Abstract
EDS1 (Enhanced Disease Susceptibility 1) plays a crucial role in both effector‐triggered immunity activation and plant basal defence. However, whether pathogen effectors can target EDS1 or an EDS1‐related pathway to manipulate immunity is rarely reported. In this study, we identified a Phytophthora capsici Avirulence Homolog (Avh) RxLR (Arg‐any amino acid‐Leu‐Arg) effector PcAvh103 that interacts with EDS1. We demonstrated that PcAvh103 can facilitate P. capsici infection and is required for pathogen virulence. Furthermore, genetic evidence showed that PcAvh103 contributes to virulence through targeting EDS1. Finally, PcAvh103 specifically interacts with the lipase domain of EDS1 and can promote the disassociation of EDS1–PAD4 (Phytoalexin Deficient 4) complex in planta. Together, our results revealed that the P. capsici RxLR effector PcAvh103 targets host EDS1 to suppress plant immunity, probably through disrupting the EDS1–PAD4 immune signalling pathway.
Keywords: EDS1, PAD4, PcAvh103, Phytophthora, plant immunity, RxLR effector
A Phytophthora capsici RxLR effector PcAvh103 targets host EDS1 to suppress plant immunity, probably through disrupting the EDS1–PAD4 immune signalling pathway.
1. INTRODUCTION
Oomycetes are a lineage of eukaryotic microorganisms phylogenetically related to diatoms and brown algae in the kingdom Stramenopila (Jiang and Tyler, 2012). They can cause devastating plant diseases, leading to enormous environmental damage and significant economic losses worldwide (Birch et al., 2006; Kamoun et al., 2015). Among them, the genus Phytophthora is the most notorious. For example, Phytophthora infestans caused potato late blight and the Great Irish Famine in history (Haas et al., 2009). P. sojae causes soybean root and stem rot, resulting in serious yield losses every year (Tyler, 2007). P. capsici, which infects a large number of agriculturally important vegetables like pepper, tomato, cucurbits, and eggplant, causes huge economic losses (Lamour et al., 2012b). In addition, P. capsici can also infect the model plants Nicotiana benthamiana and Arabidopsis thaliana, therefore it has been studied as an emerging model pathogen in plant–microbe interactions (Lamour et al., 2012b; Wang et al., 2013). However, there are still plenty of deficiencies in understanding the infection process and pathogenic mechanism of Phytophthora pathogens.
During infection, Phytophthora pathogens secrete both apoplastic and cytoplasmic effectors to target different compartments or pathways in their hosts (Birch et al., 2006; Kamoun, 2006). Among them, RxLR effectors are one class of the cytoplasmic effectors, named by their conserved Arg‐any amino acid‐Leu‐Arg (RxLR) motif at the N‐terminus (Tyler et al., 2006; Jiang et al., 2008). The RxLR motif was reported to facilitate delivery and translocation of effectors to host cells (Whisson et al., 2007; Dou et al., 2008). So far, some RxLR effectors have been reported to manipulate various aspects of plant defence (Anderson et al., 2015). Recently, P. infestans RxLR effector SFI3 (Suppressor of early Flg22‐induced Immune response 3) was shown to target the Solanum tuberosum U‐box‐kinase protein (StUBK) and suppress early transcriptional responses of the pattern‐triggered immunity (PTI) pathway (He et al., 2019). P. sojae Avh52 (Avirulence Homolog 52) recruits the cytoplasmic Glycine max transacetylase protein 1 (GmTAP1) into nuclear speckles, which acetylates histones H2A and H3, thereby enhancing plant susceptibility (Li et al., 2018). In addition, the P. capsici Avirulence (Avr) RxLR effector PcAvr3a12 can target and inhibit the FK506‐binding protein FKBP15‐2, which is required for endoplasmic reticulum (ER) stress‐mediated plant immunity (Fan et al., 2018). Furthermore, the P. sojae RxLR effector Avh238 can suppress ethylene biosynthesis and facilitate infection by destabilizing soybean 1‐aminocyclopropane‐1‐carboxylate synthase GmACSs (Yang et al., 2019).
To withstand infection of microbial pathogens, plants have evolved two layers of immune system. One is PTI, which uses pattern recognition receptors (PRRs) to perceive the conserved pathogen signatures called microbe‐associated molecular patterns (MAMPs), and the other is effector‐triggered immunity (ETI), which is activated through the recognition of effectors by nucleotide binding‐leucine rich repeat receptors (NB‐LRRs) (Jones and Dangl, 2006). PTI provides hosts with basal resistance to broad‐spectrum pathogens, including a reactive oxygen species (ROS) burst, mitogen‐activated protein kinase (MAPK) cascades phosphorylation, and callose deposition (Segonzac and Zipfel, 2011). Nevertheless, ETI brings about a robust defence response against specific pathogens, usually resulting in a hypersensitive response (HR) at the infection sites, which is also called programmed cell death (PCD) (Cui et al., 2015).
EDS1 (Enhanced Disease Susceptibility 1) was first reported as mutations in enhanced disease susceptibility1 (eds1) impair SA (salicylic acid) levels and thereby enhance susceptibility to pathogen infection (Falk et al., 1999). Subsequent studies demonstrated that both nuclear and cytoplasmic EDS1 coordinate immune responses, and nuclear EDS1 is required for reprogramming of defence gene expression and basal resistance (Garcia et al., 2010). EDS1 can form distinct protein complexes including the homomeric association with itself as well as heteromeric complexes with Phytoalexin Deficient 4 (PAD4) and SENESCENCE‐ASSOCIATED GENE 101 (SAG101) (Feys et al., 2001, 2005). In addition, PAD4 and SAG101 contact the same N‐terminal lipase domain of the EDS1 interface, and the EDS1 heterodimers respectively mediate resistance signalling (Wagner et al., 2013). EDS1–PAD4 complex works in parallel with SA in basal resistance and ETI, maintaining important SA‐related resistance genes reprogramming (Cui et al., 2017). More recently, the C‐terminal EDS1–PAD4 (EP) domain surface of EDS1 enforces timely reprogramming of resistance genes (Bhandari et al., 2019). However, a few studies reported that pathogen effectors can target or interfere with EDS1 as a virulence strategy. Considering EDS1 is a crucial component in plant immunity, we propose that Phytophthora pathogens might have evolved certain effectors that target EDS1 for virulence function.
In our study, through screening P. capsici effectors by using EDS1 as a bait, we uncovered an Avirulence Homolog (Avh) effector PcAvh103 and confirmed the interactions by yeast two‐hybrid (Y2H) and co‐immunoprecipitation (co‐IP) assays. We found that expression of PcAvh103 facilitates P. capsici infection in N. benthamiana, and silencing of PcAvh103 reduces the pathogenicity of P. capsici. Furthermore, we proved that PcAvh103 contributes to virulence through targeting EDS1 in Arabidopsis. Finally, we demonstrated that PcAvh103 specifically interacts with the lipase domain of EDS1 and can disrupt the EDS1–PAD4 complex in vivo and in vitro. Together, our results reveal that the P. capsici RxLR effector PcAvh103 targets host EDS1 for virulence, probably through disrupting the association of EDS1 and PAD4.
2. RESULTS
2.1. PcAvh103 interacts with EDS1
EDS1 plays an important role in basal resistance and ETI‐/SA‐mediated defence response, while BAK1 (BRI1‐associated kinase 1) and BIK1 (Botrytis‐induced kinase 1) are core components in the PTI signalling pathway (Lin et al., 2014; Cui et al., 2017). To investigate whether P. capsici secretes effectors that target plant EDS1, BAK1, and BIK1, 42 RxLR effectors (Lamour et al., 2012a) were separately cloned into prey vector and screened for interactors of EDS1, BAK1, and BIK1 using the Y2H approach (Li et al., 2019a, Table S1). Results showed that one effector, PcAvh103, was repeatedly identified from two independent screens (Table S1). To validate the interactions, we performed a reciprocal Y2H assay and confirmed that PcAvh103 interacted with each of the three proteins in yeast (Figure 1a). To confirm the interactions in planta, co‐IP experiments were performed by transiently expressing PcAvh103‐FLAG, EDS1‐HA, BAK1‐HA, and BIK1‐HA in Arabidopsis protoplasts. Only EDS1 was co‐immunoprecipitated with FLAG‐tagged PcAvh103; however, BAK1 or BIK1 were unable to bind to PcAvh103 (Figure 1b). We therefore focused on the interaction between PcAvh103 and EDS1 for further studies.
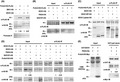
Figure 1.

PcAvh103 interacts with EDS1. (a) Interactions between EDS1, BAK1, and BIK1 with PcAvh103 in the yeast 2‐hybrid system. Yeast AH109 cells co‐transformed with bait and prey vectors were grown on QDO (SD/−Ade/−His/−Leu/−Trp) medium. The combination of pGBKT7‐53 and pGADT7‐T was used as a positive control, while pGBKT7‐Lam and pGADT7‐T was used as a negative control. (b) Interactions between EDS1, BAK1, and BIK1 with PcAvh103 in Arabidopsis. Indicated constructs were transiently co‐expressed in Arabidopsis protoplasts. The immunoprecipitated (IP) and input proteins were analysed via immunoblot assay using anti‐FLAG and anti‐HA antibodies
2.2. PcAvh103 facilitates P. capsici infection
To explore the virulence function of PcAvh103, we transiently expressed PcAvh103 in N. benthamiana and inoculated with P. capsici. N. benthamiana leaves expressing GFP‐PcAvh103 and GFP empty vector (negative control) were inoculated with P. capsici zoospores onto the infiltrated area 36 hr after infiltration. Infection lesion sizes were recorded for comparison at 36 hr post‐inoculation (hpi). As can be seen, expression of PcAvh103 significantly promoted P. capsici colonization compared to green fluorescent protein (GFP) control, with bigger lesions (Figure 2a,b). Dead cells and lesions were visualized by trypan blue staining, further confirming that PcAvh103 promotes P. capsici infection (Figure 2a). We confirmed the GFP‐PcAvh103 expression by observing the green fluorescence by confocal microscopy 48 hr after infiltration (Figure 2c). In addition, we also noticed that the GFP–PcAvh103 fluorescence signal was distributed in both the nucleus and the cytoplasm (Figure 2c).
Figure 2.
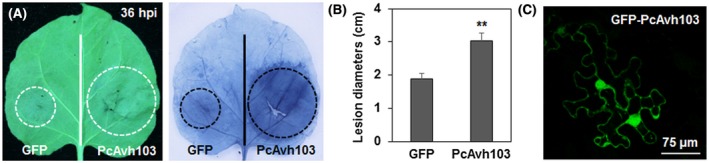
Expression of PcAvh103 can facilitate Phytophthora capsici infection. (a,b) P. capsici infection on Nicotiana benthamiana. Representative N. benthamiana leaves were inoculated by P. capsici LT263 after transient expression of green fluorescent protein (GFP) and PcAvh103, and photographed at 36 hr post‐inoculation. Dead cells and lesions were visualized by trypan blue staining. Lesion diameters were calculated from three independent biological replicates. Error bars represent + SD of at least six leaves each (**p < .01, Student's t test). (c) Expression of PcAvh103 in planta. Subcellular localization of PcAvh103 was visualized by confocal microscopy expressing in N. benthamiana epidermal cells. Scale bar represents 75 μm
2.3. PcAvh103 contributes to P. capsici virulence
To further evaluate the contribution of PcAvh103 to the virulence of P. capsici, we silenced PcAvh103 in the P. capsici strain LT263 (wild‐type, WT). Putative silenced transformants were selected and the silencing efficiency was estimated by quantitative reverse transcription PCR (RT‐qPCR). Two independently silenced transformants (T105 and T76) were obtained in which PcAvh103 transcriptional levels were reduced to approximately 0% and 20% of the WT strain. An additional transformant, T48, was selected as a control in which PcAvh103 remained unaffected (Figure 3a). To clarify whether silencing of PcAvh103 in P. capsici has an effect on its growth, we checked the growth phenotype of PcAvh103‐silenced transformants. As shown in Figure S1, PcAvh103‐silenced transformants T105 and T76 exhibited similar growth rate and mycelial morphological characteristics compared to the WT and T48 strains.
Figure 3.
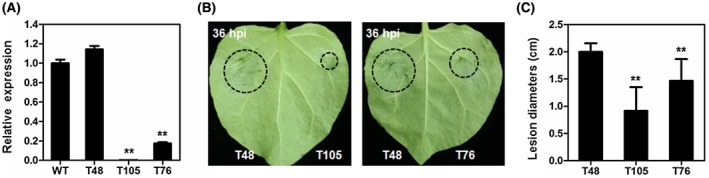
PcAvh103 is required for Phytophthora capsici virulence. (a) Relative transcript levels of PcAvh103 in transformants. The transcriptional levels were determined by quantitative reverse transcription PCR with P. capsici tubulin gene as an internal reference (**p < .01 compared with wild‐type [WT], Dunnett's test). (b) and (c) Inoculation of PcAvh103‐silenced transformants. Zoospores of T48, T105, and T76 were inoculated on Nicotiana benthamiana leaves and photographs were taken 36 hr post‐inoculation (hpi). Lesion diameters were measured at 36 hpi with at least 12 leaves in each experiment. Asterisks indicate significant differences (**p < .01 compared with T48, Dunnett's test)
The virulence of PcAvh103‐silenced transformants was determined on N. benthamiana leaves by inoculation with suspension of P. capsici zoospores. Compared to T48, T105 and T76 exhibited significantly reduced virulence with smaller lesions (Figure 3b). Statistical analysis showed that the lesion diameters in leaves inoculated with T105 and T76 were reduced to 46% and 69% relative to that inoculated with T48, respectively (Figure 3c). Together, these results suggest that PcAvh103 is required for P. capsici virulence.
2.4. PcAvh103 exhibits virulence function through EDS1
Similarly, we also checked the virulence of the PcAvh103‐silenced transformants on Arabidopsis plants (Figure 4a). Leaves from Col‐0 were inoculated with zoospore suspensions of WT, T48, and T105, respectively. Compared with WT and T48 infection leaves, we noticed the reduced colonization of pathogen in T105 infection leaves, with lighter trypan blue staining observations (Figure 4a). The quantitative assay showed the pathogen accumulation in T105 infection leaves was reduced to approximately 20% of that inoculated with WT or T48 (Figure 4a). Therefore, PcAvh103 is also required for P. capsici infection on Arabidopsis.
Figure 4.
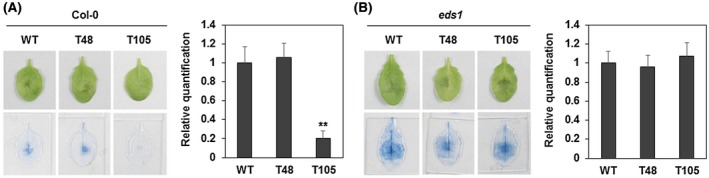
PcAvh103 contributes to Phytophthora capsici virulence through EDS1. (a) Inoculation of wild‐type (WT) and PcAvh103‐silenced P. capsici on Arabidopsis. Zoospores of LT263, T48, and T105 were inoculated on Col‐0 leaves for 36 hr. Infected leaves were stained by trypan blue. The relative biomass of P. capsici was measured by quantitative reverse transcription PCR. Experiments were repeated at least three times with at least 16 leaves used in each experiment (**p < .01 compared with T108, Dunnett's test). (b) Inoculation of WT and PcAvh103‐silenced transformants on eds1 mutant. Details same as in (a)
Considering P. capsici is known as a soilborne pathogen, we also checked whether PcAvh103 is required for P. capsici infection in soil through root inoculation. We specifically implemented root inoculation of Arabidopsis plants with corresponding zoospore suspensions. Inoculation on Col‐0 roots by WT and T48 leads to typical disease phenotypes (leaves showing yellowing or wilting with curled leaf edges), but T105 produced notably reduced disease symptoms on Col‐0 (Figure S2). In addition, the average disease indices of Col‐0 by WT and T48 inoculation are nearly 45% and significantly higher than that of T105 (Figure S2). Together, these results demonstrate PcAvh103 is also important for root colonization.
To evaluate the consequence of PcAvh103–EDS1 association in this plant–microbe interaction, we used the T‐DNA mutant eds1 to test the virulence of PcAvh103‐silenced P. capsici mutants. Similarly, eds1 mutant leaves were inoculated with zoospore suspensions of WT, T48, and T105 (Figure 4b). However, no obvious difference in disease lesions was observed after inoculation treatment, and the quantitative assay also displayed a similar amount of pathogen biomass in infected leaves for WT, T48, and T105 (Figure 4b). These results indicate that PcAvh103 contributes to P. capsici virulence by targeting EDS1.
2.5. PcAvh103 has no effect on protein accumulation of EDS1
Interfering with the stability of crucial immune components by effectors is an effective strategy that is used by a large variety of pathogens (Li et al., 2019b). To test whether PcAvh103 affects the stability of EDS1 during interaction, EDS1‐HA or BIK1‐HA was transiently co‐expressed with PcAvh103‐FLAG in Arabidopsis protoplasts, and protein levels of EDS1 and BIK1 were quantified by immunoblots (Figure 5a). The results show that neither the abundance of EDS1 nor that of non‐interacted BIK1 was significantly altered by PcAvh103, compared to expression of EDS1 or BIK1 alone (Figure 5a).
Figure 5.
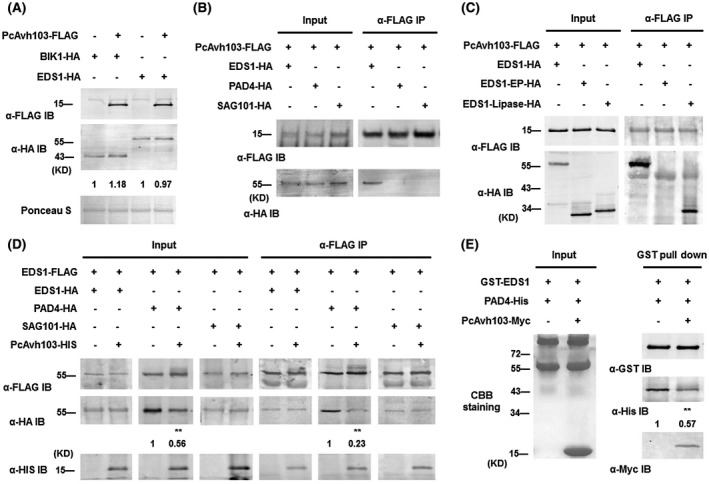
PcAvh103 can disrupt EDS1–PAD4 association. (a) Unaffected protein accumulation of EDS1 by PcAvh103. PcAvh103‐FLAG together with BIK1‐HA or EDS1‐HA were transiently co‐expressed in Arabidopsis, and protein levels were analysed by immunoblots with indicated antibodies. Numbers below represent abundances of BIK1 or EDS1 relative to solo expression. (b) and (c) Determination of the interactions in Arabidopsis. EDS1, PAD4 or SAG101 together with PcAvh103 (b) and EDS1, EDS1‐EP domain or EDS1‐Lipase domain with PcAvh103 (c) were co‐expressed in Arabidopsis protoplasts. The input and immunoprecipitated (IP) proteins were analysed via immunoblot assay using anti‐FLAG and anti‐HA antibodies. (d) Interfering with the association of EDS1 with PAD4 by PcAvh103 in vivo. Indicated constructs were co‐expressed in the presence or absence of PcAvh103 in plant cells. The immune complexes were immunoprecipitated with α‐FLAG IP, and the bound protein was detected by immunoblot with indicated antibodies. (e) Interfering with the association of EDS1 with PAD4 by PcAvh103 in vitro. Prokaryotic recombinant proteins GST‐EDS1 together with PAD4‐His were affinity purified (GST pull‐down) in the presence or absence of PcAvh103‐Myc. The gel was stained with Coomassie brilliant blue (CBB) to show equal loading of protein mixtures (input) and the amounts of bound proteins were analysed by immunoblot with indicated antibodies. Numbers below represent abundances of PAD4 relative to which in absence of PcAvh103. Asterisks indicate significant differences (**p < .01 compared with solo expression, Dunnett's test)
2.6. PcAvh103 can disrupt EDS1‐PAD4 association
Previous studies reported that EDS1, PAD4, and SAG101 have homologous lipase domain and EP domain, and form the heterodimers of EDS1–PAD4 or EDS1–SAG101 at the lipase domain of the EDS1 interface, while EDS1 also strongly interacts with itself to form homomeric associations (Feys et al., 2001; Wagner et al., 2013). To further clarify the underlying mechanism of how PcAvh103 suppresses plant defence by targeting EDS1, we tested if PcAvh103 interferes with formation of the homomeric associations as well as in heteromeric complexes of EDS1. First, PcAvh103‐FLAG was co‐expressed with EDS1‐HA, PAD4‐HA or SAG101‐HA in protoplasts and the protein extracts were processed with co‐IP assay. The results show that PcAvh103 only interacts with EDS1 (Figure 5b). Second, we demonstrated that PcAvh103 interacts with EDS1 through its lipase domain (Figure 5c). Third, we carried out in vivo co‐IP assays between EDS1‐FLAG and EDS1‐HA, PAD4‐HA or SAG101‐HA in the presence or absence of PcAvh103‐HIS. Interestingly, expression of PcAvh103 in planta only reduced the association of EDS1 with PAD4, along with the significantly lower protein levels of PAD4 in the immunoprecipitates (Figure 5d). Finally, we performed the in vitro pull‐down assay by using GST‐EDS1 together with PAD4‐His in the presence or absence of PcAvh103‐Myc. Similarly, adding extra PcAvh103 to the system resulted in decreased enrichment of PAD4 in bound resins (Figure 5e). These findings demonstrate that PcAvh103 competes with PAD4 to bind to EDS1, thus disrupting the formation of the EDS1–PAD4 complex.
3. DISCUSSION
EDS1 plays a pivotal role in plant immune systems. However, few studies have reported that pathogen effectors target EDS1 or an EDS1‐related pathway to manipulate immunity. Some impressive research reported that Pseudomonas syringae effectors AvrRps4 and HopA1 target EDS1 and alter its interactions with RPS4/6 and SRFR1g, suggesting EDS1 might be a common virulence target that is guarded by corresponding Toll‐interleukin1‐receptor (TIR)‐NB‐LRR (nucleotide binding and leucine‐rich repeat) proteins (Bhattacharjee et al., 2011). In addition, two EDS1‐like proteins in soybean (Glycine max), GmEDS1a and GmEDS1b, interacted with another P. syringae effector AvrA1 and were required for its virulence function on rpg2 (resistance to P. syringae pv. glycinea 2) plants (Wang et al., 2014). In this study, we adopted the Y2H system to screen the potential EDS1‐interacted effectors of P. capsici. Preliminarily, we identified an RxLR effector PcAvh103 that targets EDS1 and confirmed the interactions in yeast and Arabidopsis. We subsequently showed that PcAvh103 contributes to P. capsici virulence through EDS1 by using genetic approaches, indicating EDS1 indeed is the virulence target of PcAvh103. This is the first report about the Phytophthora effectors target host EDS1 for virulence, which prompted us to uncover the molecular mechanisms involved.
A variety of pathogen effectors can suppress plant immunity by using diversified strategies, termed effector‐triggered susceptibility (ETS). For example, the P. syringae effector HopAI1 inhibits plant MAPK cascades through a unique phosphothreonine lyase activity to suppress PTI (Zhang et al., 2007). Xanthomonas campestris effector XopJ interferes with SA‐dependent defence response by targeting proteasomal subunit RPT6 and inhibiting proteasome activity (Ustun et al., 2013). In addition, P. sojae effector PSR1 directly targets host PINP1, which is a previously unidentified component of RNA silencing, to promote infection (Qiao et al., 2015). More recently, P. capsici effector RxLR207 can regulate ROS‐mediated defence response to promote the transition from the biotrophic to the necrotrophic stage by targeting Arabidopsis BPA1 (binding partner of ACD11) and BPLs (BPA1‐Like proteins) (Li et al., 2019a). In our study, we found expression of PcAvh103 significantly promotes leaf colonization of P. capsici and demonstrated that PcAvh103 contributes to P. capsici virulence through targeting EDS1 in leaves. Considering P. capsici is regarded as a soilborne pathogen, it is worth mentioning that PcAvh103 is also important for root colonization. Up to now, the significance of the PcAvh103–EDS1 interaction in roots has not been validated and this will be studied in the future.
Interfering with the stability of target proteins or crucial immune components is commonly used by pathogen effectors. For instance, the P. sojae RxLR effector PsAvh262 targets and stabilizes BiPs (binding immunoglobulin proteins) to suppress ER stress‐mediated immunity and facilitate infection (Jing et al., 2016). The Xanthomonas oryzae pv. oryzae non‐TAL effector, XopK, inhibits PTI upstream of MAPK cascades by interacting with and directly ubiquitinating Oryza sativa somatic embryogenic receptor kinase 2 (OsSERK2), resulting in its degradation (Qin et al., 2018). Another P. syringae type III effector, AvrPtoB, targets NPR1 (non‐expressor of pathogenesis related‐1) and mediates the degradation of NPR1 via 26S proteasome, dependent on its E3 (ubiquitin ligase) activity (Chen et al., 2017). In our study, we co‐expressed PcAvh103 with EDS1 in planta, but found the protein levels of EDS1 were intact. Thus, we suggest that PcAvh103 has no effect on protein accumulation of EDS1 during interaction in Arabidopsis.
Previous studies reported that EDS1 can form heteromeric protein complexes with PAD4 and SAG101 (Feys et al., 2005) by PAD4 and SAG101 contacting the same N‐terminal lipase domain of the EDS1 interface, to respectively mediate resistance signalling (Wagner et al., 2013). In our study, we demonstrated that PcAvh103 specifically interacts with EDS1 through its lipase domain, implying that PcAvh103 may be involved in manipulating the homomeric or heteromeric immune complexes of EDS1. In vivo co‐IP and in vitro pull‐down assays revealed that PcAvh103 can only disrupt the EDS1–PAD4 association in plant cells. Hence, we speculated that PcAvh103 may target specific regions or sites in the N‐terminal lipase domain that play distinct roles in dimerization of EDS1–PAD4 or EDS1–SAG101. It is worth mentioning that an L262P exchange mutant in EDS1 lost interaction with PAD4, but not SAG101, reflecting a subtle difference between these two immune complexes (Rietz et al., 2011) and thus indirectly supporting our hypothesis. Additional experiments are therefore required to test our hypothesis.
Considering the functional mechanisms of the majority of effectors are still poorly understood, we advocate that interfering with the association of immune components is also a commonly used and effective virulence strategy for pathogen effectors. For example, the Phytophthora effector PsAvh23 affects the formation of the ADA2–GCN5 (Alteration/Deficiency in Activation 2‐General Control Non‐depressive 5) subcomplex to manipulate host histone acetylation and reprogramme defence gene expression (Kong et al., 2017). The P. infestans effector Pi02860 interacts with host protein NRL1 and enhances the association between NRL1 (NPH3/RPT2‐LIKE1) and SWAP70 to promote degradation of SWAP70 (He et al., 2018). Furthermore, the protein accumulation level of PAD4 was lower in the presence of PcAvh103 in protein extracts as input (Figure 5c). We hypothesized disassembly of the heteromeric interactions of EDS1–PAD4 by PcAvh103 reduced the stability of PAD4, which was also reported in previous study (Feys et al., 2005). Further studies are still needed to explore the interfering mechanisms used by PcAvh103.
It was reported that EDS1 and PAD4 are required for ETI response mediated by TIR‐NB‐LRR proteins, SA accumulation levels and SA‐related resistance genes responsiveness, and they also mediate basal resistance and PTI response (Falk et al., 1999; Feys et al., 2001). In addition, EDS1 and PAD4 are present in the nucleus and cytoplasm, and EDS1 nuclear accumulation precedes EDS1‐dependent transcriptional reprogramming (Garcia et al., 2010). Loss of the association of EDS1–PAD4 compromises basal but not TIR‐NB‐LRR‐triggered resistance (Rietz et al., 2011). In our study, we demonstrated that PcAvh103 can disrupt the EDS1–PAD4 association, probably contributing to suppression of EDS1–PAD4 immune signalling pathway‐mediated defence response. Hence, whether PcAvh103 can manipulate SA accumulation and responsiveness, TIR‐NB‐LRR‐triggered ETI, the nuclear–cytoplasm shuttle of EDS1, even PTI response will be explored in the future.
In summary, we identified a virulence essential effector PcAvh103 from P. capsici, a hemibiotrophic oomycete pathogen. PcAvh103 is required for pathogen virulence and can suppress plant defence by binding to EDS1 and disrupting the EDS1–PAD4 immune complex. This study will advance our understanding of the pathogenic mechanisms of Phytophthora pathogens.
4. EXPERIMENTAL PROCEDURES
4.1. Plant material and growth conditions
N. benthamiana plants were grown in a growth chamber at 25 °C under 16 hr light/ 8 hr dark photoperiod with a relative humidity of 60%–75%. A. thaliana plants were grown in a greenhouse at 23 °C with a photoperiod of 10 hr light/14 hr dark. The T‐DNA insertion mutant, eds1 (SALK_071051) was ordered from the Nottingham Arabidopsis Stock Center (http://arabidopsis.info). The homozygous insertion lines were verified by genomic DNA PCR with primers specific for EDS1 and the T‐DNA left border primer LB1.3 (Table S2).
4.2. Plasmid construction
For yeast two‐hybrid assay, PcAvh103 was PCR‐amplified from P. capsici LT263 and cloned into pGBKT7, and EDS1, BAK1, and BIK1 were amplified from Arabidopsis Col‐0 cDNA and cloned into pGADT7, respectively. For protoplast transfection in Arabidopsis, coding sequences of desired genes were amplified and cloned into the pUC19‐35S‐FLAG/HA‐RBS vector (Li et al., 2005). For transient expression in N. benthamiana, PcAvh103 was PCR‐amplified and inserted into pBinGFP2 vector (Song et al., 2015). For prokaryotic expression of recombinant proteins, EDS1, PAD4, and PcAvh103 were amplified and cloned into pGEX‐6P‐1, pET‐28a and pBAD/gIII, respectively. For transformation of P. capsici, PcAvh103 was cloned into pHam34, which was maintained in our laboratory. Primers used for plasmids construction are listed in Table S2.
4.3. Yeast two‐hybrid assay
Y2H assay was performed with the Matchmaker Gold yeast two‐hybrid system (Clontech), the pGBKT7 vector was used as the bait construct, and pGADT7 as the prey construct. The bait and prey vectors were co‐transformed into the yeast strain AH109 with indicated combinations. Transformants were first selected on double synthetic dropout (DDO) medium lacking leucine and histidine, growing colonies were then plated on quadruple dropout (QDO) (SD/−Leu/−Trp/−His/−Ade) selective medium to test protein interactions. pGBKT7‐53 and pGADT7‐T co‐transformant was used as the positive control, while pGBKT7‐Lam and pGADT7‐T co‐transformant was used as the negative control.
4.4. Co‐immunoprecipitation assay
Arabidopsis mesophyll protoplasts were used for co‐IP assay and 3–4‐week‐old wild‐type A. thaliana (Col‐0) leaves were used for protoplasts isolation. Protoplasts isolation, PEG (polyethylene glycol)‐mediated transfection and protoplasts cultivation were performed as previously described (Li et al., 2016). Protoplasts were transfected with 100 μg desired plasmids and incubated overnight. Total protein was extracted with extraction buffer (50 mM HEPES [N‐2‐hydroxyethylpiperazine‐N‐2‐ethane sulfonic acid]‐KOH [pH 7.5], 150 mM KCl, 1 mM EDTA, 0.3% Triton‐X 100, 1 mM DTT, protease inhibitor cocktail [Roche]). Protein was incubated with agarose‐conjugated anti‐FLAG antibody (Sigma) for 4 hr, washed seven times with washing buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 1 mM EDTA, 0.5% Triton‐X 100, 1 mM DTT) and eluted with 3 × FLAG peptide (Sigma) for 1 hr. Immunoprecipitates were separated by SDS‐PAGE gels and detected by immunoblot using the indicated antibodies.
4.5. Agrobacterium‐mediated transient expression in N. benthamiana
Agrobacterium tumefaciens GV3101 carrying indicated vectors were cultured in Luria Bertani broth with the antibiotics kanamycin and rifampicin. Cells were harvested by centrifugation, washed three times in 10 mM MgCl2 and suspended in infiltration buffer (10 mM MgCl2, 10 mM MES pH 5.6, and 150 μM acetosyringone) to a concentration of OD600 = 0.5 then incubated in an incubator in dark conditions for 3 hr. The suspensions were infiltrated into fully expanded 5–6‐week‐old N. benthamiana leaves using a needleless syringe.
4.6. Phytophthora infection assay
The P. capsici (LT263) strain used in the study was maintained routinely on 10% vegetable (V8) juice medium at 25 °C in the dark. To prepare zoospores of P. capsici, mycelial plugs were cultured in 10% (vol/vol) V8 broth at 25 °C for 3 days and washed three times with sterilized water, then incubated in 25 °C until sporangia formed. To initiate zoospore release, the water in the plates was replaced with fresh water and incubated in 4 °C for 30 min. The zoospore concentration was adjusted by dilution in sterile water and estimated with a haemocytometer. To infect N. benthamiana, 10 μl zoospore suspension (approximately 500 zoospores) was drop inoculated onto the infiltration areas of a detached leaf and incubated in a growth chamber at 25 °C in darkness for 36 hr. To infect Arabidopsis, 5 μl droplets of zoospores (100 zoospores) were inoculated for 36 hr. Relative quantification of P. capsici biomass was performed to evaluate infection severity as described (Wang et al., 2013). For root inoculation, Arabidopsis plants in pots (200 ml) were subjected to soil drench inoculation with 10 ml zoospore suspensions (105 zoospores/ml). Disease development on Arabidopsis plants was evaluated using a disease severity index as described with disease scores ranging between 0 and 4 (Liu et al., 2014). The disease index was calculated according to the formula: disease index = [(Ʃdisease grades × number of infected)/(total checked plants × 4)] × 100.
4.7. Confocal microscopy
A GFP‐fused construct of RxLR103 was transformed into A. tumefaciens GV3101. The transient expression method on N. benthamiana was described above. Images were taken in a confocal laser scanning microscope (LSM 710 META, Zeiss), with an excitation wavelength of 488 nm and a 525 nm bandpass emission filter.
4.8. Transformation of P. capsici
For P. capsici transformation, PEG‐mediated protoplast transformation was performed as described previously (Safdar et al., 2017). Putative transformants were selected on 10% V8 medium containing 30 μg/ml G418. For screening silenced transformants of PcAvh103, total RNA was extracted from mycelia and RT‐qPCR was conducted to characterize the silencing efficiency.
4.9. RNA isolation, cDNA synthesis, and RT‐qPCR
Total RNA was extracted using the RNA‐simple Total RNA Kit (Tiangen) according to the manufacturer's instructions. cDNA was synthesized using Prime Script Reverse Transcriptase (Takara). RT‐qPCR was performed using SYBR Prime‐Script RT‐PCR Kit (TaKaRa) with three technical replicates and implemented on the ABI Prism 7,500 Fast Real‐Time PCR System (Applied Biosystems Inc.). Data were analysed using the 2−ΔΔ C t method.
4.10. Trypan blue staining assay
Inoculated leaves were stained through boiling in lactophenol–trypan blue solution (10 ml lactic acid, 10 ml glycerol, 10 g phenol, 10 mg trypan blue, all dissolved in 10 ml distilled water) for 5 min. They were then destained in chloral hydrate solution (2.5 g/ml) for 12 hr with gentle shaking. Samples were photographed under natural light.
4.11. Prokaryotic expression and pull‐down assay
The recombinant proteins fused with different tags were isolated from Escherichia coli and affinity purified following the manufacturer's instructions. For glutathione S‐transferase (GST) pull‐down assay, 5 μg GST‐EDS1, PAD4‐His, and PcAvh103‐Myc (optional) were incubated at 4 °C with 30 μl glutathione agarose beads (GE Healthcare) in a buffer containing 25 mM Tris–HCl (pH 7.5), 100 mM NaCl, and 1 mM DTT for 2 hr. The bound resins were washed five times with the incubation buffer containing 0.1% Trition‐X 100. The bound proteins were eluted with 15 mM glutathione and detected by immunoblots with indicated antibodies.
CONFLICT OF INTEREST
The authors declare that no competing interests exist.
AUTHOR CONTRIBUTIONS
Q.L., M.Z., and D.D. conceived and designed the research. Q.L., J.W., T.B., M.Z., and Y.J. performed the experiments. Q.L., D.S., M.Z., and D.D. analysed the data. Q.L., J.W., and D.D. wrote the manuscript.
Supporting information
FIGURE S1 Growth of PcAvh103‐silenced transformants is similar to that of WT and control strains. Photographs were taken after 3 days of culture on the 10% (vol/vol) V8 juice medium (left panel). The colony diameters were recorded and calculated (right panel). Error bars represent +SD of at least six plates each
FIGURE S2 PcAvh103 is important for root colonization of Phytophthora capsici. Root inoculation was implemented on Arabidopsis wild‐type Col‐0 with zoospores suspensions of LT263, T48, and T105. Disease symptoms were photographed at 7 days post‐inoculation (left panel) and the disease indices were calculated from three independent biological replicates using at least 15 plants each (right panel). The values are means + SEM (**, p < .01 compared with wild‐type, Dunnett’s test)
TABLE S1 Screening of RxLR effectors in Phytophthora capsici
TABLE S2 Primers used in this study
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (31625023, 31672008, and 31721004) and the Special Fund for Agro‐scientific Research in the Public Interest (201503112). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Li Q, Wang J, Bai T, et al. A Phytophthora capsici effector suppresses plant immunity via interaction with EDS1. Molecular Plant Pathology. 2020;21:502–511. 10.1111/mpp.12912
Funding information
This work was supported by grants from the National Natural Science Foundation of China (31625023, 31672008, and 31721004) and the Special Fund for Agro‐scientific Research in the Public Interest (201503112). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Meixiang Zhang, Email: meixiangzhang@njau.edu.cn.
Daolong Dou, Email: ddou@njau.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Anderson, R.G. , Deb, D. , Fedkenheuer, K. and McDowell, J.M. (2015) Recent progress in RXLR effector research. Molecular Plant‐Microbe Interactions, 28, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Bhandari, D.D. , Lapin, D. , Kracher, B. , von Born, P. , Bautor, J. , Niefind, K. et al (2019) An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis . Nature Communications, 10, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Halane, M.K. , Kim, S.H. and Gassmann, W. (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science, 334, 1405–1408. [DOI] [PubMed] [Google Scholar]
- Birch, P.R. , Rehmany, A.P. , Pritchard, L. , Kamoun, S. and Beynon, J.L. (2006) Trafficking arms: oomycete effectors enter host plant cells. Trends in Microbiology, 14, 8–11. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Chen, J. , Li, M. , Chang, M. , Xu, K. , Shang, Z. et al (2017) A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host & Microbe, 22, e777. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Gobbato, E. , Kracher, B. , Qiu, J. , Bautor, J. and Parker, J.E. (2017) A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. The New Phytologist, 213, 1802–1817. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology, 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H. , Bruce, N.A. , Arredondo, F.D. et al (2008) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. The Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, A. , Feys, B.J. , Frost, L.N. , Jones, J.D. , Daniels, M.J. and Parker, J.E. (1999) EDS1, an essential component of R gene‐mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proceedings of the National Academy of Sciences of the United States of America, 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, G. , Yang, Y. , Li, T. , Lu, W. , Du, Y. , Qiang, X. et al (2018) A Phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum‐mediated immunity. Molecular Plant, 11, 1067–1083. [DOI] [PubMed] [Google Scholar]
- Feys, B.J. , Moisan, L.J. , Newman, M.A. and Parker, J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO Journal, 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J. , Wiermer, M. , Bhat, R.A. , Moisan, L.J. , Medina‐Escobar, N. , Neu, C. et al (2005) Arabidopsis SENESCENCE‐ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. The Plant Cell, 17, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, A.V. , Blanvillain‐Baufume, S. , Huibers, R.P. , Wiermer, M. , Li, G. , Gobbato, E. et al. (2010) Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathogens, 6, e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. et al (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- He, Q. , McLellan, H. , Hughes, R.K. , Boevink, P.C. , Armstrong, M. , Lu, Y. et al (2019) Phytophthora infestans effector SFI3 targets potato UBK to suppress early immune transcriptional responses. The New Phytologist, 222, 438–454. [DOI] [PubMed] [Google Scholar]
- He, Q. , Naqvi, S. , McLellan, H. , Boevink, P.C. , Champouret, N. , Hein, I. et al (2018) Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proceedings of the National Academy of Sciences of the United States of America, 115, E7834–E7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proceedings of the National Academy of Sciences of the United States of America, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H. and Tyler, B.M. (2012) Mechanisms and evolution of virulence in oomycetes. Annual Review of Phytopathology, 50, 295–318. [DOI] [PubMed] [Google Scholar]
- Jing, M. , Guo, B. , Li, H. , Yang, B. , Wang, H. , Kong, G. et al (2016) A Phytophthora sojae effector suppresses endoplasmic reticulum stress‐mediated immunity by stabilizing plant binding immunoglobulin proteins. Nature Communications, 7, 11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review of Phytopathology, 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Furzer, O. , Jones, J.D. , Judelson, H.S. , Ali, G.S. , Dalio, R.J. et al (2015) The top 10 oomycete pathogens in molecular plant pathology. Molecular Plant Pathology, 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L. , Qiu, X. , Kang, J. , Wang, Y. , Chen, H. , Huang, J. et al (2017) A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Current Biology, 27, 981–991. [DOI] [PubMed] [Google Scholar]
- Lamour, K.H. , Mudge, J. , Gobena, D. , Hurtado‐Gonzales, O.P. , Schmutz, J. , Kuo, A. et al (2012a) Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici . Molecular Plant‐Microbe Interactions, 25, 1350–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour, K.H. , Stam, R. , Jupe, J. and Huitema, E. (2012b) The oomycete broad‐host‐range pathogen Phytophthora capsici . Molecular Plant Pathology, 13, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Wang, H. , Jing, M. , Zhu, J. , Guo, B. , Wang, Y. et al (2018) A Phytophthora effector recruits a host cytoplasmic transacetylase into nuclear speckles to enhance plant susceptibility. Elife, 7, e40039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Ai, G. , Shen, D. , Zou, F. , Wang, J. , Bai, T. et al (2019a) A Phytophthora capsici effector targets ACD11 binding partners that regulate ROS‐mediated defense response in Arabidopsis . Molecular Plant, 12, 565–581. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Chen, Y. , Wang, J. , Zou, F. , Jia, Y. , Shen, D. et al (2019b) A Phytophthora capsici virulence effector associates with NPR1 and suppresses plant immune responses. Phytopathology Research, 1, 6. [Google Scholar]
- Li, Q. , Zhang, M. , Shen, D. , Liu, T. , Chen, Y. , Zhou, J.M. et al (2016) A Phytophthora sojae effector PsCRN63 forms homo‐/hetero‐dimers to suppress plant immunity via an inverted association manner. Scientific Reports, 6, 26951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Lin, H. , Zhang, W. , Zou, Y. , Zhang, J. , Tang, X. et al (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proceedings of the National Academy of Sciences of the United States of America, 102, 12990–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W. , Li, B. , Lu, D. , Chen, S. , Zhu, N. , He, P. et al (2014) Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proceedings of the National Academy of Sciences of the United States of America, 111, 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Song, T. , Zhang, X. , Yuan, H. , Su, L. , Li, W. et al (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nature Communications, 5, 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y.L. , Shi, J.X. , Zhai, Y. , Hou, Y.N. and Ma, W.B. (2015) Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proceedings of the National Academy of Sciences of the United States of America, 112, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, J. , Zhou, X.G. , Sun, L.F. , Wang, K.L. , Yang, F. , Liao, H.C. et al (2018) The Xanthomonas effector XopK harbours E3 ubiquitin‐ligase activity that is required for virulence. New Phytologist, 220, 219–231. [DOI] [PubMed] [Google Scholar]
- Rietz, S. , Stamm, A. , Malonek, S. , Wagner, S. , Becker, D. , Medina‐Escobar, N. et al (2011) Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. The New Phytologist, 191, 107–119. [DOI] [PubMed] [Google Scholar]
- Safdar, A. , Li, Q. , Shen, D. , Chen, L. , He, F. , Wang, R. et al (2017) An LRR receptor kinase regulates growth, development and pathogenesis in Phytophthora capsici . Microbiological Research, 198, 8–15. [DOI] [PubMed] [Google Scholar]
- Segonzac, C. and Zipfel, C. (2011) Activation of plant pattern‐recognition receptors by bacteria. Current Opinion in Microbiology, 14, 54–61. [DOI] [PubMed] [Google Scholar]
- Song, T. , Ma, Z. , Shen, D. , Li, Q. , Li, W. , Su, L. et al (2015) An oomycete CRN effector reprograms expression of plant HSP genes by targeting their promoters. PLoS Pathogens, 11, e1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, B.M. (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Molecular Plant Pathology, 8, 1–8. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H. , Aerts, A. et al (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Ustun, S. , Bartetzko, V. and Bornke, F. (2013) The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic‐acid mediated plant defence. PLoS Pathogens, 9, e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, S. , Stuttmann, J. , Rietz, S. , Guerois, R. , Brunstein, E. , Bautor, J. et al (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host & Microbe, 14, 619–630. [DOI] [PubMed] [Google Scholar]
- Wang, J.L. , Shine, M.B. , Gao, Q.M. , Navarre, D. , Jiang, W. , Liu, C.Y. et al (2014) Enhanced disease susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiology, 165, 1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Bouwmeester, K. , van de Mortel, J.E. , Shan, W. and Govers, F. (2013) A novel Arabidopsis‐oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant, Cell & Environment, 36, 1192–1203. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. et al (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Wang, Y. , Guo, B. , Jing, M. , Zhou, H. , Li, Y. et al (2019) The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. The New Phytologist, 222, 425–437. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. et al (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host & Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Growth of PcAvh103‐silenced transformants is similar to that of WT and control strains. Photographs were taken after 3 days of culture on the 10% (vol/vol) V8 juice medium (left panel). The colony diameters were recorded and calculated (right panel). Error bars represent +SD of at least six plates each
FIGURE S2 PcAvh103 is important for root colonization of Phytophthora capsici. Root inoculation was implemented on Arabidopsis wild‐type Col‐0 with zoospores suspensions of LT263, T48, and T105. Disease symptoms were photographed at 7 days post‐inoculation (left panel) and the disease indices were calculated from three independent biological replicates using at least 15 plants each (right panel). The values are means + SEM (**, p < .01 compared with wild‐type, Dunnett’s test)
TABLE S1 Screening of RxLR effectors in Phytophthora capsici
TABLE S2 Primers used in this study
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


