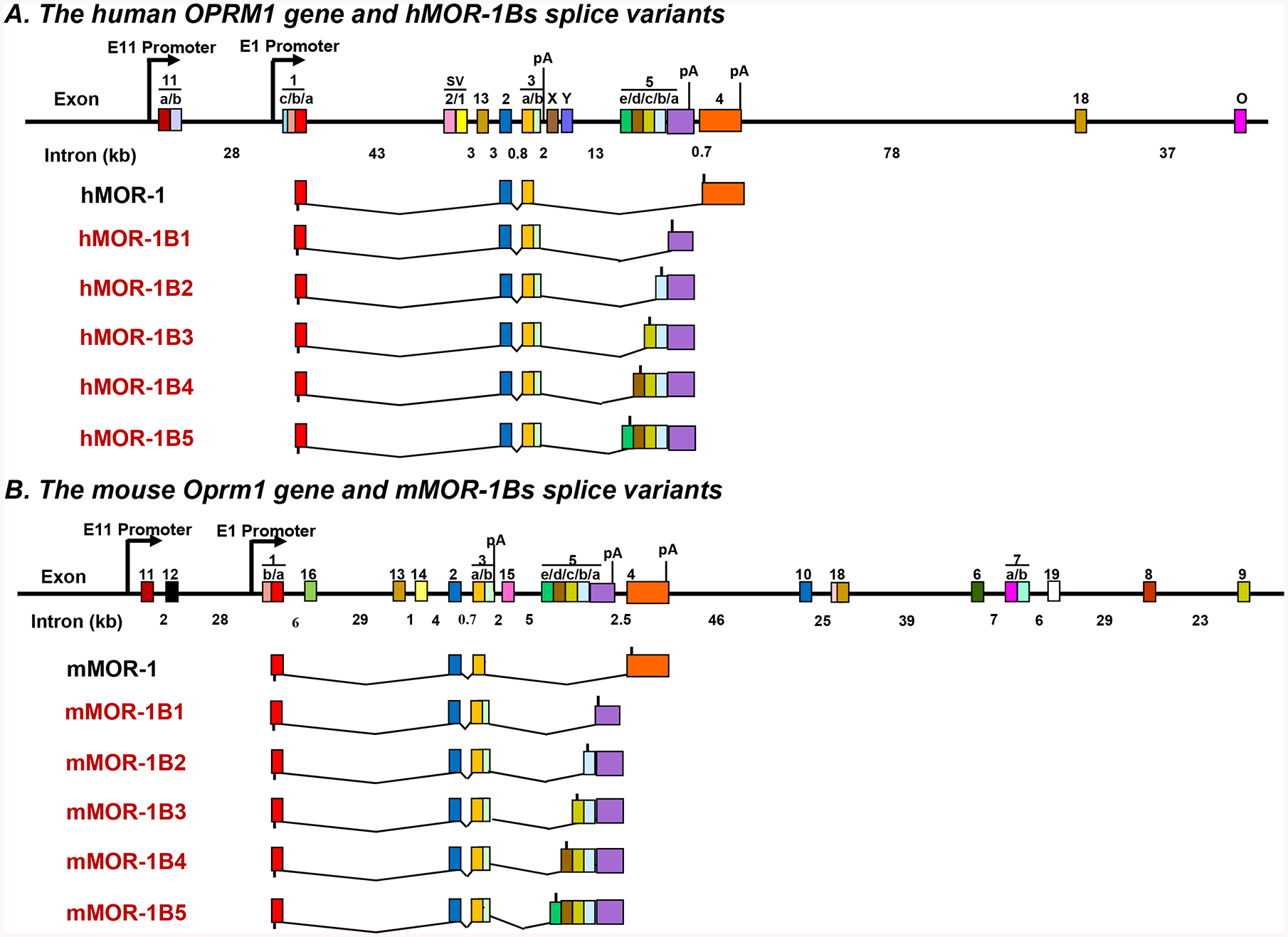
Schematic of OPRM1 gene structure and MOR-1B splice variants. MOR-1B variants from the human OPRM1 (A) and mouse (B) Oprm1 genes. Exons and introns are shown by colored boxes and black horizontal lines, respectively. Promoters are indicated by arrows. Exons are numbered in the order in which they were identified. Translation start and stop points are shown by bars below and above exon boxes, respectively. polyA sites (pA) for MOR-1A (13), MOR-1 (36, 37) and MOR-1Bs are indicated at the related exons.
miRNAs are small non-coding RNAs with 21–23 nucleotide in length that bind to complementary sequences of mRNAs, particularly located at the 3’-UTR, for degradating mRNA or inhibiting translation. Several miRNAs such as miR-23b, miR-339–3p and Let-7 regulated the expression of MOR-1 through their binding sites at the 3’-UTR of MOR-1 by destabilizing MOR-1 mRNA or suppressing MOR-1 translation (9–12). miR-103/107 modulated mouse and human MOR-1A expression through a conserved miR-103/107 binding site at the 3’-UTR of MOR-1As (13). Mu opioids including morphine and fentanyl had significant impact on the expression of a number of miRNAs, contributing to their effect on MOR-1 or MOR-1A expression. miRNA has also been associated with morphine tolerance and addiction in animal models (12–19). In the present study, we mapped a poly(A) signal sequence that defines the 3-UTRs of mouse and human MOR-1Bs and identified a conserved miR-378a-3p binding site at the 3-UTRs through which miR-378a-3p regulated expression of mouse and human MOR-1Bs at the mRNA level. We further demonstrated that chronic morphine treatment upregulated miR-378a-3p in Be(2)C cells and the brainstem of morphine tolerance mice, leading to downregulate the expression of human and mouse MOR-B3 and MOR-1B4 mRNA.
Materials and methods
Animals
C57BL/6J (B6) (stock#: 000664) mice were obtained from Jackson Laboratory. Adult (10 – 16 weeks-old) male mice were used in all experiments. All mice were housed in groups of five, maintained on a 12-h light/dark cycle and given ad libitum access to food and water. All animal studies were approved by the Institutional Animal Care and Use Committee of the Memorial Sloan-Kettering Cancer Center.
Poly(A)-rich RNA isolation, 3’-Rapid Amplification of cDNA Ends (RACE) and DNA sequencing.
Total RNA was extracted from the brain of C57BL/6J mice and Be(2)C cells, a human neuroblastoma line, with TRI Reagent following the manufacturer’s instruction (Molecular Research Center, Cincinnati, OH). Poly(A) RNA was isolated from total RNA using MicroPoly(A) Purist Kit following the manufacturer’s instruction (Ambion, Austin, TX). The 3-RACE was performed using 3’-RACE System for rapid amplification of cDNA ends Kit (Clontech) as described previously (13). In brief, the first-strand cDNA was reverse-transcribed (RT) from poly(A) RNA by SuperScript® III (Invitrogen, Carlsbad, CA) with a 3’-RACE-PCR primer (5’- CCA TCC TAA TAC GAC TCA CTA TAG GGC TCG AGC GGC TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT VN-3’), and used in nested polymerase chain reactions (PCRs). In the first-round PCR, an AP1 antisense primer (5’-TAG CTT AAC TCG GAA CTG AGT-3’) and several gene-specific sense primers (GSPn) derived from downstream of mouse or human exon 5 were used. The primers for mouse poly (A) signal identification were: mGSP1 (5’-GCA ACC GCT TGT GGG AGA ACA TTC-3’), mGSP3 (5’-GAC AAG AAG TCA AGA AAC CAG CAG AAC CAC-3’) and mGSP5 (5’-CTG AAT TCA GTT GGC AAT CAT CCA CTG TAA GC-3’); or for human poly (A) signal identification: hGSP1 (5’-CTG AGG AGT TCA GAG ATA TGG GTT CAG TGT C-3’), hGSP3 (5’-CTT ATG GTA TGC ATG GAT TGC TCC ACC-3’), hGSP5 (5’-CAC GTA GTA CTG CAA CAG CCC AGT AAA CAT G-3’) and hGSP7 (5’-GGG AGT GGT GTC TTT ATC CTA GAC-3’). The first round PCR product was then used in the second round or nested PCR with an AP2 antisense primer(5’-ACT CAC TAT AGG GCT CGA GCG GC-3’) and several gene-specific sense primers (GSPn+1) derived from downstream of mouse or human exon 5 were used. The primers for mouse poly (A) signal identification were: mGSP2 (5’-CAA TGG TTG TTG AAT GGC ATC TCT TCA TG-3’), mGSP4 (5’-GTA CAG TAT CAA GCT TAA CTG TGC ACT AC-3’) and mGSP6(5’-CA G TAG AAA AGG GGA CAG TGG TGC TTA GC-3’); or for human poly(A) identification: hGSP2(5’-GTC TCA AAA TGC CGT GGT CCT CAT GTG-3’), hGSP4 (5’-GAT ATT AAT CCA AAC AAA ACA GGA ACA GTT GGC-3’), hGSP6 (5’-GAG AAA AAT ACA TGG CCA AAA TCA AAA ATG G-3’) and hGSP8 (5’-GCC TCT AGC CTG GGT GAC AGA ACG AG-3’). PCR were performed with Platinum Taq DNA polymerase under conditions consisting of a 2-minute denaturing at 94°C and 25 cycles (in the first-round PCR) or 30 cycles in the second-round PCR) of amplification, each cycle containing a denaturing step at 94°C for 20 seconds, an annealing step at 652103 for 30 seconds, and an extension step at 72°C for 5 minutes followed by a 8-minute extension at 72°C. PCR products from the second PCRs were separated in a 1.5% agarose gel, extracted by using gel extraction with a gel extraction kit (Qiagen, Valenica, CA) and sequenced.
Reverse-transcription-polymerase chain reaction (RT-PCR).
Total RNAs extracted from Be(2)C cells and mouse brain were reverse-transcribed with Superscript® III and random hexamer. The first-strand cDNA was then used at template in PCRs using Platinum Taq DNA polymerase. We used an overlapping PCR strategy to determine if the poly(A) sites are associated with the MOR-1Bs 3’-UTRs. Based on the genomic sequences between exon 5a and the mapped poly(A)s, as well as the sequences beyond poly(A) sites, we designed five pairs of primers for human MOR-1Bs and four pairs of primers for mouse MOR-1Bs. The five pairs of primers with the predicted sizes of PCR fragments for human were: 1) hSF1 (5’-GAT TTA TTT CAA AAG TCA TCT TTA CTC AAC-3’) and hRF1 (5’-CTT TGA GTC CTT GCA ATA ATT GGC AAT GGC GCC-3’) with 30002 bp; 2) hSF2 (5’-GCC CTC TTT ACC TAG GGG ACT AAC AAA GG-3’) and hRF2 (5’-GCG GCT CCC AGG GCT GAA TGT GTA ATT GG-3’) with 1747 bp; 3) hSF3 (5’-GTG CAT GCA TCC TTC TAT CCC TCT CTA C-3’) and hRF3 (5’-CAG GCA TGA GGC ACC ACG CCC AAC TGA AGA AG-3’) with 1399 bp; 4) hSF4 (5’-GAC TTA AGA AAA TCT TCT TCA GTT GGG CGT GGT GCC-3’) and hRF4 (5’-CCC TGA CAA ATT ACA AGG TCT AAC TGT CC-3’) with 1434 bp; and 5) hSF5 (5’-GAT CAG AGT CAG AGG ACA GTT AGA CCT TG-3’) and hRF5(5’-CAG GTA AGA AAA GAG GAC TGT GG-3’) with 427 bp. The four pairs of primers with the predicted sizes of PCR fragments for mouse were: 1) mSF1 (5’-GAA GCA CAC CAA AGA TAT TTT GTT ACC ATA TGG-3’) and mRF1 (5’-GCC ACT GGG TGA CTG TGG CCA TCT GGC-3’) with 3260 bp; 2) mSF2 (5’-GAC AAG AAG TCA AGA AAC CAG CAG AAC CAC-3’) and mRF2 (5’-GAG AGA GAC TCC AGG GAG ACA CCT CCT CC-3’) with 2880 bp; 3) mSF3 (5’-CCT GCT CCT TAG GTG CAT ATA TTT GAA CTC TTG G-3’) and mRF3 (5’-CAG TCC AGA CAT CAT GCG AGG AGG CTC ATG C-3’) with 1330 bp; and 4) mSF4 (5’-CCT GAT CTT CCA AGT TCA GCA AGA AAG GC-3’ and mRF4 (5’-GTG CTG AGA CTT TGG CTT TGG ATC TGT G-3’) WITH 1466 bp. PCR products were separated in a 1.0% agarose gel, purified from the gel and sequenced. The overlapping PCRs were performed under conditions consisting of a 2-minute denaturing at 94°C and 39 cycles of amplification, each cycle containing a denaturing step at 94°C for 20 seconds, an annealing step at 65°C for 30 seconds, and an extension step at 72°C for 3 minutes followed by an 8-minute extension at 72°C.
Plasmid construction.
To generate the construct containing mMOR-1B poly(A) signal and hMOR-1B poly(A) signal in pmirGLO Dual-Luciferase miRNA Target Expression (pmir) Vector (Promega, Madison, WI), we first made a no poly(A) pmir vector (pNo-p(A)) by deleting the SV40 late poly(A) signal sequence in the pmir vector using Chang-IT™ Multiple Mutation Site Directed Mutagenesis kit (USB, Cleveland, Ohio) with a mutagenesis primer (5’-CAT AAC CCC TTG GGG CGG CCG CTT CGA GCG GCC GGC CTA TCC CGG GAA ATC GAA TTT TAA CAA AAT ATT AAC GC-3’). Then we cloned a 351 bp of mMOR-1B partial 3’-UTR PCR fragment containing the mouse poly(A) or a 743 bp of hMOR-1B partial 3’-UTR PCR fragment containing the human poly(A) into NheI/XhoI sites of pNo-p(A) vector, to generate pM-p(A) and pH-p(A) constructs, respectively. Primers used in PCRs for generating the 351 bp and 743 bp fragments were: the sense primer (5’-GGC GCT AGC AGA GCA ATT ACT CTA AGA CAG ACA AGC ATC ATT TC-3’) and the antisense primer (5’-CCG CTC GAG CTT ACA CCG ACA ATG TGC TTA TTT TAG GGC-3’) for mouse, and the sense primer (5’-GGC GCT AGC CAC TTT GTG CCA CTT TGA GAA GAA TTT CAA C-3’) and the antisense primer (5’-CCG CTC GAG GGA GAT GTT TGT ATA GGA AGC GTA AAA GGG-3’) for human.
To generate the constructs with the partial mouse or human 3’-UTR containing miR-378a-3p sequence in the pmir vector, a 359 bp (mouse) or 340 bp (human) of the partial 3’-UTR sequences were amplified by PCRs and cloned into NheI/XhoI sites of the pmir vector containing the original SV40 poly(A) as pM-wt or pH-wt, respectively. The primers used for PCRs were: the sense primer (5’-GGC GCT AGC GCA ACC GCT TGT GGG AGA ACA TTC-3’) and antisense primer (5’-CCG CTC GAG GTT CTA AAT TTG GTC AAG AAG GAG TGG-3’) for mouse, and the sense primer (5’-GGC GCT AGC CTT AGC AGG CGT GGT GGC GGG CAC C-3’) and antisense primer (5’-CCG CTC GAG CAT ATG CTT TCT TCT TCT GCT GTG AAT ATG GAC-3’) for human. The wild-type miR-378a-3p sequences in pM-wt and pH-wt constructs were mutagenized using Chang-IT Mutagenesis kit with a mutagenesis primer (5’-ATT TCT GAG TTC GGC CAA CTC CTG GTC TAC AGA GTG AGT TCT G-3’) for mouse, or a mutagenesis primer (5’-GTA TTA TAT TGG ACA AAA TAG TCT TGA ATT CAC AGC AGA AGA AAG CAT ATG-3’) for human, to generate pM-mut and pH-mut, respectively, in which only seed sequences were mutated.
To observe the effect of the identified mouse and human poly(A) signal sequences on expression of mMOR-1B1 and hMOR-1B1 cDNAs, we made mMOR-1B1 without a poly(A) (mB1-No pA) or hMOR-1B1 without a poly(A) (hB1-No pA) constructs by deleting bovine growth hormone (BGH) poly(A) in pcDNA3 constructs containing mMOR-1B1 or hMOR-1B1 previously established (3, 4) using Chang-IT Mutagenesis kit with the mutation primer (5-CGT TTA AAC CCG CTG ATC AGC CTC GGC TTC TGA GGC GGA AAG AAC CAG CTG-3). We then made mMOR-1B1 with the partial 3’-UTR containing the mouse poly(A) (mB1-mpA) or hMOR-1B1 with the partial 3’-UTR containing the human poly(A) (hB1-hpA) construct in pcDNA3 by inserting the partial 3’UTR sequences into mB1-No pA or hB1-No pA constructs using In-Fusion HD Cloning System (Clontech). The 3’-UTR sequences were amplified by PCRs using the sense primer (5’-GCT GGT GCC AAC CCC CTC TAA GAC AGA CAA GCA TCA TTT CCT AGC TC-3’) and antisense primer(5’-GCC CTC TAG ACT CGA GCT TAC ACC GAC AAT GTG CTT ATT TTA GGG C-3’) for mouse, and the sense primer (5’-GTG GAT CCG AGC TCG AAA ATA GAT TTA TTT TGA AAA GGC ATA TAC ACA GAA CTG-3’) and antisense primer (5’-TTA AAC TTA AGC TTG TTA ATT AAG GCG CGC CCT TAC ACC GAC AAT GTG CTT ATT TTA GGG C-3’) for human.
To examine the effect of miR378a-3p on expression of mMOR-1B1 cDNA, we inserted the same 359 bp of the mouse partial sequence containing miR378a-3p that was used in pmir construct into the 3’ end of mMOR-1B1 to generate the pmB1-wt construct using In-Fusion HD Cloning System. The partial sequence was amplified with the sense primer (5’-TTA AAC TTA AGC TTG TTA ATT AAG GCG CGC CGCAAC CGC TTG TGG GAG AAC ATT C-3’) and antisense primer (5’-TTA AAC TTA AGC TTG TTA ATT AAG GCG CGC CGT ACA CAT GCA TGT AGA ACT CAG AGG AC-3’). Similarly, we made the phB1-wt construct by inserting the same 340 bp of the human partial sequence containing miR378a-3p that was used in pmir construct into the 3’ end of hMOR-1B1. The partial sequence was amplified with the sense primer (5’-GTG GAT CCG AGC TCG AAA ATA GAT TTA TTT TGC TTA GCA GGC GTG GTG GCG GGC ACC-3’) and antisense primer (5’-TTA AAC TTA AGC TTG TTA ATT AAG GCG CAT ATG CTT TCT TCT GCT GTG AAT ATG GAC-3’). The wild-type miR-378a-3p sequences in pmB1-wt and the phB1-wt constructs were mutagenized by using the same mutagenesis primers and method described above to generate pmB1-mut and phB1-mut.
Cell Culture, transfection, morphine Treatment, miR378a-3p inhibitor and luciferase Assay.
HEK293T and Be(2)C cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and nonessential amino acids, and minimum Eagle’s medium supplemented with 10% fetal bovine serum and nonessential amino acids, respectively, in an atmosphere of 5% CO2 at 37°C. Plasmid constructs were transfected into HEK293T cells plated in 48- or 6-well plates using the Effectene reagent (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Antisense locked nucleic acid (LNA) oligonucleotide against miR378a-3p inhibitor (5’-CCT TCT GAC TCC AAG TCC A-3’, Exiqon, Vedbæk, Denmark) with or without plasmid constructs was transfected with Lipofactamine 2000 (Life Technologies) into HEK293T cells following the manufacturer’s protocol. A negative control LNA oligonucleotide (5’-GTG TAA CAC GTC TAT ACG CCC A-3’, Exiqon) was used as a control. After 48 hours of transfection, the cells were washed with phosphate-buffered saline and lysed with lysis buffer provided from Dual-Glo Luciferase Assay Kit (Promega). Cleared lysate was used to determine the luciferase activities by using the Dual-Glo Luciferase Assay Reagents in TD-20/20 luminometer (Promega). Luciferase activity was calculated by normalizing with Renilla luciferase activity obtained from the same assay. NeuroMag reagent (OZ Bioscience, Marseille, France) was used to transfect miR-378a-3p inhibitor into Be(2)C cells as previously described (13). Be(2)C cells were treated with 3 μM of morphine for 48 hours in the presence or absence of control LNA oligo (5 nM) or miR-378a-3p inhibitor (5 nM). Total RNAs were then extracted using miRNeasy kit (Qiagen) and used for RT-qPCR.
Chronic morphine treatment.
Chronic morphine treatment with morphine pellet (75 mg of free base) and placebo pellet (a gift from the Research Technology Branch of the National Institute on Drug Abuse, Rockville, MD) was performed as previously described (20). Briefly, the pellets were subcutaneously implanted into the back of mice under oxygen/isoflurane inhalant anesthesia. Analgesia was measured by the radiant heat tail-flick assay routinely performed in our lab. All mice implanted with a morphine pellet showed robust morphine tolerance after the 3-day implantation, whereas mice with the placebo pellet did not develop morphine tolerance (data not shown). On the fourth day, the mice were sacrificed, and brain regions were dissected for total RNA extraction.
Quantification of mouse and human MOR-1Bs mRNA miR-378a-3p by quantitative polymerase chain reaction (RT-qPCR).
Dissection of brain regions including the prefrontal cortex (Pfc), striatum (Str), thalamus (Tha) and brain stem (BS) of C57BL/6J mice and following total RNA isolation were performed as described previously (7). Total RNA isolation from Be(2) cells is described above. RNA concentration was determined by using a Qubit RNA assay in a Qubit 2.0 Fluorometer (Life Technologies), and reverse transcribed with Superscript II reverse transcriptase (Invitrogen) and random hexmers. The first-strand cDNA was then used as templates in SYBR qPCRs with Hot Start-IT SYBR Green qPCR Master Mix (Affymetrix) in a MJ Opticon 2q PCR machine (MJ Research, Waltham,MA). The following primers were used: sense primer A (5’-GTA TCC CAA CCT CTT CCA ACA TTG AGC −3’) and antisense primer A (5’-CTT TTG AAA TAA ATC TAT TTT CTG ATG ATT AGT TCT ATC-3’) for hMOR-1B1; sense primer B (5’-CAA AAT ACA GGC AAG GTT CCA TAG ATT G −3’) and antisense primer B (5’-TCT GTC TTC TTT CTC TCT GAT GAT TAG TTC TAT C −3’) for hMOR-1B2; sense primer B and antisense primer C (5’-CTT GGC TGG AGG TCC CTG ATG ATT AGT TCT ATC −3’) for hMOR-1B3; sense primer B and antisense primer D (5’-TCA TGT CAT AGT CAG CTC TGA TGA TTA GTT CTA TC −3’) for hMOR-1B4; sense primer B and antisense primer E (5’-CCA GGT TCA ATT CCA CCT GAT GAT TAG TTC TAT C-3’) for hMOR-1B5; sense primer C (5’-CAC AAA ATA CAG GCA GGG GTC CA-3’) and antisense primer F (5’-CAA AAT AAA TCT ATT TTC TGG TGG TTA GTT C-3’) for mMOR-1B1; sense primer C and antisense primer G (5’-CAC ATT AAA AGC TTC TGG TGG TTA GTT C-3’) for mMOR-1B2; sense primer D (5’-GCA CTG ATC ACG ATT CCA GAA ACC AC-3’) and antisense primer H (5’-GTG TGA GGC TTG TCT GGT GGT TAG TTC-3’) for mMOR-1B3: sense primer D and antisense primer I (5’-GTT TTT GGT GTG CCT GGT GGT TAG TTC-3’) for mMOR-1B4; sense primer C and antisense primer J (5’- GCA CTC ATA CAC ACT GGT GGT TAG TTC-3’) for mMOR-1B5; sense primer (5’-ACC ACA GTC CAT GCC ATC AC-3’) and antisense primer (5’-TCC ACC ACC CTG TTG CTG TA-3’) for for Glyceraldehyde 3-phosphate dehydrogenase (G3PDH) used as reference gene. The qPCRs were performed by initial 2 minutes at 95°C, followed by 45 cycles, each cycle consisting of 95°C for 15 seconds, 65°C for 15 seconds, and 72°C for 30 – 60 seconds. Expression levels of MOR-1BS were calculated as 2−△C(t), where △C(t)= C(t)luc2 or MOR-1BS- C(t) G3PDH).
To quantify miR-378a-3p, total RNAs were extracted with miRNeasy Kit from cells or brain regions were reverse-transcribed with a University cDNA Synthesis Kit (Exiqon). The cDNA was then used as template in SYBR green qPCR with SYBR green master mix and miR-378a-3p LNA primer set (Exiqon). U6 small nuclear RNA(snRNA) was amplified as a reference gene by using U6 snRNA LNA primer set (Exiqon) for normalization. Expression level of miR-378a-3p was calculated as 2−△C(t), where △C(t)= C(t) miR-378a-3p - C(t) U6).
Receptor binding assays.
The cells membranes were prepared from transfected HEK293T cells as described previously (21, 22). Saturation and competition binding assays were performed with 125I-3’-iodobenzoyl-6β-naltrexamide (IBNtxA)(21, 22) at 25°C for 120 min in 50 mM potassium phosphate buffer, pH 7.4, containing 5 mM magnesium sulfate. Specific binding was defined as the difference between total binding and non-specific binding, determined in the presence of 10μM levallorphan. Protein concentrations were determined by Lowry assay as previously described using bovine serum albumin as the standard.
Statistical analysis
Data are represented as the mean ± SE from at least three independent experiments. All statistical analyses were conducted using Prism 7.0 (GraphPad, San Diego, CA, USA). Comparisons of different groups were carried out by two-tailed Student t-test or one-way ANOVA with Bonferroni’s post hoc analysis for multiple comparisons or two-way ANAOVA with Fisher’s LSD’s post hoc analysis for multiple comparisons. Statistical significance was set at p < 0.05.
Results
Identification of polyadenylation (poly(A)) signal sequences in the 3’-UTR of mouse and human MOR-1BS mRNAs
Alternative 3’ splicing within exon 5 generates five MOR-1Bs, MOR-1B1, MOR-1B2, MOR-1B3, MOR-1B4 and MOR-1B5, in both mouse and human OPRM1 genes. To identify the poly(A) signal sequence for the mouse MOR-1BS, we used a 3’-RACE strategy with poly(A)-rich RNA from mouse brain (Fig. 2A). Three sets of mouse primers designed from downstream sequences of mouse exon 5a were used in the nested PCRs to identify the actual poly(A) signal sequence. Only one set of the primers in the nested PCR (mGSP5/AP1 followed by mGSP6/AP2) produced a cDNA fragment about 500 bp (Fig. 2B), while the other two sets of the primers (mGSP1/AP1 or mGSP3/AP1 followed by mGSP2/AP2 or mGSP4/AP2) did not yield any PCR product (data not shown). The sequence analysis of the fragment showed a consensus poly(A) signal sequence AAUAAA followed a 19 bp of U-rich sequence and a conserved CA dinucleotide cleavage site where the poly(A) tail was added (Fig. 2C), which is consistent with a consensus poly(A) signal sequence (23, 24). The cleavage site is located at 5,937 bp downstream of the stop codon in exon 5a for mMOR-1B1.
Fig. 2.
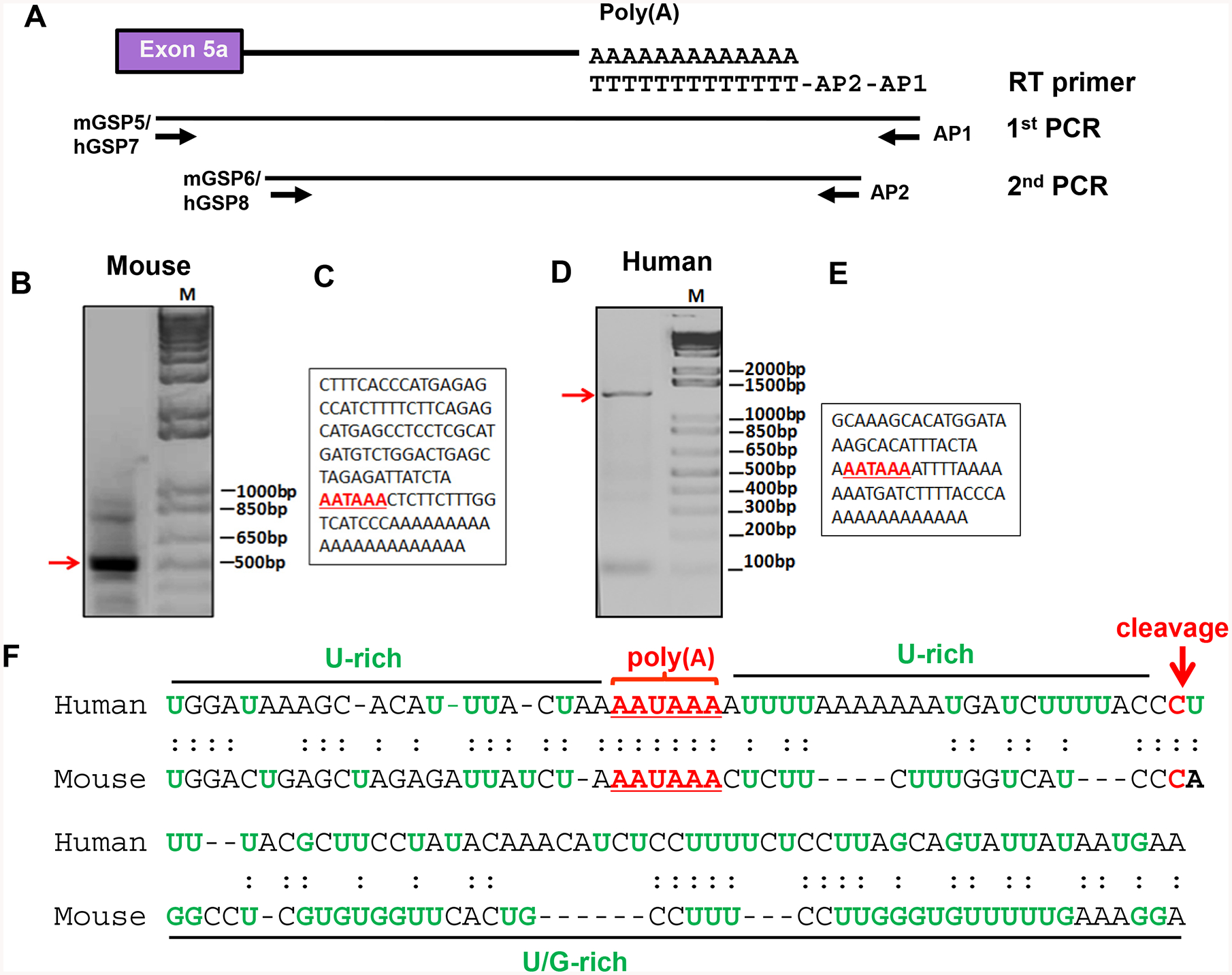
Identification of the poly(A) signal sequences of hMOR-1Bs and mMOR-1Bs. A. Schematic of the 3’-RACE strategy. The 3’-RACE was performed as described in Materials and Methods. Primers used for RT, 1st and 2nd PCRs are indicated.
B & D. Analysis of the 2nd PCR products. The 2nd PCR product of the mouse (B) or the human 3’-RACE (D) were separated on 1.5% agarose gel and stained with ethidium bromide. The gel was imaged with ChemiDoc MP system.
C & E. Partial cDNA sequences of the 2nd PCR fragments purified from mouse (B) or human (D). Poly(A) sequences are indicated by underlined red bold letters. M: 1 kb plus DNA marker.
F. Alignment of the poly(A) signal and adjacent sequences of the mouse and human MOR-1Bs. The poly(A) sequences and cleavage sites are indicated by red letter and U-rich or U/G-rich sequences and region shown by green letter and black line, respectively.
A similar 3’-RACE strategy was used to identify the poly(A) signal sequence in the 3’-UTR of human MOR-1Bs using human neuroblastoma Be(2)C cells that endogenously express all human MOR-1Bs. Among the four sets of the primers, only one set of the primers in the nested PCR (hGSP7/AP1 followed by hGSP8/AP2) yielded a 1.4 kb of cDNA fragment (Fig. 2D) that contained a consensus poly(A) signal sequence AAUAAA followed a 26 bp of U-rich sequence and a CU dinucleotide cleavage site where the poly(A) tail was added (Fig. 2E). The cleavage site is located at 7,554 bp downstream of the stop codon in exon 5a for hMOR-1B1. The other three sets of primers (hGSP1/AP1 or hGSP3/AP1 or hGSP5/AP1 followed by hGSP2/AP2 or hGSP4/AP2 or hGSP6/AP2) failed to produce any PCR product (data not shown).
We then used an overlapping PCR approach to validate whether the identified poly(A) sequences are associated with the 3’-UTR of the mouse and human MOR-1Bs transcripts. Three sets of primers for mouse and four sets of primers for human were designed based on the sequences between exon 5a and the identified poly(A) sites and used in PCR to produce three overlapping cDNA fragments for mouse (Figs. 3A & 3C) and four overlapping cDNA fragments for human (Fig3. 3B & 3D), all of which had expected sizes. The sequences of these fragments were validated by sequencing (Data not shown). We also designed one set of primers across the mapped poly(A) sites with the antisense primers located at downstream of the poly(A) sites. No PCR products were amplified in the same mouse or human RT samples (Figs. 3C & 3D). Together, these results suggest that the mapped poly(A) sites are associated with the 3’-UTR of the mouse and human MOR-1Bs transcripts.
Fig. 3.
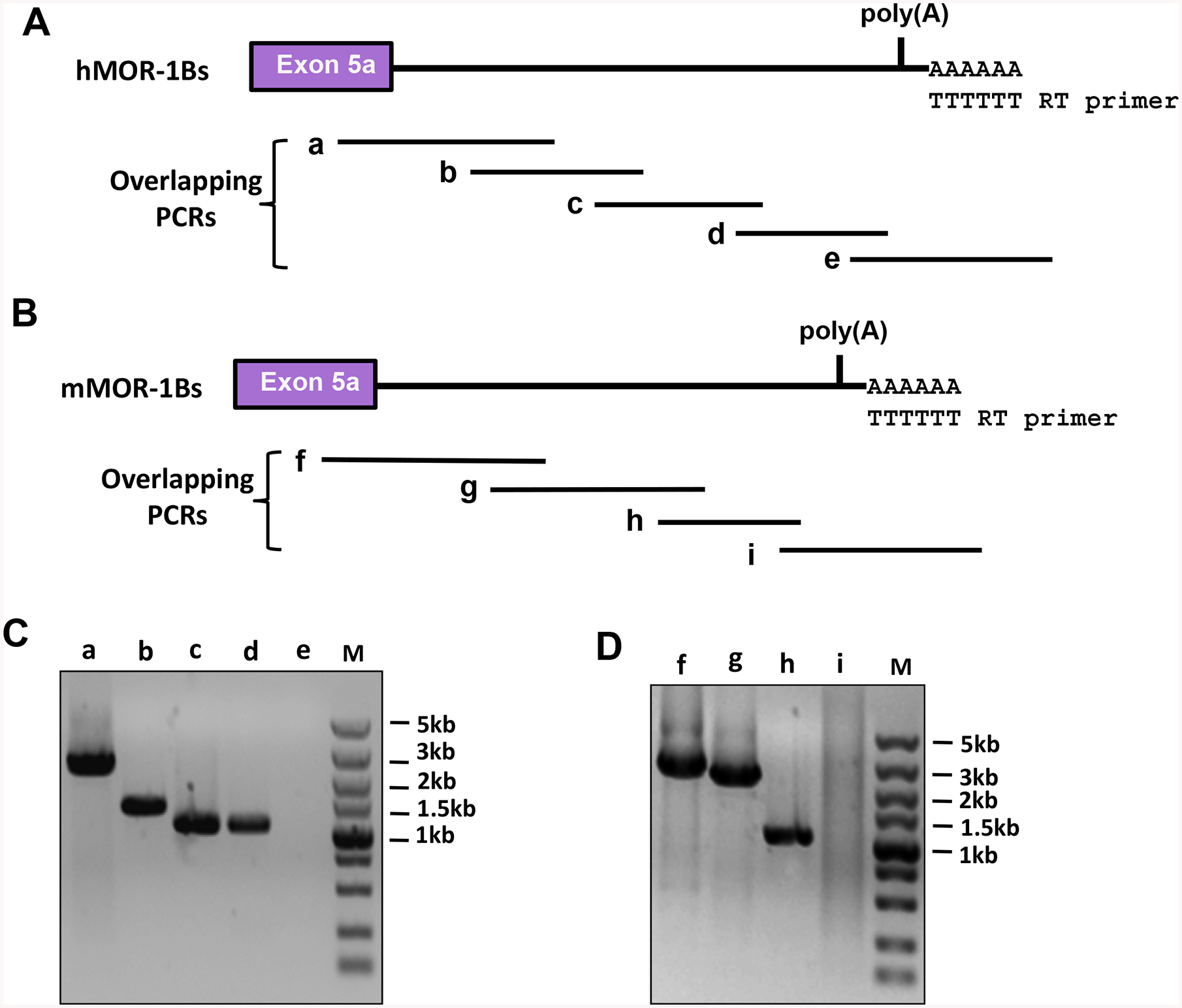
Identification of the 3’-UTR of the mouse and human MOR-1Bs using overlapping PCRs. A). Human MOR-1Bs 3’-UTR. Overlapping PCRs were performed using five pairs of primers and the first-stand cDNAs reverse-transcribed from poly(A) RNAs of Be(2)C cells to amplify overlapped PCR fragments covering the 3’-UTR, as described in Materials and Methods. a, b, c, d and e are the predicted overlapping PCR fragments.
B). Mouse MOR-1Bs 3’-UTR. Overlapping PCRs were performed using four pairs of primers and the first-stand cDNAs reverse-transcribed from total RNAs of mouse brain to amplify overlapped PCR fragments covering the 3’-UTR, as described in Materials and Methods. f, g. h and I are the predicted overlapping PCR fragments.
C & D). Analysis of the PCR products. The PCR products from Be(2)C (C) and mouse brain (D) were separated on 1% agarose gels that were stained with ethidium bromide and imaged with ChemiDoc MP System. The sizes of a – d and f – h PCR fragments were consistent with the predicted sizes (See Materials and Methods) and confirmed by DNA sequencing. PCR e and i did not yield any PCR bands.
Characterization of the mapped poly(A) signal sequences
Poly(A) sequence is crucial for mRNA stability. We then examined the role of a partial 3’-UTR containing the mapped poly(A) signal sequence in mRNA stability using a luciferase reporter assay in HEK293T cells. We made the luciferase (luc2) constructs with or without the partial MOR-1Bs 3’-UTR containing the mapped poly(A) in pmir vector (Figs. 4A). There was no apparent luciferase activity when transfected with the construct without the 3’-UTR or poly(A) signal sequence (pNo-p(A))(Fig. 4B). However, significant luciferase activities were observed when transfected constructs included either the mouse or the human 3’-UTR (pM-p(A) and pH-p(A)) at the 3’-end of luciferase (Fig. 4B), suggesting that the poly(A) signal sequences are functional in maintaining stability of luciferase activity, most likely at the mRNA level.
Fig. 4.
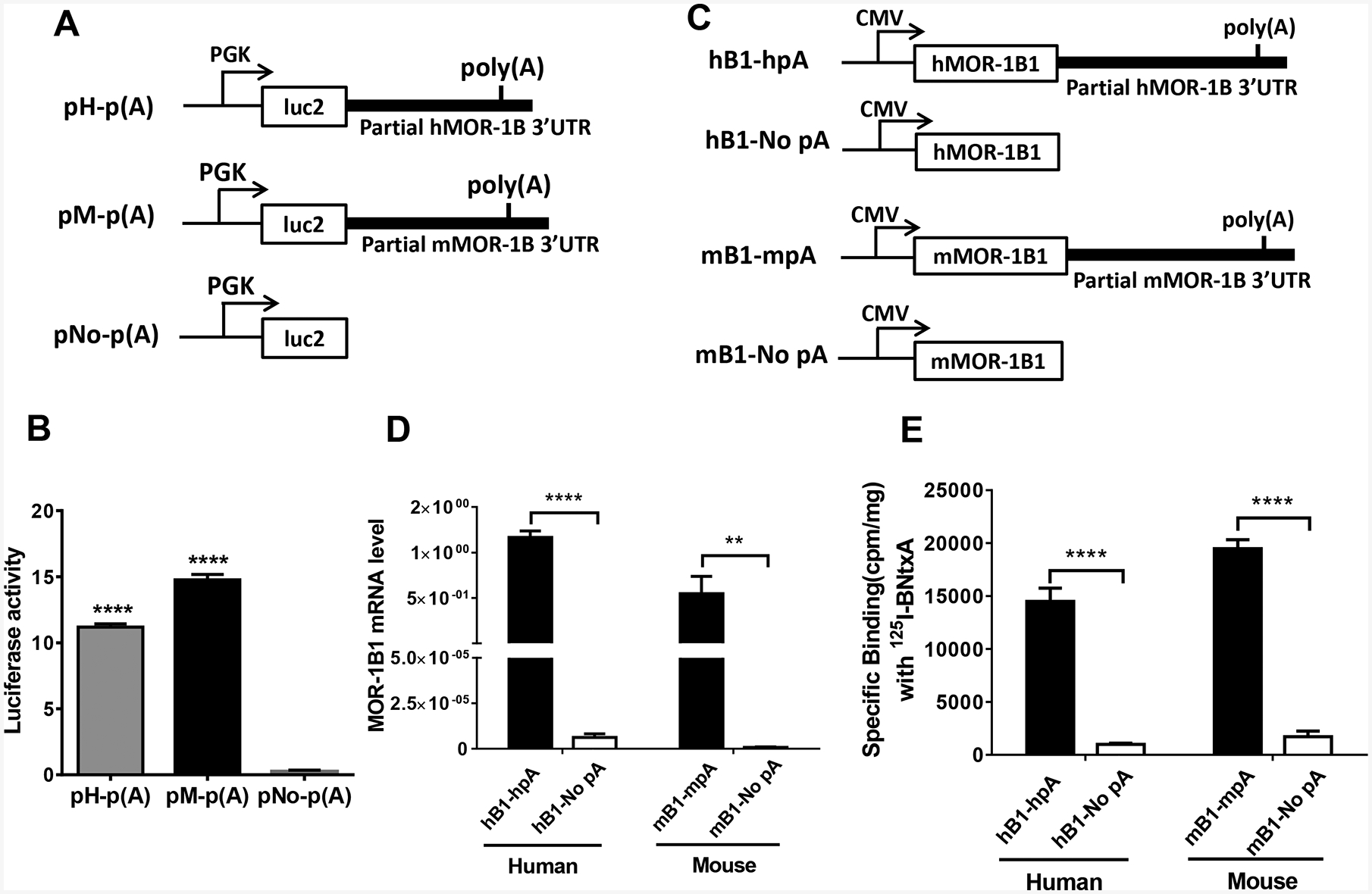
Role of the 3’-UTR of hMOR-1B and mMOR-1B in luciferase activity and MOR-1B1 expression. A). Schematic of the constructs for luciferase assay. The partial 3’-UTR of hMOR-1B and mMOR-1B containing poly(A), cleavage site and U/G-rich region was subcloned downstream of the firefly luciferase (luc2) coding region in pmir vector, as pH-p(A) and pM-p(A), respectively, as described in Materials and Methods. PGK promoter is indicated by arrows.
B). Luciferase activity. The lysates from transfected HEK293T cells with the indicated constructs (n = 4) were used for analyzing luciferase activity by using Dual-Glo Luciferase Assay, as described in Materials and Methods. Luciferase activity was calculated by normalizing with Renilla luciferase activity. Statistically significant differences were calculated by one-way ANOVA with Bonferroni’s post hoc analysis. ****: p < 0.0001 compared with pNo-p(A).
C). Schematic of the MOR-1B1 constructs. The partial 3’-UTR of hMOR-1Bs and mMOR-1Bs containing poly(A), cleavage site and U/G-rich region was subcloned downstream of hMOR-1B1 and mMOR-1B1 coding region in pcDNA3 vector, as hB1-hpA and as mB1-mpA, respectively. hB1-No pA and mB1-No pA were used as controls. CMV promoter is shown by arrows.
D). Expression of MOR-1B1 mRNAs. Total RNAs from transfected HEK293 cells with the indicated constructs (n = 4) were used in RT-qPCR to determine the expression of MOR-1B1 mRNA, as described in Materials and Methods. Two-way ANOVA with Bonferroni’s post hoc analysis was used. ****: p < 0.0001. **: p < 0.01.
E). Opioid receptor binding. Membrane proteins from transfected HEK293 cells with the indicated constructs (n = 4) were used in 125I-IBNtxA binding assay, as described in Materials and Methods. Two-way ANOVA with Bonferroni’s post hoc analysis was used. ****: p < 0.0001.
We further examined the role of the partial 3’-UTR containing the mapped poly(A)s in stability of mMOR-1B1 and hMOR-1B1 mRNA in HEK293T cells using RT-qPCR. We made MOR-1B1 constructs with or without the partial 3’-UTR sequences in pcDNA3 vector (Figs. 4C). Again, little mRNA expression was observed in either mMOR-1B1 or hMOR-1B1 construct without a poly(A) signal sequence (mB1-No pA and hB1-No pA)(Fig. 4D), while the constructs including the corresponding 3’-UTR containing the poly(A) signal sequence showed robust expression of mMOR-1B1 or hMOR-1B1 mRNA (Fig. 4D). Opioid receptor binding with 125I-IBNtxA using the same transfected cells revealed similar results to those of mRNA expression profiles (Fig. 4E), suggesting that the partial 3’-UTR with the mapped the poly(A) signal sequences can stabilize the mMOR-1B1 and hMOR-1B1 mRNAs and subsequently lead to the increased expression of mMOR-1B1 and hMOR-1B1 receptor proteins.
Reduced expression of MOR-1Bs by miR-378a-3p via its consensus Binding Site at MOR-1BS 3’-UTR.
To identify potential miRNA targets in the 3’-UTR of the hMOR-1Bs and mMOR-1Bs, we searched the sequences using several computer programs including RegRNA (http://regrna2.mbc.nctu.edu.tw/) and miRbase (http://www.mirbase.org) and identified a conserved miR-378a-3p binding site in the 3’-UTR of both mMOR-1Bs and hMOR-1Bs with a 6-mer-seed match (Fig.5A). We also found additional putative miRNA sites, most of which are not conserved (Supplemental Fig. S1). The current study focusses on this conserved miR-378a-3p site. The predicted miR-378a-3p binding site is located at 622 bp and 322 bp downstream of the stop codon in exon 5a for hMOR-1B1 and mMOR-1B1, respectively.
Fig. 5.
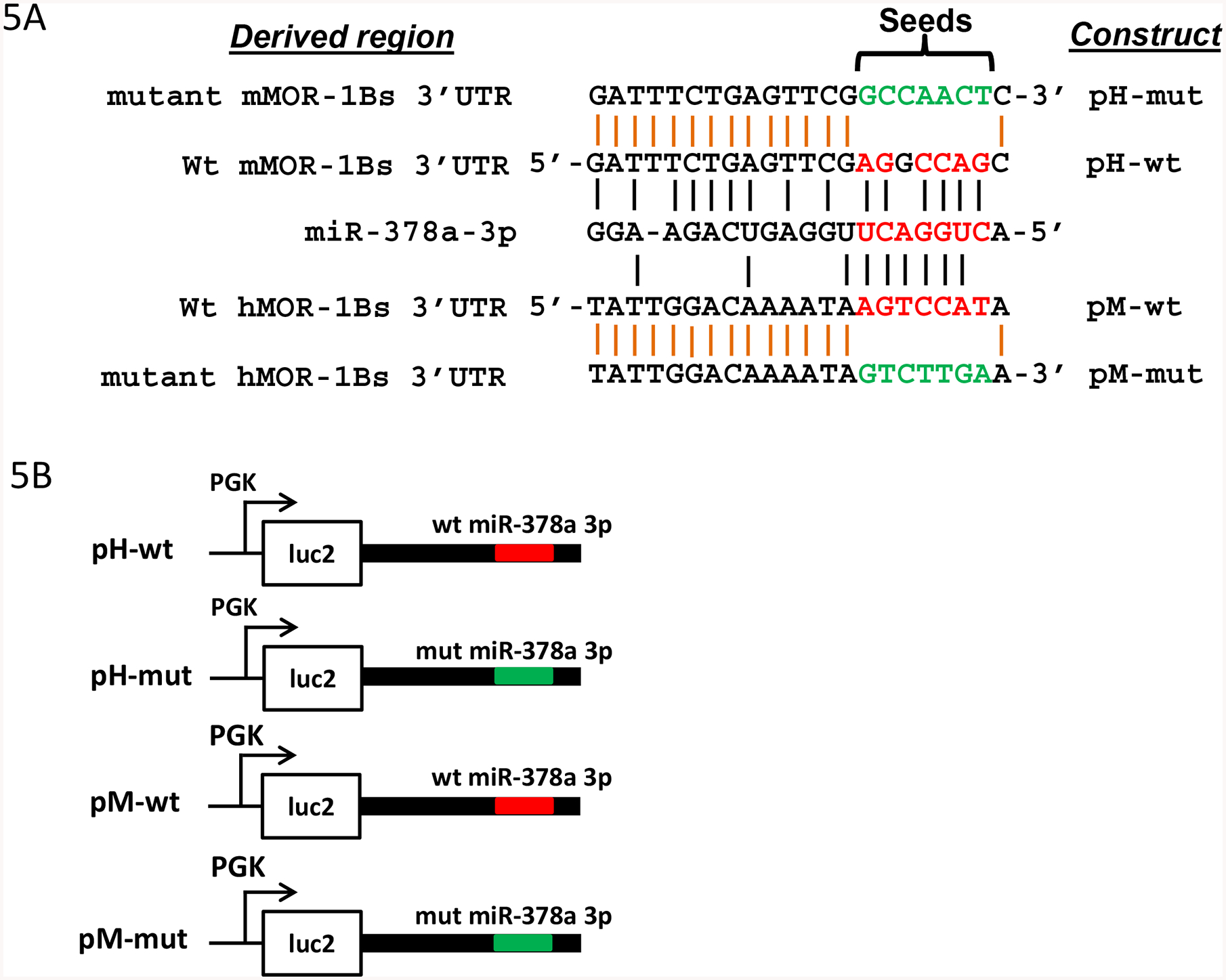
Regulation of luciferase activity by miR-378a-3p through a conserved miR-378a-3p binding site in MOR-1Bs 3’-UTRs. A). Alignment of miR-378a-3p sequences with 3’-UTR of hMOR-1Bs and mMOR-1Bs and mutagenized sequences of the miR-378a-3p sites. Red letter: the miR-378a-3p seeds and aligned 3’-UTR sequences. Green letter: mutagenized sequence. Black vertical lines show the complementary alignment and orange vertical lines indicate identical alignment.
B). Constructs in pmir vector. The 3’-UTRs of hMOR-1Bs and mMOR-1Bs containing wild-type (wt) or mutated (mut) miR-378a-3p sequences in A were subcloned into pmir as pH-wt, pM-wt, pH-mut, and pM-mut, respectively, as described in Materials and Methods. The wt and mut miR-378a-3p sequences are indicated by red and green lines, respectively.
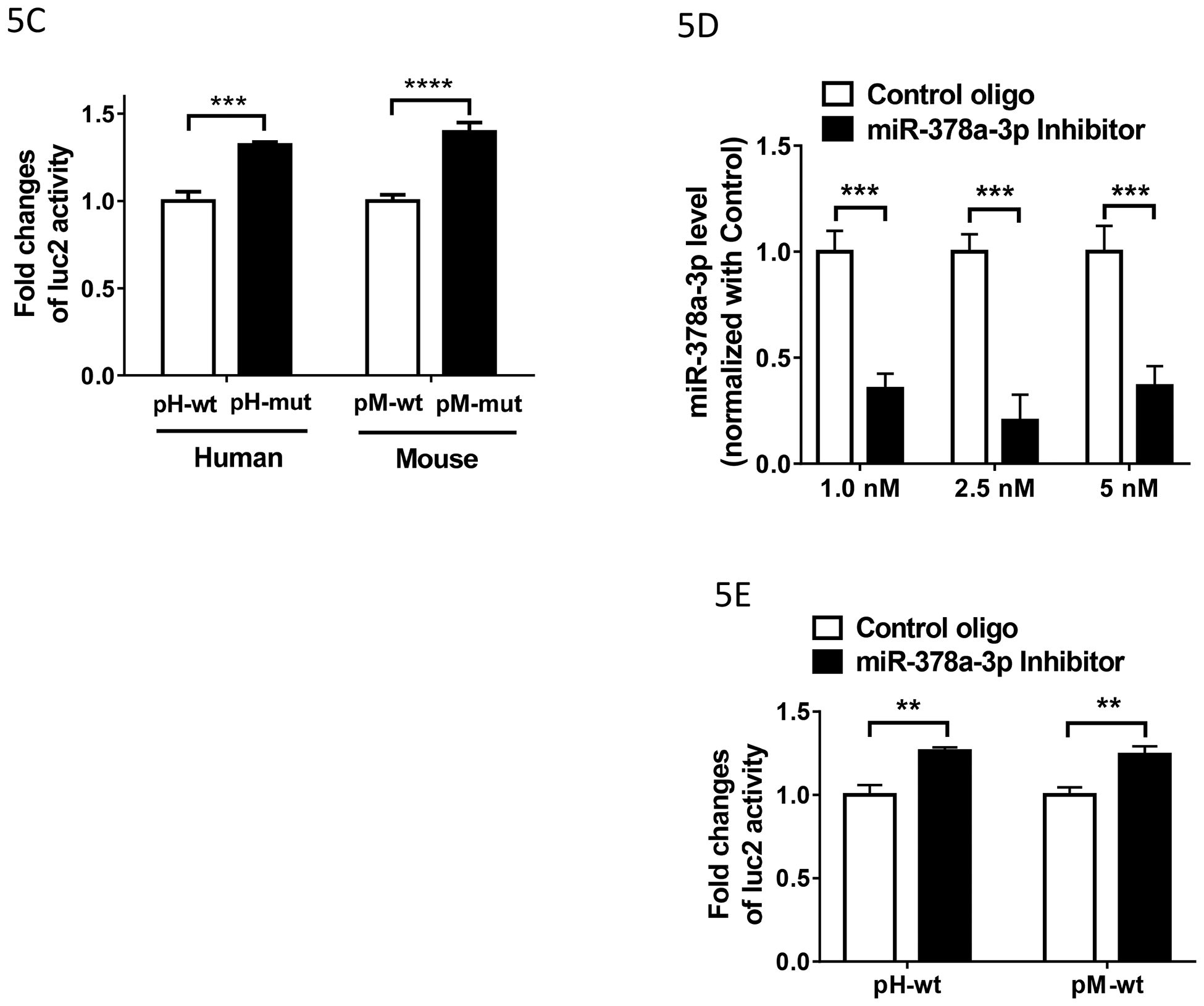
C). Effect of the miR-378a-3p mutation on the luciferase (luc2) activity in HEK293Tcells. Luciferase activity was measured using the lysate from HEK293 cells transfected with indicated constructs (n = 4), as described in Materials and Methods. Fold change of luc2 activity was calculated by normalizing the values of the mutant (mut) constructs with those of the wild-type (wt) constructs. Two ANOVA with Bonferroni’s post hoc analysis was used. ***: p < 0.001; ****: p < 0.0001.
D). Effect of miR-378a-3p inhibitor on the expression of miR-378a-3p in HEK293T cells. Total RNAs including small RNAs were isolated using miRNeasy Kit from HEK293 cells transfected with indicated concentration of Control oligo or miR-378a-3p Inhibitor (n = 3–6) and used in RT-qPCR to determine miR-378a-3p level, as described in Materials and Methods. Inhibition level by the LNA inhibitor was calculated by normalizing the levels with those of Control LNA oligo. Two-way ANOVA with Bonferroni’s post hoc analysis was used. ***: p < 0.001.
E). Effect of miR-378a-3p inhibitor on the luciferase (luc2) activity in HEK293T cells. Luciferase activity was measured using the lysate from HEK293 cells transfected with indicated constructs (n = 4). Fold change of luc2 activity was calculated by normalizing the values of the Inhibitor with those of the Control oligo. Two ANOVA with Bonferroni’s post hoc analysis was used. **: p < 0.01.
To examine whether the predicted miR-378a-3p binding sites in the 3’-UTRs can be actually targeted by miR-378a-3p, we first used a mutagenesis approach in a luciferase reporter assay. The wild-type and mutant constructs were made by subcloning the partial 3’-UTRs containing the wild-type miR-378a-3p (pH-wt and pM-wt) and mutagenized miR-378a-3p seed sequences into downstream of luciferase (pH-mut and pM-mut) and transfected into HEK293 cells (Fig. 5B). The miR-378a-3p mutation significantly increased the luciferase activities of both the human (pH-mut, ~33%) and mouse construct (pM-mut, ~40%)(Fig.5C), suggesting that these predicted miR-378a-3p seeds sites act as a repressive cis-element, likely mediated through miR-378a-3p expressed in HEK293T cells.
We next downregulate miR-378a-3p in HEK293T cells with an antisense LNA oligo (miR-378a-3p inhibitor) and investigate the effect of the downregulation on luciferase activities with the wild-type construct. We initially performed an optimization study with different concentrations of the miR-378a-3p inhibitor. The miR-378a-3p inhibitor decreased ~ 65% of miR-378a-3p at 1 nM and ~ 80% at 2.5 nM, compared to control oligo (Fig.5D). 5 nM of the inhibitor had ~ 63% of decrease. We thus used 2.5 nM of miR-378a-3p inhibitor in cotransfection studies together with the human or mouse wild-type construct in HEK293T cells. Downregulating miR-378a-3p with the inhibitor significantly enhanced luciferase activity of both the human (pH-wt, ~26%) and mouse constructs (pM-wt, 24%) (Fig.5E), compared to the control oligo. These results were consistent with the increased luciferase activities in the mutant constructs (Fig.5C), suggesting that the luciferase activity was regulated by the interplay between the predicted miR-378a-3p sites and miR-378a-3p.
To further evaluate the function of miR-378a-3p and the predicted miR-378a-3p binding sites in the context of the mouse and human MOR-1BS, we made another set of wide-type and mutant constructs by subcloning the same partial 3’-UTRs containing the wild-type and mutant miR-378a-3p as those for luciferase assay (Fig. 5B) into downstream of the human MOR-1B1 (phB1-wt and phB1-mut) or mouse MOR-1B1 (pmB1-wt and pmB1-mut) in pcDNA3 vector (Fig. 6A). When transfected into HEK293T cells, the mutant human (phB1-mut) and mouse MOR-1B1 construct (pmB1-mut) increased [125I]-IBNtxA binding over 58% and 46%, respectively (Fig. 6B), compared with the wild-type constructs (phB1-wt and pmB1-wt), consistent with the results from the luciferase constructs (Fig. 5C). Together, these data indicate that miR-378a-3p functions as a repressor to regulate luciferase activity and MOR-1B1 cDNA expression through the predicted miR-378a-3p binding sites in the mMOR-1BS and hMOR-1BS 3’-UTRs.
Fig. 6.
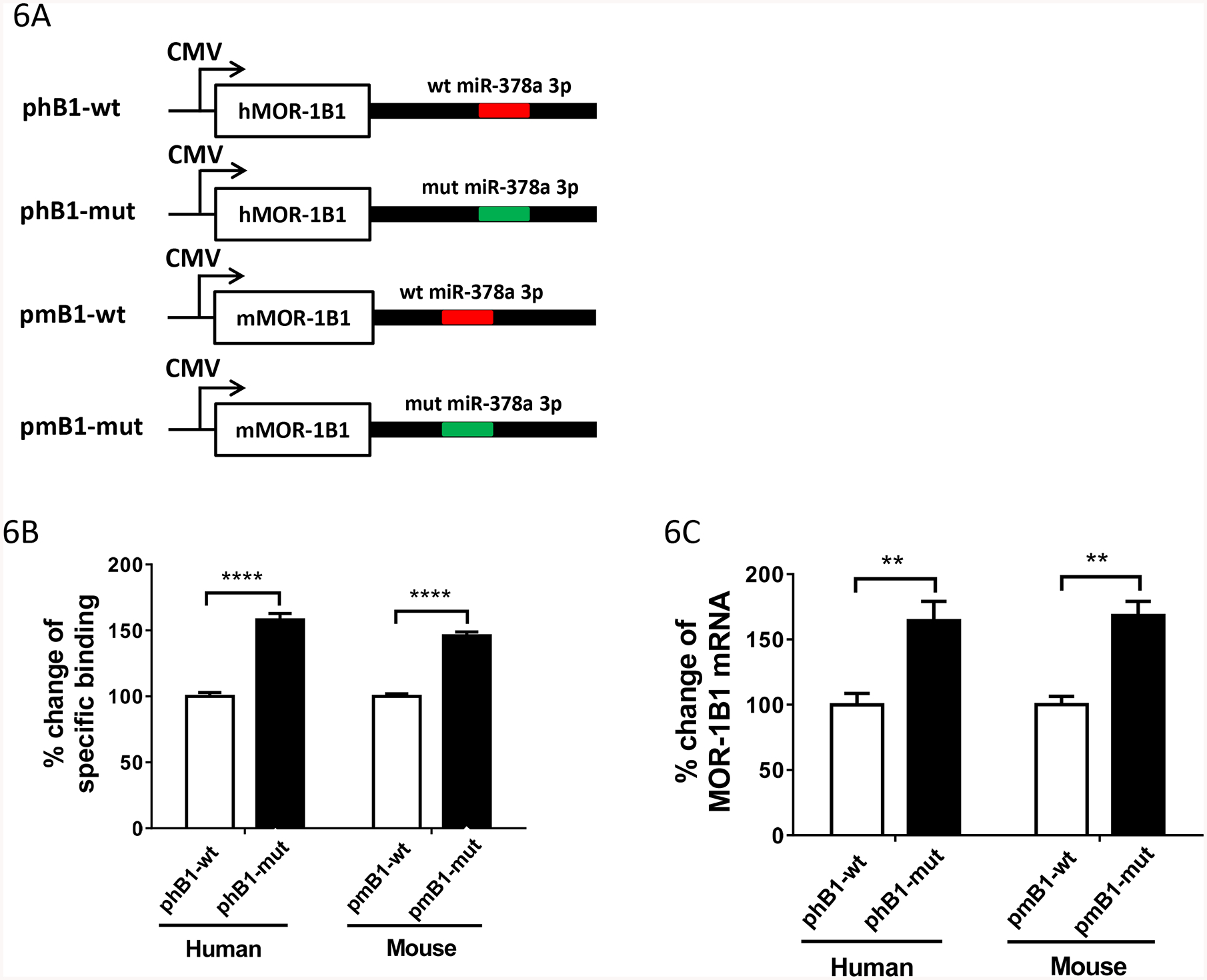
Regulation of MOR-1Bs expression by miR-378a-3p through a conserved miR-378a-3p binding site in MOR-1Bs 3’-UTR A). Constructs in pcDNA3 vector. hMOR-1B1 and mMOR-1B1 cDNAs with its partial 3’-UTRs containing wild-type (wt) or mutated (mut) miR-378a-3p sequences were subcloned into pcDNA3 vector, as described in Materials and Methods. The wt and mut miR-378a-3p sequences are indicated by red and green lines, respectively.
B). Opioid receptor binding. Membrane proteins from HEK293 cells transfected with the indicated constructs (n = 4) were used in 125I-IBNtxA binding assay, as described in Materials and Methods. Fold change of the specific binding was calculated by normalizing the values of the phB1-mut or pmB1-mut with those of the phB1-wt or pmB1-wt. Two ANOVA with Bonferroni’s post hoc analysis was used. ****: p < 0.0001.
C). Expression of MOR-1B1 mRNAs. Total RNAs from HEK293 cells transfected with indicated constructs (n = 4) were used in RT-qPCRs, as described in Materials and Methods. Percentage change of MOR-1B1 mRNA was calculated by normalizing the values of the phB1-mut or pmB1-mut with those of the phB1-wt or pmB1-wt. Two ANOVA with Bonferroni’s post hoc analysis was used. **: p < 0.01.
We then examined the effect of miR-378a-3p on the expression of the human and mouse MOR-1B1 cDNAs at the mRNA level in the same HEK293T cells transfected with the constructs with wild-type or mutant miR-378a-3p site by RT-qPCR. The human or mouse MOR-1B1 mRNA level were significantly increased in the mutant constructs (phB1-mut and pmB1-mut) by ~64% or ~68% as compared with the wild-type constructs (phB1-wt and pmB1-wt) (Fig.6C), respectively, suggesting that miR-378a-3p mainly inhibits the MOR-1B1 expression at the mRNA level. We also examined the effect of miR-378a-3p on the expression of the human and mouse MOR-1B3 and MOR-1B4 cDNAs at the mRNA level in HEK293T cells transfected with the wild-type or mutant constructs. We observed similar effect of the wild-type or mutant miR-378a-3p site on expression of MOR-1B3 and MOR-1B4 cDNAs to those seen in MOR-1B1 constructs (Supplemental Fig. S2), suggesting a common role of miR-378a-3p in the context of this in vitro system.
Upregulation of miR-378a-3p in Be(2)C cells and mouse brainstem by chronic morphine treatment.
To investigate whether morphine can regulate miR-378a-3p expression, we examined the effect of chronic morphine treatment on miR-378a-3p expression in Be(2)C Cells. Morphine (3 μM, 48 hours) markedly increased miR-378a-3p by over two-fold compared to PBS control (Fig.1A).
Fig.7.
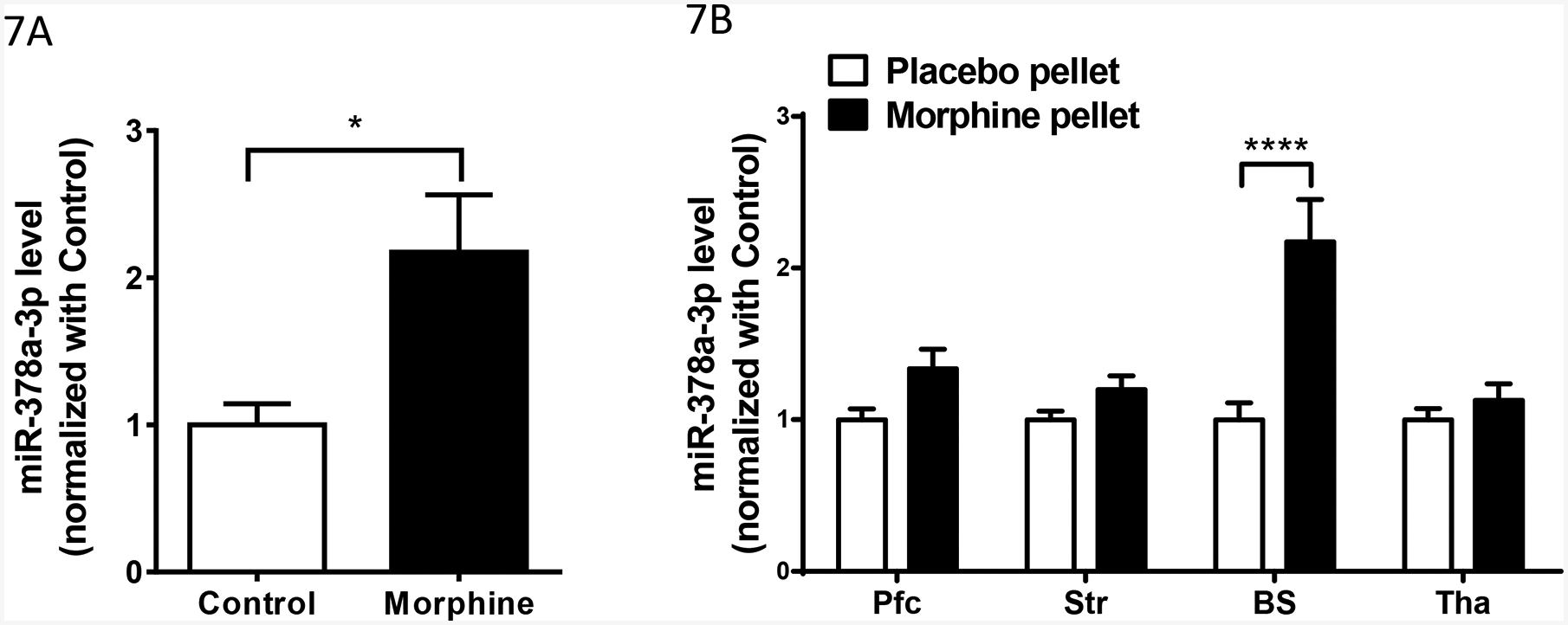
Effect of chronic morphine on miR-378a-3p expression in Be(2)C cells and brain regions of morphine tolerant mice. A). Effect of morphine on miR-378a-3p expression in Be(2)C cells. Total RNAs from Be(2)C cells treated with morphine (3 μM for 48 hours) or without morphine (Control) (n = 6) were used in RT-qPCR to determine miR-378a-3p expression, as described in Materials and Methods. Two-tailed Student t-test was used. *: p < 0.05.
B). Effect of morphine on miR-378a-3p expression in brain regions of the morphine tolerant mice. Total RNAs from the prefrontal cortex (Pfc), striatum (Str), brainstem (BS) and thalamus (Tha) of morphine tolerant mice (implanted morphine pellet (75 mg) for 3 days)(n = 10), and control mice (implanted placebo pellet for 3 days) (n = 10) were used in RT-qPCR to determine miR-378a-3p expression. Two ANOVA with Bonferroni’s post hoc analysis was used. ****: p < 0.0001.
We also examined miR-378 a-3p expression in several brain regions of a morphine tolerant mouse model. Subcutaneous implantation of the morphine pellet (75mg) for three days that produced robust morphine tolerance (data not shown) did not significantly alter miR-378a-3p expression in the PFC, striatum and thalamus, when compared to placebo pellet controls, although there was an increase trend in these regions (Fig.1B). However, the morphine pellet markedly increased miR-378a-3p level in the brainstem (Fig.1B).
Downregulation of MOR-1Bs mRNA expression by morphine via miR-378a-3p in Be(2)C cells and the mouse brainstem.
We next examined the effect of morphine on expression of endogenous human MOR-1BS mRNAs in Be(2)C cells using RT-qPCR. Morphine (3 μM, 48 hours) did not significantly alter the expression levels of hMOR-1B1, hMOR-1B2 and hMOR-1B5. However, morphine significantly reduced the expression of hMOR-1B3 and hMOR-1B4 (Fig, 8A), suggesting that morphine suppresses the expression of hMOR-1B3 and hMOR-1B4 at the mRNA level. We then downregulated miR-378a-3p in Be(2)C cells with the miR-378a-3p inhibitor to investigate whether the effect of morphine on hMOR-1B3 and hMOR-1B4 expression was mediated through miR-378a-3p.
Fig. 8.
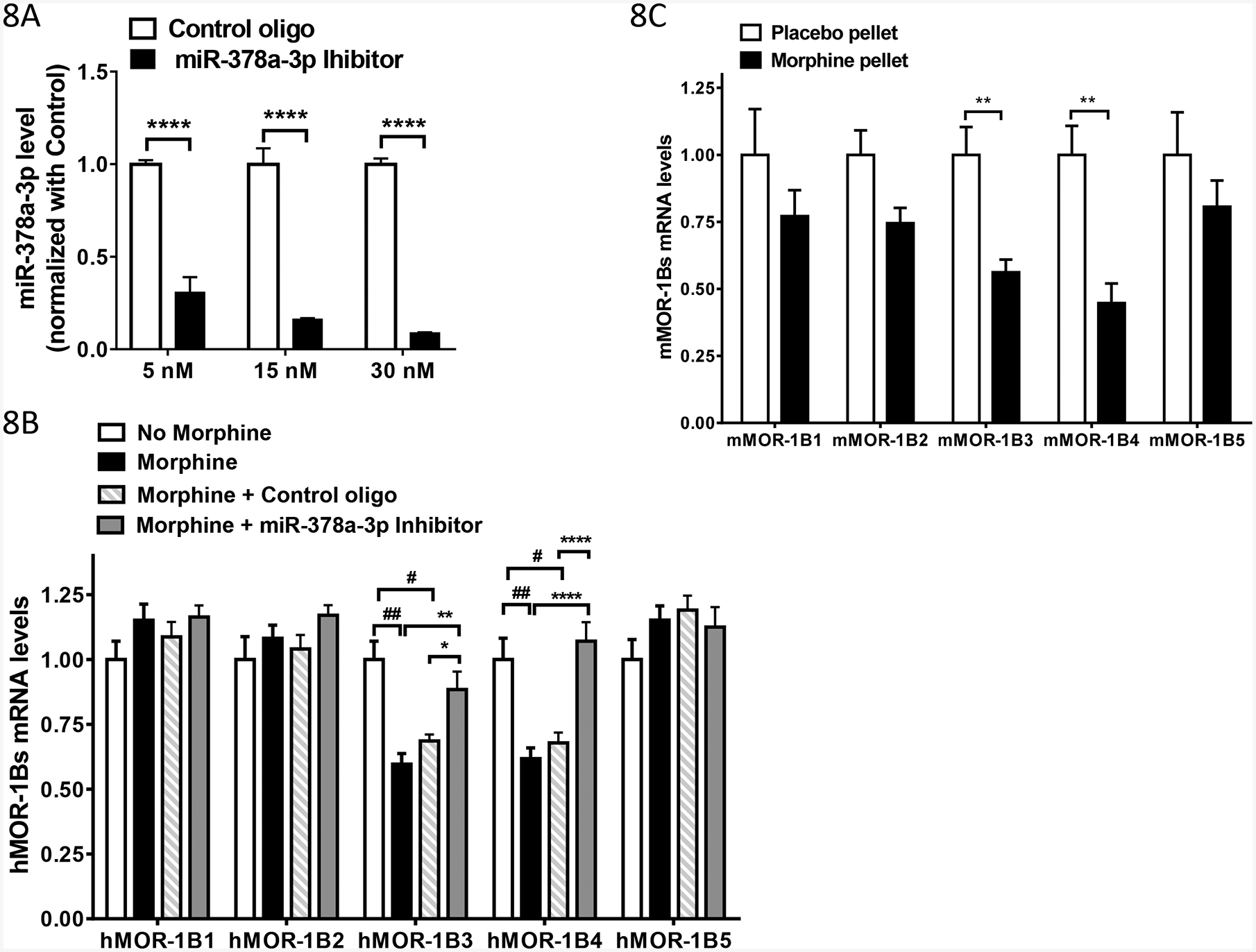
Effect of morphine on expression of MOR-1Bs via miR-378a-3p A). Effect of miR-378a-3p inhibitor on the expression of miR-378a-3p in Be(2)C cells. Total RNAs including small RNAs were isolated using miRNeasy Kit from Be(2) cells transfected with indicated concentration of Control oligo (n = 6) or miR-378a-3p Inhibitor (n = 6), and used in RT-qPCR to determine miR-378a-3p level, as described in Materials and Methods. Inhibition level by the LNA inhibitor was calculated by normalizing the levels with those of Control LNA oligo. Two-way ANOVA with Bonferroni’s post hoc analysis was used. ****: p < 0.001.
B). Effect of morphine and miR-378a-3p on the expression of hMOR-1Bs mRNAs in Be(2)C cells. Total RNAs from Be(2)C cells treated with or without morphine or with 5 nM of Control oligo or miR-378a-3p inhibitor for 48 hours (n = 4) were used in RT-qPCR to determine hMOR-1Bs levels, as described in Materials and Methods. Two-way ANOVA with Fisher’s LSD’s post hoc analysis was used. Compared to Control, ##: p < 0.0001; #: p < 0.001. Compared to Morphine + miR-378a-3p inhibitor, *: p < 0.05; **: p < 0.01; ****: p < 0.00001.
C). Effect of morphine on expression of mMOR-1Bs in the brainstem of the morphine tolerant mice. Total RNAs from the brainstem of mice implanted with morphine pellet (75 mg) or placebo pellet for 3 days (n = 4–6) were used in RT-qPCR to determine mMOR-1Bs levels, as described in Materials and Methods. Two-way ANOVA with Fisher’s LSD’s post hoc analysis was used. **: p < 0.01.
We did an optimization study using different concentration of the miR-378a-3p inhibitor. miR-378a-3p inhibitor efficiently decreased miR-378a-3p in a dose-dependent manner with inhibition of ~69%, ~84% and ~91% at 5 nM, 15 nM and 30 nM, respectively, when compared to the control LNA oligo (Fig. 8B). We choose 5 nM of miR-378a-3p inhibitor or control oligo for transfecting Be(2)C cells treated with morphine (3 μM, 48 hours). Both control oligo and miR-378a-3p inhibitor had not significant impact on the expression of hMOR-1B1, hMOR-1B2 and hMOR-1B5. Yet, miR-378a-3p inhibitor significantly increased hMOR-1B3 and hMOR-1B4 levels that were decreased by morphine treatment, while the control oligo had no such an effect (Fig. 8A). These results suggest that the decrease of hMOR-1B3 and hMOR-1B4 mRNAs by morphine in Be(2)C cells was mainly due to morphine’s action on upregulation miR-378a-3p.
Because morphine upregulates miR-378a-3p in the brainstem of morphine tolerant mice (Fig. 1B), we further investigated the effect of morphine on the expression of mMOR-1BS mRNAs in the same brainstem of morphine tolerant mice. Chronic morphine treatment using morphine pellets (75 mg) significantly reduced the expression of mMOR-1B3 and mMOR-1B4, compared to the placebo controls (Fig. 8C), while there was a trend of decrease in expression of mMOR-1B1, mMOR-1B2 and mMOR-1B5 (Fig. 8C). These results suggested that in vivo effect of morphine on regulating mMOR-1B3 and mMOR-1B4 expression in the brainstem is most likely acted through miR-378a-3p as it did in Be(2)C cells.
Discussion
Using a 3’-RACE strategy, we identified a poly(A) signal sequence and its associated cleavage site, as well as the flanking U-rich region, in both mouse and human MOR-1Bs mRNAs. These sequences are major cis-acting elements for pre-mRNA 3’ end processing, permitting to terminate transcription and define the 3’-UTR of the mouse and human MOR-1Bs mRNAs. Previous studies isolated and characterized the poly(A) sites for MOR-1 and MOR-1A in both mouse and human. The current study provides additional poly(A) site for mouse and human MOR-1Bs, confirming that alternative polyadenylation is one of mechanisms in OPRM1 alternative splicing. Interestingly, there are two poly(A) sites, one for MOR-1Bs described here and one for MOR-1A (13), is located between exon 3 and 4, raising questions regarding the role of the poly(A) sites in determining alternative splicing from exon 3 to exon 5s (producing MOR-1Bs) or to exon 4 (producing MOR-1) or intron retention (producing MOR-1A) (13).
One consensus mechanism of the poly(A) site is to maintain mRNA stability. Our results clearly demonstrated that the 3’URTs containing the poly(A) sites significantly increased the steady-state level of mMOR-1B1 and hMOR-1B1 mRNAs, leading to the increased opioid binding in HEK293 cells (Figs. 6B & 6C). We speculate that the similar role of the poly(A) sites in enhancing luciferase activity in HEK293 cells (Fig. 5C), as observed in the increased luciferase mRNA in the constructs containing the poly(A) of MOR-1A (13). Pre-mRNA 3’ end processing is tightly coupled to promoter activity. We realize that we used an exogenous phosphoglycerate kinase (PGK) promoter for luciferase or cytomegalovirus (CMV) promoter for MOR-1B1 in our constructs that were transfected into HEK293 cells. It will be interesting to investigate the interactions of the endogenous promoter with 3’ end processing in expression of these MOR-1Bs mRNAs in neuronal cells or in vivo.
One of the 3’-UTR functions is to mediate the actions of miRNAs for targeting mRNAand/or translation of the targeted mRNAs. Our study discovered that miR-378a-3p acts through a predicted and conserved miR-378a-3p binding site in the 3’-UTR of the mouse and human MOR-1Bs to regulate their expression mainly at the mRNA level. Our conclusion is supported by the observation that mutagenizing the miR-378a-3p binding site in the 3’-UTRs significantly enhances the luciferase activity (Fig. 5C) and increases mMOR-1B1 and hMOR-1B1 receptor protein expression (Fig. 6B) in HEK293 cells, and that downregulating miR-378a-3p using a miR-378a-3p inhibitor increases the luciferase activity of the 3’-UTR constructs containing the miR-378a-3p binding sites (Fig. 5E). Also, the mutation of the miR-378a-3p binding site greatly increased the expression of mMOR-1B1 and hMOR-1B1 mRNAs in HEK293 cells (Fig. 6C), suggesting that miR-378a-3p mainly function at the mRNA level.
miRNA-378a has been implicated to be involved in various biological and pathological processes, such as metabolism, angiogenesis, muscle biology, tumor biology and liver fibrosis (25). For example, miR-378a-3p regulated skeletal muscle growth and promoted the differentiation of myoblasts by targeting HDAC4 (26). miR-378a-3p suppressed activation of hepatic stellate cells by reducing Gli3 expression (27). miR-378a-3p also functioned as a tumor suppressor in several cancer models, including esophageal squamous cell carcinoma by targeting Rab10 (28) and the initial stage of carcinogenesis of colorectal cancer by suppressing IGF1R (29). Our studies offer a new function of miR-378a-3p in regulating expression of the OPRM1 splice variants, MOR-1Bs, which may contribute to development of morphine tolerance.
Increasing evidence indicates that mu opioids, such as morphine, can regulate expression of several miRNAs. Several miRNAs such as miR-219 (30), miR27a, miR-146b and miR-19b (31) were downregulated in morphine tolerant models. On the other hand, chronic morphine treatment upregulated miR-23b in mouse neuronal N2A cells (9) and let-7 in SH-SY5Y cells and in the brain of morphine tolerant mice (12). Our study provides another example in which morphine increased miR-378a-3p expression in both Be(2)C cells and the brainstem of the morphine tolerant mouse. It is interesting that morphine upregulated miR-378a-3p selectively in the brainstem, but not in the other three regions we examined, raising the possibility of the region-specific regulation by morphine and suggesting the potential role of miR-378a-3p at the brainstem in the development of morphine tolerance.
Chronic morphine treatment downregulated expression of MOR-1B3 and MOR-1B4 mRNAs in Be(2)C cells and the brainstem of morphine tolerant mice, which is correlated with the upregulation of miR-378a-3p, suggesting that morphine-induced decrease of MOR-1B3 and MOR-1B4 transcripts is mediated through miR-378a-3p. This hypothesis was further supported by observation that a miR-378a-3p inhibitor antagonized the morphine effect on reducing hMOR-1B3 and hMOR-1B4 transcripts in Be(2)C cells. Our studies suggest the potential involvement of the interplay between miR-378a-3p and MOR-1B3/B4 in the brainstem in morphine tolerance.
Our in vitro studies in HEK293 cells showed that the miR-378a-3p sites can suppress expression of both Luc2 reporter (Fig. 5) and the human and mouse MOR-1B1 mRNA and protein (Fig. 6), as well as MOR-1B3 and MOR-1B4 mRNAs (Fig. S2) by comparing the effects of the 3’UTRs containing WT or mutant miR-378a-3p, suggesting a common role of the miR-378a-3p sites under these in vitro conditions. On the other hand, in our in vitro studies using Be(2) cells with chronic morphine treatment (Fig. 8B) and in vivo studies in morphine tolerant mouse model (Fig. 8C), we observed no change in MOR-1B1 mRNA expression, but the decreased expression of MOR-1B3 and MOR-1B4 mRNAs. This discrepancy is most likely due to the differences in cell lines, constructs, 3’UTR, promoters and poly(A)s. In the in vitro studies conducted in HEK293 cells (Figs 5 and 6), we examined the expression of exogenously transfected cDNA constructs with no introns and limited 3’UTR (359 bp for mouse and 340 bp for human) with WT or mutant miR-378a-3p sites whose expression was controlled by a strong PGK (pmir) or CMV promoter (pcDNA3.1) and a SV40 poly(A) in pmir vector or BGH poly(A) in pcDNA3.1 vector. In the studies conducted in the Be(2)C cells and mouse morphine tolerant model, however, we investigated expression of endogenous MOR-1Bs mRNA in the context of intact Oprm1 gene locus including endogenous promoter and poly(A), introns and exons, the full 3’UTR regions, as well as the morphine treatment. All these elements or factors would have significant impact on expression of endogenous MOR-1Bs mRNAs.
The miR-378a-3p binding site is located at the 3’-UTR that are shared by all five MOR-1Bs. However, it appeared that only MOR-1B3 and MOR-1B4 were mainly modulated by morphine via miR-378a-3p. There are several possibilities. First, miRNA regulation and alternative splicing are two independent but closely related post-transcriptional processes (32). miRNAs can directly target some alternative splicing factors, leading to changes of alternative splicing profile. It will be interesting to further investigate whether miR-378a-3p can alter expression of the alternative splicing factors that can control splicing for specific MOR-1Bs. Second, miRNA-induced silencing complex (miRISC) involves transcription and constitutive and alternative splicing (33, 34), altering steady-state mRNA levels of the target mRNAs. It raises the possibility that miR-378a-3p-induced RISC can modulate alternative splicing of MOR-1Bs. Third, both mouse and human MOR-1Bs contain unique 3’-UTRs that may contain some distinct regulatory cis-elements for other trans-acting factors such as RNA binding proteins and other miRNAs that can be regulated by morphine besides the common 3’URT region. It could be possible that these regulatory elements and factors can antagonize the miR-378a-3p effect.
The functional relevance of MOR-1Bs has been demonstrated mainly in in vitro assays. When expressed in CHO cells, both human and mouse MOR-1B3 and MOR-1B4 displayed the differences in mu agonist-induced G protein coupling (3, 4). For example, D-Ala2,N-MePhe4,Gly5-ol]-enkephalin (DAMGO) was a full agonist against both hMOR-1B3 and hMOR-1B4 while it was a partial agonist against hMOR-1 and hMOR-1A (4). On the other hand, morphine and morphine-6β-glucuronide were more efficacious in hMOR-1 than in hMOR-1B3 and hMOR-1B4 (4). The region-specific and strain-specific expression the mouse MOR-1B3 and MOR-1B4 has been shown at the mRNA level (7, 8). The distribution of mMOR-1B4 at the protein level by immunostaining with an antisera against the C-terminus of mMOR-1B4 showed a distinct pattern when compared with that of other variants (35). It is also intriguing that mMOR-1B4 had a quite unusual binding profile. mMOR-1B4 bound [3H]diprenorphine with high affinity, but failed to bind [3H]DAMGO. Competition studies using [3H]diprenorphine revealed that most mu agonists had poor affinity toward mMOR-1B4. However, Etorphin, etonitazene and buprenorphine retained high affinity for mMOR-1B4. The current study provides another potential function of MOR-1B3 and MOR-1B4 in morphine tolerance via miR-378a-3p regulation.
In conclusion, the current study mapped a poly(A) signal sequence in both mouse and human MOR-1Bs transcripts that determined the 3’-UTR of these transcripts and maintained the stability of MOR-1Bs mRNAs. We identified a conserved miR-378a-3p binding site at the 3’-UTRs that can regulate expression of MOR-1Bs at the mRNA level. Chronic morphine treatment significantly increased miR-378–3p level in Be(2)C cells and the brainstem of the morphine tolerant mice, contributing to the decreased expression of the mouse and human MOR-1B3 and MOR-1B4. Our study provides new insights into the role of miRNAs and Oprm1 splice variants in morphine tolerance.
Supplementary Material
Acknowledgement
This work was supported by the National Institute on Drug Abuse (DA042888, DA046714 and DA007242), a National Natural Science Foundation of China grant (81673412) to Z.L., the Mayday Foundation, the Peter F. McManus Charitable Trust and a core grant from the National Cancer Institute to Memorial Sloan-Kettering Cancer Center (CA08748).
Abbreviations:
- OPRM1
mu opioid receptor gene
- 3’-RACE
Rapid Amplification of cDNA Ends
- 3’-UTR, 3’
untranslated region
- poly(A)
polyadenylation
- RT
reverse-transcription
- PCR
polymerase chain reaction
- TM
transmembrane domain
- bp
base pairs
- IBNtxA
3’-iodobenzoyl-6β-naltrexamide
- pmir
pmirGLO Dual-Luciferase miRNA Target Expression Vector
- DAMGO
D-Ala2,N-MePhe4,Gly5-ol]-enkephalin
Reference
- 1.Pan YX (2005) Diversity and complexity of the mu opioid receptor gene: alternative pre-mRNA splicing and promoters. DNA Cell Biol. 24, 736–750 [DOI] [PubMed] [Google Scholar]
- 2.Pasternak GW, and Pan YX (2013) Mu opioids and their receptors: evolution of a concept. Pharmacological reviews 65, 1257–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan YX, Xu J, Bolan E, Moskowitz HS, Xu M, and Pasternak GW (2005) Identification of four novel exon 5 splice variants of the mouse mu-opioid receptor gene: functional consequences of C-terminal splicing. Mol.Pharmacol 68, 866–875 [DOI] [PubMed] [Google Scholar]
- 4.Pan L, Xu J, Yu R, Xu MM, Pan YX, and Pasternak GW (2005) Identification and characterization of six new alternatively spliced variants of the human mu opioid receptor gene, Oprm. Neuroscience 133, 209–220 [DOI] [PubMed] [Google Scholar]
- 5.Koch T, Schulz S, Schroder H, Wolf R, Raulf E, and Hollt V (1998) Carboxyl-terminal Splicing of the Ratá¦áOpioid Receptor Modulates Agonist-mediated Internalization and Receptor Resensitization. Journal of Biological Chemistry 273, 13652–13657 [DOI] [PubMed] [Google Scholar]
- 6.Tanowitz M, Hislop JN, and von ZM (2008) Alternative splicing determines the post-endocytic sorting fate of G-protein-coupled receptors. J.Biol.Chem. 283, 35614–35621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Lu Z, Xu M, Rossi GC, Kest B, Waxman AR, Pasternak GW, and Pan YX (2014) Differential expressions of the alternatively spliced variant mRNAs of the micro opioid receptor gene, OPRM1, in brain regions of four inbred mouse strains. PloS one 9, e111267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Faskowitz AJ, Rossi GC, Xu M, Lu Z, Pan YX, and Pasternak GW (2015) Stabilization of morphine tolerance with long-term dosing: Association with selective upregulation of mu-opioid receptor splice variant mRNAs. Proceedings of the National Academy of Sciences of the United States of America 112, 279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Q, Zhang L, Law PY, Wei LN, and Loh HH (2009) Long-term morphine treatment decreases the association of mu-opioid receptor (MOR1) mRNA with polysomes through miRNA23b. Mol.Pharmacol. 75, 744–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Q, Law PY, Wei LN, and Loh HH (2008) Post-transcriptional regulation of mouse mu opioid receptor (MOR1) via its 3’ untranslated region: a role for microRNA23b. FASEB J. 22, 4085–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Hwang CK, Zheng H, Wagley Y, Lin HY, Kim DK, Law PY, Loh HH, and Wei LN (2013) MicroRNA 339 down-regulates mu-opioid receptor at the post-transcriptional level in response to opioid treatment. FASEB J 27, 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Yang C, Kirkmire CM, and Wang ZJ (2010) Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 10251–10258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z, Xu J, Xu M, Pasternak GW, and Pan YX (2014) Morphine regulates expression of mu-opioid receptor MOR-1A, an intron-retention carboxyl terminal splice variant of the mu-opioid receptor (OPRM1) gene via miR-103/miR-107. Molecular pharmacology 85, 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreyer JL (2010) New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome Med 2, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang CK, Wagley Y, Law PY, Wei LN, and Loh HH (2011) MicroRNAs in Opioid Pharmacology. J.Neuroimmune.Pharmacol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tapocik JD, Luu TV, Mayo CL, Wang BD, Doyle E, Lee AD, Lee NH, and Elmer GI (2012) Neuroplasticity, axonal guidance and micro-RNA genes are associated with morphine self-administration behavior. Addict Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, and Wang ZJ (2012) Let-7 microRNAs and Opioid Tolerance. Front Genet 3, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez RE (2012) Morphine and microRNA Activity: Is There a Relation with Addiction? Front Genet 3, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbierato M, Zusso M, Skaper SD, and Giusti P (2015) MicroRNAs: emerging role in the endogenous mu opioid system. CNS Neurol Disord Drug Targets 14, 239–250 [DOI] [PubMed] [Google Scholar]
- 20.Kolesnikov YA, Pick CG, Ciszewska G, and Pasternak GW (1993) Blockade of tolerance to morphine but not to kappa opioids by a nitric oxide synthase inhibitor. Proceedings of the National Academy of Sciences of the United States of America 90, 5162–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumdar S, Grinnell S, Le, R. V, Burgman M, Polikar L, Ansonoff M, Pintar J, Pan YX, and Pasternak GW (2011) Truncated G protein-coupled mu opioid receptor MOR-1 splice variants are targets for highly potent opioid analgesics lacking side effects. Proceedings of the National Academy of Sciences of the United States of America 108, 19778–19783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Z, Xu J, Rossi GC, Majumdar S, Pasternak GW, and Pan YX (2015) Mediation of opioid analgesia by a truncated 6-transmembrane GPCR. The Journal of clinical investigation 125, 2626–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkon R, Ugalde AP, and Agami R (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14, 496–506 [DOI] [PubMed] [Google Scholar]
- 24.Millevoi S, and Vagner S (2010) Molecular mechanisms of eukaryotic pre-mRNA 3’ end processing regulation. Nucleic Acids Res 38, 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krist B, Florczyk U, Pietraszek-Gremplewicz K, Jozkowicz A, and Dulak J (2015) The Role of miR-378a in Metabolism, Angiogenesis, and Muscle Biology. Int J Endocrinol 2015, 281756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei X, Li H, Zhang B, Li C, Dong D, Lan X, Huang Y, Bai Y, Lin F, Zhao X, and Chen H (2016) miR-378a-3p promotes differentiation and inhibits proliferation of myoblasts by targeting HDAC4 in skeletal muscle development. RNA Biol 13, 1300–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, Ha CS, Kim SW, Yun YH, and Jung Y (2016) MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun 7, 10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding N, Sun X, Wang T, Huang L, Wen J, and Zhou Y (2018) miR378a3p exerts tumor suppressive function on the tumorigenesis of esophageal squamous cell carcinoma by targeting Rab10. Int J Mol Med 42, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Dai S, Zhen T, Shi H, Zhang F, Yang Y, Kang L, Liang Y, and Han A (2014) Clinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer 50, 1207–1221 [DOI] [PubMed] [Google Scholar]
- 30.Hu XM, Cao SB, Zhang HL, Lyu DM, Chen LP, Xu H, Pan ZQ, and Shen W (2016) Downregulation of miR-219 enhances brain-derived neurotrophic factor production in mouse dorsal root ganglia to mediate morphine analgesic tolerance by upregulating CaMKIIgamma. Mol Pain 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tapocik JD, Ceniccola K, Mayo CL, Schwandt ML, Solomon M, Wang BD, Luu TV, Olender J, Harrigan T, Maynard TM, Elmer GI, and Lee NH (2016) MicroRNAs Are Involved in the Development of Morphine-Induced Analgesic Tolerance and Regulate Functionally Relevant Changes in Serpini1. Front Mol Neurosci 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kucherenko MM, and Shcherbata HR (2018) miRNA targeting and alternative splicing in the stress response - events hosted by membrane-less compartments. J Cell Sci 131 [DOI] [PubMed] [Google Scholar]
- 33.Allo M, Agirre E, Bessonov S, Bertucci P, Gomez Acuna L, Buggiano V, Bellora N, Singh B, Petrillo E, Blaustein M, Minana B, Dujardin G, Pozzi B, Pelisch F, Bechara E, Agafonov DE, Srebrow A, Luhrmann R, Valcarcel J, Eyras E, and Kornblihtt AR (2014) Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proceedings of the National Academy of Sciences of the United States of America 111, 15622–15629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Havens MA, Reich AA, and Hastings ML (2014) Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet 10, e1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Pan YX, Kolesnikov Y, and Pasternak GW (2006) Immunohistochemical labeling of the mu opioid receptor carboxy terminal splice variant mMOR-1B4 in the mouse central nervous system. Brain research 1099, 33–43 [DOI] [PubMed] [Google Scholar]
- 36.Ide S, Han W, Kasai S, Hata H, Sora I, and Ikeda K (2005) Characterization of the 3’ untranslated region of the human mu-opioid receptor (MOR-1) mRNA. Gene 364, 139–145 [DOI] [PubMed] [Google Scholar]
- 37.Wu Q, Hwang CK, Yao S, Law PY, Loh HH, and Wei LN (2005) A Major Species of Mouse {micro}-opioid Receptor mRNA and Its Promoter-Dependent Functional Polyadenylation Signal. Molecular pharmacology 68, 279–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


