Group 2 innate lymphoid cells (ILC2s) regulate inflammation and immunity in tissues1. Although ILC2s are detected in cancers of these tissues2, their role in cancer immunity and immunotherapy are unclear. Here, we identify that ILC2s infiltrate pancreatic ductal adenocarcinomas (PDAC) to activate tissue-specific tumor immunity. Interleukin-33 (IL33) activates tumor ILC2s (TILC2s) and CD8+ T cells in orthotopic pancreatic but not heterotopic skin tumors to restrict pancreas-specific tumor growth. Resting and activated TILC2s express the inhibitory checkpoint receptor PD-1, and TILC2s further expand with PD-1 blockade (αPD-1) to enhance tumor control. PD-1 blockade acts directly on TILC2s to augment anti-tumor immunity and αPD-1 immunotherapy efficacy, identifying activated TILC2s as novel targets of αPD-1. Finally, both PD-1+ TILC2s and PD-1+ T cells are present in the majority of human PDACs. Collectively, we identify ILC2s as novel anti-cancer immune cells for PDAC immunotherapy. More broadly, ILC2s emerge as tissue-specific enhancers of cancer immunity that amplify efficacy of αPD-1 immunotherapy. As ILC2s and T cells co-exist in human cancers with shared pathways of activation and inhibition, collective targeting of anti-cancer ILC2s and T cells may be a broadly applicable immunotherapeutic approach.
TILC2s infiltrate pancreatic cancers
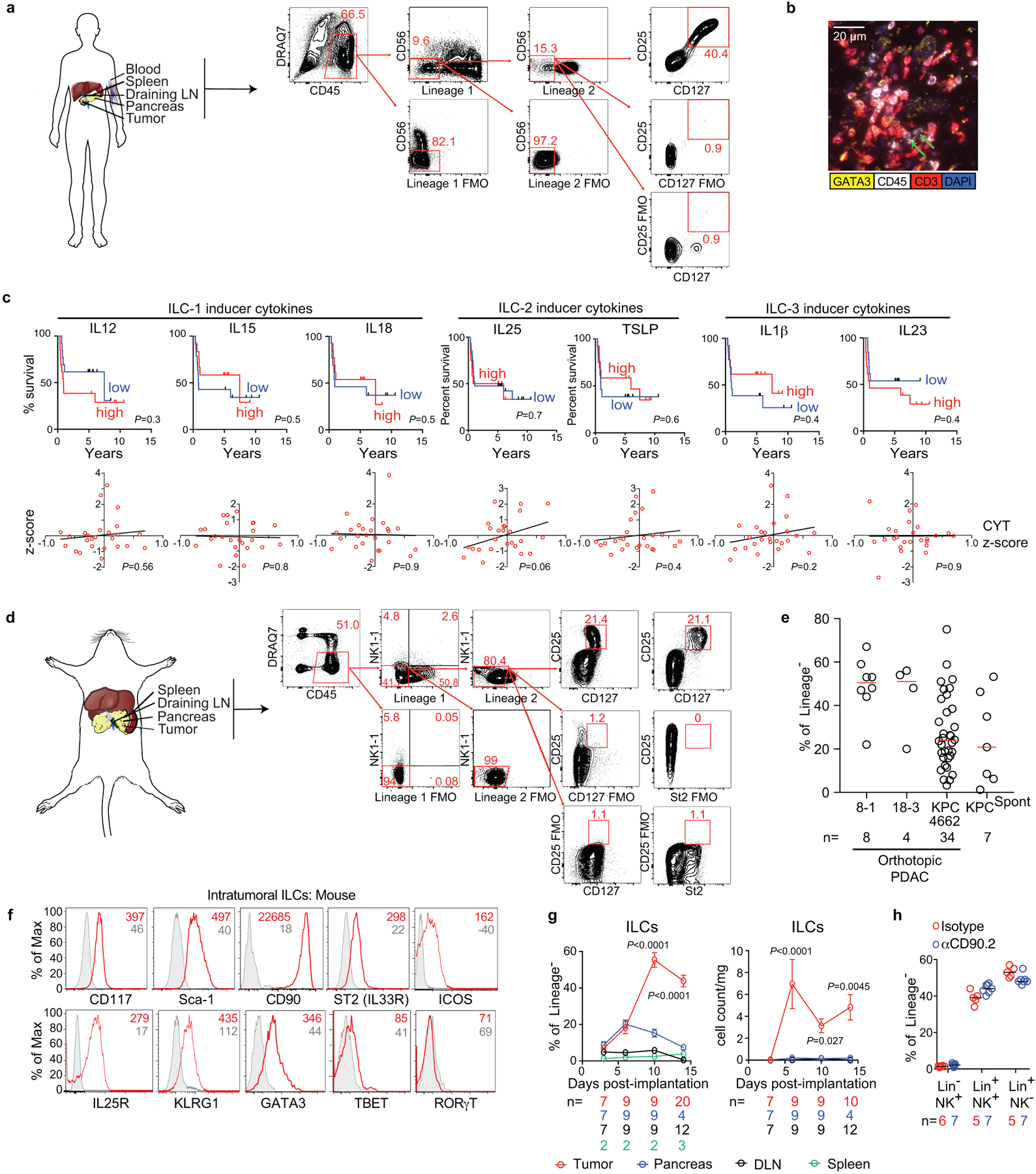
In unselected primary human PDACs, we found intratumoral cells lacking immune cell lineage markers (Lin−) but expressing ILC (CD25 and CD127)1, and ILC2 (IL33-receptor ST2/IL1RL1/IL33R, and GATA3) markers (Figure 1a, Extended Data Figure 1a, Supplementary Table 1). These putative TILC2s were enriched in rare long-term PDAC survivors with “hot” tumors (activated CD8+ T cell enriched)3 when compared to short-term survivors with cold tumors, and higher TILC2 frequencies correlated with longer survival (Figure 1b, Extended Data Figure 1b). Consistently, higher bulk tumor RNA expression of the ILC2-activating cytokine IL33, but no other ILC activating cytokine, was associated with longer survival (Figure 1c, Extended Data Figure 1c, Supplementary Table 2). Furthermore, IL33, but no other ILC-activating cytokines, correlated with higher intratumoral immune cytolytic activity (Figure 1c, Extended Data Figure 1c). Although these data assess RNA and not protein content, they suggested that IL33 and TILC2s activate anti-tumor immunity in human PDAC.
Figure 1: IL33-dependent ILC2s infiltrate human and murine pancreatic cancer.
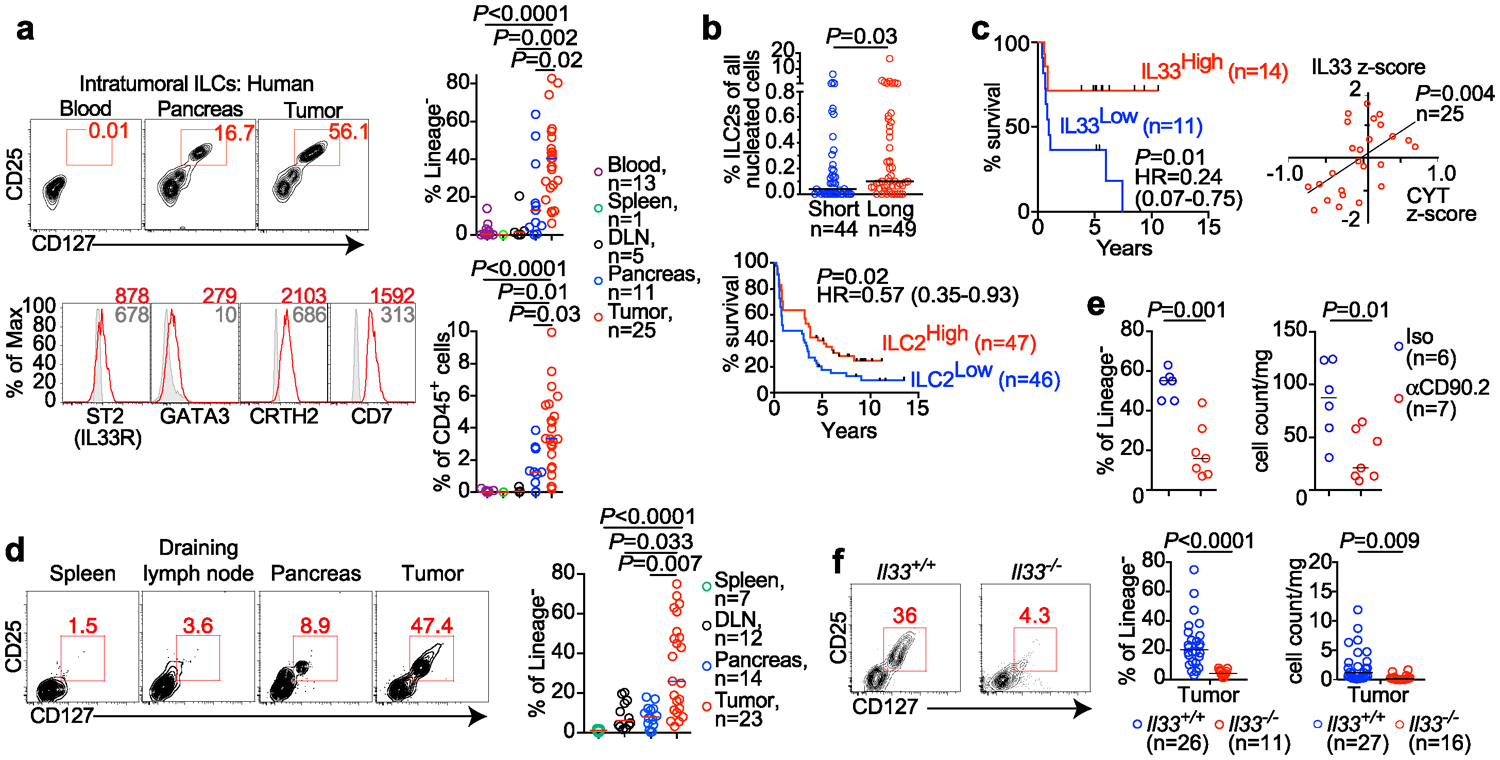
(a) Gating, frequency, and phenotype of ILCs in unselected human PDAC patients. (b) Frequency (top) and survival association (bottom) of ILC2s in tumor tissue microarrays of short-term and long-term PDAC survivors. (c) Bulk tumor IL33 mRNA associations with survival and correlation with tumor cytolytic index (CYT) in short- and long-term PDAC survivors. (d) Gating and frequency of ILCs in PDAC mice. (e) Intratumoral ILC frequency and number in Rag2−/− PDAC mice treated with αCD90.2 or isotype (Iso) antibodies. (f) Gating, frequency, and number of ILCs in Il33+/+ and Il33−/− PDAC mice. High and low in b, c defined as higher or lower, respectively, than the median for the cohort. Data were collected at 14 (d-f) post tumor implantation. n, number of tumors from individual patients or mice. Horizontal bars mark medians. Data in d-f are pooled from ≥2 independent experiments with n≥3/group; each point indicates one mouse analyzed separately. P values determined by one-way ANOVA with Tukey’s (a) and Kruskal-Wallis multiple comparison post-tests (d), two-tailed Mann-Whitney test (b, e, f), two-sided log-rank (b, c survival curves), and linear regression (c).
We next surveyed tumors in mutated Kras and p53-driven autochthonous “KPC” mice4 and orthotopic PDAC mouse models (PDAC mice5, 6) for ILCs. In both models, we detected TILC2s phenotypically similar to those in human PDAC and to murine ILC2s1,7 (Figure 1d, Extended Data Figure 1d–f). Murine TILC2s expanded in tumors, but not in adjacent organs (Figure 1d, Extended Data Figure 1g), consistent with their tissue-residency8, and were depleted in Rag2−/− mice9,10 by targeting the lymphocyte antigen CD90.2 (Figure 1e, Extended Data Fig 1h). Therefore, ILC2s are conserved cells that expand locally in mouse and human PDAC.
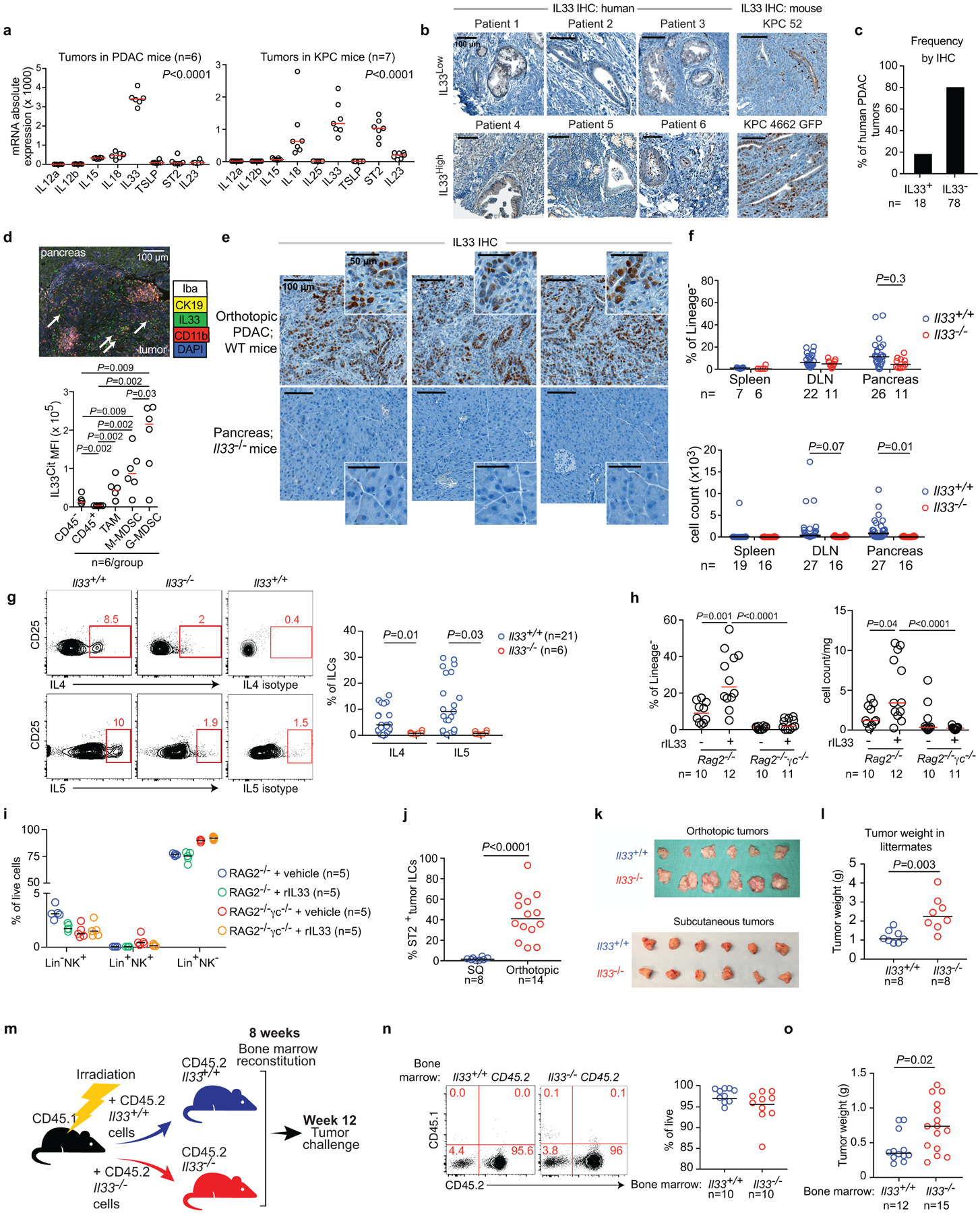
In identifying the signals inducing TILC2 expansion, we found that in both PDAC and KPC mice11 IL33 had the highest expression in tumors when compared to other ILC-inducer cytokines (Extended Data Figure 2a)6, with heterogeneous expression in both human and mouse PDACs (Extended Data Fig 2b–c) and maximal expression in intratumoral myeloid cells12,13 (Extended Data Fig 2d, e). To understand the role of IL33 and TILC2s in PDAC immunity, we studied TILC2 dependencies in IL33High PDAC mice that reflect IL33High, ILC2 enriched hot tumors in long-term human PDAC survivors. TILC2 expansion and function was IL33 dependent, as Il33−/− PDAC mice had reduced TILC2 frequency, number (Figure 1f, Extended Data Figure 2f), and cytokine production (Extended Data Figure 2g) when compared to Il33+/+ PDAC mice. Consistently, recombinant IL33 (rIL33) expanded ILCs in ILC-proficient Rag2−/− PDAC mice, but not in ILC-deficient Rag2−/−γc−/− PDAC mice (Extended Data Figure 2h, i). Collectively, these experiments demonstrated that IL33 expanded PDAC TILC2s.
TILC2s boost tumor immunity in tissues
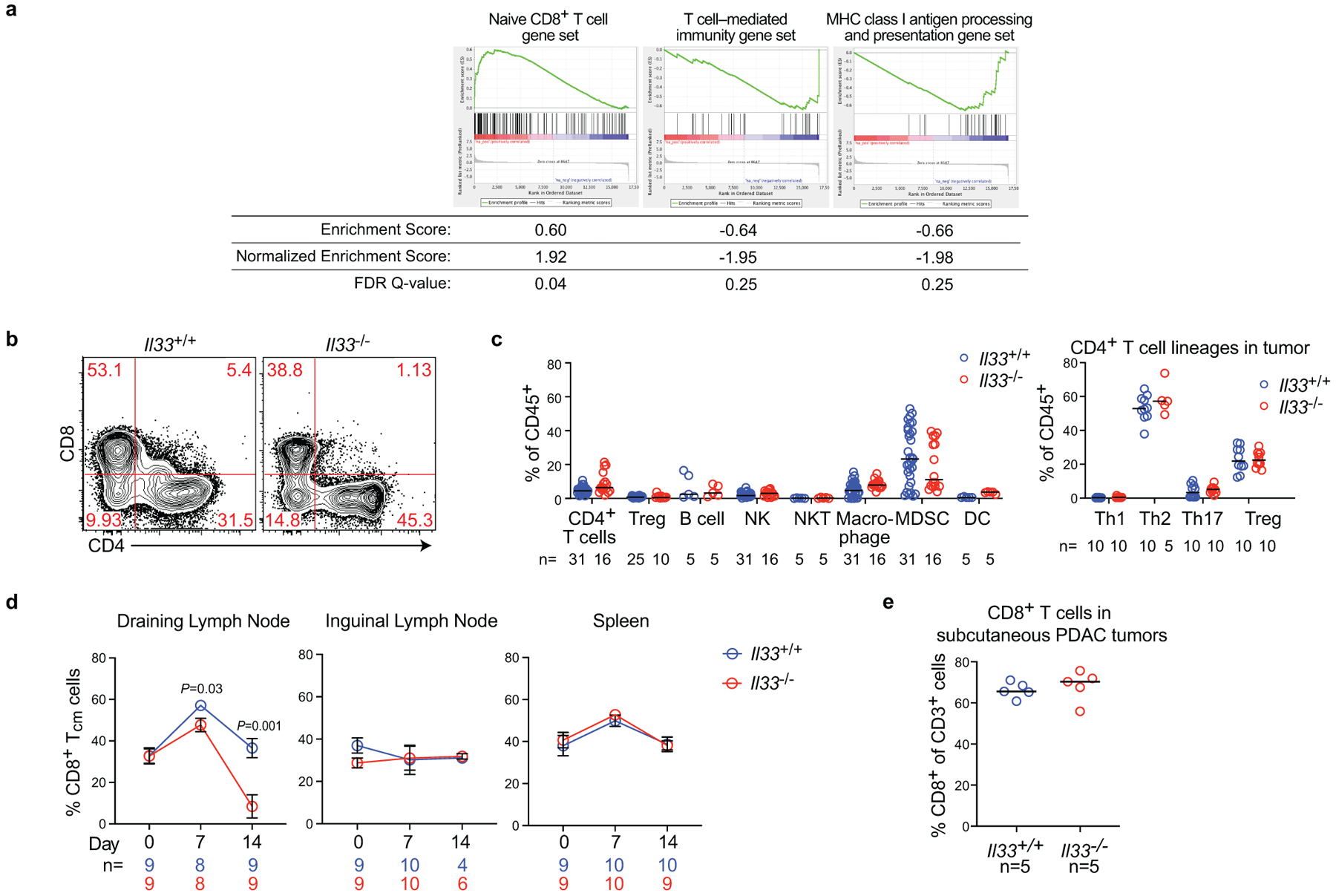
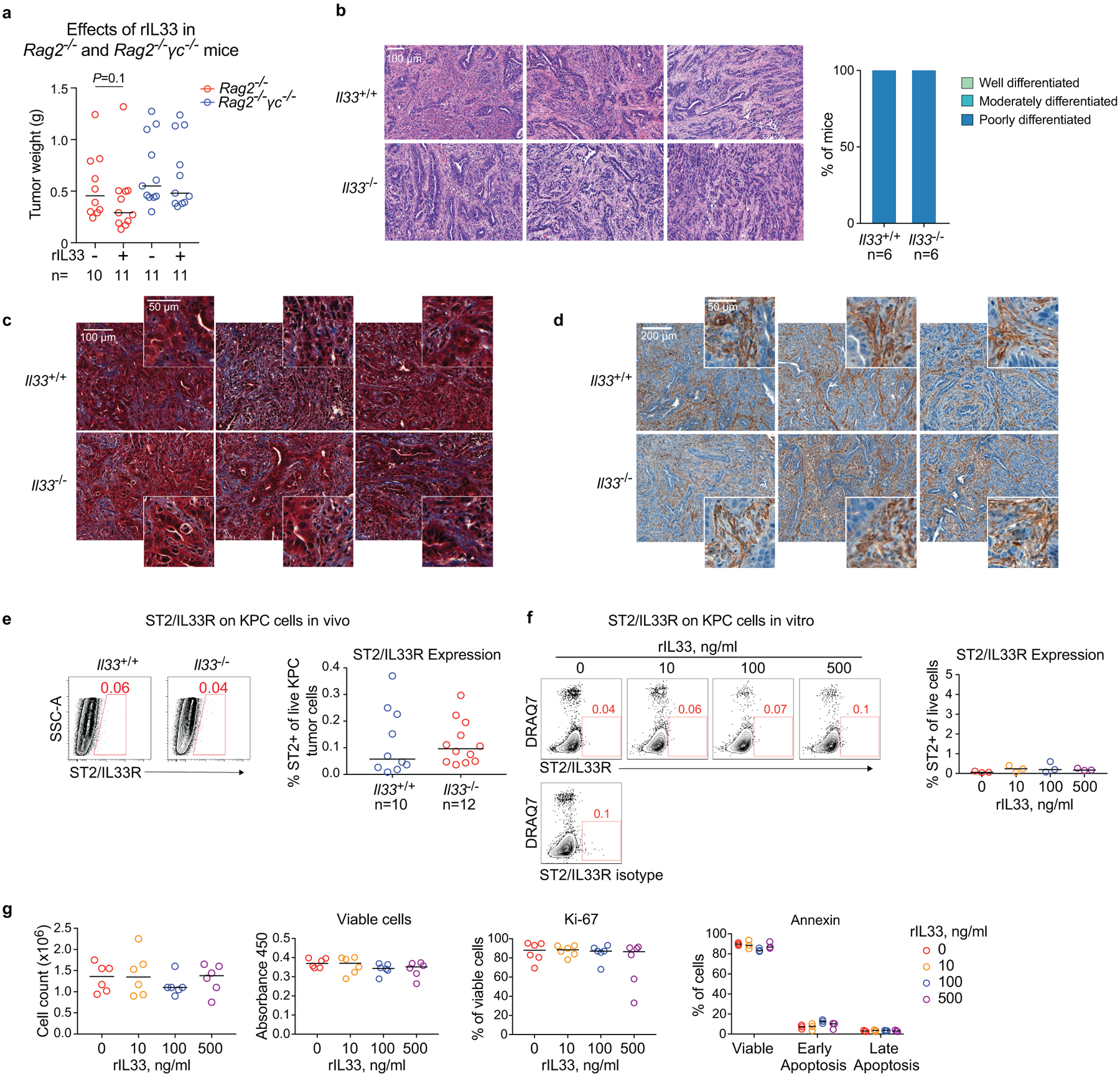
As ILC2s have tissue-specific phenotypes14, we hypothesized that TILC2 effects on PDAC immunity would be tissue-specific. To test this, we contrasted the effects of IL33 deficiency on tumor growth in the pancreas to the skin (pancreatic TILC2s express ST2, skin TILC2s do not; Extended Data Figure 2j14,15). Compared with Il33+/+ animals, Il33−/− mice with orthotopic PDAC had larger tumors, accelerated tumor growth, and worse survival (Figure 2a), in contrast to subcutaneous PDAC mice that showed no IL33-dependent phenotype (Figure 2b, Extended Data Figure 2k). Although these mice were fully backcrossed onto identical genetic backgrounds, we confirmed these differences were not due to potential minor genetic mismatches by observing larger tumors in Il33−/− vs. Il33+/+ littermates (Extended Data Figure 2l). These anti-tumor effects were host hematopoietic cell–derived IL33 dependent, as chimeric mice transplanted with Il33−/− bone marrow had larger tumors compared to control mice (Extended Data Figure 2m–o). RNA sequencing (RNA-seq) of purified CD45+ intratumoral immune cells from Il33+/+ and Il33−/− orthotopic PDAC mice revealed Il33−/− PDAC immune cells had diminished transcriptional signatures of T cell activation and MHC-I antigen processing, suggesting a T cell priming deficiency (Extended Data Figure 3a). Consistently, Il33−/− orthotopic but not subcutaneous PDAC mice had lower frequencies of global and activated tumor-infiltrating CD8+ T cells with no consistent changes in other immune cell frequencies, and reduced central memory CD8+ T cells (TCM) in draining (DLN) but not distant lymph nodes (Figure 2c, Extended Data Figure 3c–e). The increase in tumor size in Il33−/− compared to ll33+/+ mice was abrogated upon pan-T cell depletion (Figure 2d), with no differences in tumor weight in rIL33-treated Rag2−/− PDAC mice (Extended Data Figure 4a), confirming the anti-tumor effects of IL33 were T cell mediated. Orthotopic tumors from Il33−/− and Il33+/+ PDAC mice also had similar histology, collagen, and fibroblast content (Extended Data Figure 4b–d), with no effects of rIL33 on tumor cells in vitro (Extended Data Figure 4e–g), showing IL33 had no direct effects on tumor or stromal cells. Together, these data demonstrated that IL33 activated tissue-specific cancer immunity by potentially activating TILC2s to prime CD8+ T cells.
Figure 2: The IL33-ILC2 axis activates tissue-specific cancer immunity.
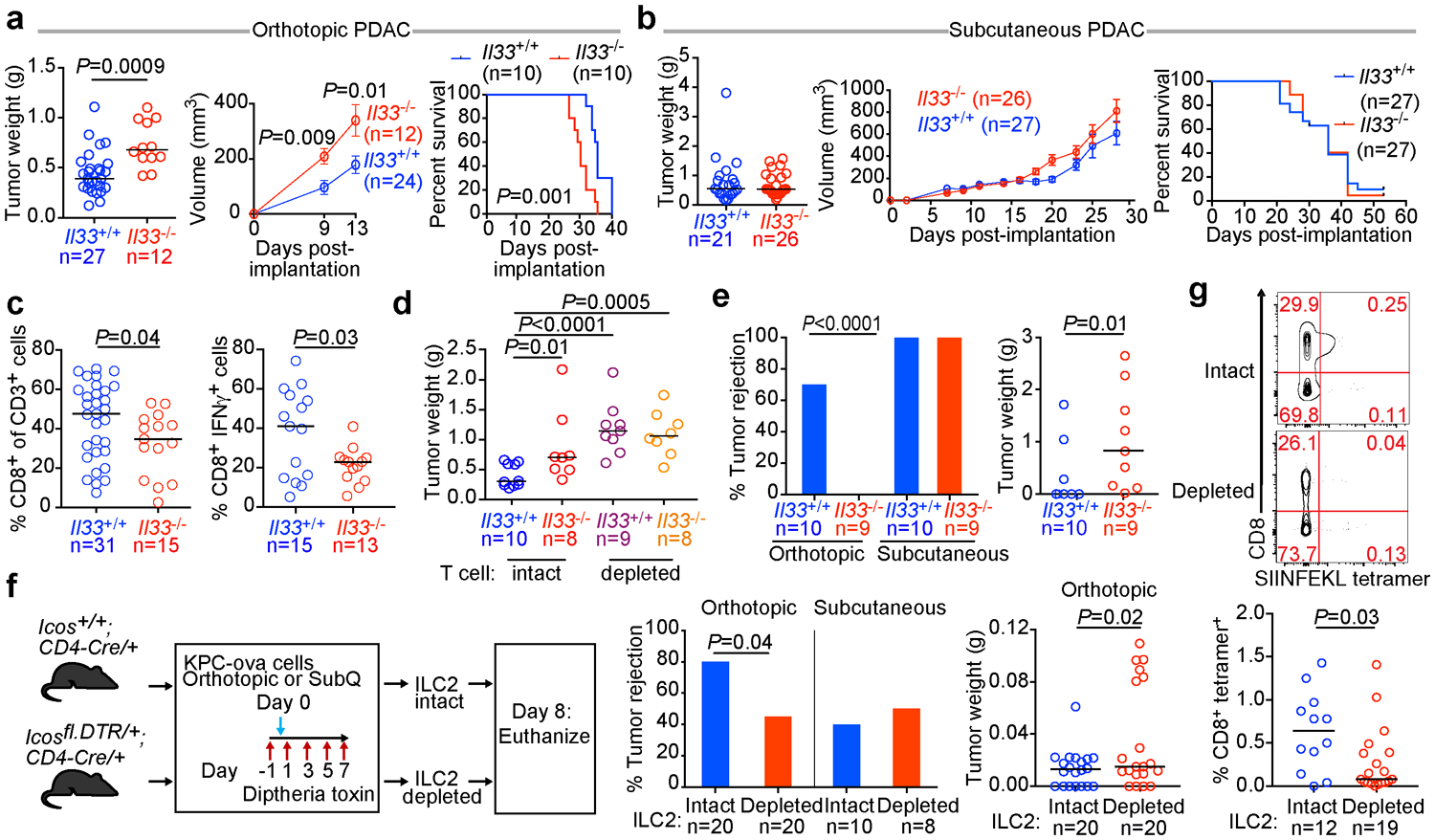
Tumor weight, volumes, and survival of Il33+/+ and Il33−/− orthotopic (a) or subcutaneous (b) PDAC mice. (c) Frequency of all (left) and IFN-γ producing (right) CD8+ T cells in orthotopic Il33+/+and Il33−/−PDAC tumors. (d) Tumor weight in T cell–depleted Il33+/+and Il33−/− orthotopic PDAC mice. (e) Frequency of tumor rejection and tumor weight in Il33+/+and Il33−/− orthotopic and subcutaneous KPC-OVA PDAC mice. (f) Experimental design (left), frequency of tumor rejection (middle), and tumor weight (right) of KPC-OVA PDAC tumors in iCOS-T mice with intact or depleted ILC2s. (g) Frequency of OVA-specific CD8+ T cells in draining lymph nodes of orthotopic KPC-OVA PDAC iCOS-T mice with intact or depleted ILC2s. Data were collected at 14 days (a, c, d), 28 days (b), 42 days (e), and 8 (f, g) days post implantation. Horizontal bars mark medians, error bars mark s.e.m. Data were pooled from ≥2 independent experiments with n≥4/group; n and data points denote individual mice analyzed separately. P values were determined by two-tailed Mann-Whitney test (a-g), two-sided log-rank test (a, b, survival curves), two-way ANOVA with Sidak’s multiple comparison test (a, b, tumor volumes), and Chi-square test (e, f % rejection).
We next investigated if the effect of IL33 on CD8+ T cells was tissue specific by contrasting the rejection phenotype of KPC cells expressing the CD8+ T cell rejection antigen ovalbumin (KPC-OVA) at different tissue sites. Interestingly, 70% of Il33+/+ mice rejected orthotopic KPC-OVA tumors, whereas 0% of Il33−/− mice did. In contrast, 100% of ll33+/+ and Il33−/− mice rejected subcutaneous KPC-OVA tumors (Figure 2e). To assess if this phenotype resulted from ILC2 deficiency and ineffective CD8+ T cell priming, we acutely depleted ILC2s and examined antigen-specific CD8+ T cells in DLNs using the iCOS-T mouse, which allows diphtheria toxin–mediated ILC2 depletion while sparing ICOS+CD4+ T cells16 (Figure 2f, Extended Data Figure 5a). ILC2 depletion recapitulated the Il33−/− phenotype, with a higher rate of tumor rejection and larger tumor size in orthotopic KPC-OVA tumors and no differences in subcutaneous tumors (Figure 2f), with anticipated variation compared to Il33−/− mice given differences in time of rejection assessment and depletion efficacy. Tetramer analysis in ILC2-depleted orthotopic KPC-OVA mice revealed reductions in OVA-specific CD8+ T cells in DLNs and spleens, and reductions in CD8+ TCM in DLNs (as seen in Il33−/− mice) (Figure 2g, Extended Data Figure 5b, c). Therefore, ILC2 deficiency partially phenocopied IL33 deficiency. Although direct effects of IL33 on CD8+ T cells cannot be ruled out, we found no ST2 expression on intratumoral CD8+ T cells (Extended Data Figure 5d). To summarize, these loss-of-function experiments suggested that the IL33-TILC2 axis primes tissue-specific CD8+ T cell PDAC immunity.
Next, to examine if rIL33 treatment had similar tissue-specific anti-tumor effects, we found rIL33 prevented tumor establishment in orthotopic PDAC mice and prolonged survival, with no effects on subcutaneous PDAC mice, leading to progressive tumor growth and ulceration requiring euthanasia (Figure 3a), with similar tissue-specific anti-tumor effects in KPC-OVA PDAC mice (Extended Data Figure 6a). Similarly, rIL18, a cytokine that preferentially activates IL18R+ skin ILC2s14, restricted the growth of subcutaneous PDACs infiltrated by IL18R+ ILCs, but not orthotopic PDACs that lack IL18R+ ILCs (Figure 3b, Extended Data Figure 6b). rIL33 selectively expanded ILC2s in DLNs and tumors of orthotopic PDAC mice (Figure 3c), with no changes in the spleen or in subcutaneous PDACs (Extended Data Figure 6c, d). ILC2 expansion was accompanied by enhanced intratumoral CD8+ T cell cytokine capacity and PD-1 upregulation (Extended Data Figure 6e), with no consistent changes in other intratumoral immune cells (Extended Data Figure 6f), although potential modulation of their function cannot be ruled out. Consistent with ILC2s priming anti-tumor CD8+ T cells indirectly, rIL33 treatment doubled intratumoral CD103+ dendritic cells (DCs) (Figure 3d, Extended Data Figure 6g) which prime and recruit CD8+ T cells into PDACs6. To determine if the effects of rIL33 depended on ILC2s, we administered rIL33 to PDAC-bearing Rorafl/flIl7rCre/+ mice constitutively deficient in ILC2s.16 ILC2 deficiency (Extended Data Figure 6h) abrogated rIL33 efficacy (Figure 3e) and attenuated increases in CD103+ DCs in tumors (Figure 3f). rIL33 also had no anti-tumor effects (Figure 3g) and failed to induce intratumoral CD8+ T cell PD-1 expression (Extended Data Figure 6i) in CD103+ DC-deficient Batf3−/− mice, establishing that CD103+ DCs were essential for rIL33-mediated tumor control. To identify if TILC2s produced chemokines to recruit DCs into tumors, we used single-cell RNA-seq (scRNA-seq) (Extended Data Figure 7a–c, Supplementary Table 3) and found activated TILC2s and DLN ILC2s retained markers of ILC2 identity but exhibited distinct transcriptional profiles (Extended Data Figure 8a–e), with rIL33-activated TILC2s selectively expressing Ccl5 (Extended Data Figure 8f), which encodes a chemokine that recruits CD103+ DCs into tumors17, and induced efficient DC migration in vitro (Figure 3h). In sum, these data suggested that rIL33 expands TILC2s to recruit CD103+ DCs into tumors, potentially through Ccl5 production, and activate CD8+ T cells to induce therapeutic tumor immunity.
Figure 3: ILC2s stimulate tissue-specific cancer immunity by recruiting intratumoral dendritic cells.
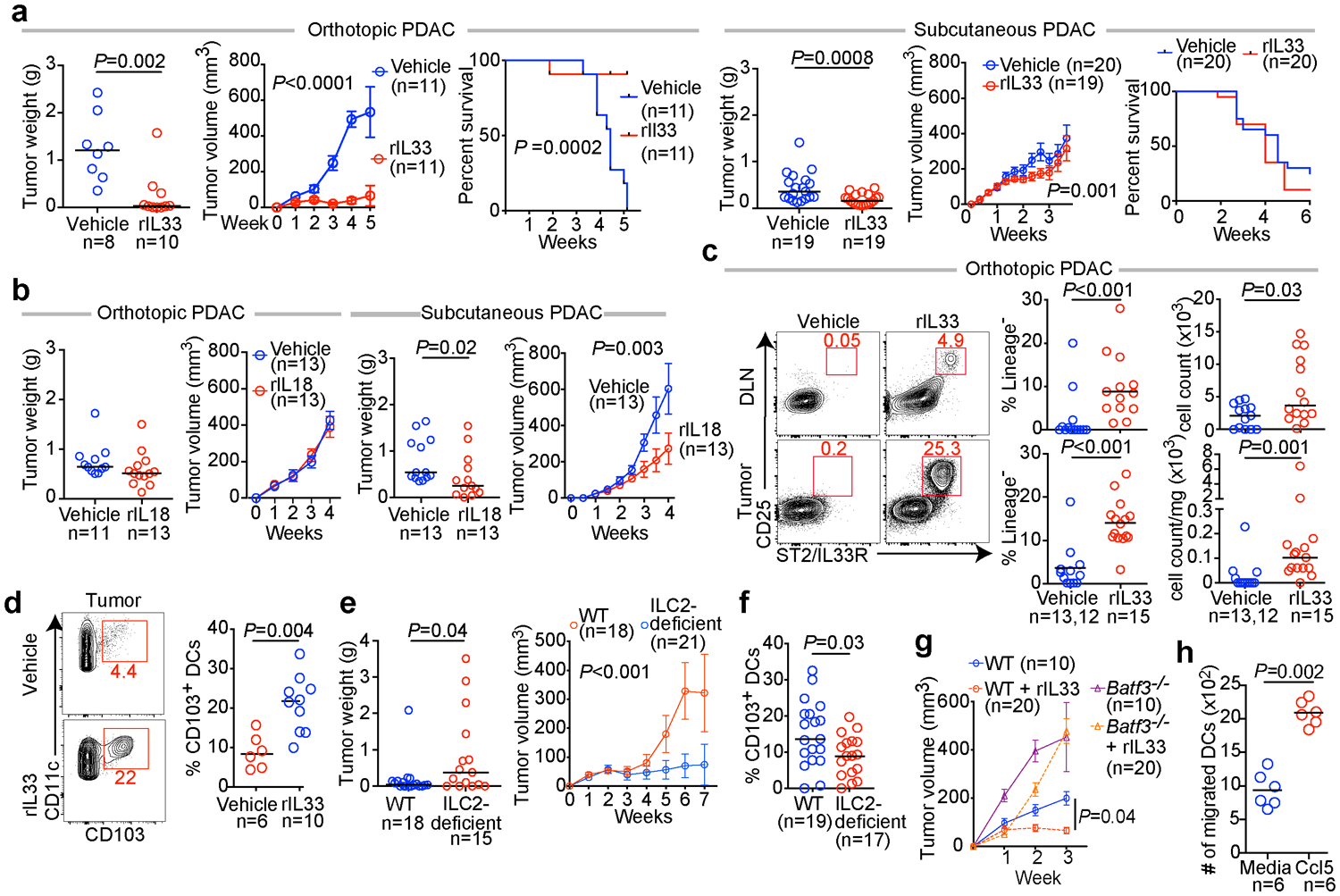
(a) Tumor weight, volume, and survival in orthotopic and subcutaneous PDAC mice treated with vehicle or recombinant IL33 (rIL33). (b) Tumor weight and volume in orthotopic and subcutaneous PDAC mice treated with vehicle or recombinant IL18 (rIL18). (c) Gating, frequency, and number of ILC2s in rIL33-treated orthotopic PDAC mice (DLN vehicle, n=13; tumor vehicle, n=12). (d) Gating and frequency of CD103+ dendritic cells (DCs) in tumors of rIL33-treated orthotopic PDAC mice. (e) Tumor weight, volume, and (f) frequency of CD103+ DCs in tumors of rIL33-treated wild-type (WT) and ILC2 deficient orthotopic PDAC mice. (g) Tumor volume in rIL33-treated WT and CD103+ DC deficient Batf3−/− orthotopic PDAC mice. (h) Migration of purified DCs towards Ccl5. Data were collected at 5 (c, d) and 7 (e, f) weeks post tumor implantation. Horizontal bars mark medians; error bars mark s.e.m. Data were pooled from ≥2 independent experiments, with n≥3/group; n and data points denote individual mice analyzed separately or (h) individual replicates. P values were determined by two-sided log-rank test (a, survival curve), two-way ANOVA (a, b, e, g, tumor volume), and two-tailed Mann-Whitney test (a-f, h).
PD-1 blockade activates TILC2s
As stimulating ILC2s with rIL33 had anti-tumor effects, we searched for strategies to further boost ILC2 activation. Recent data have shown that, like T cells, ILC2s regulate their activity through coinhibitory2,18 immune checkpoint pathways. Specifically, the immune checkpoint PD-1 regulates mouse ILC2 development19, marks effector ILCs19, and when genetically deficient or inhibited with a blocking antibody (αPD-1), IL33-activated ILC2s show greater expansion and effector function in mice and humans20. PD-1+ILC2s are also found in human tumors2. Yet, concurrent ILC2 activation and disinhibition for cancer therapy is relatively unexplored.
Using scRNA-seq (Extended Data Figure 7a–c), we found PD-1 was the only detectable coinhibitory molecule expressed at baseline by TILC2s (Extended Data Figure 9a). rIL33 treatment upregulated PD-1 on a fraction of TILC2s but not in DLN ILC2s (Extended Data Figure 9b), suggesting PD-1 may functionally restrain activated TILC2s. We therefore explored if combining rIL33 with αPD-1 could cooperatively activate TILC2s to enhance anti-tumor efficacy. Consistent with PD-1 expression only on rIL33-activated TILC2s, αPD-1 alone induced a partial response (Figure 4a) as previously reported in PDACs6 but did not appreciably alter TILC2 frequencies (Figure 4b, Extended Data Figure 9c). Combining rIL33 with αPD-1 maximally expanded ILC2s in tumors and DLNs (Figure 4b) and enhanced tumor control compared to αPD-1 alone (Figure 4a). To explore if αPD-1 was activating ILC2s by cell-intrinsic PD-1 blockade, we compared the single-cell transcriptional profiles of TILC2s and DLN ILC2s following in vivo treatment. While TILC2s retained transcriptional and cellular identities of ILC2s irrespective of treatment (Extended Data Figure 9d), TILC2s in rIL33 and αPD-1 treated PDAC mice had unique transcriptional phenotypes compared to all other conditions (Figure 4c), with increased expression of ILC2-specific markers, canonical (amphiregulin [Areg])14 and non-canonical (CXCL2)21 effector molecules, cellular activation machinery (Junb, Fosl2, Ybx1), and coinhibitory immune checkpoints (Extended Data Figure 9e–i). Finally, the anti-tumor effects of dual therapy were abrogated in ILC2-deficient mice (Figure 4d), demonstrating that ILC2s were necessary for the efficacy of dual αPD-1 and rIL33 therapy. These results suggested αPD-1 preferentially amplified activated TILC2s by possibly inhibiting the PD-1 pathway on ILC2s, and not exclusively on T cells.
Figure 4: PD-1 blockade activates TILC2s.
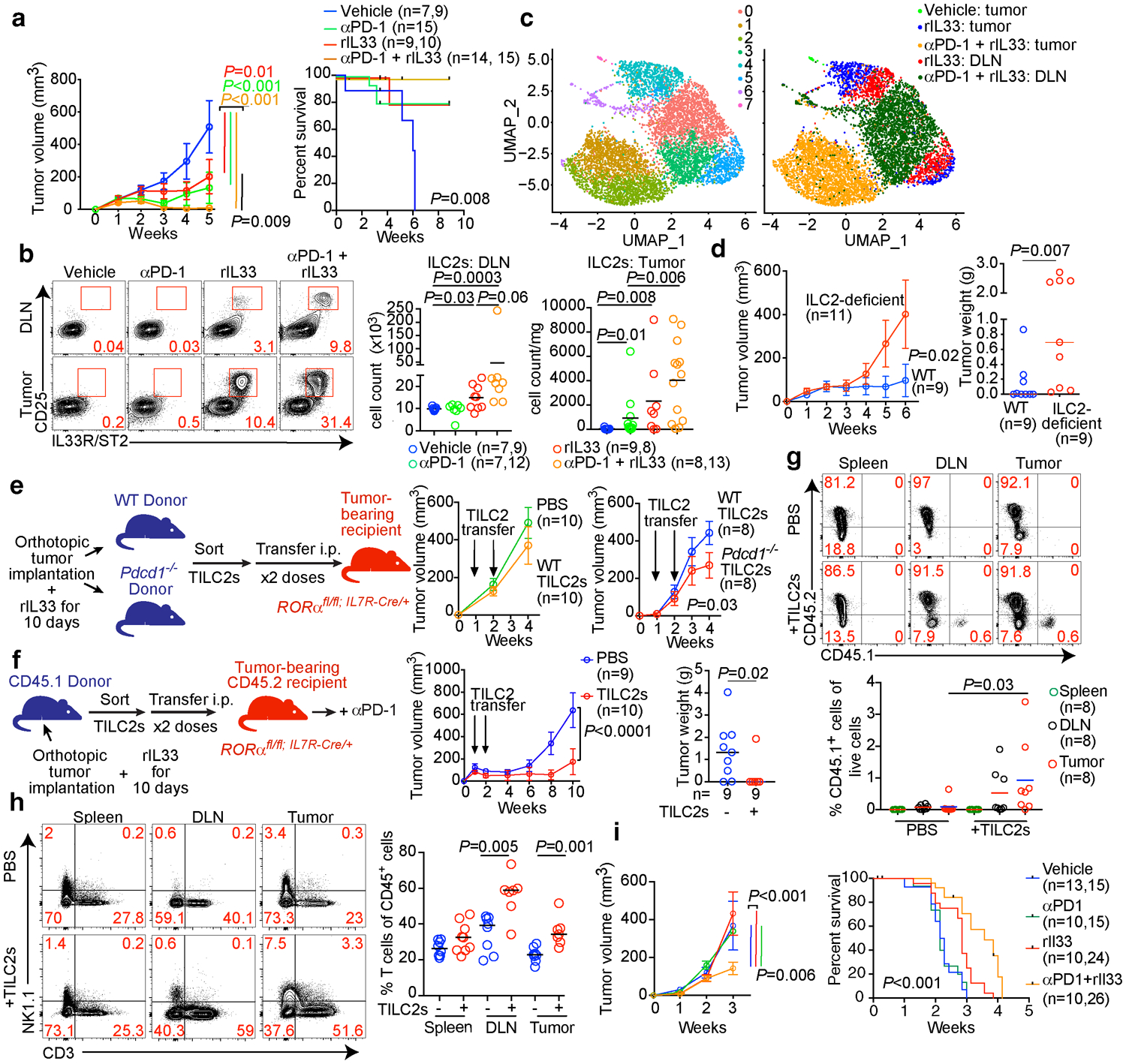
(a) Tumor volume and survival, (b) gating, frequency and number and (c) scRNA-seq (n=7,022 ILC2 single cells) in treated PDAC mice in a nonlinear representation of the top 15 principal components. Cells are colored by cluster (left) or treatment and tissue (right). In (a, b), n=n in left, right graphs respectively. (d) Tumor volume in wild-type (WT) or ILC2 deficient PDAC mice treated with αPD-1 + rIL33. (e) TILC2s were sort-purified from rIL33-treated WT or Pdcd1−/− PDAC mice, transferred into ILC2-deficient PDAC recipients, and tumor volumes measured. (f-h) TILC2s were sort-purified from rIL33-treated PDAC CD45.1 donor mice, transferred into ILC2-deficient CD45.2 PDAC recipient mice, and treated with αPD-1 post cell transfer. Tumor volume and tumor weight (f), frequency of CD45.1 and CD45.2 cells (g), and frequency of T cells (h) (TILC2s- : all groups, n=8; TILC2+ : spleen, n=9; DLN, n=7; tumor, n=7) in recipient mice 10 weeks post cell transfer. Frequencies in g = percentage of live donor- or recipient-derived immune cells. (i) Tumor volume (vehicle, n=13; other groups, n=10) and survival (vehicle and αPD-1, n=15; rIL33, n=24; rIL33+αPD-1, n=26) of treated PDAC mice (KPC 52 cells). DLN, draining lymph node. Data were collected at 5 weeks (b), 10 days (c), and 6 weeks (d) post orthotopic tumor cell implantation. Horizontal bars mark medians, error bars mark s.e.m. Data are pooled from ≥2 independent experiments with n≥3/group; n and data points denote individual mice analyzed separately. Data for scRNA-seq represent pooled purified single cells from biological replicates (vehicle n=10, rIL33 n=5, αPD-1 + rIL33 n=5). P values were determined by two-way ANOVA with Tukey’s multiple comparison post (a, d-f, i, tumor volume), two-tailed Mann-Whitney (b, d, g, h), and two-sided log-rank (a, i, survival curves) tests.
PD-1 TILC2 inhibition is cell-intrinsic
To identify if cell-intrinsic PD-1 pathway interruption on activated TILC2s contributed to the anti-tumor effects of dual therapy, we transferred sort-purified rIL33-activated PD-1-proficient (wild-type [WT]) or PD-1 deficient (Pdcd1−/−) TILC2s into tumor-bearing ILC2-deficient mice (Figure 4e, Extended Data Figure 10a). WT TILC2 transfer had no anti-tumor efficacy in established tumors, but Pdcd1−/− TILC2s restricted tumor growth, indicating that interrupting PD-1 signaling on TILC2s can enhance tumor control (Figure 4e). We next tested if rIL33-activated PD-1+ TILC2s can directly amplify the efficacy of αPD-1 therapy in established tumors. We transferred sort-purified rIL33-activated congenic CD45.1+ TILC2s into CD45.2+ ILC2-deficient mice with established tumors, and treated with αPD-1 post-transfer (Figure 4f). Transferred TILC2s were >97% PD-1+ (Extended Data Figure 10b) and accumulated in tumors and DLNs, but not spleens, of αPD-1 treated recipient mice, persisting up to 9 weeks post transfer (Figure 4g). PD-1+ TILC2 transfer augmented αPD-1 efficacy, restricted tumor growth, and increased T cell frequencies in tumors and DLNs, but not spleens, of recipient mice (Figure 4h). These data collectively demonstrated that blockade of PD-1 signaling on rIL33-activated TILC2s had direct anti-tumor effects and amplified αPD-1 efficacy.
To examine rIL33 and αPD-1 efficacy in IL33Low αPD-1–resistant tumors, we selected an aggressive “cold” PDAC model (KPC 52) that generates IL33Low tumors (Extended Data Figure 2b), has 50% fewer CD8+ T cells, and has a median survival of only 2 weeks (Extended Data Figure 10c), to mimic the immunologic and survival features of IL33Low short-term human PDAC survivors. Although KPC 52 PDAC mice do not exhibit the sequential steps of PDAC tumorigenesis from pre-invasive neoplasias to invasive PDAC as seen in spontaneous KPC mice, they recapitulated the αPD-1 resistance seen in spontaneous KPC mice and human PDAC (Figure 4i). We found that combination rIL33 and αPD-1 reduced tumor volume by nearly 40%, with a nearly 50% improvement in mouse survival (Figure 4i). Finally, to assess the potential to treat PDAC patients with dual rIL33 and αPD-1 therapy, we found nearly 60% of human PDACs had low frequencies of PD-1+ TILC2s and PD-1+ T cells, with a significant correlation between the two cell types (Extended Data Figure 10d), suggesting they frequently co-occur in human PDAC. Additionally, IL33 mRNA significantly correlated with PD-1 mRNA (Extended Data Figure 10e), which has been associated with longer survival22, suggesting the IL33-PD-1 axis may positively impact human PDAC survival. In summary, activating ILC2s with rIL33 can amplify responses to αPD-1 in both PD-1 partially sensitive and PD-1 resistant tumors.
Discussion
Our results suggest that activating ILC2s could be a broader strategy to enhance T cell priming in ILC2-infiltrated cancers (Extended Data Figure 10f). However, given the tissue-specific phenotypes of ILC2s, whether activating them will have similar effects across diverse cancers requires further study. Consistently, given the divergent effects of ILC2s on cancers in different tissue sites, our findings underscore the need for pre-clinical cancer studies to be performed in native organs to accurately reflect local immune environments.
ILC2s can express immune checkpoints, yet their ability to be harnessed by immune checkpoint blockade therapy has remained unclear. We identify that blocking PD-1 on activated ILC2s has anti-tumor effects, suggesting ILC2s may partially contribute to the efficacy of PD-1 pathway blockade in human cancers, and highlighting more broadly that differential checkpoint blockade responses may be dependent on tissue-specific factors. Refining strategies to identify ILC2s in human cancers will accelerate investigations of their prognostic and predictive potential.
The expression of several immune modulatory molecules by activated ILC2s (Extended Data Figure 10g) suggests a broader array of checkpoints can be targeted on ILC2s and T cells in tumors. As ILC2 and T cells co-exist in human cancers with shared costimulatory and coinhibitory pathways, investigation of strategies to collectively target ILC2s and T cells for cancer immunotherapy are warranted.
Methods
Mice
C57BL/6 (wild type, WT, CD45.2), C57BL/6 CD45.1, Rag2−/−, Rag2−/−γc−/−, Batf3−/−, and Pdcd1−/− mice were purchased from Jackson Labs. Il33−/−, Il33Cit/+ were a gift from M.J. Rosen. Cd4Cre/+Icosfl-Dtr/+ and Il7rCre/+Rorαfl/fl were a gift from A.N.J. McKenzie and have been previously described37,38. For all experiments, 6–12-week old mice were matched by age and sex and randomly assigned to specific treatment groups, with at least two independent experiments performed throughout. Pdx1-Cre;LSL-KrasG12D/+;LSL-Trp53R172H/+ (KPC mice) have been previously described33. Sample sizes for experiments were determined without formal power calculations. Animals were bred and maintained in a specific pathogen-free animal facility, and all experiments were conducted in accordance with an Institutional Animal Care and Use Committee (IACUC) approved protocol at Memorial Sloan Kettering Cancer Center (MSKCC) and in compliance with all relevant ethical regulations.
Cell lines and animal procedures
All tumor cell lines were derived from KPC mice. KPC 4662 cells from Pdx1-Cre;LSL-KrasG12D/+;LSL-Trp53R172H/+ (a gift of R.H. Vonderheide) were transfected with GFP and used for all experiments unless indicated otherwise. KPC 8–1, 18–3, and 52 cells derived from Ptf1a-Cre;LSL-KrasG12D/+;LSL-Trp53R172H/+ mice were a gift of C. Iacobuzio-Donahue. KPC 4662 cells engineered to express OVA were previously described39 (a gift of R.H. Vonderheide). All cell lines were authenticated as bonafide PDAC cell lines based on histopathologic verification by a dedicated pancreatic cancer pathologist. Orthotopic tumors established with KPC 4662 cells were IL33High and transiently decreased in size with αPD-1 therapy initiated at time of implantation (αPD-1 partial sensitivity). Orthotopic tumors established with KPC 52 cells were IL33Low and did not decrease in size with αPD-1 therapy initiated at time of implantation (αPD-1 resistant). All cell lines were regularly tested using MycoAlert Mycoplasma Detection Kit (Lonza). Orthotopic PDAC tumors were established as previously described34. Briefly, mice were anesthetized using a ketamine/xylazine cocktail and a small (7 mm) left abdominal side incision was made. Tumor cells (106 KPC cells/mouse; 1.25 × 105 KPC-OVA cells/mouse) were suspended in Matrigel (Becton Dickinson), diluted 1:1 with cold phosphate-buffered saline (PBS) (total volume of 50 μl), and injected into the tail of the pancreas using a 26-gauge needle. Successful injection was verified by the appearance of a fluid bubble without intraperitoneal leakage. The abdominal wall was closed with absorbable Vicryl RAPIDE sutures (Ethicon), and the skin was closed with wound clips (Roboz). For subcutaneous PDAC tumors, tumor cells (106 KPC cells/mouse; 1.25 × 105 KPC-OVA cells/mouse) were resuspended in sterile PBS (Fisher Scientific) and implanted subcutaneously. Mice were sacrificed at the indicated time points and processed for histology or flow cytometry. Autochthonous KPC mice were sacrificed when tumors were detectable by ultrasound. Tumor volumes were measured using serial ultrasound (Vevo 2100 Linear Array Imaging and Vivo LAB Version 3.1.1, Fuji Film Visual Sonics) for orthotopic tumors as previously described40. For subcutaneous tumors, tumor length and width were measured every 2–3 days by calipers, and tumor volumes were calculated as Volume = 1/2 Length × Width2. For survival analyses, survival was determined by a tumor volume of ≥500 mm3 or mouse health requiring euthanasia as defined by institutional IACUC guidelines. No mouse tumors exceeded IACUC-defined maximal tumor volumes of ≥2 cm3. No blinding was performed in experimental mouse interventions, as knowledge of the treatment groups was required.
T cell depletion
CD4 and CD8 cells were depleted by intraperitoneal (i.p.) injection of 250 μg of anti-mouse CD4 antibody (clone GK1.5, BioXcell, InVivoPlus) and 250 μg of anti-mouse CD8a antibody (clone 2.43, BioXcell, InVivoPlus). Control mice were treated with rat IgG2b isotype control (clone LTF-2, BioXcell, InVivoPlus). Mice were treated daily for 3 days prior to tumor implantation, and then every 3 days for the duration of the experiment. CD4+ and CD8+ T cell depletion were confirmed by flow cytometric analysis of tumors and secondary lymphoid organs (>85% depletion).
ILC depletion
ILCs were depleted in Rag2−/− mice by i.p. injection of 300 μg of anti-mouse CD90.2 (clone 30-H12, BioXCell) on day 0, 1, 3, 6, 9, and 13 following tumor implantation as previously described41. ILC2s were depleted in Cd4Cre/+Icosfl-DTR/+ experimental mice and Cd4Cre/+Icos+/+ control mice treated by i.p. injection of diphtheria toxin (Sigma Aldrich) at a dosage of 25 ng per gram of mouse body weight. Mice were treated the day before tumor implantation and then every other day thereafter for a total of 5 doses as previously described37. ILC2 depletion was confirmed by flow cytometric analysis of tumors (Extended Data Figure 5a).
Bone marrow chimeras
Bone marrow was harvested from CD45.2 congenically labeled donor mice, filtered through a 70-mm filter, centrifuged, and resuspended in sterile PBS to a concentration of 108 live cells per 200 μl. CD45.1 congenically labeled C57BL/6J recipient mice were irradiated (5.5 Gy × 2, 6 hours apart) 24 hours before bone marrow transplant and were maintained on endofloxacin water for 4 weeks post irradiation. A single-cell suspension of CD45.2 bone marrow chimera in sterile PBS (108 live cells per recipient mouse) was transplanted to each recipient mouse by retroorbital injection. Reconstitution was confirmed by flow cytometry of the peripheral blood at 4 and 8 weeks post transplantation. Tumor implantation experiments were performed at 12 weeks post transplantation.
Recombinant IL33, IL18, and PD-1 blockade
For rIL33, mice were treated with intraperitoneal (i.p.) injections of 500 ng of carrier-free recombinant murine IL33 (R&D Systems) in sterile PBS daily for 7 days, and then every 2 days thereafter as previously described36. For rIL18, mice were treated with i.p. injection of 2 μg of carrier-free recombinant murine IL-18 (R&D Systems) in sterile PBS at days 3, 7, 11, and 15 after tumor inoculation as previously described42. The chimeric anti-mouse PD-1 antibody (4H2) used in this study engineered as a mouse IgG1 isotype monoclonal antibody (mAb) was shown to bind to CHO transfectants expressing PD-1 and block binding of PD-L1 and PD-L2 to these cells. The affinity of 4H2 for mouse PD-1, determined by surface plasmon resonance using PD-1-Fc, was 4.68×10−9 M. The antibody was produced and purified at Bristol Myers Squibb (BMS). Each batch was certified to have <0.5 EU/mg endotoxin and be of >95% purity. All dosing solutions were prepared in PBS. Mice were treated with i.p. injection of 250 μg anti-PD1 every 2 days. Transient reduction in tumor size but subsequent regrowth while on continuous αPD-1 treatment was defined as a partial response. No reduction in tumor size while on continuous αPD-1 was defined as resistance.
Human samples
All tissues were collected at MSKCC following study protocol approval by the MSKCC Institutional Review Board. Informed consent was obtained for all patients. The study was performed in strict compliance with all institutional ethical regulations. All tumor samples were surgically resected primary PDACs.
Tissue microarray:
Tissue microarrays (TMAs) were constructed from tumor and adjacent non-tumor cores from formalin-fixed, paraffin-embedded tissue blocks from short-term survivors (n=45 tumors, 5 normal tissue) and long-term survivors (n=51 tumors, 5 normal tissue) of PDAC as previously described32. Patient subsets were randomly selected to undergo tissue microarray construction. Patients treated with neoadjuvant therapy were excluded. All tumors were subjected to pathological re-review and histological confirmation by two expert PDAC pathologists before analysis. Long-term survivors were defined as patients with overall survival of >3 years from surgery and short-term survivors as patients with survival >3 months and <1 year from surgery, to exclude perioperative mortalities. ILC2High and ILC2Low were defined as greater or lesser, respectively, than the median ILC2 frequency for the entire TMA cohort.
Tumor transcriptomic profiling:
Patient subsets were randomly selected to undergo transcriptomic profiling as previously described32. Patients in the TMA cohort with tumor tissue available for transcriptomic assessment were included in analyses in Figure 1b to allow protein confirmation of RNA expression. Extracted RNA was qualified on an Agilent BioAnalyzer and quantified by fluorometry (Ribogreen). Preparation of RNA for whole-transcriptome expression analysis was done using the WT Pico Reagent Kit (Affymetrix). Reverse transcription was initiated at the poly-A tail as well as throughout the entire length of RNA to capture both coding and multiple forms of non-coding RNA. RNA amplification was achieved using low-cycle PCR followed by linear amplification using T7 in vitro transcription technology. The cRNA was then converted to biotinylated sense-strand DNA hybridization targets. The prepared target was hybridized to GeneChip Human Transcriptome Array 2.0 (Affymetrix). Washes were performed using the GeneChip Hybridization, Wash and Stain Kit using a Fluidics Station 450/250. Arrays were scanned using the GeneChip Scanner 3000. Data analysis for the array was done using Affymetrix Expression Console Software (SST-RMA algorithm to summarize the signal from array probesets). Immune cytolytic activity was determined as previously described43.
Cell isolation
Mouse and human PDAC tumors and adjacent pancreata were mechanically dissociated and incubated in collagenase (collagenase II for murine tumors, collagenase IV for human tumors, both 5 mg/ml; Worthington Biochemical Corp., Fisher Scientific), DNAse I (0.5 mg/ml; Roche Diagnostics), and Hank’s balanced salt solution (Gibco, Fisher Scientific) for 30 minutes at 37°C. Digestion was then quenched with fetal bovine serum (FBS, Life Technologies), and cells were filtered sequentially through 100- and 40-mm nylon cell strainers (Falcon, Fisher Scientific). Tumors, adjacent pancreata, and lymph nodes were then mechanically disassociated and filtered through 100- and 40-mm nylon cell strainers (Falcon, Fisher Scientific) using PBS with 1% FBS (Life Technologies). Spleens were mechanically dissociated and filtered through 70- and 40-mm nylon cell strainers (Falcon, Fisher Scientific) using PBS with 1% FBS, followed by RBC lysis (RBC lysis buffer, ThermoFisher Scientific). Mouse Fc receptors were blocked with FcεRIII/II-specific antibody (1 μg per 1 × 106 cells; clone 2.4G2, Bio X Cell).
ILC2 adoptive transfer
CD45.1 C57Bl/6 or Pdcd1−/− orthotopic PDAC mice were treated with 500 ng of carrier-free recombinant murine IL33 (R&D Systems) in sterile PBS daily for 10 days. Live, CD45+, lineage−, CD90+, CD25+, ST2+ TILC2s were sort-purified to 98% purity at day 10 post-implantation using an Aria Cell sorter (BD Biosciences). 5 × 105 tumor ILC2s were immediately transferred to orthotopic PDAC tumor-bearing Il7rCre/+Rorαfl/fl CD45.2 mice days 7 and 14 post-tumor implantation via i.p. injection. Control mice received equivalent volumes of PBS via i.p. injection. αPD-1 treatment in recipient mice was initiated on the day of ILC2 cell transfer. Tissues were harvested at indicated time points.
Flow cytometry
Single-cell suspensions were stained using antibody cocktails in the dark at 4°C, washed, and analyzed on a FACS LSR Fortessa (BD Biosciences). Mouse ILCs were defined as live, CD45+, lineage− (CD3, CD5, NK1.1, CD11b, CD11c, CD19, FcεR1), CD25+, CD127+ cells as previously described35, 31. Mouse immune cells were defined as follows: ILC2s = live, CD45+, lineage− CD25+, ST2+ cells; central memory T cells = live, CD45+, CD3+, NK1.1−, CD8+, CD62l+, CD44+; dendritic cells = live, CD45+, CD3−, NK1.1−, Gr1−, F4/80−, CD11c+, MHC-II+; B cells = live, CD45+, CD3−, CD19+; T cells = live, CD45+, CD3+; CD4+ T cells = live, CD45+, CD3+, CD4+; CD8+ T cells = live, CD45+, CD3+, CD8+; regulatory T cells = live, CD45+, CD3+, CD4+ FoxP3+; tumor associated macrophages = live, CD45+, CD11b+, F4\80+, GR1−; myeloid derived suppressor cells (MDSCs) = live, CD45+, CD3−, CD11b+, F4\80−, GR1+. Murine cells were stained with the following antibodies: from Biolegend, CD45 (clone 30-F11, Pacific Blue), CD45.1 (clone A20, BV711), NK1.1 (clone PK136, APC), Gr-1 (clone RB6–8C5, BV605), CD103 (clone 2E7, BV711); from BD Biosciences, CD5 (clone 53–7.3, APC), CD11c (clone HL3, APC), NK1.1 (clone PK136, BV605), CD4 (clone RM4–5, BV786), CD62L (clone MEL-14, APC), CD19 (clone 1D3, BV510), Ly6C (clone AL-21, PerCP-Cy5.5), Ly6G (clone 1A8, AF700), PD1 (clone J43 BV605), TNF-α (clone MP6-XT22, BV510), IFN-γ (clone XMG1.2, APC-Cy7), CD90.2 (clone 53–2.1, BV786), Tbet (clone Q4–46, BV711), Rorγ-t (clone Q31–378, BV786), Gata3 (clone L50–823, PE-Cy7), and IL4 (clone 11B11, BV650); from ThermoFisher Scientific CD3 (clone 17A2, Alexa Fluor 700), CD11b (clone M1/70, APC), CD11b (clone M1/70, PerCP-Cy5.5), CD8 (clone 53–6.7, Alexa Fluor 700), CD19 (clone 1D3, Alexa Fluor 700), FcεR1 (clone MAR-1, APC), F4/80 (clone BM8, PE-Cy5), CD3 (clone 145–2C11, PE-Cy7), MHC-II (clone M5/114.15.2, Alexa Fluor 700), CD44 (clone IM7, PerCP-Cy5.5), CD127 (clone A7R34, FITC), CD25 (clone PC61.5, PerCP-Cy5.5), IL5 (clone TRFK5, PE), CD86 (clone GL1, PE), CD11c (clone N418, FITC), ST2 (clone RMST2–2, PE-Cy7), and FoxP3 (clone FJK-16S, APC); and from MBL international, SINFEKL tetramer (catalog # TB-5001–1, PE).
Human ILCs were defined as live CD45+, lineage− (CD3, CD5, CD56, CD11b, CD11c, CD16, CD19, TCRα/β, FcεR1), CD25+, CD127+ cells as previously described35. Human cells were stained with the following antibodies: from BD Biosciences, GATA3 (clone L50–823, BV711), TBET (clone O4–46, BV650), RORγ-T (clone Q21–559, PE); from Biolegend, CRTH2 (clone BM16, PE-Cy7), CD11b (clone ICRF44, APC), CD56 (clone NCAM16.2, BV650), CD25 (clone BC96, PerCP-Cy5.5), CD45 (clone HI30, Pacific Blue), TCRα/β (clone IP26, APC); from ThermoFisher Scientific, CD16 (clone CB16, APC), CD11c (clone 3.9, APC), CD127 (clone RDR5, FITC), CD3 (clone OKT3, Alexa Fluor 700), ST2 (clone hIL33Rcap, PE), CD5 (clone L17F12, APC), CD19 (clone HIB19, AF700), FcεR1 (clone AER-37, APC). Human-specific antibody to IL33 (clone 390412, PE) was purchased from R&D Systems. All samples for flow cytometry were from prospectively collected unselected PDAC patients.
To examine intracellular cytokine production, singe-cell suspensions of tumors were stimulated for 6 hours ex-vivo with phorbol 12-myristate (PMA, 100 ng/ml) and ionomycin (1ng/ml) in the presence of brefeldin A (10 μg/ml) (all from Sigma-Aldrich) at 37°C. Cells were then surface-stained, fixed, permeabilized, and stained for cytokine production using the Fixation and Permeabilization Buffer Kit per the manufacturer’s recommendations (Invitrogen, ThermoFisher Scientific). Appropriate isotype controls were used as indicated. Analysis was performed on FlowJo (versions 9 and 10, Tree Star).
Immunohistochemistry
Tissues were fixed in paraformaldehyde (Fisher Scientific) for 24 hours and embedded in paraffin. The tissue sections were deparaffinized with EZPrep buffer (Ventana Medical Systems), then antigen retrieval was performed with CC1 buffer (Ventana Medical Systems). Sections were blocked for 30 minutes with Background Buster solution (Innovex), followed by avidin-biotin blocking for 8 minutes (Ventana Medical Systems). Mouse IL33 (AF3626, R&D Systems), mouse smooth muscle actin (Abcam), and human IL33 (AF3625, R&D Systems) antibodies were applied, and sections were incubated for 4 hours, followed by a 60-minute incubation with biotinylated rabbit anti-goat IgG (Vector labs), or biotinylated goat anti-rabbit IgG (Vector labs) at 1:200 dilution. Detection was performed with DAB detection kit (Ventana Medical Systems) according to the manufacturer’s instructions. Any section containing cells demonstrating cytoplasmic or nuclear positivity for IL33 was designated to have positive staining. Slides were counterstained with Masson’s trichrome, or hematoxylin, and eosin,and cover-slipped with Permount (Fisher Scientific).All histologic sections were evaluated by an independent PDAC pathologist.
Immunofluorescence
Mouse
IL33/CD11b/CK19/Iba1 immunofluorescence:
Multiplex immunofluorescent staining was performed using a Discovery XT processor (Ventana Medical Systems) as described44.
IL33:
First, sections were incubated with anti-mIL33 (R&D Systems, catalog#AF3626, 1 μg/ml) for 4 hours, followed by 60 minutes incubation with biotinylated horse anti-goat IgG (Vector Laboratories) at 1:200 dilution. Detection was performed with Streptavidin-HRP D (part of DABMap kit, Ventana Medical Systems), followed by incubation with Tyramide Alexa Fluor 488 (Invitrogen) prepared according to the manufacturer’s instructions with predetermined dilutions.
CD11b:
Next, sections were incubated with anti-CD11b (Abcam, clone EPR1544) for 5 hours, followed by 60 minutes incubation with biotinylated goat anti-rabbit IgG (Vector Laboratories) at 1:200 dilution. Detection was performed with Streptavidin-HRP D (part of DABMap kit, Ventana Medical Systems), followed by incubation with Tyramide Alexa 594 (Invitrogen) prepared according to the manufacturer’s instructions with predetermined dilutions.
CK19:
Next, slides were incubated with anti-CK19 (Abcam, clone EP1580Y) for 5 hours, followed by 60 minutes incubation with biotinylated goat anti-rabbit (Vector Laboratories) at 1:200 dilution. Detection was performed with Streptavidin-HRP D (part of DABMap kit, Ventana Medical Systems), followed by incubation with Tyramide Alexa Fluor 546 (Invitrogen) prepared according to the manufacturer’s instructions with predetermined dilutions
Iba1:
Finally, sections were incubated with anti-Iba1 (Wako, catalog #019–19741) for 5 hours, followed by 60 minutes incubation with biotinylated goat anti-rabbit IgG (Vector Laboratories) at 1:200 dilution. Detection was performed with Streptavidin-HRP D (part of DABMap kit, Ventana Medical Systems), followed by incubation with Tyramide Alexa 647 (Invitrogen) prepared according to the manufacturer’s instructions with predetermined dilutions. After staining, slides were counterstained with DAPI (Sigma Aldrich) for 10 min and cover-slipped with Mowiol.
Human
The tissue sections were deparaffinized with proprietary Leica Bond buffer (Leica Biosystems), antigen retrieval was performed with Leica Bond ER2 buffer (Leica Biosystems). First, sections were incubated with anti-PD-1 antibodies (Cell Marque, clone NAT105) for 1 hour, followed by detection with Bond Polymer Refine Detection kit (Leica Biosystems) and Tyramide Alexa Fluor 488 (Invitrogen). Next, sections were incubated with anti-CD3 antibodies (DAKO, catalog#A0452) for 1 hour, followed by detection with Bond Polymer Refine Detection kit (Leica Biosystems) and Tyramide CF594 (Biotum). Next, sections were incubated with anti-GATA3 antibodies (Cell Marque, clone L50–823) for 1 hour, followed by detection with Bond Polymer Refine Detection kit (Leica Biosystems) and CF 543 (Biotum). Finally, sections were incubated with anti-CD45 antibodies (DAKO, clone 2B11 + PD7/26) for 1 hour, followed by detection with Bond Polymer Refine Detection kit (Leica Biosystems) and Tyramide Alexa Fluor 647 (Invitrogen). All detections were prepared according to manufacturer instruction with predetermined dilutions. After staining, slides were counterstained with DAPI (Sigma Aldrich) for 10 min and cover-slipped with Mowiol.
Digital image processing and analysis
The slides were digitized using Panoramic Flash 250 (3Dhistech, Budapest Hungary) using Zeiss 20x/0.8NA objective and custom filters for A488, A546, A594, and A647. Each core was exported into multi-channel tiff files and analyzed using custom macro written in FIJI/ImageJ. For quantification, each nucleus was segmented using the DAPI channel after appropriate processing and background subtraction. Then for each nucleated cell, the presence or absence of the other markers were assessed after setting appropriate thresholds for each marker. The number of cells with specific combinations of markers were tallied. ILC2s were defined as CD45+ CD3− GATA3+ nucleated cells, PD-1 expressing ILC2s were defined as CD45+ CD3− GATA3+ PD-1+ nucleated cells, and PD-1 expressing T cells were defined as CD45+ CD3+ PD-1+ nucleated cells. For each patient, the frequency of each cell type as a fraction of all nucleated cells was calculated in triplicate cores, followed by determination of the mean frequency of triplicate cores to calculate the final cellular frequency per patient.
RNA sequencing
Mouse:
Tissues from orthotopic PDAC mice (n=6) were harvested and dissociated into single-cell suspensions as described above. Tumor-infiltrating leukocytes were positively selected by magnetically activated cell sorting using mouse CD45 MicroBeads (Miltenyi Biotec). Purification of magnetically activated sorted cells was confirmed by flow cytometry and was >95%. RNA was isolated from the sorted cells using an RNeasy Plus Mini Kit (Qiagen). Poly(A) capture and paired-end RNA-seq were performed by the MSKCC Integrated Genomics Core Facility. Specifically, after RiboGreen quantification and quality control by Agilent BioAnalyzer, 500 ng of total RNA underwent polyA selection and TruSeq library preparation according to instructions provided by Illumina (TruSeq Stranded mRNA LT Kit, catalog # RS-122–2102), with 8 cycles of PCR. Samples were barcoded and ran on a HiSeq 4000 in a 100bp/100bp paired-end run, using the HiSeq 3000/4000 SBS Kit (Illumina). An average of 83 million paired reads was generated per sample. Ribosomal reads represented at most 0.03% of the total reads generated, and the percentage of mRNA bases averaged 76.6%. The expression dataset was loaded into Gene Set Enrichment Analysis (GSEA) 3.0. Gene set databases for antigen presentation and T cell mediated immunity were selected from MSIGDB v6.1, with a false discovery rate of ≤0.25 to facilitate exploratory discovery. GSEA was run with 1000 permutations. Three gene set databases met this threshold: GO 0002474 Antigen Processing And Presentation of Peptide Antigen Via MHC Class I, GO 0002711 Positive Regulation of T Cell Mediated Immunity, and GSE19825 Naïve vs Day 3 Effector CD8 T Cell Up.
Single-cell RNA sequencing
Library preparation for single-cell immune profiling, sequencing, and post-processing of the raw data was performed at the Epigenomics Core at Weill Cornell Medicine.
Single-cell RNA library preparation and sequencing
Single-cell suspensions of fluorescence activated cell (FAC)-sorted ILC2 cells from vehicle, IL33 alone, and IL33 + αPD-1 treated pancreatic KPC tumors and mesenteric DLNs were prepared as described above. scRNA-seq libraries were prepared according to 10X Genomics specifications (Chromium Single Cell V(D)J User Guide PN-1000006, 10x Genomics, Pleasanton, CA, USA). Four independent cellular suspensions (85–90% viable) at a concentration between 90–200 cells/μl, were loaded onto to the 10x Genomics Chromium platform to generate Gel Beads-in-Emulsion (GEM), targeting about 2,000 single cells per sample. After GEM generation, the samples were subjected to an incubation at 53°C for 45 min in a C1000 Touch Thermal cycler with 96-Deep Well Reaction Module (Bio-Rad, Hercules) to generate polyA cDNA barcoded at the 5’ end by the addition of a template switch oligo (TSO) linked to a cell barcode and Unique Molecular Identifiers (UMIs). GEMs were broken, and the single-strand cDNA was cleaned up with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific, Waltham, MA). The cDNA was amplified for 16 cycles (98°C for 45 s; 98°C for 20 s, 67°C for 30 s, 72°C for 1 hr). Quality of the cDNA was assessed using an Agilent Bioanalyzer 2100 (Santa Clara, CA), obtaining a product of about 1,200 bp. 50 ng of cDNA was enzymatically fragmented, end repaired, A-tailed, subjected to a double-sided size selection with SPRIselect beads (Beckman Coulter, Indianapolis, IN), and ligated to adaptors provided in the kit. A unique sample index for each library was introduced through 14 cycles of PCR amplification using the indexes provided in the kit (98°C for 45 s; 98°C for 20 s, 54°C for 30 s, and 72°C for 20 s × 14 cycles; 72°C for 1 min; held at 4°C). Indexed libraries were subjected to a second double-sided size selection, and libraries were then quantified using Qubit fluorometric quantification (Thermo Fisher Scientific, Waltham, MA). The quality was assessed on an Agilent Bioanalyzer 2100, obtaining an average library size of 450 bp. No treatment samples had concentrations below detectable limits, and cDNA amplification was done with 18 cycles and sample Index with 16 cycles. Libraries were diluted to 10 nM and clustered using a NovaSeq600 on a pair end read flow cell and sequenced for 28 cycles on R1 (10x barcode and the UMIs), followed by 8 cycles of I7 Index (sample Index), and 89 bases on R2 (transcript), obtaining about 100 million clusters per sample, except for tumors from vehicle-treated mice which was clustered at about 10 million. Primary processing of sequencing images was done using Illumina’s Real Time Analysis software (RTA). 10x Genomics Cell Ranger Single Cell Software suite v3.0.2 (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger) was used to perform sample demultiplexing, alignment to mouse genomic reference mm10, filtering, UMI counting, single-cell 5’ end gene counting, and quality control using the manufacturer parameters. Data from approximately 11,000 single cells that passed quality control were obtained with approximately 41,000 mean reads per cell (48% sequencing saturation).
scRNA-seq data processing
The Seurat R package version 3.1 pipeline was used to identify clusters on combined datasets45. First, individual datasets were read into R as count matrices and converted into Seurat objects, selecting on genes expressed in ≥3 cells and on cells with at least 200 detected genes. A standard pre-processing workflow was then used to filter cells based on excluding cells with either over 2,500 or less than 200 unique genes expressed, and cells with greater than 5% mitochondrial gene content.
Following filtering, the samples were merged, and the gene expression measurements for retained cells were log-transformed, normalized by total expression per cell, and scaled to 10,000 molecules per cell. The top 2,000 highly variable genes across the single cells were then identified, and principal components (PCs) analysis was conducted. After examining jackstraw and elbow plots, we selected the top 15 PCs for clustering using K-nearest neighbor (KNN) clustering with cluster resolution set at 0.4, identifying 6–8 clusters in all samples-combined and tumor-combined merged datasets. Non-linear dimensional reduction with UMAP was used to visualize the datasets also using the top 15 PCs. Differential gene expression for gene marker discovery across the clusters was performed using the Wilcoxon rank sum test as used in the Seurat package. Pairwise comparison using Wilcoxon rank sum test was performed with holm P value adjustment method to compare gene expression between samples.
In vitro assays
KPC 4662-GFP cells were cultured for 1 week in a 96-well flat-bottomed plate (Falcon) in complete media: RPMI-1640 with L-glutamine (Gibco, ThermoFisher) with 10% fetal bovine serum (Life Technologies), 100 units/ml of penicillin, 100 μg/ml of streptomycin, and recombinant IL33 at concentrations of 0, 10, 100, and 500 ng/ml. Culture media and cytokines were replenished every 48 hours. Viability was measured using a colorimetric tetrazolium salt assay (Cell Counting Kit, Dojindo Molecular Technologies) per the manufacturer’s instructions and read on a Synergy HT Multi-Detection Microplate Reader (Biotek). Cells were harvested and stained for Annexin V (ThermoFisher Scientific), Ki-67 (clone SolA15, ThermoFisher Scientific), and ST2 (clone RMST2, ThermoFisher Scientific). For all in vitro experiments, 2–3 technical replicates were performed per independent experiment.
In vitro dendritic cell migration assays
Murine splenic DC were isolated and enriched using a mouse pan DC isolation kit according to the manufacturer’s protocol (Miltenyi Biotech). Flow cytometry was used to assess DC purity (>70% CD11c+ of live cells). Cells were plated in complete RPMI media at 5×105 cells/ml with 50 ng/ml of recombinant mouse GM-CSF (Biolegend) overnight. Next, chemotaxis of splenic DCs was analyzed by transwell migration assays. 600 μl of RPMI with or without 100 ng/ml of recombinant mouse Ccl5 (Biolegend) was added to the lower chambers of a 6.5-mm Transwell plate with 5.0-μm pore polycarbonate membrane inserts (Sigma Aldrich). 200 μl of RPMI was also added to the upper chambers and plates were allowed to equilibrate at 37°C in 5% CO2 for 15 minutes. 1×105 splenic DCs in 100 μl of RPMI were then loaded into the upper chambers, and incubated at 37°C in 5% CO2 for 2 hours. After incubation, membrane inserts were carefully removed, and cells were harvested from the lower chambers. Migrated DCs were incubated with DAPI and CD11c antibodies for 20 minutes at 4°C, and Precision Count Beads™ (Biolegend) were added to quantify the number of live, migrated, CD11c+ cells using flow cytometry according to manufacturer’s protocol.
Statistics
Data are expressed as median. As we observed many statistically significant effects in the data without a priori sample size calculations, no statistical methods were used to determine sample size. Comparisons between two groups were performed using unpaired Mann-Whitney test with the Benjamini-Krieger-Yekutieli false discovery approach for multiple time point comparisons (2-tailed). Comparisons among multiple groups were performed using 1-way ANOVA test followed by Kruskal Wallis multiple comparison post-test. Comparisons among multiple groups across multiple time points were performed using 2-way ANOVA test. Correlations between 2 variables were calculated using linear regression. Survival curves were compared by 2-sided log-rank test. Tumor incidences were compared by Chi-square test. All alpha levels were 0.05, with P<0.05 considered a significant difference. Statistical analyses were performed using Prism 7.0 (GraphPad Software).
Extended Data
Extended Data Figure 1: Identification of IL33-dependent ILCs in pancreatic cancer.
(a) Gating strategy to identify human ILCs. The first plot was pre-gated on live (DRAQ7−) cells and singlets. Lineage (Lin) 1 cocktail: CD5, CD11b, CD11c, CD16, FcεR1. Lin 2 cocktail: CD3, CD19, TCRα/β. ILCs were identified as Lin− CD56− CD25+ CD127+ cells. FMO, fluorescence minus one. (b) Representative image of immunofluorescence of ILC2s in tumor tissue microarrays of short-term and long-term PDAC survivors (n=96). Arrows, putative ILC2s. (c) Top, overall survival of patients with greater (high) or lesser (low) than the median intratumoral mRNA level of ILC-stimulating cytokines. Bottom, correlation between expression of ILC-activating cytokines and immune cytolytic index (CYT) in long- and short-term survivors of human PDAC. Curves were fit by linear regression. n=25. (d) Gating strategy to identify murine ILCs. The first plot was pre-gated on live (DRAQ7−) cells and singlets. Lineage (Lin) 1 cocktail: CD5, CD11b, CD11c, FcεR1. Lin 2 cocktail: CD3, CD19. ILCs were identified as Lin− NK1.1− CD25+ CD127+, ILC2s as Lin− NK1.1− CD25+ St2+ cells. Gating on orthotopic PDAC mice are shown. (e) Intratumoral ILC frequency in orthotopic PDAC mice established with KPC cell lines 8–1, 18–3, and in autochthonous KPC mice with spontaneous PDAC (KPCSpont). Composite ILC frequencies from Fig. 1d and others are included for comparison (KPC 4662). (f) Phenotype of ILCs in PDAC mice. Gray curves, isotype controls; numbers, mean fluorescence intensity. (g) Expansion kinetics of ILCs in tissues of PDAC mice. (h) Changes in non-ILC cell frequency in Rag2−/− PDAC mice treated with αCD90.2 or isotype antibodies. Data were analyzed at 14 days (d-f), 10 days (h), or at the indicated time points post tumor implantation. n indicates individual mice analyzed separately in at least two independent experiments with n≥2/group. Horizontal bars mark medians, error bars mark s.e.m. P values were determined by two-sided log-rank (c, top), linear regression (c, bottom), or two-tailed Mann-Whitney test (g). P values in g indicate tumor comparisons to all other organs.
Extended Data Figure 2: Host-derived IL33 activates pancreatic ILC2s.
(a) mRNA expression of ILC1- (IL12, IL15, IL18), ILC2- (IL25, IL33, TSLP), and ILC3-inducer cytokines (IL23) and the IL33 receptor (ST2) in orthotopic PDAC tumors (left) and autochthonous PDAC tumors in KPC mice from a previously published mRNA microarray (right)11. (b) Representative IL33 immunohistochemistry (IHC) of IL33Low and IL33High human (tissue microarray, n=96) and mouse PDAC (n=3/group). (c) Frequency of human PDAC patients demonstrating IL33 positivity by IHC in a human PDAC tumor microarray. (d) Multiplexed immunofluorescence for IL33, ductal marker CK19, and myeloid markers CD11b, and Iba in mouse PDAC (top). Arrows, IL33-expressing cells. IL33 mean fluorescence intensity (MFI) in non-immune (CD45−), immune (CD45+), macrophage (TAM), and monocytic and granulocytic myeloid-derived suppressor cell (M-MDSC and G-MDSC) populations in tumors of IL33Cit reporter PDAC mice (bottom). (e) Representative IL33 protein expression by IHC in orthotopic PDAC tumors in Il33+/+ (WT) mice, and non-tumor-bearing pancreata in Il33–/– mice (n=3/group). (f) ILC frequency (top) and cell number (bottom) in organs and draining lymph nodes (DLN) of Il33+/+ and Il33−/− orthotopic PDAC mice. (g) Gating and frequency of IL4 and IL5 expression in intratumoral ILCs in Il33+/+ and Il33−/− orthotopic PDAC mice. (h) ILC2 and (i) immune cell frequencies in orthotopic Rag2–/– and Rag2–/–γc–/– PDAC mice with or without treatment with recombinant IL33 (rIL33). (j) Frequency of ST2+ tumor ILCs in mice with subcutaneous (SQ) and orthotopic PDAC. (k) Tumors in orthotopic and subcutaneous PDAC mice. (l) Tumor weight in Il33+/+ and Il33−/− littermate PDAC mice. (m) Experimental schema of bone-marrow chimeras to evaluate contribution of hematopoietic cell-derived IL33 to tumor control. (n) Hematopoietic cell reconstitution and (o) tumor weight in irradiated CD45.1 congenic mice reconstituted with either CD45.2 Il33+/+ or CD45.2 Il33−/− bone marrow. Data were collected at 14 (a, d, f, g, j, o), and 10 (h, i) days post tumor implantation. Horizontal bars mark medians. n indicates individual mice analyzed separately in at least two independent experiments with n≥2/group. P values were determined by one-way ANOVA (a) or two-tailed Mann-Whitney test (d, f-h, j, l, o).
Extended Data Figure 3: Host-derived IL33 activates pancreatic T cell immunity.
(a) Gene set enrichment analysis of bulk RNA-seq from purified CD45+ immune cells from Il33+/+ and Il33−/− PDAC mice. Enrichment plots and enrichment scores are shown for three gene sets comparing expression in Il33−/− to Il33+/+ (n=3 mice/group). FDR, false discovery rate. (b) Gating of CD8+ T cells and (c) frequencies of various immune cell types (left) and CD4+ T cell lineages (right) in Il33+/+ and Il33−/− orthotopic PDAC mice. (d) Frequency of T central memory (Tcm) cells (CD45+CD3+CD8+CD44+CD62L+) in tumor draining lymph nodes and non-tumor draining distant lymphoid organs (inguinal lymph node and spleen) in Il33+/+ and Il33−/− orthotopic PDAC mice. (e) Frequency of CD8+ T cells in subcutaneous PDAC tumors. DC, dendritic cells; MDSC, myeloid-derived suppressor cells; NK, natural killer cells; NKT, natural killer T cells; Treg, regulatory T cells. Data were analyzed 14 days post tumor implantation or at the time points indicated. Horizontal bars mark medians, error bars mark s.e.m. n indicates individual mice analyzed separately in at least two independent experiments with n≥2/group. P values determined by one-way ANOVA (d).
Extended Data Figure 4: IL33 and ILCs do not directly induce tumor cell death.
(a) Tumor weight in Rag2−/− and Rag2−/−γc−/− PDAC mice treated with vehicle or recombinant murine IL33 (rIL33). (b) Representative hematoxylin and eosin stained sections (left) with histologic tumor cell differentiation status in Il33+/+ and Il33−/− PDAC mice (right). (c) Trichrome staining in tumors of Il33+/+ and Il33−/− PDAC mice (n=3/group). (d) Immunohistochemistry for smooth muscle actin in tumors of Il33+/+ and Il33−/− PDAC mice (n=3/group). (e) Intratumoral ST2 expression on KPC cells in Il33+/+ and Il33−/− orthotopic PDAC mice. (f) ST2 expression on live KPC cells following rIL33 treatment in vitro (DRAQ7 stains dead cells) (n=3/group). (g) KPC cell number, viability, proliferation (Ki-67), and apoptosis (annexin) following rIL33 treatment in vitro (n=6/group). Horizontal bars mark medians. n in a-e indicates individual mice analyzed separately in at least two independent experiments with n≥3/group. n in f, g indicates technical replicates and is representative of at least two independent experiments. P value determined by two-tailed Mann-Whitney test (a).
Extended Data Figure 5: ILC2s induce antigen-specific CD8+ T cell priming.
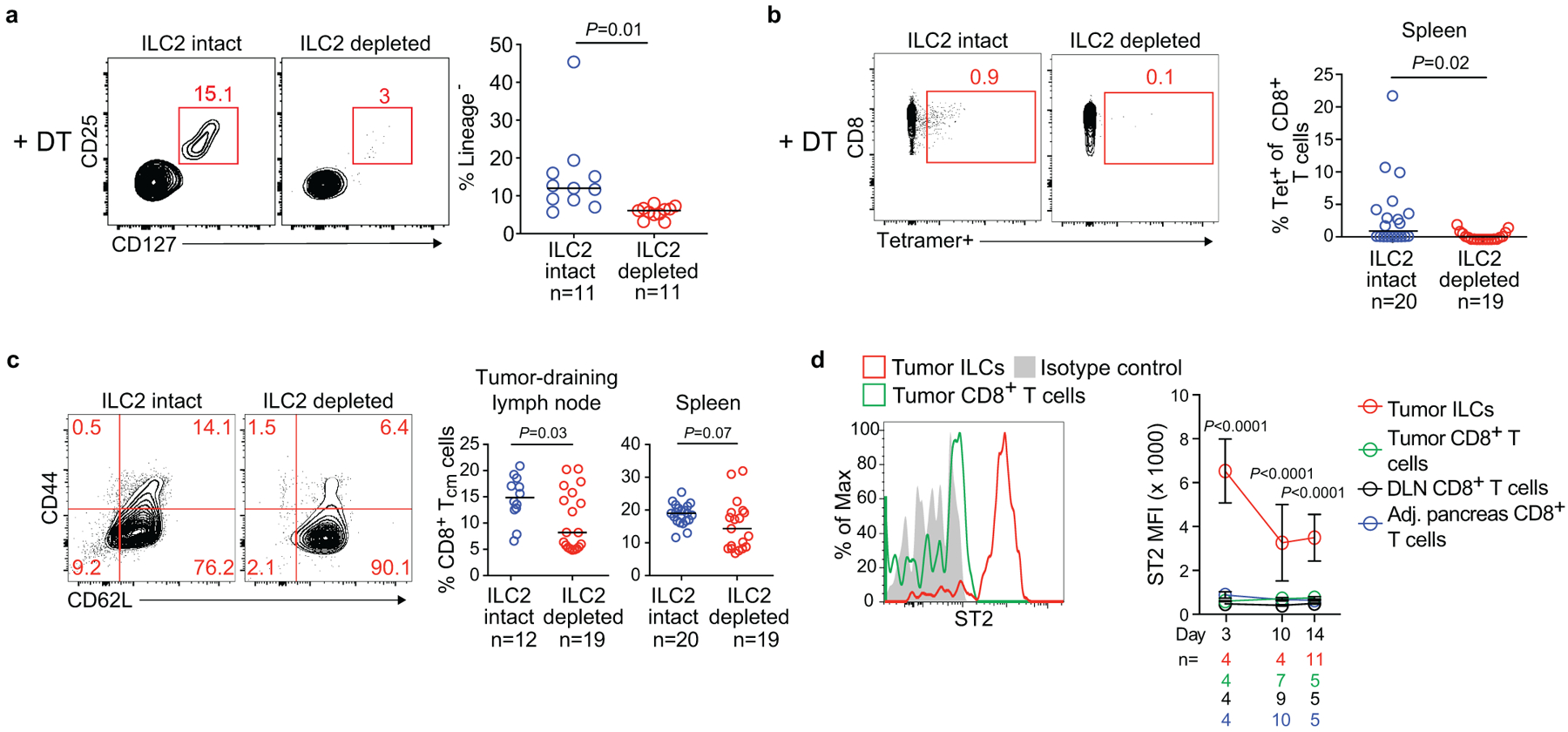
(a) Gating and frequency of intratumoral ILC2s in ILC2-intact mice (diphtheria toxin [DT]-treated Icos+/+; CD4Cre/+) and ILC2-depleted mice (DT-treated Icosfl.DTR/+; CD4Cre/+). (b) Gating and frequency of OVA-specific CD8+ T cells in spleens from ILC-intact and ILC-depleted mice. OVA-specific T cells were detected as SIINFEKL-tetramer+ cells. (c) Gating and frequency of central memory CD8+ T (TCM) cells (CD45+CD3+CD8+ CD44+CD62L+) in tumor draining lymph nodes and spleens in ILC-intact and ILC-depleted mice. (d) ST2 expression on CD45+CD3+CD8+ T cells after tumor implantation in PDAC mice. Data were collected at 14 days post tumor implantation or at the time points indicated. DLN, draining lymph node; MFI, mean fluorescence intensity. Horizontal bars mark medians; error bars mark s.e.m. n indicates individual mice analyzed separately in at least two independent experiments with n≥2/group. P values determined by two-tailed Mann-Whitney test (a-c) and two-way ANOVA with Tukey’s multiple comparison post-test (d, indicating comparison of tumor ILCs to all other groups).
Extended Data Figure 6: Immunophenotyping in rIL33-treated PDAC mice.
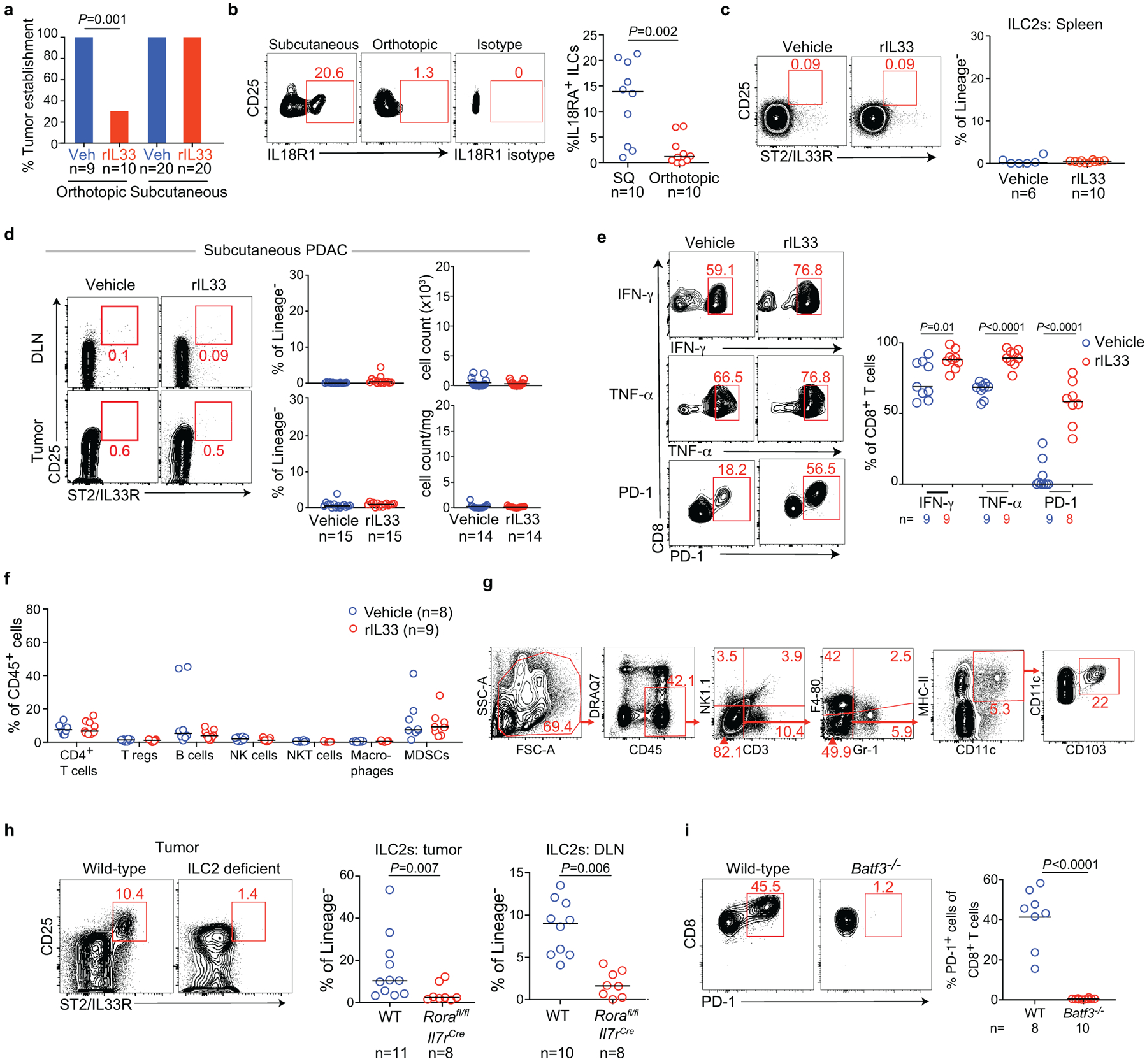
(a) Percent tumor establishment of orthotopic and subcutaneous KPC-OVA PDAC tumors in vehicle (veh) and rIL33 treated mice. (b) Gating (left) and frequency (right) of IL18R1 expression on tumor ILCs in subcutaneous (SQ) and orthotopic PDAC mice. (c) Gating (left) and frequency (right) of splenic ILC2s following rIL33 treatment in orthotopic PDAC mice. (d) Gating (left) and frequency (right) of tumor ILC2s following rIL33 treatment in subcutaneous PDAC mice. (e) Gating (left) and frequency (right) of cytokine and PD-1 expression on tumor CD8+ T cells following rIL33 treatment in orthotopic PDAC mice. (f) Frequency of immune cells in vehicle- and rIL33-treated orthotopic PDAC mice. (g) Gating strategy for identification of CD103+ dendritic cells. (h) Gating (left; tumors) and frequency (right) of ILC2s in tumors and draining lymph nodes (DLN) of wild type (WT) or Rorafl/fl IL7rCre mice (ILC2-deficient) PDAC mice following rIL33 treatment. (i) Gating (left) and frequency (right) of PD-1+ CD8+ T cells in tumors of rIL33-treated WT and Batf3−/− mice. Data were collected at 6 (a), 5 (b), and 3 (i) weeks post tumor implantation. Horizontal bars mark medians. n indicates individual mice analyzed separately in at least two independent experiments with n≥2/group. P values determined by two-tailed Mann-Whitney test (a, f, i).
Extended Data Figure 7: Single-cell RNA sequencing of tumor and draining lymph node ILC2s in PDAC mice.
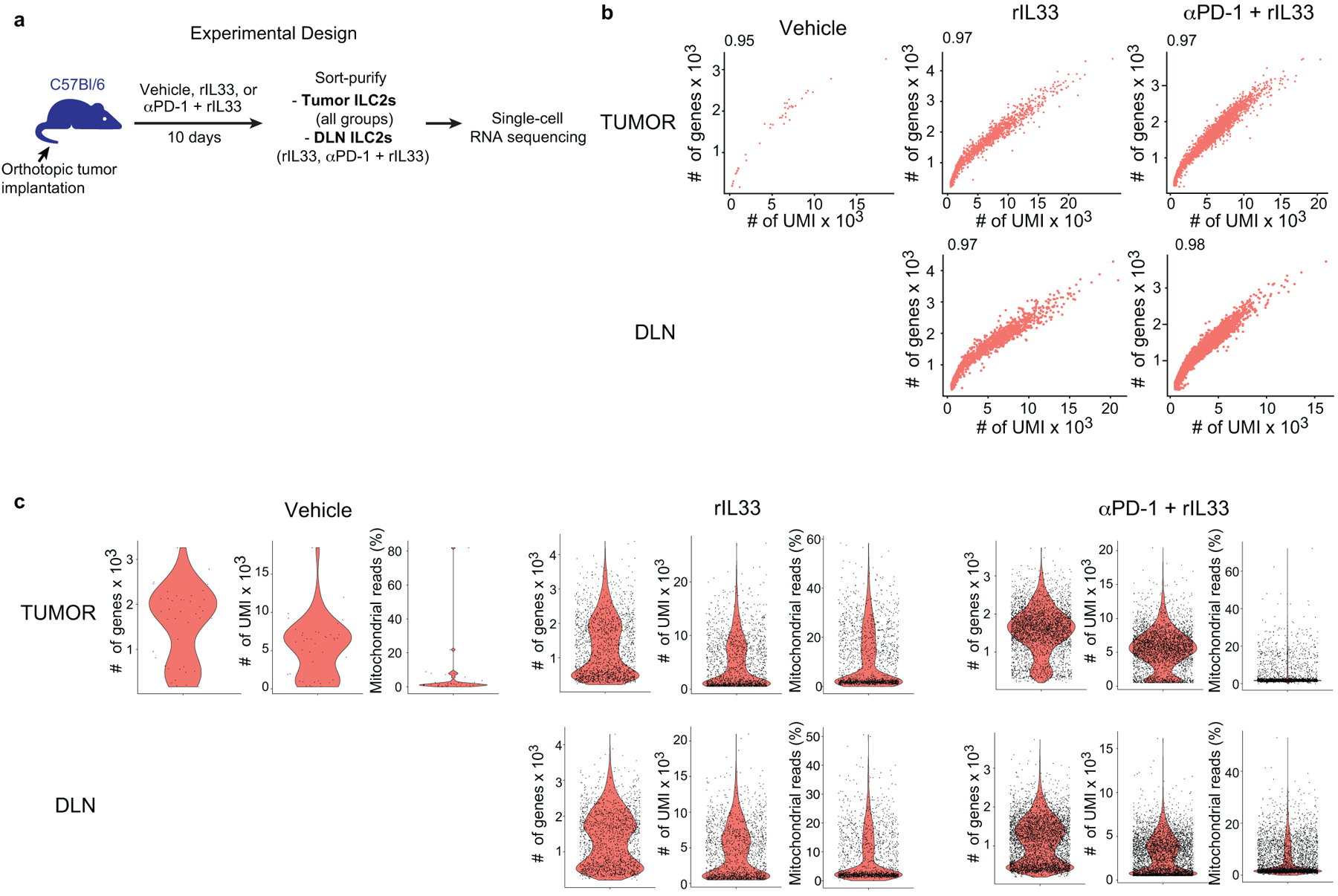
(a) Experimental design for in vivo treatment, purification, and single-cell analysis of ILC2s. (b, c) Quality metrics. (b) Scatter plots showing, for each cell, the relationship between the number of unique molecular identifiers (# of UMIs) and the number of genes (# of genes). (c) Violin plots showing the distribution of the number of genes (left), number of UMIs (middle), and percentage of normalized reads from mitochondrial genes (right) in each treatment group (columns), and each tissue (rows). Each dot represents a single cell. For each treatment group and organ, data represent pooled purified single cells from biological replicates of n=10 (vehicle), n=5 (rIL33), and n=5 (αPD-1 + rIL33) PDAC mice.
Extended Data Figure 8: Activated ILC2s from tumors and draining lymph nodes have distinct transcriptional features.
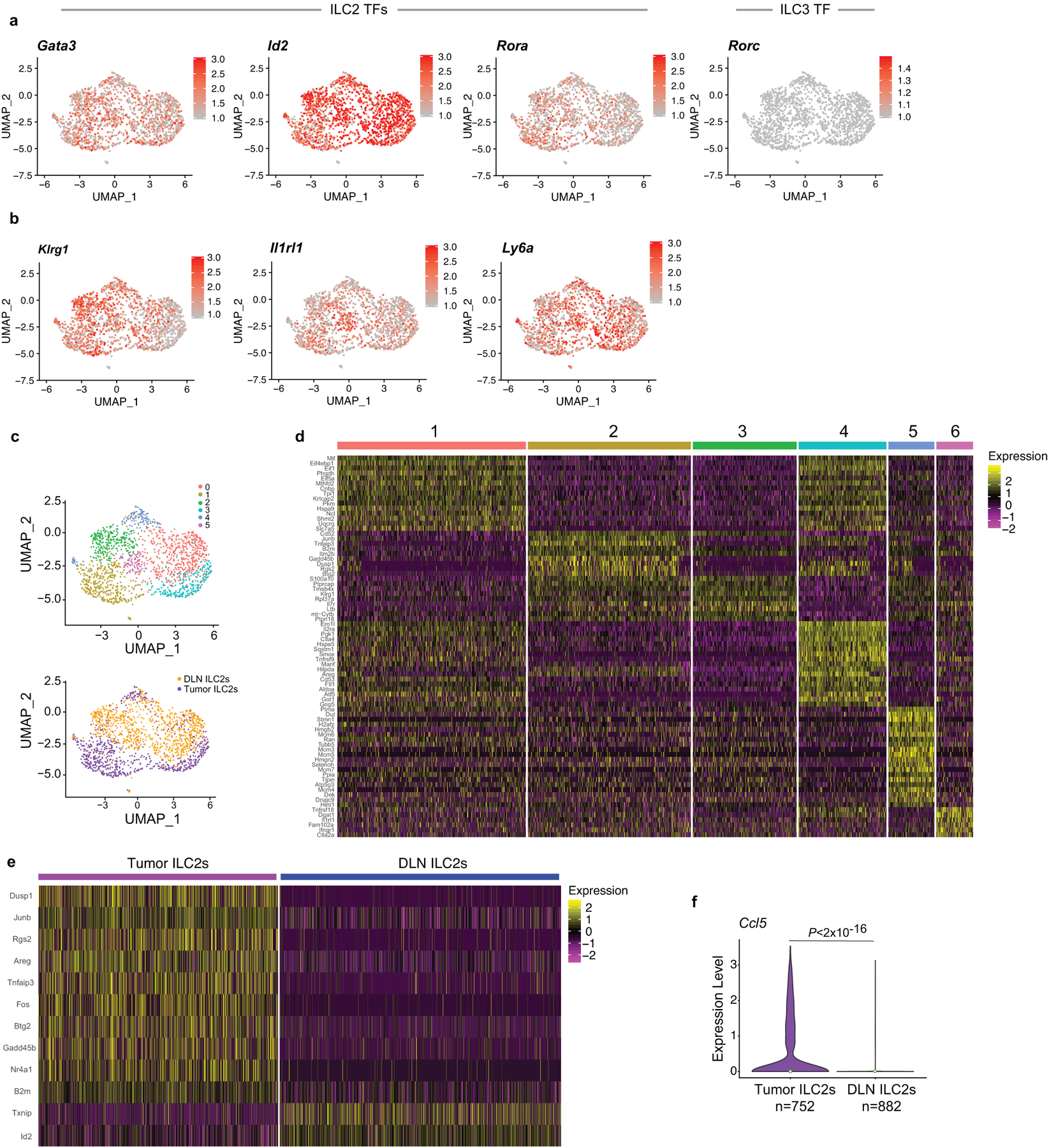
(a) Single-cell analysis of 1,634 rIL33-activated tumor and draining lymph node (DLN) ILC2s (experimental design as outlined in Extended Data Figure 7a). UMAP plots show single cells (dots) in a nonlinear representation of the top 15 principal components. Expression of (a) ILC2 (Gata3, Id2, Rora), and ILC3 (gene, Rorc; protein, Rorγt) transcription factors (TFs), (b) ILC2 surface markers, and (c) of clusters and organs. Expression of ILC-1 TF Tbx21 (T-bet) was undetectable. (d, e) Differentially expressed genes by (d) cluster and (e) organ (TILC2s and DLN ILC2s). (f) Distribution of Ccl5 expression from ILC2s in tumor and DLNs; violin plots show distribution with minima, maxima, and circle indicating median. Each dot in a and b represents a single cell. For each treatment group and organ, data represent pooled purified single cells from biological replicates of n=5 rIL33-treated PDAC mice. P values by two-sided pairwise Wilcoxon rank sum test.
Extended Data Figure 9: Combined αPD-1 and rIL33 treatment induces a unique transcriptional profile in tumor ILC2s.
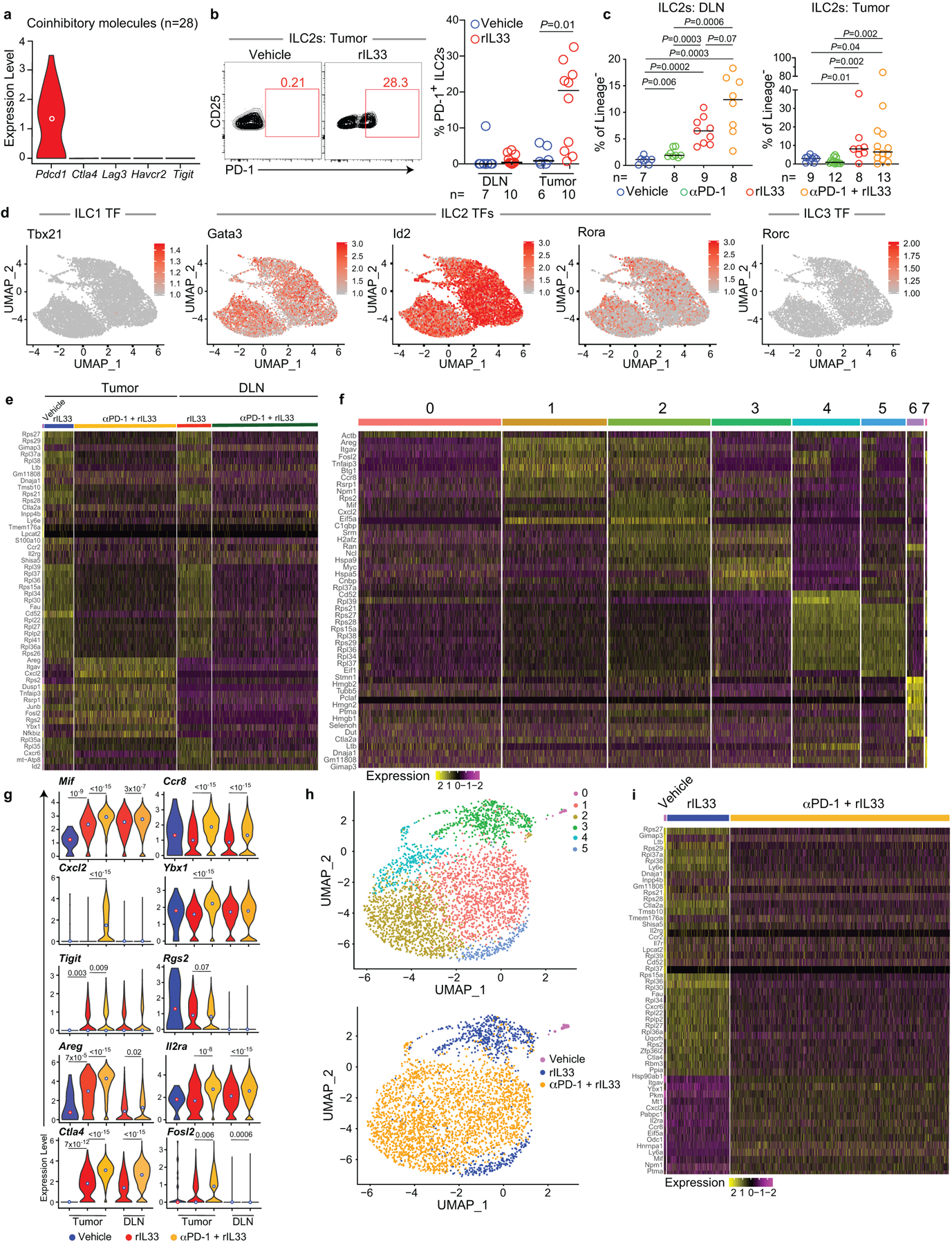
(a) Expression of coinhibitory immune checkpoints in tumor ILC2s in vehicle-treated PDAC mice by single-cell RNA sequencing (scRNA-seq). (b) Gating and frequency of PD-1+ ILC2s in vehicle- and rIL33-treated PDAC mice. DLN, draining lymph node. (c) ILC2 frequency in treated PDAC mice. Corresponding tumor volumes, weight, cell number, and scRNA-seq are shown in Figure 4a–c. (d) scRNA-seq of ILC2s from treated PDAC mice. Expression of ILC 1 (gene, Tbx21; protein, Tbet), ILC2 (Gata3, Id2, Rora), and ILC3 (gene, Rorc; protein, Rorγt) transcription factors (TFs) in purified tumor and draining lymph node (DLN) ILC2s. Corresponding UMAP plots by cluster and treatment are depicted in Figure 4c. Top differentially expressed genes by treatment and tissue (e), cluster (f), and distribution of expression for select differentially expressed genes by treatment and tissue (g) (tumor: vehicle n=28, rIL33 n=752, rIL33+PD-1 n=2,635; DLN rIL33 n=882, rIL33+PD-1 n=2,725). (h) UMAP plots of 3,387 single tumor ILC2s in a non-linear representation of the top 15 principal components. (i) Differentially expressed genes in tumor ILC2s by treatment. Each dot in d and h represents a single cell; for each treatment group and organ, data represent pooled purified single cells from biological replicates of n=10 (vehicle), n=5 (rIL33), and n=5 (αPD-1 + rIL33) PDAC mice. Violin plots show distribution with minima, maxima, and circle indicating median. Horizontal bars in b and c mark medians. P values by two-tailed Mann-Whitney test (b, c) and two-sided pairwise Wilcoxon rank sum test (g).
Extended Data Figure 10: Activated tumor ILC2s express PD-1 and co-exist with PD-1+ T cells.
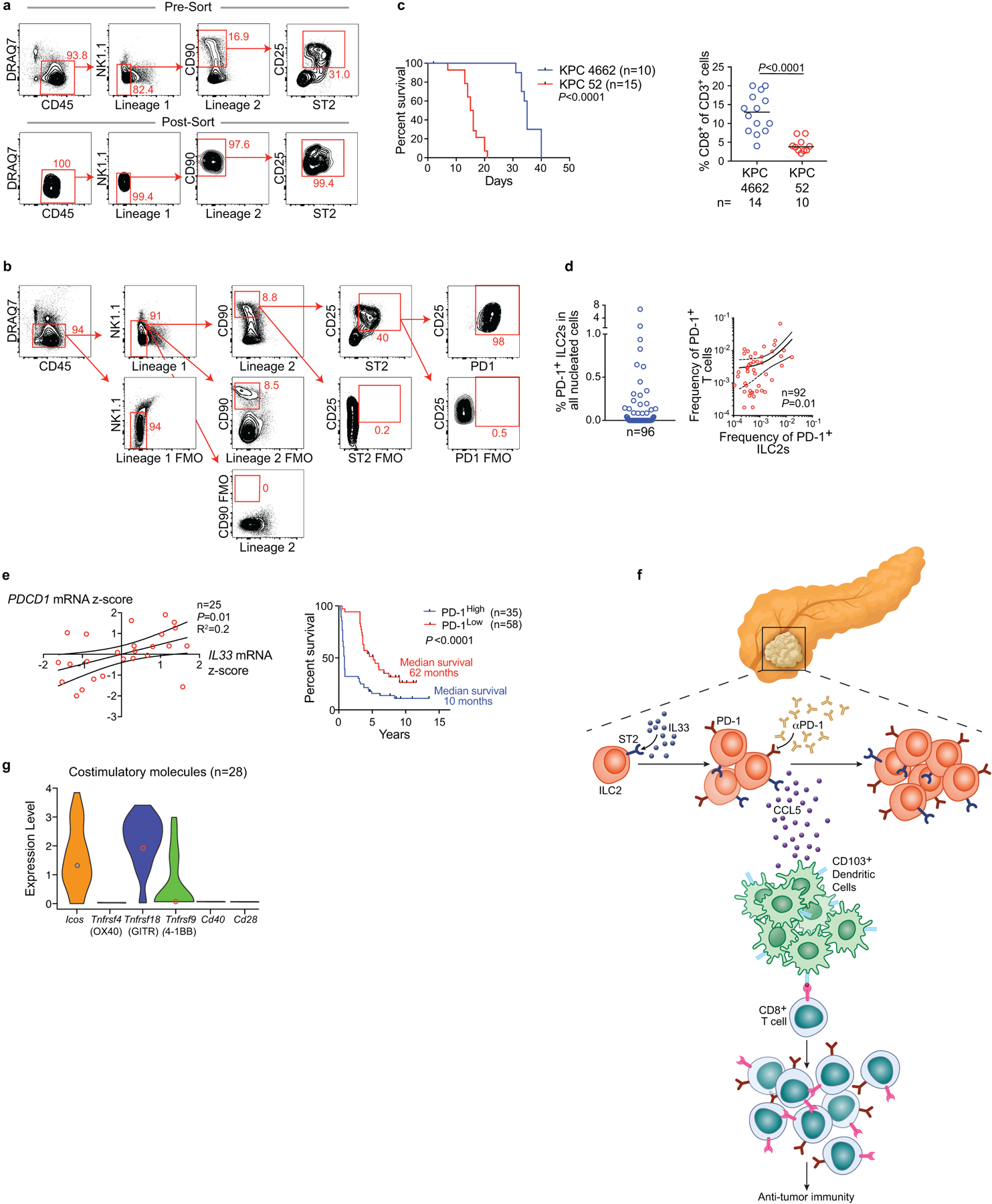
Orthotopic PDAC mice (C57Bl/6 WT, Pdcd1−/−, CD45.1) were treated with 500 ng of carrier-free recombinant murine IL33 daily for 10 days (experimental designs shown in Figure 4e, f). Live, CD45+, lineage−, CD90+, CD25+, ST2+ tumor ILC2s (TILC2s) were sort-purified to 98% purity at day 10 post-implantation. 5 × 105 TILC2s were immediately transferred to orthotopic PDAC tumor-bearing Il7rCre/+Rorafl/fl (ILC2-deficient) CD45.2 mice on days 7 and 14 post-tumor implantation via i.p. injection. Control mice received equivalent volumes of PBS via i.p. injections. (a) Representative plots for TILC2 sort-purification (top) and post-sort purity (bottom). (b) Representative plots showing PD-1 expression on sort-purified TILC2s from WT and CD45.1 mice in the experimental designs as outlined in Figure 4e, f. (c) Survival and intratumoral CD8+ T cell frequency of orthotopic KPC 4662-GFP and KPC 52 PDAC tumors; horizontal bars in c mark medians. (d) Frequency of PD-1+ ILC2s (left) and correlation with PD-1+ T cells (right) in human PDAC. (e) Linear regression analysis of IL33 and PD-1 mRNA in bulk tumor transcriptomes of short- and long-human PDAC survivors (left) and survival association of PD-1+ cells in tumor tissue microarrays of short-term and long-term PDAC survivors (right); high and low defined as higher or lower than the median for the cohort. (f) Model linking the IL33-TILC2 axis to T cell immunity in pancreatic cancer. (g) Distribution of expression of costimulatory molecules in untreated tumor ILC2s by single-cell RNA sequencing. Experimental design as shown in Extended Data Figure 7a; data represent pooled purified single cells from biological replicates of n=10 (vehicle). Data are representative of purity and PD-1 expression on sorted TILC2s in two independent experiments with n≥4/group (a, b). n and data points denote individual mice and patients analyzed separately. P values were determined by two-tailed Mann-Whitney (c), and two-sided log rank (c, e, survival curves) tests, and linear regression (d, e).
Supplementary Material
Acknowledgements
We thank J. Novak, J. Moore, and E. Patterson for editorial assistance, B. Medina, G. Vitiello, J. Zhang, S. Zeng, F. Rossi, J. Loo, N. Param, J. Maltbaek, O. Grbovic-Huezo, Y. Senbabaoglu, M. Gigoux, R. Giese, and S. Budhu for helpful discussions and technical assistance. We thank the Epigenomics Core of Weill Cornell Medical College for technical assistance with scRNA-seq. This work was supported by The V Foundation Convergence Scholar Grant (J.A.M, J.D.W., V.P.B.), the Stand Up to Cancer Convergence Award (J.D.W., V.P.B.), the National Cancer Institute K12CA184746-01A1 (V.P.B.), Damon Runyon Clinical Investigator Award (V.P.B.), the Ben and Rose Cole Pria Foundation Scholar Award (V.P.B.), The Sarah Min and Matthew Pincus Pancreatic Cancer Immunotherapy Award (V.P.B.), an administrative supplement to NIH P30-CA008748 (S.D.L., V.P.B.), NIH R01 CA204228, NIH P30CA023108 (S.D.L.), Swim Across America, and the Ludwig Institute for Cancer Research (J.D.W., T.M.), and the Parker Institute for Cancer Immunotherapy (J.D.W., T.M.). Services by the MSKCC Small-Animal Core Facility and Integrated Genomics Core were funded by the National Cancer Institute Cancer Center Support Grant (P30 CA008748-48), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Footnotes
Competing interests. V.P.B. is a recipient of an immuno-oncology translational research grant from Bristol Myers Squibb and is an inventor on a patent application related to work on neoantigen modeling. S.D.L. is a member of the scientific advisory board of Nybo Pharmaceuticals, and co-founder of Episteme Prognostics. J.D.W. is a consultant for Adaptive Biotech, Advaxis, Amgen, Apricity, Array BioPharma, Ascentage Pharma, Astellas, Bayer, Beigene, Bristol Myers Squibb, Celgene, Chugai, Elucida, Eli Lilly, F Star, Genentech, Imvaq, Janssen, Kleo Pharma, Linneaus, MedImmune, Merck, Neon Therapuetics, Ono, Polaris Pharma, Polynoma, Psioxus, Puretech, Recepta, Trieza, Sellas Life Sciences, Serametrix, Surface Oncology, and Syndax; is a recipient of research support from Bristol Myers Squibb, Medimmune, Merck Pharmaceuticals, and Genentech; and has equity in Potenza Therapeutics, Tizona Pharmaceuticals, Adaptive Biotechnologies, Elucida, Imvaq, Beigene, Trieza, and Linneaus; has received honorarium from Esanex. T.M. is a consultant for Immunos Therapeutics and Pfizer; is a co-founder with equity in IMVAQ therapeutics; receives research funding from Bristol-Myers Squibb, Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals Inc., Peregrine Pharmaceuticals Inc., Adaptive Biotechnologies, Leap Therapeutics Inc., and Aprea; is an inventor on patent applications related to work on Oncolytic Viral therapy, Alpha Virus Based Vaccine, Neo Antigen Modeling, CD40, GITR, OX40, PD-1 and CTLA-4. M.G. is an employee of Bristol Myers Squibb and has financial interest in the company.
Data availability statement
Source code for immune quantification is available in Supplementary Data 1. Bulk RNA-seq data are available under Gene Expression Omnibus (GEO) accession number GSE129388. scRNA-seq data are available under GEO accession number GSE136720. Source data are provided for all experiments. All other data are available from the corresponding author upon reasonable request.
References
- 1.Vivier E et al. Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Salimi M et al. Activated innate lymphoid cell populations accumulate in human tumour tissues. BMC Cancer 18, 341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balachandran VP et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hingorani SR et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G & Bar-Sagi D Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21, 836–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J et al. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity 49, 178–193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brestoff JR et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature 519, 242–246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasteiger G, Fan X, Dikiy S, Lee SY & Rudensky AY Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monticelli LA et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol 17, 656–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchberger S et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. Journal of Experimental Medicine 210, 917–931 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neesse A et al. CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proceedings of the National Academy of Sciences 110, 12325–12330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardman CS, Panova V & McKenzie ANJ IL‐33 citrine reporter mice reveal the temporal and spatial expression of IL‐33 during allergic lung inflammation. Eur J Immunol 43, 488–498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talabot-Ayer D et al. The mouse interleukin (Il)33 gene is expressed in a cell type- and stimulus-dependent manner from two alternative promoters. J. Leukoc. Biol 91, 119–125 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Ricardo-Gonzalez RR et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol 19, 1093–1099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalmas E et al. Interleukin-33-Activated Islet-Resident Innate Lymphoid Cells Promote Insulin Secretion through Myeloid Cell Retinoic Acid Production. Immunity 47, 928–942.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Oliphant CJ et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity 41, 283–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Böttcher JP et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto C et al. Runx/Cbfβ complexes protect group 2 innate lymphoid cells from exhausted-like hyporesponsiveness during allergic airway inflammation. Nat Commun 10, 447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y et al. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature 539, 102–106 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Taylor S et al. PD-1 regulates KLRG1(+) group 2 innate lymphoid cells. Journal of Experimental Medicine 214, 1663–1678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J et al. Intratumorally Establishing Type 2 Innate Lymphoid Cells Blocks Tumor Growth. The Journal of Immunology 196, 2410–2423 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Diana A et al. Prognostic value, localization and correlation of PD-1/PD-L1, CD8 and FOXP3 with the desmoplastic stroma in pancreatic ductal adenocarcinoma. Oncotarget 7, 40992–41004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donovan C et al. Roles for T/B lymphocytes and ILC2s in experimental chronic obstructive pulmonary disease. J. Leukoc. Biol 105, 143–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans RA et al. Lack of immunoediting in murine pancreatic cancer reversed with neoantigen. JCI Insight 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sastra SA & Olive KP Quantification of murine pancreatic tumors by high-resolution ultrasound. Methods Mol. Biol 980, 249–266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monticelli LA et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol 12, 1045–1054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Z et al. Augmentation of Immune Checkpoint Cancer Immunotherapy with IL18. Clin Cancer Res 22, 2969–2980 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarilin D et al. Machine-based method for multiplex in situ molecular characterization of tissues by immunofluorescence detection. Sci Rep 5, 9534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivier E et al. Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Balachandran VP et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hingorani SR et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G & Bar-Sagi D Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21, 836–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brestoff JR et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature 519, 242–246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalmas E et al. Interleukin-33-Activated Islet-Resident Innate Lymphoid Cells Promote Insulin Secretion through Myeloid Cell Retinoic Acid Production. Immunity 47, 928–942.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Oliphant CJ et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity 41, 283–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donovan C et al. Roles for T/B lymphocytes and ILC2s in experimental chronic obstructive pulmonary disease. J. Leukoc. Biol 105, 143–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans RA et al. Lack of immunoediting in murine pancreatic cancer reversed with neoantigen. JCI Insight 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sastra SA & Olive KP Quantification of murine pancreatic tumors by high-resolution ultrasound. Methods Mol. Biol 980, 249–266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monticelli LA et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol 12, 1045–1054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Z et al. Augmentation of Immune Checkpoint Cancer Immunotherapy with IL18. Clin Cancer Res 22, 2969–2980 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yarilin D et al. Machine-based method for multiplex in situ molecular characterization of tissues by immunofluorescence detection. Sci Rep 5, 9534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


