Abstract
The Cucuteni-Trypillia complex (CTC) flourished in eastern Europe for over two millennia (5100–2800 BCE) from the end of the Neolithic to the Early Bronze Age. Its vast distribution area encompassed modern-day eastern Romania, Moldova and western/central Ukraine. Due to a lack of existing burials throughout most of this time, only little is known about the people associated with this complex and their genetic composition. Here, we present genome-wide data generated from the skeletal remains of four females that were excavated from two Late CTC sites in Moldova (3500–3100 BCE). All individuals carried a large Neolithic-derived ancestry component and were genetically more closely related to Linear Pottery than to Anatolian farmers. Three of the specimens also showed considerable amounts of steppe-related ancestry, suggesting influx into the CTC gene-pool from people affiliated with, for instance, the Ukraine Mesolithic. The latter scenario is supported by archaeological evidence. Taken together, our results confirm that the steppe component arrived in eastern Europe farming communities maybe as early as 3500 BCE. In addition, they are in agreement with the hypothesis of ongoing contacts and gradual admixture between incoming steppe and local western populations.
Subject terms: Archaeology, Population genetics
Introduction
In the archaeological record of eastern Europe, the first evidence for an agrarian lifestyle appeared in the 6th millennium BCE, when the Neolithic societies of the Danube basin (e.g. Linear Pottery [Linearbandkeramik, LBK] and Starčevo) began to spread to the Carpathian region1–3. Following these early foundations, a new society, the Cucuteni-Trypillia complex (CTC) emerged in a vast area that encompassed modern-day eastern Romania, Moldova and western/central Ukraine (Trypillia; Fig. 1). CTC flourished in eastern Europe for about over two millennia (5100–2800 BCE) from the end of the Neolithic to the Early Bronze Age, and is commonly divided into an Early, Middle and Late period4,5 (Fig. 1). Due to its geographic location, CTC was at the nexus of several contemporaneous societies, such as the Lengyel, Funnel Beaker (FBC, also Trichterbecher TRB) and the Globular Amphorae (GAC) cultures (Fig. 1)6. CTC is characterized by a wealth of material finds, attesting to a strong farming economy, a high level of social organization and advanced metallurgy as well as by large proto-urban mega-sites that may have housed hundreds or thousands of inhabitants during the Middle period (4100–3600 BCE). However, subsequently these settlements were mostly abandoned4, and there is archaeological evidence that individuals of the Late CTC interacted with populations that lived in the vast grasslands, or steppes, of Eurasia, such as the Early Bronze Age Yamnaya pastoralists7.
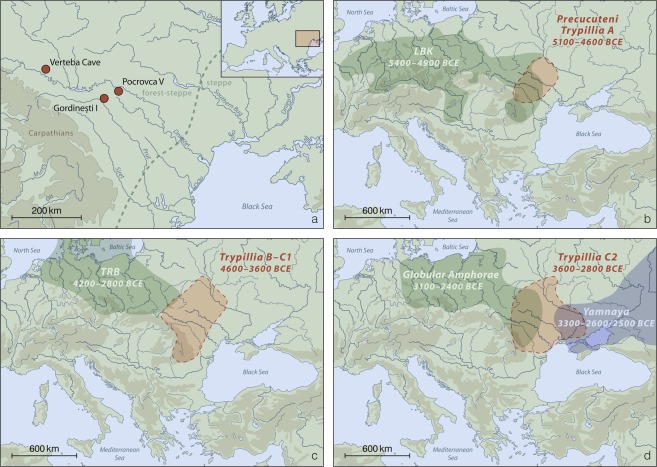
Figure 1.
(a) Map with the Moldovan sites Pocrovca V and Gordinești I from where the individuals presented in this study were recovered. Also shown is Verteba Cave in Ukraine where the CTC individuals presented in Mathieson et al.9 were discovered. (b–d) Temporal and geographic distribution of archaeological cultures mentioned in this study shown for the (b) Early (5100–4600 BCE), (c) Middle (4600–3600 BCE) and (d) Late (3600–2800 BCE) CTC. (Figure is based on a map made with Natural Earth (naturalearthdata.com) and was modified using Adobe Illustrator, Illustrator CS 6 (adobe.com)).
Despite the important role of CTC in prehistoric eastern Europe, little is known about the genetic composition of the people associated with this complex, the extent of biological contacts with their neighbors or the level of continuity with successive cultural groups. This gap in the genetic landscape can be attributed to a remarkable lack of CTC burials. Human remains have mainly been recovered from Late CTC contexts6, and ancient DNA (aDNA) studies have so far only been performed on specimens from a Trypillian site called Verteba Cave in Ukraine (Fig. 1). A mitochondrial DNA (mtDNA) study on eight Verteba individuals (3700–2900 cal. BCE) revealed in six cases maternal lineages typical of Anatolian and central European farmers. Two specimens had haplogroup U8b1 that may have been derived from European hunter-gatherers6,8. A subsequent genome-wide analysis in four males from Verteba (3900–3600 cal. BCE) confirmed the large Neolithic (~80%) and smaller hunter-gatherer (~20%) ancestry components9.
Here, we present genome-wide data generated from the human remains of four females that were excavated from two Late CTC burials in the present-day Republic of Moldova (Fig. 1). The specimens dated to 3500–3100 cal. BCE, i.e. several hundred years later than the previously investigated males from Verteba Cave9. The incorporation of these additional data sets obtained from new specimens, sites and time points allows us to draw a more nuanced picture of the population movements and dynamics during this important period in the prehistory of eastern Europe.
Results
We generated genome-wide shotgun sequencing data from three adults that were recovered from a multiple burial at the site of Pocrovca V (individuals: Pocrovca 1, Pocrovca 2, Pocrovca 3) and from a child interred in the Gordinești I flat necropolis (individual: Gordinești) in northern Moldova (Fig. 1, Supplementary Table 1). The specimens date to the Late CTC period (3500–3100 cal. BCE). Bioinformatic data analyses showed that all four individuals were females and carried the mitochondrial haplogroups U4, K1, T1 and T2, respectively (Table 1). Kinship analyses revealed no relatedness among them. When we screened the sequence data for known human blood-borne pathogens such as Yersinia pestis, Mycobacterium tuberculosis and Mycobacterium leprae, no signs of an infection were detected.
Table 1.
Summary information of the Moldova Late CTC samples analyzed in this study. Shown are the archaeological sites, anthropological data, radiocarbon dates, genetically determined sex and mitochondrial DNA haplogroup.
| Sample | Anthropological data | Calibrated years BCE | Genetic sex | mtDNA haplogroup |
|---|---|---|---|---|
| Gordinești | child 9 yo | 3482–3197 | F | U4a1 |
| Pocrovca 1 | woman 20–25 yo | 3364–3138 | F | K1a1 |
| Pocrovca 2 | woman 35–40 yo | 3366–3135 | F | T2c1d1 |
| Pocrovca 3 | woman 60–65 yo | 3341–3114 | F | T1a |
“yo”: years old.
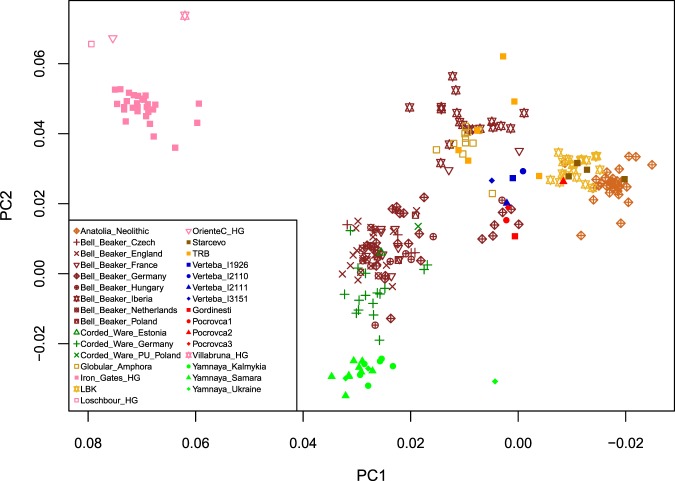
A principal component analysis of the four Moldova females together with previously published data sets of ancient Eurasians9–13 showed that Gordinești, Pocrovca 1 and Pocrovca 3 grouped with later dating Bell Beakers from Germany and Hungary close to the four CTC males from Verteba, while Pocrovca 2 fell into the LBK cluster next to Neolithic farmers from Anatolia and Starčevo individuals (Fig. 2, Supplementary Fig. 1).
Figure 2.
Principal component analysis of the CTC individuals from Moldova (Gordinești, Pocrovca 1, Pocrovca 2, Pocrovca 3) in red and the CTC individuals from Verteba Cave (I1926, I2110, I2111, I3151) in blue together with 23 selected ancient populations/individuals projected onto a basemap of 58 modern-day West Eurasian populations (not shown). HG = hunter-gatherer, LBK = Linearbandkeramik, PU = Proto-Unetice, TRB = Trichterbecher (Funnel Beaker Culture, FBC).
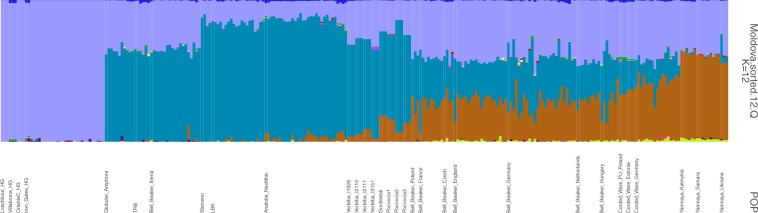
To assess the genetic diversity within the Moldovan CTC individuals, we ran ADMIXTURE analysis14. The dominant element in all Moldovan CTC females was found in Anatolian Neolithic farmers, Starčevo and LBK individuals (Fig. 3, Supplementary Fig. 2), followed by a large hunter-gatherer component. Interestingly, Gordinești, Pocrovca 1 and 3 had a considerable amount of steppe ancestry, with the Gordinești child exhibiting the highest proportion.
Figure 3.
Admixture analysis of the CTC individuals from Moldova and Ukraine together with 23 selected ancient populations/individuals. Admixture plot is shown for K = 12 ancestral genetic components. HG = hunter-gatherer, LBK = Linearbandkeramik, PU = Proto-Unetice, TRB = Trichterbecher (Funnel Beaker Culture, FBC). Farmer ancestry is illustrated in turquoise, hunter-gatherer ancestry is shown in purple and steppe-ancestry in brown.
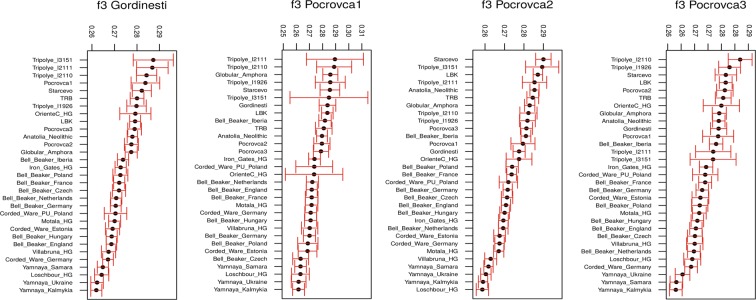
We next applied f3 outgroup statistics15 to investigate which ancient populations or individuals shared most of the genetic drift with the CTC Moldovans. All of them, except Pocrovca 2, appeared genetically close to the Verteba CTC males (Fig. 4, Supplementary Fig. 3). In contrast, Pocrovca 2 shared most of its genetic ancestry with Starčevo individuals.
Figure 4.
f3-outgroup statistics f3(Gordinești; Test, Mbuti) and f3(Pocrovca; Test, Mbuti) showing the amount of shared genetic drift between each of the four individuals analyzed in this study and selected previously published ancient populations/individuals (as used in PCA and admixture analysis). HG = hunter-gatherer, LBK = Linearbandkeramik, PU = Proto-Unetice, TRB = Trichterbecher (Funnel Beaker Culture, FBC).
We ran D statistics and qpAdm15 on the four Moldovan data sets to estimate the direction of genetic influx and the amount of the ancestry components. When the data sets from all four Moldovan CTC individuals were combined, they showed a stronger influx 1) from LBK than Anatolian Neolithic and 2) from Western hunter-gatherers than steppe-related populations. When looking at various proxies for steppe-related ancestry (Yamnaya Samara, Ukraine Mesolithic, Caucasian hunter-gatherer (CHG), Eastern hunter-gatherer (EHG), we did not observe any significant difference in genetic influx from either Yamnaya Samara, EHG or Ukraine Mesolithic. However, relative to CHG, we detected a substantial shift towards Yamnaya Samara steppe-related ancestry (Supplementary Table 2). Consequently, Yamnaya Samara, Ukraine Mesolithic and EHG appear to be equally suitable proxies for steppe-related ancestry in the Moldovan CTC individuals. This finding was confirmed by our results from the two-way admixture qpAdm models (Supplementary Table 3) for each individual separately and for the combined data.
Next, we applied a three-way admixture qpAdm model to the combined data set of our four Moldovan CTC individuals using LBK, steppe (Yamnaya Samara, EHG, Ukraine Mesolithic) as well as WHG as possible source populations, but did not obtain feasible results. We then modeled each individual separately. Interestingly, Pocrovca 1 yielded a feasible three-way admixture model suggesting the following proportions: LBK (41–60%), steppe-related ancestry (8–18%) and WHG (29–41%) (Supplementary Table 3). This finding is in accordance with the results from our admixture analysis and the D statistics. For the three individuals Gordinești, Pocrovca 2 and Pocrovca 3, the three-way admixture models were not feasible most likely due to an already achieved saturation of the hunter-gatherer ancestry in the steppe populations tested. We did not obtain feasible models when running qpAdm on the X chromosome in order to test for male-biased admixture from hunter-gatherers or individuals with steppe-related ancestry.
Discussion
CTC plays an important role in eastern European prehistory, yet owing to the scarcity of human skeletal remains and corresponding aDNA studies, the current knowledge on the origin and biological relatedness of the people associated with this cultural complex is rather limited. Up to now, genome-wide investigations have focused on only four male individuals from a single Trypillian site, Verteba Cave in Ukraine9. The vast majority of the human remains found there constituted crania and mandibles that were disposed of in the form of secondary interments. The different bone elements were found commingled and showed high levels of perimortem trauma and postmortem manipulation16,17. In contrast, the skeletal remains from the females presented in this study did not show any signs of interpersonal violence. The two sites Pocrovca and Gordinești, from which the individuals were recovered, are in close proximity to each other and a few hundred kilometers away from Verteba (Fig. 1).
Recently, it was hypothesized that due to their high population densities, the CTC mega-settlements served as a focus point for the emergence and large-scale radiation of Y. pestis lineages across Eurasia during the Neolithic18. Amongst the four Moldovan specimens, we did not detect any signals of a Y. pestis infection, although the three individuals from Pocrovca were discovered in a multiple burial (without any traces of violence), which would render death due to an epidemic event plausible.
The genome-wide data sets of the four female individuals presented in this study showed genetic ancestry common in Anatolian farmers and LBK individuals, steppe-related populations as well as Western hunter-gatherers. With a maximum of 60%, the Neolithic-derived proportion constituted the largest ancestry component. Interestingly, from our results the CTC Moldovans appeared to be genetically more likely related to LBK than to Anatolian Neolithic farmers. In the Verteba individuals, the Neolithic component was also seen in the same magnitude but had rather a northwestern Anatolian origin9. The high amount of shared genetic ancestry between CTC and LBK or Starčevo, respectively, as observed in our f3 outgroup analysis, is supported by archaeological evidence. The basis for the economic subsistence and cultural attributes of the CTC is found in the European Boian and Starčevo cultures with additional influence from the LBK19–21.
Interestingly, we detected steppe-related ancestry in the Late CTC burials from the Republic of Moldova. The presence of this component suggests moderate genetic influx from individuals affiliated with steppe cultures into the CTC-associated gene-pool as early as 3500 BCE; for this time, archaeological evidence showed an increase in Trypillia-related finds in the steppe area22,23. Thus, the steppe component had arrived in the eastern part of the continent in farmer communities well before it first appeared in the west, i.e. in the Corded Ware people around 2800 BCE9. This finding establishes eastern Europe as an old genetic contact zone between locals and incoming steppe people, which is supported by two other early dating specimens from Ukraine (from Alexandria 4045–3974 cal. BCE and Dereivka 3634–3377 cal. BCE) that also showed mixtures of steppe- and Anatolian Neolithic-related ancestry9. However, they represented individuals who still followed a hunter-gatherer subsistence, whereas the CTC females analyzed here belonged to a Neolithic agrarian culture.
One likely source population that could have introduced the steppe ancestry component into the CTC gene-pool might have been individuals associated with the eastern Eurasian Mesolithic, e.g. the Ukraine Mesolithic people, Eastern hunter-gatherers or even later-dating Yamnaya steppe pastoralists (Supplementary Table 3). Support for a mixture of Late CTC with the neighboring Early Bronze Age Yamnaya culture exists in the archaeological record (Fig. 1; 3300–2600 BCE)24. Despite the short time of overlap, artifacts found in both late CTC and Yamnaya settlements provide evidence of barter between them24. These observations and the genetic findings presented in this study (i.e. the different steppe proportions in the four Moldovans) are in agreement with ongoing contacts as well as with gradual admixture and a slow change in cultural expression, rather than total replacement. However, this hypothesis challenges a previously published scenario of Yamnaya horsemen massively migrating in war into central Europe25.
It is not surprising that Gordinești, Pocrovca 1 and Pocrovca 3 showed genetic affinities with later dating Bronze Age or Bell Beaker individuals. The common link among them is the considerable steppe-related ancestry which each group likely received independently from different parental populations26.
Our analyses (PCA, f3 outgroup analysis) also suggest a genetic relationship between the individuals from Moldova and those associated with the contemporaneous FBC/TRB and GAC, possibly indicating a common origin and/or ongoing interactions. The mtDNA study on the Verteba individuals already showed a high degree of similarity in the maternal lineage composition between CTC and FBC populations6. This connection can be explained by the geographical proximity of the FBC and CTC distribution areas (Fig. 1). An overlap of CTC and FBC settlements site has been documented and there is additional confirmation in the archaeological record for regular inter-group contacts and trade westwards and northwards from the CTC into the GAC and FBC areas27.
Overall, the different genetic makeup of the CTC individuals presented here and in Mathieson et al.9 indicates a relatively high diversity, which is surprising given that they all dated to the same Late CTC period and were buried only a few hundred kilometers apart (Fig. 1). This finding suggests population dynamics also within a culture and questions the notion of the apparently stable and uniform composition of individuals associated with a specific archaeological group.
Material and Methods
Samples
Pocrovca V: the collective burial belonging to the Trypillia C2 culture (Gordinești local group) contained three human skeletons as well as pottery and animal bones including those of three hares. The bodies of the deceased were thrown into the pit sequentially, one on top of the other. They lay in a crouched position on their right sides, which is the main inhumation type for this cultural group. Osteologically, the remains represented adult females between 20 and 65 years old (Table 1, detailed description SI).
Gordinești: the grave in the Gordinești I flat necropolis belonging to the Trypillia C2 culture (Gordinești local group) contained the incomplete skeleton of a child. The age at death estimation of 9 years ± 24 months was based on the dentition (detailed description SI).
All specimens were radiocarbon-dated to 3500–3100 cal. BCE (Table 1). Materials are stored in the National Museum of History of Moldova, Chișinău.
Ancient DNA extraction and library preparation
Petrous bones and teeth were cleaned in pure bleach solution for 5 minutes and rinsed with water. After drying overnight at 37 °C the inner ear area (cochlea and vestibule) was cut out from the petrous bone, bleached, rinsed with water and dried. The dried inner ear piece was ground in a ball mill homogenizer for 45 s at maximum speed. Fifty mg of bone/tooth powder were used for DNA extraction following a silica-based protocol28. For each sample, a double-stranded DNA sequencing library was prepared from 20 µL of extract, following partial uracil-DNA-glycosylase treatment29. Sample-specific index combinations were added to the sequencing libraries in order to allow differentiation between the individual samples after pooling and multiplex sequencing30. Sampling, aDNA extraction and the preparation of sequencing libraries were performed in clean-room facilities of the Ancient DNA Laboratory in Kiel. Negative controls were taken along for the DNA extraction and library generation steps.
Sequencing
The libraries were paired-end sequenced using 2 × 75 cycles on an Illumina HiSeq 4000. Demultiplexing was performed by sorting all the sequences according to their index combinations. Illumina sequencing adapters were removed and paired-end reads were merged. Merged reads were filtered for a minimum length of 30 bp.
Screening for pathogens
All samples were screened for their metagenomic content with the metagenome analyzer MEGAN31 and the alignment tool MALT32. MALT version V0.3.8 was used to align all pre-processed samples against a collection of available complete bacterial and viral genomes. Bacterial genomes were downloaded from the NCBI FTP server (ftp.ncbi.nlm.nih.gov/genomes/refseq/bacteria, access 12.03.2018) using a custom script. Viral genomes were downloaded from the NCBI FTP server (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/viral/, access 03.01.2018). MALT was executed in BLASTN mode with the following parameters for bacteria:
and for viruses:
where $REF is the MALT index, $IN is a clipped-and-merged FASTQ file and $OUT is the output folder for MALT. Resulting RMA files were examined for their taxonomic content using MEGAN version V6.11.4.
Mapping and aDNA damage patterns
Pre-processed sequences were mapped to the human genome build hg19 (International Human Genome Sequencing Consortium, 2001) using BWA 0.7.1233 with the reduced mapping stringency parameter “-n 0.01” to account for mismatches in aDNA. Duplicates were removed. In order to assess the authenticity of the aDNA fragments34, terminal C to T mis-incorporations were evaluated using mapDamage 2.035. After the validation of damage, the first two positions at the 5′ end of the fastq-reads were trimmed off.
Sex determination
Sex was determined based on the ratio of sequences aligning to the X and Y chromosomes compared to the autosomes36. Females are expected to have a ratio of 1 on the X chromosome and 0 on the Y chromosome, whereas males are expected to have both X and Y ratios of 0.5.
Genotyping
Alleles were drawn at random from each of the 1,233,013 SNP positions11,13 in a pseudo-haploid manner using a custom script as described in Lamnidis et al.37. 10,000 SNPs served as a minimum threshold required for a sample to be included in further analyses.
Contamination estimation and authentication
Estimation of exogenous contamination in the aDNA extracts was performed on the mitochondrial level using the software Schmutzi38.
Kinship analysis
Kin relatedness was assessed among the four Moldova CTC individuals using READ39 and lcMLkin40. READ identifies relatives based on the proportion of non-matching alleles. lcMLkin infers individual kinship from calculated genotype likelihoods.
Principal component analysis (PCA)
Genotype data sets of the Moldovan samples were merged with previously published genotypes of 5,514 modern and ancient individuals on a data set of 597,573 SNPs11–13,15. Using the software Smartpca (version 16000)41, the ancient individuals were projected on a basemap of genetic variation calculated from the following 58 West-Eurasian populations11–13: Abkhasian, Adygei, Albanian, Armenian, Balkar, Basque, Bedouin, Belarusian, Bergamo, Bulgarian, Canary Islander, Chechen, Croatian, Cypriot, Czech, Druze, English, Estonian, Finnish, French, Georgian, Greek, Hungarian, Icelandic, Iranian, South Italian, Jewish (Ashkenazi, Georgian, Iranian, Iraqi, Libyan, Moroccan, Tunisian, Turkish, Yemenite), Jordanian, Kumyk, Lebanese, Lezgin, Lithuanian, Maltese, Mordovian, North Ossetian, Norwegian, Orcadian, Palestinian, Russian, Sardinian, Saudi, Scottish, Sicilian, Spanish, North Spanish, Syrian, Turkish, Tuscan, Ukrainian.
ADMIXTURE analysis
Prior to ADMIXTURE analysis we used Plink (v1.90b3.29) to prune out SNPs in linkage disequilibrium with the parameters–indep-pairwise 200 25 0.4. ADMIXTURE (version 1.3.0)14 was run on a data set of 597,573 SNPs11–13,15 comprising 5,514 previously published ancient and modern human samples and our four Moldovan samples. We used a number of ancestral components (K) ranging from 4 to 12 in 100 bootstraps for each component, respectively.
D statistics
D statistics were run as part of the Admixtools package15 in the form of (Mbuti; Moldova; PopC; PopD), iteratively shuffling different proxies for farmer-related ancestry, hunter-gatherer-related ancestry and steppe-related ancestry, such as LBK and Anatolian Neolithic, Western, Eastern and Caucasian hunter-gatherers and Yamnaya Samara for PopC and PopD, respectively.
f3 outgroup statistics
f3 outgroup statistics were run as a part of the Admixtools package15 in the form of f3 (Moldova; Test; Mbuti) using for Test the same populations as in the ADMIXTURE and PCA analyses.
qpAdm analysis
qpAdm analysis15 was run to model the Moldovan individuals as admixture of farmers, hunter-gatherers or individuals with steppe-related ancestry, such as Yamnaya. The following populations were used as outgroups: Mbuti, Ust Ishim, Kostenki14, Mal’ta1, Han, Papuan, Onge, Chukchi and Karitiana.
Determination of mitochondrial haplogroups
Sequencing reads were mapped to the human mitochondrial genome sequence rCRS42. Consensus sequences were generated in Geneious (v. 9.1.3) using a default threshold of 85% identity among the covered positions and a minimum coverage of 3. HAPLOFIND43 was applied to determine the mitochondrial haplogroups from the consensus sequences.
Investigating male-bias in steppe-related ancestry admixture
qpAdm was used to estimate admixture proportions on the autosomes compared to the X chromosome in order to compute Z-scores for the difference between autosomes and the X chromosome as described in Mathieson et al.9, where a positive Z-score indicates a male-biased admixture through steppe populations, such as Yamnaya.
Supplementary information
Acknowledgements
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation - Projektnummer 2901391021 – SFB 1266). We are grateful to Wolfgang Haak for his advice. We thank the Institute of Clinical Molecular Biology in Kiel for providing Illumina sequencing as supported by the DFG Clusters of Excellence “Precision Medicine in Chronic Inflammation” and “ROOTS”. We thank Sören Franzenburg for support and expertise in high-throughput sequencing. We are grateful to Dr. Nicolai Telnov (Institute of Cultural Heritage, Academy of Sciences of the Republic of Moldova, Chișinău) who investigated the Gordinești burial, for providing us access to the skeletal material. We acknowledge financial support by Land Schleswig-Holstein within the funding programme Open Access Publikationsfonds.
Author contributions
B.K.-K., A.N., J.M. and S.T. conceived and designed the research. B.K.-K. generated and A.I., J.S. analyzed the ancient DNA data. A.I., S.T., A.S., J.S., O.S., G.S., R.H., J.M. A.N. and B.K.-K. interpreted the findings. A.N., A.I. and B.K.-K. wrote the manuscript with input from S.T., J.M., A.S.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Johannes Müller, Email: johannes.mueller@ufg.uni-kiel.de.
Ben Krause-Kyora, Email: b.krause-kyora@ikmb.uni-kiel.de.
Supplementary information
is available for this paper at 10.1038/s41598-020-61190-0.
References
- 1.Dergachev, V. A., Larina, O. V. Pamjatniki kul’tury Krish Moldovy (s katalogom) [Sites of Criș culture in Moldova (with a catalogue)] (in Russian). Chișinău (2015).
- 2.Saile T, et al. Zur Bandkeramik zwischen Pruth und Südlichem Bug. Praehistorische Zeitschrift. 2016;91(1):1–15. doi: 10.1515/pz-2016-0001. [DOI] [Google Scholar]
- 3.Kotova, N. S. Neolithization of Ukraine. British Archeological Reports (international series) 1109, Oxford (2003).
- 4.Müller, J., Rassmann, K. & Videiko, M. Trypillia Mega-Sites and European Prehistory: 4100–3400 BCE (Routledge, 2016).
- 5.Rassmann K, et al. High precision Tripolye settlement plans, demographic estimations and settlement organization. J Neolith Archaeol. 2014;16:63–95. [Google Scholar]
- 6.Nikitin AG, et al. Mitochondrial DNA analysis of Eneolithic Trypillians from Ukraine reveals Neolithic farming genetic roots. PLoS One. 2017;12:e0172952. doi: 10.1371/journal.pone.0172952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anthony DW. A New Approach to Language and Archaeology: The Usatovo Culture and the Separation of Pre-Germanic. J Indo-European Stud. 2008;36:1–51. [Google Scholar]
- 8.Nikitin AG, Sokhatsky MP, Kovaliukh MM, Videiko MY. Comprehensive Site Chronology and Ancient Mitochondrial DNA Analysis from Verteba Cave - a Trypillian Culture Site of Eneolithic Ukraine. Interdiscip Archaeol Nat Sci Archaeol. 2010;1:9–18. [Google Scholar]
- 9.Mathieson I, et al. The Genomic History of Southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipson M, et al. Parallel paleogenomic transects reveal complex genetic history of early European farmers. Nature. 2017;551:368–372. doi: 10.1038/nature24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haak W, et al. Massive migration from the steppe was a source of Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson N, et al. Ancient Admixture in Human History. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadrow S, Pokutta DA. The Verteba Cave: A Subterranean Sanctuary of the Cucuteni-Trypillia Culture in Western Ukraine. J Neolith Archaeol. 2016;18:1–21. [Google Scholar]
- 17.Lillie, M. C., Potekhina, I. D., Nikitin, A. G., & Sokhatsky, M. P. First evidence for interpersonal violence in Ukraine’s Trypillian farming culture: Individual 3 from Verteba Cave, Bilche Zolote, Ukraine. Gerdau-Radonić, K. & McSweeney K. (Eds.), Trends in Biological Anthropology 1, Oxbow books, 54–60 (2015).
- 18.Rascovan N, et al. Emergence and Spread of Basal Lineages of Yersinia pestis during the Neolithic Decline. Cell. 2019;176:295–305. doi: 10.1016/j.cell.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Burdo NB. Osobennosti keramicheskogo kompleksa Precucuteni – Tripolye A I problema genezisa tripol’skoj kul’tury [Peculiarities of the Precucuteni – Tripolye A ceramic complex and the problem of the origin of the Tripolye culture] (in Russian) Stratum Plus. 2002;2:141–163. [Google Scholar]
- 20.Monah, D. “Religie si arta in cultura Cucuteni” [Religion and art in Cucuteni culture], in Dumitroaia, Gheorghe, Primul muzeu Cucuteni din Romania [The first Cucuteni museum for Romania], Bibliotheca memoriae antiquitatis XV (in Romanian), Piatra-Neamţ, Romania: Editura Foton, 162–173 (2005).
- 21.Garvăn, D., Buzea, D. & Frînculeasa, A. Precucuteni. Originea unei mari civilizații [Precucuteni. The origins of a great civilization] (in Romanian). Bibliotheca Memoriae Antiquitatis XXIII. Piatra-Neamț, Editura Constantin Mătasă (2009).
- 22.Manzura I. Steps to the Steppe: Or, How the North Pontic Region Was Colonised. Oxford J Archaeol. 2005;24:313–338. doi: 10.1111/j.1468-0092.2005.00239.x. [DOI] [Google Scholar]
- 23.Manzura, I. North-Pontic steppes at the end of the 4th millennium BC: the epoch of broken borders. Zanoci, A., Kaiser, E., Kashuba, M., Izbitser, E. & Băț, M. (eds.). Man, culture and society from the Copper Age until the Early Iron Age in Northern Eurasia. Contributions in honour of the 60th anniversary of Eugen Sava. Chișinău, 53–75 (2016).
- 24.Mallory, J.P. In search of the Indo-Europeans: language, archaeology and myth. London: Thames and Hudson. ISBN 0-500-05052-X. OCLC 246601873 (1989).
- 25.Goldberg A, Günther T, Rosenberg NA, Jakobsson M. Ancient X chromosomes reveal contrasting sex bias in Neolithic and Bronze Age Eurasian migrations. Proc Natl Acad Sci USA. 2017;114:2657–2662. doi: 10.1073/pnas.1616392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olalde I, et al. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018;555:190–196. doi: 10.1038/nature25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelisiak A. The Funnel Beaker Culture Settlements Compared with Other Neolithic Cultures in the Upper and Middle Part of the Dnister Basin. Selected Issues. State of the Research. Analecta Archaeol Ressoviensia. 2007;2:23–46. [Google Scholar]
- 28.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohland N, et al. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos Trans R Soc Lond B Biol Sci. 2015;370:20130624. doi: 10.1098/rstb.2013.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huson D, et al. MEGAN Community Edition – Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput Biol. 2016;12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vågene ÅJ, et al. Salmonella enterica genomes from victims of a major sixteenth-century epidemic in Mexico. Nat Ecol Evol. 2018;2:520–528. doi: 10.1038/s41559-017-0446-6. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briggs A, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonsson H, et al. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu C, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamnidis T, et al. Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe. Nat Commun. 2018;9:5018. doi: 10.1038/s41467-018-07483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renaud G, et al. Schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 2015;16:224. doi: 10.1186/s13059-015-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn M, Jakobsson M, Günther T. Estimating genetic kin relationships in prehistoric populations. PLoS One. 2018;13:e0195491. doi: 10.1371/journal.pone.0195491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipatov, M. et al. Maximum Likelihood Estimation of Biological Relatedness from Low Coverage Sequencing Data. BioRxiv; 10.1101/023374 (2015).
- 41.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 43.Vianello D, et al. HAPLOFIND: a new method for high-throughput mtDNA haplogroup assignment. Hum Mutat. 2013;34:1189–1194. doi: 10.1002/humu.22356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.