Abstract
Silicon (Si) uptake and accumulation in plants can mitigate various biotic stresses through enhanced plant resistance against wide range of herbivores. But the role of silicon in defense molecular mechanism still remains to be elucidated in finger millet. In the present study, we identified three silicon transporter genes viz. EcLsi1, EcLsi2, and EcLsi6 involved in silicon uptake mechanism. In addition, the study also identified and characterized ten different Si transporters genes from finger millet through transcriptome assembly. The phylogenetic study revealed that EcLsi1 and EcLsi6 are homologs while EcLsi2 and EcLsi3 form another pair of homologs. EcLsi1 and EcLsi6 belong to family of NIP2s (Nod26-like major intrinsic protein), bona fide silicon transporters, whereas EcLsi2 and EcLsi3, an efflux Si transporter, belong to an uncharacterized anion transporter family having a significant identity with putative arsB transporter proteins. Further, the phylogenetic and topology analysis suggest that EcLsi1 and EcLsi2 co-evolved during evolution while, EcLsi2 and EcLsi3 are evolved from either EcLsi1 and/or EcLsi6 by fusion or duplication event. Moreover, these silicon transporters are predicted to be localized in plasma membrane, but their structural differences indicate that they might have differences in their silicon uptake ability. Silicon amendment induces the synergistic defense mechanism by significantly increasing the transcript level of silicon transporter genes (EcLsi1, EcLsi2 and EcLsi6) as well as defense hormone regulating genes (EcSAM, EcPAL and EcLOX) at 72 hpi (hours of post infestation) in both stem and roots compared to non-silicon treated plants against pink stem borer in finger millet plants. This study will help to understand the molecular defense mechanism for developing strategies for insect pest management.
Subject terms: Agricultural genetics, Plant molecular biology
Introduction
Finger millet (Eleusine coracana Gaertn.) is the most important cereal crop among small millets after sorghum and pearl millet and is grown for food and fodder in India. It is the third most widely cultivated millet crop in the semi-arid tropical and sub-tropical regions worldwide1,2. Finger millet is a sustainable food of poor people in rural areas and is also appreciated by urban populations for its nutritional quality as it provides fair amount of proteins, minerals, calcium and vitamins compared to other cereals. Still, finger millet is neglected in developing countries where food security is the major for the growing population3. Along with its nutritional quality to solve dietary issues in rural areas, finger millet is also considered as one of the hardiest crop to mitigate the effects of drought and numerous biotic stresses4. Although, it is being affected by numerous insects and pests, pink stem borer Sesamia inferens Walker (Noctuidae; Lepidoptera), is one of the major biotic constraints for crop production in finger millet growing countries5. Pink stem borer is a polyphagous pest which feeds on wide range of grasses including finger millet. It is widely recorded in India and different parts of South Asia5. Despite the economic importance of this borer, there is lack of a good pest management strategy. Currently, conventional cultural practices are preferred for suppressing the pest population during larval and pupal stages with some use of pesticides in more severe condition5. However, S.inferens has become less sensitive to many recommended pesticides in recent years6. Therefore, integrated pest management strategies recommend the combination of preventive measures using stem borer resistant/tolerant cultivars for planting, avoiding the stem borer favored conditions for planting and encouragement of indigenous parasitoids and natural enemies7. But there is an urgent need to develop effective and ecologically sound alternative method to control stem borer infestation in finger millet to maintain the high productivity. Silicon (Si)is now being recognized as a sustainable alternative to provide resistance against difference biotic and abiotic stresses which ultimately influence the crop production worldwide8,9. Although Si is not recognized as an essential element required for plant nutrition10, its beneficial effects against different insect pests has been demonstrated in many plant species8. For instance, it has been demonstrated that Si increases tolerance against all types of borers including stem borer in various cereal crops11. Hence, it was reported that Si is a “silver bullet” and a potential alternative for biotic stress management in different crop species12. It is widely known that Si creates a physical barrier by silicification of plant parts as part of defense mechanisms against insect pests. Silicon acts as an elicitor of systemic stress signals to produce the effective defense compounds against herbivores by regulating the defense related phytohormone pathways13. Still plant defense is a complex process and it can vary with feeding strategies of insect pests14. The phytohormones like salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) are the first line of defense in enhancing the plant responses to various herbivores15. JA and SA regulate defenses against herbivores, particularly JA controls cell-content-feeding and tissue-chewing insects16,17 while SA and JA together regulate the defense against phloem-feeding insects18. It has been shown recently that Si induces JA-mediated defense responses against insects and pests19, indicating that Si can enhance the plant resistance against herbivores by regulating the defense responsive phytohormone pathways. Si also induces the priming process in host plant by sensitizing and preparing the plant for quick responses against the future herbivore attacks20–22. The mechanism of uptake of Si varies amongst different plant species and apparently depends on the presence of specific Si transporters. The first report on identification of Si transporters in plants was in rice13. Genes involved in Si uptake and distribution in numbers of plant species like barley, maize, pumpkin, wheat, cucumber and horsetail millet have now been identified23,24. These transporters, namely Lsi1 and Lsi6, are Si-influx transporters belonging to the aquaporin family and are mainly involved in Si distribution in root and shoot tissues25. Lsi2 belongs to the putative anion transporter and primarily expressed in the endodermis of roots, is proton driven and it works as a Si/H + antiporter26. However, these transporters do not show polar localization in barley and maize27. Hence, the silicon uptake ability of Lsi1 and Lsi2 varies greatly in different plant species in comparison to rice28. Therefore, the aim of the present study is to isolate, identify, and characterize the silicon transporter genes in finger millet and study their expression to dissect the molecular defense mechanism and interaction with phytohormones against pink stem borer in finger millet plant.
Results
Isolation and identification of Si transporter genes
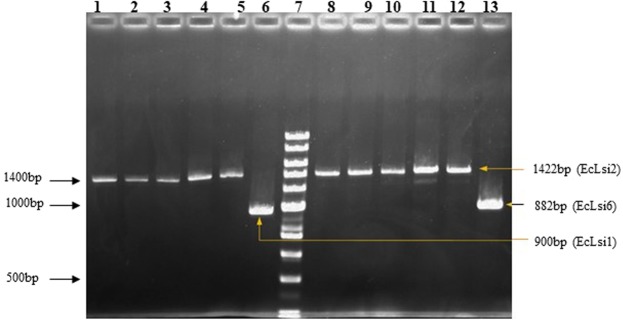
Isolation of EcLsi1, EcLsi2 and EcLsi6 CDS region from finger millet cv. Suvra employed PCR amplification using the gene specific oligonucleotides (Table S1). The PCR amplification produced sharp and bright bands of 900 bp, 1422 bp and 882bp for EcLsi1, EcLsi2 and EcLsi6 respectively (Fig. 1). The CDS regions of all three isolated genes were subjected to BLASTn hit search for identification of the genes. The EcLsi1 gene showed maximum 91.04% identity with Setaria italica aquaporin NIP2-1 mRNA (Accession no. XM_004953810.4), followed by 90.92% identity with Sorghum bicolor NOD26-like major intrinsic protein (NIP2-1) mRNA, complete CDS (Accession no. EF373651.1.). The EcLsi2 and EcLsi6 showed maximum 88.60% and 92.72% identity with Setaria italica silicon efflux transporter LSI2, mRNA (accession no. XM_004985998.2) and Setaria italica aquaporin NIP2-2, mRNA (accession number. XM_004964985.4) respectively. The blast result indicates that E.coracana silicon transporter genes are orthologous to millet family member. After identification, the CDS sequences of EcLsi1, EcLsi2 and EcLsi6 genes isolated from finger millet variety ‘Suvra’ were submitted to NCBI with the accession nos. MN517554, MN517555 and MN517556 (NCBI) respectively.
Figure 1.
Amplification of silicon transporter genes viz. EcLsi1, EcLsi2 and Eclsi6 from finger millet employing gene specific primers. 1–4: EcLsi2 gene amplification from root tissue; 6: EcLsi1 gene amplification; 7: Low range DNA ruler plus; 8–12: EcLsi2 gene amplification from stem tissue; 13: EcLsi6 gene amplification. Numbers on the right side of margin represents molecular weight marker DNA in base pairs (bps) and number on left side represents size of amplified product size in base pairs.
Transcriptome assembly
To identify the homologous silicon transporter genes, the high-throughput ILLUMINA (Next Seq 500) paired-end reads transcriptome library of finger millet cv. ML-365 was retrieved from NCBI database bearing the accession number SRR4021829. The obtained library was further assembled to mine the transcriptionally active silicon transporter genes. The raw reads of library were cleaned for adapter contamination and filtered for low-quality reads with Q20 base parameter with the Trimmomatic program. Reads less than 20 bp length were removed from libraries before assembling. De-novo assembly of these high-quality reads was performed with Trinity assembler which produced 2,64,541 unigenes after assembly. CAP3 program was then employed to reduce the overlapping length with cut off parameter > 50 and specific overlap percent identity cutoff of > 95% similarity. After CAP3 assembly, a total of 2,39,594 unigenes were generated. The unigenes were further filtered for removing sequence duplicates. The redundant unigene sequences were removed using CD-HIT-EST (v4.6.1) tool with 95% identity threshold. The CD-HIT-EST assembly yielded 1,89,632 unigenes, generating mean sequence length of 888 nt and median sequence length of 535 nt (N50 = 1736) with GC-content of 45.81% (Table S2). The largest and shortest transcript/unigene lengths were 24966 nt and 201 nt respectively after assembly. These unigenes were further used for ORF prediction for CDS identification, functional annotation and other downstream analyses. Predicted CDSs from transcriptome were subjected to BLASTx search against non-redundant (nr) protein database of NCBI with an E-value cut-off of 10e−6. Total of 1440 plant silicon transporter genes available on NCBI were queried against the predicted CDSs, total 1117 (77.57%) number of complete core genes were detected and 1272 (88.33%) complete and partial genes were detected (Table S2). After the final annotation, 10 CDSs having the significant hit with silicon transporter genes were identified as variant silicon transporter genes of finger millet. The maximum similarity of 94.26% was observed with Setaria italica silicon efflux transporter LSI2 isoform X1 (accession number XP_004985708.1) and minimum 81.22% with Panicum hallii silicon efflux transporter LSI2-like (accession number XP_025817714.1) (Table S3). Out of 10 predicted variant silicon transporter genes, 5 were Silicon-efflux transporter (EcLsi2) genes, 4 were silicon-efflux transporter (EcLsi3) genes and 1 was NOD26-like major intrinsic protein (EcLsi1) gene (Table S3).
Structural analysis of Silicon transporter protein
The nucleotide sequences were translated to amino acid sequences using Blastx tool available at NCBI (http:// blast.ncbi.nlm.nih.gov/ Blast.cgi). The basic information of physico-chemical properties including number of amino acids, molecular weight (kDa) and theoretical isoelectric point of silicon transporter (EcLsi) proteins was elucidated using Protparam tool of ExPASy (http://web.expasy.org/protparam/) (Table 1). The length of amino acid sequences of deduced silicon transporter proteins varied greatly ranging from 162 (EcLsi1_10) to 556 (EcLsi2_2) and molecular weight ranging from 17.04 (EcLsi1_10) to 60.26 kDa (EcLsi2_2) (Table 1). The highest and lowest theoretical iso-electric points (pI) of 8.52 and 5.22 were observed in EcLsi2 and EcLsi2_3 proteins respectively (Table 1). In addition, secondary structures of silicon transporter proteins were predicted by SOPMA tool and it was observed that the percentage of alpha helices (32.44 to 51.93), beta sheets (10.32 to 21.6), beta turns (3.25 to 5.56) and random coil (27.78 to 46.15) also varied greatly (Table 1). A total of six conserved motifs were identified in EcLsi2, while three conserved motifs each were identified in EcLsi1 and EcLsi6 proteins using the MEME search tool (http://meme.nbcr.net). The maximum of 15 motifs was identified in EcLsi2_2 and the minimum of 3 motifs in EcLsi1_10 protein. The unique motifs were also identified in finger millet silicon transporter proteins, EcLsi3_9 contained maximum two unique motifs i.e. motif-10 and −14, followed by EcLsi3_6 and EcLsi3_7which contained only one uniquemotif-17 (Fig. 2). But the most significant motifs possessing NPA/SPA domains, which is a feature of major intrinsic proteins, were found in 9 out of 13 silicon transporter proteins analyzed (Table 1). Particularly, NPA domain was identified in EcLsi2 and SPA domain in EcLsi1 and EcLsi6 proteins, indicating that NPA is highly conserved in finger millet Lsi2 proteins while SPA in Lsi1 and Lsi6 proteins. Further, the analysis of putative phosphorylation sites in silicon transporter proteins revealed that motifs 8 and 9 contained high number of conserved serine residues and were most probably the sites for phosphorylation (Table S4). The highest number of 22 potential serine phosphorylation sites was predicted in EcLsi2 (EcLsi2_2, EcLsi2_3, EcLsi2_4 and EcLsi2_5) and EcLsi3 (EcLsi3_9) proteins. The lowest number of 2 serine phosphorylation sites per gene were detected in EcLsi1 (EcLsi1_10) protein. EcLsi2 has an average of 18.17 potential serine phosphorylation sites per gene, which is considerably high compared to EcLsi1 and EcLsi3 proteins. Further, the sub-cellular localization of silicon transporter proteins was predicted using the Wolfpsort, CELLO2GO and TargetP servers (Table S5). The results predict that majority of silicon transporter proteins are located in the plasma membrane except EcLsi6 and EcLsi1_10 which are predicted to be located in endoplasmic reticulum and vacuole respectively. The amino acid sequences of silicon transporter proteins were used to build the tertiary structures by using Phyre2 tool. The 3-D structure of all silicon transporter protein revealed that all EcLsi proteins form the pore structure with helices, the typical characteristic of transporter proteins (Supplemental Fig. S1). Among them EcLsi2_1, EcLsi2_2 and EcLsi3_8 proteins contained maximum of 23 alpha helices each whereas the lowest of 7 alpha helices was observed in EcLsi1_10 protein. The transmembrane domain and topology of the silicon transporter protein was predicted by TMHMM2.0 and Protter tool respectively. All the identified silicon transporter proteins contained transmembrane domain indicating that all silicon transporter proteins are membrane bound and have an active role in transport across the cell. It was observed that EcLsi2 protein contained maximum number of 10 of transmembrane domains whereas EcLsi1_10 contained lowest 5 transmembrane domains (Supplemental Fig. S2). The topology prediction revealed that the EcLsi1 and EcLsi6 were the only proteins that had their N-terminal and C-terminal ends located in intracellular region (Supplemental Fig. S3), whereas, EcLsi2, EcLsi2_2, EcLsi2_4 and EcLsi3_9 showed extracellular location of N-terminal and C-terminal ends (Supplemental Fig. S3). The EcLsi2_1 was the only protein which showed intracellular N-terminal and extracellular C-terminal ends localization, while most of the silicon transporter protein showed extracellular N-terminal and intracellular C-terminal ends localization.
Table 1.
Characteristics of deduced protein of Silicon transporter genes identified.
| Gene Name | Deduced protein | Secondary Structure (%) | Number of Helices | TMHMM (TMHs) | Protter (TMHs) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A.A. Length | MW (kDa) | pI | NPA/SPA Domain | Instability index | Stable or unstable | Alpha Helix | Beta Sheet | Beta Turns | Rando m Coil | ||||
| EcLsi1 | 299 | 32.15 | 7.09 | NPA | 34.96 | Stable | 32.44 | 17.06 | 4.35 | 46.15 | 11 | 6 | 6 |
| EcLsi2 | 473 | 50.34 | 8.52 | SPA | 34.84 | Stable | 51.16 | 15.64 | 4.65 | 28.54 | 17 | 10 | 10 |
| EcLsi6 | 293 | 31.34 | 6.64 | NPA | 39.10 | Stable | 37.20 | 18.77 | 3.41 | 40.61 | 12 | 6 | 6 |
| EcLsi2_1 | 431 | 46.30 | 5.78 | — | 42.26 | Unstable | 46.87 | 13.46 | 3.25 | 36.43 | 23 | 5 | 7 |
| EcLsi2_2 | 556 | 60.26 | 5.96 | SPA | 39.05 | Stable | 47.84 | 15.65 | 4.68 | 31.83 | 23 | 9 | 10 |
| EcLsi2_3 | 436 | 47.64 | 5.22 | — | 43.02 | Unstable | 49.31 | 10.32 | 4.13 | 36.24 | 18 | 6 | 7 |
| EcLsi2_4 | 545 | 59.19 | 5.80 | SPA | 39.72 | Stable | 51.93 | 14.13 | 4.77 | 29.17 | 19 | 9 | 10 |
| EcLsi2_5 | 447 | 48.71 | 5.32 | — | 42.10 | Unstable | 48.77 | 11.86 | 4.47 | 34.90 | 19 | 6 | 7 |
| EcLsi3_6 | 518 | 55.92 | 6.01 | SPA | 36.88 | Stable | 47.88 | 12.74 | 5.02 | 34.36 | 22 | 8 | 9 |
| EcLsi3_7 | 518 | 55.92 | 6.01 | SPA | 36.88 | Stable | 47.88 | 12.74 | 5.02 | 34.36 | 22 | 8 | 9 |
| EcLsi3_8 | 519 | 55.93 | 6.01 | SPA | 37.39 | Stable | 47.40 | 14.26 | 5.20 | 33.14 | 23 | 9 | 11 |
| EcLsi3_9 | 480 | 52.05 | 5.24 | — | 39.45 | Stable | 46.46 | 14.79 | 3.54 | 35.21 | 18 | 7 | 8 |
| EcLsi1_10 | 162 | 17.04 | 6.18 | NPA | 27.91 | Stable | 45.06 | 21.60 | 5.56 | 27.78 | 7 | 5 | 5 |
(A.A. – Amino Acid, MW- Molecular Weight, pI- Isoelectric point, kDa- Kilo Dalton, TMHs- Transmembrane Helices).
Figure 2.
Organization of conserved motifs identified in finger millet silicon transporter proteins was predicted by MEME tools. The sequence and motifs identified were indicated by different coloured (1–20).
Phylogenetic analysis of silicon transporter genes
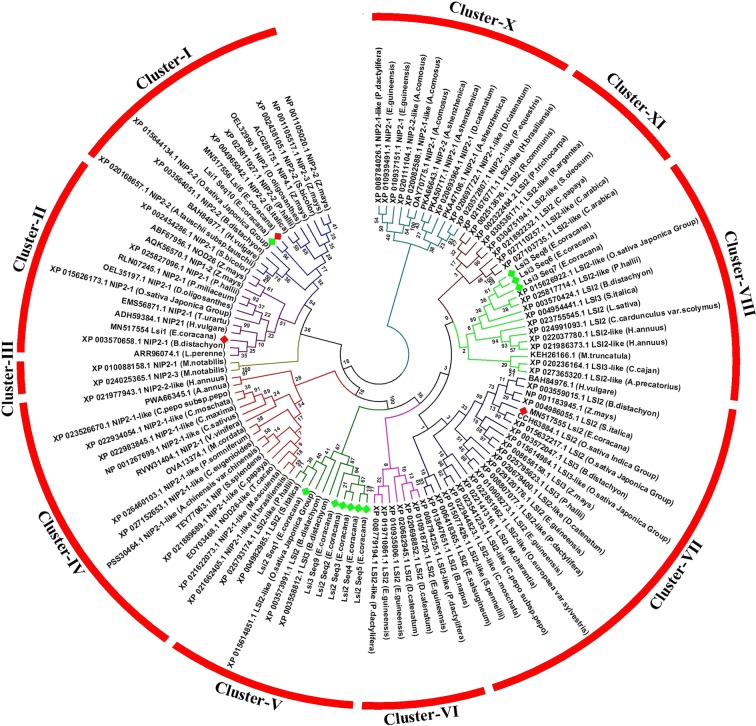
The full-length deduced protein sequences of silicon transporter genes of EcLsi1, EcLsi2 and EcLsi6 isolated from Var. Suvra and their homologs obtained through transcriptome assembly along with NCBI BLASTX hit sequences were used for phylogenetic analysis based on MSA (Multiple Sequence Alignment). The phylogenetic tree was constructed by employing bootstrap test (1000 replications) and neighbor joining method. The phylogenetic analysis revealed that silicon transporter genes from different plant species were grouped into ten main clusters (cluster-I to cluster-X). Among them cluster-VII was the largest cluster comprising 21 genes mainly Lsi2 including EcLsi2 (MN517555) while cluster-III was the smallest cluster containing only 2 genes (NIP2-1 and NIP2-3) from M. notabilis. Cluster-V contained mainly EcLsi2 genes whereas cluster-VIII contained EcLsi3 genes. The genes EcLsi1 and EcLsi6 were confined to cluster-I along with NIP2s (Nodulin 26-like intrinsic protein), a bona fide silicon transporter (Fig. 3). EcLsi1 and EcLsi6werefound to be closely related to S. italica followed by P.hallii, Z. mays and S.bicolar species with bootstrap value having 79 nodule stability (Fig. 3). It was observed that Lsi1 and Lsi6 are homologs belonging to Nod26-like major intrinsic protein (NIP2) subfamily of aquaporin protein family involved in silicon influx transport.EcLsi2 was found to be closely related to S.italica, B.distachyon and O.sativa japonica group, and EcLsi3 had more similarity with O. sativa japonica group and P.hallii (Fig. 3). TheLsi2 and Lsi3 efflux silicon transporters belong to anion transport family and have a significant identity with putative arsB transporter proteins. Furthermore, it was observed that Lsi1 might have co-evolved with Lsi6 genes in cereal species. Moreover, EcLsi2and EcLsi3 might have evolved from EcLsi1 and/or EcLsi6 and/or from both genes due to the gene duplication (Fig. 3 and Supplemental Fig. S3).
Figure 3.
Phylogenetic tree of silicon transporter proteins identified from finger millet along with different plant species generated based on homology hit. Phylogenetic tree was constructed using the neighbor-joining algorithm with 1000 replications in MEGA7 software. The proteins from finger millet are indicated with the prefixes Ec. Numbers on the tree node indicates bootstrap values in per cent. The genes isolated from finger millet are presented in red color while the genes identified by transcriptome assembly are presented in green color.
Expression analysis of silicon transporter genes
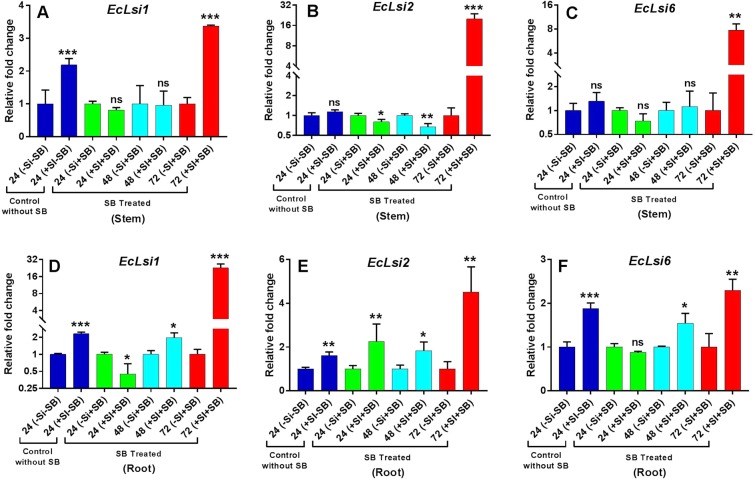
The qRT-PCR analysis is was performed to understand the expression patterns of silicon transporter genes (EcLsi1, EcLsi2 and EcLsi6) in both stem and root tissues in finger millet infested by stem borer and silicon amendment. The results showed that the expression levels of only. EcLsi1 (P = 0.001) was significantly increased in stem tissue, whereas in roots, significant up-regulation of all three genes viz. EcLsi1 (P = 0.000), EcLsi2 (P = 0.004) and EcLsi6 (P = 0.001) was observed in Si-treated plants (without SB infestation) over the non-silicon treated plants at 24 hpi (control) indicating that silicon alone was responsible for up-regulation of silicon transporter genes. Further, the expression analyses showed that all three tested genes (EcLsi1, EcLsi2 and EcLsi6) were induced by stem borer infestation in both stem and root tissues as expected. Among the three tested silicon transporter genes, EcLsi2 transcript had the highest expression observed in both stem and root tissues. However, the highest expression of EcLsi2 was observed in non-silicon treated plants in roots and stems in silicon-treated plants at 24 hpi (hours of post infestation). In roots, the t-test analysis showed a significant up-regulation of EcLsi2 (P = 0.057) and down regulation of EcLsi1 (P = 0.018) transcripts in silicon-treated and stems borer infested plants at 24 h hpi. Moreover, a significant increase in EcLsi1, EcLsi2 and EcLsi6 was observed at 48 (P = 0.020, P = 0.029 and P = 0.013) and 72 (P = 0.000, P = 0.007 and P = 0.004) hpi respectively in silicon treated plants over the non-silicon treated plants. Among all three tested genes, the highest and significant up regulation of EcLsi2 transcript was observed in root at 24hpi as compared to EcLsi1 and EcLsi6 in silicon treated plants. In stem, EcLsi2 was the only silicon transporter gene significantly down regulated at 24 (P = 0. 0.035) and 48 (P = 0.005) hpi in silicon treated plants over the non-silicon treated plants. The only significant up regulation of all three tested genes viz. EcLsi1(P = 0.000), EcLsi2 (P = 0.001) and EcLsi6 (P = 0.002) was observed in stem at 72 hpi in silicon treated plants over the non-silicon treated plants. These results indicate that stem borer infestation alone was responsible for up- regulation of EcLsi1 in root and EcLsi2 in stem in non-silicon treated plants despite of silicon availability. A considerable depression was observed up to 72 hpi which might be due to the silicon starvation or because of differential expression related to silicon availability. This major regulation of silicon transporter genes might be a feedback control in plants according to the silicon status or saturation in plants. This result indicates that the up-regulation of silicon transporter genes at 72 hpi is crucial and might have the role in defense mechanism. Compared to non-silicon treated plants, the only significant increase in relative fold change was observed for EcLsi2 transcript at all three time points viz. 24 (2.25), 48 (1.84) and 72 (4.51) hpi in root at 0.01, 0.5 and 0.01 level of significance respectively (Fig. 4E). The only significant decrease in fold change (0.45) was observed for EcLsi1 at 24 hpi (P = 0.018) in root (Fig. 4D). However, a significant decrease in fold change for EcLsi2 i.e. 0.80 and 0.68 was observed at 24 (P = 0.035) and 48 (P = 0.675) hpi in stem respectively (Fig. 4E). Moreover, the transcript levels of EcLsi1 (1.97 and 22.57) and EcLsi6 (1.54 and 2.29) were also significantly increased in root of silicon treated plants at 48 (P = 0.020 and P = 0.013) and 72 (P = 0.000 and P = 0.004) hpi respectively (Fig. 4D,F). In stem, a significant fold change for EcLsi1(3.38), EcLsi2(19.94) and EcLsi6 (7.77) was observed only at 72 hpi in silicon treated plants (Fig. 4A–C).
Figure 4.
Expression of representative genes (EcLsi1, EcLsi2 and Eclsi6) with (–Si–SB) control without Si and SB, and (–Si + SB) without silicon and with stem borer treatments at mentioned time points (24, 48 and 72 hpi) is considered as 1-fold after normalizing data with housekeeping gene (EcActin). The expression of genes in (+ Si–SB) control with Si without SB, and (+Si + SB) (with silicon and stem borer) is calculated as fold change. The fold change level of EcLsi1, EcLsi2 and Eclsi6 are presented in fig (A–C) in stem and (D—F) in root of finger millet respectively. Error bars represents ± SD (n = 3). The star above the bars graph represents different level significance obtained from paired t-test (*) at 0.05, (**) at 0.01 and (***) at 0.001 alpha levels respectively.
Expression analysis of defense hormone regulating genes
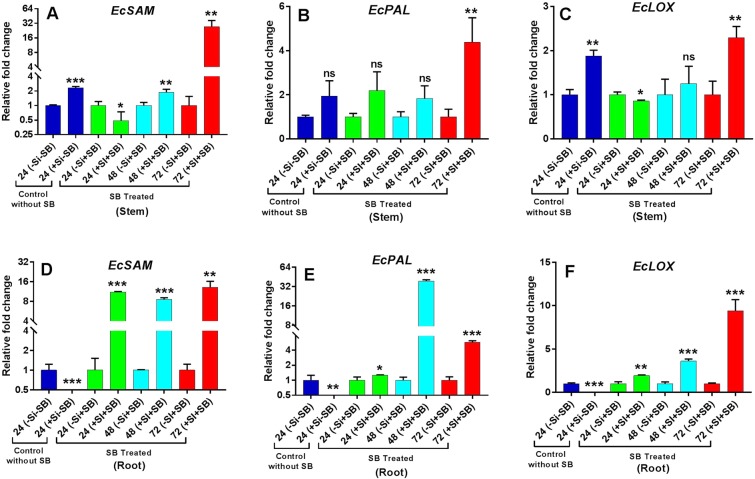
The relative expression of defense hormone regulation genes such as SAM (S-adenosyl-L- methionine), PAL (Phenyl-alanine ammonia-lyase) and LOX (Lipoxygenase) of Ethylene (ET), Salicylic acid (SA) and Jasmonic acid (JA) pathway respectively were analyzed. The expression levels of EcSAM (P = 0.000) and EcLOX (P = 0.000) genes were statistically significant at 24 hpi with mild transcriptional up-regulation in stem tissue with Si treatment (without SB infestation) indicating that the promoter(s) of these genes might be induced in response to silicon treatment, whereas down regulation of EcSAM (P = 0.001), EcPAL (P = 0.002) and EcLOX (P = 0.000) genes with no detectable transcripts were observed in root tissue. These observations indicate that these genes show spatial-temporal expression patterns. Further, the qRT-PCR analyses showed that all three tested genes were induced by stem borer infestation as expected. However, the highest significant increase of EcSAM (P = 0.053) and EcLOX(P = 0.022) was observed in non-silicon treated plants in stem at 24 hpi. T-test statistical analysis also showed that EcSAM and EcPAL transcripts were significantly increased in stem at 48 (P = 0.011 and 0.027) and 72 (P = 0.009 and 0.007) hpi in silicon treated plants over the non-silicon treated plants respectively. The up- regulation of EcLOX was found to be significant only at 72 hpi (0.004) over the non-silicon treated plants in stem. In case of roots, the stem borer infestation was not considerably responsible for induction of all three tested genes in non-silicon treated plants. But in the presence of silicon, the stem borer infestation was responsible for significantly increasing the transcript level of all three tested genes viz. EcSAM, EcPAL and EcLOX in roots at all three time periods i.e. 24 (P = 0.000, P = 0.049 and P = 0.002), 48 (P = 0.000, P = 0.000 and P = 0.000) and 72 (P = 0.002, P = 0.000 and P = 0.000) hpi over the non-silicon treated plants. In root, the highest transcript level was observed for EcSAM as compared to EcPAL and EcLOX at 24 hpi in silicon treated plants. Moreover, an antagonistic feedback control was observed in root between EcPAL (Salicylic Acid) and EcLOX (Jasmonic Acid) in silicon treated and stem borer infested plants. These results indicate that silicon was responsible for amplifying the transcripts level of all three tested genes in root under the stem borer infestation stress in finger millet plants. In stem, compared to non-silicon treated plants, significant fold change increases i.e. 27.04, 4.37 and 2.29 were observed for EcSAM, EcPAL and EcLOX transcripts at 72 hpi (P = 0.009, P = 0.007 and P = 0.004) respectively (Fig. 5A–C). In addition, EcSAM transcript was found to be increased by1.87 fold in stems at 48 hpi (P = 0.011) (Fig. 5A). Compared to non-silicon treated plants, decrease in transcript levels of EcSAM (0.48-fold change) and EcLOX (0.85-fold change) was seen in stems of silicon treated plants at 24 hpi (P = 0.053 and P = 0.022, respectively) (Fig. 5A–C). In case of root, the significant increase in expression was observed for all three tested genes at all-time points compared to non-silicon treated plants, the transcripts level of EcSAM, EcPAL and EcLOX were induced 11.11 (P = 0.000), 1.26 (P = 0.049) and 1.93 (P = 0.002) fold at 24 hpi respectively (Fig. 5D–F). At 48 hpi, the transcripts levels of EcSAM, EcPAL and EcLOX were increased by 8.53 (P = 0.000), 38.59 (P = 0.000) and 3.61 (P = 0.000) fold and at 72 hpi, they increased by 13.09 (P = 0.002), 5.69 (P = 0.000) and 9.41 (P = 0.000) fold respectively in root as compared to non-silicon treated plants after stem borer infestation (Fig. 5D–F).
Figure 5.
Expression of representative genes (EcSAM, EcPAL and EcLOX) with (–Si–SB) control without Si and SB, and (–Si + SB) without silicon and with stem borer treatments at mentioned time points (24, 48 and 72 hpi) is considered as 1 fold after normalizing data with housekeeping gene (EcActin). The expression of genes in (+Si–SB) control with Si without SB, (+Si + SB) with silicon and stem borer is calculated as fold change. The fold change level of EcSAM, EcPAL and EcLOX are presented in fig (A–C) in stem and (D–F) in root of finger millet respectively. Error bars represents ± SD (n = 3). The star above the bars graph represents different level significance obtained from paired t-test (*) at 0.05, (**) at 0.01 and (***) at 0.001 alpha levels respectively.
Inhibitory effect of Silicon content on tunnel length
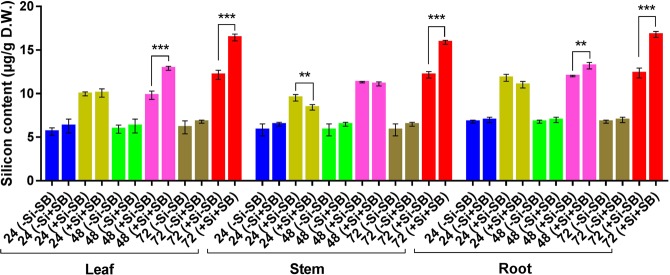
Silicon amendment with silicic acid hydrate in nutrient solution (1 mM/L) increased the silicon concentration in finger millet plants (Fig. 6). Silicon amendment alone was responsible for increasing the Si content in leaf, stem and root of Si-treated plants significantly as compared to non-treated plants at all three time-points i.e. 24, 48 and 72 hpi. The silicon content of Si-treated and stem borer infested plants was significantly increased from 9.82 to 12.90 µg/g of dry weight (P = 0.000) in leaf and from 12.00 to 13.20 µg/g of dry weight (P = 0.005) in root over non silicon treated plants at 48 hpi. The silicon content decrease was observed in stem at 24 hpi (P = 0.018) in Si-treated plants over the non-silicon treated plants. In Si-treated plants, the silicon content increased significantly in all the three plant tissues i.e. leaf (P = 0.000), stem (P = 0.000) and root (P = 0.000) at 72 hpi as compared to non-silicon treated plants (Fig. 6). The maximum silicon content was observed in roots (16.78 µg/g), followed by leaves (16.43 µg/g) and stem (15.88 µg/g) at 72 hpi in Si-treated plants.
Figure 6.
Silicon content of finger millet plant (µg/g) on dry weight basis in leaf, stem and roottissues at different time points. Error bars represents ± SD (n = 3). The star above the bars graph represents different level significance obtained from paired t-test (*) at 0.05, (**) at 0.01 and (***) at 0.001 alpha levels respectively.
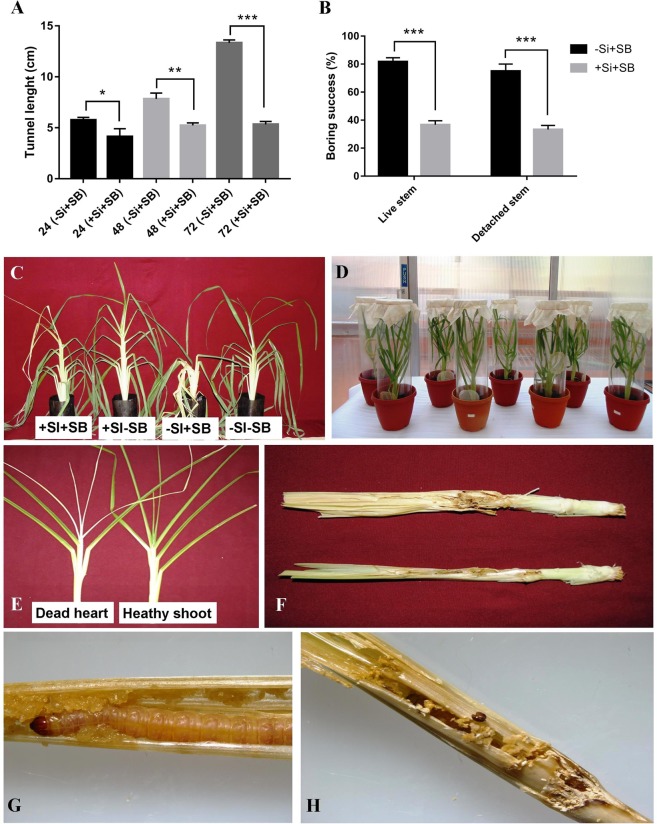
An increase in silicon concentration in plant tissue particularly in stem significantly reduced the tunnel length in Si-treated plants over the non-silicon treated plants infested with stem borer at all three time-points i.e. at 24 hpi (P = 0.026), 48 hpi (P = 0.002) and 72 hpi (P = 0.000) (Fig. 7A). The highest tunnel length (13.33 cm) was recorded at 72 hpi in non-silicon treated plant and the lowest (5.23 cm) at 48 hpi followed by 5.33 cm at 72 hpi in Si-treated plants indicating that silicon accumulation inhibits the feeding and boring ability of stem borer in finger millet. Additionally, relative silicification of leaf sheath in response to Si amendment and SB infestation was also measured by SEM-EDX analysis (Supplemental Fig. S4 and S5). The polymerized silicon dioxide in leaf sheath form dumbbell shape-like structures which are sparsely distributed (Supplemental Fig. S5 A-1 to C-1). Silicon deposition was not observed in non-silicon treated plants of both SB infested and without SB infested plants (Supplemental Fig. S4). The SB infestation and Si addition led to intensive cell silicification in ragi leaf sheath at 72 hpi (Supplemental Fig. S5 D-2). In Si-treated plants, the highest silicon deposition (0.31 weight per cent) was observed in silicon amended and SB infested plants (+Si + SB) followed by 0.12 weight percent in non-silicon infested plants (+Si-SB) at 72 hpi. The lowest silicon deposition i.e. 0.10 and 0.06 weight percent was observed in + Si–SB and + Si + SB respectively.
Figure 7.
Silicon amendment reduces stem borer infestation in finger millet plants. (A). Effect of silicon on feeding tunnel length (cm) measured with/without Si Treatment at different time points. (B). Larval boring success percentage of S. inferens larvae on Si-treated and un-treated plants of finger millet on live stem in greenhouse and on detached stem in laboratory. Error bars represents ± SD (n = 5). The star above the bars graph represents different level significance obtained from paired t-test (*) at 0.05, (**) at 0.01 and (***) at 0.001 alpha levels respectively. (C). Development of dead heart symptom in finger millet plants after one week under different treatments. (D). Experimental set up for measuring tunnel length and boring percentage on live stem in response to Si treatment. (E–H). Representative images of symptoms develop after SB infestation on finger millet plants (E) dead heart (F) feeding tunnel (G,H) frass.
Boring success on live stem
The percentage of third instar larvae that bored into the live stem of ragi plants within 72 h varied significantly under greenhouse conditions in Si-treated plants (P = 0.000) (Fig. 7B,D). Overall, the maximum percentage of larvae that bored into the stem was higher in Si-treated plants (81.67 ± 1.67%) as compared to non-silicon treated plants (37.33 ± 1.45%). The results showed that silicon accumulation in silicon treated stem reduced the boring percentage by 45.71% as compared to non-silicon treated plants.
Boring success on detached stem (‘detached stem’ assays)
Detached stem assays also showed the similar effects of Si on boring success of larvae (Fig. 7B,C,E). The results revealed that the percentage of boring success was significantly decreased in Si-treated plants (34.67 ± 2.03%) compared to non-silicon treated plants (75.67 ± 3.48%) (P = 0.000). It was observed that the success of boring percentage was reduced by 45.81% in Si- treated plants due to silicon accumulation compared to non-silicon treated plants.
Discussion
The beneficial effects of silicon in suppressing the numerous biotic and abiotic stresses by inducing the plant resistance have been evident in a wide range of plant species29. The majority of silicon accumulating crops are from the cereal family. Rice is well-known for its silicon uptake ability and can accumulate up to 10% of silicon on a dry weight basis in the shoot9. This higher accumulation of Si in different plant tissues was reported to be directly involved in physical barrier mechanism and indirectly in inducing the chemical defense mechanism against insect pests30. There is recent evidence that Si induces the defense mechanism against stem borers of economically important crops like wheat, rice, maize and sugarcane11. This induced defense mechanism depends on the silicon accumulation ability of crop plants governed by silicon transporter genes and their differential expression during stress. More recently, genes involved in Si uptake and distribution (Lsi1, Lsi2, and Lsi6) were identified in numerous plant species23,24.
In the present investigation, an attempt has been made to amplify and isolate the silicon transporter genes viz. EcLsi1, EcLsi2 and EcLsi6 from finger millet as it is not reported earlier. We isolated the three silicon transporter genes from Suvra variety of finger millet and it was observed that PCR amplification of EcLsi1, EcLsi2 and EcLsi6 produces 900 bp, 1422 bp and 882 bp long fragments respectively. After sequencing, the nucleotide sequences of all three genes were searched using BLAST tool in NCBI for identification and similarity search against different plant species. The BLAST results showed that all three isolated genes had similarity with silicon transporter genes. Recently, Sun et al.31,32 and Zellner et al.33 also isolated the silicon transporter genes from cucumber and tobacco plants. In addition, transcriptome library of finger millet was also assembled to nine homologs of silicon transporter genes. Total of ten expression transcripts of silicon transporter family were identified and named according to the BLAST hit identity such as EcLsi1 (1), EcLsi2 (5), and EcLsi3 (4). Similarly, several homologous genes OsLsi1, OsLsi2 and OsLsi6 have been isolated and functionally characterized in barley34,35, maize27,35, wheat36 and horsetail23.
Multiple sequence alignment of silicon transporter proteins amino acid residues showed that EcLsi1 and EcLsi6 were highly homologous to each other and also shared homology with other plant Lsi1s and Lsi6s. EcLsi2 and EcLsi3 shared homology with each other as well as with other Lsi2s and Lsi3s sequences of different plant species. Specific characteristics of NPA domain motifs existed in EcLsi1 and EcLsi6 while EcLsi2 and EcLsi3 contained SPA domain motifs and four amino acid residues for an ar/R selectivity filter36. Deshmukh et al.37 reported that all Nodulin 26-like intrinsic proteins (NIP2s) are bona fide silicon transporters which include Lsi1 and Lsi6 and contain NPA domain as a characteristic feature. However, EcLsi2 and EcLsi3 lack NPA domain motifs but instead contain SPA domain motif which is the characteristic feature of uncharacterized intrinsic proteins (XIPs) as reported by Deshmukh et al.37. Further, the subcellular localization prediction revealed that all silicon transporter proteins were located in plasma membrane. This subcellular localization of characterized silicon transporters is consistent with the previous studies reported in other plant species38. Similarly, Sun et al.32 found that CsLsi2 was localized at the plasma membrane which is the same as for HvLsi2 and ZmLsi2. These results indicate that EcLsi1, EcLsi2, EcLsi3 and EcLsi6 may be involved in silicon transport across the plasma membrane. However, Wolfpsort tool predicted that EcLsi6 and EcLsi1_10 may be located in endoplasmic reticulum and vacuole respectively suggesting that EcLsi6 and EcLsi1_10 play different roles in Si uptake.
Phylogenetic tree analysis revealed the evolutionary relationship between the silicon transporter genes among the different plant species. All the silicon transporter genes were grouped into ten main clusters (Cluster-I to X). Among them, cluster-V contained mainly EcLsi2 genes whereas cluster-VIII contained EcLsi3 genes, but both the clusters contain efflux silicon transporter genes belonging to anion transport family indicating that EcLsi2 and EcLsi3 are homologous to each other and also have a significant identity with putative arsB transporter proteins. Ma et al.26 also reported that Lsi2 proteins having a significant identity with arsB prokaryotic arsenic transporters are involved in anion transport. Other two silicon transporter proteins, EcLsi1 and EcLsi6, were grouped together in cluster-I along with NIP2s (Nodulin 26-like intrinsic protein), bona fide silicon transporters. It was observed that Lsi1 and Lsi6 are homologs belonging to Nod26-like major intrinsic protein (NIP2) subfamily of aquaporins involved in silicon influx transport. Ma et al.28 reported that OsLsi6 is the only homolog of OsLsi1 in rice and belongs to NIP group. Moreover, EcLsi1 and EcLsi6 were found to be closely related with other millet family member S.italica and also with other grass family members like P.hallii, Z.mays and S.bicolar. EcLsi2 was found to be closely related to S. italica, B.distachyon and O.sativa japonica group, while EcLsi3 had more similarity with O. sativa japonica group and P.hallii. Furthermore, it was observed that Lsi1 might have coevolved with Lsi6 in cereal crop species. Moreover, EcLsi2 and EcLsi3 might have evolved from EcLsi1 and/or EcLsi6 and/or from both genes due to a duplication event during evolution. The phylogenetic analysis of silicon transporter genes was found to be useful to establish the evolutionary relatedness and divergence in different plant species32,39.
The transmembrane domain and topology of all silicon transporter protein was predicted to have multi-spam helices as majority of protein structure was made up of alpha helices. It was confirmed that silicon transporter proteins are membrane-bound and involved in silicon transport across the cell. Ma and Yamaji9 have reported that the Lsi1 gene is predicted to encode a membrane protein similar to aquaporins, the water channels proteins. We also observed that both EcLsi1 and EcLsi6 proteins contained 6 transmembrane domains. Montpetit et al.36 demonstrated that silicon transporters have specific characteristics of six transmembrane domains which are well conserved in typical aquaporin proteins. However, EcLsi2 contained 10 transmembrane domains which might be due to duplication of EcLsi1 and EcLsi6 or fusion of both during evolution as evident by phylogenetic analysis. Likewise, Marron et al.40 suggested that the symmetrical 10 transmembrane domains of silicon transporter proteins (SIT) have independently evolved multiple times via duplication and fusion of 5 transmembrane domains of SIT-Ls, an extended family of related transporters present in some non-silicified organisms.
The present study revealed that the EcLsi1, EcLsi2 and EcLsi6 were differentially expressed in root and stem tissues of finger millet if infested by stem borer in silicon treated and untreated plants. The qRT-PCR analyses confirmed that EcLsi1 was primarily expressed in root followed by EcLsi6 and EcLsi1, indicating that EcLsi1is mainly involved in silicon uptake in root along with EcLsi6 and EcLsi1 genes. However, Wangkaew et al.41 showed that the Lsi1 and Lsi2 genes are highly expressed in rice root and involved in Si uptake in all growth stages. Ma and Yamaji38 also confirmed that Lsi1 and Lsi2 genes are involved in silicon uptake in rice root by knocking out Lsi1 and Lsi2 genes, resulting in significant decrease in Si uptake in the rice roots. Our study indicates that EcLsi1 is a highly expressed silicon efflux transporter involved in silicon uptake by root of finger millet, together with influx transporters, EcLsi6 and EcLsi1, during the stem borer infestation. In case of stem, the highest expression was observed for EcLsi1 followed by EcLsi6, indicating that EcLsi1 is the major silicon efflux transporter in root as well as stem during stem borer infestation and EcLsi6 is crucial in stems for silicon distribution in finger millet during stem borer attack. Similarly, Wangkaew et al.41 showed that the transcript level of Lsi6 was highly expressed in node, internode and flag leaf, especially at booting stage, resulting in increase of silicon concentration in panicle of rice plants. This result is also consistent with the suggestion by Yamaji and Ma42 that Lsi6 was involved in Si transportation to upper part of the rice plant. The expression of all three tested genes viz. EcLsi1, EcLsi2 and EcLsi6 were found to be significantly increased compared to non-silicon treated plants which indicates that the up-regulation of all these silicon transporter genes is crucial at 72 hpi against the stem borer attack. The results indicate that stem borer infestation alone was responsible for up-regulation of EcLsi1 in root and stem in non-silicon treated plants regardless of silicon availability. Thereafter, a considerable depression was observed up to 72 hpi that might be due to the silicon starvation or by a differential expression to the silicon availability. This major regulation of silicon transporter genes might be a feedback control mechanism of plants according to the silicon status or saturation in plants. This result also indicates that the up-regulation of silicon transporter genes at 72 hpi is crucial and might have a role in defense mechanism. Ye et al.43 reported that the reduced steady-state transcript levels of the Si transporters OsLsi1, OsLsi2, and OsLsi6 were observed in Si-pretreated plants after LF (Leaf Folder) attack. The common phyto-hormones salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) play primary roles in orchestrating plant defense responses15. JA is suggested to regulate defenses against both cell-content-feeding and tissue-chewing insects16,17. In the present study, the relative expression of genes encoding SAM (S-adenosyl-L-methionine), PAL (Pheny-alanine ammonia-lyase) and LOX (Lipoxygenase) involved in ethylene (ET), Salicylic acid (SA) and Jasmonic acid (JA) pathways respectively were analyzed. The qRT-PCR analyses showed that all three tested genes were induced by stem borer infestation as expected. The transcript levels of EcSAM were significantly increased at 48 and 72 hpi in stem whereas in root they were increased at all-time points i.e. at 24, 48 and 72 hpi in silicon treated plants indicating that silicon hasa direct role in triggering the ethylene-dependent defense pathway in finger millet against stem borer. Previous studies also indicate that ethylene works synergistically with jasmonic acid in regulation of plant defense responses against herbivorous insects44,45. Further, the expression analyses revealed that the transcript levels of EcPAL (Salicylic acid) were significantly increased at 48 and 72 hpi whereas EcLOX (Jasmonic acid) transcript significantly decreased and increased in silicon treated plants in stem at 24 and 72 hpi respectively. This result indicates that stem borer infestation and silicon application might have induced synergistic JA- and SA-dependent defense mechanisms in finger millet. Kahl et al.46 stated that attackers may manipulate plants for their own benefit by shutting down induced defenses by influencing the signaling network. However, an antagonistic feedback control mechanism was observed in roots between EcPAL (Salicylic Acid) and EcLOX (Jasmonic Acid) in silicon treated and stem borer infested plants. A significant increase in transcript level of EcPAL and EcLOX was observed in root indicating that silicon was responsible for amplifying the JA and SA levels under the stem borer stress in finger millet plants. Spoel et al.47 demonstrated that the SA and JA is antagonistic to each other and simultaneously induced the resistance against chewing and piercing-sucking insects.
The study was conducted to assess the effect of Si on stem borer infestation through length of the tunnel and boring on live stem under greenhouse and on detached stem (cut stem assay) in the laboratory. The results revealed that Si amendment significantly increased the silicon accumulation at 48 hpi in leaf and root and at72 hpi, silicon concentration was increased in leaf, stem and root. This increase in silicon accumulation was negatively correlated with tunnel length and boring success percentage on both live and detached stems of Si-treated plants over the non-silicon treated plants. Bandong and Litsinger48 have observed that the rice stem hardening due to deposition of lignin and cellulose on leaf sheath by silica deposition caused less penetration and reduced feeding tunnel length. Tripathy and Rath49 also observed the reduced feeding tunnel length in rice in Si-treated plants over the control against yellow stem borer. SEM-EDX analysis also revealed that silicon deposition in Si-treated leaf sheath of SB infested plants at 72 hpi was up by0.31 weight per cent indicating that accumulation of silicon subsequently reduced the tunnel length and boring success percentage significantly compared to non-silicon treated SB infested plants. The reduction in feeding tunnel length and boring success might be due to the wearing of mandibles which would prevent further penetration of larvae in silicon treated plants. Silicon is also known to reduce the digestibility of feed in the insect diet and hence an increased in silicon uptake in Si-treated plants might have inhibited the larval digestion. Studies by Ranganathan et al.50 and Chandramani et al.51 also support the findings of the present investigation.
In conclusion, the present study identified the silicon transporter genes from finger millet. The results highlighted that EcLsi1 and EcLsi6 genes are homologous silicon influx transporters while EcLsi2 and EcLsi3 genes are homologous silicon efflux transporters involved in uptake of silicon in finger millet. All silicon transporters proteins are membrane bound and predicted to be localized in plasma membrane allowing Si uptake in the finger millet plants. The phylogenetic and topology results suggest that EcLsi1 and EcLsi6 have co-evolved during evolution while EcLsi2 and EcLsi3 are evolved from either EcLsi1 and/or EcLsi6 or by fusion of both due to duplication event. The expression analyses indicate that stem borer infestation and silicon amendment increase the transcript levels of all three silicon transporter genes as well as defense phytohormone regulation genes significantly as compared to non-silicon treated plants in both stem and root at 72 hpi, indicating the crucial role of Si in inducing the defense mechanism against stem borer in finger millet. These findings also suggest that silicon transporters genes and defense phytohormone regulating genes act synergistically to mitigate the biotic stress induced by stem borer attack. The results also showed that Si amendment has a direct role in physical defense mechanism against the S.inferens due to intensified silicification in ragi leaf sheath which ultimately decreased feeding tunnel length and percent of boring success in ragi plants.
Materials and Methods
Plant materials and growth conditions
Finger millet seeds of var. Suvra susceptible to stem borer were collected from Pulses Research Centre (AICRP- All India Coordinated Research Project on Small Millets), OUAT, Berhampur, Odisha for the experiment. After germination, the germinated seeds were sown in plastic pots (27 × 19 × 4 cm) with soil less media (vermiculite 50% + Coco peat 50%) where the plants were supplemented with Hoagland solution52. After thirty days, a single seedling was transplanted to aplastic pot (50 × 40 × 15 cm) for further experimentation. The nutrient solution was prepared using deionized water (pH 6.0 ± 0.1). Si amendment (+Si) was carried out by adding silicic acid (Silicic Acid Hydrate, Himedia) to the nutrient solution at 1 mM Si/L and a control without addition of silicic acid (−Si) was included. The selected silicon concentration i.e. 1 mM Si/L was based on Nikolic et al.53 which suggested the silicon uptake rate was proportional to the solution silicon concentration in the range of 0–1 mM. At pH 6.0, nearly all the silicon is in the form of silicic acid54. The nutrient solution was replenished every five days. The plants were cultured in a greenhouse to prevent damage from rain and naturally occurring pests. Pesticides were not used throughout the experiment. Ragi plants at 60 days after transplanting (DAT) were used in the experiments.
Stem borer infestation and plants sampling
Ragi pink stem borer (SB) larvae were collected from ragi fields at AICRP-Small millet center, Berhampur, Odisha. The larvae were raised on susceptible var. Suvra until pupa formation. The pupas were collected and incubated in plastic jar covered with muslin cloth for adult emergence under the control conditions in growth chamber. After emergence, the adults were confined to caged susceptible ragi plants in the growth chamber for oviposition. A stock culture of the newly hatched first instar was maintained on ragi plants without Si addition in a growth chamber at 28 ± 1 °C, 70 ± 5% relative humidity (RH) and a 16 h photoperiod until the larvae reached the third instar stage used for infestation. The plants were treated with four treatments; (1) with Si addition and SB infestation (+Si + SB), (2) without Si addition and with SB infestation (−Si + SB), (3) with Si addition and without SB infestation (+Si-SB), and (4) without Si addition and SB infestation (-Si-SB)with three replications each (n = 3). The stems and roots were sampled at 24, 48, and 72 h post-infestation (hpi) time points and immediately frozen into liquid nitrogen and stored at −80 °C. The stem and root samples collected were used for expression pattern analysis of silicon transporter genes (EcLsi1, EcLsi2 and EcLsi6) and defense related phytohormones (ET- ethylene, JA- jasmonic acid and SA- salicylic acid) regulating genes (EcSAM- S-adenosyl-L-methionine, EcPAL- Phenylalanine ammonia-lyase and EcLOX-lipoxygenase respectively). The leaf samples were also collected for cloning of silicon transporter genes (EcLsi1, EcLsi2 and EcLsi6).
Measurement of Si content
Three plants per treatments were uprooted for collection of leaves, stems and root separately. Three samples per treatment were collected at different time points such as 24 h, 48 h and 72 h and oven dried at 70 °C for 3 days. Oven dried samples were fine-grinded and sieved through a 60 mm sieve. All the samples were further dried at 60 °C for 48 h to remove the moisture. Si content was determined by using molybdenum blue calorimetry method with slight modifications55. The absorbance was measured in Lambda 365 UV/Vis Spectrophotometer (PerkinElmer, USA) at 650 nm. The standard curved was drawn from Silicon standard solution provided by Merck Millipore (Germany). Subsequently, the Si content was determined from linear regression equation: y = 0.0067 × – 0.0121, R = 0.9977 and expressed as µg/g silicon in dry weight. In addition, scanning electron microscopy and energy dispersive X-ray analysis was performed to determine the relative Si content and silica deposition on leaf sheath surface of Si-treated and non-treated infested and non-infested plants coupled with EDX microanalysis mapping. EDX spectra of silicon were obtained using a sensitive mode with the scanned time of 3 min. The identification of the elements was done using auto identification against the built-in elemental database library.
Larval boring success study on live stem (in greenhouse)
Intact plants grown in the greenhouse was tested for larval boring success study. An additional set was also taken with Si-treated and non-treated greenhouse grown plants to assess the boring success of larvae. The no choice-study was performed by confining the third instar larvae to either Si treated or untreated control plants. The larval boring success was determined as the percentage of third instar larvae entering the stems within 72 h of being placed on plants. An experiment was set up in randomized block design (RBD) with five replications. After thirty days, the plants were infested with five third instar larvae of S. inferens per plant (Fig. 7D). The percentage of larvae bored into the stem was calculated by counting the larvae that remained outside the stems after 72 h of confining the insects with the plants. The larval boring was confirmed by observing the visible entry holes into the stem and the debris and fecal matter coming out of the stem. Based on these data, the boring success was calculated by using the following formula:
Larval boring success study on detached stem (In Laboratory)
A detached stem study was also performed in the laboratory to assess the effect of Si on boring success of S.inferens on stem. Thirty days old greenhouse grown plants treated with silicon and without silicon were sacrificed and stems were collected for the experiment. The detached stems were prepared by cutting a 20 cm long piece from the base of the tiller and placed in glass test tubes (2.5 × 20 cm). The detached stem end facing the test tube base was sealed with parafilm and the other end was sealed using a foam plug. The freshness of the stem was maintained by keeping a wet cotton plug on open end of test tube (Fig. 7G-H). The experimental design was RBD with five replications. The treatments consisted of two test tubes i.e. Si-treated and non-treated (control) and infested with five third instar S. inferens larvae per test tube. The percentage of larvae bored into the stem was calculated by counting the larvae that remained outside the stems after 72 h of confining the insects with the plants. The larval boring was confirmed by observing the visible entry holes into the stem and the frass coming out of the stem. Based on these data, the boring success was determined.
Isolation of Si transporter genes
Total RNA was isolated from stem, root and leaf tissues of finger millet variety “Suvra” amended with silicon under stem borer infestation at different time points by RNeasy Plant Mini Kit (cat. nos. 74903) as per the manual instructions. The concentration of RNA was quantified using the nanodrop spectrophotometer (Thermo scientific, USA). The absorbance and concentration were measured at 260 and 280 nm wavelength and the ratio of A260/A280 equal to 2.0 was considered as purified RNA. The total RNA was used for cDNA synthesis using Verso cDNA Synthesis Kit (Thermo Scientific, cat. nos. AB1453A) as per the manual instructions. Total RNA concentration of all samples was adjusted to 2 µg/µl. The PCR amplification of EcLsi1, EcLsi2 and EcLsi6 genes was done by taking cDNA as template and gene-specific primers (Table S1). PCR amplification was carried out in 25 μL reaction volume containing template 25–50 ng of cDNA, 2.5 μl PCR assay buffer with 2.5 mM MgCl2 (Promega), 2.0 μl of 200 μM dNTPs, 1 μl of 1U of high fidelity Pfu polymerase (M/S Emerk Bioscience, India), 1 μl of 10 pmol of each forward and reverse primers and final volume was adjusted with nuclease free water (Hi-media, India). PCR amplification was carried out in Thermal Cycler (BioRad) with PCR cycling conditions as follows: initial denaturation at 94 °C for 4 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 56–60 °C for 50 s, elongation at 72 °C for 50 s and final elongation at 72 °C for 8 min. Tris–borate–EDTA (TBE) buffer was used for electrophoresis of amplified PCR product in 1.5% (w/v) agarose gel (M/S EMerck Bioscience,India) and the gels were photographed using gel documentation system (BioRad). The single bands from agarose gel were eluted using Quick PCR purification kit (GeneiTM) as per the manual instructions. The purified single bands were sequenced using Sanger Dideoxy Sequencing method using 96 capillary high throughput sequencers (SaiGenom Labs Private Ltd, Cochin, India). The coding domain sequences (CDS) of EcLsi1, EcLsi2 and EcLsi6 genes isolated from finger millet variety Suvra has been submitted to GenBank, NCBI with the accession no. MN517554, MN517555 and MN517556 respectively.
De-novo transcriptome assembly
The silicon transporter genes were mined from assembled transcriptome library of finger millet Vr. ML-365. The high throughput ILLUMINA (Next Seq 500) sequence reads of leaf tissue (Ac. No. SRR4021829) was retrieved from NCBI database56. The sequence quality of the raw reads and trimmed reads including GC content of the sequences were checked before assembly by Fast QC program (http://www.bioinformatics.babraham.ac.uk/Projects/fastqc/). Further the Trimmomatic program was used for cleaning the raw reads and quality control (http://www.usadellab.org/cms/?page=trimmomati)57. The parameters of trimming were adjusted to remove low quality pair reads (score ≤ 20), adaptor contamination and short reads (minimum length: 20 nucleotides). The cleaned reads were then assembled using Trinity assembler (https://github.com/trinityrnaseq/trinityrnaseq/wiki) with default parameters58. All the reads derived from the trinity assembly were further used to generate one combined assembly to obtain the longer and complete sequences using the Cap3 tool (http://seq.cs.iastate.edu/cap3.html). The redundant reads from the assembly were removed using the CD-HIT-EST tool (http://www.bioinformatics.org/cd-hit/). The non-redundant sequences were annotated by comparing with all possible silicon transporter proteins available at NCBI database using BLASTx search with cut-off E-value ≤ 1.0e-5 and similarity score ≥ 60%. In addition, the reads without annotation result were separately compared with PLAZA 3.0, Uniprot and Ensemble plant protein database. This further survey of databases was to ensure the maximum annotation of putative contigs involved in silicon transport.
In-silico sequence analysis
After sequencing, the nucleotide sequences of EcLsi1, EcLsi2 and EcLsi6 were edited in BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and further translated into deduced amino acid sequence by using the BLASTx tool available at NCBI (http:// blast.ncbi.nlm.nih.gov/Blast.cgi). Along with these three deduced amino acid sequences, other putative amino acid sequences obtained through transcriptome assembly were also included in sequence analysis. The physio-chemical properties including number of amino acids, molecular weight (kDa), theoretical isoelectric point and instability index of silicon transporter proteins were elucidated in Protparam tool of ExPASy59(http://web.expasy.org/protparam/) (Table 1). The amino acid sequences of silicon transporter genes were then subjected to homology modeling search to build the protein structure by using online Phyre2 tool with default parameters60. The 3-D structures of silicon transporter protein were visualized in PyMOL software. Further the sequences were subjected to Multiple Sequence Alignment (MSA) using ClustalX program using default parameters. The MSA file was further used in MEGA7 (Molecular Evolutionary genetic analysis) software for phylogenetic analysis (version 3.1; http://www.megasoftware.net)61. The phylogenetic tree was constructed using the neighbor-joining method with bootstrap test and poisson model. Separate phylogenetic tree was constructed for three genes and combined tree was also generated to establish the evolutionary relationship between silicon transporter genes and crop species.
Expression analysis of genes
The relative expression pattern of silicon transporter genes viz. EcLsi1, EcLsi2, and EcLsi6 and defense hormone regulating genes viz. SAM (Ethylene), LOX (Salicylic acid), and PAL (Jasmonic acid) were analyzed using qPCR (CFX96TM Real-Time System, Bio-Rad, USA) using SSO Fast Eva Green Supermixes (BioRad, USA) following manufacturer’s instruction. The housekeeping gene, EcActin, was used as an internal control for normalization of data obtained. Reaction conditions for thermal cycling were 95 °C for 60 s, followed by 40 cycles of 95 °C for 20 s, 60 °C for 15 s, then 72 °C for 30 s. Fluorescence data were collected during the cycle at 60 °C. The melting curve was used as an internal check for assessing quality of the gene amplification. The experiments were repeated twice independently with three replicates each time. The threshold cycles (Ct) were used to find the relative expression level of all the genes over control using the comparative 2−ΔΔCt method62. Gene specific primers used in this study are listed in Table S6 and S7.
Supplementary information
Acknowledgements
The authors wish to acknowledge to Department of Science & Technology, Govt. of India for providing infrastructural facility under FIST grant to carry out the experiment. Authors wish to acknowledge to Department of Life Sciences and Bioinformatics, Central University, Silcher, Assam for providing laboratory facility.
Author contributions
Kundasing Rajpalsing Jadhao (K.R.J.) and Gyana R Rout (G.R.R.) designed the experiments. K.R.J. performed the experiments. K.R.J. and G.R.R. analyzed the data. K.R.J. wrote the draft paper. G.R.R. finalized the manuscript. Anuradha Bansal (A.B.) edited the whole manuscript. Authors have reviewed the manuscript and agreed to the manuscript contents.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-61182-0.
References
- 1.Rudin B, Shashidhar HE, Kulkarni RS, Hittalmani S. Genetic diversity assessment of finger millet, (Eleusine coracana (L) Gaertn), germplasm through RAPD analysis. PGR Newslett. 2004;138:50–54. [Google Scholar]
- 2.Reddy VG, Upadhyaya HD, Gowda CLL, Singh S. Characterization of Eastern African finger millet germplasms for qualitative characters at ICRISAT. J. SAT Agricultural Res. 2009;7:1–9. [Google Scholar]
- 3.Gahukar RT. Potential of minor food crops and wild plants for nutritional security in the developing world. J. Agriculture and Food Information. 2014;15:342–352. doi: 10.1080/10496505.2014.952429. [DOI] [Google Scholar]
- 4.Kumar A, Tomar V, Kaur A, Kumar V, Gupta K. Millets: a solution to agrarian and nutritional challenges. Agric. Food Secur. 2018;7:31. doi: 10.1186/s40066-018-0183-3. [DOI] [Google Scholar]
- 5.Sasmal A, Mohapatra AKB, Pradhan KC. Screening of finger millet genotypes against pink stem borer (Sesamia inferens Walker, Noctuidae, Lepidoptera) J. Entomology and Zoology Studies. 2018;6:796–799. [Google Scholar]
- 6.Li CX, Cheng X, Dai SM. Distribution and insecticide resistance of pink stem borer, Sesamia inferens (Lepidoptera: Noctuidae), in Taiwan. Formosan Entomol. 2011;31:39–50. [Google Scholar]
- 7.Gahukar RT, Reddy GVP. Management of Economically Important Insect Pests of Millet. J. Integrated Pest Management. 2019;10:28. doi: 10.1093/jipm/pmz026. [DOI] [Google Scholar]
- 8.Ma, J. F. & Takahashi, E. Soil, fertilizer and plant silicon research in Japan, Elsevier Science, Amsterdam, The Netherlands, 294 (2002).
- 9.Ma JF, Yamaji N. Silicon uptake and accumulation in higher plants. Trends in Plant Sci. 2006;11:392–397. doi: 10.1016/j.tplants.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Epstein E. Silicon. Annual Review of Plant Physiology and Plant Molecular Biol. 1999;50:641–664. doi: 10.1146/annurev.arplant.50.1.641. [DOI] [PubMed] [Google Scholar]
- 11.Debona D, Rodrigues FA, Datnoff LE. Silicon’s role in abiotic and biotic plant stresses. Ann. Rev. Phytopath. 2017;55:85–107. doi: 10.1146/annurev-phyto-080516-035312. [DOI] [PubMed] [Google Scholar]
- 12.Hou M, Han Y. Silicon-mediated rice plant resistance to the Asiatic rice borer (Lepidoptera: Crambidae): Effects of silicon amendment and rice varietal resistance. J. Econ. Entomol. 2010;103:1412–1419. doi: 10.1603/EC09341. [DOI] [PubMed] [Google Scholar]
- 13.Fauteux F, Rémus-Borel W, Menzies JG, Bélanger RR. Silicon and plant disease resistance against pathogenic fungi. FEMS MicrobiolLett. 2005;249:1–6. doi: 10.1016/j.femsle.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Ali JG, Agrawal AA. Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci. 2012;17:293–302. doi: 10.1016/j.tplants.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 15.De Vos M, et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 16.Kessler A, Baldwin IT. Plant responses to insect herbivory: The emerging molecular analysis. Ann. Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- 17.Kindt F, Joosten NN, Peters D, Tjallingii WF. Characterization of the feeding behavior of western flower thrips in terms of electrical penetration graph (EPG) waveforms. Jour Insect Physiol. 2003;49:183–191. doi: 10.1016/S0022-1910(02)00255-X. [DOI] [PubMed] [Google Scholar]
- 18.Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001;125:1074–1085. doi: 10.1104/pp.125.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, et al. Silicon supplementation alters the composition of herbivore induced plant volatiles and enhances attraction of parasitoids to infested rice plants. Front Plant Sci. 2017;8(725):1265. doi: 10.3389/fpls.2017.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds OL, Keeping MG, Meyer JH. Silicon-augmented resistance of plants to herbivorous insects: A review. Ann. Appl.Biol. 2009;155:171–186. doi: 10.1111/j.1744-7348.2009.00348.x. [DOI] [Google Scholar]
- 21.Hartley SE, DeGabriel JL. The ecology of herbivore-induced silicon defenses in grasses. Functional Ecol. 2016;30:1311–1322. doi: 10.1111/1365-2435.12706. [DOI] [Google Scholar]
- 22.Pappas ML, et al. Induced plant defences in biological control of arthropod pests: A double-edged sword. Pest Manag Sci. 2017;73:1780–1788. doi: 10.1002/ps.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grégoire C, et al. Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J. 2012;72:320–330. doi: 10.1111/j.1365-313X.2012.05082.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang HS, et al. Identification of two cucumber putative silicon transporter genes in Cucumis sativus. J. Plant Growth Regul. 2015;34:332–338. doi: 10.1007/s00344-014-9466-5. [DOI] [Google Scholar]
- 25.Mitani N, et al. Isolation and functional characterization of an influx silicon transporter in two pumpkin cultivars contrasting in silicon accumulation. Plant J. 2011;66:231–240. doi: 10.1111/j.1365-313X.2011.04483.x. [DOI] [PubMed] [Google Scholar]
- 26.Ma. JF. An efflux transporter of silicon in rice. Nature. 2007;448:209–12. doi: 10.1038/nature05964. [DOI] [PubMed] [Google Scholar]
- 27.Mitani N, Chiba Y, Yamaji N, Ma JF. Identification and characterization of maize and barley Lsi2-like silicon efflux transporters reveals a distinct silicon uptake system from that in rice. Plant Cell. 2009;21:2133–2142. doi: 10.1105/tpc.109.067884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma JF, Yamaji N, Mitani-Ueno N. Transport of silicon from roots to panicles in plants. ProcJapanAcadSer B Phys. Biol. Sci. 2011;87:377–385. doi: 10.2183/pjab.87.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma JF. Role of silicon in enhancing the resistance of plants to biotic and abiotic stresses. Soil Sci Plant Nutr. 2004;50:11–18. doi: 10.1080/00380768.2004.10408447. [DOI] [Google Scholar]
- 30.Yang L, et al. Silicon amendment is involved in the induction of plant defense responses to a phloem feeder. Scientific Reports. 2017;7:4232. doi: 10.1038/s41598-017-04571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun H, et al. Isolation and functional characterization of CsLsi1, a silicon transporter gene in Cucumissativus. Physiol Plantarum. 2017;159:201–214. doi: 10.1111/ppl.12515. [DOI] [PubMed] [Google Scholar]
- 32.Sun H, et al. Isolation and functional characterization of CsLsi2, a cucumber silicon efflux transporter gene. Annals of Bot. 2018;122:641–648. doi: 10.1093/aob/mcy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zellner W, Lutz L, Khandekar S, Leisner S. Identification of NtNIP2; 1: an Lsi1 silicon transporter in Nicotiana tabacum. Jour Plant Nutrition. 2019;42:1028–1035. doi: 10.1080/01904167.2019.1589500. [DOI] [Google Scholar]
- 34.Chiba Y, Mitani N, Yamaji N, Ma JF. HvLsi1 is a silicon influx transporter in barley. Plant Jour. 2009;57:810–818. doi: 10.1111/j.1365-313X.2008.03728.x. [DOI] [PubMed] [Google Scholar]
- 35.Mitani N, Yamaji N, Ma JF. Identification of maize silicon influx transporters. Plant Cell Physiol. 2009;50:5–12. doi: 10.1093/pcp/pcn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montpetit J, et al. Cloning, functional characterization and heterologous expression of TaLsi1, a wheat silicon transporter gene. Plant MolBiol. 2012;79:35–46. doi: 10.1007/s11103-012-9892-3. [DOI] [PubMed] [Google Scholar]
- 37.Deshmukh RK, et al. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 2013;83:303–315. doi: 10.1007/s11103-013-0087-3. [DOI] [PubMed] [Google Scholar]
- 38.Ma JF, Yamaji N. A cooperative system of silicon transport in plants. Trends in Plant Sci. 2015;20:435–442. doi: 10.1016/j.tplants.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Zhu YX, Gong HJ, Yin JL. Role of Silicon in Mediating Salt Tolerance in Plants: A Review. Plants. 2019;8:147. doi: 10.3390/plants8060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marron AO, et al. The evolution of silicon transport in eukaryotes. Molecular Biol.Evol. 2016;33:3226–3248. doi: 10.1093/molbev/msw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wangkaew B, Prom-u-thai CT, Jamjod S, Rerkasem B, Pusadee T. Silicon concentration and expression of silicon transport genes in two Thai rice Varieties. CMU J. Nat Sci. 2019;18:358–372. [Google Scholar]
- 42.Yamaji N, Ma JF. A Transporter at the node responsible for intervascular transfer of silicon in rice. The Plant Cell. 2009;21:2878–2883. doi: 10.1105/tpc.109.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye M, et al. Priming of jasmonate-mediated antiherbivore defense responses in rice by silicon. Proc. Nati. Acad. Sci. 2013;110:E3631–E3639. doi: 10.1073/pnas.1305848110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Ann. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 45.Wu J, Baldwin IT. New insights into plant responses to the attack from insect herbivores. Ann. Rev. Genetics. 2010;44:1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- 46.Kahl J, et al. Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta. 2000;210:336–342. doi: 10.1007/PL00008142. [DOI] [PubMed] [Google Scholar]
- 47.Spoel SH, et al. NPR1 modulates cross-talk between salicylate and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15:760–786. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandong JP, Litsinger JA. Rice crop stage susceptibility to the rice yellow stem borer Scirpophaga incertulas (Walker) (Lepidoptera: Pyralidae) Int. J. Pest Management. 2005;51:37–43. doi: 10.1080/09670870400028276. [DOI] [Google Scholar]
- 49.Tripathy S, Rath LK. Silicon induced resistance expression in rice to Yellow stem borer. J. Entomology and Zoology Studies. 2017;5:12–15. [Google Scholar]
- 50.Ranganathan S, et al. Effect of silicon sources on its deposition, chlorophyll content, and disease pest resistance in rice. Biol. Plantarum. 2006;50:713–716. doi: 10.1007/s10535-006-0113-2. [DOI] [Google Scholar]
- 51.Chandramani P, Rajendran R, Sivasubramanian P, Muthiah C, Chinniah C. Organic source induced silica on leaf folder, stem borer and gall midge population and rice yield. J. Biopesticides. 2010;3:423–427. [Google Scholar]
- 52.Hoagland, D. R. &Arnon, D. I. The water culture method for growing plant without soil. California Agri Exp. Sta Cir. No. 347 (University of California Berkley Press, CA, USA.) (1950).
- 53.Nikolic M, Nikolic N, Liang Y, Kirkby EA, Römheld V. Germanium-68 as an adequate tracer for silicon transport in plants. Characterization of silicon uptake in different crop species. Plant Physiol. 2007;143:495–503. doi: 10.1104/pp.106.090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi E, Hino K. Silica uptake by plant with special reference to the forms of dissolved silica. Jap J. Soil Sci and Manure. 1978;49:357–360. [Google Scholar]
- 55.Dai WM. Rapid determination of silicon content in rice. Rice Sci. 2005;12:145–147. [Google Scholar]
- 56.Hittalmani H, et al. Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics. 2017;18:465. doi: 10.1186/s12864-017-3850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gasteiger, E, et al Protein Identification and Analysis Tools on the ExPASy Server. In: Walker J.M. (ed) The Proteomics Protocols Handbook. Humana Press, New York, USA, pp 571–607 (2005).
- 60.Kelley LA, Mezulis S, Yates CM, Wass MN, Stemberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol and Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.