Abstract
Background
Human papillomavirus (HPV) infection is a primary cause of cervical cancer. Although epidemiologic study revealed that carcinogenic risk differs according to HPV genotypes, the expression patterns of HPV-derived transcripts and their dependence on HPV genotypes have not yet been fully elucidated.
Methods
In this study, 382 patients with abnormal cervical cytology were enrolled to assess the associations between HPV-derived transcripts and cervical intraepithelial neoplasia (CIN) grades and/or HPV genotypes. Specifically, four HPV-derived transcripts, namely, oncogenes E6 and E6*, E1^E4, and viral capsid protein L1 in four major HPV genotypes—HPV 16, 18, 52, and 58—were investigated.
Results
The detection rate of E6/E6* increased with CIN progression, whereas there was no significant change in the detection rate of E1^E4 or L1 among CIN grades. In addition, we found that L1 gene expression was HPV type-dependent. Almost all HPV 52-positive specimens, approximately 50% of HPV 58-positive specimens, around 33% of HPV 16-positive specimens, and only one HPV18-positive specimen expressed L1.
Conclusions
We demonstrated that HPV-derived transcripts are HPV genotype-dependent. Especially, expression patterns of L1 gene expression might reflect HPV genotype-dependent patterns of carcinogenesis.
Keywords: Human papillomavirus, Cervical intraepithelial neoplasia, Viral transcripts
Background
Human papillomavirus (HPV) infection is a common sexually transmitted disease, with approximately 50–80% of sexually active adolescents being infected within 2–3 years of initiating intercourse [1]. Most HPV infections are latent by immune regression, while about 10% of the infections are proliferative, which is associated with cervical cancer development [2]. The International Agency for Research on Cancer divided the HPV genotypes into the following groups according to their carcinogenesis: the highly carcinogenic Group 1 (HPVs 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59); the probably carcinogenic Group 2A (HPV 68); and the possibly carcinogenic Group 2B (HPVs 26, 30, 34, 53, 66, 67, 69, 70, 73, 82, 85, and 97) [3]. Continuous expression of HPV E6 and E7 oncogenes, mainly caused by integration of the HPV genome into the human genome, is critical in cervical cancer progression [4]. High-risk HPV E6 and E7 are likely to transform CIN lesion to cancer. Especially, HPV 16 and 18 are the most carcinogenic. The prevalence of HPV 16 and 18 in cervical cancer and cervical intraepithelial neoplasia (CIN) are quite different from other high-risk HPV. About 50 and 15% of cervical cancer are positive for HPV 16 and 18, whereas about 40% and 3–7% of high-grade CIN (CIN2/3) are positive, respectively [5, 6]. Other data show that the rate of progression of HPV 16- or 18-infected cervical epithelium to CIN3 or more is around 15% at 10 years post-infection, which is much higher compared to other HR-HPV [7]. Further, HPV 18 is likely to integrate the viral genome into the host genome compared to HPV 16 [8]. Furthermore, there are HPV type-dependent features among cancer histological types. Most HPV 16-positive cancers are squamous cell carcinomas, whereas around 50% of HPV 18-positive cancers are adenocarcinomas [5].
HPV generates numerous viral transcripts via differential RNA splicing. For example, at least 13 transcripts are derived from eight HPV genes in HPV 16-infected W12E cells [9]. There are six genes (E6, E7, E1, E2, E4, and E5) located in the early region of the HPV genome, and two genes (L1 and L2) in the late region. Expression of these genes is altered during epithelial differentiation and/or CIN progression. E6 and E7 are oncogenes encoding proteins that suppress p53 and pRb activation, respectively [10]. E6* is a splicing isoform of E6, which is the main E6 isoform in cervical cancer, and might facilitate E7 expression [11]. Although the roles of E1^E4 are not explicitly defined, E1^E4 is considered to be associated with viral replication [12]. The L1 and L2 proteins are components of the viral capsid and are associated with HPV infection [13].
The expression patterns of HPV-derived transcripts vary depending on CIN grade. For example, the expression of E6 and E7 is higher in high-grade squamous intraepithelial lesions (high-grade SILs) than in low-grade SILs [14]. In contrast, expression of the L1 protein is lower in high-grade SILs [15, 16]. The expression patterns of HPV-derived transcripts also differ among HPV genotypes. For example, among E6 isoforms, HPV 18 cancers exhibit significantly higher ratios of the non-spliced isoform of E6 oncoprotein than HPV 16 cancers [17]. Furthermore, Griffin et al. demonstrated that CIN3 with HPV 18 exhibited no E4 protein expression [18].
In this study, we analyzed each HPV-derived transcript to gain a better understanding of HPV genotype-dependent carcinogenesis. To represent the viral life cycle, we focused on the expression levels of HPV-derived transcripts E6/E6*, E1^E4, and L1. E6/E6* are oncogenes regulated by the early promoter, the E1^E4 splicing site relates to both early and late gene expression and can contribute to viral replication, and L1 expression is observed in the late phase of viral differentiation, which is regulated by the late promoter [19]. In addition to HPV 16 and 18, we focused on HPV 52 and 58, which are highly prevalent in East Asia [20].
Methods
Patients and sample collection
All experimental procedures were approved by the institutional review boards of The University of Tokyo (approval number: G10082), Keio University (approval number: 2015–388), Chiba University (approval number: 560), Akita University (approval number: 2174), the National Institute of Infectious Diseases (approval number: 659), and Nihon University (approval number: 234–0), and signed informed consent for the use of tissues was obtained from each participant.
In total, 382 patients with cervical cytological abnormality who were admitted to the University of Tokyo, Chiba University, or Keio University between February 2016 and December 2017 were enrolled. Cervical tissues were obtained from biopsy under colposcopic examination. Samples were stored at − 80 °C until analysis.
Variables
Clinical data, such as age, gravidity, smoking history, usage of steroids or immunosuppressants, and time from first detection of abnormal cytology, were obtained by a medical interview. Histological results were classified into three CIN grades: CIN1, CIN2, and CIN3. Diagnosis was confirmed by a pathologist at Akita University.
The results of the HPV genotyping in cervical samples were recorded. It was permitted to assign multiple genotypes to a single patient. In this study, on the basis of the classification of the International Agency for Research on Cancer, we defined HPVs classified in Group 1 (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59) as “high-risk HPVs (hrHPVs)” [3]. Of these, HPVs 16, 18, 52, and 58 were separately categorized. hrHPVs other than HPV 16, 18, 52, and 58 were classified as “other hrHPVs.”
HPV genotyping
DNA was extracted from cervical specimens using the Tissue Genomic DNA Extraction Mini Kit (Favorgen Biotech Corp., Ping-Tung, Taiwan) at The University of Tokyo. HPV genotyping was performed at the National Institute of Infectious Diseases using the PGMY-CHUV assay method as described previously [21]. Briefly, standard PCR was conducted using the PGMY09/11 L1 consensus primer set and human leukocyte antigen-DQ (HLADQ) primer sets. Subsequently, reverse blotting hybridization was performed. Heat-denatured PCR amplicons were hybridized to probes specific for 31 HPV genotypes and HLA-DQ references [22].
Primer design and standard plasmid
PCR primers were designed using Primer-Blast (NCBI) in reference to PaVE, the papilloma virus genome database. The following criteria were considered when designing the primer pairs: (1) each primer should be 19–23 bp in length, and (2) the amplicon should be between 70 and 260 bp in length. The primer design is shown in S1 Fig. Plasmid standards (Eurofin Scientific, Luxembourg City, Luxembourg) were used to derive standard curves for absolute quantification.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cervical specimens using an miRNeasy Mini Kit (Qiagen, Hilden, Germany) after DNase treatment using the RNase-Free DNase Set (Qiagen, Hilden, Germany) at The University of Tokyo. Extracted RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. To assess mRNA expression levels, qRT-PCR was performed using a Light Cycler 480 system (Roche Diagnostics GmbH, Mannheim, Germany) with 1 μL of cDNA. Expression of HPV-derived transcripts was normalized to that of GAPDH mRNA as an internal standard. The normalized copy number was calculated as follows: normalized copy number = copy number/2^[30 - GAPDH Cp value]. Primer pairs for amplification of GAPDH and each HPV-derived transcript are shown in Table 1. PCR conditions were as follows: 45 cycles at 95 °C for 10 s, 62 °C for 10 s, and 72 °C for 18 s. All PCR reactions were assessed using melting curve analysis.
Table 1.
Primer pairs used for qRT-PCR
| Target | Direction | Sequence | Product size (bp) | Genome position |
|---|---|---|---|---|
| GAPDH | Forward | GAAAGGTGAAGGTCGGAGTC | 227 | |
| Reverse | GAAGATGGTGATGGGATTTC | |||
| HPV 16 E6 | Forward | AGCGACCCAGAAAGTTACCAC | 260 | 123–143 |
| Reverse | GTTGTATTGCTGTTCTAATGTTG | 382–360 | ||
| HPV 16 E6* | Forward | AGCGACCCAGAAAGTTACCAC | 114 | 123–143 |
| Reverse | TTAATACACCTCACGTCGC | 418–409 + 226–217 | ||
| HPV 16 E1^4 | Forward | CCTGCAGCAGCAACGAAGTATC | 218 | 874–880 + 3358–3372 |
| Reverse | TTGGTCGCTGGATAGTCGTC | 3479–3460 | ||
| HPV 16 L1 | Forward | GTCTCTTTGGCTGCCTAGTG | 89 | 5641–5660 |
| Reverse | TGCGTGCAACATATTCATCCG | 5729–5709 | ||
| HPV 18 E6 | Forward | AACACGGCGACCCTACAAG | 248 | 125–143 |
| Reverse | ATGTGTCTCCATACACAGAGTC | 372–351 | ||
| HPV 18 E6* | Forward | AACACGGCGACCCTACAAG | 120 | 125–143 |
| Reverse | ACCGCAGGCACCTCTGTAAG | 426–416 + 233–225 | ||
| HPV 18 E1^4 | Forward | GATCCAGAAGTACCAGTGAC | 194 | 920–929 + 3434–3443 |
| Reverse | GAGAAGTGGGTTGACAGGTC | 3617–3598 | ||
| HPV 18 L1 | Forward | TCCTTCTGTGGCAAGAGTTGT | 123 | 5657–5677 |
| Reverse | CCACCTGCAGGAACCCTAAAA | 5779–5759 | ||
| HPV 52 E6 | Forward | TTTGAGGATCCAGCAACAC | 197 | 105–123 |
| Reverse | TAGGCACATAATACACACGCC | 302–282 | ||
| HPV 52 E6* | Forward | TTTGAGGATCCAGCAACAC | 128 | 105–123 |
| Reverse | GACAAATTATACATCTCTCTTCG | 510–502 + 216–224 | ||
| HPV 52 E1^4 | Forward | AGGACCCTGAAGTAACGAAG | 150 | 868–879 + 3345–3352 |
| Reverse | CTGGAGTCTGTGACGTCTGG | 3482–3463 | ||
| HPV 52 L1 | Forward | ACTGTGTACCTGCCTCCTGTA | 72 | 5670–5690 |
| Reverse | GATGCTTGTGCGAGACACAT | 5741–5722 | ||
| HPV 58 E6 | Forward | GAAACCACGGACATTGCATG | 254 | 130–149 |
| Reverse | GTGTTTGTTCTAATGTGTCTCC | 383–362 | ||
| HPV 58 E6* | Forward | GAAACCACGGACATTGCATG | 109 | 130–149 |
| Reverse | CAAATAATACATCTCAGATCGC | 515–510 + 232–223 | ||
| HPV 58 E1^4 | Forward | GACCCTGAAGTGATCAAATATC | 127 | 889–898 + 3358–3372 |
| Reverse | GTGTTGTCTCTGGAGTCTGG | 3471–3452 | ||
| HPV 58 L1 | Forward | CCTCCTGTGCCTGTGTCTAA | 104 | 5682–5700 |
| Reverse | GGATTGCCAACAGCCAAAAGT | 5785–5765 |
Primer information, such as sequence, product size, and genome position of the primer pairs was summarized
Statistical analysis
Categorized clinical features such as gravidity, smoking history, usage of steroids or immunosuppressants according to HPV categories and CIN grades were evaluated using the Analysis of Variance. Other clinical features such as age and time from first detection of abnormal cytology and the expression levels of each transcriptome according to HPV types and CIN grades were analyzed using a Steel-Dwass test. The relationship between each HPV infection and positive ratio of each transcriptome was evaluated using the Cochran-Armitage trend test. Statistical analyses were performed with the JMP Pro software (13.0.0). p < 0.05 was considered significant. If the viral gene copy number was greater than 10 copies/L, the sample was considered positive for gene expression.
Results
HPV prevalence of four major genotypes
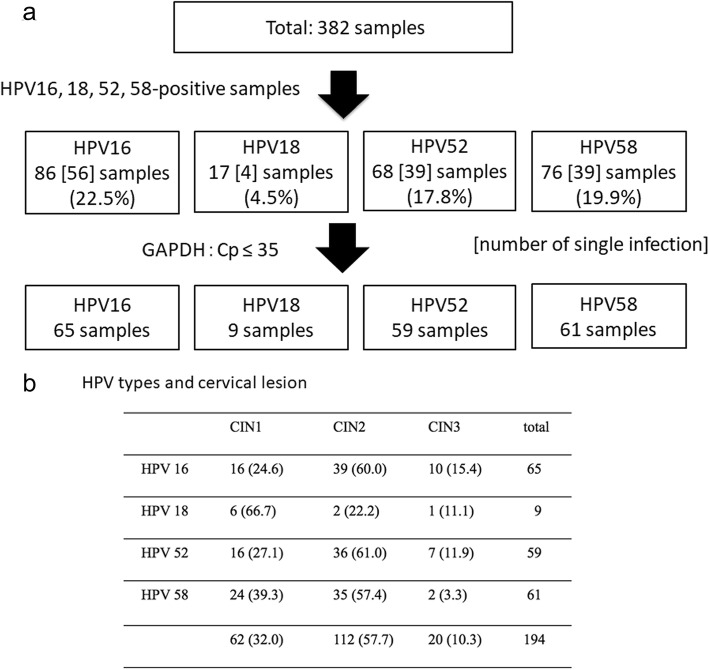
For the 382 patients with cervical cytological abnormality enrolled in the study, CIN grades and infected HPV types are summarized in Fig. 1. HPV 16 was detected in 86 (22.5%) samples, HPV 18 was detected in 17 (4.5%) samples, HPV 52 was detected in 68 (17.8%) samples, HPV 58 was detected in 76 (19.9%) samples, and other hrHPVs were detected in 83 (21.7%) samples. Samples infected with a single genotype included 56 (65.1%) HPV 16-positive samples, 4 (23.5%) HPV 18-positive samples, 39 (57.3%) HPV 52-positive samples, and 39 (51.3%) HPV 58-positive samples (Fig. 1). The ages of patients with each HPV genotype were significantly different, whereas no significant differences were found in gravidity, smoking history, usage of steroids or immunosuppressants, or time from first detection of abnormal cytology among patients with each HPV genotype (Tables 2 and 3).
Fig. 1.
CIN grades and prevalence of four major HPV genotypes. a Flow chart of patient enrollment; b CIN grades and HPV infection genotypes of the patients enrolled in this study
Table 2.
Clinical features according to HPV type
| HPV 16 | HPV 18 | HPV 52 | HPV 58 | Other hrHPVs | Negative | p-value | |
|---|---|---|---|---|---|---|---|
| Age (years) | 37 ± 0.9 | 39 ± 3.6 | 38 ± 0.8 | 39 ± 1.0 | 44 ± 1.2 | 40 ± 1.7 | 0.001*1 |
| Months since diagnosis | 40 ± 5.2 | 38 ± 10.0 | 36 ± 4.4 | 39 ± 5.1 | 37 ± 7.2 | 37 ± 7.0 | 0.95*1 |
| Smoking (%) | 15/55 (27.3) | 3/10 (30.0) | 14/56 (25.0) | 19/56 (28.4) | 16/72 (22.2) | 7/39 (18.0) | 0.57*2 |
| Parity ≥1 (%) | 23/57 (40.3) | 3/10 (30.0) | 23/56 (41.1) | 22/57 (38.6) | 26/74 (35.1) | 15/40 (37.5) | 0.97*2 |
| Steroid use (%) | 0/59 (0.0) | 0/10 (0.0) | 2/57 (3.5) | 2/58 (3.5) | 2/74 (2.7) | 1/36 (2.8) | 0.56*2 |
Clinical features were summarized according to HPV categorize. HPVs 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 were classified as hrHPVs. Of these, HPV 16, 18, 52, and 58 were categorized separately. hrHPVs other than HPVs 16, 18, 52, and 58 were classified as “other hrHPVs.” Patients who were not infected with any hrHPVs were referred to as “no hrHPVs” patients. Statistical analysis was performed using a Steel-Dwass test (*1) and the Analysis of Variance (*2)
HPV human papillomavirus
Table 3.
Clinical features according to cervical lesion grade
| CIN1 | CIN2 | CIN3 | p-value | |
|---|---|---|---|---|
| Age (years) | 36 ± 1.1 | 38 ± 0.6 | 32.5 ± 2.0 | 0.28*1 |
| Months since diagnosis | 23 ± 4.3 | 26 ± 3.9 | 9 ± 9.9 | 0.07*1 |
| Smoking (%) | 10/55 (18) | 31/93 (33) | 6/19 (32) | 0.12*2 |
| Parity ≥1 (%) | 21/56 (38) | 36/96 (38) | 10/19 (53) | 0.36*2 |
| Steroid use (%) | 2/57 (3.5) | 2/98 (2.0) | 0/19 (0.0) | 0.67*2 |
Clinical features were summarized according to cervical lesion grade. Statistical analysis was performed using a Steel-Dwass test (*1) and the Analysis of Variance (*2)
CIN cervical intraepithelial neoplasia
Differential E6/E6* expression in HPV-positive specimens
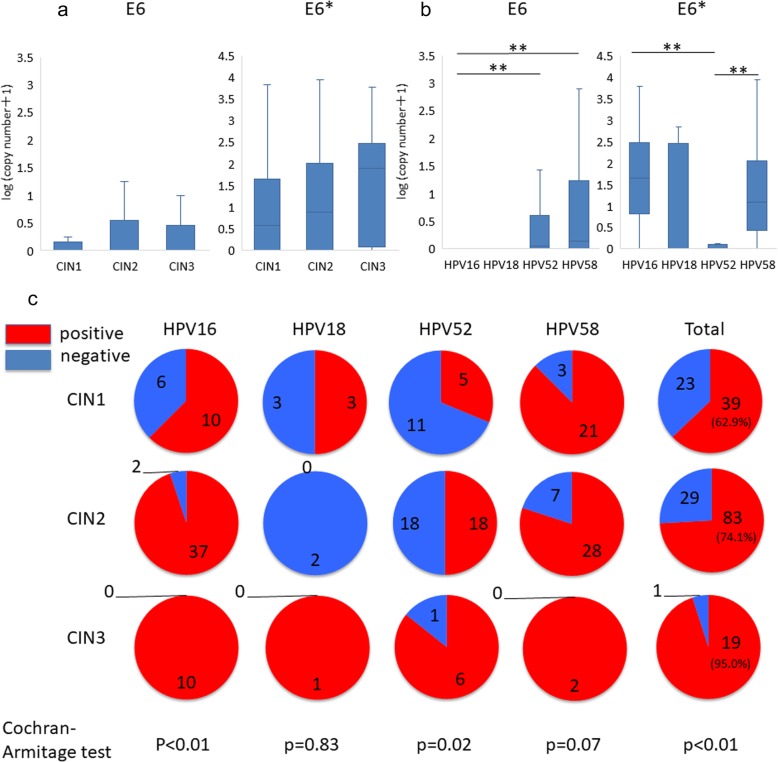
Expression levels of oncogene E6 and its isoform E6* were evaluated in each sample. Although there was no difference in the expression levels of E6 or E6* among CIN grades (Fig. 2a and b), the detection rate of E6 and/or E6* increased with severity of the CIN grade (Cochran-Armitage test, p < 0.01, Fig. 2c). In terms of HPV genotype-dependent analysis, E6 expression was lowest and E6* expression was highest in HPV 16 positive-specimens among the four HPV genotypes. In HPV 18-positive specimens, although no significant differences compared to other HPV genotypes were observed, possibly due to the small sample size, E6 and E6* expression patterns were similar to HPV 16-positive specimens. Conversely, in HPV 52-positive specimens, E6 expression was higher than E6*, while comparable expression levels of both E6 and E6* were observed in HPV 58-positive specimens (Fig. 2b).
Fig. 2.
Expression of HPV E6 and/or E6* genes. a Copy number of E6 and E6* genes in specimens with different CIN grades. Statistical analysis was performed using a Steel-Dwass test. ** indicates p < 0.01. b Copy number of E6 and E6* genes in specimens with HPV16, 18, 52, and 58 infection. Statistical analysis was performed using a Steel-Dwass test. ** indicates p < 0.01. c Detection rate of E6 and/or E6* gene in each genotype stratified by CIN grade. Statistical analysis was performed using a Cochran-Armitage test
Differential E1^E4 expression in HPV-positive specimens
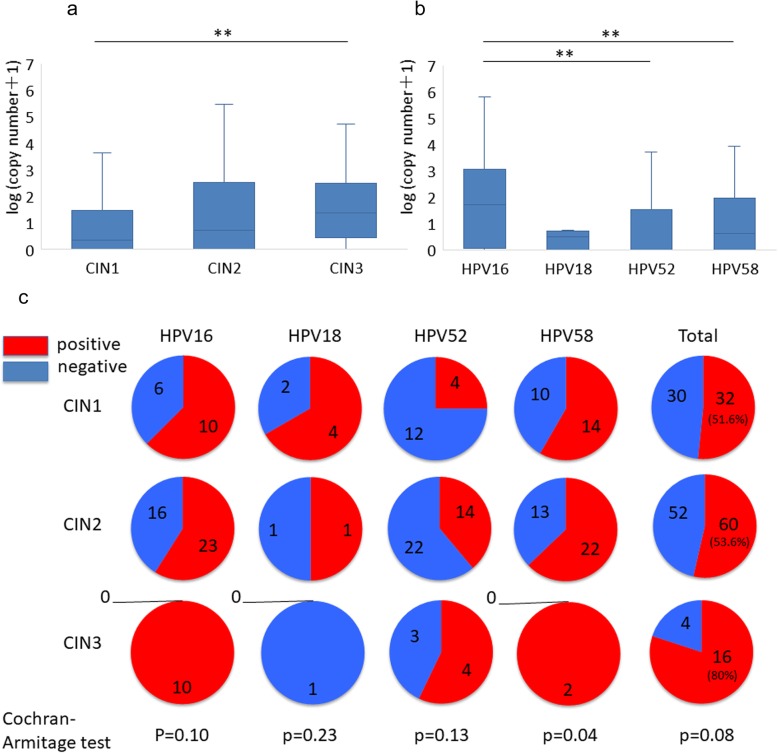
Subsequently, expression of the E1^E4 splicing site was evaluated. Positivity of this site is related to both early and late gene expression [19]. First, we compared E1^E4 expression levels across CIN grades and found its highest expression in CIN3 (Fig. 3a). The detection rate of E1^E4 tended to increase with CIN progression in HPV 16-, 52-, and 58-positive specimens (Cochran-Armitage test, p = 0.10, p = 0.13, and p = 0.04, respectively) (Fig. 3c). HPV genotype-dependent analysis revealed that E1^E4 expression levels were highest in HPV 16-positive specimens and lowest in HPV 18-positive specimens (Fig. 3b). Further, the detection rate of E1^E4 was higher in HPV 16- and 58-positive specimens than HPV 52-positive specimens (Fig. 3c).
Fig. 3.
Expression of HPV E1^E4 gene. a Copy number of the E1^E4 gene in specimens with different CIN grades. Statistical analysis was performed using a Steel–Dwass test. ** indicates p < 0.01. b Copy number of the E1^E4 gene in specimens with HPV16, 18, 52, and 58 infection. Statistical analysis was performed using a Steel–Dwass test. * indicates normalized copy number. ** indicates p < 0.01. c Detection rate of E1^E4 gene in each genotype stratified by CIN grade. Statistical analysis was performed using a Cochran-Armitage test
HPV type-dependent expression of major capsid protein L1
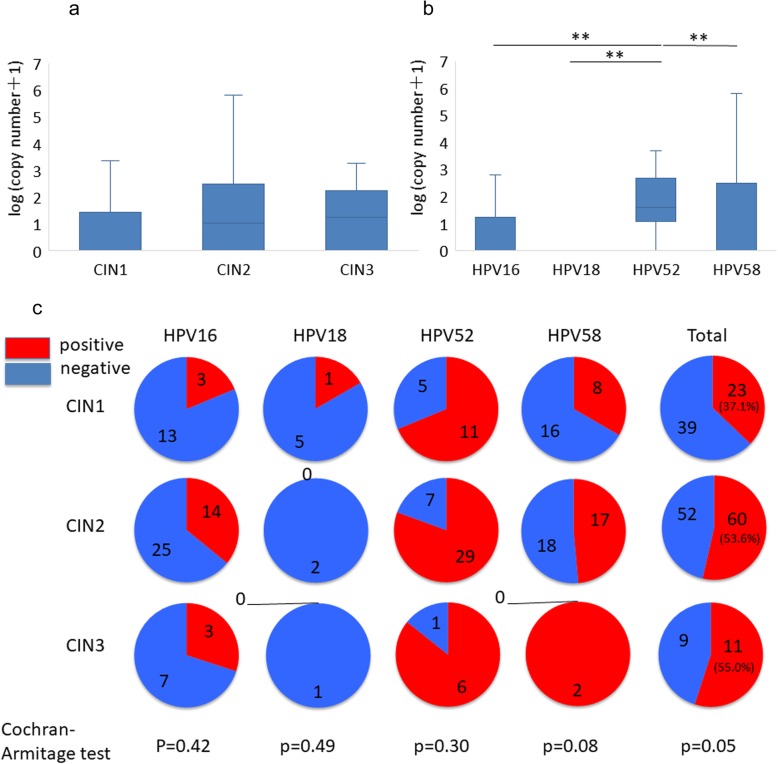
Further, expression levels of the L1 gene, which encodes a major capsid protein, was assessed. There was no difference in L1 expression among CIN grades (Fig. 4a). Comparison of L1 gene expression among HPV genotypes revealed the highest L1 expression in HPV 52-positive specimens, followed by HPV 58-positive specimens, and there was almost no L1 expression in HPV 18-positive specimens (Fig. 4b). Additionally, L1 expression was HPV type-dependent, in which nearly 100% of HPV 52-positive specimens, around 50% of HPV 58-positive specimens, approximately 33% of HPV 16-positive specimens, and almost 0% of HPV 18-positive specimens expressed L1 (Fig. 4c).
Fig. 4.
Expression of the HPV L1 gene. a Copy number of the L1 gene in specimens with different CIN grades. Statistical analysis was performed using a Steel–Dwass test. ** indicates p < 0.01. b Copy number of L1 gene in specimens with HPV16, 18, 52, and 58 infection. Statistical analysis was performed using a Steel–Dwass test. ** indicates p < 0.01. c Detection rate of L1 gene in each genotype stratified by CIN grade. Statistical analysis was performed using a Cochran-Armitage test
Since L1 expression is a hall mark of viral production and viral production is typically accompanied by epithelial differentiation, we performed immunohistochemistry analysis of KRT10 and KRT13 to investigate epithelial differentiation. However, there was no difference in KRT10 and KRT13 expression among samples with each HPV genotype (S2 Fig).
Discussion
We performed an in-depth analysis of HPV-derived transcript levels according to HPV genotype and CIN grade. The detection rate of E6/E6* increased with CIN progression, which is consistent with a previous study [23], whereas there was no significant change in the detection rate of E1^E4 or L1 among CIN grades. Furthermore, type-dependent analysis revealed that expression patterns of HPV-derived transcripts were HPV genotype-dependent.
Interestingly, L1 expression level was lowest in HPV 18-positive specimens among the four genotype groups. HPV 18 is one of the most carcinogenic genotypes among HR-HPV [24], and frequently observed in young aged cervical cancer [1]. In addition, around 40% of cervical adenocarcinoma is caused by HPV 18 infection [5]. L1 protein, a major component of the viral capsid, is a hallmark of viral production accompanied with cellular differentiation. Therefore, low level or lack of L1 expression in HPV 18-positive specimens may be associated with the loss of cellular differentiation and non-proliferative HPV infection, suggesting that stratified epithelium differentiation is not necessary for the HPV 18 genome replication and maintenance of HPV 18-related carcinogenesis. Our results added a new insight on HPV 18-related carcinogenesis from the aspect of HPV-derived transcripts. Other than loss of cellular differentiation, expression of the HPV L1 capsid protein disappears when HPV DNA is integrated into the host genome. Viral genome integration occurs earlier in HPV 18-positive cervical cells than in HPV 16-positive cells [8]. Loss of cell differentiation and viral genome integration from the early stage of CIN might be associated with rapid cancer development of HPV 18-infected CINs.
Conversely, L1 expression was highest in HPV 52, even in high-grade SIL (CIN2 and CIN3). Usually, L1 gene expression decreases as the CIN grades progress due to the lack of cellular differentiation [15, 16]. In contrast to HPV 18, the high expression of the L1 gene in HPV 52-positive specimens, even in high-grade SILs, may indicate that proliferative HPV infection accompanied with cellular differentiation may be maintained in HPV 52-positive lesions. In this study, we could not identify the human differentiation markers reflecting HPV 52-positive lesions; further studies are needed to identify human gene expression profiles that can distinguish the expression patterns of HPV-derived transcriptomes. Furthermore, the high level of L1 gene expression in HPV 52-positive CIN3 suggests that viral genome integration occurs in the late stage of CIN progression in these samples. Combined with the previously reported epidemiological findings, i.e. frequent observation of HPV 52 in CIN lesions compared to cancer lesions [5], high L1 expression represents the long-term persistence of HPV 52-related CINs.
This study has several limitations. First, regarding the analysis of E1^E4 gene expression, we only evaluated the expression level of the E1^E4 splicing site. Therefore, it is difficult to precisely demonstrate the significance of E1^E4 expression on the biology of viral replication or cancer development. Second, the sample size of the HPV 18-positive specimen was small. Therefore, further study using a large sample size is warranted to confirm our results. Third, this study is a cross-sectional study and does not investigate prognosis of CIN patients. As such, a prospective cohort study is needed to investigate whether expression of these HPV-derived transcripts can be biomarkers of CIN progression or regression.
Conclusions
In this study, we investigated the expression of three HPV-derived transcripts downstream of early, early/late, and late promoters in CIN lesions. Their expression patterns differed among HPV genotypes. In particular, L1 gene expression levels were lowest in HPV 18, while highest in HPV 52, suggesting HPV type dependence of HPV-derived carcinogenesis and viral maintenance in the cervical epithelium.
Supplementary information
Additional file 1: Figure S1. Primer designs for each transcriptome. The primer designs for E6, E6*, E1^E4, and L1 were summarized.
Additional file 2: Figure S2. Keratin (KRT)10 and KRT13 immunohistochemistry of cervical lesions. (a) Expression of KRT10 and KRT13 in CIN1 and 3. Specimens were stained with anti-KRT10 antibody (GNT, Irvine, CA, USA) and anti-KRT13 antibody (GNT, Irvine, CA, USA) according to the manufacturer’s instructions. Bars indicate 100 μm. (b) Detection rate of KRT10 and KRT13 in each genotype stratified by CIN grade.
Acknowledgments
We thank Takae Shimada for excellent technical support.
Abbreviations
- CIN
Cervical intraepithelial neoplasia
- HLA-DQ
Human leukocyte antigen-DQ
- HPV
Human papillomavirus
- PaVE
Papillomavirus episteme
- SILs
Squamous intraepithelial lesions
Authors’ contributions
Conception and design: Ayumi Taguchi, Kei Kawana. Acquisition of data: Satoshi Baba, Ayumi Taguchi, Akira Kawata, Satoko Eguchi, Mayuyo Mori, Katsuyuki Adachi, Seichiro Mori, Takashi Iwata, Akira Mitsuhashi. Analysis and interpretation of data: Satoshi Baba, Ayumi Taguchi, Konan Hara, Daichi Maeda, Iwao Kukimoto. Writing, review, and/or revision of the manuscript: Satoshi Baba, Ayumi Taguchi, Iwao Kukimoto, Kei Kawana. Study supervision: Atsushi Komatsu, Takeshi Nagamatsu, Katsutoshi Oda, Yutaka Osuga, Tomoyuki Fujii, Iwao Kukimoto, Kei Kawana. The author(s) read and approved the final manuscript.
Funding
This study was supported by Practical Research for Innovative Cancer Control (KK) and J-PRIDE (AT) under Grant Number 19fm0208013h0003 from the Japan Agency for Medical Research and Development (AMED).
Availability of data and materials
Ethics approval and consent to participate
All experimental procedures were approved by the institutional review boards of The University of Tokyo (approval number: G10082), Keio University (approval number: 2015–388), Chiba University (approval number: 560), Akita University (approval number: 2174), the National Institute of Infectious Diseases (approval number: 659), and Nihon University (approval number: 234–0), and signed informed consent for the use of tissues was obtained from each participant.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12985-020-01306-0.
References
- 1.Moscicki A-B. HPV infections in adolescents. Dis Markers. 2007;23:229–234. doi: 10.1155/2007/136906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 3.Bzhalava D, Guan P, Franceschi S, Dillner J, Clifford G. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology. 2013;445:224–231. doi: 10.1016/j.virol.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Mittal S, Banks L. Molecular mechanisms underlying human papillomavirus E6 and E7 oncoprotein-induced cell transformation. Mutat Res Rev Mutat Res. 2017;772:23–35. doi: 10.1016/j.mrrev.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Azuma YR, Takeuchi F, Uenoyama A, Kondo K, Tsunoda H, Nagasaka K, et al. Human papillomavirus genotype distribution in cervical intraepithelial neoplasia grade 2/3 and invasive cervical cancer in Japanese women. Jpn J Clin Oncol. 2014;44:910–917. doi: 10.1093/jjco/hyu112. [DOI] [PubMed] [Google Scholar]
- 6.Bulk S, Berkhof J, Rozendaal L, Daalmeijer NF, Gök M, de Schipper FA, et al. The contribution of HPV18 to cervical cancer is underestimated using high-grade CIN as a measure of screening efficiency. Br J Cancer. 2007;96:1234–1236. doi: 10.1038/sj.bjc.6603693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005;97:1072–1079. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]
- 8.Collins SI, Constandinou-Williams C, Wen K, Young LS, Roberts S, Murray PG, et al. Disruption of the E2 gene is a common and early event in the natural history of cervical human papillomavirus infection: a longitudinal cohort study. Cancer Res. 2009;69:3828–3832. doi: 10.1158/0008-5472.CAN-08-3099. [DOI] [PubMed] [Google Scholar]
- 9.Milligan SG, Veerapraditsin T, Ahamet B, Mole S, Graham SV. Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virology. 2007;360:172–181. doi: 10.1016/j.virol.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang S, Tao M, McCoy JP, Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papilloma virus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biryukov J, Myers JC, McLaughlin-Drubin ME, Griffin HM, Milici J, Doorbar J, et al. Mutations in HPV18 E1^E4 impact virus capsid assembly, infectivity competence, and maturation. Viruses. 2017;9:385. doi: 10.3390/v9120385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi YS, Kang WD, Kim SM, Choi YD, Nam JH, Park CS, et al. Human papillomavirus L1 capsid protein and human papillomavirus type 16 as prognostic markers in cervical intraepithelial neoplasia 1. Int J Gynecol Cancer. 2010;20:288–293. doi: 10.1111/IGC.0b013e3181cd184c. [DOI] [PubMed] [Google Scholar]
- 14.Ho CM, Lee BH, Chang SF, Chien TY, Huang SH, Yan CC, et al. Type-specific human papillomavirus oncogene messenger RNA levels correlate with the severity of cervical neoplasia. Int J Cancer. 2010;127:622–632. doi: 10.1002/ijc.25078. [DOI] [PubMed] [Google Scholar]
- 15.Xiao W, Bian M, Ma L, Liu J, Chen Y, Yang B, et al. Immunochemical analysis of human papillomavirus L1 capsid protein in liquid-based cytology samples from cervical lesions. Acta Cytol. 2010;54:661–667. doi: 10.1159/000325229. [DOI] [PubMed] [Google Scholar]
- 16.Stemberger-Papić S, Vrdoljak-Mozetic D, Ostojić DV, Rubesa-Mihaljević R, Manestar M. Evaluation of the HPV L1 capsid protein in prognosis of mild and moderate dysplasia of the cervix uteri. Coll Antropol. 2010;34:419–423. [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin H, Wu Z, Marnane R, Dewar V, Molijn A, Quint W, et al. E4 antibodies facilitate detection and type-assignment of active HPV infection in cervical disease. PLoS One. 2012;7:e49974. doi: 10.1371/journal.pone.0049974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham SV. Keratinocyte differentiation-dependent human papillomavirus gene regulation. Viruses. 2017;9:245. doi: 10.3390/v9090245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasagawa T, Maehama T, Ideta K, Irie T, Fujiko I. Population-based study for human papillomavirus (HPV) infection in young women in Japan: a multicenter study by the Japanese human papillomavirus disease education research survey group (J-HERS) J Med Virol. 2016;88:324–335. doi: 10.1002/jmv.24323. [DOI] [PubMed] [Google Scholar]
- 21.Kojima S, Kawana K, Tomio K, Yamashita A, Taguchi A, Miura S, et al. The prevalence of cervical regulatory T cells in HPV-related cervical intraepithelial neoplasia (CIN) correlates inversely with spontaneous regression of CIN. Am J Reprod Immunol. 2013;69:134–141. doi: 10.1111/aji.12030. [DOI] [PubMed] [Google Scholar]
- 22.Estrade C, Menoud PA, Nardelli-Haefliger D, Sahli R. Validation of a low-cost human papillomavirus genotyping assay based on PGMY PCR and reverse blotting hybridization with reusable membranes. J Clin Microbiol. 2011;49:3474–3481. doi: 10.1128/JCM.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JJ, Cao XC, Zheng XY, Wang HY, Li YW. Feasibility study of a human papillomavirus E6 and E7 oncoprotein test for the diagnosis of cervical precancer and cancer. J Int Med Res. 2018;1:300060517736913. doi: 10.1177/0300060517736913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Primer designs for each transcriptome. The primer designs for E6, E6*, E1^E4, and L1 were summarized.
Additional file 2: Figure S2. Keratin (KRT)10 and KRT13 immunohistochemistry of cervical lesions. (a) Expression of KRT10 and KRT13 in CIN1 and 3. Specimens were stained with anti-KRT10 antibody (GNT, Irvine, CA, USA) and anti-KRT13 antibody (GNT, Irvine, CA, USA) according to the manufacturer’s instructions. Bars indicate 100 μm. (b) Detection rate of KRT10 and KRT13 in each genotype stratified by CIN grade.