Abstract
Begomoviruses of the Geminiviridae are usually transmitted by whiteflies and rarely by mechanical inoculation. We used tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus, to address this issue. Most ToLCNDV isolates are not mechanically transmissible to their natural hosts. The ToLCNDV‐OM isolate, originally identified from a diseased oriental melon plant, is mechanically transmissible, while the ToLCNDV‐CB isolate, from a diseased cucumber plant, is not. Genetic swapping and pathological tests were performed to identify the molecular determinants involved in mechanical transmission. Various viral infectious clones were constructed and successfully introduced into Nicotiana benthamiana, oriental melon, and cucumber plants by Agrobacterium‐mediated inoculation. Mechanical transmissibility was assessed via direct rub inoculation with sap prepared from infected N. benthamiana. The presence or absence of viral DNA in plants was validated by PCR, Southern blotting, and in situ hybridization. The results reveal that mechanical transmissibility is associated with the movement protein (MP) of viral DNA‐B in ToLCNDV‐OM. However, the nuclear shuttle protein of DNA‐B plays no role in mechanical transmission. Analyses of infectious clones carrying a single amino acid substitution reveal that the glutamate at amino acid position 19 of MP in ToLCNDV‐OM is critical for mechanical transmissibility. The substitution of glutamate with glycine at this position in the MP of ToLCNDV‐OM abolishes mechanical transmissibility. In contrast, the substitution of glycine with glutamate at the 19th amino acid position in the MP of ToLCNDV‐CB enables mechanical transmission. This is the first time that a specific geminiviral movement protein has been identified as a determinant of mechanical transmissibility.
Keywords: begomovirus, mechanical transmissibility, movement protein, ToLCNDV oriental melon isolate, tomato leaf curl New Delhi virus
Begomoviruses are usually transmitted by whiteflies and rarely by mechanical inoculation. The movement protein of tomato leaf curl New Delhi virus is identified as a determinant of mechanical transmissibility.

1. INTRODUCTION
Begomoviruses cause epidemics in many economically important crops worldwide, including cassava, cucurbits, eggplant, legumes, okra, pepper, potato, and tomato (Green et al., 2003; Varma and Malathi, 2003; Shih et al., 2007). The infection of cassava by African cassava mosaic virus (ACMV) causes annual economic losses of $1.9–2.7 billion in Africa (Legg and Fauquet, 2004). The infection of sugar beet by bean golden mosaic virus (BGMV) and tomato by tomato yellow leaf curl virus (TYLCV) also leads to a severe reduction in crop yields (Varma and Malathi, 2003).
One of the most important factors contributing to viral epidemics is the mode of transmission. Comprehensive knowledge of viral transmission mechanisms and their potential modifications during viral evolution may lead to the development of more effective preventive measures. Begomoviruses are usually transmitted by whiteflies (Bemisia tabaci). Similar to many phloem‐limited viruses, including other geminiviruses and closteroviruses, begomoviruses are rarely mechanically transmissible by rub or sap inoculation (Garnsey et al., 1977; Wege and Pohl, 2007). Thus far, only approximately 20 out of 409 known begomovirus species have been reported to be mechanically transmissible to their natural hosts (Bock and Guthrie, 1978; Morales et al., 1990; Gilbertson et al., 1991; Garrido‐Ramirez et al., 2000; Chatchawankanphanich and Maxwell, 2002; Usharani et al., 2004; Ajlan et al., 2007; Tsai et al., 2011).
The mechanisms underlying mechanical or nonmechanical transmissibility among related viruses of the same genus are of significant interest from the point of view of evolution and epidemics. The identification of the viral determinants associated with mechanical transmissibility is a critical step towards mechanistic studies. Begomoviruses have a single‐stranded circular monopartite (DNA‐A like) or bipartite (DNA‐A and DNA‐B) genome (Fauquet et al., 2008). The DNA‐A genome encodes proteins required for virus replication and encapsidation, whereas DNA‐B encodes proteins responsible for viral movement between subcellular and intercellular compartments as well as pathogenicity (Krenz et al., 2012). Although the DNA‐B genomes of some begomoviruses have been implicated in mechanical transmission (Levy and Czosenk, 2003; Wege and Pohl, 2007), the specific viral proteins involved in this aspect remain largely unknown.
Tomato leaf curl New Delhi virus (ToLCNDV) is a begomovirus with a bipartite genome. This virus could cause severe damage to many economically important crops in the families of Solanaceae and Cucurbitaceae, including tomato, pepper, potato, cucumber, melon, sponge gourd, and pumpkin (Padidam et al., 1995; Samretwanich et al., 2000; Usharani et al., 2004; Hussain et al., 2005; Ito et al., 2008; Khan et al., 2012; Lopez et al., 2015). ToLCNDV DNA‐A has two open reading frames (ORFs; AV1 and AV2) on the viral‐sense strand and four ORFs (AC1 to AC4) on the antisense strand. AV1 encodes a coat protein (CP) and AV2 encodes a pre‐coat protein. AC1, AC2, and AC3 encode a replication‐associated protein (Rep), a transcriptional activator protein (TrAP), and a replication enhancer (REn), respectively. AC4 encodes a protein required for symptom development (Fondong, 2013). DNA‐B has two ORFs: BV1 of the viral‐sense strand encodes a nuclear shuttle protein (NSP) and BC1 of the antisense strand encodes a cell‐to‐cell movement protein (MP) (Jeske, 2009).
ToLCNDV is transmitted by whiteflies in nature. However, greenhouse tests have revealed that some ToLCNDV isolates can be mechanically transmitted by rub inoculation while others cannot. The ToLCNDV‐OM isolate, originally identified from a diseased oriental melon plant, can infect oriental melon, pickling melon, bottle gourd, cucumber, zucchini, and luffa via mechanical sap inoculation (Chang et al., 2010). The ToLCNDV‐potato isolate can also be transmitted to host plants via mechanical inoculation (Usharani et al., 2004). In contrast, the ToLCNDV‐severe and ToLCNDV‐cucumber isolates, which share a very similar genome organization and size with oriental melon and potato isolates, cannot be mechanically transmitted to their hosts (Padidam et al., 1995; Samretwanich et al., 2000). The divergent modes of mechanical transmissibility among ToLCNDV isolates may be due to the high genomic recombination and mutation rates of geminiviruses, which could lead to the emergence of a new isolate with different transmission abilities or host ranges (Chatchawankanphanich and Maxwell, 2002). New viral variants are often more pathogenic or can exhibit a wider host range than previously existing variants (Arguello‐Astorga et al., 2007; Duffy and Holmes, 2008). For example, tomato yellow leaf curl Thailand virus (TYLCTHV) is mechanically transmissible and has spread widely to become a predominant strain in Taiwan (Tsai et al., 2011). In contrast, an endogenous tomato leaf curl Taiwan virus (ToLCTWV), which is not mechanically transmissible, has become less common in the region.
The mechanisms of the mechanical transmission of begomoviruses have not received much attention, because many of them are transmitted by whiteflies. The new ToLCNDV‐CB isolate, recently identified from a cucumber plant with symptoms, shares high genome sequence similarity with the ToLCNDV‐OM isolate. Unlike ToLCNDV‐OM, the ToLCNDV‐CB isolate cannot be transmitted to host plants by sap inoculation. In this study, genetic and pathological approaches were employed to identify the molecular determinants responsible for the mechanical transmission of ToLCNDV‐OM. Gene swapping and point mutations revealed that the 19th amino acid residue at the N‐terminus of the MP of DNA‐B plays a critical role in mechanical transmission. The results also showed that the NSP‐coding region plays no role in mechanical transmission.
2. RESULTS
2.1. ToLCNDV isolates share high sequence similarity
The ToLCNDV‐OM isolate, originally obtained from a diseased oriental melon, could be mechanically transmitted to its host plants. In contrast, the closely related ToLCNDV‐CB isolate, originally collected from cucumber with symptoms, failed to infect Nicotiana benthamiana, oriental melon, or cucumber plants by mechanical sap inoculation. Similar to ToLCNDV‐OM, the ToLCNDV‐CB isolate was found to have a bipartite genome consisting of DNA‐A (2,738 nt) and DNA‐B (2,695 nt). Pairwise sequence comparisons of the ToLCNDV‐CB genome with the ToLCNDV‐OM, ToLCNDV‐severe (nonmechanically transmissible), and ToLCNDV‐potato (mechanically transmissible) genome sequences available in GenBank were conducted to determine their genetic relationships. The whole‐DNA‐A‐genome sequences of the ToLCNDV isolates shared from 92.8% to 96.5% nucleotide identity, and those of DNA‐B shared from 84.4% to 92.3% nucleotide identity. The amino acid sequences encoded by DNA‐A or DNA‐B were highly similar, with the lowest similarity found among AC4 (88.1%–91.5%) of DNA‐A and BC1 (88.7%–97.2%) of DNA‐B (Table 1). ToLCNDV‐OM and ToLCNDV‐CB showed 96.5% identity of their DNA‐A sequences and 92.3% of their DNA‐B genomes. In addition to their high sequence similarity, the two isolates were found to induce similar symptoms and present very similar host ranges (Figure S1).
Table 1.
Sequence similarity of full‐length DNA‐A and DNA‐B and individual genes of the tomato leaf curl New Delhi virus (ToLCNDV)‐OM with those of other ToLCNDV isolates
| Isolate | Sequence identity and similarity (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA‐A | AV1 | AV2 | AC1 | AC2 | AC3 | AC4 | DNA‐B | BV1 | BC1 | |
| CB | 96.5a | 99.0 (99.6)b | 97.6 (99.1) | 95.1 (96.1) | 98.3 (96.3) | 97.3 (97.8) | 97.2 (91.5) | 92.3 | 94.3 (95.2) | 95.4 (97.2) |
| Severe | 93.4 | 95.2 (97.7) | 94.4 (95.6) | 93.2 (93.9) | 95.6 (92.6) | 95.9 (97.1) | 94.9 (88.1) | 85.1 | 87.5 (89.6) | 89.7 (88.7) |
| Potato | 92.8 | 94.0 (95.7) | 94.4 (93.8) | 93.2 (94.8) | 95.3 (90.4) | 93.4 (91.2) | 95.5 (89.8) | 84.4 | 87.7 (90.0) | 90.2 (94.0) |
The sequences used in this study can be retrieved from the GenBank database: ToLCNDV‐OM isolate (DNA‐A, accession number GU180095; DNA‐B, accession number MK883714); ToLCNDV‐CB isolate (DNA‐A, accession number MK883715; DNA‐B, accession number MK883716); ToLCNDV‐severe isolate (DNA‐A, accession number U15015; DNA‐B, accession number U15017), and ToLCNDV‐potato isolate (DNA‐A, accession number AY286316; DNA‐B, accession number AY158080).
The identity of nucleotide sequences.
The similarity of amino acids.
2.2. The DNA‐B genome of ToLCNDV‐OM is required for mechanical transmissibility
Because ToLCNDV‐CB was unable to infect oriental melon and N. benthamiana by mechanical sap inoculation, infectious clones of ToLCNDV‐CB DNA‐A and DNA‐B amplified via the rolling circle amplification (RCA) method were independently constructed in the binary vector pCAMBIA1304 (Figure 1) for further tests. Each clone contained two copies of the respective DNA sequences, and they were thus designated pCB2A (DNA‐A) and pCB2B (DNA‐B). The pCB2A and pCB2B infectious clones were successfully introduced into N. benthamiana, oriental melon, and cucumber by agroinoculation, resulting in visible symptoms (Table 2). Viral symptoms including curling, puckering, and mosaic appeared on N. benthamiana leaves 5–7 days post‐inoculation (dpi) and on oriental melon and cucumber leaves 7–10 dpi. However, sap prepared from diseased N. benthamiana leaves inoculated with pCB2A and pCB2B failed to induce symptoms in any of the test plants after rub inoculation. The ToLCNDV‐OM infectious clones pOM2A and pOM2B, constructed in our previous study (Chang et al., 2010), were also introduced into N. benthamiana, oriental melon, and cucumber by agroinoculation and induced symptoms at similar rates and with similar magnitudes to those induced by the ToLCNDV‐CB clones. Sap prepared from the ToLCNDV‐O‐infected N. benthamiana leaves also resulted in symptoms on the test plants following rub inoculation at incidence rates ranging from 40% to 85% (Table 2). Viral DNA‐A and DNA‐B in the plant leaves with symptoms were monitored by PCR amplification using genome‐specific primers, resulting in products with similar intensities.
Figure 1.

The DNA‐B genome of the tomato leaf curl New Delhi virus (ToLCNDV)‐OM isolate is required for mechanical transmissibility. (a) Physical maps of DNA infectious clones (pCB2A, pCB2B, pOM2A, and pOM2B) carrying the duplicated DNA‐A or DNA‐B genome of the ToLCNDV‐CB or ToLCNDV‐OM isolate. Genes encoded by DNA‐A or DNA‐B and the location of the intergenic region (IR) sequence are also indicated. The arrow indicates the direction of the transcription of each gene. (b) Images of Nicotiana benthamiana (10 days post‐inoculation, dpi), oriental melon, and cucumber plants (12 dpi) after mechanical inoculation with the wild‐type virus (pCB2A + pCB2B and pOM2A + pOM2B) or pseudorecombinant viruses (pCB2A + pOM2B and pOM2A + pCB2B) developing viral symptoms (O) or exhibiting no symptoms (X). The viral inoculum used for mechanical sap inoculation was prepared from diseased N. benthamiana after agroinoculation with an appropriate combination of the clones. (c) PCR detection of virus accumulation in the leaves after mechanical inoculation with pCB2A + pCB2B (lanes 1, 5, and 9), pOM2A + pOM2B (lanes 2, 6, and 10), pCB2A + pOM2B (lanes 3, 7, and 11), and pOM2A + pCB2B (lanes 4, 8, and 12). The specific DNA fragments amplified by PCR are indicated on the left. The pseudoinfectious clones leading to successful mechanical sap inoculation are indicated by asterisks
Table 2.
Summary of infection results for the tomato leaf curl New Delhi virus (ToLCNDV) isolates, pseudorecombinants, and mutants
| DNA‐Aa | DNA‐B | Agroinoculation | Mechanical inoculationc | ||||
|---|---|---|---|---|---|---|---|
| Nicotiana benthamiana | Oriental melon | Cucumber | Nicotiana benthamiana | Oriental melon | Cucumber | ||
| pCB2A | pCB2B | 10/10 | 11/12 (91.7%) | 11/12 (91.7%) | 0/26 | 0/25 | 0/24 |
| pOM2A | pOM2B | 10/10 | 11/12 (91.7%) | 10/12 (83.3%) | 22/26 (84.6%) | 10/25 (40.0%) | 11/25 (44.0%) |
| pCB2A | pOM2B | 10/10 | 11/12 (91.7%) | 12/12 | 19/26 (73.1%) | 11/26 (42.3%) | 10/27 (37.0%) |
| pOM2A | pCB2B | 9/10 | 10/12 (83.3%) | 11/12 (91.7%) | 0/26 | 0/25 | 0/25 |
| pOM2A | pOM1B::NSP ins39 | 5/5 | 2/5 (40.0%) | ‐ | 6/6 | 5/8 (62.5%) | ‐ |
| pCB2A | pCB1B | 10/10 | 15/17 (88.2%) | 10/11 (90.9%) | 0/26 | 0/30 | 0/28 |
| pOM2A | pOM1B | 10/10 | 11/13 (84.6%) | 13/13 | 21/26 (80.8%) | 17/29 (58.6%) | 14/27 (51.9%) |
| pCB2A | pCB1B::OMNSP | 9/10 (90.0%)b | 11/13 (84.6%) | 10/12 (83.3%) | 0/26 | 0/29 | 0/27 |
| pOM2A | pOM1B::CBNSP | 10/10 | 12/13 (92.3%) | 12/13 (92.3%) | 22/26 (84.6%) | 16/29 (55.2%) | 12/27 (44.4%) |
| pCB2A | pCB1B::OMMP | 12/12 | 17/17 | 10/11 (90.9%) | 23/26 (88.5%) | 14/30 (46.7%) | 12/30 (40.0%) |
| pOM2A | pOM1B::CBMP | 10/10 | 12/13 (92.3%) | 13/13 | 0/26 | 0/29 | 0/27 |
| pCB2A | pCB1B::OM5′ MP | 8/8 | –d | – | 7/8 (87.5%) | 4/8 (50.0%) | 3/8 (37.5%) |
| pCB2A | pCB1B::OM3′ MP | 8/8 | – | – | 0/8 | 0/8 | 0/8 |
| pOM2A | pOM1B::CB5′ MP | 8/8 | – | – | 0/8 | 0/8 | 0/8 |
| pOM2A | pOM1B::CB3′ MP | 8/8 | – | – | 6/8 (75.0%) | 3/8 (37.5%) | 4/8 (50.0%) |
| pCB2A | pCB1BMP (3I→T) | 8/8 | – | – | 0/8 | 0/9 | 0/5 |
| pCB2A | pCB1BMP (6D→E) | 8/8 | – | – | 0/8 | 0/12 | 0/12 |
| pCB2A | pCB1BMP (8V→M) | 8/8 | – | – | 0/8 | 0/9 | 0/5 |
| pCB2A | pCB1BMP (19G→E) | 8/8 | – | – | 7/8 (87.5%) | 6/9 (66.7%) | 1/5 (20.0%) |
| pOM2A | pOM1BMP (3T→I) | 8/8 | – | – | 8/8 | 7/9 (77.8%) | 1/5 (20.0%) |
| pOM2A | pOM1BMP (6E→D) | 8/8 | – | – | 7/8 (87.5%) | 3/12 (25.0%) | 9/12 (75.0%) |
| pOM2A | pOM1BMP (8M→V) | 8/8 | – | – | 7/8 (87.5%) | 6/9 (66.7%) | 1/5 (20.0%) |
| pOM2A | pOM1BMP (19E→G) | 8/8 | – | – | 0/8 | 0/9 | 0/5 |
Abbreviations for amino acids: G, glycine; E, glutamate; I, isoleucine; T, threonine; D, aspartic acid; V, valine; M, methionine.
Detailed construction maps of the infectious DNA‐A and ‐B are illustrated in Figures 1a, 3a, 4a and 5.
Disease incidence (%) is indicated by number of infected plants/total number of plants inoculated.
The sap was prepared from the agroinoculated N. benthamiana and used as inoculum for mechanical inoculation.
The symbol “–“ indicates that infectivity was not determined.
To determine whether DNA‐A or DNA‐B plays a role in mechanical transmission, a combination of DNA‐A and DNA‐B from ToLCNDV‐CB and ToLCNDV‐OM was tested for infectivity via mechanical sap inoculation. When the test plants were infected with the pCB2A + pOM2B or pOM2A + pCB2B pseudorecombinants by agroinoculation, most developed symptoms at rates ranging from 83.3% to 100% (Table 2). When sap extracted from diseased N. benthamiana leaves infected with pCB2A + pOM2B was rub inoculated onto the test plants, many of them also developed symptoms at rates (37% to 73%) comparable to those in plants inoculated with pOM2A + pOM2B. In contrast, sap extracted from N. benthamiana infected with pOM2A + pCB2B failed to induce symptoms on the test plants after rub inoculation (Figure 1b), indicating a role of DNA‐B from the ToLCNDV‐OM isolate in mechanical transmissibility. The presence of both DNA‐A and DNA‐B in sap‐inoculated plants was further confirmed by PCR using genome‐specific primers (Figure 1c).
2.3. The NSP gene of ToLCNDV‐OM plays no role in mechanical transmissibility
Sequence alignment has shown that the NSP genes of ToLCNDV‐OM and ToLCNDV‐potato (both mechanically transmissible isolates) are 39 nt shorter than that of the ToLCNDV‐severe isolate, which is not mechanically transmissible (Chang et al., 2010). To determine whether the addition of 39 nt at the 5′ end of the NSP gene of ToLCNDV‐severe plays a negative role in mechanical transmission, the pOM1B::NSP ins39 clone (Figure 2a), containing an extra 39‐nt fragment from the NSP gene of the ToLCNDV‐severe isolate, was constructed in the DNA‐B genome of ToLCNDV‐OM. The agroinoculation of test plants with pOM2A + pOM1B::NSP ins39 resulted in a high level of infectivity in N. benthamiana and oriental melon (Table 2). Sap prepared from N. benthamiana leaves with symptoms infected with pOM2A + pOM1B::NSP ins39 produced visible symptoms on two plant species following rub inoculation (Figure 2b), indicating that the addition of the 39 nt had no negative effect on mechanical transmissibility. The presence of ToLCNDV‐OM DNA‐A and DNA‐B in the mechanically infected plants was also confirmed by PCR using DNA‐specific primers. The DNA‐B primers amplified the expected 772‐bp fragment from pOM2B and 811‐bp fragment from pOM1B::NSP ins39 (Figure 2c).
Figure 2.

The addition of 39 nucleotides of the NSP gene of the tomato leaf curl New Delhi virus (ToLCNDV)‐OM has no effect on mechanical transmissibility. (a) Physical map of the pOM1B::NSP ins39 infectious clone constructed in the ToLCNDV‐OM DNA‐B genome (pOM1B) by adding an extra 39‐nt fragment (indicated by a pink box) from the NSP gene of the ToLCNDV‐severe isolate by PCR using two overlapping primers (FJJ2009‐26 and FJJ2009‐27). A recognition site for the NcoI endonuclease was incorporated into FJJ2009‐26 to facilitate cloning. Other primers used for cloning and the location of the intergenic region (IR) are also indicated. (b) Symptoms of Nicotiana benthamiana and oriental melon after sap inoculation with wild‐type ToLCNDV‐OM (pOM2A + pOM2B) and the NSP mutant (pOM2A + pOM1B::NSP ins39) at 10 days post‐inoculation. The viral inoculum used for mechanical sap inoculation was prepared from N. benthamiana with symptoms after agroinoculation with an appropriate combination of the clones. (c) Detection of viral DNA using PCR. Specific primers (Table S1) were used to amplify DNA‐A (upper panel) and DNA‐B (lower panel). An 772‐bp fragment representing the DNA‐B genome of the wild‐type virus was amplified from plants mechanically inoculated with pOM2A + pOM2B (lanes 1 and 3). A 811‐bp fragment representing the DNA‐B genome of pOM1B::NSP ins39 was amplified from plants inoculated with pOM2A + pOM1B::NSP ins39 (lanes 2 and 4)
To determine the roles of NSP and MP in mechanical transmission, two infectious clones, pCB1B and pOM1B, containing a single copy of the DNA‐B genomes of the respective ToLCNDV isolates and 832‐bp intergenic region (IR) tandem repeats, were generated (Figure 3a). As controls, the three test species were agroinoculated with pCB2A + pCB1B or pOM2A + pOM1B, which resulted in a high disease incidence. Sap extracted from N. benthamiana infected with pOM2A + pOM1B, but not with pCB2A + pCB1B, also induced symptoms on the test plants (Table 2 and Figure 3b).
Figure 3.
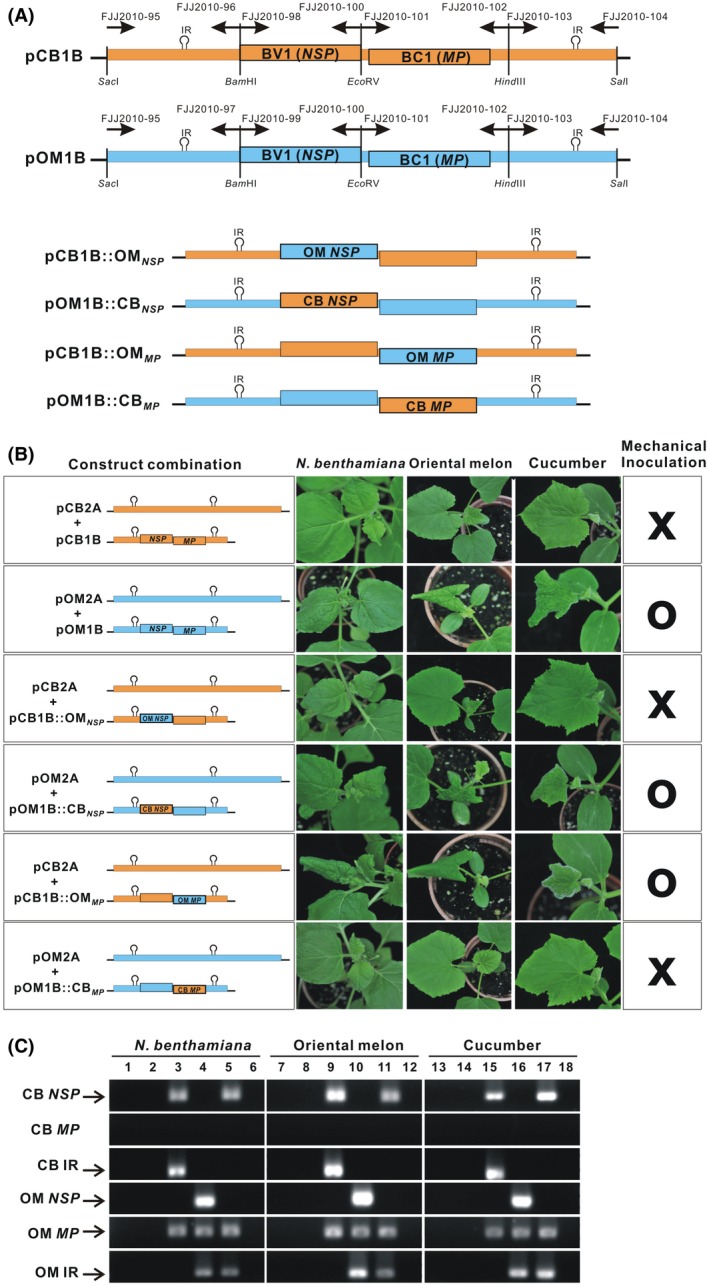
The tomato leaf curl New Delhi virus (ToLCNDV)‐OM movement protein (MP), but not nuclear shuttle protein (NSP), is required for mechanical transmission. (a) Schematic illustration of the pCB1B and pOM1B infectious clones carrying a single copy of the DNA‐B genome of the CB or OM isolate, respectively. Four additional constructs, pCB1B::OMNSP, pOM1B::CBNSP, pCB1B::OMMP, and pOM1B::CBMP, were produced by swapping the BV1 (encoding NSP) or BC1 (encoding MP) gene between two ToLCNDV isolates. The primers used for construction are indicated above each genome. (b) Nicotiana benthamiana, oriental melon, and cucumber plants after mechanical inoculation with pCB2A + pCB1B, pOM2A + pOM1B, or other infectious clones, as indicated, developed viral symptoms (O) or exhibited no symptoms (X) at 10–12 days post‐inoculation. The viral inoculum used for mechanical sap inoculation was prepared from N. benthamiana with symptoms after agroinoculation with an appropriate combination of the clones. Symptoms were observed exclusively in plants inoculated with the constructs carrying the ToLCNDV‐OM MP gene. (c) PCR detection of viral DNA in plants after mechanical inoculation with pCB2A + pCB1B (lanes 1, 7, and 13), pCB2A + pCB1B::OMNSP (lanes 2, 8, and 14), pCB2A + pCB1B::OMMP (lanes 3, 9, and 15), pOM2A + pOM1B (lanes 4, 10, and 16), pOM2A + pOM1B::CBNSP (lanes 5, 11, and 17), or pOM2A + pOM1B::CBMP (lanes 6, 12, and 18). Specific primers were used to amplify the NSP, MP, or intergenic region (IR) fragment from the OM or CB isolate. Amplicons were obtained only from plants inoculated with the constructs carrying the ToLCNDV‐OM MP gene
To confirm the role of NSP in mechanical transmission, two infectious clones, pCB1B::OMNSP and pOM1B::CBNSP, were generated by exchanging the NSP‐coding genes of two ToLCNDV isolates and tested for infectivity. The agroinoculation of the test plants with pCB2A + pCB1B::OMNSP or pOM2A + pOM1B::CBNSP resulted in symptoms on all three plant species (Table 2). Sap extracted from N. benthamiana plants infected with pOM2A + pOM1B::CBNSP mechanically infected all three plant species, and viral DNA was detectable by PCR using gene‐specific primers (Figure 3b,c). However, sap prepared from N. benthamiana leaves infected with pCB2A + pCB1B::OMNSP failed to induce symptoms, and viral DNA was not detected in the test plants after rub inoculation. Thus, a role of NSP in mechanical transmissibility was ruled out in this study.
2.4. The ToLCNDV‐OM MP is required for mechanical transmission
To determine the role of the ToLCNDV‐OM movement protein in mechanical transmission, two infectious clones were generated. The pCB1B::OMMP clone was generated by replacing ToLCNDV‐CB MP with ToLCNDV‐OM MP. The other clone, designated pOM1B::CBMP, was generated by replacing ToLCNDV‐OM MP with ToLCNDV‐CB MP. Agroinoculation of three plant species with pCB2A + pCB1B::OMMP or pOM2A + pOM1B::CBMP resulted in symptoms at 7 dpi. Sap extracted from N. benthamiana plants infected with pCB2A + pCB1B::OMMP mechanically infected the test plants (Figure 3b) and viral DNA propagated, as demonstrated by PCR using gene‐specific primers for NSP, MP or IR (Figure 3c). However, sap prepared from N. benthamiana leaves infected with pOM2A + pOM1B::CBMP failed to induce symptoms, and the virus was not detected after rub inoculation, indicating the involvement of the ToLCNDV‐OM MP gene in mechanical transmission.
2.5. The 5′ end of the MP of ToLCNDV‐OM is required for mechanical transmissibility
The comparison of MP amino acid sequences between ToLCNDV‐CB and ToLCNDV‐OM (Figure S2) revealed that the two MPs were highly similar, differing only by four amino acid residues in the N‐terminal region and another four in the C‐terminal region (Figure 4a). To identify the critical amino acid residue(s) involved in mechanical transmissibility, chimeric MP genes were constructed through the combination of portions of two viral MP‐encoding genes and tested for infectivity (Figure 4a). The pCB1B::OM5′ MP clone was constructed in the DNA‐B backbone of ToLCNDV‐CB by substituting the 5′ fragment of the MP gene with its ToLCNDV‐OM counterpart. pCB1B::OM3′ MP was generated by replacing the 3′ end of ToLCNDV‐CB MP with its ToLCNDV‐OM counterpart. Similar approaches were used to construct pOM1B::CB5′ MP and pOM1B::CB3′ MP. Combined with the corresponding DNA‐A infectious clone, all four constructs induced symptoms in N. benthamiana after agroinoculation (Table 2).
Figure 4.

The 5′‐end of the movement protein (MP) of the tomato leaf curl New Delhi virus (ToLCNDV)‐OM is required for mechanical transmissibility. (a) Schematic illustration of the pCB1B and pOM1B infectious clones showing eight different amino acid residues in MP between the ToLCNDV‐OM and CB isolates. Four different amino acids (3rd, 6th, 8th, and 19th) were found at the N terminus, and four other amino acids (193rd, 225th, 233rd, and 262nd) were found at the C terminus of MPs. Two clusters of different amino acids were separated via the PstI restriction enzyme recognition site. Four recombinant constructs, pCB1B::OM5′ MP, pCB1B::OM3′ MP, pOM1B::CB5′ MP, and pOM1B::CB3′ MP, were produced by exchanging the 5′MP‐intergenic region (IR) fragment (455 bp) and the 3′‐end MP fragment (817 bp) between two isolates using EcoRV, PstI, and/or HindIII cleavage. (b) Nicotiana benthamiana, oriental melon, and cucumber plants after mechanical inoculation with the infectious clones, either developed viral symptoms (O) or exhibited no symptoms (X) at 10–12 days post‐inoculation. The viral inoculum used for mechanical sap inoculation was prepared from N. benthamiana with symptoms after agroinoculation with an appropriate combination of the clones. Symptoms were observed only in plants inoculated with the constructs carrying the 5′ region of the ToLCNDV‐OM MP gene. (c) PCR detection of the NSP‐MP DNA fragment in plants after mechanical inoculation with pCB2A + pCB1B (lanes 1, 7, and 13), pCB2A + pCB1B::OM5′ MP (lanes 2, 8, and 14), pCB2A + pCB1B::OM3′ MP (lanes 3, 9, and 15), pOM2A + pOM1B (lanes 4, 10, and 16), pOM2A + pOM1B::CB5′ MP (lanes 5, 11, and 17), and pOM2A + pOM1B::CB3′ MP (lanes 6, 12, and 18). Amplicons were obtained only from plants inoculated with the constructs carrying the 5′ end of the MP of ToLCNDV‐OM
Mechanical transmission assays using sap extracted from N. benthamiana plants infected with pCB2A + pCB1B::OM5′ MP or pOM2A + pOM1B::CB3′ MP resulted in symptoms on N. benthamiana, oriental melon, and cucumber (Figure 4b). In contrast, sap from N. benthamiana infected with pOM2A + pOM1B::CB5′ MP or pCB2A + pCB1B::OM3′ MP failed to induce symptoms on the test plants after rub inoculation, indicating an important role of the 5′ end of the MP of ToLCNDV‐OM in mechanical transmissibility. The presence of chimeric NSP‐MP sequences after mechanical inoculation was identified by PCR only in plants inoculated with pCB2A + pCB1B::OM5′ MP or pOM2A + pOM1B::CB3′ MP (Figure 4c). The identity of the MP fragments in the mechanically infected N. benthamiana was further verified by restriction enzyme digestion and gel analysis. The digestion of NSP‐MP amplicons with DdeI revealed polymorphisms of the expected sizes from pCB2A + pCB1B::OM5′ MP (271, 431, 643, and 794 bp) and pOM2A + pOM1B::CB3′ MP (233, 431, 643, and 794 bp), which were clearly different from those of pOM2A + pOM1B (233, 431, and 1,437 bp). Because pCB1B::OM5′ MP contained a BsrGI recognition site, which was not present in pOM1B::CB3′ MP, BsrGI was used to differentiate the two genomes (Figure S3). The results indicate that the 5′ but not the 3′ end of the MP of ToLCNDV‐OM was required for mechanical transmissibility.
2.6. A single amino acid in the ToLCNDV‐OM MP determines mechanical transmissibility
Sequence alignment of ToLCNDV‐OM and ToLCNDV‐CB MPs revealed four amino acid differences (3rd, 6th, 8th, and 19th) in the N‐terminal region (Figure 4a). Site‐directed mutagenesis was performed to identify the specific amino acid residue(s) that are essential for mechanical transmissibility. Eight infectious clones, each containing a specific mutation of these four amino acids, were constructed and tested for infectivity (Figure 5). Agroinoculation and rub inoculation assays revealed that infectious clones pCB1BMP (3I→T), pCB1BMP (6D→E), pCB1BMP (8V→M), pOM1BMP (3T→I), pOM1BMP (6E→D), and pOM1BMP (8M→V), carrying substitution mutations at the 3rd, 6th, and 8th amino acid residues of either CB or OM, did not alter mechanical transmissibility when paired with the corresponding DNA‐A clone. However, ToLCNDV‐CB became mechanically transmissible when the 19th amino acid was changed from glycine (G) to glutamate (E), as demonstrated by the pairing of pCB1BMP (19G→E) with pCB2A (Figure 5). Moreover, the mechanical transmissibility of ToLCNDV‐OM was abolished by a substitution mutation at the 19th amino acid causing a change from glutamate to glycine, as demonstrated by the pairing of pOM1BMP (19E→G) with pOM2A. The presence of the viral variants in the mechanically infected plants was confirmed by PCR and sequencing analysis (data not shown).
Figure 5.

A single glutamate residue in the tomato leaf curl New Delhi virus (ToLCNDV)‐OM movement protein (MP) determines mechanical transmissibility. Nicotiana benthamiana, oriental melon, and cucumber plants after mechanical inoculation with pCB2A paired with pCB1BMP (3I→T), pCB1BMP (6D→E), or pCB1BMP (8V→M) carrying a single amino acid substitution at the 3rd, 6th or 8th position of the MP of the CB isolate did not develop visible symptoms (X). However, plants that were mechanically inoculated with pCB2A and pCB1BMP (19G→E) carrying a substitution mutation at the 19th amino acid, resulting in a change from glycine to glutamate, developed viral symptoms (O). Plants that were mechanically inoculated with pOM2A and pOM1BMP (3T→I), pOM1BMP (6E→D), or pOM1BMP (8M→V) also developed visible symptoms. In contrast, plants that were mechanically inoculated with pOM2A and pOM1BMP (19E→G), carrying a single amino acid substitution at the 19th amino acid, resulting in a change from glutamate to glycine, failed to develop symptoms. The viral inoculum used for mechanical sap inoculation was prepared from N. benthamiana with symptoms after agroinoculation with an appropriate combination of the clones. Abbreviations for amino acids: G, glycine; E, glutamate; I, isoleucine; T, threonine; D, aspartic acid; V, valine; M, methionine
2.7. Nonmechanically transmissible viruses fail to accumulate at detectable levels in plants after sap inoculation
Oriental melon plants that were inoculated with ToLCNDV infectious clones developed visible symptoms at 7–10 dpi. After mechanical transmission, Southern blot analysis was performed to determine the viral titres present in oriental melon cotyledons at 0, 5, 10, and 14 dpi using a ToLCNDV‐OM MP probe (Figure 6a). Viral DNA in the form of open circular (oc) and supercoiled (sc) double‐stranded DNA as well as single‐stranded DNA (ss) could be detected in all plants immediately after sap inoculation (0 dpi). Surprisingly, viral DNA was barely detectable or undetectable at 5 dpi in all oriental melon plants except those inoculated with pCB2A + pCB1BMP (19G→E). At 10 and 14 dpi, viral DNA could be detected in all plants inoculated with infectious clones pOM2A + pOM1B, pCB2A + pCB1B::OMMP, pCB2A + pCB1B::OM5′ MP, and pCB2A + pCB1BMP (19G→E), which have been shown to be mechanically transmittable. However, no viral DNA was detected in plants inoculated with clones pCB2A + pCB1B, pOM2A + pOM1B::CBMP, pOM2A + pOM1B::CB5′ MP, and pOM2A + pOM1BMP (19E→G), which were not mechanically transmissible. PCR analysis of samples prepared from apical leaves also identified the expected amplicons only in plants inoculated with the infectious constructs that were mechanically transmissible (data not shown). Further analysis via in situ hybridization identified ToLCNDV‐OM viruses in N. benthamiana leaves inoculated with pOM2A + pOM1B or pCB2A + pCB1BMP (19G→E) at 5 dpi. No signal was detected in leaf samples inoculated with pCB2A + pCB1B or pOM2A + pOM1BMP (19E→G) (Figure 6b). The results indicate the critical role of glutamate as the 19th residue of MP in the mechanical transmissibility of the ToLCNDV OM isolate.
Figure 6.
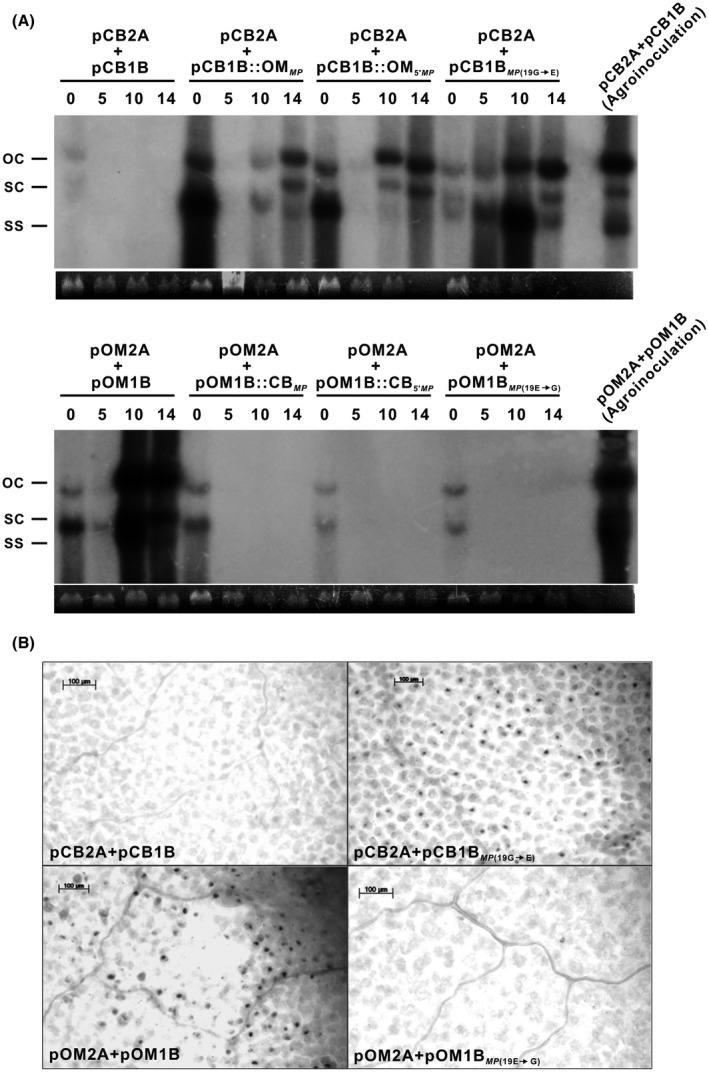
Nonmechanically transmissible viruses fail to accumulate to detectable levels in plants after sap inoculation. (a) Southern blot analysis of viral DNA in the open circular (oc), supercoiled (sc) double‐stranded DNA, and single‐stranded DNA (ss) forms in the cotyledons of oriental melon mechanically inoculated with infectious clones as indicated over time (0, 5, 10, and 14 days post‐inoculation, dpi using the tomato leaf curl New Delhi virus (ToLCNDV)‐OM MP probe. DNA prepared from oriental melon inoculated with pCB2A + pCB1B (upper panel) or pOM2A + pOM1B (lower panel) by using Agrobacterium was used as a positive control. DNA stained with ethidium bromide was used as the loading control. (b) In situ hybridization of Nicotiana benthamiana leaves with the ToLCNDV‐OM MP probe at 5 dpi with the infectious clones, as indicated. The dark spots observed in samples inoculated with pCB2A + p pCB1BMP (19G→E) and pOM2A + pOM1B represent viral hybridization signals. Bar = 100 μm. Only representative examples are shown
2.8. The glutamate residue is related to the tertiary structure of the ToLCNDV‐OM MP
Protein sequence analysis using the predictor of natural disordered regions (PONDR) revealed that the N terminus of the MP of ToLCNDV‐OM was probably a disordered region and that the counterpart in ToLCNDV‐CB was an ordered region (Figure 7). In contrast, the deduced pOM1BMP (19E→G) amino acid sequence (ToLCNDV‐OM mutant), which differed only at the 19th (glycine, G) residue from that of ToLCNDV‐OM, was an ordered region. The deduced pCB1BMP (19G→E) amino acid sequence (ToLCNDV‐CB mutant), containing glutamate (E) instead of glycine at the 19th position, was a disordered region. The results suggest a close relationship between the 19th glutamate and the tertiary structure of MP in the ToLCNDV OM isolate. However, sequence alignment of MP sequences available in the database revealed that glutamate as the 19th residue was not conserved among ToLCNDV isolates. Both the ToLCNDV‐severe isolate (nonmechanically transmissible) and the ToLCNDV‐potato isolate (mechanically transmissible) exhibited glutamate as the 19th amino acid residue, indicating that glutamate alone is not sufficient to determine mechanical transmissibility.
Figure 7.
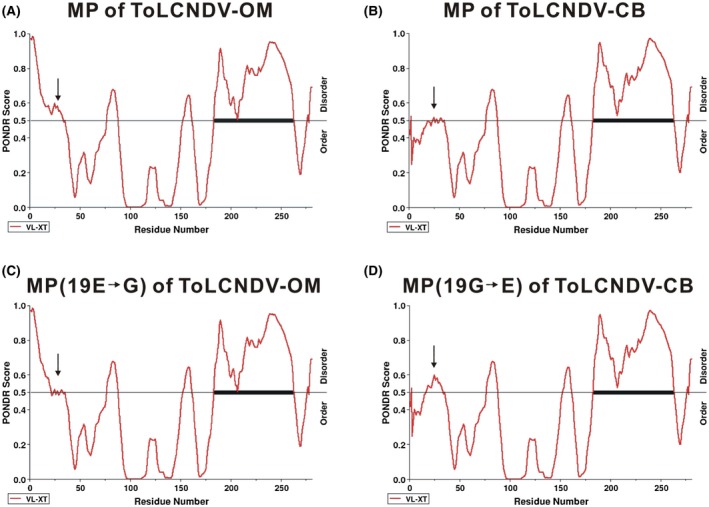
Prediction of natural disordered regions in movement proteins (MPs) using VL‐XT PONDR software. PONDR plot of the disorder score for each of the MP amino acid residue of the tomato leaf curl New Delhi virus (ToLCNDV)‐OM (a), ToLCNDV‐CB (b), ToLCNDV‐OMMP (19E→G) (c), and ToLCNDV‐CBMP (19G→E) (d). The horizontal lines indicate the threshold for disorder prediction. The 19th amino acid is indicated by an arrow
3. DISCUSSION
Begomoviruses are often transmitted by whiteflies and are rarely mechanically transmissible by rub or sap inoculation (Wege and Pohl, 2007). Only a few known species in the Begomovirus genus have been reported to be mechanically transmissible to their natural hosts (Gilbertson et al., 1991; Usharani et al., 2004; Tsai et al., 2011). ToLCNDV is a begomovirus with a bipartite (DNA‐A and DNA‐B) genome (Padidam et al., 1995). In the present study, genetic and pathological approaches were applied to identify the molecular determinants involved in mechanical transmissibility in a ToLCNDV‐OM isolate originally collected from a diseased oriental melon. Unlike ToLCNDV‐OM, the closely related ToLCNDV‐CB isolate is not mechanically transmissible to oriental melon, cucumber or N. benthamiana via sap inoculation. The two ToLCNDV isolates show strong nucleotide sequence identity for both DNA‐A (96.5%) and DNA‐B (92.2%), and exhibit very similar host ranges. Genetic evidence indicates that DNA‐B of ToLCNDV‐OM plays a critical role in mechanical transmissibility, consistent with findings for TYLCTHV and abutilon mosaic virus (AbMV) (Wege and Pohl, 2007; Tsai et al., 2011).
DNA‐B contains both the NSP and MP genes. NSP encodes a protein that is presumably responsible for the transportation of single‐stranded or double‐stranded DNA between the cytoplasm and the nucleus. MP encodes a protein required for cell‐to‐cell movement. Although NSP has been reported to be required for begomovirus virulence (Hussain et al., 2005; Zhou et al., 2007), a role of this gene in mechanical transmissibility was ruled out by testing the infectious clone pOM1B::NSP ins39 in the present study. This clone carries an extra 39‐nt fragment from a nonmechanically transmissible isolate (ToLCNDV‐severe) at the 5′ end of ToLCNDV‐OM NSP, and together with pOM2A it failed to infect N. benthamiana and oriental melon via sap inoculation. Further substitution of NSP between ToLCNDV‐OM and ToLCNDV‐CB also ruled out the possible involvement of NSP in mechanical transmission because swapping NSP between these two isolates did not alter the mode of transmissibility.
Genetic evidence derived from a series of mutations has revealed that MP in DNA‐B contains the key determinant required for mechanical transmission in ToLCNDV‐OM. The exchange of MPs between ToLCNDV‐OM and ToLCNDV‐CB revealed that an infectious clone carrying ToLCNDV‐OM MP, but not ToLCNDV‐CB MP, was able to induce symptoms on three test plant species after sap inoculation. Moreover, it appears that the N terminus of DNA‐B MP contains the elements required for mechanical transmissibility. Sequence alignment revealed that ToLCNDV‐OM and ToLCNDV‐CB MPs share 97.2% similarity, differing in only eight amino acids, four of which are located in the N terminus, while the other four are located at the C terminus. The involvement of the C terminus was ruled out by DNA‐B swapping assays. The evaluation of a series of point mutations revealed that glutamate at the 19th amino acid position at the N‐terminal end of MP is critical for the mechanical transmission of ToLCNDV‐OM. Point mutations affecting the 3rd, 6th, or 8th amino acid residue of ToLCNDV‐CB or ToLCNDV‐OM did not affect the mechanical transmissibility of either isolate. Moreover, ToLCNDV‐CB, which is not mechanically transmissible, became mechanically transmissible when a point mutation at the 19th amino acid of MP caused a change from glycine (G) to glutamate (E). In contrast, changing the 19th amino acid from glutamate to glycine in ToLCNDV‐OM rendered the virus nonmechanically transmissible, which might be due to the characteristics of the amino acids, as glycine is nonpolar and glutamate is negatively charged.
Although the glutamate residue at the 19th position of the ToLCNDV‐OM MP plays a critical role in mechanical transmission, glutamate alone is seemingly not sufficient to determine mechanical transmissibility. Sequence alignment revealed that the MPs of both the ToLCNDV‐severe (nonmechanically transmissible) and ToLCNDV‐potato (mechanically transmissible) isolates also exhibited glutamate at the 19th position, suggesting the presence of other factors or determinants that may contribute to the mechanical transmission of ToLCNDV‐OM. This assumption is supported by findings obtained using PONDR VL‐XT. PONDR plots can be used to visualize regions within disordered regions and to determine potential binding sites in proteins (Oldfield et al., 2005; Cheng et al., 2007). Disordered protein regions, characterized by the lack of a fixed tertiary structure, have been proposed to be involved in many biochemical functions, including protein binding and recognition (Dunker et al., 2002; Xue et al., 2010). PONDR analyses suggested that the N terminus of ToLCNDV‐OM MP is a disordered region; a switch from glutamate to glycine at the 19th amino acid position changes the prediction to an ordered region. Moreover, the N terminus of ToLCNDV‐CB MP is predicted to be an ordered region; however, the sequence is predicted to be a disordered region after the substitution of glycine with glutamate at the 19th amino acid.
In addition to MP, CP may be involved in mechanical transmissibility in ToLCNDV‐OM as demonstrated in BGMV, another member of the Begomovirus genus. The BGMV CP has been demonstrated to be required for both mechanical and whitefly transmissions (Azzam et al., 1994). A BGMV mutant with a defective CP fails to induce symptoms on sap inoculation, but an infection develops after the mutant is biolistically bombarded into leaf cells because biolistic inoculation can deliver the virus directly into the host nucleus. Thus, CP is required for the localization of BGMV to the nucleus. Intriguingly, the CP of two geminiviruses, mungbean yellow mosaic virus (MYMV) and ACMV, have been reported to contain a nuclear localization signal (NLS) (Unseld et al., 2001; Guerra‐Peraza et al., 2005), suggesting the involvement of CP in nuclear localization. Therefore, the difference in mechanical transmissibility between ToLCNDV‐OM and ToLCNDV‐CB may result from the interactions of CP and MP, which was shown not to be required for the mechanical transmission of ToLCNDV in the present study.
MP has also been implicated in intra‐ and intercellular trafficking by interacting with host proteins (Jeske, 2009), which could lead to different modes of transmission as seen in different ToLCNDV isolates. AbMV MP can interact with host kinases and affect the development of symptoms and the accumulation of viral DNA in host cells (Kleinow et al., 2009). Studies have identified histone H3 (Zhou et al., 2011), synaptotagmin (SYTA) (Lewis and Lazarowitz, 2010), and the 70 kDa heat shock protein cpHSC70‐1 (Krenz et al., 2010) as proteins that can interact with the MP of begomoviruses. Histone H3 is probably required for the assembly of the MP‐nucleoprotein complex in the nucleus. SYTA is required for cell‐to‐cell trafficking and systemic infection. The cpHSC70‐1 protein regulates the MP conformation at the cell periphery and localization to chloroplasts.
Viral MPs are required for both cell‐to‐cell and long‐distance movements. Studies on AbMV, another geminivirus, have suggested that a so‐called “pilot domain” (amino acid residues from 1 to 49) of MP is responsible for the transportation of the virus to cell periphery or nucleus (Zhang et al., 2002). Two models have been proposed to explain the functions of MP in geminiviruses (Rojas et al., 2005; Jeske, 2009). For phloem‐limited begomoviruses such as AbMV and cabbage leaf curl virus (CaLCuV), MP is thought to attach to the NSP‐bound viral DNA and form a complex that localizes along the plasma membrane or is transferred to adjacent cells through plasmodesmata. For mesophyll‐invading begomoviruses, MP is thought to take over viral DNA from NSP after being exported from the nucleus to the cytoplasm and deliver it to adjacent cells. Most nonmechanically transmissible begomoviruses have been reported to be phloem limited because they can be introduced into the phloem translocation stream through the stylets of insects and become systemic (Czosnek et al., 2017). One of the reasons that ToLCNDV‐CB fails to be mechanically transmissible and to initiate systemic infection after sap inoculation may be its inability to travel beyond the site of inoculation. ToLCNDV‐CB is probably a phloem‐limited virus because this isolate is unable to propagate in the mesophyll after sap inoculation, as demonstrated by Southern blot and in situ hybridization analyses. In contrast, ToLCNDV‐OM is capable of replicating in mesophyll after sap inoculation.
For systemic infection, ToLCNDV must be loaded into the phloem sieve tube system for long‐distance movement. ToLCNDV‐OM is able to propagate and travel through mesophyll cells, the bundle sheath, and eventually the phloem after sap inoculation, which is very rare for a geminivirus (Wang et al., 1996). In contrast, ToLCNDV‐CB apparently has to rely on the stylet of its insect vector to reach the phloem. After sap inoculation, ToLCNDV‐CB is unable to move beyond its inoculation foci, for which there may be many possible explanations. ToLCNDV‐CB may trigger plant defence, fail to manipulate plasmodesmata, and be unable to replicate. A time‐course study of viral accumulation by Southern blotting (Figure 6) revealed that ToLCNDV‐OM DNA is barely detectable or undetectable at 5 dpi in oriental melon plants and can be detected again at 10 and 14 dpi, suggesting the suppression of plant defence during the early stages of virus–plant interactions. ToLCNDV‐OM, but not ToLCNDV‐CB, might be able to overcome host defence reactions and, thus, infect the host systemically after sap inoculation.
In conclusion, genetic experiments have clearly demonstrated that the glutamate residue at the 19th position in the N terminus of the MP of the ToLCNDV‐OM DNA‐B genome is required for mechanical transmission. Our genetic evidence also ruled out the involvement of NSP in mechanical transmission. It is very likely that MP interacts with host proteins to enable the mechanical transmission of ToLCNDV‐OM. Understanding the molecular mechanisms involved in the mechanical transmission of viruses could contribute to the development of more efficient strategies to manage diseases caused by begomoviruses.
4. EXPERIMENTAL PROCEDURES
4.1. Characteristics and origins of ToLCNDV isolates
ToLCNDV‐OM was originally isolated from a diseased oriental melon (Cucumis melo “Silver Light”) collected from Yilan, Taiwan in 2007 (Chang et al., 2010). ToLCNDV‐CB (accession numbers MK883715 and MK883716) was originally isolated from a diseased cucumber in Thailand. The sequences of ToLCNDV‐OM (accession numbers GU180095 and MK883714), ToLCNDV‐severe (accession numbers U15015 and U15017) and ToLCNDV‐potato (accession numbers AY286316 and AY158080) were retrieved from the GenBank database of NCBI. Sequence alignment was conducted with the ClustalW algorithm of the MegAlign program available in Lasergene 7 software (DNASTAR).
4.2. Construction of infectious viral DNAs
Viral infectious clones of ToLCNDV‐OM and ToLCNDV‐CB were constructed via the rolling circle amplification (RCA) method (Wu et al., 2008). The infectious clones pOM2A (formerly pGPhi‐ToLCNDV‐2A) and pOM2B (formerly pGPhi‐ToLCNDV‐2B) containing tandem repeated sequences of ToLCNDV‐OM DNA‐A and DNA‐B, respectively, were constructed in the binary vector pGANP (Lin et al., 2011) in a previous study (Chang et al., 2010). The infectious clones, pCB2A and pCB2B of ToLCNDV‐CB, were constructed in the binary vector pCAMBIA1304 (Cambia). The oligonucleotide primers (FJJ2007‐13 and FJJ2007‐15) used for the construction of infectious clones are listed in Table S2.
A 39‐nt fragment was amplified by PCR using the primers FJJ2009‐26, FJJ2009‐27, and FJJ2009‐32, corresponding to sequences from the NSP gene of the ToLCNDV‐severe isolate (Hussain et al., 2005). The amplicon was fused with the ToLCNDV‐OM DNA‐B fragment digested with NcoI and NheI, and the resultant fragment was cloned into NcoI and NheI‐digested pGANP to create pOM2B::NSP ins39.
An NSP‐MP fragment was amplified either from pOM2B, with the primers FJJ2010‐99 and FJJ2010‐100 for NSP or FJJ2010‐101 and FJJ2010‐102 for MP, or from pCB2B, with the primers FJJ2010‐98 and FJJ2010‐100 for NSP and FJJ2010‐101 and FJJ2010‐102 for MP (Figure 3a and Table S2). The IR fragment was amplified from either pOM2B, with the primers FJJ2010‐95, FJJ2010‐97, FJJ2010‐103, and FJJ2010‐104, or from pCB2B, with the primers FJJ2010‐95, FJJ2010‐96, FJJ2010‐103, and FJJ2010‐104 (Figure 3 and Table S2). A recognition site for restriction endonuclease (SacI, BamHI, EcoRV, HindIII or SalI) was incorporated into the primer as appropriate to facilitate cloning. NSP‐MP and IR were fused to form an IR‐NSP‐MP‐IR fragment. The resultant fragment was digested with SacI and SalI and cloned into pCAMBIA1304 to generate pOM1B or pCB1B.
4.3. Recombination of partial 5′ or 3′ MP sequences between ToLCNDV‐CB and ToLCNDV‐OM
Four infectious clones, each containing a chimeric MP, were constructed in the backbone of pOM1B or pCB1B. The HindIII‐PstI fragment (partial IR and 5′ end MP, 455 bp) of pCB1B was replaced with its counterpart from pOM1B to generate pCB1B::OM5′ MP. The PstI‐EcoRV fragment (3′ end MP, 817 bp) of pCB1B was replaced with its counterpart from pOM1B to generate pCB1B::OM3′ MP. Similarly, the HindIII‐PstI fragment and PstI‐EcoRV of pOM1B were replaced with the counterparts from pCB1B to create pOM1B::CB5′ MP and pOM1B::CB3′ MP, respectively.
4.4. Single amino acid substitutions in MP
Point mutations at each of four amino acid residues (3rd, 6th, 8th, and 19th) of MP (Figure 4a) were individually generated in pCB1B and pOM1B using a QuickChange XL Site‐Directed Mutagenesis Kit following the manufacturer's recommendations (Stratagene). In total, eight clones (pCB1BMP (3I→T), pCB1BMP (6D→E), pCB1BMP (8V→M), pCB1BMP (19G→E), pOM1BMP (3T→I), pOM1BMP (6E→D), pOM1BMP (8M→V), and pOM1BMP (19E→G)) were generated.
4.5. Agroinoculation and mechanical inoculation
Infectious clones were individually transformed into Agrobacterium tumefaciens C58 by using a GenePulser II electroporator (Bio‐Rad). Agroinoculation was conducted according to the procedure described by Llave et al. (2000) with some modifications. Briefly, 0.5 ml of an overnight bacterial culture was recultured in 10 ml of Luria Bertani (LB) medium (pH 5.6) containing kanamycin (50 μg/ml), streptomycin (50 μg/ml), 2‐N‐morpholino‐ethanesulfonic acid (MES, 10 mM), and acetosyringone (AS, 0.04 mM) at 28 °C for 16 hr on a shaker set at 200 rpm. The bacterial cells were pelleted by centrifugation at 5,000 × g for 10 min and resuspended in 10 ml of an infiltration solution (10 mM MgCl2, 0.15 mM AS, pH 5.6). Agrobacteria carrying the infectious DNA‐A or DNA‐B constructs were co‐injected in equal amounts into the leaves of N. benthamiana, oriental melon (C. melo “Silver Light”) or cucumber (C. sativus “Vantage”). After agroinoculation, N. benthamiana leaves with symptoms (c.1 g) were collected and ground in 0.01 M potassium phosphate buffer (pH 7) (1:20, wt/vol). The resultant sap was inoculated onto N. benthamiana leaves or cotyledon explants of oriental melon and cucumber plants by rubbing with carborundum powder. All inoculated plants were kept in a greenhouse at 25–28 °C to observe symptom development.
4.6. Detection of viral DNA by PCR
DNA was extracted from plant leaves following the method described by Lin et al. (2011). PCR was performed in a 25 μl reaction containing 100 ng DNA, 20 each of the primers at nM, 0.05 mM dNTPs, 0.25 U Pro Taq Plus DNA polymerase (Protech Technology) and buffer. The conditions used for amplification were as follows: one cycle at 95 °C for 5 min followed by 35 cycles of 95 °C for 1 min, 50–67 °C depending on the specific primer pairs used (Table S1) for 1 min and 72 °C for 1 min, with a final cycle at 72 °C for 10 min. The PCR products were subjected to electrophoresis in a 0.8% agarose gel. Sequencing was performed using an ABI PRISM 3730 automatic DNA sequencer (Applied Biosystems) available at the Biotechnology Center, National Chung Hsing University (Taichung, Taiwan).
4.7. Southern blot hybridization
After mechanical inoculation, the cotyledons of oriental melon were harvested at 0, 5, 10, and 14 dpi and subjected to DNA isolation. DNA (5 μg) was subjected to electrophoresis in a 1% agarose gel and transferred to a nylon membrane (PerkinElmer) via the alkaline transfer method, and hybridization was performed on the full‐length MP of ToLCNDV‐OM. The probe was labelled by α‐[32P]dATP. The hybridization and detection of the probe were performed according to the methods described by Kon et al. (2003). DNA isolated from the apical leaves of oriental melon agroinoculated with ToLCNDV‐OM (pOM2A + pOM1B) or ToLCNDV‐CB (pCB2A + pCB1B) was used as a positive control.
4.8. In situ hybridization
Tissue fixation and in situ hybridization were performed following the protocol described by Takeda et al. (2011) with some modifications. N. benthamiana leaves at 5 dpi were collected, cut into small pieces, and fixed in solution containing 50% ethanol, 5% acetic acid, and 5% formaldehyde (FAA) for 2 hr at room temperature. Leaf samples were soaked in 30% ethanol for 20 min and then in sterile water twice for 20 min each. The samples were soaked in Tris‐EDTA (TE) buffer (100 mM Tris‐HCl, 50 mM EDTA, pH 8.0) for 30 min and treated with 2 μg/ml proteinase K for 30 min at 37 °C. The samples were next washed with phosphate‐buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4) and fixed in 4% paraformaldehyde for 20 min at 4 °C. The samples were washed with PBS again and incubated in hybridization solution for 20 min. The DNA probe was labelled and detected using a Digoxigenin (DIG)‐High Prime DNA Labeling and Detection Starter Kit I (Roche). The samples were incubated in hybridization solution containing a DIG‐labelled MP probe (200 bp) at 50 °C overnight. The detection of the probe was performed via an immunological assay using alkaline phosphatase‐conjugated DIG antibodies followed by nitroblue tetrazolium and 5‐bromo‐4‐chloro‐3‐indolyl phosphate (NBT/BCIP) colour development. Samples were photographed under a Zeiss light microscope equipped with AxioVision Rel 4.8 software (Carl Zeiss).
4.9. Predictor of natural disordered regions
The PONDR within the movement protein sequences of ToLCNDV‐OM and ToLCNDV‐CB was performed by using the PONDR VL‐XT predictor available at http://www.pondr.com/.
Supporting information
FIGURE S1 Symptoms of Nicotiana benthamiana, oriental melon, and cucumber plants after agroinoculation with the infectious clones at 10–12 days post‐inoculation
FIGURE S2 Alignment of the amino acid residues of the movement proteins of the tomato leaf curl New Delhi virus (ToLCNDV)‐CB and ToLCNDV‐OM
FIGURE S3 Confirmation of the presence of the 5′ or 3′ MP fragment in Nicotiana benthamiana, oriental melon, and cucumber plants after mechanical inoculation with the tomato leaf curl New Delhi virus (ToLCNDV)‐CB or ToLCNDV‐OM mutant
TABLE S1 Characteristics of oligonucleotide primers used to amplify the viral DNA of the tomato leaf curl New Delhi virus (ToLCNDV)‐CB and ToLCNDV‐OM isolates in this study
TABLE S2 Characteristics of the oligonucleotide primers used to construct infectious clones of tomato leaf curl New Delhi virus
ACKNOWLEDGEMENTS
We are grateful to Dr Kuang‐Ren Chung for his critical review of this manuscript. We also thank Dr Shi‐Dong Yeh for valuable discussion. This work was supported by grants from the Ministry of Science and Technology (NSC 101‐2313‐B‐005‐039‐MY3 and MOST 105‐2313‐B‐005‐019‐MY3), Executive Yuan, Taiwan. The authors have no conflicts of interest to declare.
Lee C‐H, Zheng Y‐X, Chan C‐H, Ku H‐M, Chang C‐J, Jan F‐J. A single amino acid substitution in the movement protein enables the mechanical transmission of a geminivirus. Molecular Plant Pathology. 2020;21:571–588. 10.1111/mpp.12917
Chia‐Hwa Lee and You‐Xiu Zheng contributed equally to this work.
Funding information
This work was supported by grants from the Ministry of Science and Technology (NSC 101‐2313‐B‐005‐039‐MY3 and MOST 105‐2313‐B‐005‐019‐MY3), Executive Yuan, Taiwan.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Ajlan, A.M. , Ghanem, G.A.M. and Abdulsalam, K.S. (2007) Tomato yellow leaf curl virus (TYLCV) in Saudi Arabia: identification, partial characterization and virus–vector relationship. Arab Journal of Biotechnology, 10, 179–192. [Google Scholar]
- Arguello‐Astorga, G. , Ascencio‐lbanez, J.T. , Dallas, M.B. , Orozco, B.M. and Hanley‐Bowdoin, L. (2007) High‐frequency reversion of geminivirus replication protein mutants during infection. Journal of Virology, 81, 11005–11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam, O. , Frazer, J. , de la Rosa, D. , Beaver, J.S. , Ahlquist, P. and Maxwell, D.P. (1994) Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology, 204, 289–296. [DOI] [PubMed] [Google Scholar]
- Bock, K.R. and Guthrie, E.J. (1978) Transmission of African cassava mosaic by mechanical inoculation. Plant Disease Reporter, 62, 580–581. [Google Scholar]
- Chang, H.H. , Ku, H.M. , Tsai, W.S. , Chien, R.C. and Jan, F.J. (2010) Identification and characterization of a mechanical transmissible begomovirus causing leaf curl on oriental melon. European Journal of Plant Pathology, 127, 219–228. [Google Scholar]
- Chatchawankanphanich, O. and Maxwell, D.P. (2002) Tomato leaf curl Karnataka virus from Bangalore, India, appears to be a recombinant begomovirus. Phytopathology, 92, 637–645. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Oldfield, C.J. , Meng, J. , Romero, P. , Uversky, V.N. and Dunker, A.K. (2007) Mining a‐helic‐forming molecular recognition features with cross species sequence alignments. Biochemistry, 46, 13468–13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czosnek, H. , Hariton‐Shalev, A. , Sobol, I. , Gorovits, R. and Ghanim, M. (2017) The incredible journey of begomoviruses in their whitefly vector. Viruses, 9, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, S. and Holmes, E.C. (2008) Phylogenetic evidence for rapid rates of molecular evolution in the single‐stranded DNA begomovirus Tomato yellow leaf curl virus . Journal of Virology, 82, 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker, A.K. , Brown, C.J. , Lawson, J.D. , Iakoucheva, L.M. and Obradović, Z. (2002) Intrinsic disorder and protein function. Biochemistry, 41, 6573–6582. [DOI] [PubMed] [Google Scholar]
- Fauquet, C.M. , Briddon, R.W. , Brown, J.K. , Moriones, E. , Stanley, J. , Zerbini, M. et al (2008) Geminivirus strain demarcation and nomenclature. Archives of Virology, 153, 783–821. [DOI] [PubMed] [Google Scholar]
- Fondong, V.N. (2013) Geminivirus protein structure and function. Molecular Plant Pathology, 14, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnsey, S.M. , Gonsalves, D. and Purcifull, D.E. (1977) Mechanical transmission of Citrus tristeza virus . Phytopathology, 67, 965–968. [Google Scholar]
- Garrido‐Ramirez, E.R. , Sudarshana, M.R. and Gilbertson, R.L. (2000) Bean golden yellow mosaic virus from Chiapas, Mexico: characterization, pseudorecombination with other bean‐infecting geminiviruses and germ plasm screening. Phytopathology, 90, 1224–1232. [DOI] [PubMed] [Google Scholar]
- Gilbertson, R.L. , Hidayat, S.H. , Martinez, R.T. , Leong, S.A. , Faria, J.C. and Morales, F. (1991) Differentiation of bean‐infecting geminiviruses by nucleic acid hybridization probes and aspects of bean golden mosaic in Brazil. Plant Disease, 75, 336–342. [Google Scholar]
- Green, S.K. , Tsai, W.S. , Shih, S.L. , Rezaian, M.A. and Duangsong, U. (2003) Molecular characterization of a new begomovirus associated with tomato yellow leaf curl and eggplant yellow mosaic diseases in Thailand. Plant Disease, 87, 446. [DOI] [PubMed] [Google Scholar]
- Guerra‐Peraza, O. , Kirk, D. , Seltzer, V. , Veluthambi, K. , Schmit, A.C. , Hohn, T. et al (2005) Coat proteins of Rice tungro bacilliform virus and Mungbean yellow mosaic virus contain multiple nuclear‐localization signals and interact with importin alpha. Journal of General Virology, 86, 1815–1826. [DOI] [PubMed] [Google Scholar]
- Hussain, M. , Mansoor, S. , Iram, S. , Fatima, A.N. and Zafar, Y. (2005) The nuclear shuttle protein of Tomato leaf curl New Delhi virus is a pathogenicity determinant. Journal of Virology, 79, 4434–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T. , Sharma, P. , Kittipakorn, K. and Ikegami, M. (2008) Complete nucleotide sequence of a new isolate of tomato leaf curl New Delhi virus infecting cucumber, bottle gourd and muskmelon in Thailand. Archives of Virology, 153, 611–613. [DOI] [PubMed] [Google Scholar]
- Jeske, H. (2009) Geminiviruses. Current Topics in Microbiology and Immunology, 331, 185–226. [DOI] [PubMed] [Google Scholar]
- Khan, M.S. , Ji, S.H. and Chun, S.C. (2012) Begomoviruses and their emerging threats in South Korea: a review. Plant Pathology Journal, 28, 123–136. [Google Scholar]
- Kleinow, T. , Tanwir, F. , Kocher, C. , Krenz, B. , Wege, C. and Jeske, H. (2009) Expression dynamics and ultrastructural localization of epitope‐tagged Abutilon mosaic virus nuclear shuttle and movement proteins in Nicotiana benthamiana cells. Virology, 391, 212–220. [DOI] [PubMed] [Google Scholar]
- Kon, T. , Dolores, L.M. , Bajet, N.B. , Hase, S. , Takahashi, H. and Ikegami, M. (2003) Molecular characterization of a strain of Squash leaf curl china virus from Philippines. Journal of Phytopathology, 151, 535–539. [Google Scholar]
- Krenz, B. , Jeske, H. and Kleinow, T. (2012) The induction of stromule formation by a plant DNA‐virus in epidermal leaf tissues suggests a novel intra‐ and intercellular macromolecular trafficking route. Frontiers in Plant Science, 3, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenz, B. , Windeisen, V. , Wege, C. , Jeske, H. and Kleinow, T. (2010) A plastid‐targeted heat shock cognate 70 kDa protein interacts with the Abutilon mosaic virus movement protein. Virology, 401, 6–17. [DOI] [PubMed] [Google Scholar]
- Legg, J.P. and Fauquet, C.M. (2004) Cassava mosaic geminiviruses in Africa. Plant Molecular Biology, 56, 585–599. [DOI] [PubMed] [Google Scholar]
- Levy, A. and Czosnek, H. (2003) The DNA‐B of the non‐phloem‐limited Bean dwarf mosaic virus (BDMV) is able to move the phloem‐limited Abutilon mosaic virus (AbMV) out of the phloem, but DNA‐B of AbMV is unable to confine BDMV to the phloem. Plant Molecular Biology, 53, 789–803. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D. and Lazarowitz, S.G. (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell‐to‐cell transport. Proceedings of the National Academy of Sciences of the United States of America, 107, 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.Y. , Ku, H.M. , Tan, C.W. , Yeh, S.D. and Jan, F.J. (2011) Construction of binary vectors with bi‐selectable markers for generating marker‐free transgenic plants. Botanical Studies, 52, 239–248. [Google Scholar]
- Llave, C. , Kasschau, K.D. and Carrington, J.C. (2000) Virus‐encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proceedings of the National Academy of Sciences of the United States of America, 97, 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, C. , Ferriol, M. and Pico, M.B. (2015) Mechanical transmission of Tomato leaf curl New Delhi virus to cucurbit germplasm: selection of tolerance sources in Cucumis melo . Euphytica, 204, 679–691. [Google Scholar]
- Morales, F. , Niessen, A. , Ramirez, B. and Castano, M. (1990) Isolation and partial characterization of a geminivirus causing bean dwarf mosaic. Phytopathology, 80, 96–101. [Google Scholar]
- Oldfield, C.J. , Cheng, Y. , Cortese, M.S. , Romero, P. , Uversky, V.N. and Dunker, A.K. (2005) Coupled folding and binding with alpha‐helix‐forming molecular recognition elements. Biochemistry, 44, 12454–12470. [DOI] [PubMed] [Google Scholar]
- Padidam, M. , Beachy, R.N. and Fauquet, C.M. (1995) Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. Journal of General Virology, 76, 25–35. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R. , Hagen, C. , Lucas, W.J. and Gilbertson, R.L. (2005) Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annual Review of Phytopathology, 43, 361–394. [DOI] [PubMed] [Google Scholar]
- Samretwanich, K. , Chiemsombat, P. , Kittipakorn, K. and Ikegami, M. (2000) Tomato leaf curl geminivirus associated with cucumber yellow leaf disease in Thailand. Journal of Phytopathology, 148, 615–617. [Google Scholar]
- Shih, S.L. , Green, S.K. , Tsai, W.S. , Lee, L.M. and Levasseur, V. (2007) First report of a distinct begomovirus associated with okra yellow crinkle disease in Mali. Plant Pathology, 56, 718. [Google Scholar]
- Takeda, R. , Petrov, A.I. , Leontis, N.B. and Ding, B. (2011) A three‐dimensional RNA motif in Potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana . The Plant Cell, 23, 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, W.S. , Shih, S.L. , Kenyon, L. , Green, S.K. and Jan, F.J. (2011) Temporal distribution and pathogenicity of the predominant tomato‐infecting begomoviruses in Taiwan. Plant Pathology, 60, 787–799. [Google Scholar]
- Unseld, S. , Hohnle, M. , Ringel, M. and Frishmuth, T. (2001) Subcellular targeting of the coat protein of African cassava mosaic geminiviruses. Virology, 286, 373–383. [DOI] [PubMed] [Google Scholar]
- Usharani, K.S. , Surendranath, B. , Paul‐Khurana, S.M. , Garg, I.D. and Malathi, V.G. (2004) Potato leaf curl‐a new disease of potato in northern India caused by a strain of Tomato leaf curl New Delhi virus . Plant Pathology, 53, 235. [Google Scholar]
- Varma, A. and Malathi, V.G. (2003) Emerging geminivirus problems: a serious threat to crop production. The Annals of Applied Biology, 142, 145–164. [Google Scholar]
- Wang, H.L. , Gilbertson, R.L. and Lucas, W.J. (1996) Spatial and temporal distribution of bean dwarf mosaic geminivirus in Phaseolus vulgaris and Nicotiana benthamiana . Phytopathology, 86, 1204–1214. [Google Scholar]
- Wege, C. and Pohl, D. (2007) Abutilon mosaic virus DNA B component supports mechanical virus transmission, but does not counteract begomoviral phloem limitation in transgenic plants. Virology, 365, 173–186. [DOI] [PubMed] [Google Scholar]
- Wu, C.Y. , Lai, Y.C. , Lin, N.S. , Hsu, Y.H. , Tsai, H.T. , Liao, J.Y. et al (2008) A simplified method of constructing infectious clones of begomovirus employing limited restriction enzyme digestion of products of rolling circle amplification. Journal of Virological Methods, 147, 355–359. [DOI] [PubMed] [Google Scholar]
- Xue, B. , Dunbrack, R.L. , Williams, R.W. , Dunker, A.K. and Uversky, V.N. (2010) PONDR‐FIT: a meta‐predictor of intrinsically disordered amino acids. Biochimica et Biophysica Acta, 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.C. , Ghosh, R. and Jeske, H. (2002) Subcellular targeting domains of Abutilon mosaic geminivirus movment protein BC1. Archives of Virology, 147, 2349–2363. [DOI] [PubMed] [Google Scholar]
- Zhou, Y.C. , Garrido‐Ramirez, E.R. , Sudarshana, M.R. , Yendluri, S. and Gilbertson, R.L. (2007) The N‐terminus of the Begomovirus nuclear shuttle protein (BV1) determines virulence or avirulence in Phaseolus vulgaris . Molecular Plant‐Microbe Interactions, 20, 1523–1534. [DOI] [PubMed] [Google Scholar]
- Zhou, Y.C. , Rojas, M.R. , Park, M. , Seo, Y.S. , Lucas, W.J. and Gilbertson, R.L. (2011) Histone H3 interacts and colocalizes with the nuclear shuttle protein and the movement protein of a geminivirus. Journal of Virology, 85, 11821–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Symptoms of Nicotiana benthamiana, oriental melon, and cucumber plants after agroinoculation with the infectious clones at 10–12 days post‐inoculation
FIGURE S2 Alignment of the amino acid residues of the movement proteins of the tomato leaf curl New Delhi virus (ToLCNDV)‐CB and ToLCNDV‐OM
FIGURE S3 Confirmation of the presence of the 5′ or 3′ MP fragment in Nicotiana benthamiana, oriental melon, and cucumber plants after mechanical inoculation with the tomato leaf curl New Delhi virus (ToLCNDV)‐CB or ToLCNDV‐OM mutant
TABLE S1 Characteristics of oligonucleotide primers used to amplify the viral DNA of the tomato leaf curl New Delhi virus (ToLCNDV)‐CB and ToLCNDV‐OM isolates in this study
TABLE S2 Characteristics of the oligonucleotide primers used to construct infectious clones of tomato leaf curl New Delhi virus
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


