Summary
The biotrophic fungal pathogen Ustilaginoidea virens causes rice false smut, a newly emerging plant disease that has become epidemic worldwide in recent years. The U. virens genome encodes many putative effector proteins that, based on the study of other pathosystems, could play an essential role in fungal virulence. However, few studies have been reported on virulence functions of individual U. virens effectors. Here, we report our identification and characterization of the secreted cysteine‐rich protein SCRE1, which is an essential virulence effector in U. virens. When SCRE1 was heterologously expressed in Magnaporthe oryzae, the protein was secreted and translocated into plant cells during infection. SCRE1 suppresses the immunity‐associated hypersensitive response in the nonhost plant Nicotiana benthamiana. Induced expression of SCRE1 in rice also inhibits pattern‐triggered immunity and enhances disease susceptibility to rice bacterial and fungal pathogens. The immunosuppressive activity is localized to a small peptide region that contains an important ‘cysteine‐proline‐alanine‐arginine‐serine’ motif. Furthermore, the scre1 knockout mutant generated using the CRISPR/Cas9 system is attenuated in U. virens virulence to rice, which is greatly complemented by the full‐length SCRE1 gene. Collectively, this study indicates that the effector SCRE1 is able to inhibit host immunity and is required for full virulence of U. virens.
Keywords: fungal effector, immunosuppressive peptide, rice immunity, SCRE1, Ustilaginoidea virens
The Ustilaginoidea virens effector SCRE1 can suppress plant immunity and promote disease development in rice and a novel small peptide region is identified to be essential for SCRE1 immunosuppressive ability.

INTRODUCTION
Ustilaginoidea virens (telemorph Villosiclava virens) is an important fungus that causes false smut of rice (Oryza sativa). The disease spreads rapidly throughout the world and poses a significant economic impact (Rush et al., 2000). Particularly in China and in South Asia, false smut has become one of the most important rice diseases (Tang et al., 2013). U. virens infects rice flowers through stamen filaments at the late booting stage and develops false smut balls on the panicles at the heading stage (Han et al., 2015; Hu et al., 2014; Zhang et al., 2014). In the formation of false smut balls during infection, the pathogen produces two major types of mycotoxins: ustiloxins and ustilaginoidins (Koiso et al., 1994; Koyama and Natori, 1988). These mycotoxins inhibit cell division and impose a health threat to humans and animals (Koiso et al., 1994). Because false smut is a newly emerging disease, little is known about the virulence mechanisms of U. virens, which makes this fungal disease difficult to predict and manage (Fang et al., 2016; Han et al., 2015; Zhang et al., 2014).
Phytopathogenic fungi and oomycetes secrete a large number of effector proteins into the extracellular matrix (Alfano and Collmer, 2004; Galán and Collmer, 1999; Lo Presti et al., 2015). Some of these effectors remain and function in the apoplast and are called apoplastic effectors, while others, named cytoplasmic effectors, enter the host cells (Giraldo and Valent, 2013). During host–pathogen coevolution, some pathogen effectors are recognized by host immune receptors to trigger plant immunity in resistant plants. In the absence of such recognition (i.e. in susceptible plants), many effectors subvert plant immunity and other host cellular processes to promote infection (Cui et al., 2015; Oliveira‐Garcia and Valent, 2015). Studies have shown that fungal effectors play virulence functions using a variety of molecular mechanisms. Multiple apoplastic effectors, such as Avr4 and Ecp6 from Cladosporium fulvum, Slp1 from Magnaporthe oryzae and Mg3LysM from Mycosphaerella graminicola, block the elicitation of chitin‐triggered immunity (van Esse et al., 2007; de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012). Avr4, Mg1LysM and Mg3LysM, as well as the effector SnTox1 from the necrotrophic fungus Parastagonospora nodorum bind to chitin in the fungal cell wall and protect them against hydrolysis by plant chitinases (van Esse et al., 2007; Liu et al., 2012; Marshall et al., 2011; Mentlak et al., 2012). On the other hand, Ecp6 and Slp1 sequester chitin oligosaccharides that are released from the cell walls of invading hyphae to prevent activation of chitin‐induced immunity (Chen et al., 2014; de Jonge et al., 2010; Mentlak et al., 2012). The cytoplasmic effector Cmu1 of Ustilago maydis counteracts salicylic acid‐induced immunity by functioning as a chorismate mutase to reduce the level of chorismate, a precursor for salicylic acid synthesis (Djamei et al., 2011). In addition, the fungal effector candidate PpEC23 from Phakopsora pachyrhizi suppresses plant immunity and interacts with the transcription factor GmSPL12l, a negative immune regulator in soybean (Qi et al., 2016a). Fungal effectors can also target host proteins residing in subcellular organelles. For example, the small cysteine‐rich effector SsSSVP1 from Sclerotinia sclerotiorum interacts with the host mitochondrial protein QCR8 and facilitates S. sclerotiorum infection, probably through disrupting the host cell respiratory chain (Lyu et al., 2016).
In addition, pathogen effectors often suppress plant immunity through targeting various types of catalytic enzymes. The Pep1 effector from U. maydis interacts with the secreted maize peroxidase POX12 and inhibits its activity, thereby suppressing the reactive oxygen species (ROS) burst in plants (Hemetsberger et al., 2012). Another U. maydis effector, Pit2, inhibits a set of maize apoplastic cysteine proteases whose activities promote salicylic acid‐associated plant defences (Mueller et al., 2013). The C. fulvum apoplastic effector Avr2 also inhibits several cysteine proteases that contribute to plant basal defences (Shabab et al., 2008). The rust transferred protein 1, a cytoplasmic effector, represents yet another family of fungal effectors that exhibit inhibitory activity toward plant proteases (Pretsch et al., 2013). In addition, several effectors have been identified to target the host ubiquitin‐proteasome system and inhibit plant immunity. For example, AvrPiz‐t in M. oryzae binds to the rice RING E3 ligase APIP6 and inhibits its ubiquitination activity (Park et al., 2012). APIP6 is required for ROS generation and defence against M. oryzae (Park et al., 2012). APIP10, another E3 ligase that functions as a positive regulator of plant immunity in the non‐Piz‐t background in rice, is also a target of AvrPiz‐t (Park et al., 2016). In addition, PEC6 from the wheat stripe rust pathogen Puccinia striiformis interacts with adenosine kinases and suppresses pattern‐triggered immunity (Liu et al., 2016a).
Comparative transcriptome analyses suggest that a set of U. virens secreted proteins could be putative effectors (Han et al., 2015; Zhang et al., 2014). In a large‐scale screening, eight U. virens putative effectors were identified to induce the immunity‐associated hypersensitive responses when transiently overexpressed in the nonhost plant Nicotiana benthamiana (Fang et al., 2016). On the other hand, when delivered by the type III secretion system of the bacterium Burkholderia glumae, many putative U. virens effectors were found to suppress B. glumae‐induced hypersensitive cell death in N. benthamiana (Zhang et al., 2014). The U. virens secreted cysteine‐rich effector (SCRE) SCRE2 (UV_1261) has been recently demonstrated to be an essential virulence factor (Fan et al., 2019; Fang et al., 2019).
In this study, we demonstrate that the small SCRE candidate 1 (SCRE1) from U. virens not only inhibits hypersensitive cell death in the nonhost N. benthamiana but also suppresses pattern‐triggered immunity and enhances disease susceptibility in the host rice. When ectopically expressed in M. oryzae, SCRE1 is secreted and accumulated in the biotrophic interfacial complexes during infection, suggesting that SCRE1 is a cytoplasmic effector. The study identifies SCRE1 as an essential virulence effector in U. virens.
RESULTS
SCRE1 is a secreted protein in U. virens
It was predicted that UV_2799 encodes a putative secreted cysteine‐rich effector (named thereafter SCRE1) in U. virens (Zhang et al., 2014). We first used a yeast secretion system to verify the functionality of the SCRE1 signal peptide (Fang et al., 2016; Jacobs et al., 1997). The predicted signal peptide‐encoding sequence of SCRE1 was fused in frame with a truncated SUC2 gene that encodes invertase but lacks its own signal peptide. When the fusion construct was transformed into the YTK12 strain, the yeast strain restored the ability to grow on YPRAA medium (Fig. 1A). The N‐terminal sequences of Avr1b and Mg87 were used as positive and negative controls, respectively. The results indicate that the predicted SCRE1 signal peptide is functional in yeast and can guide the secretion of invertase. Furthermore, pCAMBIA1301‐RP27::SCRE1‐HA was transformed into the U. virens strain P1 to express SCRE1‐HA under the fungal RP27 promoter (Bruno et al., 2004). HA‐tagged β‐tubulin (UV_1410‐HA), which is a major component of the eukaryotic cytoskeleton, was also expressed in U. virens. The results from immunoblotting showed that SCRE1‐HA was detected in total cell lysate and in the culture medium, while β‐tubulin‐HA was only detected in total cell lysate, but not in the culture medium (Fig. 1B). More convincingly, the native β‐actin was also only detectable in total cell lysates, but not in the culture medium of the transformed strains, indicating that no cytoplasmic protein was leaking into the culture medium (Fig. 1B). These results confirmed that SCRE1 is a secreted protein in U. virens.
Figure 1.
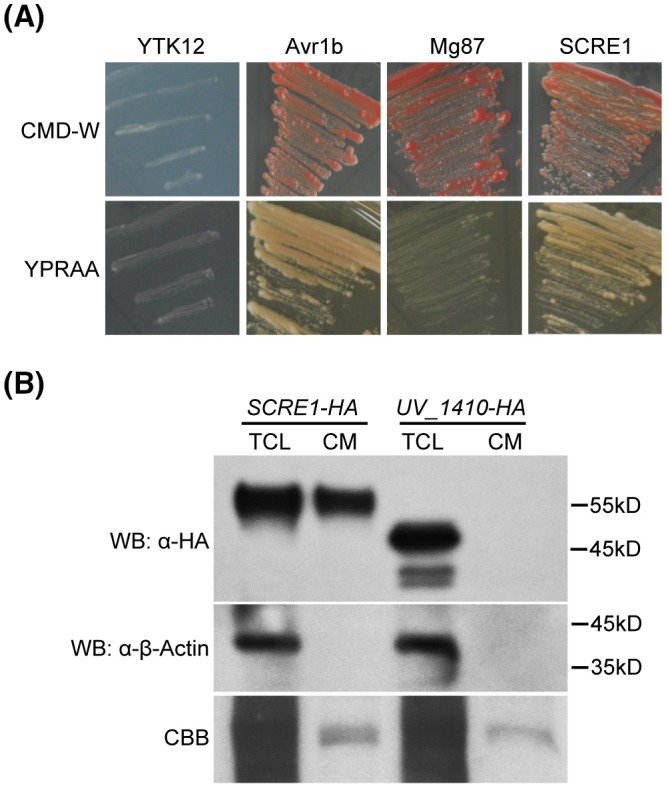
SCRE1 is a secreted protein in Ustilaginoidea virens. (A) The putative signal peptide of SCRE1 is functional to guide invertase secretion into culture medium (CM). YTK12 is an invertase secretion‐deficient yeast strain. The secretion of invertase is indicated by the growth of YTK12 on YPRAA plates with raffinose as sole carbon source. The signal peptide of Avr1b and the first 25 amino acids of Mg87 were used for positive and negative controls, respectively. (B) SCRE1‐HA was detected in CM and in the total cell lysate (TCL) via western blot (WB) analysis. The non‐secreted β‐tubulin protein UV_1410‐HA and β‐actin, as negative controls, were only detected in the TCL, but not in the CM. α‐HA, anti‐haemagglutinin antibody; α‐β‐actin, anti‐β‐actin antibody. CCB, Coomassie brilliant blue staining; SCRE1‐HA, U. virens transformed with pCAMBIA1301‐RP27::SCRE1‐HA; UV_1410‐HA, U. virens transformed with pCAMBIA1301‐RP27::UV_1410‐HA.
Ectopically expressed SCRE1 in M. oryzae is translocated into plant cells during infection
Generally, effector genes are transcriptionally regulated during pathogen infection. To investigate the expression pattern of SCRE1, the transcript level of SCRE1 was quantified at different time points after inoculation via quantitative reverse transcription‐PCR (RT‐qPCR) analyses. SCRE1 exhibited a strong up‐regulated expression pattern during U. virens infection in the susceptible rice variety LYP9, with a peak of expression at 5 days after inoculation and a gradual decrease thereafter (Fig. 2A).
Figure 2.
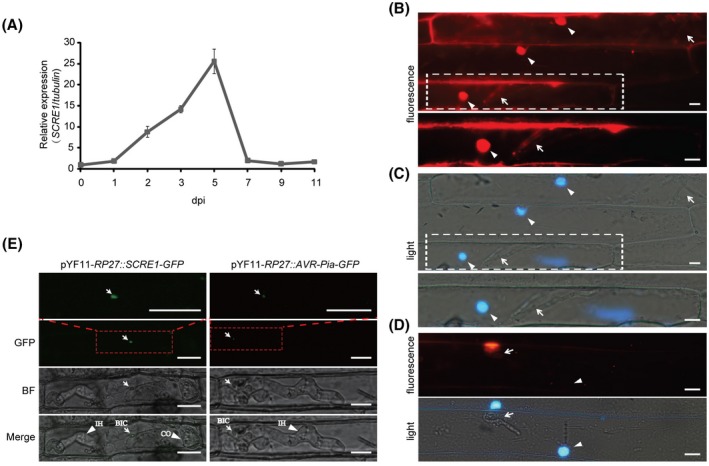
SCRE1 is an effector in Ustilaginoidea virens as revealed by expression and translocation assays. (A) SCRE1 expression was dramatically induced and peaked at 5 days post‐inoculation (dpi) and gradually decreased thereafter during U. virens infection. SCRE1 expression was normalized to the internal reference tubulin gene. (B), (C) Red fluorescence was clearly observed in barley cell nuclei and invasive hyphae of Magnaporthe oryzae. The fluorescence (B) and light (C) microscopy images were taken at 30 h after barley leaves were inoculated with M. oryzae carrying pKS‐RP27::SCRE1‐mCherry‐NLS. The barley nuclei stained with DAPI are indicated by arrowheads, while M. oryzae invasive hyphae are indicated by arrows. The images in lower panels are enlarged from the regions in broken squares in upper panels. (D) Red fluorescence was only observed in M. oryzae hyphae, but not in barley nuclei. The fluorescence (upper panel) and light (lower panel) microscopy images were taken 30 h after barley leaves were inoculated with M. oryzae carrying pKS‐RP27::mCherry‐NLS. The nuclei stained with DAPI are indicated by arrowheads, while M. oryzae hyphae are indicated by arrows. (E) Green fluorescence was clearly observed in biotrophic interfacial complexes during the infection of M. oryzae strains transformed with pYF11‐RP27::SCRE1‐GFP and pYF11‐RP27::AVR‐Pia‐GFP. The transformed M. oryzae Guy11 strains were inoculated into rice sheaths. The images were captured by confocal microscopy 30 h after inoculation. The images in the uppermost panels are enlarged from the regions in broken squares in the GFP panels. GFP, green fluorescent protein; BF, bright field; Merge, the overlay of GFP and BF images; IH, invasive hyphae; CO, conidia; BIC, biotrophic interfacial complex. Scale bar: 10 μM.
Next, living cell imaging was used to investigate whether SCRE1 is secreted and translocated into plant cells using a heterologous system. SCRE1 carrying a nuclear localization signal (NLS) from simian virus large T‐antigen (Ai et al., 2007) was ectopically expressed as a fusion with mCherry in M. oryzae driven by the RP27 promoter. The conventional NLS is sufficient for nuclear localization of fusion proteins and thus facilitates visualization of mCherry fluorescence (Park et al., 2012). The engineered M. oryzae strains were inoculated onto detached barley leaves. Red fluorescence was detectable inside the invasive hyphae of mCherry‐labelled M. oryzae by microscopy at 30 h after inoculation (Fig. 2B,C). The majority of barley epidermal cells infected by SCRE1‐mCherry‐NLS‐transformed M. oryzae exhibited red fluorescence in the nuclei at 30 h after inoculation (Fig. 2B,C). However, red fluorescence was only observed in the hyphae of mCherry‐NLS expressing M. oryzae during infection (Fig. 2D).
Cytoplasmic effectors in M. oryzae are generally accumulated in a novel structure called the biotrophic interfacial complex (BIC), where some effectors are translocated into rice cytoplasm during infection (Khang et al., 2010). To further investigate SCRE1 translocation in M. oryzae, the Guy11 strain was transformed with pYF11‐RP27::SCRE1‐GFP and then inoculated into detached rice sheaths. At 30 h after inoculation of M. oryzae, green fluorescence from SCRE1‐GFP was clearly observable in BICs by confocal microscopy (Fig. 2E). As a positive control, green fluorescence was visible in the BICs after the M. oryzae strain expressing AVR‐Pia‐GFP was inoculated into rice sheaths (Fig. 2E). Collectively, these data indicate that SCRE1, when expressed in M. oryzae, is secreted and translocated through BICs during infection, and thus is most likely a cytoplasmic effector.
SCRE1 is required for full virulence of U. virens to rice
To investigate whether SCRE1 plays a role in U. virens virulence, we generated the scre1 mutants using a modified CRISPR/Cas9 system. Five, six and five gene replacement mutants were preliminarily identified out of 32, 18 and 18 transformants via PCR screening using three single guide (sg) RNAs targeting different regions of SCRE1, respectively. Four independent mutants, scre1‐1‐10, scre1‐1‐26, scre1‐2‐4 and scre1‐3‐23, were confirmed by Southern blot analysis (Fig. S1A). The scre1‐1‐10, scre1‐2‐4 and scre1‐3‐23 mutants generated from different sgRNAs were subsequently injected into rice panicles before heading for virulence assays. The disease incidence rate of panicles inoculated with the wild‐type P1 strain was much higher than those inoculated with the three mutants. Consistently, the number of diseased grains per P1‐inoculated panicle was significantly greater than that caused by these mutants (Figs 3 and S1B). Furthermore, the complementary strain was created by introducing the plasmid‐borne SCRE1 gene into the scre1‐10 mutant. Inoculation assays showed that the virulence of the complementary strain was partially restored (Fig. 3). These data indicate that SCRE1 is required for the virulence of U. virens.
Figure 3.
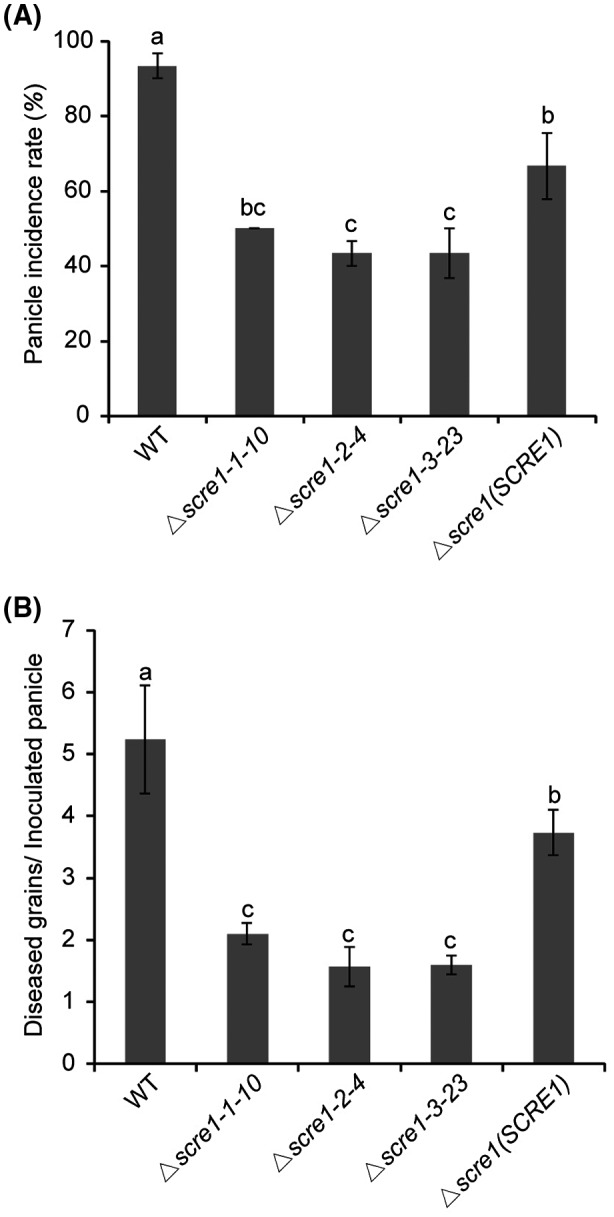
SCRE1 is required for full virulence of Ustilaginoidea virens to rice. The panicle incidence rate (%) (A) and average diseased grains per inoculated panicle (B) after injection inoculation of the wild‐type, different scre1 mutant and complementary strains in U. virens. The complementary strain was generated through transforming plasmid‐borne full‐length SCRE1 gene into the scre1‐1‐10 mutant. Different letters above error bars indicate statistically significant differences among the wild‐type, scre1 mutant and complementary strains of U. virens. Data shown are combined from three independent inoculation experiments. Error bars represent means ± standard error (SE).
SCRE1 suppresses immunity‐associated hypersensitive cell death in N. benthamiana
To determine the immunosuppressive ability of SCRE1, we demonstrated here that B. glumae carrying pEDV‐SCRE1‐HA caused a considerably reduced nonhost cell death response in N. benthamiana leaves compared with the wild‐type B. glumae (Fig. S2A). Immunoblotting assays showed that SCRE1‐HA was expressed in B. glumae and secreted into culture medium when bacteria were cultured in the type III secretion‐inducing minimal medium (Fig. S2B). This result suggests that the delivery of SCRE1 via the bacterial type III secretion system into plant cells can suppress B. glumae‐triggered hypersensitive responses (HR) in N. benthamiana. To elaborate this result, we demonstrated that transient expression of SCRE1 in N. benthamiana inhibited hypersensitive cell death triggered by the Phytophthora infestans elicitor INF1 (Fig. 4A) (Kamoun et al., 1998). Similarly, SCRE1 also suppressed cell death in N. benthamiana induced by the mammalian protein BAX, which has been shown to cause HR in numerous plant species (Lam et al., 2001; Oltval et al., 1993) (Fig. 4A). Furthermore, co‐expression of SCRE1 without signal peptide also suppressed INF1‐ and BAX‐triggered HR in N. benthamiana (Fig. 4A). Electrolyte leakage in N. benthamiana triggered by INF1 and BAX was significantly attenuated in the presence of SCRE1‐FLAG compared with co‐expression with FLAG‐tagged green fluorescent protein (GFP‐FLAG) (Fig. 4B). Western blot analysis showed that BAX‐FLAG was well produced when co‐expressed with SCRE1‐FLAG and GFP‐FLAG in N. benthamiana leaves (Fig. 4C). We further investigated subcellular localization of SCRE1/SCRE1‐SP when mCherry‐labelled proteins were transiently expressed in N. benthamiana cells. After plasmolysis, neither SCRE1‐mCherry nor SCRE1‐SP‐mCherry was visible in periplasmic spaces (Fig. S3). As controls, SCRE2‐mCherry was clearly observed in periplasmic spaces of plasmolysed cells, while SCRE2‐SP‐mCherry was not (Fig. S3; Fang et al., 2019). Collectively, these data suggest that SCRE1, as a secreted protein, is taken up inside plant cells and functions as a suppressor of immunity‐associated cell death in plants.
Figure 4.
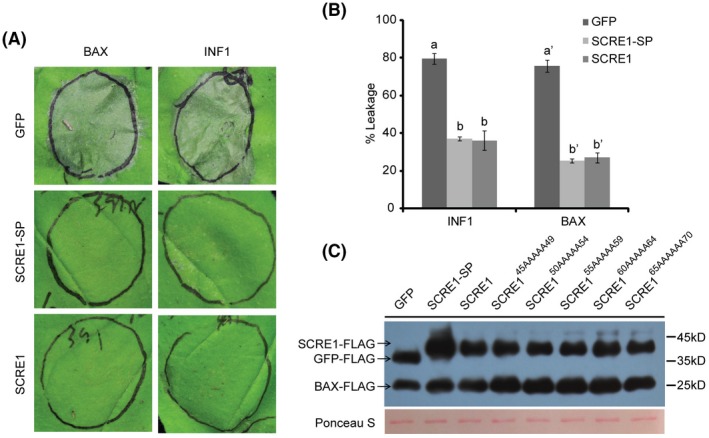
SCRE1 suppresses different types of immunity‐associated hypersensitive cell death in Nicotiana benthamiana. (A) Co‐expression of SCRE1 or truncated SCRE1 without signal peptide (SCRE1‐SP) suppressed BAX‐ (left panel) and INF1‐ (right panel) triggered hypersensitive cell death in N. benthamiana. (B) SCRE1/SCRE1‐SP expression, in contrast to green fluorescent protein (GFP) expression, significantly suppressed BAX‐ and INF1‐induced electrolyte leakage in N. benthamiana. Different letters over error bars indicate significant difference in BAX‐ and INF1‐induced electrolyte leakage of N. benthamiana leaves between SCRE1/SCRE1‐SP co‐expression and GFP co‐expression (P < 0.05). (C) SCRE1‐FLAG and GFP‐FLAG co‐expression imposed no evident effect on BAX‐FLAG expression in N. benthamiana leaves. Upper panel: the proteins isolated from the agroinfiltrated leaves were detected via immunoblotting with anti‐FLAG antibody (α‐FLAG). The bands of SCRE1‐FLAG, GFP‐FLAG and BAX‐FLAG are indicated by arrows. Lower panel: protein loading is indicated by Ponceau S staining.
SCRE1 suppresses pattern‐triggered immunity and promotes pathogen infection in rice
To investigate whether SCRE1 suppresses rice immunity, we generated 22 transgenic rice lines with dexamethasone (DEX)‐inducible expression of SCRE1. Three transgenic lines, Z178, Z195 and Z196, with strong DEX‐induced expression and no detectable basal expression of SCRE1, were chosen for further studies (Fig. S4A). Chitin and flg22 triggered a strong and rapid generation of ROS in the wild‐type, Z195 and Z196 transgenic rice plants without DEX induction. The induced oxidative burst was significantly suppressed in these transgenic plants after DEX treatment. In contrast, DEX treatment had no effect on flg22‐ and chitin‐triggered ROS burst in the wild‐type plants (Figs 5A,B and S4B–E), indicating that SCRE1 inhibits microbe‐associated molecular pattern (MAMP)‐triggered oxidative burst. Quantitative RT‐PCR assays showed that expression of the defence marker genes OsPR10a and OsPR1b was strongly up‐regulated by flg22 and chitin in the wild‐type, Z178 and Z196 transgenic seedlings without DEX treatment. By contrast, flg22‐ and chitin‐induced expression of these defence‐related genes was significantly suppressed in the DEX‐treated transgenic lines Z178 and Z196 (Figs 5C,D and S4F‐G). Thus, transgenically expressed SCRE1 suppresses rice pattern‐triggered immunity.
Figure 5.
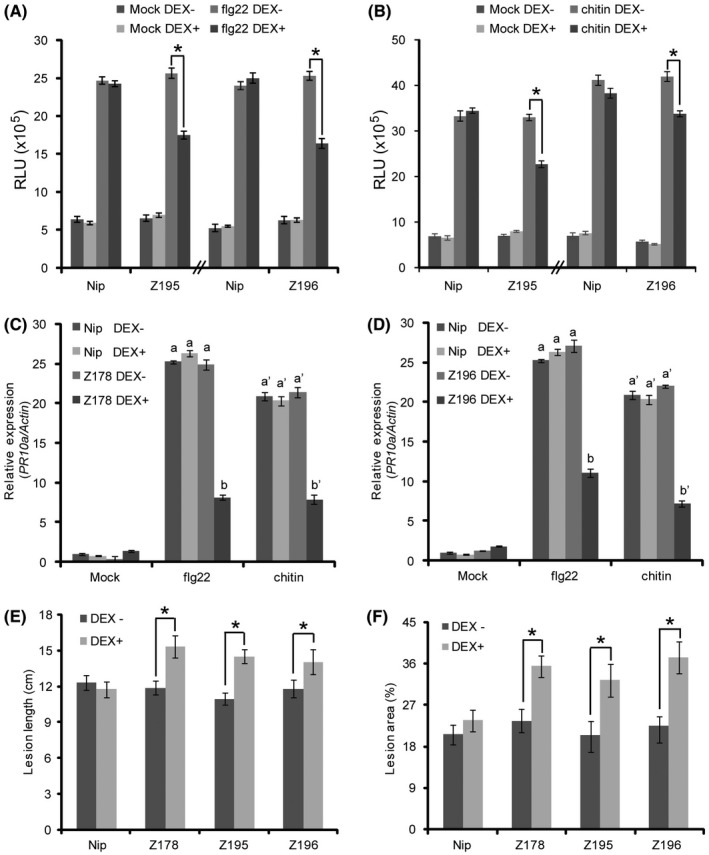
Ectopic expression of SCRE1 suppresses pattern‐triggered immunity and promotes pathogen infection in rice. (A), (B) Dexamethasone (DEX)‐induced expression of SCRE1 inhibited the oxidative burst triggered by flg22 (A) and chitin (B) in the Z195 and Z196 transgenic rice plants. Leaf disks collected from the wild‐type, Z195 and Z196 transgenic plants were immersed in DEX (10 µM) and mock solution for 16 h followed by treatment of flg22 (10 µM) and hexa‐N‐acetyl‐chitohexaose (8 µM). The area under the curve for 30‐min oxidative burst (relative luminescence units × time, see Fig. S4B–E) was determined for the wild‐type and transgenic plants after different treatments. Asterisks indicate statistical significance in oxidative burst from flg22‐ and chitin‐treated transgenic rice leaves with and without DEX treatment (P < 0.05). Histogram shows means ± SE for nine leaf disks from different plants. RLU, relative luminescence unit. (C), (D) DEX‐induced expression of SCRE1 significantly inhibited OsPR10a expression triggered by flg22 and chitin in the Z178 (C) and Z196 (D) transgenic lines. The wild‐type and transgenic seedlings were treated with DEX (10 µM) and mock solution for 24 h followed by flg22 (1 µM) and chitin (10 µg/mL). OsPR10a expression was detected by RT‐qPCR and normalized to the reference gene OsActin. Different letters above error bars indicate statistically significant differences in the OsPR10a transcript levels in the wild‐type and transgenic lines under different treatments (P < 0.05). (E), (F) SCRE1 expression in the transgenic lines significantly promoted pathogen infection after inoculation of rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae (E) and rice blast fungus Magnaporthe oryzae (F). The wild‐type and transgenic plants were challenged with pathogens at 24 h after spraying with DEX (10 µM) and mock solution. At least 12 leaves were measured for lesion lengths at 12 days after bacterial inoculation. Lesion areas on 20 inoculated leaves were photographed at 6 days after fungal inoculation and calculated using Abode Photoshop. Asterisks indicate statistically significant differences in disease lesions on the inoculated leaves with and without DEX treatment. Error bars represent means ± SE. Nip, Nipponbare; Z178, Z195 and Z196, three independent transgenic lines; DEX−, no dexamethasone treatment; DEX+, with dexamethasone treatment.
To investigate whether SCRE1 affects disease susceptibility in rice, the transgenic lines Z178, Z195 and Z196 were infected by Xanthomonas oryzae pv. oryzae PXO99 (a major bacterial pathogen of rice) or M. oryzae strain S5 (a major fungal pathogen of rice). The three transgenic lines after DEX treatment all exhibited significantly longer disease lesions on PXO99‐inoculated leaves than the same transgenic lines without DEX treatment and the wild‐type plants (Figs 5E and S4H). Similarly, M. oryzae inoculation caused significantly greater disease lesions on the leaves of DEX‐treated transgenic lines than on the mock‐treated wild‐type and transgenic plant leaves (Figs 5F and S4I). These results demonstrated that transgenic expression of SCRE1 compromises rice basal defences against both bacterial and fungal pathogens.
SCRE145‐70 is an essential fragment for the virulence function of SCRE1
SCRE1 contains no known conserved functional domain but two types of tandem and almost identical repeats, each repeat containing two Cys residues (Fig. S5A). To delineate the region in SCRE1 that is essential for its virulence ability, a series of truncated SCRE1 variants were expressed in N. benthamiana (Fig. S5B). A small peptide that contains the 45–70th amino acid residues (SCRE145‐70) and the truncated SCRE1 proteins including this fragment had a similar ability as full‐length SCRE1 to suppress INF1‐ or BAX‐induced hypersensitive responses and/or electrolyte leakage in N. benthamiana (Figs 6A and S5B). In contrast, other small peptides that contain only part of this fragment, such as SCRE150‐70 and SCRE145‐65, largely lost the HR‐suppressive ability in N. benthamiana. Furthermore, alanine‐scanning mutagenesis was performed to generate five multiple‐point mutations in the 45–70 amino acid region of SCRE1 (Fig. 6B). Transient expression of SCRE145AAAAA49 (the 45CPARS49 motif replaced by AAAAA) lost the ability to suppress BAX‐ and INF1‐induced ion leakage, while the other four mutant proteins retained the immunosuppressive ability in N. benthamiana (Fig. 6C,D). These mutant proteins were well expressed in N. benthamiana (Fig. 4C). To further confirm these findings, we purified the truncated SCRE11‐179 from Escherichia coli (full‐length SCRE1 was not well expressed in E. coli). Infiltration of purified SCRE11‐179 into N. benthamiana leaves was able to suppress BAX‐ or INF1‐triggered cell death, while purified SCRE11‐179(45AAAAA49) did not (Fig. 6E). Consistently, SCRE11‐179 infiltration significantly alleviated BAX‐ or INF1‐triggered electrolyte leakage from N. benthamiana leaves, while SCRE11‐179(45AAAAA49) did not (Fig. 6F). Taken together, these data define the 45–70 amino acid residues of SCRE1, particularly the 45CPARS49 motif, as the essential domain for the virulence function of SCRE1.
Figure 6.
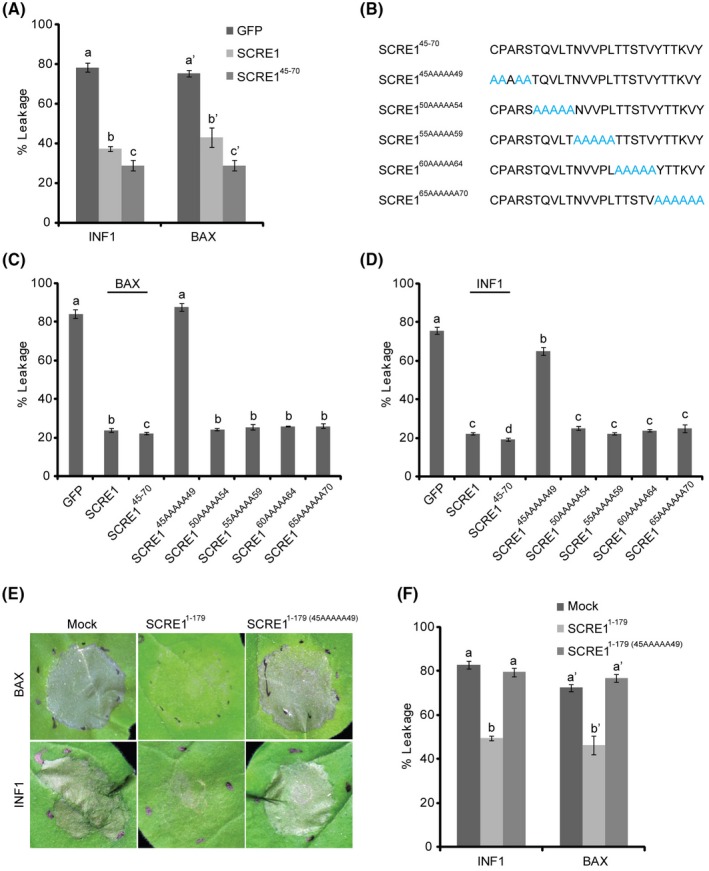
SCRE145‐70 is an essential region that suppresses BAX‐ and INF1‐triggered cell death in Nicotiana benthamiana. (A) Transient expression of SCRE145‐70 had an even stronger ability than full‐length SCRE1 to inhibit BAX‐ and INF1‐triggered electrolyte leakage in N. benthamiana. BAX and INF1 were transiently expressed at 6 h after expression of green fluorescent protein (GFP), SCRE1 and SCRE145‐70 in N. benthamiana. Letters above error bars indicate statistically significant differences in BAX‐ and INF1‐triggered electrolyte leakage when co‐expressing with different proteins in N. benthamiana. (B) Diagrams of alanine‐scanning mutagenesis in the 45–70th residues of full‐length SCRE1. (C), (D) SCRE145AAAAA49, in which the 45–49th residues of SCRE1 were all replaced with Ala, significantly lost the ability to suppress BAX‐ (C) and INF1‐ (D) induced ion leakage in N. benthamiana. Different letters (a–d) indicate statistically significant differences in BAX‐ and INF1‐triggered electrolyte leakage in N. benthamiana. BAX and INF1 were expressed at 6 h after expression of the indicated proteins. (E), (F) The Ala replacement in the 45‐49th residues of SCRE11‐179 greatly disrupted the ability of in vitro‐purified SCRE11‐179 to suppress BAX‐ and INF1‐induced hypersensitive response symptoms (E) and ion leakage (F). In vitro‐purified proteins (1 µM) were co‐infiltrated with Agrobacterium strains carrying pGR107‐BAX and pGR107‐INF1 constructs into N. benthamiana leaves. Hypersensitive response‐like symptoms and electrolyte leakage in the infiltrated leaves were investigated at 5 days post‐infiltration.
To investigate whether purified SCRE1 has the ability to suppress rice immunity, flg22‐ and chitin‐triggered immunity was examined in SCRE11‐179‐pretreated rice seedlings. As expected, expression of OsPR10a and OsPR1b was induced by flg22 and chitin in mock‐pretreated rice seedlings. In contrast, flg22‐ and chitin‐triggered expression of OsPR10a and OsPR1b was significantly attenuated in SCRE11‐179‐pretreated seedlings, while SCRE11‐179(45AAAAA49) pretreatment had no effect on flg22‐ or chitin‐induced expression of OsPR10a and OsPR1b (Fig. 7A,B). Furthermore, flg22‐ and chitin‐triggered MAPK activation in rice seedlings was also greatly dampened with SCRE11‐179 pretreatment, but not with SCRE11‐179(45AAAAA49) (Fig. 7C). Collectively, these results showed that purified SCRE11‐179 suppresses pattern‐triggered rice immunity and that the 45CPARS49 motif is critical for its suppression activity.
Figure 7.
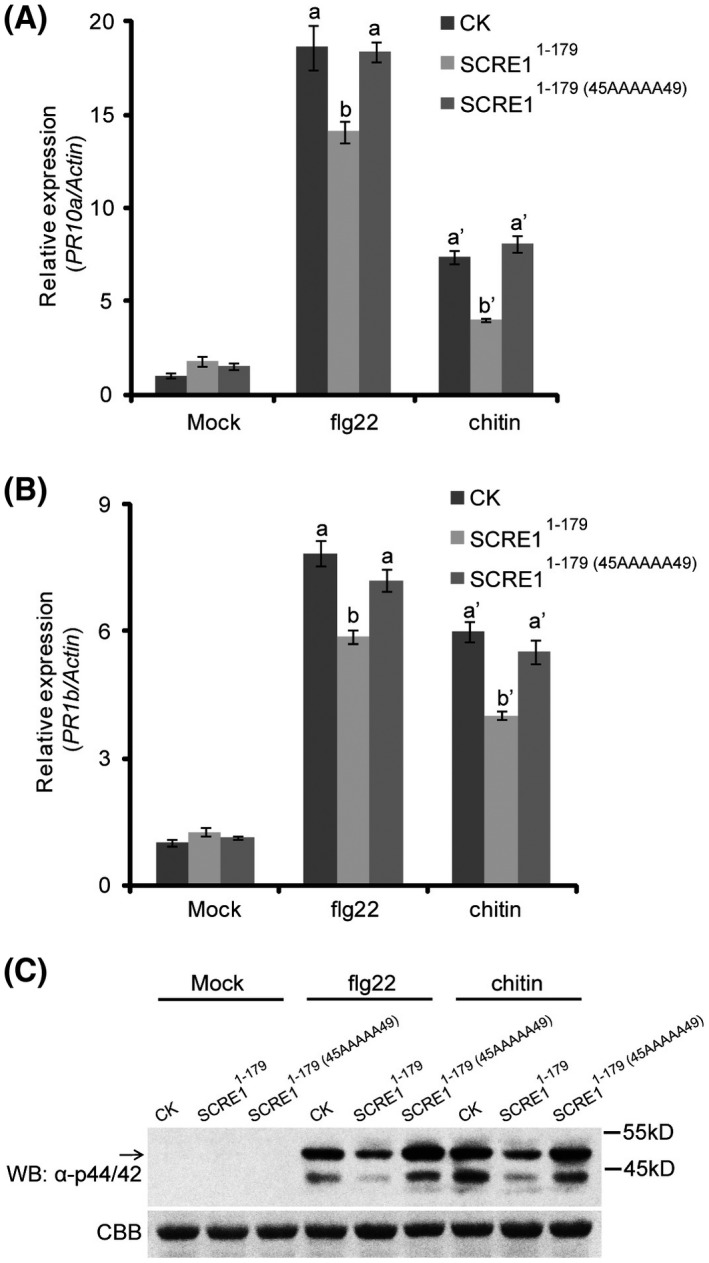
SCRE1 compromises pattern‐triggered immunity in rice. (A), (B) The flg22‐ and chitin‐induced expression of OsPR10a (A) and OsPR1b (B) was significantly attenuated in SCRE11‐179‐pretreated rice seedlings, but not in mock‐ and SCRE11‐179(45AAAAA49)‐pretreated seedlings. Rice seedlings were incubated with 1 µM of SCRE11‐179, SCRE11‐179(45AAAAA49) or mock solution for c. 12 h followed by treatment with flg22 (1 µM) or chitin (10 µg/mL) for 6 h. The expression level of OsPR10a and OsPR1b was detected by quantitative RT‐PCR and normalized to that of the reference gene OsActin. Different letters above error bars indicate significant differences in the transcript levels of PR genes among SCRE11‐179, SCRE11‐179(45AAAAA49) and mock treatments. (C) The flg22‐ and chitin‐induced MAPK phosphorylation was greatly inhibited in the SCRE11‐179‐pretreated rice seedlings, but not in the SCRE11‐179(45AAAAA49)‐pretreated seedlings. The SCRE11‐179‐pretreated seedlings were treated by flg22 and chitin for 15 min and were then collected for protein extraction. MAPK activation was detected through immunoblotting with anti‐Phospho‐p44/42 MAPK antibody (α‐p44/42). Lower panel: protein loading is indicated by Coomassie brilliant blue (CBB) staining.
DISCUSSION
Rice false smut is an emerging global disease and poses significant threats to rice production and human health. Currently, little is known about virulence functions of U. virens effectors (Rush et al., 2000; Tang et al., 2013; Zhang et al., 2014). Effectors play essential roles in fungal virulence (Lo Presti et al., 2015; Oliveira‐Garcia and Valent, 2015). However, despite genomics‐based predictions, few effectors have been functionally characterized in U. virens (Fang et al., 2016). In this study, we identified an essential virulence effector, SCRE1, in U. virens and provided evidence that this effector suppresses rice immunity.
In U. virens, 193 secreted proteins have been predicted to be potential effectors based on sequence and structural features (Zhang et al., 2014). Through a large‐scale high‐throughput screen, many putative U. virens effectors, including SCRE1, were previously shown to suppress B. glumae‐induced HR‐like cell death in the nonhost plant N. benthamiana (Zhang et al., 2014). In this study, we showed that SCRE1 also has the ability to inhibit HR‐like cell death induced by BAX and INF1 in N. benthamiana (Fig. 4). Furthermore, transgenically expressed SCRE1 compromises the expression of defence‐marker genes and the oxidative burst triggered by flg22 and chitin in rice plants (Fig. 5A–D) and the SCRE1 transgenic plants were more susceptible to both bacterial and fungal pathogens (Fig. 5E,F). These results are consistent with the hypothesis that SCRE1 is a fungal effector that is involved in immunosuppression. SCRE1 has other features that are characteristic of a genuine fungal effector protein: it is transcriptionally induced during infection and secreted by U. virens (Figs 1 and 2). In addition, red fluorescence from SCRE1‐mCherry‐NLS was clearly visible in barley nuclei after infection by M. oryzae with heterologous expression of SCRE1‐mCherry‐NLS, indicating that SCRE1 is translocated into plant cells after secretion (Fig. 2B,C). Many cytoplasmic effectors in M. oryzae, such as BAS1, PWL2 and AVR‐Pia, have been demonstrated to be secreted into the BIC structure and translocated to rice cytoplasm during M. oryzae infection (Khang et al., 2010; Mosquera et al., 2009; Sornkom et al., 2017; Yoshida et al., 2009). Here, we showed that ectopically expressed SCRE1‐GFP in M. oryzae was accumulated in the BIC structure during infection of rice cells (Fig. 2E), indicating that SCRE1 is a cytoplasmic effector. In oomycetes, the RxLR‐dEER motifs are sufficient for transit of cytoplasmic effectors into host cells (Dou et al., 2008). However, autonomous translocation motifs for fungal effectors are still mysterious, although some cytoplasmic fungal effectors have been demonstrated to be transported into plant cells (Kale et al., 2010; Manning and Ciuffetti, 2005; Rafiqi et al., 2010). Furthermore, multiple independent scre1 mutants of U. virens all exhibited a significant reduced virulence to rice and the plasmid‐borne full‐length SCRE1 gene significantly restored the virulence of the mutant (Fig. 3), indicating that SCRE1 is required for the virulence of U. virens. Taken together, we conclude that SCRE1 is an important cytoplasmic effector with virulence functions in U. virens.
Interestingly, we revealed that the truncated SCRE1 without its signal peptide retained HR‐suppressive ability (Fig. 4A). In contrast, our previous study demonstrated that signal peptides of the U. virens secreted proteins that trigger cell death were all required for their HR‐inducing ability. The difference is probably because the HR‐inducing and immunosuppressive effectors might function in different locations. Similar to the HR‐inducing effectors in M. oryzae (Chen et al., 2013), HR‐inducing effectors in U. virens likely function in apoplastic spaces while cytoplasmic effectors including SCRE1 might be transported inside host cells after they are secreted into periplasmic spaces by fungi. This speculation is supported by the subcellular localization assay of SCRE1‐mCherry, demonstrating that red fluorescence from SCRE1‐mCherry was localized predominantly inside the cells and was hardly visible in periplasmic spaces of plasmolysed N. benthamiana cells (Fig. S3). The purified SCRE11‐179 had the ability to inhibit BAX‐ and INF1‐triggered HR‐like symptoms in nonhost N. benthamiana and to compromise pattern‐triggered immunity in host rice seedlings (Figs 6E,F and 7). This finding indicates that SCRE1 in the extracellular matrix might be translocated inside the cells through an unknown mechanism. However, we cannot completely rule out the possibility of SCRE1 functioning as an apoplastic effector. Although no known conserved functional domain or motif was found in SCRE1, we discovered that SCRE1 possesses two types of Cys‐containing tandem repeats (Fig. S5A). Through a series of truncations of SCRE1, we were able to define the key immunosuppressive region of SCRE1. Remarkably, a small 26 amino acid peptide containing amino acid residues 45–70, SCRE145‐70, was found to be sufficient for the HR‐inhibitory ability in N. benthamiana (Figs 6 and S5). It is notable that transient expression of the small peptide SCRE145‐70 had an even stronger inhibition effect on INF1‐ and BAX‐induced cell death than full‐length SCRE1 (Fig. 6A). The flg22 peptide functions more effectively as an elicitor of defence responses than flagellin (Felix et al., 1999). We propose two explanations for this phenomenon: (i) small peptides are more easily accessible to plant cells, and (ii) functional peptides are somewhat covered by other fragments of full‐length proteins. It is well known that small peptides can induce plant immunity (Yamaguchi and Huffaker, 2011). However, to our knowledge, few studies have reported that a small peptide is able to disarm plant immunity. Furthermore, using site‐directed mutagenesis, we identified a 5‐amino‐acid motif, 45CPARS49, that is critical for the immunosuppressive function of SCRE1 (Fig. 6). Interestingly, the HR‐suppressive region (i.e. 45–70th amino acids) of SCRE1 encompasses mostly the sequence between the first tandem repeats (Fig. S5A). The finding suggests that the tandem repeats themselves in SCRE1 are not directly associated with its immunosuppressive activity. The role of these repeats, if any (e.g. in stabilizing SCRE1 protein in vivo), needs to be further investigated. Most recently, the conserved 14‐amino‐acid motif (PID14) in U. maydis Pit2 has been shown to function as an inhibitor of papain‐like cysteine proteases to suppress plant immunity (Villamil et al., 2019). In future studies, it would be worthwhile identifying host targets of SCRE1 and clarifying whether the immunosuppressive peptide in SCRE1 functions in a similar way to PID14 as an enzyme inhibitor.
EXPERIMENTAL PROCEDURES
Plant materials, bacterial and fungal strains
Oryza sativa subsp. japonica 'Nipponbare', N. benthamiana and Hordeum sativa plants were grown in the greenhouse or in the growth chamber. Bacterial and fungal strains are listed in Table S1. E. coli strains were grown at 37 °C in Luria–Bertani (LB) medium. The other bacteria and fungi used were grown at 28 °C. B. glumae and A. tumefaciens strains were cultured in LB medium, X. oryzae pv. oryzae PXO99 in nutrient broth, M. oryzae in oatmeal‐tomato paste medium, U. virens isolates P1 in potato sucrose medium, and yeast strains in yeast extract‐peptone‐dextrose medium, unless specifically noted. The PCR primers used in this study are listed in Table S2.
Agrobacterium‐mediated transient assays in N. benthamiana
SCRE1 fragments were amplified using the primers in Table S2. The pGR107 constructs (Jones et al., 1999) were transformed into A. tumefaciens strains using the freeze‐thaw method (An et al., 1988). Overnight‐cultured A. tumefaciens cells were collected and resuspended in the infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, 150 μM acetosyringone). After being incubated at room temperature for 2 h, A. tumefaciens cells were pressure‐infiltrated into the leaves of 4–6‐week‐old N. benthamiana plants at an OD600 of 0.2 for INF1 and BAX expression or OD600 = 0.5 for other protein expression. A. tumefaciens strains carrying pGR107‐INF1 or pGR107‐BAX were infiltrated together with different truncated constructs of SCRE1 or at 6 h after infiltration of different SCRE1 constructs. Subcellular localization of SCRE1‐mCherry and SCRE1‐SP‐mCherry was performed as described previously (Fang et al., 2019).
Electrolyte leakage measurement
Electrolyte leakage in the inoculated N. benthamiana leaves was measured as described previously (Fang et al., 2016).
The yeast secretion assay
The yeast invertase secretion assay was performed using the SP trap vector pSUC2 as previously described (Jacobs et al., 1997). Briefly, the signal sequence of SCRE1 was PCR‐amplified and subcloned into pSUC2. The pSUC2‐SCRE1(SP) construct was transformed into the invertase‐deficient yeast strain YTK12 (SUC2−) as described previously (Jacobs et al., 1997). The transformants were cultured on CMD‐W (0.67% yeast N base, 0.075% tryptophan drop‐out supplement, 2% sucrose, 0.1% glucose, 2% agar) or YPRAA (1% yeast extract, 2% peptone, 2% raffinose, 2 μg/L antimycin A, 2% agar) plates for invertase secretion assays.
Agrobacterium‐mediated transformation of U. virens
The fungal promoter RP27 (Bruno et al., 2004) was PCR‐amplified and cloned into pCAMBIA1301. The 3 × HA coding sequence was amplified from pGWB14 and cloned into pCAMBIA1301‐RP27. The SCRE1 coding sequence was amplified and ligated into pCAMBIA1301‐RP27‐3 × HA. The construct was then transformed into A. tumefaciens strain AGL1. Agrobacterium‐mediated U. virens transformation was performed as described with some modifications (Mullins et al., 2001). Briefly, the U. virens P1 culture was filtered through lens paper. The conidia were collected from the filtrate by centrifugation. A. tumefaciens was cultured in LB medium at 28 °C for 12 h with shaking. A. tumefaciens cells were collected and resuspended in co‐cultivation medium to a cell density of OD600 ≈ 0.15, and were further cultured for 6 h. The culture was mixed with equal volume of P1 conidial suspension (106 spores/mL) and spread onto nitrocellulose membrane (Whatman, GE Healthcare, Amersham, UK) placed on co‐cultivation medium plates. After the plates had been incubated for 3 days, the nitrocellulose membrane was transferred to TB3 (3 g/L yeast extract, 3 g/L casamino acids, 2% sucrose, 1.2% agar) plates containing 100 μg/mL of hygromycin B for transformant selection. Individual colonies appearing after 5–6 days of incubation were transferred to TB3 for further identification.
SCRE1 secretion assays
B. glumae transformed with pEDV‐SCRE1‐HA was cultured in the hrp‐inducing SMMXC medium (Sun et al., 2011). The wild‐type U. virens and transformants carrying pCAMBIA1301‐RP27::SCRE1‐HA and pCAMBIA1301‐RP27::UV_1410‐HA were cultured in potato‐sucrose medium for 5–7 days with shaking. The culture medium were collected by consecutive filtrations through filter paper and a 0.45 µm filter. Proteins in the medium were precipitated by 10% trichloroacetic acid on ice overnight and then spun down (13,800 g, 30 min). The pellet was washed with acetone and air‐dried. The protein precipitants were dissolved in 1 × loading buffer and separated on SDS‐PAGE gels (12%). The proteins were electrophoretically blotted on Immobolin‐P membranes (Merck Millipore, Darmstadt, Germany), which were then probed with anti‐HA (Roche Diagnostics, Basel, Switzerland) and anti‐β‐actin (CWBio, Beijing, China) antibodies.
Generation of gene knockout and complementary strains in U. virens
To create gene‐replacement mutants in U. virens, polyethylene glycol (PEG)‐mediated protoplast transformation was performed in combination with a modified CRISPR/Cas9 system (Zheng et al., 2016). Briefly, the protoplasts were prepared from the mycelia after being incubated in lysis buffer (25 g/L driselase, 10 g/L lysing enzymes and 0.05 g/L lyticase in 1.2 M KCl). The protoplasts were rinsed and then resuspended in STC buffer (20% sucrose, 50 mM Tris‐Cl, pH 8.0, 50 mM CaCl2) to 108 protoplasts/mL. The upstream and downstream sequences (c. 1 kb) of SCRE1 were PCR‐amplified and were then fused to the 5ʹ and 3ʹ termini of hygromycin resistance gene by fusion PCR, respectively. Three pairs of SCRE1‐specific spacer primers for different sgRNA target sequences (Table S2) were designed following the instructions (http://www.rgenome.net/). These primers were annealed and inserted into the BsmBI‐digested entry vector pmCAS9‐tRp‐gRNA. Individual plasmid constructs and PCR products (5 µg each) were transformed into U. virens protoplasts as described (Li et al., 2019). The protoplasts were cultured for 12–14 days to screen transformants. In addition, the full‐length SCRE1 gene with the native promoter was amplified and cloned into pYP102 vector (Zhou et al., 2018). The vector was transformed into the scre1‐1‐10 mutant to construct the complementary strain.
Southern blot analysis
Southern blot analysis was performed using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) following the manufacturer’s instructions (Li et al., 2019).
The translocation assay
The pYF11‐RP27::SCRE1‐GFP construct was generated through homologous recombination in Saccharomyces cerevisiae (Qi et al., 2016b). The recombined plasmid was isolated using a yeast plasmid extraction kit (Zomanbio, Beijing, China) and was then transformed into E. coli DH5ɑ. After being confirmed by sequencing, the plasmid was transformed into M. oryzae Guy11 via PEG‐mediated protoplast transformation as described (Yang et al., 2010). M. oryzae conidial suspension (100 µL, 105 spores/mL) was inoculated on the inner leaf sheath cuticle cells. After 30–36 h incubation under humid conditions at 28 °C, the leaf sheaths were observed under confocal microscopy.
Plant transformation
For conditional expression of SCRE1, the SCRE1 open reading frame was amplified and cloned into pUC19‐RBS‐3 × FLAG. The open reading frame was released together with the 3 × FLAG coding sequence by XhoI and SpeI digestion, and was then ligated into pTA7001 (Aoyama and Chua, 1997). These constructs were transformed into A. tumefaciens EHA105. Agrobacterium‐mediated rice transformation was performed as described previously (Li et al., 2015; Lu et al., 2015).
The oxidative burst assay
The oxidative burst after MAMP treatment was detected in the leaf disks using luminol chemiluminescence assay with minor modifications (Schwacke and Hager, 1992; Park et al., 2012). Briefly, leaf disks were punched from 4‐week‐old wild‐type and transgenic plants and were then pre‐incubated in 10 µM DEX or mock solution for 16 h. To detect ROS generation, three leaf discs were soaked in 100 μL solution with luminol substrate, peroxidase and different MAMPs [10 µM flg22, 8 µM hexa‐N‐acetyl‐chitohexaose (Toronto Research Chemicals H290750), 10 µg/mL chitin (Sigma C9752), or sterile H2O as control]. Luminescence was recorded continuously at 1‐min intervals for 30 min with a GloMax 20/20 luminometer (Promega).
Rice defence response assays
Seven‐day‐old rice seedlings were pretreated with 10 μM DEX and mock solution overnight, and were then treated with 1 µM flg22, 10 µg/mL chitin or sterile H2O plus 0.01% Silwet L‐77 (GE Healthcare, Amersham, UK). The seedlings were collected at 6 h after treatment for detection of defence‐marker gene expression and at 15 min for MAPK activation assay. Total RNAs were isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) from the leaves of treated seedlings following the manufacturer’s instructions. The synthesis of cDNA was performed with the Reverse Transcription System (Takara, Dalian, China) using oligo‐d(T)18 as primers. The transcript levels of defence‐marker genes were analysed using the SYBR PrimeScript RT‐PCR kit (Takara) on an ABI PRISM 7500 system and normalized to the reference gene OsActin. The proteins were isolated from rice seedlings and MAPK activation was detected as described (Wang et al., 2015). Phospho‐serine/‐threonine was detected using anti‐Phospho‐p44/42 MAPK antibody (1:2000 dilution) (Cell Signaling Technology, Danvers, MA, USA) in 4 °C overnight, followed by anti‐rabbit‐HRP conjugated secondary antibody (CWBio) at 1:5000 dilution.
Bacterial inoculation assays
Resistance to bacterial blight was evaluated on 6‐week‐old rice plants using the leaf clipping method (Kauffman et al., 1973; Wang et al., 2018). The wild‐type and transgenic plants were sprayed with 10 µM DEX at 24 h before inoculation. The X. oryzae pv. oryzae strain PXO99A was cultured overnight and then resuspended with 10 mM MgCl2 to an OD600 of 0.8 for inoculation. Lesion lengths of the inoculated leaves (≥12 leaves, each line) were measured at 12–14 days after inoculation.
Fungal inoculation assays
The U. virens isolate P1 and mutant strains were inoculated into rice panicles of the false smut susceptible cv. LYP9 at the booting stage by injection (Han et al., 2015; Wang et al., 2008). M. oryzae S5 inoculation was performed as described previously (Lu et al., 2015; Peng and Shishiyama, 1988). At least 20 inoculated leaves were photographed at 5–6 days post‐inoculation and disease severity was scored by estimating lesion areas using Abode Photoshop (Adobe Systems, San Jose, CA, USA).
Recombinant protein expression and purification
SCRE11‐179 and its variants were generated via PCR using the primers in Table S2 and were subcloned into pET32b (His6‐tag). E. coli BL21 (DE3) cells (OD600 ≈ 0.6) transformed with individual constructs were incubated with 100 µM isopropyl β‐d‐thiogalactopyranoside for 3–5 h at 25 °C to induce protein expression. The His6‐tagged proteins were purified using Ni‐NTA beads (Novagen, Billerica, MA, USA) according to the manufacturer’s instructions. The protein concentration was determined by Pierce BCA Protein Assay Kit (Thermo, Rockford, IL, USA) and was also confirmed by SDS‐PAGE gels after staining with Coomassie brilliant blue.
Protein extraction and immunoblotting
Total proteins were isolated from the inoculated N. benthamiana leaves and the transfected protoplasts following the described procedure (Liu et al., 2016b; Sun et al., 2006). Briefly, the ground powder and the collected protoplasts were incubated in 1 mL of extraction buffer [50 mM Tris‐Cl, pH 7.4, 5 mM EDTA, 50 mM NaCl, 10% glycerol, 0.1% Triton X‐100, 20 mM dithiothreitol and 1% protease inhibitor cocktail (Sigma‐Aldrich, St Louis, MO, USA)] on ice for 1 h. The extracts were separated in a 12% polyacrylamide gel and then electrophoretically transferred onto Immobilon‐P membrane (Millipore, Bedford, MA, USA) as described (Wang et al., 2015). The membranes were subjected to immunoblotting with different antibodies.
Statistical analysis
All experiments were independently repeated at least three times with similar results unless specifically noted. Statistical significance was determined by one‐way ANOVA followed by Duncan’s multiple range test using SPSS software. Pairwise comparisons were performed by Student’s t‐test using Microsoft Excel.
Competing Interests
The authors declare no competing financial interests.
Supporting information
Fig. S1 Virulence assay to rice of the wild‐type, scre1 mutant and complementary strains. (A) The scre1 mutants were confirmed via Southern blot analyses. Genomic DNA isolated from the wild‐type (WT) strain, scre1‐3‐23, scre1‐2‐4, scre1‐1‐26 and scre1‐1‐10 mutant candidates was digested with SphI. After separation by gel electrophoresis, the digested DNA was blotted on Hybond‐N membrane. The membrane was hybridized with the probe that was PCR‐amplified and DIG‐labelled. The molecular markers are shown in bp on the left. (B) Disease symptoms on rice panicles after injection inoculation of the wild‐type P1, scre1‐1‐10 mutant and scre1(SCRE1) complementary strains. The images were taken at 4 weeks after inoculation.
Fig. S2 SCRE1 expressed and secreted from Burkholderia glumae suppresses nonhost cell death in Nicotiana benthamiana. (A) Infiltration of the rice bacterial pathogen B. glumae caused nonhost cell death in N. benthamiana leaves while the engineered B. glumae transformed with pEDV‐SCRE1‐HA did not induce cell death. B.g., B. glumae; B.g. (SCRE1), B. glumae transformed with pEDV‐SCRE1‐HA. (B) SCRE1‐HA was secreted by B. glumae in the SMMXC hrp‐inducing minimal medium under different pH conditions. Upper panel: SCRE1‐HA was detected in the cell pellet and culture medium by western blotting. SCRE1‐HA secretion was dramatically induced at pH 6.5 in the SMMXC medium. Lower panel: protein loading is indicated by Ponceau S staining. CM, culture medium; TCL, total cell lysate; WB, western blot; α‐HA, anti‐haemagglutinin antibody.
Fig. S3 Subcellular localization of SCRE1 and SCRE1‐SP in Nicotiana benthamiana cells. Red fluorescence was observed in the plasmolysed N. benthamiana cells transiently expressing mCherry, SCRE1/2‐mCherry and SCRE1/2‐SP‐mCherry via laser scanning confocal microscopy. Neither SCRE1‐mCherry nor SCRE1‐SP‐mCherry was visible in periplasmic spaces of plasmolysed N. benthamiana cells. In contrast, SCRE2‐mCherry was clearly observed in periplasmic spaces, while SCRE2‐SP‐mCherry was not. SCRE1/2‐SP, SCRE1/2 without signal peptides; mCherry panels, red fluorescence; zoom‐in panels, enlarged images from broken square areas in mCherry panels. Arrows indicate the periplasmic spaces after N. benthamiana cells were plasmolysed. Scale bar: 75 μm.
Fig. S4 Induced expression of SCRE1 suppresses microbe‐associated molecular pattern (MAMP)‐triggered immunity and promotes pathogen infection in transgenic rice plants. (A) SCRE1‐FLAG was conditionally expressed after dezamethasone (DEX) induction in three independent transgenic lines, Z178, Z195 and Z196, as detected by immunoblotting with anti‐FLAG antibody (α‐FLAG). Lower panel: protein loading is indicated by Coomassie brilliant blue (CBB) staining. Nip, Nipponbare; DEX, dexamethasone; –, no DEX treatment; +, with DEX treatment. (B–E) SCRE1 expression induced by DEX significantly suppressed reactive oxygen species (ROS) burst triggered by flg22 (B, D) and chitin (C, E) in the Z195 (B, C) and Z196 (D, E) transgenic lines. Leaf discs were collected from the wild‐type and transgenic lines at 24 h after spraying with 10 µM DEX and mock solution, and were treated with 10 µM flg22 and 8 µM hexa‐N‐acetyl‐chitohexaose. (F, G) DEX‐induced expression of SCRE1 significantly inhibited OsPR1b expression triggered by flg22 and chitin in the Z178 (F) and Z196 (G) transgenic lines. The wild‐type and transgenic lines were treated with 10 µM DEX and mock solution for 24 h followed by 1 µM flg22 and 10 µg/mL chitin. The expression of OsPR1b was detected by quantitative RT‐PCR and normalized to the reference gene OsActin. Different letters above error bars indicate significant differences in the transcript levels of OsPR1b in the wild‐type and transgenic lines under different treatments (P < 0.05). Error bars represent means ± SE. (H, I) Disease symptoms on the leaves of the wild‐type, Z178, Z195 and Z196 transgenic rice lines with and without DEX treatment after Xanthomonas oryzae pv. oryzae PXO99 inoculation (H) and after Magnaporthe oryzae inoculation (I). The images in H and I were photographed at 12 and 6 days after inoculation, respectively.
Fig. S5 The SCRE1 fragment including the 45–70th amino acid residues retains the ability to suppress BAX‐triggered cell death. (A) The derived protein sequences and structural features of SCRE1. The predicted signal peptide is highlighted in orange. The two types of tandem and almost identical repeats are shown in yellow and purple, respectively. The Cys residues are in bold. The 45–70th amino acid residues are underlined and highlighted in blue. (B) Different truncated SCRE1 proteins have distinct abilities to suppress BAX1‐triggered hypersensitive responses in Nicotiana benthamiana. The truncated proteins including the 45–70th residues all have hypersensitive response (HR)‐suppressive ability similar to the full‐length SCRE1. +, having the ability to suppress BAX‐induced cell death; –, having no HR suppressive ability. BAX was transiently expressed at 6 h after expression of GFP, SCRE1 and different truncated SCRE1 proteins in N. benthamiana. HR symptoms in the infiltrated leaves were observed and photographed at 5 days post‐infiltration.
Table S1 The strains used in this study
Table S2 The primers used in this study
Acknowledgments
We thank Li‐Jia Qu and Genji Qin at Peking University for the pOS‐sgRNA and pOS‐Cas9 vectors; Zhengguang Zhang at Nanjing Agricultural University for the yeast strain XK‐125, M. oryzae Guy11 and pYF11 vector; Zhaoxi Luo at Huazhong Agricultural University for the JS60‐2 isolate; Jinrong Xu at Purdue University for the pmCAS9‐tRp‐gRNA vector. The work is supported by National Natural Science Foundation of China grants 31630064 to W.S., the National Key Research and Development Program of China 2016YFD0300700 and the 111 project B13006 to Y.P.
References
- Ai, H. , Shaner, N.C. , Cheng, Z. and Tsien, R.Y . and Campbell, R.E. (2007) Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry, 46, 5904–5910. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- An, G. , Ebert, P.R. , Mitra, A. and Ha, S.B. (1988) Binary Vectors In: Plant Molecular Biology Manual (Gelvin S.B. and Schilperoort R.A., eds), pp. A3, 1–19. Dordrecht, Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Aoyama, T. and Chua, N.H. (1997) A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Bruno, K.S. , Tenjo, F. , Li, L. , Hamer, J.E. and Xu, J. (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea . Eukaryot. Cell, 3, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Songkumarn, P. , Venu, R.C. , Gowda, M. , Bellizzi, M. , Hu, J. , Liu, W. , Ebbole, D. , Meyers, B. , Mitchell, T. and Wang, G.L. (2013) Identification and characterization of in planta‐expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Mol. Plant‐Microbe Interact. 26, 191–202. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Shi, T. , Yang, J. , Shi, W. , Gao, X. , Chen, D. , Xu, X. , Xu, J. , Talbot, N.J. and Peng, Y. (2014) N‐glycosylation of effector proteins by an α‐1,3‐mannosyltransferase is required for the rice blast fungus to evade host innate immunity. Plant Cell, 26, 1360–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Djamei, A. , Schipper, K. , Rabe, F. , Ghosh, A. , Vincon, V. , Kahnt, J. , Osorio, S. , Tohge, T. , Fernie, A.R. , Feussner, I. , Feussner, K. , Meinicke, P. , Stierhof, Y.‐D. , Schwarz, H. , Macek, B. , Mann, M. and Kahmann, R. (2011) Metabolic priming by a secreted fungal effector. Nature, 478, 395–398. [DOI] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, H.P. , Bolton, M.D. , Stergiopoulos, I. , de Wit, P.J. and Thomma, B.P. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant‐Microbe Interact. 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Fan, J. , Du, N. , Li, L. , Li, G. , Wang, Y. , Zhou, Y. , Hu, X. , Liu, J. , Zhao, J. , Li, Y. , Huang, F. and Wang, W.M. (2019) A core effector UV_1261 promotes Ustilaginoidea virens infection via spatiotemporally suppressing plant defense. Phytopathol. Res. 1, 11. [Google Scholar]
- Fang, A. , Han, Y. , Zhang, N. , Zhang, M. , Liu, L. , Li, S. , Lu, F. and Sun, W. (2016) Identification and characterization of plant cell death–inducing secreted proteins from Ustilaginoidea virens . Mol. Plant‐Microbe Interact. 29, 405–416. [DOI] [PubMed] [Google Scholar]
- Fang, A. , Gao, H. , Zhang, N. , Zheng, X. , Qiu, S. , Li, Y. , Zhou, S. , Cui, F. and Sun, W. (2019) A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice. Front. Microbiol. 10, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Galán, J.E. and Collmer, A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Giraldo, M.C. and Valent, B. (2013) Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. [DOI] [PubMed] [Google Scholar]
- Han, Y. , Zhang, K. , Yang, J. , Zhang, N. , Fang, A. , Zhang, Y. , Liu, Y. , Chen, Z. , Hsiang, T. and Sun, W. (2015) Differential expression profiling of the early response to Ustilaginoidea virens between false smut resistant and susceptible rice varieties. BMC Genom. 16, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemetsberger, C. , Herrberger, C. , Zechmann, B. , Hillmer, M. and Doehlemann, G. (2012) The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8, e1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M. , Luo, L. , Wang, S. , Liu, Y. and Li, J. (2014) Infection processes of Ustilaginoidea virens during artificial inoculation of rice panicles. Eur. J. Plant Pathol. 139, 67–77. [Google Scholar]
- Jacobs, K.A. , Collins‐Racie, L.A. , Colbert, M. , Duckett, M. , Golden‐Fleet, M. , Kelleher, K. , Kriz, R. , LaVallie, E.R. , Merberg, D. , Spaulding, V. , Stover, J. , Williamson, M.J. and McCoy, J.M. (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene, 198, 289–296. [DOI] [PubMed] [Google Scholar]
- Jones, L. , Hamilton, A.J. , Voinnet, O. , Thomas, C.L. , Maule, A.J. and Baulcombe, D.C. (1999) RNA‐DNA interactions and DNA methylation in post‐transcriptional gene silencing. Plant Cell, 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Kombrink, A. , Shinya, T. , Desaki, Y. , Bours, R. , van der Krol, S. , Shibuya, N. , Joosten, M.H. and Thomma, B.P. (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. [DOI] [PubMed] [Google Scholar]
- Kale, S.D. , Gu, B. , Capelluto, D.G.S. , Dou, D. , Feldman, E. , Rumore, A. , Arredondo, F.D. , Hanlon, R. , Fudal, I. , Rouxel, T. , Lawrence, C.B. , Shan, W. and Tyler, B.M. (2010) External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell, 142, 284–295. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , van West, P. , Vleeshouwers, V.G. , de Groot, K.E. and Govers, F. (1998) Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell, 10, 1413–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, H.E. , Reddy, A.P.K. , Hsieh, S.P.Y. and Merca, S.D. (1973) Improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae . Plant Dis. Rep. 57, 537–541. [Google Scholar]
- Khang, C.H. , Berruyer, R. , Giraldo, M.C. , Kankanala, P. , Park, S. , Czymmek, K. , Kang, S. and Valent, B. (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell‐to‐cell movement. Plant Cell, 22, 1388–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiso, Y. , Li, Y. , Iwasaki, S. , HANAKA, K. , Kobayashi, T. , Sonoda, R. , Fujita, Y. , Yaegashi, H. and Sato, Z. (1994) Ustiloxins, antimitotic cyclic peptides from false smut balls on rice panicles caused by Ustilaginoidea virens . J. Antibiot. (Tokyo), 47, 765–773. [DOI] [PubMed] [Google Scholar]
- Koyama, K. and Natori, S. (1988) Further characterization of seven bis(naphtho‐γ‐pyrone) congeners of ustilaginoidins, coloring matters of Claviceps virens (Ustilaginoidea virens). Chem. Pharm. Bull. 36, 146–152. [Google Scholar]
- Lam, E. , Kato, N. and Lawton, M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature, 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Li, S. , Wang, Y. , Wang, S. , Fang, A. , Wang, J. , Liu, L. , Zhang, K. , Mao, Y. and Sun, W. (2015) The type III effector AvrBs2 in Xanthomonas oryzae pv. oryzicola suppresses rice immunity and promotes disease development. Mol. Plant‐Microbe Interact. 28, 869–880. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, M. , Liu, Z. , Zhang, K. , Cui, F. and Sun, W. (2019) Towards understanding the biosynthetic pathway for ustilaginoidin mycotoxins in Ustilaginoidea virens . Environ. Microbiol. 10.1111/1462-2920.14572. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Zhang, Z. , Faris, J.D. , Oliver, R.P. , Syme, R. , McDonald, M.C. , McDonald, B.A. , Solomon, P.S. , Lu, S. , Shelver, W.L. , Xu, S. and Friesen, T.L. (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1 . PLoS Pathog. 8, e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Pedersen, C. , Schultz Larsen, T. , Aguilar, G.B. , Madriz Ordeñana, K. , Hovmøller, M.S. and Thordal Christensen, H. (2016a) The stripe rust fungal effector PEC6 suppresses pattern‐triggered immunity in a host species‐independent manner and interacts with adenosine kinases. New Phytol. 211, 1052–1064. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Wang, Y. , Cui, F. , Fang, A. , Wang, S. , Wang, J. , Wei, C. , Li, S. and Sun, W. (2016b) The type III effector AvrXccB in Xanthomonas campestris pv. campestris targets putative methyltransferases and suppresses innate immunity in Arabidopsis. Mol. Plant Pathol. 18, 768–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti, L. , Lanver, D. , Schweizer, G. , Tanaka, S. , Liang, L. , Tollot, M. , Zuccaro, A. , Reissmann, S. and Kahmann, R. (2015) Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. [DOI] [PubMed] [Google Scholar]
- Lu, F. , Wang, H. , Wang, S. , Jiang, W. , Shan, C. , Li, B. , Yang, J. , Zhang, S. and Sun, W. (2015) Enhancement of innate immune system in monocot rice by transferring the dicotyledonous elongation factor Tu receptor EFR. J. Integr. Plant Biol. 57, 641–652. [DOI] [PubMed] [Google Scholar]
- Lyu, X. , Shen, C. , Fu, Y. , Xie, J. , Jiang, D. , Li, G. and Cheng, J. (2016) A small secreted virulence‐related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 12, e1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, V.A. and Ciuffetti, L.M. (2005) Localization of Ptr ToxA produced by Pyrenophora tritici‐repentis reveals protein import into wheat mesophyll cells. Plant Cell, 17, 3203–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, R. , Kombrink, A. , Motteram, J. , Loza‐Reyes, E. , Lucas, J. , Hammond‐Kosack, K.E. , Thomma, B.P. and Rudd, J.J. (2011) Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlak, T.A. , Kombrink, A. , Shinya, T. , Ryder, L.S. , Otomo, I. , Saitoh, H. , Terauchi, R. , Nishizawa, Y. , Shibuya, N. , Thomma, B.P. and Talbot, N.J. (2012) Effector‐mediated suppression of chitin‐triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell, 24, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera, G. , Giraldo, M.C. , Khang, C.H. , Coughlan, S. and Valent, B. (2009) Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1‐4 as biotrophy‐associated secreted proteins in rice blast disease. Plant Cell, 21, 1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, A.N. , Ziemann, S. , Treitschke, S. , Aßmann, D. and Doehlemann, G. (2013) Compatibility in the Ustilago maydis–maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9, e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, E.D. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D.M. and Kang, S. (2001) Agrobacterium‐mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Oliveira‐Garcia, E. and Valent, B. (2015) How eukaryotic filamentous pathogens evade plant recognition. Curr. Opin. Microbiol. 26, 92–101. [DOI] [PubMed] [Google Scholar]
- Oltval, Z.N. , Milliman, C.L. and Korsmeyer, S.J. (1993) Bcl‐2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programed cell death. Cell, 74, 609–619. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Shirsekar, G. , Bellizzi, M. , Chen, S. , Songkumarn, P. , Xie, X. , Shi, X. , Ning, Y. , Zhou, B. , Suttiviriya, P. , Wang, M. , Umemura, K. and Wang, G.‐L. (2016) The E3 ligase APIP10 connects the effector AvrPiz‐t to the NLR receptor Piz‐t in rice. PLoS Pathog. 12, e1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. , Bellizzi, M. , Valent, B. and Wang, G.L. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. and Shishiyama, J. (1988) Temporal sequence of cytological events in rice leaves infected with Pyricularia oryzae . Can. J. Bot. 66, 730–735. [Google Scholar]
- Pretsch, K. , Kemen, A. , Kemen, E. , Geiger, M. , Mendgen, K. and Voegele, R. (2013) The rust transferred proteins‐a new family of effector proteins exhibiting protease inhibitor function. Mol. Plant Pathol. 14, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, M. , Link, T.I. , Müller, M. , Hirschburger, D. , Pudake, R.N. , Pedley, K.F. , Braun, E. , Voegele, R.T. , Baum, T.J. and Whitham, S.A. (2016a) A small cysteine‐rich protein from the Asian soybean rust fungus, Phakopsora pachyrhizi, suppresses plant immunity. PLoS Pathog. 12, e1005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Z. , Liu, M. , Dong, Y. , Zhu, Q. , Li, L. , Li, B. , Yang, J. , Li, Y. , Ru, Y. and Zhang, H. (2016b) The syntaxin protein (MoSyn8) mediates intracellular trafficking to regulate conidiogenesis and pathogenicity of rice blast fungus. New Phytol. 209, 1655–1667. [DOI] [PubMed] [Google Scholar]
- Rafiqi, M. , Gan, P.H. , Ravensdale, M. , Lawrence, G.J. , Ellis, J.G. , Jones, D.A. , Hardham, A.R. and Dodds, P.N. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell, 22, 2017–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush, M.C. , Shahjahan, A. , Jones, J.P. and Groth, D.E. (2000) Outbreak of false smut of rice in Louisiana. Plant Dis. 84, 100. [DOI] [PubMed] [Google Scholar]
- Schwacke, R. and Hager, A. (1992) Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca2+ and protein‐kinase activity. Planta, 187, 136–141. [DOI] [PubMed] [Google Scholar]
- Shabab, M. , Shindo, T. , Gu, C. , Kaschani, F. , Pansuriya, T. , Chintha, R. , Harzen, A. , Colby, T. , Kamoun, S. and van der Hoorn, R.A. (2008) Fungal effector protein AVR2 targets diversifying defense‐related cys proteases of tomato. Plant Cell, 20, 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sornkom, W. , Miki, S. , Takeuchi, S. , Abe, A. , Asano, K. and Sone, T. (2017) Fluorescent reporter analysis revealed the timing and localization of AVR‐Pia expression, an avirulence effector of Magnaporthe oryzae . Mol. Plant Pathol. 18, 1138–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W. , Dunning, F.M. , Pfund, C. , Weingarten, R. and Bent, A.F. (2006) Within‐species flagellin polymorphism in Xanthomonas campestris pv. campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2‐dependent defenses. Plant Cell, 18, 764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W. , Liu, L. and Bent, A.F. (2011) Type III secretion‐dependent host defence elicitation and type III secretion‐independent growth within leaves by Xanthomonas campestris pv. campestris . Mol. Plant Pathol. 12, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y.X. , Jin, J. , Hu, D.W. , Yong, M.L. , Xu, Y. and He, L.P. (2013) Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 62, 1–8. [Google Scholar]
- Villamil, J.C.M. , Mueller, A.N. , Demir, F. , Meyer, U. , Ökmen, B. , Hüynck, J.S. , Breuer, M. , Dauben, H. , Win, J. , Huesgen, P.F. and Doehlemann, G. (2019) A fungal substrate mimicking molecule suppresses plant immunity via an inter‐kingdom conserved motif. Nat. Commun. 10, 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Li, M. , Dong, H. , Liu, X.Z. , Bai, Y.J. and Yang, H. (2008) Sporulation, inoculation methods and pathogenicity of Ustilaginoidea albicans, the cause of white rice false smut in China. J. Phytopathol. 156, 755–757. [Google Scholar]
- Wang, S. , Sun, Z. , Wang, H. , Liu, L. , Lu, F. , Yang, J. , Zhang, M. , Zhang, S. , Guo, Z. , Bent, A.F. and Sun, W. (2015) OsFLS2‐mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola . Mol. Plant, 8, 1024–1037. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wang, S. , Hu, K. , Yang, J. , Xin, X. , Zhou, W. , Fan, J. , Cui, F. , Mou, B. , Zhang, S. , Wang, G. and Sun, W. (2018) The kinase OsCPK4 regulates a buffering mechanism that fine‐tunes innate immunity. Plant Physiol. 176, 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y. and Huffaker, A. (2011) Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14, 351–357. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Zhao, X. , Sun, J. , Kang, Z. , Ding, S. , Xu, J. and Peng, Y. (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae . Mol. Plant‐Microbe Interact. 23, 112–123. [DOI] [PubMed] [Google Scholar]
- Yoshida, K. , Saitoh, H. , Fujisawa, S. , Kanzaki, H. , Matsumura, H. , Yoshida, K. , Tosa, Y. , Chuma, I. , Takano, Y. , Win, J. , Kamoun, S. and Terauchi, R. (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae . Plant Cell, 21, 1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, K. , Fang, A. , Han, Y. , Yang, J. , Xue, M. , Bao, J. , Hu, D. , Zhou, B. , Sun, X. , Li, S. , Wen, M. , Yao, N. , Ma, L.‐J. , Liu, Y. , Zhang, M. , Huang, F. , Luo, C. , Zhou, L. , Li, J. , Chen, Z. , Miao, J. , Wang, S. , Lai, J. , Xu, J.‐R. , Hsiang, T. , Peng, Y.‐L. and Sun, W. (2014) Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat. Commun. 5, 4849. [DOI] [PubMed] [Google Scholar]
- Zheng, D. , Wang, Y. , Han, Y. , Xu, J. and Wang, C. (2016) UvHOG1 is important for hyphal growth and stress responses in the rice false smut fungus Ustilaginoidea virens . Sci. Rep. 6, 24824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. , Shi, W. , Xu, X.W. , Li, Z.G. , Yin, C.F. , Peng, J.B. , Pan, S. , Chen, X.L. , Zhao, W.S. , Zhang, Y. , Yang, J. and Peng, Y. (2018) Glutamate synthase MoGlt1‐mediated glutamate homeostasis is important for autophagy, virulence and conidiation in the rice blast fungus. Mol. Plant Pathol. 19, 564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Virulence assay to rice of the wild‐type, scre1 mutant and complementary strains. (A) The scre1 mutants were confirmed via Southern blot analyses. Genomic DNA isolated from the wild‐type (WT) strain, scre1‐3‐23, scre1‐2‐4, scre1‐1‐26 and scre1‐1‐10 mutant candidates was digested with SphI. After separation by gel electrophoresis, the digested DNA was blotted on Hybond‐N membrane. The membrane was hybridized with the probe that was PCR‐amplified and DIG‐labelled. The molecular markers are shown in bp on the left. (B) Disease symptoms on rice panicles after injection inoculation of the wild‐type P1, scre1‐1‐10 mutant and scre1(SCRE1) complementary strains. The images were taken at 4 weeks after inoculation.
Fig. S2 SCRE1 expressed and secreted from Burkholderia glumae suppresses nonhost cell death in Nicotiana benthamiana. (A) Infiltration of the rice bacterial pathogen B. glumae caused nonhost cell death in N. benthamiana leaves while the engineered B. glumae transformed with pEDV‐SCRE1‐HA did not induce cell death. B.g., B. glumae; B.g. (SCRE1), B. glumae transformed with pEDV‐SCRE1‐HA. (B) SCRE1‐HA was secreted by B. glumae in the SMMXC hrp‐inducing minimal medium under different pH conditions. Upper panel: SCRE1‐HA was detected in the cell pellet and culture medium by western blotting. SCRE1‐HA secretion was dramatically induced at pH 6.5 in the SMMXC medium. Lower panel: protein loading is indicated by Ponceau S staining. CM, culture medium; TCL, total cell lysate; WB, western blot; α‐HA, anti‐haemagglutinin antibody.
Fig. S3 Subcellular localization of SCRE1 and SCRE1‐SP in Nicotiana benthamiana cells. Red fluorescence was observed in the plasmolysed N. benthamiana cells transiently expressing mCherry, SCRE1/2‐mCherry and SCRE1/2‐SP‐mCherry via laser scanning confocal microscopy. Neither SCRE1‐mCherry nor SCRE1‐SP‐mCherry was visible in periplasmic spaces of plasmolysed N. benthamiana cells. In contrast, SCRE2‐mCherry was clearly observed in periplasmic spaces, while SCRE2‐SP‐mCherry was not. SCRE1/2‐SP, SCRE1/2 without signal peptides; mCherry panels, red fluorescence; zoom‐in panels, enlarged images from broken square areas in mCherry panels. Arrows indicate the periplasmic spaces after N. benthamiana cells were plasmolysed. Scale bar: 75 μm.
Fig. S4 Induced expression of SCRE1 suppresses microbe‐associated molecular pattern (MAMP)‐triggered immunity and promotes pathogen infection in transgenic rice plants. (A) SCRE1‐FLAG was conditionally expressed after dezamethasone (DEX) induction in three independent transgenic lines, Z178, Z195 and Z196, as detected by immunoblotting with anti‐FLAG antibody (α‐FLAG). Lower panel: protein loading is indicated by Coomassie brilliant blue (CBB) staining. Nip, Nipponbare; DEX, dexamethasone; –, no DEX treatment; +, with DEX treatment. (B–E) SCRE1 expression induced by DEX significantly suppressed reactive oxygen species (ROS) burst triggered by flg22 (B, D) and chitin (C, E) in the Z195 (B, C) and Z196 (D, E) transgenic lines. Leaf discs were collected from the wild‐type and transgenic lines at 24 h after spraying with 10 µM DEX and mock solution, and were treated with 10 µM flg22 and 8 µM hexa‐N‐acetyl‐chitohexaose. (F, G) DEX‐induced expression of SCRE1 significantly inhibited OsPR1b expression triggered by flg22 and chitin in the Z178 (F) and Z196 (G) transgenic lines. The wild‐type and transgenic lines were treated with 10 µM DEX and mock solution for 24 h followed by 1 µM flg22 and 10 µg/mL chitin. The expression of OsPR1b was detected by quantitative RT‐PCR and normalized to the reference gene OsActin. Different letters above error bars indicate significant differences in the transcript levels of OsPR1b in the wild‐type and transgenic lines under different treatments (P < 0.05). Error bars represent means ± SE. (H, I) Disease symptoms on the leaves of the wild‐type, Z178, Z195 and Z196 transgenic rice lines with and without DEX treatment after Xanthomonas oryzae pv. oryzae PXO99 inoculation (H) and after Magnaporthe oryzae inoculation (I). The images in H and I were photographed at 12 and 6 days after inoculation, respectively.
Fig. S5 The SCRE1 fragment including the 45–70th amino acid residues retains the ability to suppress BAX‐triggered cell death. (A) The derived protein sequences and structural features of SCRE1. The predicted signal peptide is highlighted in orange. The two types of tandem and almost identical repeats are shown in yellow and purple, respectively. The Cys residues are in bold. The 45–70th amino acid residues are underlined and highlighted in blue. (B) Different truncated SCRE1 proteins have distinct abilities to suppress BAX1‐triggered hypersensitive responses in Nicotiana benthamiana. The truncated proteins including the 45–70th residues all have hypersensitive response (HR)‐suppressive ability similar to the full‐length SCRE1. +, having the ability to suppress BAX‐induced cell death; –, having no HR suppressive ability. BAX was transiently expressed at 6 h after expression of GFP, SCRE1 and different truncated SCRE1 proteins in N. benthamiana. HR symptoms in the infiltrated leaves were observed and photographed at 5 days post‐infiltration.
Table S1 The strains used in this study
Table S2 The primers used in this study


