Summary
Despite being an autosomal dominant disorder caused by a known coding mutation in the gene HTT, Huntington’s disease (HD) patients with similar trinucleotide repeat mutations can have an age of onset that varies by decades. One likely contributing factor is the genetic heterogeneity of patients that might modify their vulnerability to disease. We report that although the heterozygous depletion of the autophagy adaptor protein Alfy/Wdfy3 has no consequence in control mice, it significantly accelerates age of onset and progression of HD pathogenesis. Alfy is required in the adult brain for the autophagy-dependent clearance of proteinaceous deposits, and its depletion in mice and neurons derived from patient fibroblasts accelerates the aberrant accumulation of this pathological hallmark shared across adult onset neurodegenerative diseases. These findings indicate that selectively compromising the ability to eliminate aggregated proteins is a pathogenic driver, and the selective elimination of aggregates may confer disease resistance.
eTOC Blurb
Fox et al. demonstrate in a Huntington’s disease (HD) mouse model and in HD patient-derived neurons that the autophagy adaptor protein Alfy/WDFY3 is required for removal of aggregated protein and that depletion of Alfy hastens pathogenic onset of HD.
Introduction
Protein accumulation is a pathogenic hallmark across adult onset neurodegenerative diseases, but to what extent accumulation actually contributes to neurodegeneration is unclear. In HD, the characteristic intracellular proteinaceous deposits are the byproduct of a heritable mutation causing expansion of a polymorphic CAG repeat region within exon 1 of the HTT gene (HDCRG, 1993). The mutation, in a length dependent manner, results in an expanded polyglutamine (polyQ) tract in the Huntingtin protein (Htt) that causes this otherwise soluble protein to aggregate. Although the exact relevance of these deposits remains unclear, the correlation between mutation length and aggregation echoes the correlation between mutation length and an earlier age of disease onset (MacDonald and Gusella, 1996; Perutz and Windle, 2001). In addition, given that aggregate-clearance upon gene suppression in HD mice is strongly correlated with therapeutic reversal (Kordasiewicz et al., 2012; Stanek et al., 2014; Yamamoto et al., 2000), further interrogation of the relationship between these two observations would clarify whether protein accumulation impacts pathogenesis.
Cell-based systems have demonstrated that aggregated proteins are eliminated by macroautophagy (Iwata et al., 2005a; Iwata et al., 2005b; Ravikumar et al., 2002; Yamamoto et al., 2006), and their recognition by adaptor proteins promotes their selective degradation (Klionsky, 2007). In cell lines, we have previously characterized the large PI3P-binding protein autophagy-linked FYVE protein (Alfy/Wdfy3) as an adaptor required for the degradation of preformed, detergent-insoluble, aggregated proteins by selective macroautophagy (Eenjes et al., 2016; Filimonenko et al., 2010). Consistent with its role as a selectivity adaptor, Alfy is not required for starvation-mediated or basal macroautophagy in either cells or the mammalian brain (Dragich et al., 2016; Filimonenko et al., 2010). In addition to mutant Htt, Alfy has been implicated in the turnover of other disease-relevant protein aggregates of α-synuclein, as well as protein complexes such as the midbody ring (Clausen et al., 2010; Hocking et al., 2010; Isakson et al., 2013; Kadir et al., 2016; Park et al., 2014). Whether selective macroautophagy eliminates aggregates in the adult brain, however, remains unclear.
Given the impact of genetic modifiers to contribute to disease onset and progression of HD (Genetic Modifiers of Huntington’s Disease, 2015; Gusella and MacDonald, 2009; Wexler et al., 2004), we hypothesized that the pathological relevance of proteinaceous inclusions would be revealed by modifying the dosage of genes involved in selective macroautophagy. We therefore used a combination of mouse genetics, biochemistry, and cell biology to establish if aggregation modifies disease pathogenesis. Upon establishing that Alfy-mediated selective autophagy is required in the adult brain to clear accumulated proteins, we demonstrate that depleting levels of Alfy globally has no detectable impact on normal adult mice, but significantly accelerates aggregation and symptomatic-onset in a mouse model of HD. Using a novel, patient-derived, medium spiny neuron model of HD, Alfy depletion was also sufficient to increase significantly aggregation of endogenous mutant Htt, suggesting that heterogeneity of Alfy levels in patients might similarly impact HD pathogenesis. Taken together, our findings imply that modulating the proteinopathy can directly impact disease.
Results
Alfy is required for aggregate clearance in the adult brain
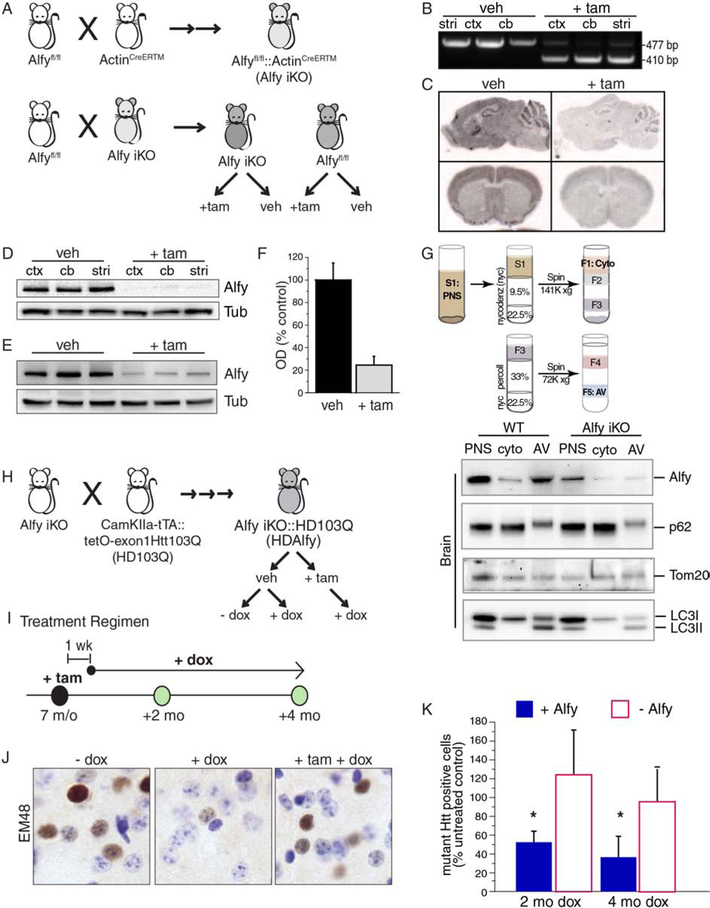
To establish firmly the importance of Alfy in aggregate-clearance in the adult brain, we generated a genetic model in which temporally regulated deletion of Alfy would occur together with conditional expression of a fragment model of HD. To do this, we interbred a mouse model that permits tamoxifen (tam)-inducible Cre-mediated elimination of Alfy expression (Alfy iKO) (Figure 1A–G, S1A–D) with a newly created model expressing an exon 1 fragment of the HD gene with 103 glutamines (encoded by CAGCAA alternating repeats, exon1Htt103Q) in a tet-regulated manner (HD103Q) (Figure 1H–K, S1E–I). Our combined analyses of induced-deletion of Alfy in 7 month old (m/o) mice revealed that maximal loss of Alfy, on average 80%, was achieved 7 d after the induction of Cre (Figure 1F). Similarly to cell lines (Filimonenko et al., 2010), the loss of Alfy had no measurable impact on autophagosome biogenesis under basal conditions; 12 mos after Alfy KO, autophagosomes from Alfy iKO brains were isolated and similar levels of p62 and the mitochondrial protein Tom20 was present in the AV fraction.
Figure 1. Alfy is required for the clearance of aggregated mHtt in the adult brain.
(A) Schematic representation of the creation of Alfy iKO mice. Two rounds of breeding create the iKO mice, which are homozygous for the conditional Alfy allele (Alfyfl/fl) and heterozygous for ActinCreERTM. Experimental cohorts are created by using these iKO mice, and crossing back to AlfyloxP/loxP mice. Offspring are then treated with veh or +tam. See also Figure S1.
(B) PCR genotyping of veh vs. +tam Alfy iKO mice. Genomic DNA from cortex (ctx), cerebellum (cb) and striatum (str) of 11 m/o iKO mice that were treated at 7 m/o. Cre-mediated excision of exon 5 is detected by a change in PCR product size (477 vs. 410 bp). n=3 mice/treatment.
(C) In situ hybridization using a probe complementary to exon 5 of Alfy 1 wk after the final injection in 6 m/o Alfy iKO mice. n=3 mice/treatment.
(D-F) Western blot analyses of detergent-soluble brain lysates from 11 m/o Alfy iKO mice in (D) different brain regions or (E) whole brain lysates of mice, quantified in (F). n=3 mice/treatment.
(G) Tissue fractionation for autophagosome enrichment reveals no notable alteration in basal macroautophagy up to 1 year after the induced-deletion of Alfy. Fractionation from brain and liver shown. Fractions representing the post-nuclear supernatant (PNS), cytosol (cyto), and autophagic vacuoles (AV) are probed for Alfy, p62, Tom20 and LC3. Note the lack of p62 accumulation despite the loss of Alfy. n=6–7 mice/genotype.
(H) An abbreviated schematic of the genetic crosses and treatment regimens to create HDAlfy mice. Alfy iKO and HD103Q mice are intercrossed to introduce the six genetic modifications that are necessary to create the breeders and ultimately the HDAlfy mice. HDAlfy mice are treated with +tam to modify Alfy levels and dox to regulate expression of the exon1Htt103Q transgene. See also Figure S1.
(I) Treatment regimen for HDAlfy mice. 7 m/o mice are injected with tam, followed by treatment with dox or control solution for up to 4 mos, one wk after the final injection. Mice are collected for analyses at the time points indicated.
(J, K) Alfy is required for aggregate clearance. (J) EM48 staining of HDAlfy striatum, before treatment and 4 mos post-treatment. Dox treated mice are compared to HD gene suppressed mice lacking Alfy (+tam +dox). (K) Stereological assessment of EM48-positive Htt deposits at 2 and 4 mos post dox with or without Alfy. Data is shown as a percentage of 7 m/o HDAlfy mice (no treatment). ANOVA revealed a significant overall effect of the presence of Alfy (F(1,20)=7.032, p=0.0153) and a significant interaction between Alfy and length of dox treatment (F(1,2)=4.001, p=0.0346)(n=4–5 mice/genotype/age). Although after 2 and 4 mo of dox led to a significant decrease in aggregate load (2 mo: F(1,6)=8.632, p=0.0260; 4 mo: F(1,8)=6.347, p=0.0358), deletion of Alfy impeded this clearance.
To determine the impact of Alfy iKO on the selective clearance of mHtt deposits in the adult brain, aggregate clearance in the HDAlfy mice was monitored in the presence or absence of Alfy (Figure 1I). Stereologic quantification of aggregate load revealed that although a time-dependent decrease in mHtt deposits was observed in Alfy+ mice after eliminating exon1Htt103Q expression (Figure 1J, K), the decrease was significantly impeded in mice lacking Alfy even after 4 months of exon1Htt103Q suppression (Figure 1J, K). Taken together, these data indicate that although the presence of Alfy is not essential for macroautophagy to occur, it is required for neurons to efficiently eliminate aggregated protein in the adult brain.
Heterozygous depletion of Alfy accelerates aggregation and behavioral dysfunction in a mouse model of HD
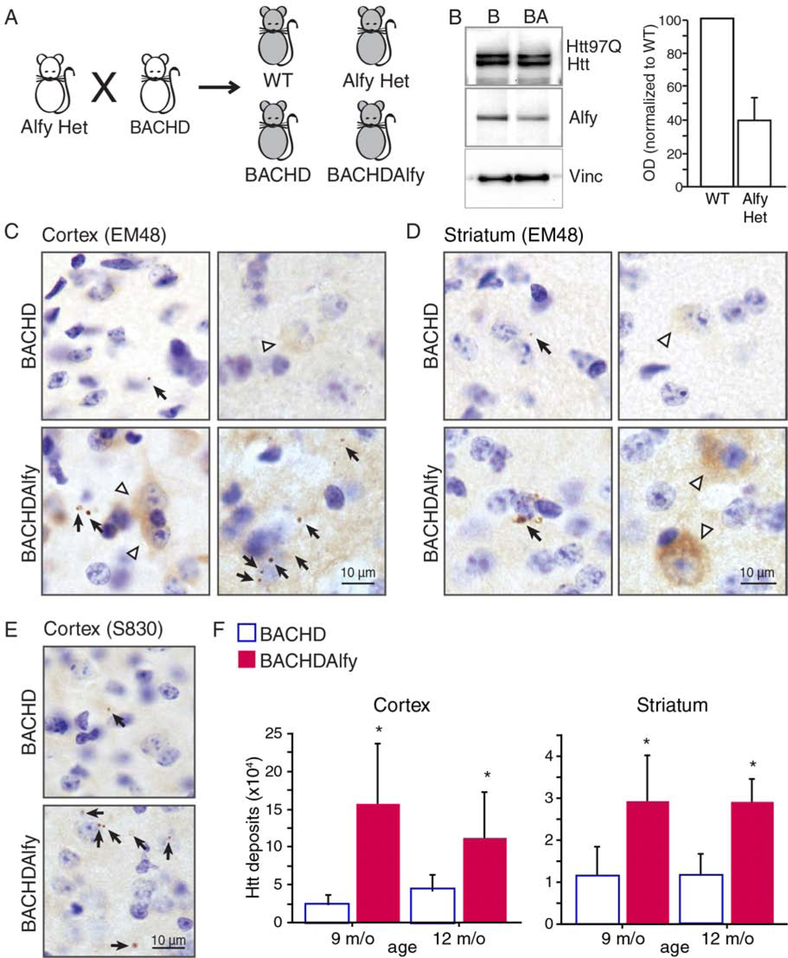
The bigenetic design of the HD mouse model led to an unexpectedly restricted expression pattern of mutant Htt into the lateral striatum, thereby leading to no progressive decline on motor tasks such as the rotarod (Figure S1H) as previously predicted (Gu et al., 2005). Thus, to determine if aggregation impacts the age of onset in a mouse model of HD, we crossed the BACHD mouse model (Gray et al., 2008), which expresses the full length human HTT genomic sequence with 97 repeats, to mice heterozygous for the gene encoding Alfy Wdfy3 (AlfyHet), to create BACHDAlfy mice (Figure 2A). Lowering Alfy levels by 50% (Figure 2B) was sufficient to accelerate significantly the appearance of S830- or EM48-positive mutant Htt (mHtt) inclusions in cortex and striatum compared to BACHD mice (Figure 2C–E). Anti-EM48 staining revealed the appearance of cytoplasmic and perinuclear punctae as well as diffuse cytoplasmic staining as early as 9 m/o in BACHDAlfy cortex and striatum, whereas only sparse EM48-positive puncta and minimal cytoplasmic accumulation was apparent in 12 m/o BACHD (Figure 2C and D). Stereological quantification of aggregate-load revealed by S830 immunoreactivity was similarly higher in the BACHDAlfy brains compared to BACHD mice at these ages (Figure 2E). In addition, biochemical fractionation and immunoblotting of the lysate against the toxic mHtt epitopes 3B5H10 (Miller et al., 2011) and 1C2 (Trottier et al., 1995) revealed that Alfy-depletion slowed the elimination of detergent-insoluble mHtt but had no impact on detergent-soluble mHtt levels (Figure S2A and B), consistent with Alfy’s role as an adaptor protein for aggregated proteins. Overall, depleting Alfy in BACHD leads to accelerated accumulation of detergent-insoluble, aggregated mHtt, further supporting that aggregate-clearance by selective autophagy in the adult brain requires Alfy.
Figure 2. Heterozygous depletion of Alfy accelerates the accumulation of aggregates in a mouse model of HD.
(A) Creation of BACHDAlfy mice and the experimental littermates, wild-type (WT), AlfyHet and BACHD.
(B) Representative immunoblot of BACHD (B) and BACHDAlfy (BA) mice probed for Htt (3B5H10) and Alfy. Alfy levels corrected by vinculin (vinc) at right.
(C-F) Depletion of Alfy accelerates accumulation of mHtt in BACHD mice. (C, D) Immunohistochemistry against the EM48 epitope in 12 m/o (C) cortex and (D) striatum. Black arrows indicate cytoplasmic and perinuclear punctate structures; white arrowheads indicate diffuse cytoplasmic or neuritic staining. Scale bar=10 m. (E, F) S830-positive mHtt puncta in the cortex and striatum. (E) S830-positive structures in the cortex of 12 m/o mice. Similar structures were revealed in the striatum (not shown). (F) Stereological quantification of aggregate load. ANOVA revealed a significant increase of aggregates in the BACHDAlfy brain compared to BACHD (Cortex: F(1,11)=15.920, p=0.0021; Striatum: F(1,11)=9.058, p=0.0119), and no significant effect of age (Cortex: F(1,11)=0.242, p=0.6324; Striatum: F(1,11)=0.520, p=0.4857). Data are shown as mean ± SD. Unlike aggregated proteins, detergent soluble proteins are unaffected (See Figure S2).
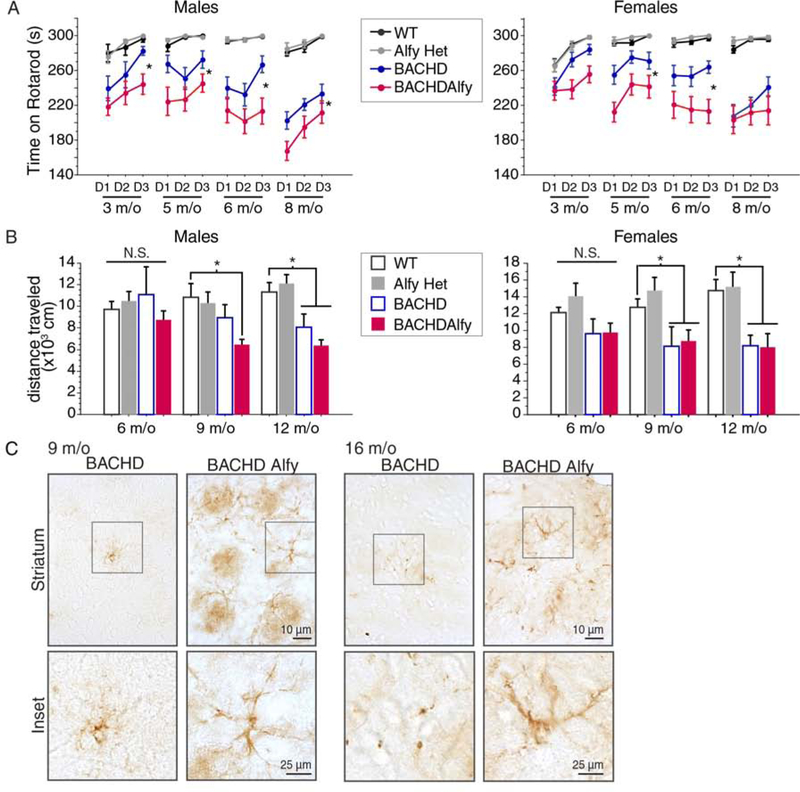
To determine if aggregate-turnover affected phenotypic onset, we performed two behavioral paradigms (Figure 3). The repeated-accelerated rotarod paradigm revealed that although AlfyHet mice were indistinguishable from WT littermates, BACHDAlfy onset was faster and performed significantly worse than BACHD mice (Figure 3A). These data indicate that lowering Alfy levels by 50% increased the vulnerability of the mice in the context of disease, but did not otherwise affect normal function. Habituated locomotor activity in the open field maze across time revealed similar results (Figure 3B): In males, BACHD mice became hypolocomotive at 12 m/o, as previously reported (Wang et al., 2014), whereas BACHDAlfy mice onset at 9 m/o. Unlike the rotarod behavior, the hypolocomotor response did not progressively decline, possibly due to a floor effect in habituated animals. This was particularly evident in the female BACHD and BACHDAlfy cohorts, which showed diminished locomotion, but there was no change across time. Given that hypothalamic accumulation of mutant Htt had previously been implicated to affect body weight in BACHD mice (Hult et al., 2011), we also monitored body weight and food consumption upon Alfy depletion to determine if the locomotor differences were independent of body weight (Figure S3A and B): Although BACHD are heavier than control mice, the depletion of Alfy had no measureable impact on body weight across time. Moreover, the amount of food consumed over a single 24hr period was similar across all genotypes tested. Thus, a decrease in Alfy levels that accelerates aggregation significantly accelerates the onset of motoric dysfunction in BACHD mice.
Figure 3: Accelerated aggregation correlates with an accelerated onset of HD symptoms.
(A) Latency to fall from the accelerating rotarod. Data are shown as mean±SD. In males and females, RM-ANOVA revealed a significant overall effect of genotype (Males: F(3,188)=88.171, p<0.0001; Females: F(3,171)=80.182, p<0.0001) and age (Males: F(3,188)=6.768, p=0.0002; Females: F(3,171)=4.283, p=0.0061). In only females an interaction between genotype and age (F(3,171)=3.017, p=0.0023) was also found. Fisher PLSD revealed that BACHDAlfy showed significantly poorer performance compared to BACHD at all ages in males (3 m/o, p=0.0446; 4 m/o, p=0.0097; 5 m/o, p=0.0033; 8 m/o, p=0.0119) and at 4 and 5 m/o in females (3 m/o, p=0.0538; 4 m/o, p=0.0006; 5 m/o, p=0.0008; 8 m/o, p=0.2809). No significant difference between WT and AlfyHet mice were ever found (Male: 3 m/o, p=0.8627; 4 m/o, p=0.8629; 5 m/o, p=0.9630; 8 m/o, p=0.7559; Female: 3 m/o, p=0.9556; 4 m/o, p=0.7554; 5 m/o, p=0.7554; 8 m/o, p=0.6852).
(B) Distance traveled in the open field arena. Data are shown as mean±SD. ANOVA revealed a significant overall effect of genotype in both males and females at 9 m/o (males: F(3,37)=3.660, p=0.0209; females: F(3,43)=3.953, p=0.0141) and 12 m/o (males: F(3,39)=9.025, p=0.0001; females: F(3,34)=6.724, p=0.0011). In males Fisher’s PLSD confirmed that BACHD mice become hypolocomotive at 12 m/o (9 m/o, p=0.1930; 12 m/o p=0.0141), whereas BACHDAlfy become hypolocomotive at 9 m/o (9 m/o, p=0.0142; 12 m/o, p=0.0002). In contrast, both BACHD (9 m/o, p=0.0064; 12 m/o p=0.0025) and BACHDAlfy females (9 m/o, p=0.0098; 12 m/o p=0.0034) are hypolocomotive at these ages and do not differ from each other (9 m/o, p=0.7780; 12 m/o p = 0.9202). WT and AlfyHet are also indistinguishable (Males: 9 m/o, p=0.7136; 12 m/o, p=0.5596; Females: 9 m/o, p=0.3830; 12 m/o, p=0.8289).
(C) GFAP staining reveals widespread reactivity in BACHDAlfy compared to BACHD. Scale bar=10μm, inset=25μm. Images are representative of n=3–4 mice/genotype. Also see Figure S3.
Reactive astrocytosis in regions such as the dorsal striatum is a neuropathological phenotype observed in HD patient brains indicative of neuronal stress (Vonsattel and DiFiglia, 1998). GFAP staining in 9 and 16 m/o mouse striatum (Figure 3C and S3C) revealed widespread staining of GFAP+ reactive astrocytes extending throughout the striatum of BACHDAlfy mice. This was in contrast to AlfyHet and BACHD mice, for which we observed occasional isolated instances of GFAP reactivity but did not observe the typical morphologies associated with astrocytosis. Despite the increased astrocytosis, the loss of Alfy did not accelerate cell loss or tissue volume, as assessed stereologically (Figure S3D). Given that the Alfy depletion is required to drive the appearance of these structures in the striata of BACHD mice, this suggests that the inability to clear aggregates efficiently allows inclusions to drive secondary events that impact homeostasis. Taken together, these data imply that Alfy can modify HD pathogenesis by impacting protein aggregation, and that protein aggregation affects neuronal stress and function but does not overtly impact neuronal death.
Depletion of Alfy accelerates aggregation in a HD patient-derived neuronal model
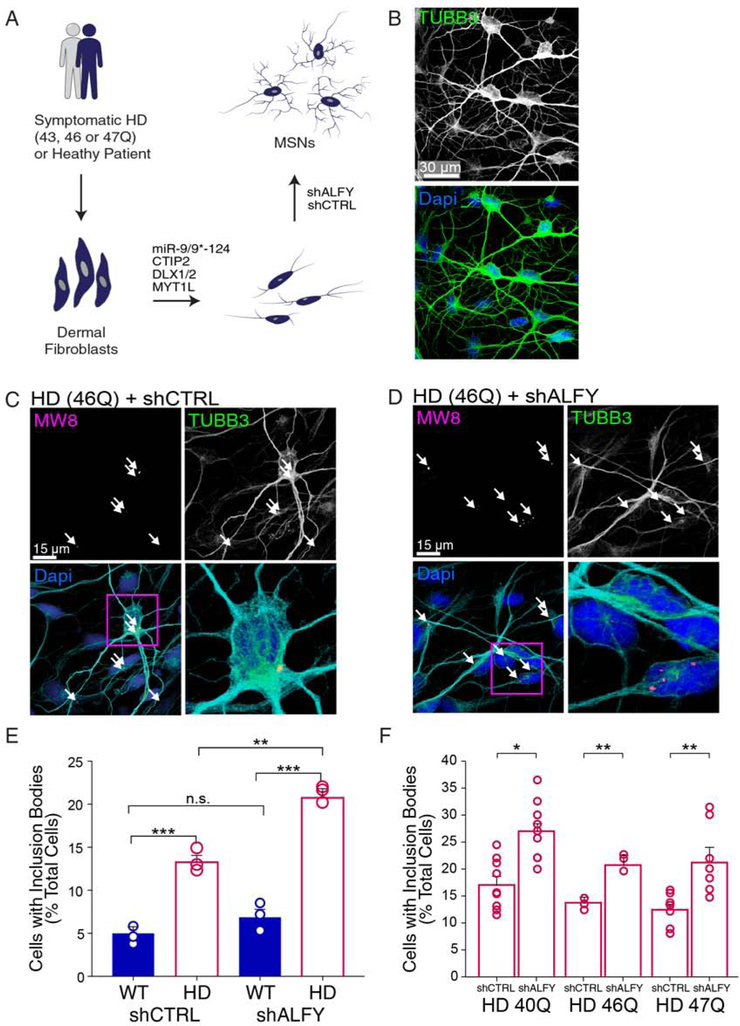
We next sought to determine if modifying Alfy levels can impact cellular events that more closely replicates what might occur in adult HD. Recently, an HD patient fibroblast model from which medium spiny neurons (MSNs) are generated through direct neuronal conversion (Huh et al., 2016; Mertens et al., 2015) was shown to demonstrate an aggregation phenotype of endogenous mHtt, in the absence of extreme polyQ expansions that are rarely observed in the patient population (Victor et al., 2018). Importantly, much like patient brains (Gutekunst et al., 1999; Vonsattel, 2008), less than 20% of the neurons demonstrated predominantly neuropil aggregates of Htt carrying CAG lengths typical of adult onset. Using this model system, we next determined if Alfy depletion augmented aggregation. Patient fibroblasts with a polyQ length of 40, 46 or 47 repeats were converted to MSNs following a previously established protocol (Figure 4A, B)(Victor et al., 2018), then treated with an Alfy shRNA sequence which depleted Alfy levels to about 60% (Figure S4A). In contrast to control MSNs (Figure S4B), shALFY led to a significant increase in aggregation in the patient-derived MSNs (Figure 4C–F), indicating that Alfy also can modify aggregation in this model without affecting autophagy overall (Figure S4C, D). Notably, despite increasing aggregation, Alfy depletion was not sufficient to augment cell death, although little cell death was observed overall in the HD line at the time point chosen (Figure S4E). These data suggest that increasing aggregation provokes circuitry dysfunction, but cell autonomous toxicity of mHtt is independent of aggregation.
Figure 4. ALFY depletion increases endogenous mHtt aggregation in human neurons directly reprogrammed from HD patient fibroblasts.
(A) Schematic depicting the conversion of human fibroblasts isolated into cells analogous to medium spiny neurons (MSNs) using brain-enriched microRNAs, miR-9/9*−124, combined with striatum-enriched factors. shRNA against Alfy (shALFY) or a control sequence (shCTRL) was administered after replating.
(B) Representative image from converted fibroblasts at post-induction day (PID) 19, immunostained for TUBB3 (green) and the nuclear stain DAPI (blue).
(C-F) The percentage of MSNs positive for mHtt inclusions. HD-MSNs treated with (C) shCTRL or (D) shALFY immunostained with MW8 antibody at PID 19. (E) Quantification of MSNs positive for mHtt inclusion bodies relative to unaffected patient fibroblasts. Data shown as mean±sem, with individual data points. ANOVA revealed a significant difference of shALFY(F(5,12)= 19.88, P<0.0001). Post hoc Tukey’s test comparison outcomes indicated. N=average of 400 cells/genotype/shRNA. (F) Quantification of percentage of MSNs positive for mHtt inclusion bodies across different patient fibroblast lines. Pairwise t-tests are performed within each fibroblast line. n=219–450 cells/cell line/shRNA. ***, p<0.001; **, p<0.01; *, p<0.05; n.s.=not significant. See also Figure S4.
Discussion
From patient-derived neurons to mice, we demonstrate that Alfy, an adaptor of selective autophagy, is required for the clearance of aggregated proteins in the adult brain, and reduction of its levels can accelerate disease pathogenesis. Notably, using the HD103Q and the BACHD models, which differ in expression strategy, aggregation profile, and behavioral phenotype, we show that Alfy is required in vivo for the effective turnover of aggregated Htt, and demonstrate a specific need for Alfy-mediated aggregate clearance in order to resist neurodegenerative and behavioral symptomatology.
It is increasingly clear that the study of protein aggregation and turnover is a complex one, and that from yeast to mammalian cells, all aggregates are not considered equal. For example, in a HeLa cell based model, we had found that although Alfy promoted the turnover of preformed inclusions, there was a pool of inclusions that was more susceptible to degradation than others (Eenjes et al., 2016). These findings echoed a model proposed by Frydman and colleagues, in which there are two distinct compartments for misfolded proteins, a soluble pool (JUNQ) and an insoluble pool (IPOD), the latter of which is targeted for autophagic degradation (Kaganovich et al., 2008). Our biochemical studies indicate that consistent with our original cell based findings (Eenjes et al., 2016; Filimonenko et al., 2010), Alfy-mediated turnover in the adult brain preferentially effects the detergent-insoluble pool (Figure S2). Although it still remains unexplored whether proteins are similarly sequestered in the postmitotic and highly compartmentalized adult neurons, these data suggest that an IPOD-like pool may indeed exist.
Our results strongly suggest that genetic modifications that impact protein aggregation can modify the age of onset of HD, accelerating the appearance of motor dysfunction. Accelerating aggregation did not impact cytotoxicity outright in both the BACHD mice (Figure S3) and the HD MSN model (Figure 4), but instead exerted its deleterious effects by impacting neural circuitry and thereby accelerating behavioral dysfunction (Figure 3). This suggests that disease toxicity is influenced by two factors, with the non-aggregating mHtt driving disease-specific pathogenesis, and aggregating mHtt modifying this toxicity (Figure S4E). In light of the in vitro evidence that the rate of aggregation and accumulation is directly related to the polyQ length, our work suggests that it is the rate of aggregation that contributes to the earlier age of onset often associated with a longer polyQ length. Thus, if accumulation can be slowed, our data indicate that age of onset might be significantly delayed and quality of life preserved. Importantly, Alfy is not specific to the autophagic elimination of mHtt, but generally to proteins in either an aggregated state or a large protein complex, including other disease-relevant deposits such as alpha-synuclein. Future studies establishing how broadly these observations apply across the many proteinopathic diseases will allow us to gain better insight on how protein aggregation contributes to disease, and whether the appearance of a proteinopathy is indeed a unifying feature of pathogenesis.
STAR Methods text
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ai Yamamoto (ai.yamamoto@columbia.edu). All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
In vivo animal studies
All experiments were reviewed and approved by the Columbia University Medical Center’s Institutional Animal Care and Use Committee (IACUC). Experimental mice were bred and housed in facilities at the William Black Medical Research Building. Up to five mice per cage were maintained sex-segregated in a temperature-controlled environment on a 12 hour light/dark cycle with access to food and water ad libitum. For breeding and housing pre-experiment, mice were maintained on a cycle with the lights on between 6 am and 6 pm. Mice were randomly assigned to experimental groups, and experiments were all performed blind to genotype, by using either AVID chips or earpunching for identification. The respective age at which the experiments were performed are noted below.
Mouse strains
BACHD were obtained from William Yang at UCLA (Gray et al., 2008). They are maintained by crossing onto a wild-type FVB/J strain. Creation of the Alfy gene trap mice and conditional Wdfy3 allele are described previously (Dragich et al, 2016), and are maintained on the CBAXC57Bl6 F1 backgrounds. Actin-CreER™ mice (Hayashi and McMahon, 2002) were obtained from the Jackson Laboratory (Bar Harbor, Maine). Actin-CreER™ mice were maintained as heterozygotes on a C57BL/6 background by crossing to wild-type C57BL/6 mice every 3–4 months. CamKIIa-tTA +/− mice were obtained from Jackson labs and are originally described by Mayford et al. (1996). They were maintained as heterozygotes on a C57BL/6 background.
Alfy iKO (Alfy flox/flox::Actin-CreER™) were created as follows: Mice homozygous for the conditional Wdfy3 allele (Alfyfl/fl) were outcrossed to Actin-CreER™. Resulting Cre+Alfyfl/+ F1 offspring are recrossed to the original Alfyfl/fl lines to maintain strain.
HD103Q mice (CamKIIa-tTA::tetO exon1Htt103Q) were created as follows: An NheI/EcoRV insert containing a polyglutamine-expanded (103Q) exon I fragment of HTT using an alternating CAGCAA sequence (Kazantsev et al., 1999) was subcloned into pBi5 downstream of a bidirectional TetO operator sequence (Baron et al., 1995) (pBi5 from Dr. Herman Bujard). Site-directed mutagenesis was used to introduce a stop codon downstream in exon I of HTT (Quickchange mutagenesis, Promega). Successful mutagenesis of the insert was confirmed by Sanger DNA sequencing. Pronuclear injection was carried out by the Transgenic Mouse Shared Resource Center at Columbia University Medical Center using 50 μg of pBi2-HTT103Q which was linearized using AseI; transgenic mice were produced on a CBAXC57Bl6 F1 background. For colony maintenance, tetO-exon1Htt103Q was bred to homozygosity, with no observed toxicity. HD103Q mice were created by outcrossing tetO-exon1Htt103Q+/+ with CamKIIa-tTA+/−. Resulting F1 offspring were used for subsequent experiments.
HDAlfy mice were created as follows: To create regulatable HD mice with the potential for Alfy iKO, several generations of backcrosses were required to bring Alfyfl/fl and tetO-exon1Htt103Q+/+ to homozygosity with simultaneous heterozygous expression of CamKIIa-tTA and Actin-CreER™. The resulting offspring for each subsequent cross were used to ultimately bring together the 5 different transgenes onto the same mouse line. In the experimental mice in Figures 1 and S1, Alfy knockout and exon1Htt103Q expression are controlled by administering tamoxifen and dox, respectively.
BACHDAlfy mice were created as follows: BACHD mice were outcrossed to AlfyGT/+ mice. All 4 resulting F1 offspring were used as described in the main text.
Cell lines
Patient fibroblasts were obtained from Coriell. Their CAG lengths were verified and maintained as previously described (Victor et al., 2018). The description of each patient line is provided in the main figures.
METHOD DETAILS
Genotyping
PCR to detect genes of interest was performed with 50–200 ng of DNA phenol:chloroform extracted from earpunch biopsies, using 5 PRIME Hot Mastermix (5-PRIME) in the Eppendorf AG 22331 Hamburg Mastercycler. BACHD (Gray et al., 2008) and Alfy GT mice (Dragich et al., 2016) were genotyped as previously described. PCR for Alfy excision was performed using the forward primer from the first loxP site (P1) and the reverse primer from the second loxP site (P2). The primers were as follows: Alfy lox P1 forward GAA ACG AAG CTC GTT TAC GG, reverse TGC AGT GAC ATT TCC TCT GG; Alfy lox P2 forward ACT TGG GAA GAG GGA AGC TC, reverse AGG TTA CCA GCC ACA ACC AG; Actin CreERTM forward GCG GTC TGG CAG TAA AAA CTA TC, reverse GTG AAA CAG CAT TGC TGT CAC TT; Internal control for Actin CreERTM forward CTA GGC CAC AGA ATT GAA AGA TCT, reverse GTA GGT GGA AAT TCT AGC ATC ATC C; tetO-exon1Htt103Q forward TCC TCT GAC ACA TAA TTC GCC, reverse GTT GTT CCA TTC CAT CAC GG; CamKIIa tTA forward GTG ATT AAC AGC GCA TTA GAG C, reverse GAA GGC TGG CTC TGC ACC TTG GTG; and DNA Loading control for transgenics (Terd) forward CAA ATG TTG CTT GTC TGG TG, reverse GTC AGT CGA GTG CAC AGT TT.
Preparation and administration of drugs:
Tamoxifen (Sigma) was prepared at 10 mg/mL in a solution containing 5% ethanol in corn oil, by heating at 37°C for 4 h and vortexing frequently. Vehicle control was prepared by omitting tamoxifen from the solution. Tam or vehicle control was administered intraperitoneally at 2 mg/26 g BW (or 200 μL solution/26 g BW) daily for five consecutive days. Doxycycline (Sigma) was prepared at 2 mg/mL in 5% sucrose solution, in red tinted water bottles to protect against light sensitivity. Non-doxycycline-treated control mice received control water containing 5% sucrose. Sucrose water was autoclaved and cooled prior to the addition of doxycycline and was replaced 1–2 times weekly. Mice received dox beginning at 7 months of age for durations varying between 2 weeks and 4 months, as indicated in the text.
Behavioral Testing
All animals were drug naïve prior to treatment. Mice were maintained in group housing, and experiments were therefore performed by cage. Littermates of the same sex were randomly assigned to experimental groups at weaning. Two weeks prior to the start of behavioral testing, experimental cohorts were relocated to a reverse-cycle core facility with lights off between 10 am and 10 pm, so that longitudinal experiments could be performed during dark (active) hours. For testing on the Accelerated Rotarod, mice were first trained on the rotarod apparatus with grip (Ugo Basile, Varese, Italy) on an initial exposure day, with one trial at a constant speed of 5 rpm for 5 min, a second trial with speed accelerating from 5 rpm to 15 rpm over 5 min, and a third trial with speed accelerating from 5 to 25 rpm. The number of training days was dependent upon the strain, which was established by running only control animals across multiple days on this training regimen such that 2-month-old mice can successfully complete the task. 3 (BACHD) or 4 (HD103Q) days of testing with 3 trials daily of 5 minutes each, with speeds accelerating from 5 to 40 rpm and lasting 300 seconds per trial. Resting periods in between trials were always 35–60 min. Mice were allowed 1 fall from the rotarod and on the second fall, the latency in seconds was recorded. For Open Field, BACHD and littermate controls were given a one-hour exposure to the open field arena (43.2 cm × 43.2 cm × 30.5 cm) at intervals of two months. Distance traveled was recorded using equipment and software from Med Associates.
Tissue preparation for detergent soluble and insoluble fractions
For the preparation of tissue lysates, mice were euthanized with CO2 and decapitated. Brains were removed and placed into ice-cold 0.1 M Sorensen’s phosphate buffer pH 7.4 (1X PB). Samples were flash-frozen on powdered dry ice and stored at −80°C until processing. Where noted, brains were dissected prior to freezing. Frozen tissue (50–500 mg) was placed in a glass dounce homogenizer with an equal volume of 1X phosphate buffer saline (PBS) containing 2X Halt protease inhibitor cocktail (ThermoFisher) and disaggregated with 40 pumps of a pestle. The suspension was transferred to a tube and an equal volume of detergent (2% TritonX-100 [Tx-100] in PBS) was added to bring the final detergent concentration to 1%Tx-100 in PBS (PBS-Tx), and incubated for 30 minutes on ice, then spun in an Eppendorf 5417R centrifuge at 1000 rpm for 5 minutes. To segregate the Tx-100 soluble versus insoluble fraction, the supernatant (S1) was centrifuged at 14,000 rpm for 5 minutes. The resulting supernatant (S2) represents the detergent soluble fraction, whereas the pellet (P2) the detergent insoluble fraction. P2 was suspended in 1 PBS-Tx in 8 M urea and incubated on ice for 15 minutes. This was then centrifuged at 14,000 rpm for 5 minutes, and if a small viscous pellet (nucleic acid) was detected, the pellet was carefully removed leaving the solubilized supernatant (S3). The protein concentration of relevant suspensions was quantified using the BioRad DC protein kit according to manufacturer’s instructions. Detergent soluble and insoluble fractions were probed for 1C2, 3B5H10, Huntingtin (C-term) gamma tubulin and vinculin. S1 was also probed for Nbr1, Optineurin, Atg5 ,vinculin and Alfy
Fractionation for autophagic vacuoles
Cell fractionation was performed with 10 g of brain tissue as previously described (Filimonenko et al., 2010; Stromhaug and Seglen, 1993). Briefly, brain homogenates were incubated with 0.5 mM GPN (to break lysosomes) then spun at 2000 x g for 2 min at 4°C to pellet nuclei. The post-nuclear supernatant was fractionated using step gradients of Nycodenz (Sigma), followed by Nycodenz and Percoll (Sigma) and spun at 141,000 and 72,000 x g respectively at 4 °C. Percoll was removed from the AV-enriched fraction with 30% Optiprep (Sigma) at 71,000 x g at 4°C for 30 min. Fractions were collected at each step, quantified by Bradford (Biorad) and run on Western blots. Blots were probed for LC3B, p62, Alfy, and Tom20.
Western blot (SDS-PAGE)
5–20 μg of protein was loaded onto a NuPAGE 4–12% bis-tris gel (ThermoFisher), or a 3–12% tris-acetate gel (ThermoFisher, for Alfy and mHtt) and transferred to PVDF or PVDF invitrolon membranes (ThermoFisher). Membranes were blocked in 3% BSA at room temperature for 1 hour and incubated in primary antibody overnight. Primary antibody dilutions contained 0.01% tween in PBS with 3% BSA. Blots were incubated for 1–2 hours with horseradish peroxidase-conjugated secondary antibodies (ThermoFisher Pierce) at 1:2000 to 1:5000 for most antibodies (1:1000 for anti-Alfy (Lystad et al., 2014)) in 0.01% 1X PBS-Tween containing 3% BSA, and developed using Clarity Western ECL substrate (BioRad) and detected using the BioRad VersaDoc imaging system. Band intensities were analyzed using QuantityOne software (Biorad) or ImageJ and normalized to a loading control.
In situ hybridization:
Mice were deeply anesthetized with isoflurane and decapitated. Brains were removed and halved sagitally, then frozen in −20°C isopropane and embedded in Tissue-Tek (Torrance, CA) optimal cutting temperature (OCT) embedding medium over dry ice. 20 μm sections were cut using a Leica CM 1950 cryostat and thaw-mounted onto untreated Fisher Superfrost plus slides at intervals of 200 μm. Slides were stored at −80°C until required. Prior to probing, sections were dried at RT for 30 minutes and transferred into cold 4% paraformaldehyde (PFA) in 1X PBS. Slides were washed once in 1X PBS for 5 minutes, followed by dehydration in 70% EtOH for 5 minutes and storage in 100% EtOH at 4°C until use. To create the hybridization probe, a 45-base oligonucleotide probe was designed complementary to bases 766–810 of Wdfy3 mRNA (GenBank accession no. NM_172882). 150 ng of oligonucleotide was radiolabeled with α-[32P]-dATP using the Roche terminal transferase 3′-end labeling kit, according to manufacturer’s instructions. The labeled probe was extracted using phenol-chloroform-isoamyl alcohol and unincorporated radionucleotides were removed by centrifugation through an illustra Microspin-G25 column (GE Healthcare, Chicago, IL). Radioactivity was measured by liquid scintillation counter. Probes were diluted in minimal hybridization buffer containing 50% formamide, 4X SSC, and 10% dextran sulfate. Prior to probe application sections were air dried for 15 minutes. Hybridization was performed overnight at 42°C in a humidified chamber. Slides were washed first at 60°C for 30 minutes, then with 1X SSC and 0.1X SSC at room temperature for 5 min. Slides were briefly dehydrated in 70 and 95% ethanol, air dried, and exposed to film for 2 weeks.
Immunohistochemistry
Mice were deeply anesthetized (as confirmed with multiple toe pinches) with isoflurane and transcardially perfused with 0.9% saline for 2 minutes followed by 4% paraformaldehyde (PFA) for 3 minutes. Brains were removed and post-fixed in 4% PFA for 4 hours, then incubated in 30% sucrose in 1X PB for 48–72 h. Brains were snap-frozen in powdered dry ice then cryoprotected in OCT embedding medium and stored at −80°C. Tissue was sectioned at 30 μm using a Leica CM 1950 cryostat and stored at 4°C in phosphate buffer containing 0.02% sodium azide as a preservative. Representative sections throughout the forebrain, spaced at 240 μm, were stained with antibodies against huntingtin and GFAP. Briefly, sections were washed in PBS containing 0.02% Triton-X100, followed by antigen retrieval in sodium citrate, pH 9.0 at 80°C for 30 minutes. After three more washes, endogenous peroxidases were blocked with 1% hydrogen peroxide, and again washed three times. Sections were blocked in 0.4% Triton-X for 1 hour (for EM48 staining, blocking solution also contained 3% BSA and 2% NGS), then incubated overnight for up to 3 days in primary antibody. Sections were incubated in biotinylated secondary antibody at room temperature for 1 hour or overnight, and signal was amplified using the Vectastain ABC Kit (Vector labs). For S830 staining only (Sathasivam et al., 2010), this was followed by an additional 10 minute tyramide amplification step (Perkin Elmer) followed by washes and a second ABC treatment. Signal was detected using diaminobenzidine (DAB) (10 mg/25 mL) in phosphate buffer containing 0.00001% H2O2. Following the DAB reaction, sections were mounted on glass slides and air-dried, counterstained with thionin, dehydrated in ascending grades of ethanol, cleared in xylene, and coverslipped with Permount mounting medium (Fisher).
Stereology
Serially-cut coronal brain sections (every 240 μm) were used for all volumetric, neuronal, and huntingtin aggregate analyses. Tissue was matched across brains during sectioning, by designating “set 1” as the representative set containing the section with the initial crossing of the anterior commissure. For each immunostain, a number between 1 and 8 was selected at random, and this set number was used for staining that epitope across all brains. For assessment of HD103Q striatal accumulation of Htt epitope EM48, analysis of the right hemisphere began at the anterior tip of the genu of the corpus callosum (interaural 4.98 mm/bregma 1.18 mm, Franklin and Paxinos 2001) and extended through the anterior emergence of CA3 (interaural 2.86 mm/bregma −0.94 mm, Franklin and Paxinos 2001) These rostrocaudal bounds resulted in 8–10 sections per brain spaced 240 microns apart. The neostriatum was defined within the rostral-caudal plane as being bounded dorsally by the corpus callosum, dorsolaterally by the external capsule, and medially by the lateral ventricle. Ventrolaterally the neostriatum continues in an arc bounded ventrolaterally by the claustrum, the dorsal endopiriform nucleus, the lateral striatal stripe, the lateral accumbens shell (or more ventrally, the interstitial nucleus of the posterior limb of the anterior commissure), through the anterior commissure and/or internal capsule, and to the ventral tip of the lateral ventricle. For assessment of BACHD cortical accumulation of Htt epitope S830, anatomical bounds were the same as above for the striatum, with additional analysis of the left dorsolateral cortex. The extent of the cortical region analyzed was dorsal to the corpus callosum arcing ventrolaterally and extending to the secondary somatosensory cortex, as bounded by tracing horizontally from the lateral edge of the external capsule to the edge of the tissue. Unbiased stereological counts were obtained from the striatum or cortex using Stereo Investigator Software (MBF Bioscience). The optical dissector method was used to count profiles (neurons or Htt-positive aggregates) in an unbiased random selection of serial sections in a defined volume of the cortex or striatum. Based on pilot studies, the counting parameters selected involved a 300 × 300 grid with a 50 × 50 counting frame, and a fixed tissue thickness of 30 μm. In all mice, Gundersen coefficients of error (CE) for Nissl-positive neuronal counts in all mice were on average (0.02).
Direct conversion
Lentiviral preparation of a doxycycline-responsive synthetic cluster of miR-9/9* and miR-124 (Yoo et al., 2011) as well as transcription factors cloned downstream of the EF1α promoter were used to transduce these cells as previously reported to generate cells analogous to human medium spiny neurons (Richner et al., 2015; Victor et al., 2014; Victor et al., 2018). Infected human fibroblasts were maintained in 15%FBS DMEM media for 5 days with doxycycline before re-plating onto coated coverslips. Cells were then selected with appropriate antibiotics in neuronal media (ScienCell) or Neurobasal −B27plus (Invitrogen) supplemented with valproic acid (1 mM), dibutyryl cAMP (200 μM), BDNF (10 ng/mL), NT-3 (10 ng/mL), RA (1 μM), and RevitaCell (100x). Doxycycline was replenished every two days and media were changed every 4 days. Knockdown of ALFY was achieved by transducing shRNAs into human fibroblasts at the origin of microRNA-mediated reprogramming. Briefly, single-stranded oligos designed for human ALFY or a non-specific target (Control shRNA) were annealed into hairpins and ligated into the PLKO.1 puro shRNA vector ((Stewart et al., 2003)Addgene vector #8453) which was modified in-house to carry blasticidin resistance rather than puromycin, as to not interfere with the vector carrying the synthetic cluster of miR-9/9* and miR-124. At day 19 of reprogramming, cells were analyzed by immunofluorescence. Cells were fixed using 4% paraformaldehyde for 20 min at room temperature (RT) followed by permeabilization with 0.2% Triton X-100 for 10 min. One percent goat serum was used for blocking followed by incubation of primary antibodies overnight at 4°C. Secondary antibodies conjugated to Alexa-488, or −594 were applied for 1 hour at RT. Images were captured using a Leica SP5X white light laser confocal system with Leica Application Suite (LAS) Advanced Fluorescence 2.7.3.9723. Staining quantification in direct converted neurons was performed by counting number of cells bearing inclusions (MW8 or MAB5492 positive) over DAPI signal. LC3B was quantified with ImageJ particle analyzer tool and averaged over total DAPI-positive count. Antibodies were validated by staining fibroblasts as negative controls, and exhibited low background.
QUANTIFICATION AND STATISTICAL ANALYSIS
All images were prepared using Adobe Photoshop CS5 and Adobe Illustrator CS5. Statistical analyses of behavioral data, stereologically-obtained counts and volumes (obtained using MicroBrightfield StereoInvestigator), puncta (ImageJ) and Western blot band intensities (obtained with ImageJ) were performed using Statview (Scientific Computing), Matlab or R. Power analyses for all in vivo testing were performed using GPower3.1, for which the variance was established in pilot studies or previously published data. Statistical approaches and complete statistical information are provided in the appropriate figure legend. Significance was accepted at the 95% probability level. Behavioral data are represented as mean +/− SEM or SD as noted in figures. The n values are defined in the respective figure legends, but are briefly as follows: Figure 1, n represents number of mice; Figure 2, n represents number of brains; Figure 3, n represents number of mice; and Figure 4, n represents number of cells.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code
Supplementary Material
Highlights.
Autophagy adaptor protein Alfy/WDFY3 is required to clear mutant huntingtin in vivo. (84 c)
Alfy depletion accelerates aggregation, neuronal stress, and pathogenesis in HD mice. (85 c)
Reduction of Alfy increases aggregation but not cell death in patient-derived MSNs. (83 c)
Genetic modifications impacting protein aggregation may modify age of disease onset. (84 c)
Acknowledgements
The authors would like to acknowledge and thank Drs. Robert Burke and Christoph Kellendonk and their lab members for technical guidance and support, and Drs. Dritan Agalliu, Robert Burke, Tyler Cutforth, Lisa Ellerby, Lloyd Greene, Christoph Kellendonk and members of the Yamamoto lab for helpful discussions. A special thanks to Katharine Abbot and William Jackson Lovejoy for their contributions, and Drs. Gillian Bates and Masaaki Komatsu for use of antibodies created in their labs. This work was supported by the HDF (LMF, KRC, AY), NINDS R01 NS063973 (JRB, EE, CWJ, AY), NINDS R01 NS077111 (JMD, LMF, AY), NINDS R01 NS107488 (SC, ASY), HDSA Don King Summer Fellowship (KK), Ruth L. Kirschstein NRSA Predoctoral Fellowship (LMF), T32 NS064928 (LMF) and NSF Predoctoral Fellowship (KRC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Uncategorized References
- Baron U, Freundlieb S, Gossen M, and Bujard H (1995). Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res 23, 3605–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, et al. (2010). p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344. [DOI] [PubMed] [Google Scholar]
- Dragich JM, Kuwajima T, Hirose-Ikeda M, Yoon MS, Eenjes E, Bosco JR, Fox LM, Lystad AH, Oo TF, Yarygina O, et al. (2016). Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain.Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eenjes E, Dragich JM, Kampinga HH, and Yamamoto A (2016). Distinguishing aggregate formation and aggregate clearance using cell based assays. J Cell Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. (2010). The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Molecular cell 38, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Modifiers of Huntington’s Disease, C. (2015). Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell 162, 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al. (2008). Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci 28, 6182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Li C, Wei W, Lo V, Gong S, Li SH, Iwasato T, Itohara S, Li XJ, Mody I, et al. (2005). Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron 46, 433–444. [DOI] [PubMed] [Google Scholar]
- Gusella JF, and MacDonald ME (2009). Huntington’s disease: the case for genetic modifiers. Genome Med 1, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, and Li XJ (1999). Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci 19, 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, and McMahon AP (2002). Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Developmental biology 244, 305–318. [DOI] [PubMed] [Google Scholar]
- HDCRG (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72, 971–983. [DOI] [PubMed] [Google Scholar]
- Hocking LJ, Mellis DJ, McCabe PS, Helfrich MH, and Rogers MJ (2010). Functional interaction between sequestosome-1/p62 and autophagy-linked FYVE-containing protein WDFY3 in human osteoclasts. Biochem Biophys Res Commun 402, 543–548. [DOI] [PubMed] [Google Scholar]
- Huh CJ, Zhang B, Victor MB, Dahiya S, Batista LF, Horvath S, and Yoo AS (2016). Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hult S, Soylu R, Bjorklund T, Belgardt BF, Mauer J, Bruning JC, Kirik D, and Petersen A (2011). Mutant huntingtin causes metabolic imbalance by disruption of hypothalamic neurocircuits. Cell metabolism 13, 428–439. [DOI] [PubMed] [Google Scholar]
- Isakson P, Lystad AH, Breen K, Koster G, Stenmark H, and Simonsen A (2013). TRAF6 mediates ubiquitination of KIF23/MKLP1 and is required for midbody ring degradation by selective autophagy. Autophagy 9, 1955–1964. [DOI] [PubMed] [Google Scholar]
- Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, and Kopito RR (2005a). Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci U S A 102, 13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, and Kopito RR (2005b). HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 280, 40282–40292. [DOI] [PubMed] [Google Scholar]
- Kadir R, Harel T, Markus B, Perez Y, Bakhrat A, Cohen I, Volodarsky M, Feintsein-Linial M, Chervinski E, Zlotogora J, et al. (2016). ALFY-Controlled DVL3 Autophagy Regulates Wnt Signaling, Determining Human Brain Size. PLoS Genet 12, e1005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, and Frydman J (2008). Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, and Housman D (1999). Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci U S A 96, 11404–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8, 931–937. [DOI] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, Shihabuddin LS, et al. (2012). Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lystad AH, Ichimura Y, Takagi K, Yang Y, Pankiv S, Kanegae Y, Kageyama S, Suzuki M, Saito I, Mizushima T, et al. (2014). Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep 15, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ME, and Gusella JF (1996). Huntington’s disease: translating a CAG repeat into a pathogenic mechanism. Curr Opin Neurobiol 6, 638–643. [DOI] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, and Kandel ER (1996). Control of memory formation through regulated expression of a CaMKII transgene. Science 274, 1678–1683. [DOI] [PubMed] [Google Scholar]
- Mertens J, Paquola ACM, Ku M, Hatch E, Bohnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. (2015). Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 17, 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Arrasate M, Brooks E, Libeu CP, Legleiter J, Hatters D, Curtis J, Cheung K, Krishnan P, Mitra S, et al. (2011). Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nature chemical biology 7, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Jang I, Zuber C, Lee Y, Cho JW, Matsuo I, Ito Y, and Roth J (2014). ERADication of EDEM1 occurs by selective autophagy and requires deglycosylation by cytoplasmic peptide N-glycanase. Histochem Cell Biol 142, 153–169. [DOI] [PubMed] [Google Scholar]
- Perutz MF, and Windle AH (2001). Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature 412, 143–144. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, and Rubinsztein DC (2002). Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11, 1107–1117. [DOI] [PubMed] [Google Scholar]
- Richner M, Victor MB, Liu Y, Abernathy D, and Yoo AS (2015). MicroRNA-based conversion of human fibroblasts into striatal medium spiny neurons. Nature protocols 10, 1543–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K, Lane A, Legleiter J, Warley A, Woodman B, Finkbeiner S, Paganetti P, Muchowski PJ, Wilson S, and Bates GP (2010). Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington’s disease. Hum Mol Genet 19, 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek LM, Sardi SP, Mastis B, Richards AR, Treleaven CM, Taksir T, Misra K, Cheng SH, and Shihabuddin LS (2014). Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington’s disease. Hum Gene Ther 25, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug PE, and Seglen PO (1993). Evidence for acidity of prelysosomal autophagic/endocytic vacuoles (amphisomes). Biochem J 291 (Pt 1), 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier Y, Devys D, Imbert G, Saudou F, An I, Lutz Y, Weber C, Agid Y, Hirsch EC, and Mandel JL (1995). Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nat Genet 10, 104–110. [DOI] [PubMed] [Google Scholar]
- Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng PY, Klyachko VA, Nerbonne JM, and Yoo AS (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, Huh CJ, Zhang B, Davidson BL, Yang XW, et al. (2018). Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat Neurosci 21, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP (2008). Huntington disease models and human neuropathology: similarities and differences. Acta neuropathologica 115, 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, and DiFiglia M (1998). Huntington disease. J Neuropathol Exp Neurol 57, 369–384. [DOI] [PubMed] [Google Scholar]
- Wang N, Gray M, Lu XH, Cantle JP, Holley SM, Greiner E, Gu X, Shirasaki D, Cepeda C, Li Y, et al. (2014). Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington’s disease. Nat Med 20, 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts SA, Gayan J, et al. (2004). Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc Natl Acad Sci U S A 101, 3498–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, and Rothman JE (2006). Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol 172, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, and Hen R (2000). Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell 101, 57–66. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, and Crabtree GR (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code