Abstract
TRIM44 has oncogenic roles in various cancers. However, TRIM44 expression and its function in renal cell carcinoma (RCC) are still unknown. Here in this study, we investigated the clinical significance of TRIM44 and its biological function in RCC. TRIM44 overexpression was significantly associated with clinical M stage, histologic type (clear cell) and presence of lymphatic invasion (P = .047, P = .005, and P = .028, respectively). Moreover, TRIM44 overexpression was significantly associated with poor prognosis in terms of cancer‐specific survival (P = .019). Gain‐of‐function and loss‐of‐function studies using TRIM44 and siTRIM44 transfection showed that TRIM44 promotes cell proliferation and cell migration in two RCC cell lines, Caki1 and 769P. To further investigate the role of TRIM44 in RCC, we performed integrated microarray analysis in Caki1 and 769P cells and explored the data in the Oncomine database. Interestingly, FRK was identified as a promising candidate target gene of TRIM44, which was downregulated in RCC compared with normal renal tissues. We found that cell proliferation was inhibited by TRIM44 knockdown and then recovered by siFRK treatment. Taken together, the present study revealed the association between high expression of TRIM44 and poor prognosis in RCC patients and that TRIM44 promotes cell proliferation by regulating FRK.
Keywords: FRK, immunohistochemistry, microarray, renal cell carcinoma, TRIM
Based on the results of immunohistochemistry and functional experiments, TRIM44 promotes cell proliferation and migration by inhibiting FRK in renal cell carcinoma.
1. INTRODUCTION
Renal cell carcinoma (RCC) is the most common type of histology in kidney cancer and represents the 6th and 10th most frequently diagnosed cancer in men and women, respectively.1, 2 Despite new systemic approaches in RCC treatment including tyrosine kinase inhibitors,3, 4 mTOR inhibitors5 and immune checkpoint inhibitors,6 survival still depends on the success of surgical treatment.7, 8 The median overall survival of metastatic RCC treated with a combination of cytoreductive nephrectomy and vascular endothelial growth factor targeted therapy was 19.8 months compared to 9.4 months for patients who were not treated with cytoreductive therapy.9 Discovery of new therapeutic targets is required to improve prognosis in metastatic RCC patients.
Tripartite motif (TRIM) family proteins are a group of proteins possessing unique domains, such as the Really Interesting New Gene (RING) finger, B‐box and coiled‐coil domain. TRIM proteins are subclassified into C‐I to C‐XI and one unclassified (UC) subgroup according to their types of structure.10 TRIM proteins that belong to subgroup C‐I to C‐XI all have a RING finger domain, which is responsible for E3 ubiquitin activity.10, 11 Therefore, most of the TRIM proteins are directly involved in ubiquitination, which is a common process in post‐translational modification.11 These proteins are likely to be involved in innate immunity and cancer.10, 12
TRIM44 is one of the proteins belonging to the UC subgroup and does not possess the RING finger domain.11 Instead, it contains a zinc‐finger ubiquitin binding domain (ZF UBP) that is also found in ubiquitin specific protease (USP) USP7/HAUSP, USP20/VDU2 and USP5/IsoT, which are members of the USP family.13, 14, 15, 16 USP are involved in the deubiquitination process.14, 15, 16
The oncogenic effects of TRIM44 have been reported in a number of studies in various types of cancers.17, 18, 19, 20, 21, 22, 23, 24, 25, 26 We have previously shown that TRIM44 is involved in carcinogenesis of testicular germ cell tumor and breast cancer.21, 22 Other reports have suggested its involvement with various pathways, including mTOR,23, 24 NFκB25 and MAPK.26 However, the mechanism of TRIM44 involvement in carcinogenesis of RCC remains unknown.
In the present study, we aimed to investigate the clinical significance and function of TRIM44 in RCC and to further identify targeted genes of TRIM44.
2. MATERIALS AND METHODS
2.1. Patient characteristics and tissue preparation
Tissue samples were obtained from 102 patients with renal cell carcinoma who underwent surgical treatment either by partial nephrectomy or radical nephrectomy between 1989 and 2015. No patients received neoadjuvant cancer treatment including immunotherapy and molecular‐target therapy. Staging was performed according to the AJCC staging system (https://www.cancer.org/cancer/kidney-cancer/detection-diagnosis-staging/staging.html). The present study was carried out in accordance with the Helsinki declaration, and was approved by our institutional ethical committee (#2283). All patients provided written informed consent.
2.2. Immunostaining and immunohistochemical assessment
A Histofine Kit (Nichirei, Bioscience Tokyo, Japan), which employs the streptavidin–biotin amplification method, was used for immunohistochemistry in our study. As primary antibody, we applied 1:200 diluted rabbit polyclonal antibody. Human breast carcinoma tissue was used as the positive control for TRIM44 immunostaining,21 while rabbit immunoglobulin G (IgG) was used as the negative control. Immunoreactivity (IR) of TRIM44 was evaluated by intensity score of immunostaining, which was rated from 0 to 2+ (0: none, 1+: weak, 2+: moderate to strong) based on a report by Yamada et al22 with a modification. Positive IR was defined as having intensity score ≥1+. ST (pathologist who was blind to the clinical data) evaluated the slides.
2.3. Antibodies
Anti–TRIM44 polyclonal antibody was raised in our laboratory as described in a previous report.22 Briefly, this antibody is an affinity purified rabbit polyclonal antibody raised by immunizing rabbits with a glutathione S‐transferase (GST) fusion protein with amino acids of full‐length mouse TRIM44 protein as an antigen. GST‐bound resin was used to remove anti‐GST antibody. The cross‐reactivity to human TRIM44 protein and quality of anti–TRIM44 antibody were confirmed by western blotting analysis in previous literature.22
2.4. Plasmid construction and transfection
TRIM44 cDNA was amplified by PCR with specific primers. TRIM44 cDNA was then subcloned into pcDNA3, a mammalian expression vector (Invitrogen). Transfection with expression vectors was performed 24 hours after cell seeding. Lipofectamine 3000 (Invitrogen) was used according to the manufacturer's protocol. Halo‐tagged FRK clone (FRK‐Halo Tag human ORF in pFN21A; catalog # FHC08658) was purchased from PROMEGA KK. Double induction of TRIM44 and FRK in Caki‐1 cells was carried out by performing transfection with TRIM44 and FRK expression vectors simultaneously with the same protocol as single induction of expression vector.
2.5. Cell culture
We used human RCC‐originated cells (Caki‐1 and 769P). These cells were cultured at 37°C in a humidified air and 5% CO2 atmosphere. DMEM and RPMI with 10% FBS and 1% penicillin‐streptomycin were used for media to cultivate Caki1 and 769P cells, respectively (Sigma‐Aldrich Japan).
2.6. Western blot analysis
Cell lysis, gel electrophoresis and western blot analysis were performed as previously described.22 Cells were lysed with NP40 buffer (50 mmol/L Tris, pH 8.0, 150 mmol/L NaCl, 1% NP‐40) containing proteinase inhibitor. SDS sample buffer was added to these cell lysates and used for gel electrophoresis. Anti–TRIM44 and anti–β‐actin from Sigma were used for primary antibodies. An enhanced chemiluminescence system (GE Healthcare UK) was used for western blot analysis.
2.7. RNA extraction and quantitative reverse transcription‐polymerase chain reaction
Quantitative RT‐PCR (qRT‐PCR) was performed as previously described.27 Total RNA extraction from cell lines was performed using ISOGEN reagent (Nippon Gene, Tokyo, Japan). First‐strand cDNA was synthesized using PrimeScript (Takara). The Applied Biosystems 7300 Real‐Time PCR System and SYBR Green fluorescence were used for qRT‐PCR. The following primers were designed and used for qRT‐PCR:
GAPDH forward: 5′‐GGTGGTCTCCTCTGACTTCAACA‐3′
GAPDH reverse: 5′‐GTGGTCGTTGAGGGCAATG‐3′
TRIM44 forward: 5′‐GTGGACATCCAAGAGGCAAT‐3′
TRIM44 reverse: 5′‐AGCAAGCCTTCATGTGTCCT‐3′
FRK forward: 5′‐CGGACTGCTGAGGACTTGAG‐3′
FRK reverse: 5′‐TTCGCCAAACTGACCAGATCC‐3′.
2.8. Small interfering RNA transfection
TRIM44 and FRK knockdown was performed using siRNA transfection. Two siRNA that specifically target TRIM44 and one non–targeting siRNA (siRNA control) were purchased from RNAi Inc (Tokyo, Japan). siFRK (Silencer Select Pre‐designed siRNA, siFRK #1: siRNA ID s5363, Catalog #4390824; siFRK #2: siRNA ID s5364, Catalog # 4392420) were purchased from Thermo Fisher Scientific. These siRNA were used for transfection in RCC cells by using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Double knockdown of TRIM44 and FRK was performed simultaneously with the same protocol as single gene knockdown. Downregulation of TRIM44 and/or FRK was confirmed by performing qRT‐PCR and/or western blot analysis. The sequences of the siRNAs were as follows:
siControl
Sense: 5′‐GUACCGCACGUCAUUCGUAUC‐3′
Antisense: 5′‐UACGAAUGACGUGCGGUACGU‐3′
siTRIM44‐A
Sense: 5′‐GAAUCAGUCGGAUACUCAUAG‐3′
Antisense: 5′‐AUGAGUAUCCGACUGAUUCUG‐3′
siTRIM44‐B
Sense: 5′‐CCGCUAUGAUCGAAUUGGUGG‐3′
Antisense: 5′‐ACCAAUUCGAUCAUAGCGGCC‐3′.
2.9. Cell proliferation assay
Cells were seeded in 96‐well plates (4.0 × 103 cells/well) and transfected with TRIM44 plasmids or siRNA (siTRIM44, siFRK) after 24 hours. MTS assay was performed at 24 and 48 hours after transfection using The Cell Titer 96 Aqueous One Solution Cell Proliferation Assay (Promega KK) according to the manufacturer's instructions. Assays were performed in quintuplicate, and data are presented as mean value ± SD.
2.10. Cell migration assay
Cell migration assay was carried out as previously described.22 Cell culture inserts with an 8.0‐µm‐pore‐sized PET filter (Becton Dickinson) were used in the assay. Medium without FBS was added to the lower chamber. The RCC cells on the upper surface of the filter were carefully removed 48 hours after transfection. The filters were dipped in methanol for 30 minutes, washed with PBS, and stained with Giemsa for 30 seconds. After washing three times with fresh PBS, filters were mounted on glass slides. The cells migrated on the lower surface and were counted in five randomly selected fields microscopically at a magnification of ×200. Data are presented as mean value ± SD.
2.11. Microarray analysis
TRIM44 knockdown was performed on 769P cells by using siTRIM44‐A or siTRIM44‐B. In addition, TRIM44 knockdown (siTRIM44‐A) and TRIM44 overexpression were performed on Caki‐1 cells. Forty‐eight hours after transfection, total RNA from these RCC cell lines were extracted using the Qiagen RNeasy Micro Kit according to the manufacturer's instructions. RNA integrity numbers (RIN) were above 9.0 in all RNA samples. GeneChip Human Exon 1.0 ST Array (Affymetrix) was used in microarray analysis according to the manufacturer's protocol. Fold changes of gene expressions were log2 transformed. Cutoff values were set at 0.3 (upregulated) or −0.3 (downregulated). We then used Oncomine datasets (https://www.oncomine.org) and qRT‐PCR to validate and confirm our microarray results.
2.12. Statistical analyses
JMP Pro version 14.1.0 (SAS Institute) was used for data analyses. Pearson's χ2 test and Fisher's test were used (when frequency was <5) to analyze association between TRIM44 IR and clinicopathological parameters. Student's t test was used in analyzing data of qRT‐PCR, MTS assay and migration assay. The log‐rank test was used in analyzing the statistical difference of cancer‐specific and overall survival. Univariate and multiple hazard risk models were used to evaluate independent predictors of cancer‐specific mortality in RCC patients. P‐value < .05 was defined as statistically significant.
3. RESULTS
3.1. TRIM44 is overexpressed in renal cell carcinoma and is associated with poor prognosis
Analysis of the ONCOMINE datasets (https://www.oncomine.org) showed that mRNA expression of TRIM44 was higher in kidney cancer than in normal kidney tissue (Figure 1A,B). Immunohistochemistry of TRIM44 protein in RCC revealed that TRIM44 immunoreactivity (IR) was detected in the cytoplasm of RCC cells (Figure 1C,D). We found that overexpression of TRIM44 was significantly associated with clinical M stage, histologic type (clear cell type) and presence of lymphatic invasion (P = .047, P = .005 and P = .028, respectively; Tables 1 and 2). Multivariate analysis also revealed that TRIM44 overexpression was a significant risk factor of cancer‐specific mortality in patients with RCC (Table 3). Kaplan‐Meier curves showed that cancer‐specific survival was significantly shorter in patients with TRIM44 overexpression (Figure 1H).
Figure 1.
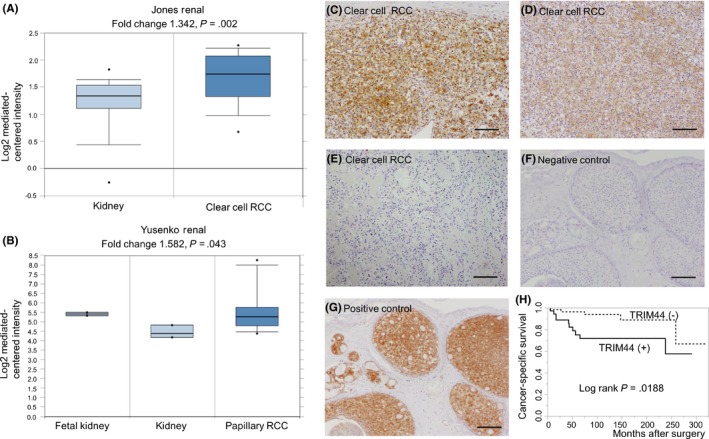
Overexpression of TRIM44 was associated with cancer‐specific and overall survival in patients with renal cell carcinoma (RCC). A, B, Oncomine data showing TRIM44 overexpression in RCC samples. C‐F, Representative images of immunohistochemistry. C, Anti–TRIM44 immunostaining of clear cell carcinoma showing intensity score 2+. D, Anti–TRIM44 immunostaining of clear cell carcinoma showing intensity score 1+. E, Anti–TRIM44 immunostaining of clear cell carcinoma showing intensity score 0. F, Negative control (breast cancer tissue, rabbit immunogloblin G antibody). G, Positive control (breast cancer tissue). Scale bar = 100 μm, respectively. H, Cancer‐specific survival of patients with RCC. Patients with positive TRIM44 immunoreactivity showed poor prognosis (log‐rank test; P = .019)
Table 1.
Patient characteristics (N = 102)
| TRIM44 IR | |||
|---|---|---|---|
| Negative (N = 61) | Positive (N = 41) | P‐value | |
| Age | 59.2 ± 10.5 | 56.8 ± 14.9 | .343 |
| Sex | |||
| Female | 16 | 8 | .433 |
| Male | 45 | 33 | |
| Clinical stage | |||
| T stage | |||
| cT1 | 41 | 22 | .167 |
| cT2‐T4 | 20 | 19 | |
| N stage | |||
| N0 | 59 | 39 | 1.000 |
| N1‐2 | 2 | 2 | |
| M stagea | |||
| M0 | 58 | 33 | .047 |
| M1 | 3 | 7 | |
| Surgical typea | |||
| Partial nephrectomy | 11 | 4 | .393 |
| Radical nephrectomy | 50 | 37 | |
“IR positive” was defined as intensity score ≥1. Student's t test was used for continuous values and Pearson's χ2 tests were used for categorical values.
Abbreviations: IR, immunoreactivity; TRIM44, tripartite motif 44.
M stage was unknown in 1 patient. Fisher's test was used when categorical values were under 5.
Table 2.
Relationships between TRIM44 IR and pathological parameters in patients with renal cell carcinoma (N = 102)
| Clinical parameters | TRIM44 IR | ||
|---|---|---|---|
| Negative (N = 61) | Positive (N = 41) | P‐value | |
| Histology type | |||
| Clear cell type | 53 | 26 | .005 |
| Others | 8 | 15 | |
| Gradea | |||
| Grade <2 | 17 | 5 | .059 |
| Grade ≥2 | 44 | 36 | |
| Lymphatic invasiona | |||
| Negative | 59 | 34 | .028 |
| Positive | 2 | 7 | |
| Venous invasion | |||
| Negative | 48 | 27 | .149 |
| Positive | 13 | 14 | |
| Pathological T stage | |||
| pT1 | 41 | 23 | .254 |
| pT2‐T4 | 20 | 18 | |
Pearson's χ2 tests were used for statistical analysis.
Abbreviations: IR, immunoreactivity; TRIM44, tripartite motif 44. “IR positive” was defined as intensity score ≥1.
Fisher's test was used when categorical values were under 5.
Table 3.
Risk analysis of cancer‐specific mortality in patients with renal cell carcinoma (N = 102)
| Variable | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% index | P‐value | hazard ratio | 95% index | P‐value | |
| Histologic type (clear cell vs others) | 0.3 | 0.1‐0.9 | .037 | 0.4 | 0.1‐1.4 | .192 |
| Lymph node or distant metastasis (present vs absent) | 18.3 | 5.6‐59.2 | <.001 | 17.2 | 5.0‐58.5 | <.001 |
| TRIM44 IR (positive vs negative) | 3.3 | 1.1‐9.9 | .026 | 3.4 | 1.0‐11.3 | .042 |
Abbreviations: IR, immunoreactivity; TRIM, tripartite motif.
3.2. TRIM44 overexpression promoted proliferation and migration and vice versa in TRIM44 knockdown in renal cell carcinoma cells
Overexpression of TRIM44 in Caki1 and 769P cells was confirmed by qRT‐PCR and western blot analysis (Figure 2A,B and Figure S1A,B). Cell proliferation and migration were markedly promoted in RCC cells treated with TRIM44 overexpression (Figures 2C‐E and S1C‐E).
Figure 2.
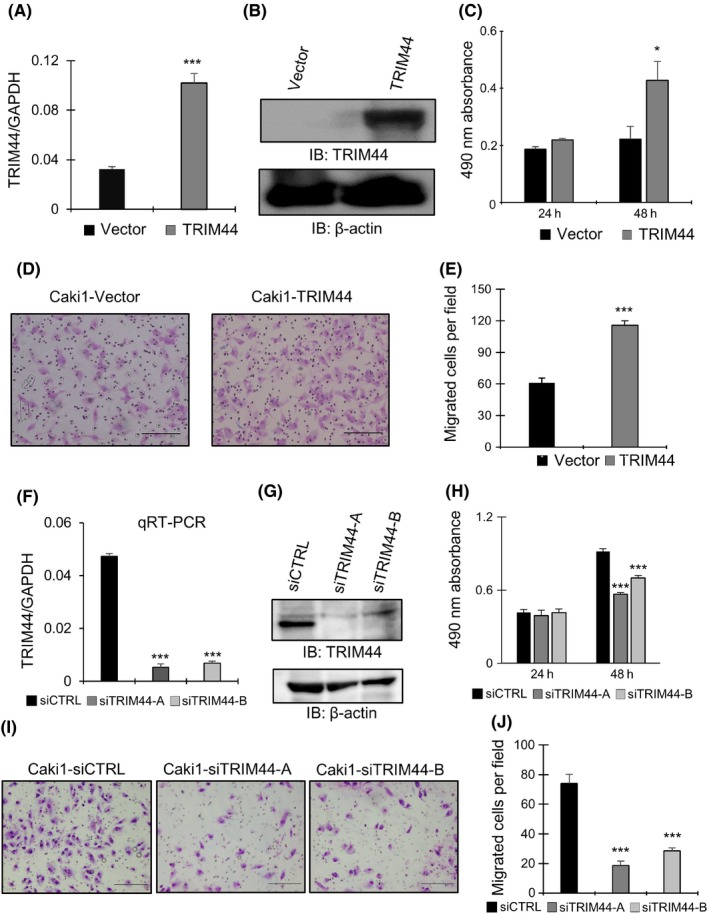
Functional in vitro effects of TRIM44 in Caki1 cells. A, mRNA expression level of TRIM44 was analyzed by quantitative RT‐PCR (qRT‐PCR) in TRIM44 transfected cells (***P < .001, Student's t test). B, Western blotting analysis showing protein level of TRIM44 in TRIM44 overexpression Caki1 cells. C, MTS assay of TRIM44 transfected Caki1 cells. Cell proliferation was promoted in Caki1 cells treated with TRIM44 overexpression (*P < .05, Student's t test). D, Representative pictures of migration assay. Migrated cells of Caki1 cells treated with TRIM44 overexpression are shown. Scale bars indicate 200 μm. E, Migrated cells of Caki1 cells treated with TRIM44 overexpression were counted and analyzed. Results are shown as mean + SD (N = 5, ***P < .001, Student's t test). F, mRNA levels of TRIM44 in TRIM44 knockdown Caki1 cells were measured by qRT‐PCR (***P < .001, vs siCTRL, Student's t test). G, Protein levels of TRIM44 in siTRIM44‐treated Caki1 cells. TRIM44 knockdown was confirmed in both siTRIM44‐A‐treated and siTRIM44‐B‐treated Caki1 cells. H, MTS assay of Caki1 cells treated with TRIM44 knockdown (***P < .001, vs siCTRL, Student's t test). Results are shown as mean + SD (N = 5). I, Representative pictures of migration assay. Migrated cells of Caki1 cells treated with siTRIM44 are shown. J, Migrated cells of Caki1 cells treated with TRIM44 knockdown were counted and analyzed (***P < .001, vs siCTRL, Student's t test)
Loss‐of‐function studies in RCC cell lines were performed using two types of siTRIM44. Caki1 and 769P cells were treated with siTRIM44‐A and siTRIM4‐B. TRIM44 knockdown in these siTRIM44‐treated RCC cells were confirmed by qRT‐PCR and western blotting (Figures 2F,G and S1F,G). Cell proliferation and migration were significantly repressed by TRIM44 knockdown (Figures 2H‐J and S1H‐J).
3.3. FRK was identified as the target gene of TRIM44 in renal cell carcinoma
We conducted a microarray analysis to investigate TRIM44‐regulated signals that were responsible for RCC progression by identifying differentially expressed genes in RCC cell lines treated with either TRIM44 knockdown or overexpression.
Integrated analysis containing microarray data of 769P and Caki1 cells revealed 12 candidate genes that were potentially downregulated by TRIM44 (Figure 3A, Table S1). Of the 12 genes that were downregulated by TRIM44, three genes were found to possess tumor suppressive functions (Figure 3A; FRK, EGR1 and BCL2L14). 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42 We then used the Oncomine dataset to analyze the expression level of these three genes in tumors. FRK was the only gene that showed low expression levels in cancer compared with benign tissues (Figures 3B,C and S2). In addition, validation of FRK regulation by TRIM44 was performed using qRT‐PCR in RCC cells (Figure 3D‐G). mRNA expression levels of FRK were elevated by TRIM44 knockdown in both Caki‐1 and 769P cells (Figure 3D‐G). Taken together, these results led us to focus on FRK as the key gene among genes downregulated by TRIM44.
Figure 3.

Integrative analysis of microarray data was performed to identify genes downregulated by TRIM44 in renal cell carcinoma (RCC) cells. A, Microarray analysis was performed to identify TRIM44regulated genes. For 769P cells, expression levels were analyzed in TRIM44 knockdown cells (siTRIM44‐A vs siCTRL and siTRIM44‐B vs siCTRL). Expression levels were also analyzed in TRIM44 knockdown and TRIM44 overexpression Caki1 cells (siTRIM44‐A vs siCTRL and TRIM44 overexpression vs Vector). To analyze genes showing tumor suppressive effect, upregulated genes by TRIM44 knockdown or downregulated genes by overexpression were identified in each microarray dataset, as shown in the flowchart. These data were integrated to identify the overlapped TRIM44‐regulated genes that possess a tumor suppressive effect (three genes). B, C, Oncomine data showing expression level of FRK in RCC samples. Expression levels of FRK showed low levels in cancer compared with benign renal tissues. D‐G, mRNA expression levels of FRK normalized by GAPDH are shown (*P < .05, **P < .01, ***P < .001; Student's t test)
We also explored upregulated genes by TRIM44 using the same microarray data (Figure S3, Table S2). Fourteen candidate genes were identified by integrating four sets of microarray data (Figure S3). Of these 14 genes, GRK5 was the only gene known to possess oncogenic effect (Figure S3). In fact, the oncogenic effect of GRK5 in renal cell carcinoma was previously reported.43
3.4. TRIM44 promotes cell proliferation by regulating FRK in renal cell carcinoma
Knockdown of FRK in Caki1 cells was confirmed by qRT‐PCR (Figure 4A). Double knockdown of TRIM44 and FRK was also performed in Caki1 cells by using siTRIM44 + siFRK and was confirmed by qRT‐PCR (Figure 4B,C). MTS assay of Caki1 cells treated with siCTRL, siFRK, siTRIM44 + siCTRL or siTRIM44 + siFRK revealed that cell proliferation was inhibited by TRIM44 knockdown and then promoted by siFRK supplement (Figure 4D,E).
Figure 4.
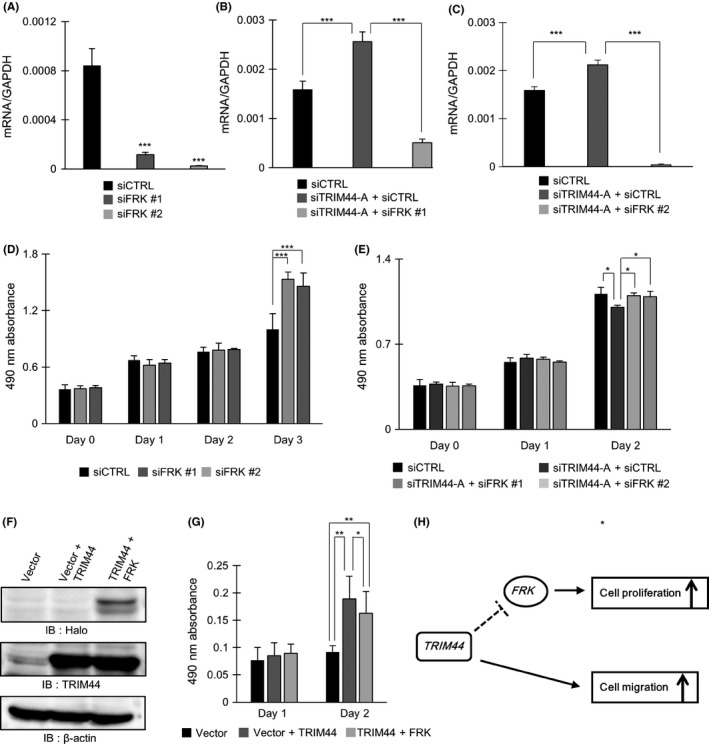
TRIM44 inhibits cell proliferation through regulating FRK. A, Quantitative RT‐PCR showing FRK mRNA expression levels in Caki1 cells treated with FRK knockdown (***P < .001; Student's t test). B, C, qRT‐PCR showing mRNA expression levels of FRK in siFRK‐treated Caki1 cells (***P < .001; Student's t test). D, E, MTS assay of siTRIM44 and siFRK‐treated renal cell carcinoma (RCC) cells (*P < .05, ***P < .001; Student's t test). F, Western blot analysis of Vector, Vector + TRIM44, and TRIM44 + FRK in Caki1 cells. Halo Tag protein was expressed in Caki1 cells treated with TRIM44 + FRK overexpression. TRIM44 protein was expressed in both Vector + TRIM44 and TRIM44 + FRK‐treated Caki1 cells. G, MTS assay of Vector + TRIM44 and TRIM44 + FRK treated Caki1 cells. Cell proliferation was elevated in Caki1 cells treated with Vector + TRIM44. Furthermore, cell proliferation was suppressed in cells treated with TRIM44 + FRK. H, Proposed schema of mechanism regarding TRIM44 involvement in RCC carcinogenesis
Double induction of TRIM44 and FRK in Caki1 cells was carried out by performing transfection with TRIM44 and FRK simultaneously. Transfection of Halo‐FRK was confirmed by western blot analysis (Figure 4F). Overexpression of TRIM44 was also confirmed in Caki1 cells transfected with TRIM44 alone or both TRIM44 and Halo‐FRK (Figure 4F). Then, MTS assay was performed in Caki1 cells treated with Vector, Vector + TRIM44 and TRIM44 + Halo‐FRK (Figure 4G). We observed increased cell proliferation through TRIM44 overexpression that was then attenuated by overexpressing Halo‐FRK with TRIM44 (Figure 4G).
3.5. Proposed mechanism of TRIM44 involvement in renal cell carcinoma carcinogenesis
The present study revealed that TRIM44 promotes cell proliferation and migration in RCC cells. Furthermore, TRIM44 may promote cell proliferation of RCC cells by inhibiting FRK (Figure 4H).
4. DISCUSSION
TRIM44 is genetically located at chromosome 11p13 and belongs to the UC subgroup of the TRIM family, which do not possess the RING finger domain.10, 11 Therefore, TRIM44 itself may not have the E3 ligase activity. From the molecular structural perspective, this suggests a couple of hypotheses of how TRIM44 is involved in the ubiquitination process. One theory is that TRIM44 may function as a promoter of deubiquitination. The N‐terminal region of TRIM44 contains the ZF UBP that binds to ubiquitin with high affinity.14 ZF UBP are found in the deubiquitinating enzymes containing USP.13 USP typically remove ubiquitin from substrates, leading to the inhibition of substrate degradation.15, 16 For example, USP7/HAUSP deubiquitinates and stabilizes p53(Li M).15 USP20/VDU2 deubiquitinates and stabilizes HIF1α.16 Previously, we reported that TRIM44 binds to TRIM17 and stabilizes it to activate the function of TRIM17.13 TRIM44 also targets virus‐induced signaling adaptor (VISA) and stabilizes it by preventing VISA ubiquitination.44
Another theory can be projected by reflecting on mechanisms of other TRIM proteins. TRIM29 is one of the proteins that also do not possess the RING finger.11 This protein is suggested to be involved in DNA repair response by serving as a scaffold protein to assemble DNA repair proteins into damaged chromatin.45 Xing et al46 show that the E3 ligase activity, which is usually the distinct function of the RING finger domain, could be functionally compensated by B‐box2 domain. Therefore, the B‐box2 of TRIM44 could have a similar function that may directly affect the ubiquitination process. In theory, TRIM44 is capable of targeting proteins directly or indirectly for degradation by ubiquitination despite the structural absence of RING finger domain.
Accumulating studies have shown that TRIM family proteins may positively or negatively regulate carcinogenesis depending on the types of TRIM or the origin of cancer.10, 11 Specifically, TRIM44 has been reported in a number of studies showing oncogenic effect in various cancers.17, 18, 19, 20, 21, 22, 23, 24, 25, 26 Ong et al20 have reported that TRIM44 overexpression resulted from genomic amplification in 16.1% of all epithelial cancers. From a clinicopathological point of view, we previously showed that TRIM44 overexpression was associated with poor prognosis in testicular germ cell tumors.22 Other studies include an immunohistochemical (IHC) analysis that demonstrated that overexpression of TRIM44 was an independent predictor of poor prognosis in a series of 109 patients with epithelial ovarian cancer.47 Another IHC study revealed that TRIM44 overexpression was significantly associated with histological grade, depth of myometrial invasion and lymph node metastasis in endometrial carcinoma.48 In esophageal squamous cell carcinoma, overexpression of TRIM44 was associated with the status of lymph node metastasis and poor overall survival.49
As described, the role of TRIM44 in cancer is likely to be oncogenic, although the involved mechanism seems to differ depending on types of cancer. TRIM44 facilitated the cell growth and motility of non–small cell lung cancer cells through the mTOR signaling pathway.24 In non–small cell lung cancer cells and breast cancers, TRIM44 promoted migration of cancer cells via the NF‐κβ signaling pathway.21, 25 Zhou et al18 have revealed that TRIM44 knockdown inhibited cell proliferation, migration and the epithelial‐mesenchymal transition (EMT) process through suppression of Wnt/beta‐catenin pathway in papillary thyroid cancer cells. An in vitro study in cholangiocarcinoma has indicated that TRIM44 overexpression activated MAPK signaling and reversed cell EMT and apoptosis.26 Despite the known TRIM44‐regulated pathways mentioned, these pathways were not enriched in the RCC cells according to the microarray results in the present study. In contrast, the present study identified FRK, a novel tumor suppressive gene,40, 50 as a direct or indirect target of TRIM44.
The fyn‐related kinase (FRK) is one of the tumor suppressive genes that belong to a subfamily of non–receptor tyrosine kinases.40, 50 FRK positively regulated PTEN by phosphorylating PTEN at tyrosine residue 336, which led to a stabilization of PTEN protein in breast cancer cells.50 This mutation of the tyrosine residue seemed to prevent the NEDD4‐1 mediated ubiquitination of PTEN.50 Another study revealed that FRK inhibited cell migration of breast cancer cells through suppressing EMT.39 FRK is also involved in large cell lymphoma and glioma as a tumor suppressor.38, 40, 41, 42 FRK overexpression caused G1 phase arrest that led to apoptosis in glioma cells.41In the present study, TRIM44 downregulated FRK which showed a tumor suppressive effect in RCC cells. However, the specific regulatory mechanism of FRK by TRIM44 remains unclear, because our results cannot distinguish whether TRIM44 suppresses FRK directly or indirectly. It is possible that TRIM44 may indirectly suppress FRK through regulating other transcriptional factors.
In conclusion, our study shows that overexpression of TRIM44 is associated with poor prognosis in patients with RCC and that TRIM44 targets FRK and promotes cell proliferation and migration.
DISCLOSURE
The authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
We thank A. Saitoh and N. Sasaki for their technical assistance. This study was supported by the Takeda Science Foundation (SI, KT) and the Japan Agency for Medical Research and Development (SI, P‐CREATE, JP18cm0106144), and by grants from the Japan Society for the Promotion of Science (YY, grant 17K16781; KT, grant 15K15581; SI, grant 15K15353).
Yamada Y, Kimura N, Takayama K‐I, et al. TRIM44 promotes cell proliferation and migration by inhibiting FRK in renal cell carcinoma. Cancer Sci. 2020;111:881–890. 10.1111/cas.14295
YY and NK contributed equally to the work
REFERENCES
- 1. Weikert S, Ljunberg B. Contemporary epidemiology of renal cell carcinoma: perspectives of primary prevention. World J Urol. 2010;28:247‐252. [DOI] [PubMed] [Google Scholar]
- 2. Capitanio U, Bensalah K, Bex A. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75:74‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gore ME, Szczylik C, Porta C, et al. Safety and efficacy of sunitinib for metastatic renal cell carcinoma: an expanded‐access trial. Lancet Oncol. 2009;10:757‐763. [DOI] [PubMed] [Google Scholar]
- 4. Motzer RJ, Hutson TE, Cella D. Pazopanib versus sunitinib in metastatic renal‐cell carcinoma. N Engl J Med. 2013;369:722‐731. [DOI] [PubMed] [Google Scholar]
- 5. Motzer RJ, Escudier B, Oudard S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double‐blind, randomized, placebo‐controlled phase III trial. Lancet. 2008;9:449‐456. [DOI] [PubMed] [Google Scholar]
- 6. Motzer RJ, Rini BI, McDermott DF, et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33:1430‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dabestani S, Marconi L, Hofmann F, et al. Local treatments for metastases of renal cell carcinoma: a systematic review. Lancet Oncol. 2014;15:e549‐e561. [DOI] [PubMed] [Google Scholar]
- 8. Yu X, Wang B, Li X, et al. The significance of metastasectomy in patients with metastatic renal cell carcinoma in the era of targeted therapy. Biomed Res Int. 2015;2015:176373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choueiri TK, Xie W, Kollmannsberger C, et al. The impact of cytoreductive nephrectomy on survival of patients with metastatic renal cell carcinoma receiving vascular endothelial growth factor targeted therapy. J Urol. 2011;185:60‐66. [DOI] [PubMed] [Google Scholar]
- 10. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792‐804. [DOI] [PubMed] [Google Scholar]
- 11. Watanabe M, Hatakeyama S. TRIM proteins and diseases. J Biochem. 2017;161:135‐144. [DOI] [PubMed] [Google Scholar]
- 12. Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol. 2012;770:27‐37. [DOI] [PubMed] [Google Scholar]
- 13. Urano T, Usui T, Takeda S, et al. TRIM44 interacts with and stabilizes terf, a TRIM ubiquitin E3 ligase. Biochem Biophys Res Commun. 2009;383:263‐268. [DOI] [PubMed] [Google Scholar]
- 14. Reyes‐Turcu FE, Horton JR, Mullally JE, et al. The ubiquitin binding domain ZnF UBP recognizes the C‐terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197‐1208. [DOI] [PubMed] [Google Scholar]
- 15. Li M, Chen D, Shiloh A, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648‐653. [DOI] [PubMed] [Google Scholar]
- 16. Li Z, Wang D, Messing EM, et al. VHL protein‐interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF‐1 alpha. EMBO Rep. 2005;6:373‐378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan Y, Yao H, Hu J, et al. Knockdown of TRIM44 inhibits the proliferation and invasion in prostate cancer cells. Oncol Res. 2017;25:1253‐1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Z, Liu Y, Ma M, et al. Knockdown of TRIM44 inhibits the proliferation and invasion in papillary thyroid cancer cells through suppressing the Wnt/β‐catenin signaling pathway. Biomed Pharmacother. 2017;96:98‐103. [DOI] [PubMed] [Google Scholar]
- 19. Kashimoto K, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci. 2012;103:2021‐2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ong CA, Shannon NB, Ross‐Innes CS, et al. Amplification of TRIM44: pairing a prognostic target with potential therapeutic strategy. J Natl Cancer Inst. 2014;106:pii:dju050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawabata H, Azuma K, Ikeda K, et al. TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF‐κβ Signaling. Int J Mol Sci. 2017;18:pii:E1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamada Y, Takayama KI, Fujimura T, et al. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci. 2017;108:32‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiong D, Jin C, Ye X, et al. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci. 2018;109:3080‐3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xing Y, Meng Q, Chen X, et al. TRIM44 promotes proliferation and metastasis in non–small lung cancer via mTOR signaling pathway. Oncotarget. 2016;7:30479‐30491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo Q, Lin H, Ye X, Huang J, Lu S, Xu L. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF‐κβ signaling pathway. Int J Clin Oncol. 2015;20:508‐517. [DOI] [PubMed] [Google Scholar]
- 26. Peng R, Zhang PF, Zhang C, et al. Elevated TRIM44 promotes intrahepatic cholangiocarcinoma progression by inducing EMT via MAPK signaling. Cancer Med. 2018;7:796‐808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamada Y, Fujimura T, Takahashi S, et al. Clinical significance of amyloid precursor protein in patients with testicular germ cell tumor. Adv Urol. 2013;2013:348438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo B, Godzik A, Reed JC. Bcl‐G, a novel pro‐apoptotic member of the Bcl‐2 family. J Biol Chem. 2001;276:2780‐2785. [DOI] [PubMed] [Google Scholar]
- 29. Jiang P, Wang X, Chen X, et al. A potential molecular model for studying apoptosis enhanced by the interaction of BCL‐G with JAB1 in swine. Oncotarget. 2016;7:62912‐62924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pickard MR, Mourtada‐Maarabouni M, Williams GT. Candidate tumour suppressor Fau regulates apoptosis in human cells: an essential role for Bcl‐G. Biochem Biophys Acta. 2011;1812:1146‐1153. [DOI] [PubMed] [Google Scholar]
- 31. Inoue K, Fry EA. Tumor suppression by the EGR1, DMP1, ARF, p53, and PTEN network. Cancer Invest. 2018;36:520–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun M, Nie FQ, Zang C, et al. The peudogene DUXAP8 promotes non–small‐cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol Ther. 2017;25:739‐751. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Choi EJ, Yoo NJ, Kim MS, et al. Putative tumor suppressor genes EGR1 and BRSK1 are mutated in gastric and colorectal cancers. Oncology. 2016;91:289‐294. [DOI] [PubMed] [Google Scholar]
- 34. Yan L, Wang Y, Liang J, et al. MiR‐301b promotes the proliferation, motility, and epithelial‐to‐mesenchymal transition of bladder cancer cells by targeting EGR1. Biochem Cell Biol. 2017;95:571‐577. [DOI] [PubMed] [Google Scholar]
- 35. Ju JH, Yang W, Lee KM, et al. Regulation of cell proliferation and migration by Keratin 19‐induced nuclear import of early growth response‐1 in breast cancer cells. Clin Cancer Res. 2013;19:4335‐4346. [DOI] [PubMed] [Google Scholar]
- 36. Yang M, Teng W, Qu Y, Wang H, Yuan Q. Sulforaphene inhibits triple negative breast cancer through activating tumor suppressor Egr1. Breast Cancer Res Treat. 2016;158:277‐286. [DOI] [PubMed] [Google Scholar]
- 37. Lee SH, Bahn JH, Choi CK, et al. ESE‐1/EGR‐1 pathway plays a role in tolfenamic acid‐induced apoptosis in colorectal cancer cells. Mol Cancer Ther. 2008;7:3739‐3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu G, Dasari S, Asmann YW, et al. Targetable fusions of the FRK tyrosine kinase in ALK‐negative anaplastic large cell lymphoma. Leukemia. 2018;32:565‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogunbolude Y, Dai C, Bagu ET, et al. FRK inhibits breast cancer cell migration and invasion by suppressing epithelial‐mesenchymal transition. Oncotarget. 2017;8:113034‐113065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brauer PM, Tyner AL. RAKing in AKT: A tumor suppressor function for the intracellular tyrosine kinase FRK. Cell Cycle. 2009;8:2728‐2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hua L, Zhu M, Song X, et al. FRK suppresses the proliferation of human glioma cells by inhibiting cyclin D1 nuclear accumulation. J Neurooncol. 2014;119:49‐58. [DOI] [PubMed] [Google Scholar]
- 42. Shi Q, Song X, Wang J, et al. FRK inhibits migration and invasion of human glioma cells by promoting N‐cadherin/β‐catenin complex formation. J Mol Neurosci. 2015;55:32‐41. [DOI] [PubMed] [Google Scholar]
- 43. Zhao TL, Gan XX, Bao Y, et al. GRK promotes tumor progression in renal cell carcinoma. Neoplasma. 2019;66:261‐270. [DOI] [PubMed] [Google Scholar]
- 44. Yang B, Wang Y, Zhou H, et al. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol. 2013;190:3613‐3619. [DOI] [PubMed] [Google Scholar]
- 45. Masuda Y, Takahashi H, Sato S, et al. TRIM29 regulates the assembly of DNA repair proteins into damaged chromatin. Nat Commun. 2015;6:7299. [DOI] [PubMed] [Google Scholar]
- 46. Xing J, Weng L, Yuan B, et al. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat Immunol. 2016;17:1373‐1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu S, Yin H, Ji H, et al. Overexpression of TRIM44 is an independent marker for predicting poor prognosis in epithelial ovarian cancer. Exp Ther Med. 2018;16:3034‐3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li P, Yin H, Meng F, et al. High TRIM44 expression in endometrial carcinoma is associated with a poorer patient outcome. Pathol Res Pract. 2018;214:727‐731. [DOI] [PubMed] [Google Scholar]
- 49. Kawaguchi T, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 is related to invasive potential and malignant outcomes in esophageal squamous cell carcinoma. Tumour Biol. 2017;39:1010428317700409. [DOI] [PubMed] [Google Scholar]
- 50. Yim EK, Peng G, Dai H, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


