Abstract
With the explosion of high-throughput data, effective integrative analyses are needed to decipher the knowledge accumulated in biological databases. Existing meta-analysis approaches in systems biology often focus on hypothesis testing and neglect real expression changes, i.e. effect sizes, across independent studies. In addition, most integrative tools completely ignore the topological order of gene regulatory networks that hold key characteristics in understanding biological processes. Here we introduce a novel meta-analysis framework, Network-Based Integrative Analysis (NBIA), that transforms the challenging meta-analysis problem into a set of standard pathway analysis problems that have been solved efficiently. NBIA utilizes techniques from classical and modern meta-analysis, as well as a network-based analysis, in order to identify patterns of genes and networks that are consistently impacted across multiple studies. We assess the performance of NBIA by comparing it with nine meta-analysis approaches: Impact Analysis, GSA, and GSEA combined with classical meta-analysis methods (Fisher’s and the additive method), plus the three MetaPath approaches that employ multiple datasets. The 10 approaches have been tested on 1,737 samples from 27 expression datasets related to Alzheimer’s disease, acute myeloid leukemia (AML), and influenza. For all of the three diseases, NBIA consistently identifies biological pathways relevant to the underlying diseases while the other 9 methods fail to capture the key phenomena. The identified AML signature is also validated on a completely independent cohort of 167 AML patients. In this independent cohort, the proposed signature identifies two groups of patients that have significantly different survival profiles (Cox p-value 2 × 10−6). The NBIA framework will be included in the next release of BLMA Bioconductor package (http://bioconductor.org/packages/release/bioc/html/BLMA.html).
Subject terms: Microarrays, Statistical methods
Introduction
Microarray and sequencing technologies have transformed biological and medical research by allowing us to monitor the biological systems at the molecular level. Enormous volumes of molecular data have accumulated in public repositories, including Gene Expression Omnibus (GEO)1, cBioPortal2, and TCGA (http://cancergenome.nih.gov). Regardless of the high-throughput platforms being used, a standard comparative analysis of expression data usually produces a set of differentially expressed (DE) genes, which are often regarded as potential biological markers. These genes are important in classifying and subtyping patients, as well as in identifying entities that may involve in biological processes of the underlying diseases3–6. However, taken alone, gene biomarkers are insufficient to reveal biological mechanisms. In order to translate the differential expression to biological knowledge, researchers have been developing knowledge bases7,8 that map genes and gene products to known functional modules and regulatory networks. Concurrently, computational approaches have been developed for the identification of biomarkers at the systems level from differential expression9–14.
Remarkably, reproducibility poses big challenges in biomarker identification. Due to measurement errors and inherent study bias, analyses of independent datasets studying the same condition often result in distinctively different sets of DE genes15,16 and pathways17. Therefore, effective data integration is needed to integrate such similar studies to obtain reliable and consistent findings. For this purpose, meta-analyses have been performed at both gene18–21 and systems levels22–24. These approaches typically analyze individual studies independently to assess the significance of differential expression, either at gene or pathway level. The results from individual studies are then combined using p-value-based meta-analysis methods such as Fisher’s25, Stouffer’s26, maxP27, minP28, and addCLT29. One of the critical pitfalls of these p-value-based meta-analysis methods is that they neglect the actual expression changes, i.e. effect sizes. This might result in information loss. Although p-value is influenced by effect size, it is also greatly affected by sample size30. For datasets with large sample size, a test for differential expression will almost always result in a significant p-value, unless the effect size is exactly zero, which is very unlikely in reality. Simply combining the p-values would likely produce varying degree of false discoveries. In addition, most integrative approaches do not take into consideration the topological order of genes that hold key characteristics in understanding biological processes.
Here we propose Network-Based Integrative Analysis (NBIA), a network-based approach that utilizes techniques from both p-values-based and effect-sizes-based methods to reliably identify genes and pathways that are likely to be impacted by the underlying disease. The meta-analysis of effect sizes accurately estimates the central tendency of expression change for individual genes. The estimated genome-scale expression change allows for topology-aware analysis, in which gene interaction and signal propagation are taken into consideration. This approach transforms the meta-analysis problem into a standard topology-aware pathway analysis problem that has been solved efficiently. We illustrate the performance of NBIA using 1,737 samples from 27 studies related to Alzheimer’s disease, influenza, and acute myeloid leukemia (AML). We compared NBIA with 9 other approaches: Impact Analysis (IA), GSEA, and GSA combined with Fisher’s25 and the addCLT method29, plus 3 MetaPath approaches23. NBIA outperforms existing approaches in identifying biological processes relevant to the disease.
Methods
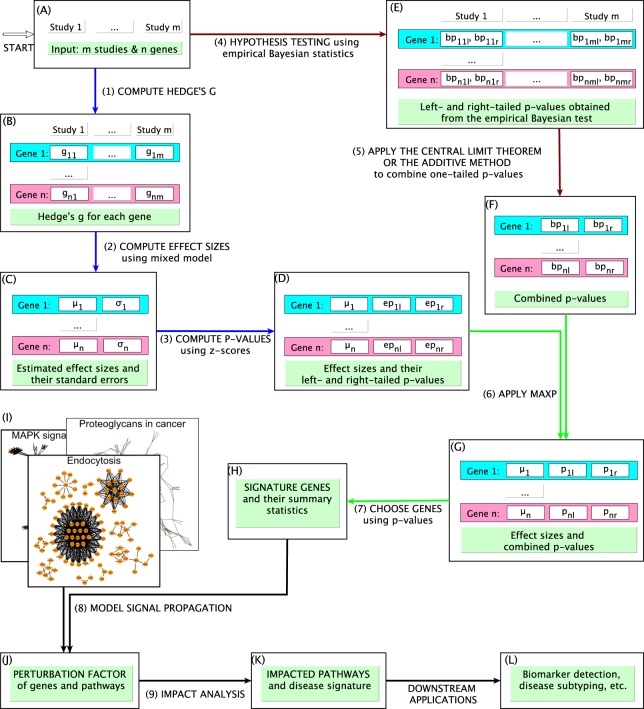
The overall pipeline consists of four main modules: (i) estimating the expression changes (i.e. standardized mean difference), standard errors, and their p-values, (ii) computing the p-values obtained from standard hypothesis testing, (iii) combining the two types of evidence to identify impacted genes and their summary statistics, and finally (iv) performing a network-based pathway analysis. The output is a set of impacted pathways and gene patterns that are consistently impacted across independent studies. These can serve as the disease signature for other downstream analyses. In Fig. 1, the brown arrows show the steps of the first module while the blue and green arrows display the steps of the second and third modules, respectively. The black arrows show the steps of the fourth module, which integrates the computed statistics and the pathway knowledge to identify the biological processes that are impacted or disrupted by the disease.
Figure 1.
The overall pipeline of NBIA. The input consists of m independent datasets and n genes. Step (1): calculate effect size (Hedge’s g) for each gene in each study. Step (2): combine effect sizes for each gene across multiple studies using the REstricted Maximum Likelihood (REML) algorithm. Step (3): compute the z-score () and calculate the left- and right-tailed p-values (epil and epir) using the standard normal distribution. This ends the first module. Step (4): perform hypothesis testing at gene level using empirical Bayesian statistics. For gene ith and dataset jth, the left- and right-tailed p-values obtained from the Bayesian test are bpijl and bpijr. Step (5): combine the one-tailed p-values for each gene, i.e., bpil = addCLT(bpi1l, …, bpiml) and bpir = addCLT(bpi1r, …, bpimr). This ends the second module. Step (6): combine Bayesian p-values with the p-values of the effect size using maxP, i.e. pil = max(epil, bpil) and pir = max(epir, bpir). Step (7): choose genes that are significantly impacted from both hypothesis testing and effect size perspectives using FDR-adjusted p-values (1% threshold by default). This ends the third module. Step (8): compute the perturbation factors for NBIA-prioritized genes and pathways. Step (9): identify impacted pathways using impact analysis.
To estimate the effect sizes of genes across all studies, we first compute standardized mean difference (SMD) for each gene in individual studies. We next estimate the overall effect size and standard error using the random-effects model31. This overall effect size represents the gene’s expression change under the effect of the condition. We then calculate the z-scores and the p-values of observing such effect sizes. Concurrently, we also calculate the p-values obtained from classical hypothesis testing. By default, we apply the linear model and empirical Bayesian testing provided by limma32 to compute the p-values for differential expression. The two-tailed p-values are converted to one-tailed p-values (left- and right-tailed). For each gene, the one-tailed p-values across all datasets are then combined using the addCLT method29. These p-values represent how likely the differential expression is observed by chance.
In the third module, we combine the two types of evidence (one p-value from empirical Bayesian statistics, and one p-value from effect size and standard error). We want that if a p-value is found significant, then it should be significant from classical hypothesis testing point of view, and the expression change should be well beyond the range of the standard error. Finally, the impacted genes and their summary statistics (p-values and effect sizes) are used to compute perturbation factors (detailed below) for the NBIA-prioritized genes and pathways. These perturbation factors are formulated to take into account gene interactions and signal propagation. Through permutation, we construct the null distribution of each pathway, and then compute the p-values of pathways as the fractions that are more extreme than the observed perturbation factors. The identified impacted pathways can be considered as the signature of the disease. This signature can be used for other downstream analyses.
Effect size and standard error (in Module 1)
Since the datasets are obtained from independent studies, it is reasonable to expect that the expression values are scaled differently in each study. Therefore, it is more reasonable to use standardized mean difference (SMD) as metrics to measure effect sizes, instead of raw mean difference. By default, we use Hedge’s g33 as the metric to measure expression change between two conditions (see Supplemental Section 1). However, this metric can be substituted by any existing metrics designed for the same purpose.
The central tendency of effect sizes for a gene can be estimated either using a fixed-effects model or a random-effects model21. The fix-effects model assumes that there is only one true effect size that underlies all of the studies, and the variability among studies is due to sampling error. This assumption, however, is unlikely to be correct when analyzing multiple independent datasets, since it cannot account for batch effects and heterogeneity between studies34,35. In contrast, the random-effects model explicitly takes into consideration the batch effects and data heterogeneity. This model decomposes the variability of effect size estimates into two variance components35,36. The first component represents batch effects and data heterogeneity across studies, while the second component represents the variability within each study. In other words, this model includes batch effects and data heterogeneity as a covariate in the designated formula. That is the main reason we favor the random-effects model over the fixed-effects model. See Supplementary Section 3.3, Figs. S5–S8, and Table S5 for more discussion regarding batch effects.
Consider one specific gene and denote y1, y2, …, ym as Hedge’s g values computed for m studies. We can write the random-effects model as yi = μ + τi + ϵi with τi ~ N(0, σ2) and . In this formula, μ is the central tendency of the effect size, τi represents the term by which the effect size in the ith study differs from the central tendency, and ϵi represents within-study variability. The τi variables represent batch effects and data heterogeneity among datasets. The overall effect size μ of the gene and its standard error σ are estimated iteratively, as described in the literature35,37–39. The algorithm stops when further iterations do not change the values of μ and σ. After the REML algorithm stops, we compute the z-score using the formula and then calculate the left- and right-tailed p-values of observing such z-score. The obtained μ and p-values (epl and epr where ep stands for “effect size p-value”) represent the overall expression change of the gene and the reliability of the estimated effect size.
Classical hypothesis testing and meta-analysis (in Module 2)
In this work, we apply the linear model and empirical Bayesian test provided by limma32 to calculate the two-tailed p-values. We then convert these p-values into one-tailed p-values. We note that this step can be substituted by any other hypothesis testing methods. We favor this empirical approach to avoid relying on strong assumption about the distributions of the expression values. For each gene, the one-tailed p-values are independent and uniformly distributed under the null. We next combine the individual p-values of the m studies to obtain one left- and one right-tailed p-value for each gene.
p-value aggregation (in Module 3)
To combine the p-values obtained from each study, we use the addCLT method that is built on the Central Limit Theorem29. This method uses the average of p-values as the test statistic; therefore, it is robust against extreme p-values. Denoting the individual p-values to be combined as P1, P2, …, Pm, and , the probability density function (pdf) is derived from a linear transformation of the Irwin-Hall distribution40,41: . When m is large, the computation of the Irwin-Hall distribution becomes unstable due to underflow/overflow of memory29. In this case, we use the Central Limit Theorem42 to estimate this distribution. From the Central Limit Theorem, the average of such m independently and identically distributed variables follows a normal distribution with mean and variance , i.e. for large values of m. The method is named “addCLT” for “additive-Central Limit Theorem”29. See Supplemental Section 1 for details.
Impacted genes (in Module 3)
After performing effect-size-based meta-analysis and classical hypothesis testing, we have the following statistics for a gene with index i: (1) the central tendency μi of effect sizes, (2) the left- and right-tailed p-values, epil and epir, obtained from the z-score ( where σi is the standard error), and (3) the left and right-tailed p-values obtained from Bayesian statistics, bpil and bpir. We further combine the two types of p-values as follows:
The intuition behind using maxP27 to combine the two types of p-values is to reduce the number of potential false positives. We want to make sure that the selected DE genes are significant from the classical hypothesis testing perspective, as well as have the effect size that is outside the range of standard error. After this, we correct the p-values for multiple comparisons using Benjamini-Hochberg’s False Discovery Rate (FDR)43. By default, genes with FDR ≤ 1% are considered as genes that are significantly impacted under the effects of the disease. We note that to have a p-value of 1%, the absolute z-score must be at least 2. Therefore, with a cutoff of 1% we choose genes that are not only statistically significant using the empirical Bayesian test, but also have the absolute effect size at least twice the standard error (see Supplementary Sections 3.1 and 3.4 and Figs. S3 and S9 for more discussion about the contribution of each type of p-values and their impact on false positive rate). These p-values and the effect sizes calculated above serve as the input of the Impact Analysis to identify impacted signaling pathways.
Perturbation factors of genes and pathways (in Module 4)
To identify the biological processes that are impacted by the disease, the Impact Analysis (IA) method44 combines two types of evidence: (i) the over-representation of significantly impacted genes in a given pathway, and (ii) the perturbation of the pathway, as measured by propagation expression changes through the network. These two aspects are represented by two p-values: pde and ppert. The first p-value, pde, is calculated using the hypergeometric model — this probability quantified the over-representation of DE genes in a pathway, compared to the rest of the transcriptome. The second term, ppert, is obtained from an empirical hypothesis testing in which we take into account both the identity of DE genes and their known interactions. It is calculated based on the perturbation factor in each pathway. The perturbation factor (PF) of each gene is defined as: . The first term, ΔE(g), captures the signed normalized expression change of the gene, i.e. standardized mean difference (SMD). In the context of meta-analysis, we use the central tendency of effect sizes μ to represent ΔE(g). This value is estimated from multiple studies and is expected to be more robust against noise and bias than the SMD obtained from any single study. The second term is the sum of all PFs of upstream genes, normalized by the number of downstream genes. The value of βug quantifies the strength of interaction between u and g. By default, βug = 1 for activation and βug = −1 for repression. The total perturbation in the pathway is then computed as: .
For each pathway Pi, we construct the null distribution of PF(Pi) by permuting both sample and gene labels. The p-value ppert is calculated by the fraction of the null distribution of Pi that is more extreme than the observed value. The two p-values, pde and ppert, are then combined using Fisher’s method to obtain one single p-value for the pathway. This combined p-value represents how likely the pathway is impacted under the effects of the condition44. See Supplementary Section 3.2 and Fig. S4 for more discussion.
Results
Here we analyze 1,737 samples from 27 independent datasets related to Alzheimer’s disease, influenza, and AML. We selected these conditions for our analysis due to two main reasons. First, we were able to find multiple datasets/experiments in public repositories for each of the three diseases. Second, for each disease, there is pathway that was created in KEGG7 to describe the known biology and mechanisms of the underlying disease. We use these KEGG pathways to validate the methods and refer to them as target pathways. We expect that a good analysis method to identify these target pathways as significant. Supplemental Table S1 shows the details of each dataset, including the number of samples, platforms, and tissues. For graphical representation of biological processes, we use the KEGG database version 76, which includes 182 signaling pathways.
We compare NBIA with 4 other pathway analysis approaches: Impact Analysis (IA)44, GSA45, GSEA9, and MetaPath23. IA is a topology-aware method while GSEA and GSA are enrichment-based methods. Since IA, GSEA, and GSA are not able to perform meta-analysis, we use addCLT29 and Fisher’s method25 to combine individual p-values. MetaPath, on the other hand, is a stand-alone meta-analysis method, which performs pathway analysis without the need of any external analysis tool. There are three MetaPath methods: (i) MetaPath_G which performs meta-analysis at the gene level, (ii) MetaPath_P which performs meta-analysis at the pathway level, and (iii) MetaPath_I which combines the results obtained from MetaPath_G and MetaPath_P. In summary, we compare NBIA with 9 different integrative approaches: 6 GSEA-, GSA-, and IA-based approaches, plus 3 MetaPath methods. We consistently set the significance threshold at 5% for all approaches. Pathways with FDR-adjusted p-values smaller than the threshold are consider significantly impacted.
The experimental study consists of two parts. In the first part, we use NBIA for each of the diseases to identify the genes that are consistently differentially expressed. The signature genes and their effect sizes are then used to identify the biological processes at the systems level. We show that NBIA outperforms other approaches: GSEA9, GSA45, and Impact Analysis44 and the MetaPath methods23. In the second part, we use the pathway signature identified by NBIA as biomarkers to cluster RNA-Seq data obtained from TCGA for 167 AML patients. We show that the discovered subtypes have significantly different survival profiles using 4 different clustering methods. The Cox p-values obtained from the discovered subtypes equal to 2 × 10−4, 3 × 10−4, 4 × 10−5, and 2 × 10−6 for consensus clustering, hierarchical clustering, local shrinkage, and cluster ensemble, respectively. We also show that this would not be possible without knowing the NBIA signature.
Alzheimer’s disease
There is a target pathway in KEGG, Alzheimer’s disease, that describes the known mechanisms and biological processes involved in this disease. However, it is well known that the pathways Parkinson’s disease and Huntington’s disease share many genes and mechanisms with Alzheimer’s disease46–49. Therefore, we expect that good analysis methods to identify all of the three neurological disorder pathways as statistically significant and rank them on top.
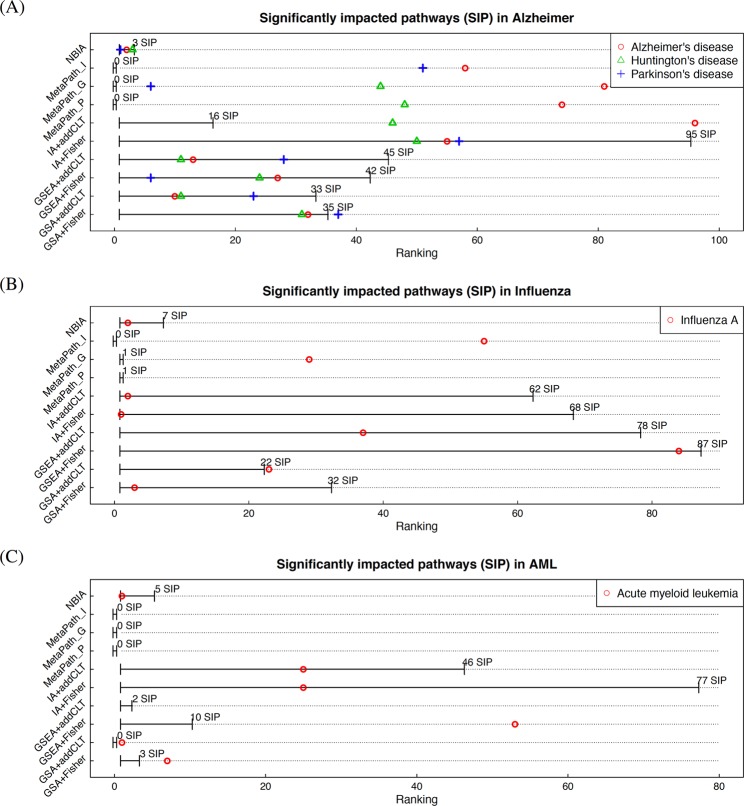
Each of the 10 meta-analysis methods (NBIA, three MetaPath methods, and six GSA-, GSEA-, and IA-based approaches) produces a list of KEGG pathways ranked according to their p-values. Table 1 shows the 10 top ranked pathways and FDR-corrected p-values for NBIA while Supplementary Table S2 shows the 20 top ranked pathways for the other nine methods. Pathways with FDR-corrected p-values less than 5% are considered significant. Figure 2A summarizes the results by showing the number of significant pathways and the ranking of the three neurological disorder pathways for the 10 methods. The horizontal axis shows the ranking of the pathways while the vertical axis shows the 10 methods. For each method, we draw a segment that represents the range of the significant pathways. For example, using NBIA, we identified three significant pathways (Table 1), which are exactly the three neurological disorder pathways. Therefore, the segment for NBIA ranges from 1 to 3 and the three neurological disorders pathways fall onto this segment (top row in Fig. 2A). In another example, using IA + addCLT, we identified 16 pathways as significant (third column in Table S2). Therefore, the segment for IA + addCLT ranges from 1 to 16 in Fig. 2A. The pathway Alzheimer’s disease is ranked 96th (red circle) and thus falls outside of the segment. Similarly, using GSA + Fisher, we identified 35 significant pathways. The three neurological disorder pathways, Alzheimer’s disease (red circle), Huntington’s disease (green triangle), and Parkinson’s disease (blue plus sign), are ranked at the positions 32nd, 31st, and 37th, respectively. The pathway Parkinson’s disease is not significant and thus does not fall onto the segment of significant pathways.
Table 1.
The top 10 ranked pathways and FDR-corrected p-values obtained by combining Alzheimer’s data using NBIA. The horizontal line represents the cutoff of 5%. All of the three target pathways are ranked on top with FDR-adjusted p-values smaller than 5%.
| NBIA | ||
|---|---|---|
| Pathway | p.fdr | |
| 1 | Parkinson’s disease | 7e-07 |
| 2 | Alzheimer’s disease | 0.0024 |
| 3 | Huntington’s disease | 0.0086 |
| 4 | Glutamatergic synapse | 0.1008 |
| 5 | Amyotrophic lateral sclerosis (ALS) | 0.1750 |
| 6 | Sphingolipid signaling pathway | 0.2137 |
| 7 | Regulation of actin cytoskeleton | 0.2137 |
| 8 | Synaptic vesicle cycle | 0.3868 |
| 9 | Retrograde endocannabinoid signaling | 0.6446 |
| 10 | Fc gamma R-mediated phagocytosis | 0.6446 |
Figure 2.
Number of significant pathways and their ranking in Alzheimer’s disease (panel A), influenza (panel B), and AML (panel C) using 10 meta-analysis approaches. The horizontal axis shows the ranking of the pathways while the vertical axis shows the 10 methods. The significance threshold is consistently set to 5% for all approaches. For each method, we draw a segment that represents the range of the significant pathways. For all of the three diseases, MetaPath finds no significant pathway at the significance cutoff of FDR = 5%. The 6 GSA-, GSEA-, and IA-based methods generally identify large sets of significant pathways, among which many are likely to be false positives. NBIA consistently identifies the target pathways as significant and ranks them on top in each of the three conditions.
The three MetaPath methods fail to identify the three neurological disorder pathways as the most significant ones. MetaPath_P identifies no significant pathway. The three pathways Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease are ranked at positions 74th, 48th, and 121st, respectively. Similarly, MetaPath_G and MetaPath_I also fail to identify the three neurological disorder pathways as significant. MetaPath_G produces no significant pathway and ranks the three pathways at positions 81st, 6th, and 44th, respectively. In consequences, MetaPath_I also fails to identify the three neurological disorder pathways as significant (adjusted p-values 0.85, 0.87, and 0.85 with rankings 58th, 83rd, and 51st, respectively). IA + addCLT and IA + Fisher, which are topology-aware methods, rank the target pathways very low (not in top 40). IA + addCLT fails to identify any of the three neurological disorder pathways as significant. The GSA-based and GSEA-based methods appear to perform well for this disease. These methods identify the target pathways as significant. However, the two methods also identify a large number of significant pathways, among which many are likely to be false positives.
Finally, we apply NBIA to combine the 10 studies (Table 1). NBIA identifies all of the three neurological disorder pathways as significant and ranks them at the very top. The pathway Alzheimer’s disease is ranked 2nd with adjusted p = 0.002.
Influenza
There is a dedicated pathway Influenza A that was created in order to describe the known mechanisms involved in the influenza disease. We expect that a good meta-analysis method to identify this target pathway as significant and ranks it among the top impacted pathways. The number of significant pathways and the ranking of the target pathway for the 10 methods are shown in Fig. 2B. Supplemental Table S3 shows the details of top ranked pathways of the competing methods.
MetaPath_P, MetaPath_G and MetaPath_I fail to identify the target pathway as significant and ranks it at the positions 167th, 29th and 55th, respectively. The two topology-aware methods, IA combined with addCLT and Fisher’s method, identify the pathway Influenza A as significant and rank it on top at positions 1st and 2nd, respectively. However, these methods also provide a large set of significant pathways (62 and 68 pathways). Similarly, GSA + Fisher and GSEA + addCLT identify the target pathway as significant but likely to include many false positives as well.
Table 2 shows the 10 top ranked pathways using NBIA. NBIA finds 7 signifiant pathways with the threshold FDR = 5%. The target pathway Influenza A is ranked 2nd with FDR = 8 × 10−5. The other significant pathways, Herpes simplex infection, Systemic lupus erythematosus, Viral carcinogenesis, Pertussis, Measles, and NOD-like receptor signaling pathway, are also known to share common mechanisms with influenza and closely associated with immune response of the body50–53.
Table 2.
The top 10 ranked pathways and FDR-corrected p-values obtained by combining influenza data using NBIA. The horizontal line represents the cutoff of 5%. The target pathway Influenza A is ranked 2nd with an FDR-adjusted p-value of 8 × 10−5.
| NBIA | ||
|---|---|---|
| Pathway | p.fdr | |
| 1 | Herpes simplex infection | 4e-06 |
| 2 | Influenza A | 8e-05 |
| 3 | Systemic lupus erythematosus | 0.0002 |
| 4 | Viral carcinogenesis | 0.0052 |
| 5 | Pertussis | 0.0052 |
| 6 | Measles | 0.0179 |
| 7 | NOD-like receptor signaling pathway | 0.0441 |
| 8 | Staphylococcus aureus infection | 0.0642 |
| 9 | Cytosolic DNA-sensing pathway | 0.0642 |
| 10 | Alcoholism | 0.0642 |
Acute myeloid leukemia
For this disease, the target pathway is Acute myeloid leukemia. Again, we use the 10 methods to combine the 8 AML datasets. The ranking and the number of significant pathways are shown in Fig. 2C. The top pathways of the 9 other methods are shown in Supplemental Table S4. Again, the three MetaPath methods identify no significant pathways at the cutoff of 5%. The four GSA- and GSEA-based methods fail to identify the pathway Acute myeloid leukemia as significant. IA + addCLT and IA + Fisher succeed in identifying the target pathway as significant but rank it at a relatively low position, 25th. The 10 top pathways of NBIA are shown in Table 3. The target pathway Acute myeloid leukemia is ranked on top with FDR = 0.0066.
Table 3.
The top 10 ranked pathways and FDR-corrected p-values obtained by combining AML data using NBIA. The horizontal line represents the cutoff of 5%. The target pathway Acute myeloid leukemia is ranked on top with an FDR-adjusted p-value of 0.0066.
| NBIA | ||
|---|---|---|
| Pathway | p.fdr | |
| 1 | Acute myeloid leukemia | 0.0066 |
| 2 | Neurotrophin signaling pathway | 0.0178 |
| 3 | Non-small cell lung cancer | 0.0353 |
| 4 | Renal cell carcinoma | 0.0353 |
| 5 | Transcriptional misregulation in cancer | 0.0384 |
| 6 | ErbB signaling pathway | 0.0628 |
| 7 | Non-alcoholic fatty liver disease (NAFLD) | 0.1461 |
| 8 | Colorectal cancer | 0.1913 |
| 9 | Insulin resistance | 0.2792 |
| 10 | Endometrial cancer | 0.2792 |
Subtyping AML data
To further validate the signature identified for AML, we downloaded RNA-Seq data for 167 AML patients. The raw TCGA data was sequenced using Illumina GASeq. The processed data and the overall survival information were downloaded from the Broad Institute’s website http://gdac.broadinstitute.org/.
As we reported above, NBIA identified 5 pathways that are significantly impacted in AML. The total number of genes belonging to these pathways are 364. We simply use these genes as selected features in order to refine the partitioning of the 167 AML patients. The comparison between the partitioning with and without feature selection show that the selected pathways and genes play a crucial role in identifying subtypes with significantly different survival.
Here we use three existing methods, consensus clustering54,55 (CC), hierarchical clustering (HC), and local shrinkage56, as well as one newly developed cluster ensemble approach to cluster the gene expression data. We show that using each of the three clustering methods, we discovered subtypes that have significantly different survival profiles. Figure 3 shows the Kaplan-Meier survival analysis57 of the discovered subtypes using the four clustering methods. The heatmaps that visualize different subtypes of AML patients on all genes and NBIA signature are shown in Supplementary Fig. S2.
Figure 3.
Kaplan-Meier survival analysis of AML subtypes discovered by consensus clustering (A panels), hierarchical clustering (B panels), local shrinkage (C panels), and cluster ensemble (D panels). The top panels show AML subtypes discovered using genome-wide expression values (all genes) while the bottom panels shows the subtypes discovered using genes selected by NBIA. In each panel, the colored curves represent the survival probability of different subtypes. For any of the four methods, we are not able to find subtypes with significantly different profiles when using the genome-wide expression values. In contrast, when applied in conjunction with the pathway signature obtained from NBIA, any of the four methods identifies subtypes with very significant survival profiles. Interestingly, there is one group of patients that are always grouped together in the high-survival group using CC, HC, and local shrinkage. When performing an ensemble of the three partitioning, we are able to separate this group from the rest (panel D). The cluster ensemble algorithm identifies two groups of patients that have very different survival profiles (Cox p-value 2 × 10−6). Among the high-survival group, almost 60% of the patients survived at the end of the study (more than 8 years). In contrast, only approximately 10% of the other group survived at the end.
Without feature selection, we are unable to identify subtypes with significant survival differences by using genome-wide expression values. With feature selection, CC is able to find 4 subtypes with Cox p-value = 2 × 10−4 while HC finds 2 subtypes with p-value = 3 × 10−4. Similarly, the local shrinkage finds 4 subtypes with p-value = 4 × 10−5. We note that there is a group of patients that always belongs to the highest-survival group in the three partitionings. The cluster ensemble approach that is designed to look for common pattern between the partitionings is able to separate this group of patients from the rest. This approach identifies two groups of patients with very different survival profiles (Cox p-value = 2 × 10−6). Among the high-survival group, almost 60% of the patients survived at the end of the study (more than 8 years). In contrast, only approximately 10% of the other group survived at the end.
We also perform subtyping using the pathway signatures identified by the other meta-analysis methods. The four methods, MetaPath_I, MetaPath_G, MetaPath_P and GSA+addCLT, yield no significant pathway and thus have no pathway signture. The other five methods, IA + addCLT, IA + Fisher, GSEA + addCLT, GSEA + Fisher, and GSA + Fisher, identify 46, 77, 2, 10, and 3 pathways as significant, respectively. We use the pathway signatures of these five methods to subtype AML patients. The Kaplan-Meier survival analysis of the discovered subtypes is shown in Supplementary Fig. S1. The Cox p-values obtained for each analysis are shown in Table 4. Using any of the clustering methods, NBIA has the most significant p-values. In addition, it is the only method that provides significant p-values across all four clustering methods.
Table 4.
Cox p-values obtained from four clustering methods (consensus clustering, hierarchical clustering, local shrinkage, and cluster ensemble) using seven sets of genes: all genes and the signatures obtained from IA + addCLT, IA + Fisher, GSEA + addCLT, GSEA + Fisher, GSA + Fisher, and NBIA. Cells with emboldening text have p-values smaller than 5%. Using any of the clustering methods, NBIA has the most significant p-values. In addition, it is the only method that provides significant p-values across all four clustering methods.
| All genes | IA + addCLT | IA + Fisher | GSEA + addCLT | GSEA + Fisher | GSA + Fisher | NBIA | |
|---|---|---|---|---|---|---|---|
| Consensus clustering | 0.359 | 0.016 | 0.001 | 0.145 | 0.089 | 0.072 | 1e-04 |
| Hierarchical clustering | 0.742 | 0.004 | 0.002 | 0.009 | 0.896 | 0.345 | 3e-04 |
| Local shrinkage | 0.066 | 0.002 | 0.09 | 0.788 | 0.02 | 0.24 | 4e-05 |
| Cluster ensemble | 0.591 | 0.902 | 0.048 | 0.916 | 0.068 | 0.132 | 2e-06 |
Conclusion
In this article, we present a novel network-based meta-analysis that is able to combine multiple studies and identify the signaling pathways that are significantly impacted in a given phenotype. The main innovation of NBIA is that it transforms the challenging meta-analysis problem into a set of standard analysis problems that can be solved efficiently. This approach utilizes techniques from both p-value-based and effect-size-based meta-analysis techniques in order to reliably identify a robust set of impacted genes. This set of genes serves as the input of the impact analysis (IA) approach to identify the biological processes that are significantly impacted under the effect of the disease.
To evaluate this framework, we examined 1,737 samples from 27 independent datasets related to Alzheimer’s disease, acute myeloid leukemia (AML), and influenza. NBIA was compared against 9 different approaches, GSA, GSEA, and IA combined with Fisher’s method and addCLT, plus three MetaPath approaches. We demonstrated that NBIA outperforms existing approaches to consistently identify the target pathways as significant and top ranked. We also assessed NBIA’s performance in simulation studies, including Monte Carlo evaluations of batch effects, false positive rates, and discuss the relative contributions of the different quantification steps in the NBIA workflow.
To further validate the framework, we also used the identified signature to cluster RNA-Seq data of 167 AML patients obtained from TCGA. For any of the 4 clustering methods tested, consensus clustering, hierarchical clustering, local shrinkage, and cluster ensemble, the discovered subtypes have significant survival differences with Cox p-value as small as 2 × 10−6. Even though our analysis stops at disease subtyping, NBIA can be used for many other applications, such as biomarker detection, drug repurposing, drug synergy, and anti-aging. In each of these areas, identifying the correct set of biological processes that are impacted by the disease/drug is the key for success.
Supplementary information
Acknowledgements
This work was partially supported by NASA under grant number 80NSSC19M0170. This work has also been supported by Startup Fund and Research Enhancement Grant at the University of Nevada Reno to Tin Nguyen. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of any of the funding agencies.
Author contributions
T.N. conceived and designed the approach. A.S., T.M.N., A.G.S. and S.D. helped with data analysis and interpretation. All authors revised and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60981-9.
References
- 1.Barrett T, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Research. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proceedings of the National Academy of Sciences of the Unites States of America. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of Clinical Oncology. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen T, Tagett R, Diaz D, Draghici S. A novel approach for data integration and disease subtyping. Genome Research. 2017;27:2025–2039. doi: 10.1101/gr.215129.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen H, Shrestha S, Draghici S, Nguyen T. PINSPlus: A tool for tumor subtype discovery in integrated genomic data. Bioinformatics. 2019;35:2843–2846. doi: 10.1093/bioinformatics/bty1049. [DOI] [PubMed] [Google Scholar]
- 7.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Research. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceeding of The National Academy of Sciences of the Unites States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozerov IV, et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nature Communications. 2016;7:13427. doi: 10.1038/ncomms13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doungpan N, Engchuan W, Chan JH, Meechai A. GSNFS: Gene subnetwork biomarker identification of lung cancer expression data. BMC Medical Genomics. 2016;9:70. doi: 10.1186/s12920-016-0231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen T, Mitrea C, Draghici S. Network-based approaches for pathway level analysis. Current Protocols in Bioinformatics. 2018;61:8–25. doi: 10.1002/cpbi.42. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen H, et al. A comprehensive survey of tools and software for active subnetwork identification. Frontiers in Genetics. 2019;10:155. doi: 10.3389/fgene.2019.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen T-M, Shafi A, Nguyen T, Draghici S. Identifying significantly impacted pathways: a comprehensive review and assessment. Genome Biology. 2019;20:203. doi: 10.1186/s13059-019-1790-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan PK, et al. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Research. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen T, Mitrea C, Tagett R, Draghici S. DANUBE: Data-driven meta-ANalysis using UnBiased Empirical distributions - applied to biological pathway analysis. Proceedings of the IEEE. 2017;105:496–515. doi: 10.1109/JPROC.2015.2507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shafi, A., Nguyen, T., Peyvandipour, A. & Draghici, S. GSMA: an approach to identify robust global and test Gene Signatures using Meta-Analysis. Bioinformatics btz561 (2019). [DOI] [PMC free article] [PubMed]
- 19.Rhodes DR, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Tseng GC. An adaptively weighted statistic for detecting differential gene expression when combining multiple transcriptomic studies. The Annals of Applied Statistics. 2011;5:994–1019. doi: 10.1214/10-AOAS393. [DOI] [Google Scholar]
- 21.Nguyen, T., Diaz, D. & Draghici, S. TOMAS: A novel TOpology-aware Meta-Analysis approach applied to System biology. In Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, 13–22 (ACM, 2016).
- 22.Kaever A, et al. Meta-analysis of pathway enrichment: combining independent and dependent omics data sets. PLoS One. 2014;9:e89297. doi: 10.1371/journal.pone.0089297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen K, Tseng GC. Meta-analysis for pathway enrichment analysis when combining multiple genomic studies. Bioinformatics. 2010;26:1316–1323. doi: 10.1093/bioinformatics/btq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen, T., Diaz, D., Tagett, R. & Draghici, S. Overcoming the matched-sample bottleneck: an orthogonal approach to integrate omic data. Scientific Reports6, 29251 (2016). [DOI] [PMC free article] [PubMed]
- 25.Fisher, R. A. Statistical methods for research workers (Oliver & Boyd, Edinburgh, 1925).
- 26.Stouffer, S., Suchman, E., DeVinney, L., Star, S. & Williams, R. M. Jr. The American Soldier: Adjustment during army life, vol. 1 (Princeton University Press, Princeton, 1949).
- 27.Wilkinson B. A statistical consideration in psychological research. Psychological Bulletin. 1951;48:156. doi: 10.1037/h0059111. [DOI] [PubMed] [Google Scholar]
- 28.Tippett, L. H. C. The methods of statistics (Williams & Norgate, London, 1931).
- 29.Nguyen T, Tagett R, Donato M, Mitrea C, Draghici S. A novel bi-level meta-analysis approach-applied to biological pathway analysis. Bioinformatics. 2016;32:409–416. doi: 10.1093/bioinformatics/btv588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan GM, Feinn R. Using effect size-or why the p value is not enough. Journal of Graduate Medical Education. 2012;4:279–282. doi: 10.4300/JGME-D-12-00156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viechtbauer W. Conducting meta-analyses in R with the metafor package. Journal of Statistical Software. 2010;36:1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 32.Smyth, G. K. Limma: linear models for microarray data. In Gentleman, R., Carey, V., Dudoit, S., Irizarry, R. & Huber, W. (eds) Bioinformatics and Computational Biology Solutions Using R and Bioconductor, 397–420 (Springer, New York, 2005).
- 33.Hedges, L. V. & Olkin, I. Statistical method for meta-analysis (Academic Press, London, 2014).
- 34.Milliken, G. A. & Johnson, D. E. Analysis of messy data volume 1: designed experiments, vol. 1 (Chapman & Hall/CRC, London, 2009).
- 35.Viechtbauer W. Bias and efficiency of meta-analytic variance estimators in the random-effects model. Journal of Educational and Behavioral Statistics. 2005;30:261–293. doi: 10.3102/10769986030003261. [DOI] [Google Scholar]
- 36.Goldstein, H. Multilevel statistical models, vol. 922 (John Wiley & Sons, New York, 2011).
- 37.Harville DA. Maximum likelihood approaches to variance component estimation and to related problems. Journal of the American Statistical Association. 1977;72:320–338. doi: 10.1080/01621459.1977.10480998. [DOI] [Google Scholar]
- 38.Corbeil RR, Searle SR. Restricted maximum likelihood (REML) estimation of variance components in the mixed model. Technometrics. 1976;18:31–38. doi: 10.2307/1267913. [DOI] [Google Scholar]
- 39.Patterson, H. & Thompson, R. Maximum likelihood estimation of components of variance. In Proceedings of the 8th International Biometric Conference, 197–207 (1975).
- 40.Hall P. The distribution of means for samples of size n drawn from a population in which the variate takes values between 0 and 1, all such values being equally probable. Biometrika. 1927;19:240–244. doi: 10.2307/2331961. [DOI] [Google Scholar]
- 41.Irwin JO. On the frequency distribution of the means of samples from a population having any law of frequency with finite moments, with special reference to Pearsonas Type II. Biometrika. 1927;19:225–239. doi: 10.1093/biomet/19.3-4.225. [DOI] [Google Scholar]
- 42.Kallenberg, O. Foundations of modern probability (Springer-Verlag, New York, 2002).
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of The Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 44.Draghici S, et al. A systems biology approach for pathway level analysis. Genome Research. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efron B, Tibshirani R. On testing the significance of sets of genes. The Annals of Applied Statistics. 2007;1:107–129. doi: 10.1214/07-AOAS101. [DOI] [Google Scholar]
- 46.Swerdlow RH. Brain aging, Alzheimer’s disease, and mitochondria. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2011;1812:1630–1639. doi: 10.1016/j.bbadis.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maruszak A, Żekanowski C. Mitochondrial dysfunction and Alzheimer’s disease. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2011;35:320–330. doi: 10.1016/j.pnpbp.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Zhu X, Perry G, Smith MA, Wang X. Abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. Journal of Alzheimer’s Disease. 2013;33:S253–S262. doi: 10.3233/JAD-2012-129005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Querfurth HW, LaFerla FM. Mechanisms of disease. New England Journal of Medicine. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 50.Carter C. Schizophrenia susceptibility genes directly implicated in the life cycles of pathogens: cytomegalovirus, influenza, herpes simplex, rubella, and Toxoplasma gondii. Schizophrenia Bulletin. 2008;35:1163–1182. doi: 10.1093/schbul/sbn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Djeu J, et al. Positive self regulation of cytotoxicity in human natural killer cells by production of interferon upon exposure to influenza and herpes viruses. Journal of Experimental Medicine. 1982;156:1222–1234. doi: 10.1084/jem.156.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abu-Shakra M, et al. Specific antibody response after influenza immunization in systemic lupus erythematosus. The Journal of Rheumatology. 2002;29:2555–2557. [PubMed] [Google Scholar]
- 53.Cliff A, Haggett P. Statistical modelling of measles and influenza outbreaks. Statistical Methods in Medical Research. 1993;2:43–73. doi: 10.1177/096228029300200104. [DOI] [PubMed] [Google Scholar]
- 54.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. doi: 10.1023/A:1023949509487. [DOI] [Google Scholar]
- 56.Chang F, Qiu W, Zamar RH, Lazarus R, Wang X. Clues: an R package for nonparametric clustering based on local shrinking. Journal of Statistical Software. 2010;33:1–16. doi: 10.18637/jss.v033.i04. [DOI] [Google Scholar]
- 57.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of the American Statistical Association. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.