Abstract
Recent studies have highlighted the value of microRNA-21 (miR-21) as a prognostic biomarker in gliomas. However, the role of miR-21 in predicting prognosis remains controversial. We performed a comprehensive study based upon a meta-analysis and The Cancer Genome Atlas (TCGA) glioma dataset validation to clarify the prognostic significance of miR-21 in glioma patients. In this study, we searched Embase, PubMed, Web of science, CNKI, SinoMed, and Wanfang databases for records up to May 2018. Relevant data were extracted to assess the correlation between miR-21 expression and survival in glioma patients. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) were used to describe association strength. We further used multivariate Cox regression analysis to assess miR-21 expression in the TCGA glioma dataset to validate the relationship between miR-21 expression and survival. Nine studies were included in the meta-analysis. Among them, eight studies provided data on overall survival (OS) with a pooled HR of 1.91 (95% CI: 1.34, 2.73), indicating that higher expression of miR-21 was significantly associated with worse OS in glioma patients; for the other study, which provided data on progression-free survival (PFS), no statistically significant HR was reported for PFS in the glioma patients (HR = 1.23, 95% CI: 0.41, 3.72). A multivariate Cox regression analysis of the miR-21 expression in the TCGA glioma dataset revealed that overexpression of miR-21 was a potential independent prognostic biomarker of poorer OS (HR = 1.27, 95% CI: 1.01, 1.59) and poorer PFS (HR = 1.46, 95% CI: 1.17, 1.82). Our findings suggest that higher expression of miR-21 is correlated with poorer glioma prognosis.
Subject terms: CNS cancer, CNS cancer
Introduction
Gliomas are one of the most common central nervous system (CNS) glial neoplasms, accounting for 30% of all CNS tumors and 80% of malignant brain tumors1, respectively. According to the World Health Organization (WHO) classification, gliomas are classified as grade I through grade IV2. Low-grade gliomas (LGG, grades I–II) are well-differentiated and grow slowly, while high-grade gliomas (HGG, grades III–IV) are characterized by poor differentiation and rapid progression, and the prognosis of HGG is usually poor. Grade IV glioma, known as glioblastoma multiforme (GBM), is extremely aggressive, and comprises 55.1% of all gliomas3. Although recent progress has been made in surgical and radiotherapy techniques, the prognoses of GBM patients have not significantly improved4,5, and GBM remains one of the most incurable cancers6, with a 5-year survival rate of only 5.1%3. Some clinical factors impact the prognosis of glioma patients, such as age at diagnosis, WHO grade, duration of symptoms, tumor size, postoperative treatment, and karnofsky performance score (KPS)7,8. However, studies have shown that biological alterations in specific genes or molecules can affect the prognosis of glioma patients7,8. Therefore, clinical factors alone are not sufficient to evaluate the prognoses of glioma patients, and further research is needed to search for better prognostic biomarkers.
Micro-RNAs (miRNAs) are a class of small non-coding RNAs of 18–25 nucleotides with the ability to regulate gene expression at a post-transcriptional level by targeting messenger RNA (mRNA);9,10 miRNAs serve as key regulatory components in various biological processes, including cell proliferation, differentiation, angiogenesis, and apoptosis11,12. Dysregulation of miRNAs may lead to certain pathological states, such as cancer13. Recently, many studies have revealed that miRNAs can function as oncogenes or tumor suppressors in cancer, affecting the clinical outcome14,15. Studies have shown that miRNA expression is related to prognosis in patients with gliomas; for example, elevated expressions of microRNA-21 (miR-21), microRNA-10b (miR-10b), and microRNA-221/222 (miR-221/222) in patients with gliomas are correlated with shorter overall survival (OS) time16–18. This suggests that for the patients with gliomas, survival outcome can be predicted; therefore, miRNA expression may be clinically useful for management and prognosis.
Overexpressions of miR-21, also known as oncomiR, has been reported in various cancers, including gliomas19–22. Previous studies have shown that overexpression of miR-21 is associated with poor survival of glioma patients23–29. However, two studies reported insignificant correlations between miR-21 and prognosis in glioma patients30,31.
Meta-analysis is a method that allows quantitative analysis of the results of multiple independent studies, and has been used extensively to analyze the relationship between specific genes and prognosis in cancer patients32–34. Because of the inconsistent findings on the prognostic value of miR-21 in gliomas, a literature-based meta-analysis would be beneficial. A meta-analysis concerning the association between miR-21 expression and OS in glioma patients was published in 2016;35 however, several new studies on miR-21 expression and OS in glioma patients have been published since then. The Cancer Genome Atlas (TCGA) dataset has a large sample size of glioma patients, and the data have been standardized. Therefore, we performed an updated literature-based meta-analysis and also analyzed the miR-21 levels obtained from the TCGA database (https://portal.gdc.cancer.gov/) to elucidate the prognostic significance of miR-21 expression in glioma patients.
Results
Meta-analysis. study selection
A total of 465 articles were identified from the literature databases according to the inclusion criteria. Of these articles, 440 articles were removed because they were reviews, or were overlapped and irrelevant studies. Of the remaining 25 candidates, 16 articles were excluded because of one of the following: conference abstract (n = 4), lacking sufficient data to calculate hazard ratios (HRs) (n = 3), duplicated data (n = 5), and studying a set of microRNAs but not miR-21 alone (n = 4). Finally, nine articles were enrolled in our evaluation of the association between miR-21 expression and glioma prognosis. A flow chart of the eligible study selection process is shown in Fig. 1.
Figure 1.

Flow chart of the study selection process.
Characteristics of enrolled studies
The nine included studies for this meta-analysis were cohort studies, and the main characteristics of these nine studies are summarized in Table 1. The enrolled nine studies from China (n = 5), USA (n = 2), Denmark (n = 1), and Austria (n = 1) were published between 2010 and 2017, and included a total of 1,059 glioma patients. Of the nine studies, the tumor grades of the glioma patients were heterogeneous. Three studies included patients with grades I–IV, two studies with grades II–IV, one study with grades III–IV, and three studies with grade IV only. The expressions of miR-21 were all examined in glioma tissues. The miR-21 expressions were verified by quantitative real-time polymerase chain reaction (qRT-PCR) in eight studies, and by in situ hybridization (ISH) in one study. The follow-up periods of the nine studies ranged from 44 months–120 months. The cut-off values used to define the high- or low-expression of miR-21, the adopted therapeutic regimen for the glioma patients, and the reference of miR-21 quantified in the tumor tissues in the nine included studies were different. Among the nine included studies, one study31 provided data on progression-free survival (PFS), and the other eight studied OS. The values of HR and 95% confidence interval (95% CI) were extracted from original data in six studies, and in the other three studies, the values were estimated using Kaplan-Meier curves (K-M curves) (n = 2) or original data (n = 1). A multivariate Cox regression analysis was performed in five studies23–26,29, and the adjustment variables were age, sex, KPS and the malignancy grade of glioma, among others. The Newcastle-Ottawa scales (NOSs) for all the eligible studies were assigned more than five stars, indicating a high methodological quality in the included studies.
Table 1.
Characteristics of the enrolled studies.
| First Author, Publication Year | Location of Sample Collection | n | miRNA Source | miRNA Assay | plain housekeeping miRNAs | Cut-off*(Actual Value) | Grade | Follow-up (months) | Outcome | HR (95% CI) | Extracting Method | Adjustment Variables | NOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang28 | China | (Asian) | 92 | tissue | qRT-PCR | U6 | Mean (—) | III-IV | 96 | OS | 3.401 (1.296, 8.922) | Reported | — | 7 |
| Qu27 | China | (Asian) | 35 | tissue | qRT-PCR | RNU6B | 1.5-fold (—) | II-IV | 72 | OS | 2.66 (1.02, 6.92) | K-M curve | — | 5 |
| Shi23 | China | (Asian) | 198 | tissue | qRT-PCR | GAPDH | median (—) | II-IV | 102 | OS | 1.634 (1.083, 2.467) | Reported | Age, Sex, KPS, WHO grade | 7 |
| Sathyan30 | USA | (Non-Asian) | 69 | tissue | qRT-PCR | EEF1A | median (—) | IV | 90 | OS | 1.63 (0.82, 3.22) | K-M curve | — | 5 |
| Barbano26 | USA | (Non-Asian) | 185 | tissue | qRT-PCR | RNU48 | mean (—) | IV | 120 | OS | 1.19 (1.01, 1.41) | Reported | MGMT unmethylated, IDH1 mutation, Treatment, Recurrence | 7 |
| Hermansen29 | Denmark | (Non-Asian) | 189 | tissue | ISH | U6 | mean (—) | I-IV | 70 | OS | 1.545 (1.002, 2.381) | Reported | Age, WHO grade | 9 |
| Wu24 | China | (Asian) | 152 | tissue | qRT-PCR | U6 | median (20.99) | I-IV | 60 | OS | 3.17 (2.39, 4.179) | Reported | Age, Gender, WHO grade, KPS | 8 |
| Zh25 | China | (Asian) | 124 | tissue | qRT-PCR | Has-miR-16 | median (—) | I-IV | 98 | OS | 1.882 (1.07, 3.308) | Reported | Gender, Age, WHO grade, hsa-miR-106a, hsa-miR-181b | 9 |
| Ilhan-Mutlu31 | Austria | (Non-Asian) | 15 | tissue | qRT-PCR | RNU6B | median (—) | IV | 44 | PFS | 1.23 (0.41, 3.72) | Data-extrapolation | — | 5 |
*The cut-off values to define the high- or low-expression of miR-21.
Correlation between miR-21 expression and OS
Among the nine studies enrolled in evaluating the association of miR-21 expression and glioma prognosis, eight studies provided data on OS, and one study provided data on PFS31. For the eight studies with OS data, the random-effect model was adopted to calculated the pooled HR and 95% CI because of the high heterogeneity among studies (I2 = 82.0%, P < 0.001). The pooled result (HR = 1.91, 95% CI: 1.34, 2.73) showed that a higher miR-21 expression significantly predicted a poorer OS in the patients with gliomas (Fig. 2).
Figure 2.
The association between miR-21 expression and OS in eight studies.
Subgroup analyses
Subgroup analyses were conducted to explore the causes of heterogeneity according to locations of the sample collection, methods of the miRNA assay, cut-off values of miR-21 expression, follow-up periods, and adjustment variables (Table 2). The subgroup analysis by location of sample collection showed that a higher expression of miR-21 predicted poorer prognoses in Asian glioma patients (HR = 2.37, 95% CI: 1.68, 3.35) compared with that in Non-Asian patients (HR = 1.25, 95% CI: 1.07, 1.45). The subgroup analysis by different miRNA test methods indicated that the qRT-PCR group (HR = 1.99, 95% CI: 1.31, 3.01) predicted worse prognoses with higher expressions of miR-21 in the glioma patients. Concerning subgroups by different cut-off values, a significant HR was found in the median group (pooled HR = 2.09, 95% CI: 1.41, 3.11), while the mean group showed an insignificant pooled HR (HR = 1.51, 95% CI: 1.00, 2.27). According to the subgroups of follow-up time, a follow-up time of > 60 months group showed a significant HR (pooled HR = 1.36, 95% CI: 1.19, 1.56), indicating that higher miR-21 expression was associated with a poorer OS in glioma patients. The subgroup analysis by the adjustment variables of the multivariate Cox regression analysis showed that higher expression of miR-21 predicted poorer prognoses (HR = 1.78, 95% CI: 1.15, 2.75).
Table 2.
Subgroup analyses of associations between miR-21 expression and OS.
| Subgroup | Number of Studies | Number of Patients | Pooled model | Pooled HR (95% CI) | Heterogeneity | Publication bias (P-value) | ||
|---|---|---|---|---|---|---|---|---|
| I2 (%) | P-value | Begg’s test | Egger’s test | |||||
| All | 8 | 1059 | random | 1.91 (1.34, 2.73) | 82 | <0.001 | 0.216 | 0.236 |
| Locations | ||||||||
| Asian | 5 | 509 | random | 2.37 (1.68, 3.35) | 52 | 0.083 | 0.624 | 0.719 |
| Non-Asian | 3 | 640 | fixed | 1.25 (1.07, 1.45) | 0 | 0.400 | 0.602 | 0.164 |
| Method | ||||||||
| qRT-PCR | 7 | 960 | random | 1.99 (1.31, 3.01) | 85 | <0.001 | 0.293 | 0.262 |
| ISH | 1 | 189 | — | 1.55 (1.00, 2.38) | — | — | — | — |
| Cut-off value | ||||||||
| Median | 4 | 705 | random | 2.09 (1.41, 3.11) | 67 | 0.028 | 0.497 | 0.195 |
| Mean | 3 | 409 | random | 1.51 (1.00, 2.27) | 63 | 0.068 | 0.117 | 0.087 |
| 1.5-fold | 1 | 35 | — | 2.66 (1.02, 6.93) | — | — | — | — |
| Follow-up (month) | ||||||||
| ≤60 | 1 | 152 | — | 3.17 (2.40, 4.19) | — | — | — | — |
| >60 | 7 | 997 | fixed | 1.36 (1.19, 1.56) | 43 | 0.107 | 0.051 | 0.000 |
| Adjustment variables | ||||||||
| Yes | 5 | 848 | random | 1.78 (1.15, 2.75) | 89 | <0.001 | 0.624 | 0.447 |
| No | 3 | 211 | fixed | 2.22 (1.37, 3.59) | 0 | 0.433 | 0.117 | 0.165 |
Sensitivity analysis
A sensitivity analysis showed that the removal of individual studies, in turn, did not change the HR effect of the combined effect (range of pooled HRs: 1.36–2.10, all lower limits of the 95% CIs > 1.0) (Table 3), indicating that the result was stable in the meta-analysis.
Table 3.
Sensitivity analysis of pooled HRs of higher miR-21 expression for OS.
| Study Omitted | Pooled HR | 95% CI | Heterogeneity | |
|---|---|---|---|---|
| I2 (%) | P-value | |||
| Zhang28 | 1.82 | (1.26, 2.63) | 84 | <0.001 |
| Qu27 | 1.86 | (1.28, 2.70) | 84 | <0.001 |
| Shi23 | 1.97 | (1.30, 2.99) | 85 | <0.001 |
| Sathyan30 | 1.95 | (1.32, 2.89) | 85 | <0.001 |
| Barbano26 | 2.10 | (1.56, 2.82) | 54 | 0.042 |
| Hermansen29 | 1.99 | (1.31, 3.01) | 85 | <0.001 |
| Wu24 | 1.36 | (1.19, 1.56) | 43 | 0.107 |
| Zhi25 | 1.92 | (1.29, 2.86) | 84 | <0.001 |
Publication bias
Publication bias was evaluated by Begg’s test and Egger’s test. For the eight studies, the Begg’s test (P = 0.216), and Egger’s test (P = 0.236) provided no evidence of publication bias (Table 2).
Correlation between miR-21 expression and PFS
In this meta-analysis, only one study exploring the relationship between miR-21 expression and the survival outcome on PFS in glioma patients was included. No significant HR was reported in the study (HR = 1.23, 95% CI: 0.41, 3.72).
TCGA data extraction
Of the glioma patients in TCGA dataset, 641 glioma patients were selected according to the selection criteria to verify the prognostic significant of miR-21, and the clinical features of the 641 patients are summarized in Table 4.
Table 4.
Clinical information of glioma patients from TCGA dataset (n = 641).
| Variables | Overall (n = 641) | high miR-21 expression (n = 330) | low miR-21 expression (n = 311) | P-value* |
|---|---|---|---|---|
| Average age at diagnosis (mean ± standard deviation, year) | 51.4 ± 15.2 | 55.1 ± 13.7 | 47.6 ± 15.7 | <0.001 |
| Grade (n, II/III/IV) | 110/136/395 | 23/54/253 | 87/82/142 | <0.001 |
| Gender (n, male/female) | 364/277 | 199/131 | 165/146 | 0.064 |
| KPS (n, <80/≥80) | 123/518 | 78/252 | 45/266 | 0.003 |
| Median OS time (day) | 714 | 550 | 1315 | <0.001 |
| Median PFS time (day) | 463 | 11.2 | 32.7 | <0.001 |
*Comparison of high miR-21 expression group and low miR-21 expression group.
TCGA data validation
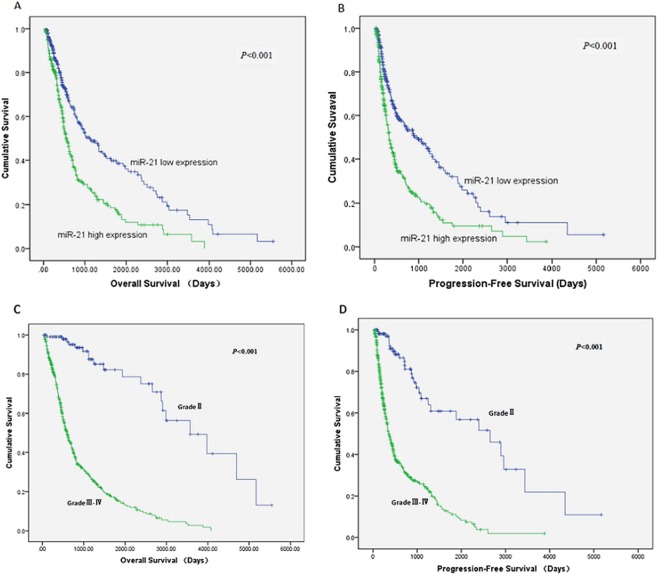
To validate the results of the meta-analysis, the TCGA glioma dataset was used to analyze the relationship between miR-21 expression and survival in the patients with gliomas. Figure 3 shows the Kaplan-Meier estimates for the high miR-21 expression group and the low miR-21 expression group. The results of a log-rank test showed that the glioma patients with high levels of miR-21 expression had a poorer OS (P < 0.001) (Fig. 3A) and a poorer PFS (P < 0.001) (Fig. 3B), and the results of a log-rank test showed that the glioma patients with grade III–IV had a poorer OS (P < 0.001) (Fig. 3C) and a poorer PFS (P < 0.001) (Fig. 3D). A multivariate Cox regression analysis indicated that the OS-related variables were miR-21 expression, tumor grade, age at diagnosis, gender, and KPS (Table 5), while the PFS-related variables were miR-21 expression, tumor grade, age at diagnosis and gender (Table 6). Meanwhile, the results of the multivariate Cox regression analysis showed that high miR-21 expression was an independent prognostic biomarker for a poorer OS (HR = 1.27, 95% CI: 1.01, 1.59) and poorer PFS (HR = 1.46, 95% CI: 1.17, 1.82) in patients with gliomas (Tables 5 and 6).
Figure 3.
Kaplan-Meier estimates for glioma patients from the TCGA glioma dataset. (A) Kaplan-Meier estimates of OS for groups with high and low miR-21 expression; (B) Kaplan-Meier estimates of PFS for groups with high and low miR-21 expression; (C) Kaplan-Meier estimates of OS for groups with grade II and grade III–IV; (D) Kaplan-Meier estimates of PFS for groups with grade II and grade III–IV.
Table 5.
Multivariate Cox regression analysis for OS in glioma patients.
| Variables | HR | 95% CI | P-value |
|---|---|---|---|
| miR-21 (high/low) | 1.27 | (1.01, 1.59) | 0.042 |
| Grade (III–IV/II) | 6.83 | (4.06, 11.50) | <0.001 |
| Age at diagnosis (≥50/<50) | 2.55 | (1.98, 3.29) | <0.001 |
| Gender (male/female) | 1.32 | (1.06, 1.65) | 0.014 |
| KPS (<80/≥80) | 2.11 | (1.62, 2.75) | <0.001 |
Table 6.
Multivariate Cox regression analysis for PFS in glioma patients.
| Variables | HR | 95% CI | P-value |
|---|---|---|---|
| miR-21 (high/low) | 1.46 | (1.17, 1.82) | 0.001 |
| Grade (III–IV/II) | 3.96 | (2.64, 5.96) | <0.001 |
| Age at diagnosis (≥50/<50) | 1.84 | (1.46, 2.32) | <0.001 |
| Gender (male/female) | 1.29 | (1.04, 1.60) | 0.023 |
| KPS (<80/≥80) | 0.97 | (0.71, 1.31) | 0.828 |
Discussion
Alterations in the expression levels of specific miRNAs can be easily and stably detected in tumor tissues17–20, plasma and serum36,37 and cerebrospinal fluid (CSF)38,39. Therefore, miRNAs are potential tumor biomarkers40–43. In recent years, an increasing number of studies have shown that miRNAs are valuable for predicting tumor prognosis20,22,40–43. Previous studies have shown that the role of miR-21 in predicting the prognoses of glioma patients remains controversial, possibly because small sample sizes could have led to an inadequate statistical ability to detect certain relationships in individual studies. Therefore, our findings in this meta-analysis and the multivariate Cox regression analysis of the TCGA glioma data on miR-21 expression to assess the prognostic value of miR-21 in glioma patients are more believable.
A meta-analysis to assess the relationship between miR-21 expression and OS in glioma patients was first performed in 201635. Compared with that study, our current meta-analysis included four new eligible studies, and we also analyzed the relationship between miR-21 expression and PFS in glioma patients. In our study, we found that the expression level of miR-21 is associated with prognosis, with a pooled HR of 1.91 (95% CI: 1.34, 2.73). This suggests patients with higher miR-21 expression have shorter OS.
Gliomas are clinically complex tumors with various manifestations. There are many factors affecting the prognoses of glioma patients. In analyzing the relationship between miR-21 and prognosis, it is important to consider confounding effects caused by the degree of malignancy as well as other factors23–31. In this meta-analysis, five included studies used a multivariate Cox regression analysis to study the relationship between miR-21 expression and OS, and in each of these five studies, the covariate confounding effect was adjusted by performing a multivariate Cox regression analysis. The pooled HR for the five included articles was 1.78 (95% CI: 1.15, 2.75), indicating that glioma patients with higher miR-21 expression have shorter OS.
In addition, the multivariate Cox regression analysis of covariates (the miR-21 expression, grade, age at diagnosis, gender, and KPS) in the TCGA glioma datasets revealed that overexpression of miR-21 is a potential independent prognostic biomarker of poorer OS (HR = 1.29, 95% CI: 1.01, 1.59) and poorer PFS (HR = 1.46, 95% CI: 1.17, 1.82) in glioma patients. Therefore, based on the meta-analysis results and the validation results by the TCGA dataset, we believe that miR-21 is a significant and independent prognostic biomarker for a glioma patient survival.
The associations between elevated miR-21 expression and poor survival can partly be explained by its role in the cascade of tumorigenesis and progression. The gene miR-21 is up-regulated in gliomas, and its oncogenic effect may be mediated through regulation of certain transcriptional targets and downstream signaling pathways. Currently, some tumor suppressor genes have been identified as targets of miR-21, such as programmed cell death 4 (PDCD4)44,45 and phosphatase and tensin homolog (PTEN)46. Furthermore, cellular pathways such as p53 and the PI3K-Akt pathway, are also part of the miR-21 regulatory network47,48. By attenuating or inhibiting these tumor suppressor genes, miR-21 can promote tumor proliferation, invasion, and metastasis, and reduce sensitivity to chemotherapy, thereby affecting the prognosis of glioma patients.
Because of the existence of heterogeneity among included studies, subgroup analyses were conducted according to locations of sample collection, methods of the miRNA assays, cut-off values of miR-21 expression, and follow-up periods. The results showed that a higher expression of miR-21 was predictive of poorer prognoses in Asian glioma patients (HR = 2.37, 95% CI: 1.68, 3.35); and the prognostic effect of miR-21 in the glioma patients could be influenced by the follow-up periods, test methods, and cut-off values of miR-21 expression. The different adopted therapeutic regimen for the glioma patients and the different reference of miR-21 quantified in the tumor tissues may lead to heterogeneity between the included studies. There were three included studies conducted in non-Asian subjects, one of which29 employed an ISH technique and the other two of which26,30 employed qRT-PCR technique. Although the methodological approach to quantify miR-21 were different, the heterogeneity among the three studies was not significant (I2 = 0%, P = 0.400). In the included studies, the different cut-off values were used to define the high- or low-expression of miR-21, which may affect the power of miR-21 as a prognostic biomarker in glioma. The random effect model and subgroup analyses conducted according to the cut-off values of miR-21 expression were performed to weaken the influence of heterogeneity on the conclusion of miR-21 as a predictive biomarker in glioma. There was no significant publication bias identified in this meta-analysis, and the result of the sensitivity analyses also showed the robustness of our results.
The literature review for this updated meta-analysis was thorough and adequately chosen. Concurrently, the TCGA data used to verify the meta-analysis results of this study are standardized data, and the sample size of the TCGA data set is large, so the results of this study are reliable. However, it should be noted that there were some limitations existed in our study. Firstly, there was heterogeneity in the included studies. Although we performed subgroup meta-analyses and adopted a random-effect model to minimize the effects of the heterogeneity, the heterogeneity among the studies may still affect the reliability of the combined results. For example, in a subgroup analysis of different miR-21 detection methods, the expression of miR-21 was verified by qRT-PCR in seven studies, and heterogeneity existed in these seven studies (I2 = 85%, P < 0.001). A possible reason is that the subjects came from different countries (Asian and non-Asian), and the tumor grades varied by study (IV, III–IV, II–IV, I–IV). Secondly, the cut-off values to define the high- or low-expression of miR-21 were different in the included studies. Thirdly, a few studies only provided results calculated by a univariate analysis, which did not adjust for the impacts of certain variables on the prognosis, such as patients’ age, gender, treatment received, and tumor grade. Therefore, the authentic prognostic value of miR-21 may be affected in the glioma patients. Fourthly, because there were relatively few original studies that met the inclusion criteria, the total sample size of this meta-analysis (n = 1,059) is small, which may reduce the statistical power of the summarizing results. Fifthly, most of the included studies were based on Chinese patients, while only three included studies were conducted in non-Asian subjects. Sixthly, there may be some possible confounding parameters influencing the prognostic role of miR-21, while the confounding effects of the clinical features were only considered in this study of the relationship between miR-21 and the prognosis of glioma patients. Considering the limitations of the included studies in this meta-analysis, we will continue to focus on relevant studies and improve the limitations of this study to get a more reliable conclusion.
In summary, elevated miR-21 expressions could indeed predict worse OS in glioma patients, and the prognostic effect of miR-21 was more prominent in the Asian group. There are still some detailed issues that need to be addressed for the clinical application of miR-21 as a prognostic marker in gliomas. For examples, it is necessary to establish a standard cut-off value defining the high and low levels of miR-21 expression, and a standard method for detecting the expression of miR-21 to ensure accuracy and comparability in glioma patients. We will track the research reports of the prognostic value of miR-21 in gliomas, and explore the influence of the difference between the detection of miR-21 in tumor tissue and liquid biopsy on the prognostic value of miR-21 in gliomas.
Material and Methods
Meta-analysis
Search strategy
We conducted a systematic search for available literature in the electronic databases Embase, PubMed, Web of Science, Chinese national knowledge infrastructure (CNKI), China biomedical literature service system (SinoMed), and Wanfang up to May 2018. The search terms used were “miR-21 OR microRNA-21”, “glioma OR glioblastoma OR GBM OR astrocytoma”, and “prognosis OR prognostic OR survival OR recurrence”. These three search terms were combined by the Boolean operator “AND”. In addition, we sought eligible studies by conducting a manual search of references from relevant articles and reviews to avoid missing potentially related articles.
Inclusion criteria
Two investigators independently determined the eligibility for each included article in the meta-analysis according to the inclusion criteria and exclusion criteria. Any disagreements during the selection process were resolved by discussion with a third author.
Eligible studies: (1) described the correlation between miR-21 expression and survival in glioma patients; (2) provided HRs with 95% CI directly, or key information to calculate HR indirectly, such as K-M curves and original survival data; (3) described a case-control study or cohort study; (4) categorized glioma patients into low- and high-expression groups only based on the miR-21 expression; and (5) were written in English or Chinese.
The exclusion criteria were as follows: (1) reviews, basic laboratory research, and conference abstracts; and (2) duplicated or overlapped studies.
Quality assessment
The quality of all included studies was assessed using the NOS49. The NOS contains eight items that are categorized into three groups (selection, comparability, and outcome or exposure). A star system is employed to assess the quality of the included study, so that the highest quality study is assigned to a maximum of one star per item, except for the item related to the comparability that allows for two stars. NOS ranges from zero to nine stars, and the highest quality study was assigned nine stars. Each included study with more than five stars was considered to be high quality50.
Data extraction
For each included study, the following data were extracted: (1) first author’s name, publication year, and location of sample collection; (2) characteristics of the studied population (sample size, follow-up period, tumor grade, sampling type); (3) miRNA test method, cut-off value to define high- or low-expression of miRNA, extracting method of HRs (95% CIs), outcome (OS or PFS), and NOS; and (4) HRs of miR-21 expression associated with survival of glioma patients in terms of OS and PFS with 95% CIs. If without HRs (95% CIs), and only the raw data were provided in the study, the HRs (95% CIs) were calculated from the raw data; or, if without HRs (95% CIs), and only the K-M curves were provided in the study, the HRs (95% CIs) were extracted using the method provided by Tierney and Parmar51,52.
Quantitative data synthesis
HR with 95% CI was used to evaluate the effect size for the OS or PFS. HRs from individual studies were transformed to their logarithms to stabilize the variances and normalize the distributions. To assess the heterogeneity of HRs across the included studies, the Cochran Q’s statistic and Higgins I2 statistic were calculated. A fixed-effect model (Mantel-Haenszel method) was adopted to calculate the pooled HRs (95% CIs) if the heterogeneity was absent among studies (P > 0.10 and I2 < 50%), while if the heterogeneity was observed among studies, the random-effect model (DerSimonian-Laird method) was selected to calculate the pooled HRs (95% CIs), and the subgroup meta-analyses were performed (P ≤ 0.10 or I2 ≥ 50%). A sensitivity analysis was used to evaluate the stability and reliability of the results, by omitting one individual study at a time and analyzing the remaining studies to detect whether the results were influenced excessively by any single study.
Begg’s test and Egger’s test were both used to test the significance of publication bias, with a P-value ≤ 0.10 considered significant. All P-values were two-sided. All calculations were carried out using R (version 3.2.3, R Foundation for Statistical Computing, Vienna, Austria).
TCGA dataset analysis
The miR-21 expression data and the clinical data of the LGG patients and the GBM patients were obtained from the TCGA database. MiR-21 expression was measured by Illumina Hi-Seq platform in the LGG dataset and Agilent 8 × 15 K Human microRNA platform in the GBM dataset. The inclusion criteria for the patients in the LGG and GBM datasets were as follows: (a) miR-21 expression level data and the corresponding follow-up data were available; (b) the OS or PFS were ≥ 30 days; and (c) clinical data of the patients, such as age at diagnosis, gender, tumor grade, and KPS, were available. The high- and low-expression groups were distinguished by the median value of the miR-21 expressions. Unpaired t test and chi-square test were used for the comparison of high miR-21 expression group and low miR-21 expression group. The survival curves were estimated by the Kaplan–Meier method. Survival differences between the high-expression group and the low-expression group were assessed by a log-rank test. A multivariate Cox regression analysis was used to identify miR-21 expression as an independent prognostic biomarker. All the statistical analyses were performed using PASW Statistics Version 18.0 (SPSS Inc., Chicago, Illinois, USA).
Acknowledgements
This project was supported by the National Natural Science Foundation of China (Grant No. 81472860). We also thank Katrina Krogh, MD, from essayreview.co.kr for editing a draft of this manuscript. All authors contributed to the drafting of this manuscript, and have read and approved the final version of the manuscript.
Author contributions
Xiaomin Zeng conceived of the study idea and led this study. Guli Jiang, Jing Mu, Xing Liu, Xiangni Peng, Feiya Zhong, Fang Deng and Sihua Peng performed the meta-analysis. Guli Jiang, Jing Mu, Wenliang Yuan, Feiya Zhong and Xiaoning Peng analyzed the TCGA data. Xiaomin Zeng, Xiaoning Peng and Sihua Peng ultimately finished the paper. All authors contributed to the drafting of this manuscript, and have read and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guli Jiang and Jing Mu.
Contributor Information
Xiaoning Peng, Email: pxiaoning@hunnu.edu.cn.
Sihua Peng, Email: shpeng@shou.edu.cn.
Xiaomin Zeng, Email: zxiaomin@csu.edu.cn.
References
- 1.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–21. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrom QT, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the united states in 2008-2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho VK, et al. Changing incidence and improved survival of gliomas. Eur. J. Cancer. 2014;50(13):2309–18. doi: 10.1016/j.ejca.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10)):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Sci. 2008;321(5897)):1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu X, et al. Prognostic factors for the survival outcome of bilateral thalamic glioma: an integrated survival analysis. World Neurosurg. 2018;110:e222–e230. doi: 10.1016/j.wneu.2017.10.132. [DOI] [PubMed] [Google Scholar]
- 8.Pan IW, Ferguson SD, Lam S. Patient and treatment factors associated with survival among adult glioblastoma patients: a USA population-based study from 2000-2010. J. Clin. Neurosci. 2015;22(10)):1575–81. doi: 10.1016/j.jocn.2015.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros V. The functions of animal microRNAs. Nat. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 12.Gaur A, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–68. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–4. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Luo YP. MicroRNAs in breast cancer: oncogene and tumor suppressors with clinical potential. J. Zhejiang Univ. Sci. B. 2015;16(1):18–31. doi: 10.1631/jzus.B1400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nohata N, Hanazawa T, Kinoshita T, Okamoto Y, Seki N. MicroRNAs function as tumor suppressors or oncogenes: aberrant expression of microRNAs in head and neck squamous cell carcinoma. Auris Nasus Larynx. 2013;40(2):143–9. doi: 10.1016/j.anl.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Zhang XX, Cheng J, Fu L, Li QS. Overexpression of tissue microRNA10b may help predict glioma prognosis. J. Clin. Neurosci. 2016;29:59–63. doi: 10.1016/j.jocn.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 2012;10:119. doi: 10.1186/1479-5876-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakomy R, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 2011;102(12):2186–90. doi: 10.1111/j.1349-7006.2011.02092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piwecka M, et al. Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol. Oncol. 2015;9(7):1324–40. doi: 10.1016/j.molonc.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang WY, et al. MiR-21 expression predicts prognosis in hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2014;38(6):715–9. doi: 10.1016/j.clinre.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Li B, et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer. 2014;83(2):146–53. doi: 10.1016/j.lungcan.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Liu XG, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med. Oncol. 2012;29(2):618–26. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 23.Shi R, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6(29):26971–81. doi: 10.18632/oncotarget.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, et al. MicroRNA-21 expression is associated with overall survival in patients with glioma. Diagn. Pathol. 2013;8:200. doi: 10.1186/1746-1596-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhi F, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur. J. Cancer. 2010;46(9):1640–9. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Barbano R, et al. A miRNA signature for defining aggressive phenotype and prognosis in gliomas. PLoS One. 2014;9(10):e108950. doi: 10.1371/journal.pone.0108950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu K, et al. Extracellular miRNA-21 as a novel biomarker in glioma: Evidence from meta-analysis, clinical validation and experimental investigations. Oncotarget. 2016;7(23):33994–4010. doi: 10.18632/oncotarget.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z. The Correlation Study between the Expression for E GFR, VEGF and miR-21 and the prognosis on Hum an Glioma. Xinjiang Medical University, http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CMFD&dbname=CMFD201801&filename=1017172699.nh&uid=WEEvREcwSlJHSldRa1FhdXNXa0d1ZzEzRUIydDk4RDB4TDVNZ1JoaVZhYz0=$9A4hF_YAuvQ5obgVAqNKPCYcEjKensW4IQMovwHtwkF4VYPoHbKxJw!!&v=MDgzNjIxTHV4WVM3RGgxVDNxVHJXTTFGckNVUkxLZVplZHFGeW5nVWI3QVZGMjZHYksvSE5mRnBwRWJQSVI4ZVg= (2017).
- 29.Hermansen SK, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW. MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J. Neurooncol. 2013;111(1):71–81. doi: 10.1007/s11060-012-0992-3. [DOI] [PubMed] [Google Scholar]
- 30.Sathyan P, et al. Mir-21-Sox2 axis delineates glioblastoma subtypes with prognostic impact. J. Neurosci. 2015;35(45):15097–112. doi: 10.1523/JNEUROSCI.1265-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilhan-Mutlu A, et al. Comparison of microRNA expression levels between initial and recurrent glioblastoma specimens. J. Neurooncol. 2013;112(3):347–54. doi: 10.1007/s11060-013-1078-6. [DOI] [PubMed] [Google Scholar]
- 32.Lei YY, et al. The clinicopathological parameters and prognostic significance of HER2 expression in gastric cancer patients: a meta-analysis of literature. World J. Surg. Oncol. 2017;15(1):68. doi: 10.1186/s12957-017-1132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, et al. Clinicopathological characteristics and prognostic value of cancer stem cell marker CD133 in breast cancer: a meta-analysis. Onco Targets Ther. 2017;10:859–70. doi: 10.2147/OTT.S124733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan, F., Mao, H., Deng, L., Li, G. & Geng, P. Prognostic and clinicopathological significance of microRNA-21 overexpression in breast cancer: a meta-analysis. Int J Clin Exp Pathol7 (9), 5622–33, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4203174/ (2014). [PMC free article] [PubMed]
- 35.Li C, et al. Prognostic role of microRNA-21 expression in gliomas: a meta-analysis. J. Neurooncol. 2016;130(1):11–7. doi: 10.1007/s11060-016-2233-7. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, et al. Plasma microRNA-based signatures to predict 3-year postoperative recurrence risk for stage II and III gastric cancer. Int. J. Cancer. 2017;141(10):2093–102. doi: 10.1002/ijc.30895. [DOI] [PubMed] [Google Scholar]
- 37.Wang N, et al. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem. Biophys. Res. Commun. 2017;493(3):1322–8. doi: 10.1016/j.bbrc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Marques TM, et al. MicroRNAs in cerebrospinal fluid as potential biomarkers for parkinson’s disease and multiple system atrophy. Mol. Neurobiol. 2017;54(10):7736–45. doi: 10.1007/s12035-016-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akers JC, et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget. 2017;8(40):68769–79. doi: 10.18632/oncotarget.18332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J, et al. Potential value of miR-221/222 as diagnostic, prognostic, and therapeutic biomarkers for diseases. Front. Immunol. 2017;8:56. doi: 10.3389/fimmu.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuberi M, et al. Utility of serum miR-125b as a diagnostic and prognostic indicator and its alliance with a panel of tumor suppressor genes in epithelial ovarian cancer. PLoS One. 2016;11(4):e0153902. doi: 10.1371/journal.pone.0153902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu S, et al. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J. Transl. Med. 2013;11:10. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JA, Lee HY, Lee ES, Kim I, Bae JW. Prognostic implications of microRNA-21 overexpression in invasive ductal carcinomas of the breast. J. Breast Cancer. 2011;14(4):269–75. doi: 10.4048/jbc.2011.14.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Wang JJ, Tang HM, To SS. Targeting strategies on miRNA-21 and PDCD4 for glioblastoma. Arch. Biochem. Biophys. 2015;580:64–74. doi: 10.1016/j.abb.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Li Y, Wang X, Jiang C. Ursolic acid inhibits proliferation and induces apoptosis in human glioblastoma cell lines U251 by suppressing TGF-beta 1/miR-21/PDCD4 pathway. Basic. Clin. Pharmacol. Toxicol. 2012;111(2):106–12. doi: 10.1111/j.1742-7843.2012.00870.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab. Invest. 2010;90(2):144–55. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 47.Gwak HS, et al. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7(10):e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X, Choudhury SN, Hua X, Dai Z, Li Y. Interaction of the oncogenic miR-21 microRNA and the p53 tumor suppressor pathway. Carcinogenesis. 2013;34(6):1216–23. doi: 10.1093/carcin/bgt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells G I. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses, http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (2018).
- 50.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25(9):603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 51.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parmar Mahesh K. B., Torri Valter, Stewart Lesley. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in Medicine. 1998;17(24):2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]