Abstract
Accumulating evidence suggests that our ability to predict chemical effects on breast cancer is limited by a lack of physiologically relevant in vitro models; the typical in vitro breast cancer model consists of the cancer cell and excludes the mammary microenvironment. As the effects of the microenvironment on cancer cell behavior becomes more understood, researchers have called for the integration of the microenvironment into in vitro chemical testing systems. However, given the complexity of the microenvironment and the variety of platforms to choose from, identifying the essential parameters to include in a chemical testing platform is challenging. This review discusses the need for more complex in vitro breast cancer models and outlines different approaches used to model breast cancer in vitro. We provide examples of the microenvironment modulating breast cancer cell responses to chemicals and discuss strategies to help pinpoint what components should be included in a model.
Keywords: breast cancer, drugs, environmental chemicals, microenvironment, 3D
Insight Box
This review provides insight into the challenges of predicting the effects of chemicals on breast cancer. It discusses the need for more complex in vitro breast cancer models and reviews the different approaches used to model breast cancer in vitro. The review is innovative as it integrates concepts from cancer biology, tissue engineering, and cellular biology to guide researchers with the design and validation of model systems.
DEFINING THE NEED FOR IMPROVED SYSTEMS TO TEST CHEMICAL EFFECTS ON BREAST CANCER
A large fraction of breast cancer research is devoted to identifying and understanding drugs, drug candidates, and environmental chemicals (together herein referred to as ‘chemicals’) that reduce or increase cancerous behaviors of breast epithelial cells. These studies have identified multiple pathways that are effective targets for breast cancer therapies [1] and uncovered several chemicals present in the environment and consumer products that increase breast cancer risk [2]. However, breast cancer remains the most common non-cutaneous cancer in women and the second leading cause of cancer-related deaths among women [3]. Understanding the effects that chemicals have on breast cancer risk and progression will improve our ability to reduce cancer incidence and mortality.
The existing framework used to evaluate the effect of chemicals on human breast cancer relies on the model systems that are on two ends of a wide spectrum: simple, inexpensive in vitro models, and resource intensive, physiologically complex rodent models. The simplicity and low cost/time requirements of in vitro models are useful for uncovering the innerworkings of signaling pathways and screening for the molecular targets of chemicals, such as the affinity for a receptor. These kinds of experiments are difficult to perform in vivo, as animal models are too complex to easily dissect molecular interactions. Unfortunately, in vitro platforms do not reliably predict the toxic or therapeutic potential of a chemical. Rodent models are valuable as they incorporate the systemic and local effects of chemicals and enable the assessment of chronic chemical exposures and the importance of life stages. However, the biological differences between humans and rodents are problematic when extrapolating data between the species; humans and rodents differ in the development of breast cancer [4, 5], mammary gland physiology [6], and chemical metabolism [7]. Both in vitro and in vivo models are indispensable to the breast cancer field, but each harbors shortcomings that limit their utility.
To increase the efficiency of chemical testing, leaders in pharmacology and toxicology have called for the development and implementation of in vitro models that better recapitulate human physiology [8, 9]. These new systems would help bridge the gap between in vitro and in vivo systems and improve the ability to predict chemical effects on breast cancer risk and progression. While the approaches differ, the prevailing hypothesis is that inclusion of the mammary microenvironment will enhance the ability of in vitro models to predict in vivo outcomes. When integrating the microenvironment into in vitro models, it is important to find a balance between the simplicity and complexity of the system. If a platform becomes too complex, many of the advantages of being in vitro are lost: chemical mechanisms become difficult to decipher, and costs and variability increase. Therefore, to maintain a level of simplicity, it is critical that only the components needed to recapitulate in vivo responses are included in a platform. As the mammary microenvironment is complex and contains many different cell types and proteins, a major challenge is identifying which components are needed to predict chemical responses.
This review describes the normal and cancerous mammary microenvironment and the various strategies used to model breast cancer in vitro. We provide examples in which the microenvironment regulates chemical responses in breast cancer cells and discuss strategies to develop and validate in vitro models.
THE NORMAL AND CANCEROUS MAMMARY GLAND
As illustrated in Fig. 1, the human mammary gland is a complex tissue that evolves as cancer is initiated and progresses. The mammary gland is composed of a series of ducts that collect into the nipple. Mammary ducts are bilayered: the milk-secreting luminal epithelial cells line the ductal lumens while the basal myoepithelial cells face the basement membrane. The basement membrane, a specialized form of extracellular matrix (ECM) that is rich in laminin and collagen IV, provides mechanical support and separates the ducts from the surrounding stroma. The stroma is composed of an ECM rich in fibrous collagen, glycoproteins, and proteoglycans as well as different stromal cells including fibroblasts, adipose stromal cells, adipocytes, and immune, neural, and endothelial cells. The cross talk between the epithelium and stroma tightly regulates the development and maintenance of the mammary gland [10]. Here, we will briefly review the changes that occur within the mammary microenvironment as cancer progresses.
Figure 1.
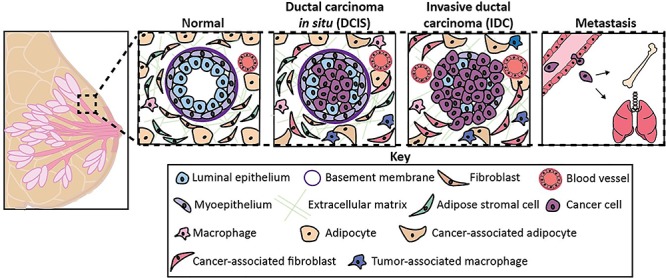
The normal and cancerous mammary microenvironment. The transition from a normal to cancerous mammary gland is characterized by changes in both the epithelium and stroma. The ductal epithelium becomes hyperplastic, the myoepithelial layer is lost, and the basement membrane is degraded. The stromal cell types become activated, immune cells infiltrate to the area, and angiogenesis is increased.
Epithelium
Ductal carcinoma in situ (DCIS) is considered the earliest form of breast cancer and occurs when breast cancer cells have proliferated to fill the breast ducts with cells [11]. When lumens exhibit this high-grade hyperplasia, the intraductal microenvironment becomes hypoxic, acidic, and deprived of nutrients [12, 13]. The transition from DCIS to invasive ductal carcinoma is not well understood; however, the loss of basement membrane, epithelial polarity, and myoepithelial layer are defining features [14].
Extracellular matrix
The ECM of breast cancer tissue is stiffened due to increased collagen cross-linking and increased deposition of ECM proteins such as fibronectin, collagen I, and tenascin-C [15, 16]. Cancer and stromal cells secrete enzymes such as lysyl oxidase, cathepsins, fibroblast activation protein, and matrix metalloproteases that catalyze cross-linking and/or degradation and so remodel the ECM [17–21]. In contrast to the normal mammary gland in which the collagen fibers are curved and randomly organized, the collagen fibers of breast cancer tissue become linear and thick. These aligned collagen fibers facilitate cancer cell invasion into the vasculature [22]. Keely and her colleagues identified three collagen signatures, which correlate with prognosis. Tumor-associated collagen signature-3 (TACS-3), defined by collagen >60° perpendicular to the tumor boundary, predicts worse overall survival of breast cancer patients [23, 24].
Fibroblasts
Fibroblasts are stromal cells that are critical to maintaining the ECM and overall homeostasis of the mammary gland. The fibroblasts of breast cancer tissue acquire an activated phenotype, upregulating expression of proteins such as smooth muscle-alpha actin and fibroblast activation protein [25]. The activated fibroblasts of breast cancer tissue, referred to as cancer-associated fibroblasts, secrete elevated levels of ECM proteins and pro-tumorigenic factors that contribute to the fibrosis and collagen-associated signatures observed in late-stage breast cancers [24]. Fibroblasts can also be activated by exogenous factors, which can contribute to cancer development. In the 1990s, Barcellos-Hoff and colleagues discovered that ionizing radiation damages and activates fibroblasts, stimulating the irradiated fibroblasts to secrete elevated levels of growth factors and hormones [26, 27]. Her group later found that mammary epithelial cells grafted into irradiated mammary gland stroma form more aggressive breast cancers at a faster rate, compared to cells grafted into sham-irradiated hosts [28].
Adipose tissue
Adipocytes (i.e. fat cells) are a major component of the normal mammary gland, where they store energy and secrete a collection of growth factors and pro-inflammatory cytokines collectively referred to as adipokines. In contrast to the normal mammary gland where adipocytes are abundant, the stroma of breast cancer tissue contains mostly immune cells and fibroblasts [29]. Experimental studies have found that adipocytes decrease in size, become delipidated, and acquire a fibroblast-like morphology when exposed to secreted factors from cancer cells, suggesting that a portion of the cancer-associated fibroblasts observed in breast cancer are derived from adipocytes [30]. Cancer-associated adipocytes secrete adipokines such as TNFα, IL-6, and leptin, stimulating cancer cell invasion and proliferation [31, 32].
In addition to mature adipocytes, breast adipose tissue contains adipose stromal cells, a term that refers to a heterogenous mixture of fibroblast-like cells encompassing adipose fibroblasts and adipose stem cells [33]. Similar to fibroblasts, the soluble factors secreted by cancer cells can transform adipose stromal cells into cancer-associated fibroblasts, stimulating the cells to produce elevated levels of cytokines, growth factors, and ECM proteins [34].
Vasculature
Cancer cells and activated stromal cells secrete pro-angiogenic factors such as vascular endothelial growth factor (VEGF) that stimulate the endothelial cells that line blood and lymphatic vessels, thereby inducing angiogenesis and lymphangiogenesis [35]. The new blood vessels provide cancer cells with oxygen and metabolites, which is critical for the progression of high-grade tumors as their cores become hypoxic, acidic, and nutrient deprived [36]. Pericytes, which provide structural support to blood vessels, are decreased in late-stage breast cancers. Blood vessels that exhibit poor pericyte coverage are more prone to leakiness and excessive sprouting [37, 38]. While lymphatic and blood vessels are both routes of metastasis for breast cancer, breast cancer cells more commonly utilize the lymphatic system [39, 40].
Immune cells
Macrophages are recruited to tumors in response to elevated levels of CSF1, CCL2, and CCL8 and can comprise up to 50% of a breast tumor’s volume [41]. The tumor-associated macrophages (TAMs) observed in breast cancer exhibit an M2-like phenotype, secreting immunosuppressive factors which prevent the recruitment of T cells and other immune cells [42, 43]. TAMs also secrete factors that promote the metastasis and proliferation of cancer cells, as well as angiogenic factors [44]. Other immune cell subpopulations, such as natural killer cells [45, 46], neutrophils [47], and T cells [48, 49], also exist within the breast cancer microenvironment and are important to breast cancer progression but are beyond the scope of this review.
Breast cancer is influenced by the ratio of stromal cell types
Due to the impact that each cell type has on cancer progression, it is unsurprising that the ratios of different mammary cell types correlates with cancer outcome. For example, breast cancer patients whose tumors contain a high density of TAMs have a worse prognosis [50]. Decreased pericyte coverage is correlated with poor patient survival [37], and the number of nerve fibers is increased in invasive breast cancers [51]. Costa and colleagues identified four subtypes of cancer-associated fibroblasts in breast cancer tissue and identified correlations between the CAF subtypes and cancer subtype and histological grade [52]. Furthermore, the cellular composition of normal mammary tissue influences breast cancer risk: women whose breast tissue is composed mostly of adipocytes have a lower risk of breast cancer compared to women with dense breast tissue or breast tissue that has a large proportion of fibroblasts and epithelial cells [53].
Summary
As illustrated above, there is a complex interplay between cancer cells and each component of the microenvironment: cancer cells induce an activated phenotype in stromal cells, and cancer-associated stromal cells send signals back to cancer cells to fuel their growth and invasiveness [54]. Similarly, cancer cells secrete factors that remodel the ECM, and the ECM regulates cancer cell invasion [22]. Components of the microenvironment also modify the behavior of one another; both fibroblasts and cancer cells secrete proteases that degrade the ECM, release pro-inflammatory signals that recruit macrophages, and stimulate angiogenesis by secreting angiogenic factors such as VEGF [55–58]. The dynamic cross talk between the cancer cell and each microenvironmental component, as well among components of the microenvironment, is integral to the development and progression of breast cancer (Fig. 2). With this in mind, it is unsurprising that breast cancer cells cultured in a traditional in vitro culture model respond to chemicals differently than those grown in their native microenvironment [59].
Figure 2.
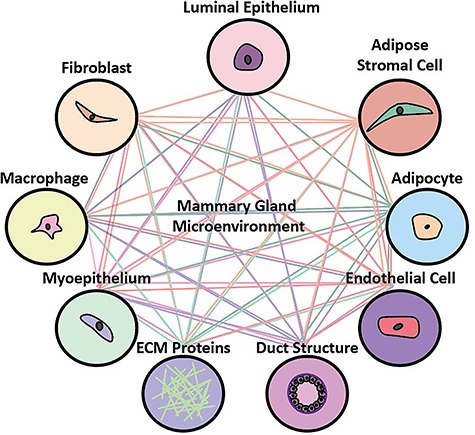
The mammary gland interactome. Each component of the microenvironment communicates with the breast cancer cell, as well as with one another.
TOOLBOX OF IN VITRO CULTURE PLATFORMS
A major advantage of in vitro platforms over in vivo platforms is the ability to precisely control experimental conditions. For this reason, it is easy to understand why most in vitro studies use a simple platform consisting of an immortalized cell line cultured on a flat plastic surface: experimental results are easy to interpret, as the variables affecting cell interactions are well defined. However, due to the growing recognition that in vitro platforms often fail to predict in vivo responses, a variety of strategies have been developed to better recapitulate in vivo biology [60]. Here, we discuss approaches used to improve in vitro breast cancer models, such as the inclusion of primary cells, tissue architecture, and stromal cells. We also summarize the advantages offered by microphysiological models and ex vivo systems. Table 1 provides examples of these different tools and some of their advantages and disadvantages.
Table 1.
Culture strategies that move beyond standard 2D monocultures.
| Cell input | Advantages | Disadvantages | Examples |
|---|---|---|---|
| Primary cells | -Recapitulates heterogeneity observed in tumors and between patients | -Dependent on the availability of primary tissue -Challenging to maintain phenotype in long-term culture |
Hopkinson et al., 2017 Costa et al., 2018 Morgan et al., 2019 |
| Stromal cells | -Paracrine signals induce biological pathways to behave more similar to in vivo Example: fibroblasts encourage differentiation of epithelium and cell polarity |
-Can be challenging to separate different cell types -Media must be compatible with all cell types |
Sung et al., 2013 Carter et al., 2017 Nash et al., 2015 |
| Culture systems | Advantages | Disadvantages | Examples |
| 3D matrices | -Incorporates mechanical and biological signals from the extracellular matrix -Better represents differences between normal and cancerous cells |
-Can be difficult to isolate and analyze cells -Natural matrices can vary batch to batch |
Gudjonsson et al., 2002 Nelson et al., 2006 Nguyen et al., 2017 |
| Microphysiological systems | -Less reagents and cells needed per sample -Can incorporate defined culture areas, geometry, gradients, and structure -High cell to media volume increases sensitivity of paracrine signaling |
-Can be technically challenging to fabricate and use -Small number of cells used per device can lead to a poor reflection of tumor heterogeneity |
Chen et al., 2016 Ayuso et al., 2018 Domenech et al., 2009 |
| Ex vivo systems | -Extracellular matrix, microenvironment, and/or tissue structure is preserved and is native to the tissue of interest | -Dependent on availability of primary tissue -Less control over experimental variables -Generally measured via low-throughput techniques such as microscopy |
Dunne et al., 2014 Vaira et al., 2010 Dean et al., 2012 |
Immortalized and primary cells
Most in vitro breast cancer studies are conducted using immortalized cell lines because they replicate indefinitely and are easy to culture. However, while the genetic profiles of breast cancer cell lines generally reflect the profiles of breast tumors, immortalized cell lines poorly recapitulate the heterogeneity observed within tumors and between different patients [61]. Most breast cancer cell lines are derived from metastatic tumors, so cell lines that model early-stage breast cancer are limited. In addition, while estrogen receptor (ER)-positive breast cancer accounts for the majority of breast cancer cases, most breast cancer cell lines are ER-negative [61, 62]. We expect that immortalized stromal cells suffer from similar issues, and do not recapitulate the heterogeneity of stromal cells seen in vivo [52, 63]. Furthermore, as stromal and cancer cells vary dramatically from patient-to-patient, co-culturing primary tumor and stromal cells from the same patient may better represent patient-specific drug responses, in comparison to culturing the primary tumor cells alone or with stromal cell types derived from a different individual. While there are clear advantages to utilizing primary cells, primary cells are difficult to acquire, and they lose their in vivo phenotypes after a few passages [64]. Consequently, the goal of the experiment should dictate whether immortalized cells or primary cells are more appropriate. For instance, immortalized cell lines may be more useful for hypothesis-generating studies or chemical screens, while primary cells may be more valuable for validation or experiments that study patient-specific responses.
Tissue architecture
While researchers originally overlooked the impact of the structural environment on cell behavior, landmark studies by Mina Bissell’s lab and others revealed that culturing cells in 3D uncovers striking differences in the phenotypes of normal and cancerous breast epithelial cells. When grown as 2D monolayers, non-tumorigenic and cancerous mammary epithelial cell lines appear morphologically similar. However, when grown in a reconstituted basement membrane (rBM, e.g. Matrigel), non-tumorigenic cell lines organize into growth-arrested polarized lumens, while cancerous cell lines form disorganized proliferative colonies [65]. This led to a multitude of publications that reported striking differences in cell behavior when cells are grown in matrices compared to when cultured on plastic [66–68].
To increase the structural relevance of platforms, researchers culture cells on top of matrices (2.5D cultures), independent of matrices (anchorage-independent), or embedded in matrices (3D cultures) [69]. In these platforms, breast epithelial cells self-organize into ductal structures or solid spheres (e.g. spheroids), collectively referred to as organoids, and thus provide a more structurally relevant environment to model preinvasive disease. Multiple groups have generated patient-derived mammary organoids to test potential cancer therapeutics in vitro and have thereby more closely recapitulated the heterogeneity in patient responses that is observed in the clinic [70–72].
Further studies revealed that researchers should carefully consider the type and composition of the matrix used in their in vitro model. When non-tumorigenic mammary epithelial cells are grown in rBM, they form polarized, hollow lumens. However, when the same cells are grown in collagen, they exhibit reverse polarity or no polarity [73]. In addition, changing the stiffness or geometry of the matrix can change cell behavior [74–76]. For example, mesenchymal stromal cells differentiate into cancer-associated fibroblasts when cultured on a stiff collagen matrix [77]. Other studies that compared hydrogels of varying stiffnesses found that plasticity influences breast cancer cell proliferation, invasion [78], and metabolism [79]. This demonstrates similarities to in vivo studies that show a correlation between increased stromal collagen and tumor formation and metastasis in mice [80–83].
Unfortunately, incorporating matrix proteins into in vitro models introduces technical challenges. Cells that are embedded in matrices are less accessible to antibodies and are harder to image and isolate, making qPCR, microscopy, and flow cytometry difficult. To make cells more accessible for these analyses, researchers have designed systems in which cells are cultured on top of matrices rather than within matrices [82]. Another point of difficulty is that natural matrices such as rBM can vary considerably lot to lot, which can negatively impact experimental reproducibility. As such, researchers have turned to synthetic matrixes, such as polyethylene glycol (PEG), because they have well-defined chemistries and are highly reproducible. While biologically inert, synthetic matrices can be modified to better mimic the biological and mechanical properties of natural matrices [84, 85]. For instance, PEG hydrogels can be designed to include the RGD motif that is present in some ECM proteins [86]. The utility of synthetic vs. natural hydrogels has been demonstrated in the context of drug screening: Murphy and colleagues demonstrated that endothelial cells grown on PEG hydrogels are more sensitive to anti-angiogenic chemicals and have more reproducible responses compared to those grown on rBM [83]. While synthetic matrices offer enhanced reproducibility, the trade-off is that they lack the biological complexity offered by natural matrices. Therefore, synthetic hydrogels may be a better choice for studies that need high reproducibility such as high-throughput drug screening experiments, while natural matrices may be more beneficial for studies that utilize biologically sensitive samples, such as primary cells.
Stromal cells
As described above, each stromal cell type of the mammary microenvironment regulates cancer progression. Moreover, they modulate cancer cell function by multiple mechanisms, including secreted proteins and direct contact. Several strategies have been developed to study the effect of stromal cells on cancer cell behavior, largely through the use of conditioned media, compartmentalized co-culture systems, and direct co-culture systems [69]. As there are trade-offs with each type of co-culture method, the preferred strategy varies based on the experimental question.
Researchers sometimes co-culture cells embedded in or on top of matrix proteins and in doing so have revealed dramatic differences in cancer cell behavior when co-cultured with stromal cells in 2D compared to 3D. Multiple studies have found that breast cancer cell invasion is increased when co-cultured with fibroblasts in 3D as compared to 2D, as fibroblasts in 3D rearrange the ECM and secrete higher levels of cytokines [87]. Stromal cells have also been shown to direct the organization of mammary epithelial cells grown in 3D. As described above, benign breast epithelial cell lines are polarized when grown in rBM but fail to polarize when grown in collagen. As stromal cells are important to mammary gland homeostasis, Gudjonsson et al. hypothesized that myoepithelial cells would be sufficient to induce polarity in collagen organoids. When myoepithelial cells were included in the collagen matrix that housed the luminal epithelial cells, the epithelium organized into a polarized structure that appeared morphologically similar to cells grown in rBM [73]. A later paper compared the organization of mixed luminal/myoepithelial cultures when grown in a 3D collagen gel or rBM. Myoepithelial and luminal epithelial cells grown in collagen organized into a bilayered structure, where myoepithelial cells laid against the basement membrane and luminal epithelial cells lined the inner lumen. However, the two cell types did not self-organize when grown in rBM; instead, luminal and epithelial cells were randomly organized throughout the organoid [88]. Several other groups have found that stromal cell types regulate the tissue morphogenesis of mammary epithelial cells grown in 3D; in many cases the structures developed in co-culture were more complex than structures developed in monoculture [89–91].
Microphysiological models
One challenge with increasing the complexity of in vitro models is that standard culture platforms (e.g. well plates) offer limited control over the culture environment. For example, the majority of 3D breast cancer models rely on cancer cells to self-assemble into luminal structures, which can be inefficient as in some scenarios only a small percentage of samples will undergo structural morphogenesis and/or the structures vary dramatically in shape or size [92]. In addition, when studying stromal–epithelial interactions, cells are typically mixed together or layered on top of one another, while in the mammary gland, the spatial organization of cells is tightly regulated. Consequently, microfluidic platforms are increasingly popular due to enhanced control over the culture environment [93].
Microfluidic platforms culture cells and tissues at the physiological scale, where the dimensions of microfluidic devices range from micrometers to millimeters. Microfluidic platforms are highly modular as they are fabricated using cheap and easy to modify materials; microfluidic platforms can incorporate multiple culture areas, diffusion channels, gradients, and/or structures [94]. Our lab uses a microfluidic platform to culture a biomimetic mammary duct surrounded in a collagen matrix containing stromal cells, which is similar to the organization of a normal or premalignant mammary gland [95, 96]. We found that this system could model the gradients of hypoxia and nutrient deprivation that are seen in DCIS tissues in vivo; the metabolic phenotypes of the cells varied from the lumen perimeter to the center, and tirapazamine, a drug that induces DNA damage under hypoxic conditions, was highly toxic to the ductal core and had little effect on the edges of the ducts [95]. The temporal control enabled by microphysiological models also eases the study of paracrine signaling that would be challenging to evaluate in traditional in vitro models; for example, our lab has developed microfluidic models that cultures cells in nearby compartments, allowing different cell types to be added and/or removed at different distances and times [97, 98]. Kamm and colleagues have developed several microphysiological models that culture metastatic microenvironments adjacent to a biomimetic vasculature; cancer cells can be added to the vasculature and monitored for extravasation into the bone microenvironment [99, 100]. In addition to increased spatial and temporal control, microfluidic platforms require small amounts of cells and reagents, which can be advantageous when working with primary cells or expensive matrix proteins. The high surface area to volume ratio in microfluidic platforms also increases the sensitivity of paracrine signaling: there is less medium volume per cell, which results in a higher concentration of secreted factors [101].
While there are clear advantages to utilizing microfluidic models, most of these platforms are in their infancy, and future studies are needed to confirm if they recapitulate human responses more faithfully than traditional culture platforms. In addition, microfluidic devices can be technically challenging to learn, are relatively low throughput to build, are usually constructed by academic labs, and, as such, are not commercially available [102]. Further, the small amounts of cells used per device can lead to a poor reflection of tumor heterogeneity. The emergence of bioprinting may overcome some of these concerns. Bioprinting eases the construction and use of complex microphysiological models, as multiple types of materials and cell types can be printed simultaneously [103]. Altogether, commercializing platforms to decrease variability and increase usability will be essential for the widespread adoption of microscale platforms.
Ex vivo systems
The above strategies involve combining cells or matrix proteins from different sources to create an in vitro model. However, the organization and composition of a tissue is complex, and it is consequently challenging to accurately recapitulate tissues in vitro. To combat this issue, some researchers perform in vitro studies in ex vivo systems that preserve the native tissue architecture: tumor explants or decellularized tissues.
Tumor explants are tumors that are removed from patients, sectioned, and then cultured in vitro. The cells of these ex vivo models can be evaluated in a system where the tissue architecture, cellular composition, and microenvironment of the tumors remain intact [104, 105]. A different approach is to decellularize, or remove cells from, a tissue or organ and then use the tissue scaffold as the base model for cell seeding. For instance, one group decellularized subcutaneous adipose tissue and seeded breast cancer cells in the scaffold to create a physiologically relevant drug testing model [106]. Utilizing decellularized tissues is advantageous because extracellular matrices differ by disease state and by cancer, which creates challenges when mimicking native microenvironments in vitro. The major drawbacks of these ex vivo systems are that they are limited by the availability of patient samples, experimental parameters are more difficult to control, and their responses are generally measured via low-throughput techniques such as microscopy.
EXAMPLES OF CHEMICAL INTERACTIONS THAT ARE REGULATED BY THE MICROENVIRONMENT
Due to the myriad of effects that the microenvironment has on cancer cell behavior, the responses of cancer cells to chemicals can differ dramatically depending on the surrounding microenvironment. In addition, some chemicals that target the microenvironment directly, such as immunotherapies or aromatase inhibitors, cannot be evaluated when cancer cells are grown alone. To illustrate this point, we provide examples of chemical interactions in the mammary gland that are regulated by the microenvironment.
Stromal cell modulation of targeted therapy resistance
A number of studies have found that stromal cells regulate the expression of ER alpha, a nuclear receptor protein that controls breast cancer cell proliferation in approximately 75% of breast cancer cases [96, 107]. One study found that stromal cells that lack the cell surface marker CD146 enhance ER-independent growth in breast cancer cells via activation of the EGFR, HER2, and IGF1R pathways. By inducing ER-independent growth, CD146− cells confer resistance to tamoxifen, an ER antagonist used to treat breast cancer. Interestingly, the authors showed that an epithelial gene signature indicative of a high percentage of CD146- fibroblasts is predictive of decreased recurrence-free survival in breast cancer patients treated with tamoxifen [108].
Stromal cell modulation of apoptosis in breast cancer cells
Triple-negative breast cancer (TNBC) accounts for approximately 15% of all breast cancers and can be organized into different subtypes. The mesenchymal subtype (M) and the basal-like subtypes (BLS) of TNBC differ in chemotherapeutic resistance: the mesenchymal subtype of TNBC exhibits higher resistance to chemotherapies in comparison to the BLS of TNBC [109]. Interestingly, when M and BLS cell lines were grown as a monoculture and exposed to breast cancer chemotherapeutics, they did not segregate by chemotherapeutic resistance. However, when the cancer cell subtypes were co-cultured with fibroblasts, they responded to chemotherapeutics similarly to what is observed clinically. While the mechanism is not entirely understood, the authors found that the fibroblasts differentially altered apoptotic priming depending on the TNBC subtype [110].
Chemotherapeutic-induced activation of fibroblasts
Recently researchers found that exposure to doxorubicin, 4-hydroxy-cyclophosphamide, and paclitaxel induce an activated phenotype in human mammary fibroblasts. When co-cultured with chemotherapy-treated fibroblasts, breast cancer cells exhibited more proliferative and invasive behavior compared to those co-cultured with vehicle-treated fibroblasts [111]. This finding has been observed by other breast cancer researchers [112] as well as in other types of cancer [113, 114]. Importantly, adjusting the dosing paradigm was sufficient to prevent fibroblast activation: treatment of fibroblasts with a low dose of chemotherapy for 10 days rather than a high dose for 1 day prevented fibroblast activation [111]. This data has prompted some authors to hypothesize that therapeutics that prevent fibroblast activation would decrease the incidence of chemotherapeutic resistance [58].
Matrix proteins mediate drug responses
It is well-accepted that cells grown in 3D show different drug responses as compared to those grown on plastic. For instance, primary breast cancer cells are dramatically less sensitive to the breast cancer therapeutic trastuzumab when cultured on plastic, compared to when grown on collagen I, collagen III, or rBM [115]. In addition, the composition and stiffness of the matrices used in in vitro models influence drug responses. Breast cancer cells grown in high-density collagen are more resistant to the ER antagonist 4-hydroxytamoxifen, compared to when grown in low-density collagen [76]. HER2+ breast cancer cells grown on the basement membrane protein laminin-5 are less sensitive to trastuzumab, compared to when grown on plastic, collagen I, or fibronectin [116]. These findings underscore the importance of the ECM composition in in vitro drug testing systems.
Microenvironmental regulation of hepcidin expression
Hepcidin is a hormone that contributes to breast cancer progression via the regulation of iron metabolism [117]. A recent study found that hepcidin expression differs dramatically depending on the culture condition. Compared to 2D cultures, breast cancer cells grown as spheroids exhibited markedly higher levels of hepcidin mRNA and protein and increased iron retention. Through a series of mechanistic studies, the authors concluded that the BMP signaling pathway was activated when breast cancer cells were grown as spheroids, which caused hepcidin to be regulated by growth differentiation factor 15 (GDF-15). The expression of hepcidin and GDF-15 were correlated in human breast cancer tissue, while no relationship was observed between hepcidin and GDF-15 when breast cancer cells were cultured in 2D. The authors also found that breast cancer cells grown as spheroids exhibited an even higher hepcidin expression when co-cultured with fibroblasts, as fibroblasts secreted IL-6 which induces hepcidin expression in breast cancer cells. Interestingly, IL-6 had no effect on hepcidin expression when breast cancer cells were cultured in 2D [118]. This study provides a clear example of a signaling pathway that is differentially activated when breast cancer cells are cultured in 3D as compared to 2D.
Environmental obesogens target adipocytes
Some environmental chemicals have been found to alter the differentiation and/or proliferation of adipocytes, primarily by interacting with the master regulators of lipogenesis PPARy and RXR. Named after their ability to alter body weight, obesogens are an emerging area of research in toxicology [119]. Several types of environmental chemicals have been implicated as obesogens, including organotin compounds, phthalates, perfluoroalkyl compounds, and bisphenols [120]. Obesogens are relevant to the breast cancer field because obesity is associated with an increased risk of developing breast cancer. While the mechanisms are not understood, signals from the adipose microenvironment are thought to be primarily responsible: adipocytes secrete hormones and cytokines, coined adipokines, which are upregulated in obese individuals and are mitogenic to breast cancer cells [121, 122]. Consequently, some authors have hypothesized that obesogens can indirectly increase breast cancer risk or progression by altering the adipose stroma [123].
Modulation of aromatase in adipose stromal cells
The primary source of estrogen in the mammary gland of postmenopausal women is adipose stromal cells. Mammary adipose stromal cells produce the enzyme aromatase, which metabolizes androgens to estrogens [124]. Like many potential targets in the cancer microenvironment, aromatase is a target of both breast cancer therapies and environmental chemicals: aromatase inhibitors letrozole and anastrozole are the first-line treatment for postmenopausal women with ER+ breast cancer [125], and several chemicals present in pesticides and plasticizers modulate aromatase expression [126]. Some environmental chemicals that target ER also modulate aromatase expression [127]; since aromatase controls estrogen production, these chemicals could lead to synergistic or neutralizing effects on ER signaling. While aromatase studies have mostly been limited to in vivo studies, the inclusion of adipose stromal cells in in vitro breast cancer models would provide a more tractable approach. We recently co-cultured adipose stromal cells with breast cancer cells in a microphysiological breast model to study the mechanisms responsible for the increased risk of aromatase inhibitor resistance in obese women, and we showed that the type of co-culture platform determines the response of cells to the aromatase inhibitor anastrozole. When incorporated into the microphysiological model, mammary adipose stromal cells derived from obese women increased resistance to anastrozole as compared to lean-derived stromal cells. In contrast, no difference in anastrozole resistance was detected between the two groups when they were evaluated in a 2D co-culture platform [128].
Another factor to consider when evaluating aromatase modulators is inflammatory cells. Under chronic states of inflammation such as obesity, the pro-inflammatory cytokines secreted by immune cells are hypothesized to enhance aromatase expression in mammary stromal cells, leading to aromatase inhibitor resistance [129]. In support of this hypothesis, in vitro studies have found the conditioned media from macrophages increases aromatase expression in adipose stromal cells [130]. Another study found that the frequency of crown-like structures formed by macrophages correlates with aromatase expression in adipose stromal cells of human mammary tissue, further supporting the importance of macrophages on aromatase expression in breast cancer [131].
Immune-targeted therapies
In response to an increased understanding of the interactions between the immune system and cancer progression, scientists have developed a variety of therapeutic strategies that target immune cell types such as T cells, natural killer cells, and/or macrophages. As TAMs typically exhibit an anti-inflammatory phenotype that deters the recruitment and activation of T cells [43, 132], researchers have developed therapies that either prevent the infiltration of TAMs or repolarize TAMs to a pro-inflammatory phenotype. For example, anti-CCL2 antibodies prevent the recruitment of inflammatory monocytes, inhibit metastasis, and prolong the survival of mice with breast cancer [133]. A more recent study showed that PI3Kγ inhibition promoted a pro-inflammatory phenotype in macrophages and enhanced T-cell-mediated cytotoxicity, which led to a decrease in tumor growth and metastasis of murine breast tumors [134]. Other immune therapies directly target T cells to increase their cytotoxicity. For example, in CAR-T therapy, T cells are harvested from a patient and genetically engineered to express chimeric antigen receptors (CARs), which improves the T cells’ ability to recognize cancer cells. These CAR-T cells are then infused back into the patient [135]. While tremendous progress has been made in the immune therapy field, a major challenge with assessing immune therapies is that studies are largely limited to humans or animal models, as traditional in vitro systems are incapable of modeling these complex multicellular interactions. However, recent advances in engineering and cellular biology have enabled researchers to co-culture immune cells and cancer cells in vitro. For example, Wallstabe et al. [136] utilized a microphysiological breast cancer model to evaluate the therapeutic effect of CAR-T cells on breast cancer cells in vitro. The development of in vitro models that can model immune–cancer cell interactions will certainly improve understanding of the effects that immune therapies have on breast cancer.
Stromal mediation of fibroblast growth factor receptor
Fibroblast growth factor receptor (FGFR) is frequently dysregulated in several types of cancer, where activation of FGFR increases the proliferation of cancer cells and stimulates resistance to several types of cancer therapies. While the mechanisms can vary, cancer-associated stromal cells secrete elevated FGFs, which activates FGFR increasing drug resistance [137]. For example, FGF2 secreted by adipocytes and cancer-associated fibroblasts increases resistance to anti-VEGF therapy, and inhibition of FGF signaling restores drug sensitivity [138]. A different study found that cancer-associated fibroblasts secrete FGF5, which increases the number of cancer stem cells and induces resistance to chemotherapies [139]. These findings have increased interest in co-administrating FGFR inhibitors with standard cancer drugs to prolong therapeutic sensitivity. As FGFs secreted by macrophages and fibroblasts enhance resistance to FGFR inhibitors in vitro [140], co-culture systems that utilize patient-matched stromal and cancer cells may be useful for predicting if an individual is a good candidate for FGFR inhibitors.
CONSIDERATIONS FOR THE DESIGN OF IN VITRO MODELS
There is a delicate balance between the complexity and simplicity of any model system. As additional components are integrated into a platform, the model becomes more resource intensive, and responses become difficult to interpret. Ideally, a researcher would use the simplest possible model system that predicts human responses to chemicals. However, there are many strategies to build an in vitro model, and the ideal approach is often unclear. Consider a situation in which a researcher wants to develop an in vitro model and is deciding between a platform that contains no matrix proteins, collagen, or rBM, as well as a monoculture or co-culture; in this scenario there are six combinations to choose from. If the researcher also considers a macroscale or microscale platform, there are 12 options. When the variety of cancer subtypes, cell types, and life stages are included in the decision, there are an infinite number of possibilities. Pinpointing the best components to include in a model and which responses will best predict human physiology is an ongoing challenge. In this section, we discuss the factors to consider when choosing an in vitro system for chemical testing.
The significance of the microenvironment ultimately depends on the question being asked. Breast cancer is a heterogeneous disease encompassing multiple subtypes that differ in signaling and progression [141], and chemicals vary in the pathways that they target. Therefore, the importance of the microenvironment should be evaluated case by case as the significance depends on the context, such as the chemical and endpoint being evaluated, and the cancer subtype and stage. For instance, a simple 2D monoculture platform is an ideal system to test if a chemical activates a receptor in a given cell type or to gain a basic understanding of a chemical’s cytotoxicity. However, in vitro models that incorporate microenvironmental components are often needed when deciphering the more complex effects of a chemical, such as if the chemical alters metabolism in addition to receptor binding or if the chemical targets multiple cell types [128]. For instance, co-culture systems may be better equipped for understanding the efficacy of drugs that target paracrine interactions (e.g. aromatase inhibitors, FGF inhibitors) or are influenced by paracrine interactions (chemotherapeutics, ER antagonists).
This point is underscored in ‘Examples of chemical interactions that are regulated by the microenvironment’ where we provided examples of different scenarios where models of varying degrees of complexity were needed to recapitulate in vivo responses. For example, in ‘Stromal cell modulation of targeted therapy resistance’ and ‘Stromal cell modulation of apoptosis in breast cancer cells’, a co-culture (rather than a monoculture) was needed to mimic breast cancer cell responses to ER antagonists and chemotherapies, respectively. In ‘Stromal cell modulation of apoptosis in breast cancer cells’, the authors did not observe dramatic differences in apoptosis when the cells were co-cultured in 2D compared to in 3D; a 2D co-culture was sufficient for modeling the clinical outcome. However, in ‘Modulation of aromatase in adipose stromal cells’, we discussed a case where a 3D co-culture better recapitulated in vivo biology than a 2D co-culture: while cancer cells responded similarly to the aromatase inhibitor anastrozole when co-cultured with adipose stromal cells derived from lean and obese women in 2D, co-culturing the same cells in an organotypic co-culture system segregated the anastrozole responses of the lean and obese donors. The take-home message from these case studies is that the perfect model system will vary depending on the experimental question.
To inform the design of future model systems and help define the scenarios in which the microenvironment is needed, we must increase understanding of the influence that the microenvironment has on breast cancer development and chemical responses. Studies must assess if more complex in vitro models respond to chemicals differently than traditional in vitro models and whether the response is predictive of what is seen clinically in humans. Because failure to compare to standard culture models stunts the ability to determine if a more complex microenvironment is needed, researchers that evaluate chemicals in a complex in vitro platform should concurrently evaluate chemicals in a traditional in vitro model.
In vitro responses must also be compared directly to human data to validate that the responses observed in vitro are reflective of human responses. Models for pharmaceutical research can be validated by conducting in vitro studies using clinical samples alongside, or retrospectively, with clinical trials. For example, Hans Clevers’ group validated their organoid models by comparing chemical responses observed in patient-derived organoids to clinical data, which revealed that the organoids responded to tamoxifen similarly to their respective donors [70]. Comparison of responses of human and experimental models is more difficult when evaluating chemicals for their ability to increase cancer risk. While results can be compared to animal studies, critical differences between the physiology of animals and humans present uncertainties when extrapolating these data across species. Consequently, validating in vitro data regarding chemical risk factors is limited to studying accidental or occupational exposures, which can be challenging to analyze since the dose, exposure duration, and other factors are usually unclear. Comparing data about unknown chemicals to well-characterized chemicals, such as pharmaceuticals, is one strategy to validate chemical hits.
In summary, the ideal chemical testing system is the simplest model that can accurately predict human responses to chemicals. Known information about the experimental parameters can be harnessed to help decide if a more complex in vitro model is needed to predict the effects of a chemical. The chemical responses observed in the complex in vitro model should be compared to a simple in vitro model, as well as to human data, to help determine which model system should be used for chemical testing.
SUMMARY
Accumulating evidence suggests that the in vitro tools used to study breast cancer are often too simple to recapitulate in vivo responses; this hinders our ability to predict the effects of chemicals on breast cancer risk and progression. Advances in tissue engineering and cancer biology have enabled researchers to integrate components of the microenvironment into in vitro platforms, which has shown that the in vitro microenvironment strongly influences how cancer cells respond to chemicals. Future studies are needed to determine the effect of the microenvironment on xenobiotic responses and to define the scenarios where complex in vitro models are needed to better understand chemical actions. We expect that further work in elucidating the relevant factors in in vitro systems will undoubtedly provide great strides in breast cancer research.
Author contribution
M.M.M. conducted the literature search, wrote the manuscript, and drew the figure. L.A.S. and J.C.C. edited the review. D.J.B., E.T.A., B.P.J., and L.A.S. supervised the preparation of the manuscript and helped construct the idea. All authors approved the final version of the manuscript.
Conflict of interest statement
Three of the authors have conflicts of interests. D.J.B. holds equity in Bellbrook Labs, L.L.C.; Tasso, Inc.; Stacks to the Future, L.L.C.; Salus Discovery, L.L.C.; Lynx Biosciences, Inc.; and Onexio Biosystems L.L.C. B.P.J. holds equity in Onexio Biosystems L.L.C. M.M.M. is an employee of Salus Discovery where she has received compensation.
Funding
This work was supported by the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520, Environmental Protection Agency's Science to Achieve Results (STAR) grant number 83573701, NIH R01 CA186134, National Institute of Health (NIH) National Cancer Institute T32 CA157322 to D.J.B., K99ES028744 to B.P.J., NIH National Institute of Environmental Health Sciences T32 ES007015-39 to M.M.M. and J.C.C., NIH R01 CA179556 to L.A.S., and funding from the University of Wisconsin Carbone Cancer Center and the McArdle Laboratory for Cancer Research to E.T.A.
Contributor Information
Molly M Morgan, Email: mmemorgan5@gmail.com.
Linda A Schuler, Email: linda.schuler@wisc.edu.
Jordan C Ciciliano, Email: jordanciciliano@gmail.com.
Brian P Johnson, Email: bpjohnson5@wisc.edu.
Elaine T Alarid, Email: alarid@oncology.wisc.edu.
David J Beebe, Email: djbeebe@wisc.edu.
References
- 1. Masoud V, Pagès G. Targeted therapies in breast cancer: new challenges to fight against resistance. World J Clin Oncol. 2017;8:120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodgers KM, Udesky JO, Rudel RA et al. Environmental chemicals and breast cancer: an updated review of epidemiological literature informed by biological mechanisms. Environ Res. 2018;160:152–82. [DOI] [PubMed] [Google Scholar]
- 3. NIH Cancer of the breast (female) - SEER stat fact sheets, 2016. http://seer.cancer.gov/statfacts/html/breast.html.
- 4. Holen I, Speirs V, Morrissey B et al. In vivo models in breast cancer research: progress challenges and future directions. Dis Model Mech. 2017;10:359–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sflomos G, Dormoy V, Metsalu T et al. A preclinical model for ERalpha-positive breast cancer points to the epithelial microenvironment as determinant of luminal phenotype and hormone response. Cancer Cell. 2016;29:407–22. [DOI] [PubMed] [Google Scholar]
- 6. McNally S, Stein T. Overview of mammary gland development: a comparison of mouse and human. Methods Mol Biol. 2017;1501:1–17. [DOI] [PubMed] [Google Scholar]
- 7. Zhao H, Zhou L, Shangguan AJ et al. Aromatase expression and regulation in breast and endometrial cancer. J Mol Endocrinol. 2016;57:R19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pamies D, Hartung T. 21st century cell culture for 21st century toxicology. Chem Res Toxicol. 2017;30:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vidi PA, Leary J, Lelièvre SA. Building risk-on-a-chip models to improve breast cancer risk assessment and prevention. Integr Biol (Camb). 2013;5:1110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inman JL, Robertson C, Mott JD et al. Mammary gland development: cell fate specification stem cells and the microenvironment. Development 2015;142:1028–42. [DOI] [PubMed] [Google Scholar]
- 11. Nelson AC, Machado HL, Schwertfeger KL. Breaking through to the other side: microenvironment contributions to DCIS initiation and progression. J Mammary Gland Biol Neoplasia. 2018;23:207–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Espina V, Liotta LA. What is the malignant nature of human ductal carcinoma in situ? Nat Rev Cancer. 2011;11:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 14. Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Acerbi I, Cassereau L, Dean I et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb). 2015;7:1120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ioachim E, Charchanti A, Briasoulis E et al. Immunohistochemical expression of extracellular matrix components tenascin, fibronectin, collagen type IV and laminin in breast cancer: their prognostic value and role in tumour invasion and progression. Eur J Cancer. 2002;38:2362–70. [DOI] [PubMed] [Google Scholar]
- 17. Salvador F, Martin A, Lopez-Menendez C et al. Lysyl oxidase-like protein LOXL2 promotes lung metastasis of breast cancer. Cancer Res. 2017;77:5846–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malik R, Lelkes PI, Cukierman E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 2015;33:230–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kai F, Drain AP, Weaver VM. The extracellular matrix modulates the metastatic journey. Dev Cell. 2019;49:332–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gocheva V, Wang HW, Gadea BB et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koczorowska MM, Tholen S, Bucher F et al. Fibroblast activation protein-alpha, a stromal cell surface protease, shapes key features of cancer associated fibroblasts through proteome and degradome alterations. Mol Oncol. 2016;10:40–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han W, Chen S, Yuan W et al. Oriented collagen fibers direct tumor cell intravasation. Proc Natl Acad Sci U S A. 2016;113:11208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Esbona K, Yi Y, Saha S et al. The presence of cyclooxygenase 2, tumor-associated macrophages, and collagen alignment as prognostic markers for invasive breast carcinoma patients. Am J Pathol. 2018;188:559–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conklin MW, Eickhoff JC, Riching KM et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol. 2011;178:1221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng JD, Valianou M, Canutescu AA et al. Abrogation of fibroblast activation protein enzymatic activity attenuates tumor growth. Mol Cancer Ther. 2005;4:351–60. [DOI] [PubMed] [Google Scholar]
- 26. Barcellos-Hoff MH. The potential influence of radiation-induced microenvironments in neoplastic progression. J Mammary Gland Biol Neoplasia. 1998;3:165–75. [DOI] [PubMed] [Google Scholar]
- 27. Barcellos-Hoff MH, Park C, Wright EG. Radiation and the microenvironment - tumorigenesis and therapy. Nat Rev Cancer. 2005;5:867–75. [DOI] [PubMed] [Google Scholar]
- 28. Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer Res. 2000;60:1254–60. [PubMed] [Google Scholar]
- 29. Tan J, Buache E, Chenard MP et al. Adipocyte is a non-trivial, dynamic partner of breast cancer cells. Int J Dev Biol. 2011;55:851–9. [DOI] [PubMed] [Google Scholar]
- 30. Wang YY, Attane C, Milhas D et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight. 2017;2:e87489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dirat B, Bochet L, Dabek M et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011;71:2455–65. [DOI] [PubMed] [Google Scholar]
- 32. Arendt LM, McCready J, Keller PJ et al. Obesity promotes breast cancer by CCL2-mediated macrophage recruitment and angiogenesis. Cancer Res. 2013;73:6080–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bourin P, Bunnell BA, Casteilla L et al. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy. 2013;15:641–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sakurai M, Miki Y, Takagi K et al. Interaction with adipocyte stromal cells induces breast cancer malignancy via S100A7 upregulation in breast cancer microenvironment. Breast Cancer Res. 2017;19:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jussila L, Alitalo K. Vascular growth factors and lymphangiogenesis. Physiol Rev. 2002;82:673–700. [DOI] [PubMed] [Google Scholar]
- 36. Boudreau N, Myers C. Breast cancer-induced angiogenesis: multiple mechanisms and the role of the microenvironment. Breast Cancer Res. 2003;5:140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cooke VG, LeBleu VS, Keskin D et al. Pericyte depletion results in hypoxia-associated epithelial-to-mesenchymal transition and metastasis mediated by met signaling pathway. Cancer Cell. 2012;21:66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas HM, Cowin AJ, Mills SJ. The importance of pericytes in healing: wounds and other pathologies. Int J Mol Sci 182017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mohammed RA, Martin SG, Gill MS et al. Improved methods of detection of lymphovascular invasion demonstrate that it is the predominant method of vascular invasion in breast cancer and has important clinical consequences. Am J Surg Pathol. 2007;31:1825–33. [DOI] [PubMed] [Google Scholar]
- 40. Lee E, Pandey NB, Popel AS. Crosstalk between cancer cells and blood endothelial and lymphatic endothelial cells in tumour and organ microenvironment. Expert Rev Mol Med. 2015;17:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cassetta L, Fragkogianni S, Sims AH et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, bio-markers, and therapeutic targets. Cancer Cell 2019;35:588–602.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Linde N, Casanova-Acebes M, Sosa MS et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun. 2018;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pathria P, Louis TL, Varner JA. Targeting tumor-associated macrophages in cancer. Trends Immunol. 2019;40:310–27. [DOI] [PubMed] [Google Scholar]
- 44. Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17:887–904. [DOI] [PubMed] [Google Scholar]
- 45. Kim R, Kawai A, Wakisaka M et al. A potential role for peripheral natural killer cell activity induced by preoperative chemotherapy in breast cancer patients. Cancer Immunol Immunother. 2019;68:577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mamessier E, Sylvain A, Thibult ML et al. Human breast cancer cells enhance self-tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest. 2011;121:3609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu L, Saxena S, Awaji M et al. Tumor-associated neutrophils in cancer: going pro. Cancers (Basel) 2019;11:564–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu J, Sun C, Bernatchez C et al. T-cell homing therapy for reducing regulatory T cells and preserving effector T-cell function in large solid Tumors. Clin Cancer Res. 2018;24:2920–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mahmoud SM, Paish EC, Powe DG et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–55. [DOI] [PubMed] [Google Scholar]
- 50. Qiu SQ, Waaijer SJH, Zwager MC et al. Tumor-associated macrophages in breast cancer: innocent bystander or important player? Cancer Treat Rev. 2018;70:178–89. [DOI] [PubMed] [Google Scholar]
- 51. Pundavela J, Roselli S, Faulkner S et al. Nerve fibers infiltrate the tumor microenvironment and are associated with nerve growth factor production and lymph node invasion in breast cancer. Mol Oncol. 2015;9:1626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Costa A, Kieffer Y, Scholer-Dahirel A et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 2018;33:463–79.e10. [DOI] [PubMed] [Google Scholar]
- 53. Nazari SS, Mukherjee P. An overview of mammographic density and its association with breast cancer. Breast Cancer. 2018;25:259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eiro N, Gonzalez L, Martinez-Ordonez A et al. Cancer-associated fibroblasts affect breast cancer cell gene expression, invasion and angiogenesis. Cell Oncol (Dordr) 2018;41:369–78. [DOI] [PubMed] [Google Scholar]
- 55. Li X, Gao Y, Li J et al. FOXP3 inhibits angiogenesis by downregulating VEGF in breast cancer. Cell Death Dis. 2018;9:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Low-Marchelli JM, Ardi VC, Vizcarra EA et al. Twist1 induces CCL2 and recruits macrophages to promote angiogenesis. Cancer Res. 2013;73:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lebeau A, Nerlich AG, Sauer U et al. Tissue distribution of major matrix metalloproteinases and their transcripts in human breast carcinomas. Anticancer Res. 1999;19:4257–64. [PubMed] [Google Scholar]
- 58. LeBleu VS, Kalluri R. A peek into cancer-associated fibroblasts: origins, functions and translational impact. Dis Model Mech 112018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morgan MM, Johnson BP, Livingston MK et al. Personalized in vitro cancer models to predict therapeutic response: challenges and a framework for improvement. Pharmacol Ther. 2016;165:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brock EJ, Ji K, Shah S et al. In vitro models for studying invasive transitions of ductal carcinoma in situ. J Mammary Gland Biol Neoplasia. 2019;24:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vincent KM, Findlay SD, Postovit LM. Assessing breast cancer cell lines as tumour models by comparison of mRNA expression profiles. Breast Cancer Res. 2015;17:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dai X, Cheng H, Bai Z et al. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 2017;8:3131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang M, Li Z, Ren M et al. Stromal infiltration of tumor-associated macrophages conferring poor prognosis of patients with basal-like breast carcinoma. J Cancer. 2018;9:2308–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hopkinson BM, Klitgaard MC, Petersen OW et al. Establishment of a normal-derived estrogen receptor-positive cell line comparable to the prevailing human breast cancer subtype. Oncotarget. 2017;8:10580–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Petersen OW, Rønnov-Jessen L, Howlett AR et al. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Muthuswamy SK, Li D, Lelievre S et al. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Debnath J, Mills KR, Collins NL et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. [DOI] [PubMed] [Google Scholar]
- 68. Marchese S, Silva E. Disruption of 3D MCF-12A breast cell cultures by estrogens--an in vitro model for ER-mediated changes indicative of hormonal carcinogenesis. PLoS One. 2012;7:e45767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15:647–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sachs N, Ligt J, Kopper O et al. A living biobank of breast cancer Organoids captures disease heterogeneity. Cell 2018;172:373–86.e10. [DOI] [PubMed] [Google Scholar]
- 71. Pauli C, Hopkins BD, Prandi D et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Walsh AJ, Cook RS, Sanders ME et al. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 2014;74:5184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gudjonsson T, Ronnov-Jessen L, Villadsen R et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nelson CM, VanDuijn MM, Inman JL et al. Tissue geometry determines sites of mammary branching morphogenesis in Organotypic cultures. Science. 2006;314:298–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zaman MH, Trapani LM, Sieminski AL et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103:10889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Barcus CE, Holt EC, Keely PJ et al. Dense collagen-I matrices enhance pro-tumorigenic estrogen-prolactin crosstalk in MCF-7 and T47D breast cancer cells. PLoS One. 2015;10:e0116891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ishihara S, Inman DR, Li WJ et al. Mechano-signal transduction in mesenchymal stem cells induces prosaposin secretion to drive the proliferation of breast cancer cells. Cancer Res. 2017;77:6179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cavo M, Fato M, Penuela L et al. Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci Rep. 2016;6:35367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Morris BA, Burkel B, Ponik SM et al. Collagen matrix density drives the metabolic shift in breast cancer cells. EBioMedicine 2016;13:146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Provenzano PP, Inman DR, Eliceiri KW et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Barcus CE, O'Leary KA, Brockman JL et al. Elevated collagen-I augments tumor progressive signals, intravasation and metastasis of prolactin-induced estrogen receptor alpha positive mammary tumor cells. Breast Cancer Res. 2017;19:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lee GY, Kenny PA, Lee EH et al. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nguyen EH, Daly WT, Le NNT et al. Versatile synthetic alternatives to Matrigel for vascular toxicity screening and stem cell expansion. Nat Biomed Eng. 2017;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li Y, Kumacheva E. Hydrogel microenvironments for cancer spheroid growth and drug screening. Sci Adv 2018;4:eaas8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li J, Mooney DJ. Designing hydrogels for controlled drug delivery. Nat Rev Mater. 2016;1:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Loessner D, Stok KS, Lutolf MP et al. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials. 2010;31:8494–506. [DOI] [PubMed] [Google Scholar]
- 87. Sung KE, Su X, Berthier E et al. Understanding the impact of 2D and 3D fibroblast cultures on in vitro breast cancer models. PLoS One 2013;8:e76373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Carter EP, Gopsill JA, Gomm JJ et al. A 3D in vitro model of the human breast duct: a method to unravel myoepithelial-luminal interactions in the progression of breast cancer. Breast Cancer Res. 2017;19:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lühr I, Friedl A, Overath T et al. Mammary fibroblasts regulate morphogenesis of normal and tumorigenic breast epithelial cells by mechanical and paracrine signals. Cancer Lett. 2012;325:175–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Koledova Z, Lu P. A 3D fibroblast-epithelium co-culture model for understanding microenvironmental role in branching morphogenesis of the mammary gland. Methods Mol Biol. 2017;1501:217–31. [DOI] [PubMed] [Google Scholar]
- 91. Nash CE, Mavria G, Baxter EW et al. Development and characterisation of a 3D multi-cellular in vitro model of normal human breast: a tool for cancer initiation studies. Oncotarget 2015;6:13731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sokol ES, Miller DH, Breggia A et al. Growth of human breast tissues from patient cells in 3D hydrogel scaffolds. Breast Cancer Res. 2016;18:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 2015;14:248–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014;507:181–9. [DOI] [PubMed] [Google Scholar]
- 95. Ayuso JM, Gillette A, Lugo-Cintron K et al. Organotypic microfluidic breast cancer model reveals starvation-induced spatial-temporal metabolic adaptations. EBioMedicine 2018;37:144–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Morgan MM, Livingston MK, Warrick JW et al. Mammary fibroblasts reduce apoptosis and speed estrogen-induced hyperplasia in an organotypic MCF7-derived duct model. Sci Rep. 2018;8:7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sung KE, Yang N, Pehlke C et al. Transition to invasion in breast cancer: a microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol (Camb). 2011;3:439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yu J, Berthier E, Craig A et al. Reconfigurable open microfluidics for studying the spatiotemporal dynamics of paracrine signalling. Nat Biomed Eng. 2019;3:830–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chen MB, Lamar JM, Li R et al. Elucidation of the roles of tumor integrin beta1 in the extravasation stage of the metastasis Cascade. Cancer Res. 2016;76:2513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jeon JS, Bersini S, Gilardi M et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc Natl Acad Sci U S A. 2015;112:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Domenech M, Yu H, Warrick J et al. Cellular observations enabled by microculture: paracrine signaling and population demographics. Integr Biol (Camb). 2009;1:267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Paguirigan AL, Beebe DJ. Microfluidics meet cell biology: bridging the gap by validation and application of microscale techniques for cell biological assays. Bioessays. 2008;30:811–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ma X, Liu J, Zhu W et al. 3D bioprinting of functional tissue models for personalized drug screening and in vitro disease modeling. Adv Drug Deliv Rev. 2018;132:235–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Vaira V, Fedele G, Pyne S et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci U S A. 2010;107:8352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Dean JL, McClendon AK, Hickey TE et al. Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors. Cell Cycle. 112012:2756–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dunne LW, Huang Z, Meng W et al. Human decellularized adipose tissue scaffold as a model for breast cancer cell growth and drug treatments. Biomaterials. 2014;35:4940–9. [DOI] [PubMed] [Google Scholar]
- 107. Huang J, Woods P, Normolle D et al. Downregulation of estrogen receptor and modulation of growth of breast cancer cell lines mediated by paracrine stromal cell signals. Breast Cancer Res Treat. 2017;161:229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Brechbuhl HM, Finlay-Schultz J, Yamamoto TM et al. Fibroblast subtypes regulate responsiveness of luminal breast cancer to Estrogen. Clin Cancer Res. 2017;23:1710–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lehmann BD, Jovanovic B, Chen X et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Landry BD, Leete T, Richards R et al. Tumor-stroma interactions differentially alter drug sensitivity based on the origin of stromal cells. Mol Syst Biol. 2018;14:e8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chan TS, Hsu CC, Pai VC et al. Metronomic chemotherapy prevents therapy-induced stromal activation and induction of tumor-initiating cells. J Exp Med. 2016;213:2967–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Peiris-Pages M, Sotgia F, Lisanti MP. Chemotherapy induces the cancer-associated fibroblast phenotype, activating paracrine hedgehog-GLI signalling in breast cancer cells. Oncotarget. 2015;6:10728–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sun Y, Campisi J, Higano C et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18:1359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yang M, Liu P, Wang K et al. Chemotherapy induces tumor immune evasion by upregulation of programmed cell death ligand 1 expression in bone marrow stromal cells. Mol Oncol. 2017;11:358–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hanker AB, Estrada MV, Bianchini G et al. Extracellular matrix/integrin Signaling promotes resistance to combined inhibition of HER2 and PI3K in HER2(+) breast cancer. Cancer Res. 2017;77:3280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yang XH, Flores LM, Li Q et al. Disruption of Laminin-integrin-CD151-FAK Axis sensitizes breast cancer cells to ErbB2 antagonists. Cancer Res. 2010;70:2256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Vela D, Vela-Gaxha Z. Differential regulation of hepcidin in cancer and non-cancer tissues and its clinical implications. Exp Mol Med. 2018;50:e436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Blanchette-Farra N, Kita D, Konstorum A et al. Contribution of three-dimensional architecture and tumor-associated fibroblasts to hepcidin regulation in breast cancer. Oncogene. 2018;37:4013–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Heindel JJ, Blumberg B. Environmental obesogens: mechanisms and controversies. Annu Rev Pharmacol Toxicol. 2019;59:89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Grün F, Blumberg B. Minireview: the case for obesogens. Mol Endocrinol 232009:1127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bougaret L, Delort L, Billard H et al. Adipocyte/breast cancer cell crosstalk in obesity interferes with the anti-proliferative efficacy of tamoxifen. PLoS One. 2018;13:e0191571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gui Y, Pan Q, Chen X et al. The association between obesity related adipokines and risk of breast cancer: a meta-analysis. Oncotarget. 82017:75389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Burks H, Pashos N, Martin E et al. Endocrine disruptors and the tumor microenvironment: a new paradigm in breast cancer biology. Mol Cell Endocrinol. 2017;457:13–9. [DOI] [PubMed] [Google Scholar]
- 124. Yaghjyan L, Colditz GA. Estrogens in the breast tissue: a systematic review. Cancer Causes Control. 2011;22:529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. EBCTCG Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386:1341–52. [DOI] [PubMed] [Google Scholar]
- 126. Whitehead SA, Rice S. Endocrine-disrupting chemicals as modulators of sex steroid synthesis. Best Pract Res Clin Endocrinol Metab. 2006;20:45–61. [DOI] [PubMed] [Google Scholar]
- 127. Duursen MB, Nijmeijer SM, Morree ES et al. Genistein induces breast cancer-associated aromatase and stimulates estrogen-dependent tumor cell growth in in vitro breast cancer model. Toxicology. 2011;289:67–73. [DOI] [PubMed] [Google Scholar]
- 128. Morgan MM, Arendt LM, Alarid ET et al. Mammary adipose stromal cells derived from obese women reduce sensitivity to the aromatase inhibitor anastrazole in an organotypic breast model. FASEB J. 2019;33:8623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Iyengar NM, Brown KA, Zhou XK et al. Metabolic obesity, adipose inflammation and elevated breast aromatase in women with normal body mass index. Cancer Prev Res (Phila). 2017;10:235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Au CC, Docanto MM, Zahid H et al. Des-acyl ghrelin inhibits the capacity of macrophages to stimulate the expression of aromatase in breast adipose stromal cells. J Steroid Biochem Mol Biol. 2017;170:49–53. [DOI] [PubMed] [Google Scholar]
- 131. Subbaramaiah K, Morris PG, Zhou XK et al. Increased levels of COX-2 and prostaglandin E2 contribute to elevated aromatase expression in inflamed breast tissue of obese women. Cancer Discov. 2012;2:356–65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132. Hung CH, Chen FM, Lin YC et al. Altered monocyte differentiation and macrophage polarization patterns in patients with breast cancer. BMC Cancer. 2018;18:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Qian BZ, Li J, Zhang H et al. CCL2 recruits inflammatory monocytes to facilitate breast tumor metastasis. Nature. 2011;475:222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kaneda MM, Messer KS, Ralainirina N et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature. 2016;539:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wilkie S, Schalkwyk MC, Hobbs S et al. Dual targeting of ErbB2 and MUC1 in breast cancer using chimeric antigen receptors engineered to provide complementary signaling. J Clin Immunol. 2012;32:1059–70. [DOI] [PubMed] [Google Scholar]
- 136. Wallstabe L, Gottlich C, Nelke LC et al. ROR1-CAR T cells are effective against lung and breast cancer in advanced microphysiologic 3D tumor models. JCI Insight 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Piasecka D, Braun M, Kitowska K et al. FGFs/FGFRs-dependent signalling in regulation of steroid hormone receptors - implications for therapy of luminal breast cancer. J Exp Clin Cancer Res. 2019;38:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Incio J, Ligibel JA, McManus DT et al. Obesity promotes resistance to anti-VEGF therapy in breast cancer by up-regulating IL-6 and potentially FGF-2. Sci Transl Med. 2018;10:432–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Cazet AS, Hui MN, Elsworth BL et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat Commun. 2018;9:2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang X, Ai J, Liu H et al. The Secretome engages STAT3 to favor a cytokine-rich microenvironment in mediating acquired resistance to FGFR inhibitors. Mol Cancer Ther. 2019;18:667–79. [DOI] [PubMed] [Google Scholar]
- 141. Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7:659–72. [DOI] [PubMed] [Google Scholar]


