Abstract
Background
Major latex proteins (MLPs) belong to the MLP subfamily in Bet v 1 protein family and respond to both biotic and abiotic stresses, which play critical roles in plant disease resistance. As the type species of widely distributed and economically devastating Potyvirus, Potato virus Y (PVY) is one of the major constraints to important crop plants including tobacco (Nicotiana benthamiana) worldwide. Despite the great losses owing to PVY infection in tobacco, there is no previous study investigating the potential role of MLPs in developing resistance to viral infection.
Results
In this study, for the first time we have identified and functionally analyzed the MLP-like protein 28 from N. benthamiana, denoted as NbMLP28 and investigated its role in conferring resistance to N. benthamiana against PVY infection. NbMLP28 was localized to the plasmalemma and nucleus, with the highest level in the root. NbMLP28 gene was hypothesized to be triggered by PVY infection and was highly expressed in jasmonic acid (JA) signaling pathway. Further validation was achieved through silencing of NbMLP28 through virus-induced gene silencing (VIGS) that rendered N. benthamiana plants more vulnerable to PVY infection, contrary to overexpression that enhanced resistance.
Conclusions
Taken together, this is the first study describing the role of NbMLP28 in tobacco against PVY infection and provide a pivotal point towards obtaining pathogen-resistant tobacco varieties through constructing new candidate genes of MLP subfamily.
Keywords: Potato virus Y, Resistance, N. Benthamian, MLP-like proteins, NbMLP28, Jasmonic acid, Overexpression, VIGS, Gene silencing
Background
Potato virus Y (PVY) is highly destructive plant virus with worldwide distribution and pose serious economic losses to tobacco production [1–3]. PVY is mainly transmitted systemically by aphids [4], and can lead to mosaic, mottle, dwarfism, deformity, and necrosis in tobacco plants, seriously damaging yield and quality [5]. Current control measures of PVY in tobacco rely heavily on aphid prevention, agronomic practices, and PVY-resistant tobacco varieties [6, 7]. To date, PVY-resistant tobacco varieties are rare, while the resistance of most of the tobacco germplasm is not achieved yet [8]. Plants employ multiple strategies to cope with virus infection. Such as, plant hormones trigger the defense response and enhance stress resistance upon infection [9]. Moreover, ethylene (ET), salicylic acid (SA), and jasmonic acid (JA) signaling participate in plant defense [10]. ET and JA cooperatively regulate induced systemic resistance (ISR) in plants in the presence of non-pathogenic microbes such as rhizobacteria. Ryu et al. reported that in Arabidopsis, JA induced by rhizobacterium could alleviate the symptoms caused by Cucumber mosaic virus (CMV) infection [11]. Furthermore, JA pretreatment followed by SA confers strong resistance against the Tobacco mosaic virus (TMV) in N. benthamiana [12].
The major latex protein (MLP) was first identified from the latex of opium poppy (Papaver somniferum) [13, 14]. MLP proteins are members of MLP subfamily in the Bet v 1 family and exist in many plant species [15] and the orthologues of MLP, the MLP-like proteins, are also found in various plant species including Arabidopsis, soybean and tobacco [16, 17]. Most of the MLP/RRP subfamily members in wild strawberry and cucumber were expressed during the fruit ripening [18, 19], and also by wounding in immature bell peppers [20]. As revealed by the microarray analyses of Arabidopsis, the expression of three paralogous MLP gene pairs was significantly downregulated upon oxidative stress, indicating that MLP may participate in stress response [21]. In addition, many studies have demonstrated the necessity of MLP function against pathogen infection. For instance, Arabidopsis MLP28 (AT1G70830) and MLP3 were induced by Alternaria and Plasmodiophora brassicae, respectively [22, 23], and MLP expression was detected in stem phloem sap of melon plants infected by CMV [24]. Despite the importance of MLPs in biotic and abiotic stress responses, no systematic study on the relationship between MLP family members and PVY infection has been conducted.
In this study, for the first time we have identified and cloned the MLP-like protein 28 (NbMLP28) gene from N. benthamiana. The expression profile of this gene revealed that it was responsive to PVY infection and defense-related signaling molecules including ET, JA, and SA. Furthermore, virus-induced silencing of NbMLP28 rendered N. benthamiana plants more susceptible to PVY infection, whereas transient and constitutive overexpression of NbMLP28 enhanced resistance in tobacco plants against PVY. In addition, we identified the pathway that modulates the expression of NbMLP28 in N. benthamiana. The promoter sequence of NbMLP28 was amplified and analyzed to contain cis-acting elements in response to JA, light, drought, auxin, endosperm expression, etc. Conclusively, this is the first identification of NbMLP28 in tobacco and also the first detailed study describing its importance as a contributor to plant defense against PVY infection and provide strong bases to obtain pathogen-resistant tobacco varieties through constructing new candidate genes of MLP subfamily.
Results
Identification of the NbMLP28 gene and Phylogenetic analysis
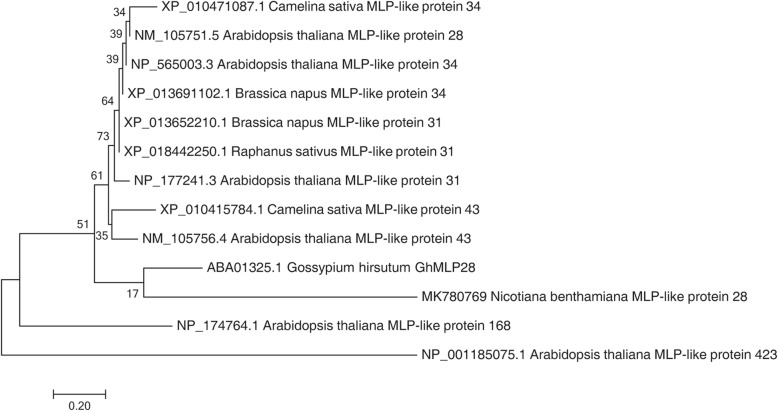
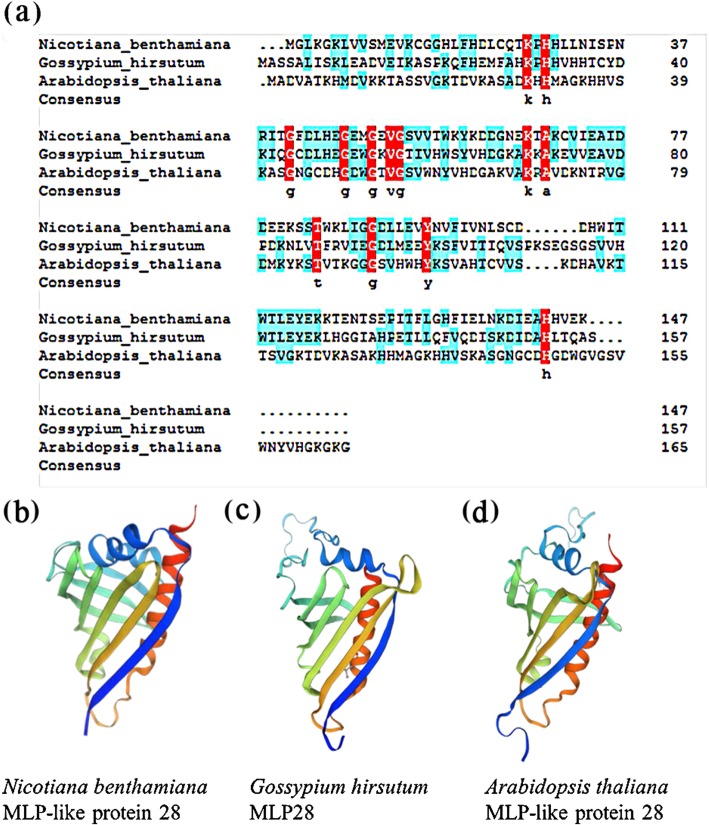
We amplified the ORF of NbMLP28 from N. benthamiana using primers, and the ORF of NbMLP28 was aligned with the predicted ORF sequence of NbMLP28 in the N. benthamiana database (https://solgenomics.net/organism/Nicotiana_benthamiana/genome). The ORF of NbMLP28 was submitted to NCBI under accession number MK780769. We constructed a phylogenetic tree of NbMLP28 and members of the MLP family in related species (Fig. 1). The result showed that the NbMLP28 shares the highest sequence similarity with Cossypium hirsutum MLP28, which is a putative defense-related protein [25]. The multiple alignment analysis revealed 31.16% similarity between Gossypium hirsutum MLP28 and Arabidopsis thaliana MLP28, They all contain a Gly-rich loop whose sequence is GxxxxxG (Fig. 2a). The structure of NbMLP28 protein predicted by SWISS-MODEL exhibits properties similar to those of the Gossypium hirsutum and Arabidopsis MLP28 (Fig. 2b-d). We cloned and analyzed the 3000 bp NbMLP28 promoter region and identified several potential cis-acting elements involved in Me-JA and light responses, one MYB binding site involved in drought-inducibility, one auxin-responsive element, one element involved in the abscisic acid (ABA) response, and one enhancer-like element involved in anoxic-specific induction (Table 1).
Fig. 1.
Phylogenetic analysis between MLP-like protein 28 of Nicotiana benthamiana and MLPs in other plants
Fig. 2.
Sequence analysis of N. benthamiana MLP-like protein28. a Sequence alignment analysis of N. benthamiana MLP-like protein28 with Arabidopsis thaliana MLP28 (NM_105751.5) and Gossypium hirsutum MLP28 (ABA01325.1) using DNAMAN. b-d The 3D-structure of N. benthamiana MLP-like protein 28 with Gossypium hirsutum MLP28 and Arabidopsis thaliana MLP-like protein 28
Table 1.
Cis-acting regulatory element analysis of the promoter of NbMLP28 gene
| Number | Site name | Amount | Sequence | Function of site |
|---|---|---|---|---|
| 1 | CGTCA-motif | 2 | CGTCA | cis-acting regulatory element involved in the MeJA-response |
| 2 | Gap-box | 1 | CAAATGAA(A/G)A | part of a light responsive element |
| 3 | I-box | 1 | GTATAAGGCC | part of a light responsive element |
| 4 | Box 4 | 3 | ATTAAT | part of a conserved DNA module involved in light response |
| 5 | circadian | 2 | CAAAGATATC | cis-acting regulatory element involved in Circadian control |
| 6 | TGACG-motif | 2 | TGACG | cis-acting regulatory element involved in the MeJA-response |
| 7 | 3-AF3 binding site | 1 | CACTATCTAAC | part of a conserved DNA module array (CMA3) |
| 8 | LAMP-element | 1 | CTTTATCA | part of a light responsive element |
| 9 | CCAAT-box | 2 | CAACGG | MYB Hv1 binding site |
| 10 | chs-CMA1a | 1 | TTACTTAA | part of a light responsive element |
| 11 | GCN4_motif | 1 | TGAGTCA | element involved in endosperm expression |
| 12 | CAAT-box | 44 | CCAAT | cis-acting element in promoter and enhancer regions |
| 13 | G-Box | 1 | CACGTT | regulatory element involved in light responsiveness |
| 14 | GATA-motif | 3 | AAGGATAAGG | part of a light responsive element |
| 15 | O2-site | 2 | GATGATGTGG | regulatory element involved in zein metabolism regulation |
| 16 | G-box | 2 | CACGTC | regulatory element involved in light responsiveness |
| 17 | MRE | 1 | AACCTAA | MYB binding site involved in light responsiveness |
| 18 | ARE | 3 | AAACCA | regulatory element essential for the anaerobic induction |
| 29 | MBS | 1 | CAACTG | MYB binding site involved in drought-inducibility |
| 20 | GC-motif | 1 | CCCCCG | enhancer-like element involved in anoxic specific inducibility |
| 21 | TGA-element | 2 | AACGAC | auxin-responsive element |
| 22 | ABRE | 3 | ACGTG | element involved in the abscisic acid responsiveness |
Subcellular localization of NbMLP28
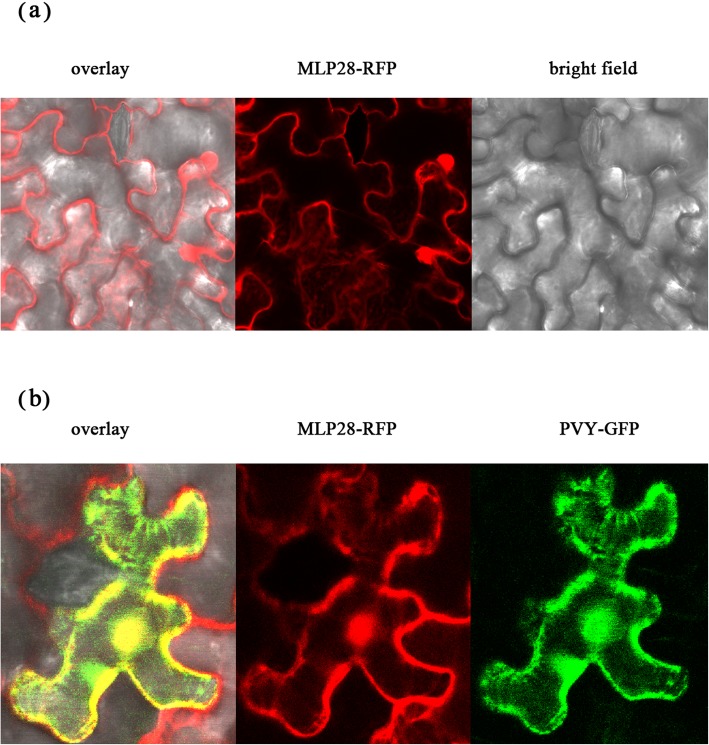
We predicted the subcellular localization of NbMLP28 using an online Plant-mPLo tool (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). The results suggested that the protein is localized in the cytoplasm. In addition, the protein contains a nuclear localization signal peptide (GLKGKLVVSMEVKCGGHLFHDLCQTKPHHLL) with a score of 4.2, as predicted by the NLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi#opennewwindow). Confocal results revealed that NbMLP28 is mainly localized to the plasmalemma and nucleus with or without virus infection (Fig. 3).
Fig. 3.
The subcellular localization of MLP-like protein 28. a The subcellular localization of MLP-like protein 28 in healthy N. benthamiana. b The subcellular localization of MLP-like protein during viral infection
Expression profiling of NbMLP28
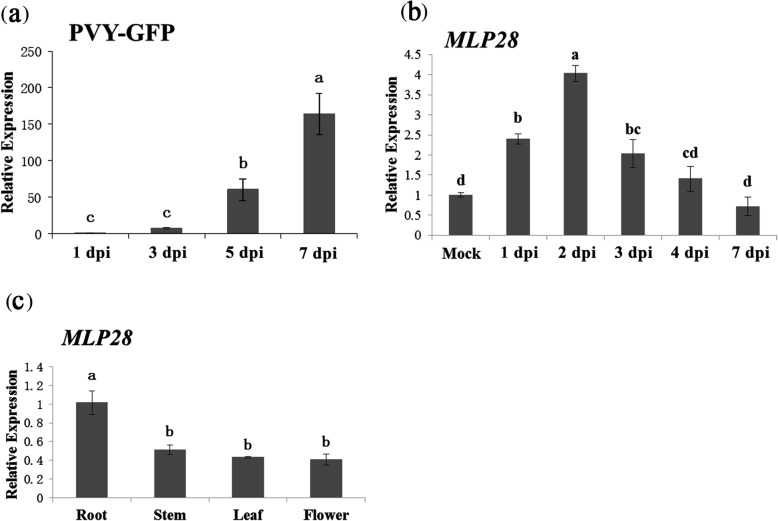
The accumulation of virus showed an upward trend after 1, 3, 5, and 7 days post inoculation (dpi) of PVY, and reached the peak of at seven dpi (Fig. 4a). Likewise, the expression of NbMLP28 was induced 1 day after PVY-GFP infection and maximized at 2 dpi (Fig. 4b). The qRT-PCR analysis detected uniformly NbMLP28 expression in various tissues in healthy N. benthamiana plants and the root exhibited a relatively highest level of NbMLP28 transcripts than other tissues investigated (Fig. 4c).
Fig. 4.
Gene expression pattern of NbMLP28 in wildtype plants. a PVY-GFP expression trend at 1.3.5.7 days of inoculation. bNbMLP28 expression trend after PVY inoculation. cNbMLP28 expression trends in root, stem, leaf and flower of healthy plants. The data were analyzed by Duncan’s multiple range tests in the ANOVA program of SPSS, different letters indicate that values of the four treatments were significantly different at P < 0.05
Silencing of NbMLP28 Renders N. benthamiana plants more susceptible to PVY infection
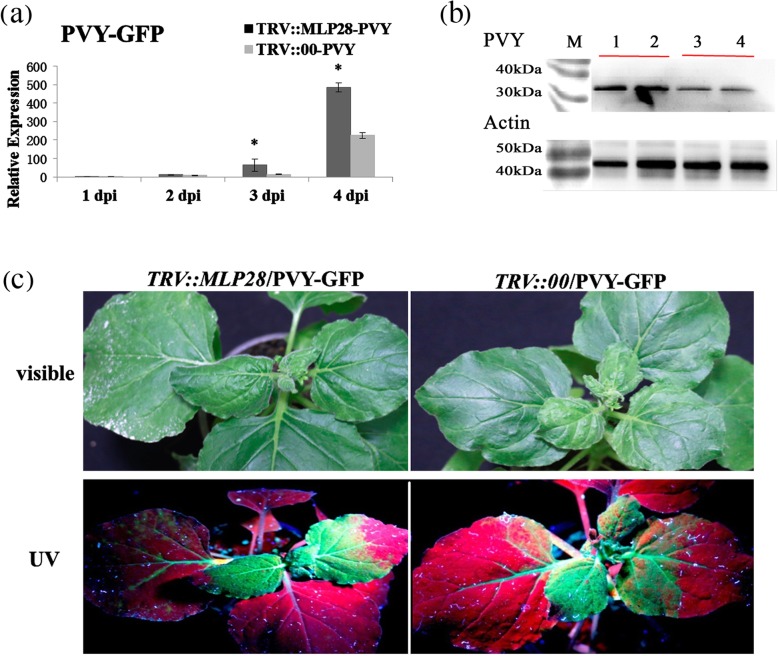
To further investigate the role of NbMLP28 in plant defense, we silenced the NbMLP28 gene using VIGS. Silencing efficiency was tested by comparing the expression levels of NbMLP28 in TRV::MLP28 plants versus TRV::00 control plants. Efficiency of the VIGS of NbMLP28 was 87% and no phenotypic difference was observed between the TRV::MLP28 and control plants (Supplementary Figure 1). Next, we infected the TRV::MLP28 and control plants with PVY-GFP and monitored virus infection for at least 1 week. The results showed that virus infection in TRV::MLP28 was significantly higher than that in the TRV::00 control group one to 4 days following inoculation, the treatment was 3.6 times and 1.2 times higher than the control at 3 and 4 dpi, respectively. (Fig. 5a). Western blotting detected a relatively higher level of viral coat protein in TRV::MLP28 than in TRV::00 3 days after virus inoculation (Fig. 5b, Supplementary Figure 4). Consistently, strong and far-ranging GFP signal was observed in TRV::MLP28 leaves, whereas only fewer fluorescent spots were observed in TRV::00 leaves. Especially, GFP signal was detected throughout the whole TRV::MLP28 plant nine dpi but was only observable in the leaf vein, petiole and lower leaves in TRV::00 individuals. The number and size of the infected areas in the systematic leaves of TRV::MLP28 were significantly greater than those of TRV::00 (Fig. 5c). Moreover, the TRV::MLP28 plants showed severe malformation of emerging leaves as compared to the control at nine dpi, indicating the severity of PVY in the absence of this protein (Fig. 5c). Taken together, these results indicated that the silencing of NbMLP28 rendered N. benthamiana plants highly susceptible to PVY infection.
Fig. 5.
Effects of silencing NbMLP28 on PVY infection. a PVY was inoculated after silencing NbMLP28 14 days, and the virus expression was higher compared to the control TRV::00 inoculated PVY at the RNA level. b Samples inoculated with PVY for 3 days were used to detect viral protein expression differences by Western blotting. 1–2 is TRV::MLP28 inoculated with PVY, 3–4 is TRV::00 inoculated with PVY. c The fluorescence difference of these two treatments
Transient NbMLP28 overexpression enhances PVY resistance
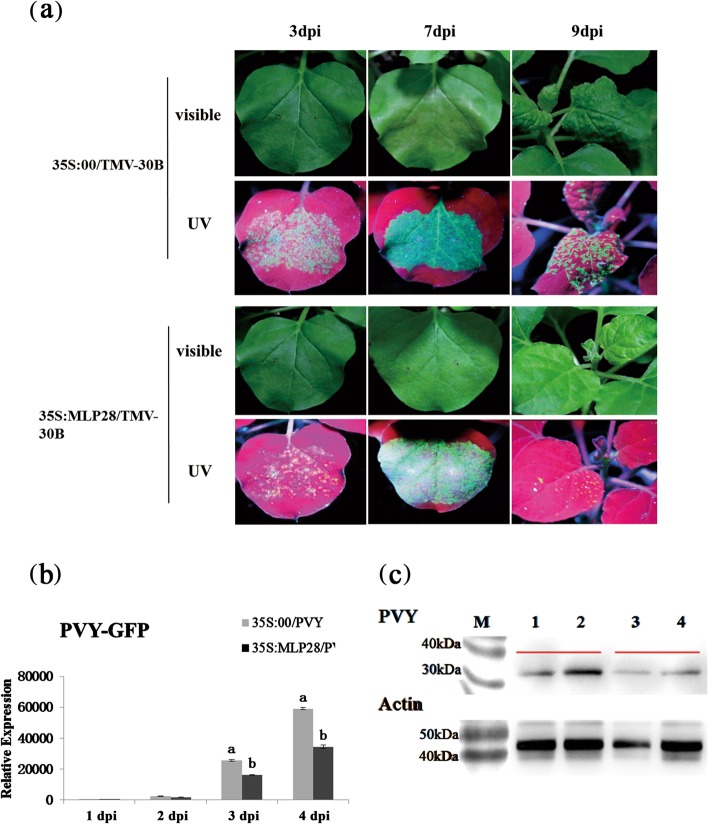
NbMLP28 was transiently overexpressed in N. benthamiana to further determine its role in response to PVY infection. N. benthamiana leaves were infiltrated with Agrobacterium carrying the 35S::MLP28 construct or the empty vector 35S::00 (negative control). The differences in viral accumulation between PVY-GFP-infected 35S::MLP28 and 35S::00 leaves were assessed by examining the intensity of GFP signals. We continued to observe virus fluorescence differences of inoculated leaves and system leaves at 3dpi, 7dpi and 9dpi. Lower PVY accumulation was observed in 35S::MLP28 plants compared with the 35S::00 control three, seven, and 9 days after inoculation (Fig. 6a). For example, the number and size of infected areas in the systematic leaves of 35S::MLP28 were significantly reduced compared with those of the empty vector control 9 days after inoculation (Fig. 6a). There was no significant malformation of systematic leaves in 35S::MLP28 plants comparing with the control at 9 dpi (Fig. 6a). Moreover, the results of subsequent qRT-PCR and western blotting analyses were in line with the severity of PVY infection. Specifically, the PVY level in 35S::MLP28 leaves was reduced approximately 37% at 3 dpi and 42% at 4 dpi than 35S::00 leaves (Fig. 6b), and western blotting confirmed the lower PVY protein level in 35S::MLP28 than in 35S::00 at 3 dpi (Fig. 6c, Supplementary Figure 5). We also verified the results using 35S::MLP28 transgenic plants (Supplementary Figure 2). Taken together, these results supported the notion that NbMLP28 overexpression enhanced PVY tolerance in N. benthamiana plants.
Fig. 6.
Effects of overexpression NbMLP28 on PVY infection. a Transiently infiltrating tobacco 35::MLP28/PVY-GFP and 35::00/PVY-GFP, respectively, observed UV fluorescence difference at 3, 7 and 9 days of treatment. b Real-time PCR detected virus expression difference at 1, 2, 3, 4 days after inoculation. 35S::MLP28/PVY was the treatment, 35S::00/PVY was the control group. The data were analyzed with independent sample T test using SPSS Statistics v.21 software, different letters indicated that values of the two treatments were significantly different at P < 0.05. c Virus expression was differentially detected 3 days after PVY inoculation. 1–2 is 35S::00 inoculated with PVY, 3–4 is 35S::MLP28 inoculated with PVY
NbMLP28 overexpression in N. benthamiana promotes germination and root growth
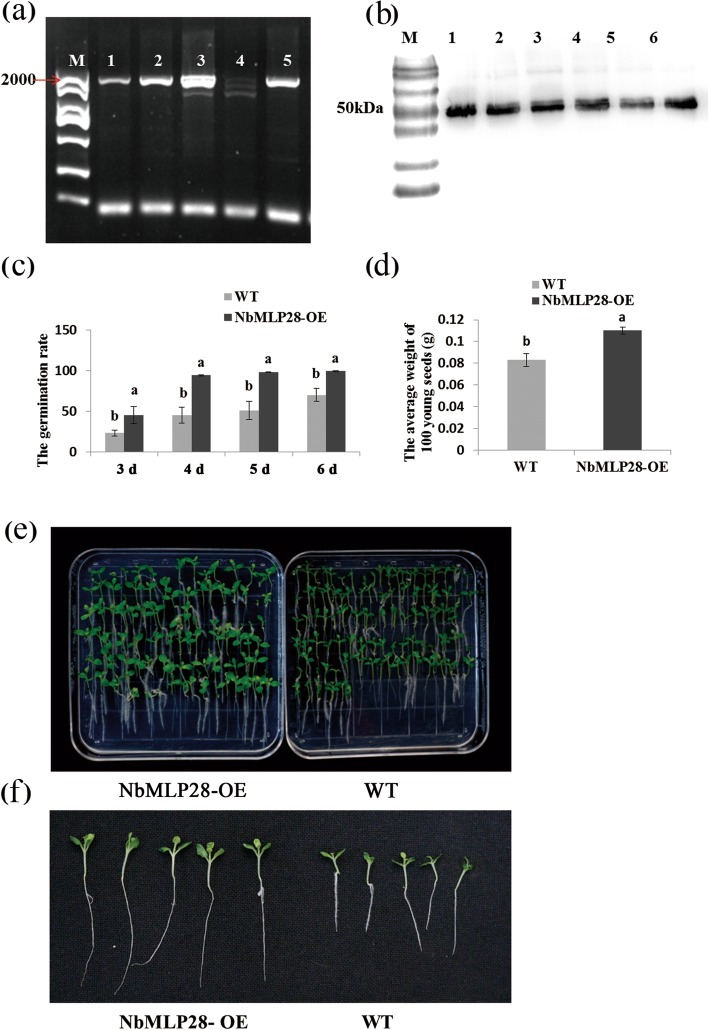
Next, MLP28 was constitutively expressed in frame with a RFP tag under the 35S promoter (35S::MLP28::RFP) in wild-type N. benthamiana. Positive 35S::MLP28::RFP transgenic plants were screened by PCR using primers E100F and E100R (Fig. 7a, Supplementary Figure 6). Through the antibiotic screening test, we obtained the T3 generation homozygous transgenic seeds and were used for subsequent experiments. The T4 plants of 35S::MLP28::RFP were confirmed by western blotting using RFP antibody (Fig. 7b, Supplementary Figure 6). The 35S::MLP28::RFP seeds started to germinate 2 days after sowing when the wild-type control showed no sign of germination. On day three, 45% germination of the 35S::MLP28::RFP seeds was achieved, whereas only 23% of the wild-type seeds germinated. The roots of 12-day-old 35S::MLP28::RFP plants were significantly longer than those of the wild-type seedlings and the two lines differed substantially in root morphology (Fig. 7e). The root length of the 35S::MLP28::RFP plants averaged 2.8 cm, and the root length of the wild type control averaged 2.0 cm (Fig. 7f). Detailed statistical analyses revealed significant difference in germination rate between the 35S::MLP28::RFP and control seeds three to 6 days after sowing (Fig. 7c). In addition, the fresh weight of 35S::MLP28::RFP seedlings was significantly higher than that of the wild-type control (Fig. 7d). Collectively, these data revealed that MLP28 overexpression promotes seed germination and root growth in tobacco.
Fig. 7.
Overexpression of NbMLP28 in transgenic N. benthamiana has germination and rooting promoting effect. a PCR amplification to selected highly overexpressed transgenic plants, sample 3 was a transgenic tobacco that strongly expresses NbMLP28.b Protein detection of overexpressing plants T4 generation stably expressing RFP-tagged MLP28. c Statistics on germination rate of over-expressed NbMLP28 transgenic tobacco and wild type at 3, 4, 5, and 6 days after seeding. d The fresh weight statistics of over-expression of NbMLP28 transgenic tobacco and wild-type at 12 days after seeding. e Differences in growth status of overexpressed NbMLP28 transgenic tobacco and wild type at 12 days after seeding. f Detection root length of over-expressed NbMLP28 transgenic tobacco and wild-type at 12 days after seeding
NbMLP28 is highly responsive to JA signaling in N. benthamiana
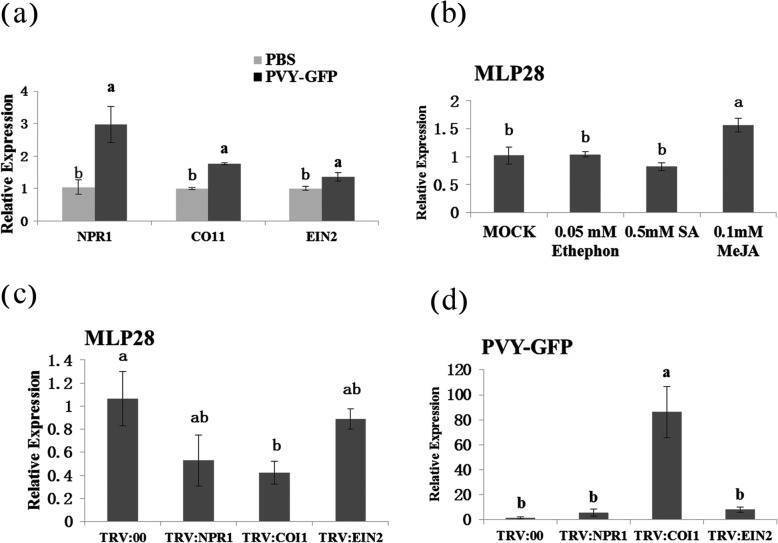
To explore the molecular basis of NbMLP28 in conferring PVY tolerance, we measured the relative expression levels of NPR1, COI1 and EIN2, key genes in SA, JA and ET signaling, respectively, in 4-week-old N. benthamiana following PVY-GFP infiltration. Our data show that the expression levels of NPR1, COI1 and EIN2 were significantly influenced upon PVY-GFP infiltration (Fig. 8a), which is consistent with a previous finding that the SA, JA, and ET signaling pathways are involved in plant pathogens resistance [26]. To test the effects of SA, JA and ET on NbMLP28 expression, we measured NbMLP28 transcript levels in N. benthamiana plants treated with 0.5 mM SA, 0.1 mM Me-JA or 0.05 mM Ethephon. Notably, Me-JA treatment significantly boosted the transcript level of NbMLP28 by approximately 0.5-fold in N. benthamiana leaves (Fig. 8b). SA treatment slightly decreased the transcript level of NbMLP28 by 0.2-fold in N. benthamiana plants, and 0.05 mM Ethephon had a minor effect on NbMLP28 expression (Fig. 8b).
Fig. 8.
Identification of NbMLP28 response to the hormonal pathway in N. benthamiana. a Expression of signal transduction key genes NPR1, COI1 and EIN2 of SA, JA and ET after 3 days of virus infection. The data were analyzed with independent sample T test using SPSS Statistics v.21 software, different letters indicate that values of the two treatments were significantly different at P < 0.05 (b) The expression of NbMLP28 after spraying with 0.5 mM SA, 0.1 mM Me JA or 0.05 mM Ethephon, the MOCK was sprayed with water with 0.02% Tween 20. The data were analyzed by Duncan’s multiple range tests in the ANOVA program of SPSS, different letters indicate that values of the four treatments were significantly different at P < 0.05, the same as C and D. c The expression of NbMLP28 after silencing NPR1, COI1 and EIN2. d The expression of PVY-GFP after silencing NPR1, COI1 and EIN2
We then silenced the NPR1, COI1 or EIN2 genes in wild-type N. benthamiana plants using VIGS to further dissect the interactions between NbMLP28 expression and SA, JA, and ET signaling [27].
The silencing efficiencies of NPR1, COI1 and EIN2 were 72, 70 and 75%, respectively (Supplementary Figure 7). We examined the expression pattern of NbMLP28 under these three treatments and observed a strong NbMLP28 reduction upon NPR1 and COI1 silencing—relative NbMLP28 expression was down-regulated by 50 and 60% in TRV::NPR1 and TRV::COI1 individuals than TRV::00, respectively. By contrast, EIN2 silencing only slightly reduced NbMLP28 expression by 16% (Fig. 8c). Meanwhile, boosted PVY-GFP expression was observed in TRV::NPR1, TRV::COI1 and TRV::EIN2 transgenic lines compared with that of the control group, with TRV::COI1 individuals showing the highest level of PVY-GFP expression (Fig. 8d). Taken together, these findings suggested the responsiveness of NbMLP28 to JA signaling.
Discussion
Despite the importance of MLP proteins in biotic and abiotic stress responses, their role in PVY-tobacco interaction remain unclear. In this study, we identified NbMLP28, a novel MLP-like protein 28 and investigated its functional profile in response to PVY infection in N. benthamiana. NbMLP28 was localized in both, the plasmalemma and nucleus. It was expressed uniformly in tobacco plants with the root exhibiting the highest level. Furthermore, the expression level of NbMLP28 peaked 2 days after PVY infection in tobacco plants. Whilst, transient and stable transgenic plants overexpressing NbMLP28 were more resistant to PVY infection, whereas silencing of this gene facilitated the viral infection.
The isolated ORF of MLP28 from N. benthamiana encoded a protein of 147 amino acids with a predicted conserved Bet v 1 domain, which was named after Bet v 1, a ribonuclease-active birch pollen allergen PR-10 protein [28]. PR-10 accumulation could be induced by pathogen infection, abiotic stress, related signaling molecules, hypersensitive response (HR), and systemic acquired resistance (SAR) [29], which was important for plant’s defense against biotic and abiotic stresses [30]. The presence of this Bet v 1 domain in NbMLP28 strongly indicated that it might be involved in plant defense. An MLP gene (At4g14060) has been reported to be down-regulated to a significant level during infection of Arabidopsis by Plum pox virus (PPV) – like PVY, an important member of the genus Potyvirus [31]. While, MLP induction has been a common observation following Verticillium dahliae (V. dahlia) attack in cotton [32–34], and ectopic overexpression of GhMLP28 in tobacco leads to improved V. dahliae tolerance [35]. In line with these reports, we have also observed an enhanced PVY resistance at both the mRNA and protein levels in N. benthamiana overexpressing NbMLP28.
Plant hormones are known to play essential roles in biotic and abiotic stress responses. Previous studies have identified the Arabidopsis MLP43 as a positive regulator of drought response, which modulated water loss efficiency, electrolyte leakage, ROS levels, and the expression levels of genes involved in ABA signaling [36], cotton MLP28 induced ethylene responsive factor 6 upon V. dahlia infection [35]. In accordance with these findings, our results showed that the NbMLP28 at transcriptomic level significantly elevated in tobacco after the exogenous application of Me-JA, as compared to SA and ET. We concluded that NbMLP28 responds to PVY infection via the JA signaling pathway. Additionally, the silencing of NPR1, COI1 and EIN2, the key genes involved in hormone signaling, downregulated NbMLP28 expression in N. benthamiana, where silencing of COI1 significantly decreased the NbMLP28 expression and enhanced PVY accumulation as compared to NPR1and EIN2. Similar findings have indicated that NPR1 and COI1 silencing in tobacco led to increased TMV susceptibility [12], suggesting that reduced NbMLP28 expression in COI1-silenced plants might hampered systemic resistance in N. benthamiana against PVY infection. Meanwhile, through analyzing the 3000 bp NbMLP28 promoter sequence, we identified two cis-regulatory elements (with the CGTCA and TGACG motifs, respectively) involved in Me-JA-response. However, the detailed mechanism by which NbMLP28 induced these responses still require further studies.
No phenotypic difference was observed between the 35S::MLP28 and control plants, indicating that NbMLP28 overexpression did not affect the growth and development of transgenic tobacco plants (Supplementary Figure 3). Conclusively, the higher NbMLP28 expression level is associated with increased PVY resistance in N. benthamiana. To our knowledge, this is the first mechanistic study of how NbMLP28 modulates the resistance of N. benthamiana against PVY infection. However, the detailed molecular mechanism by which this protein affected the defense pathway warrant future research.
Conclusions
This is the first ever identification and functional analysis of NbMLP28 in PVY-infected N. benthamiana. Additionally, we have analysed its defensive role upon PVY infection, that will further provide strong bases for constructing new candidate genes of MLP family to develop disease-resistant varieties of tobacco.
Methods
Plant materials
Two sets of N. benthamiana plants were used: (a) wild-type (seeds were obtained from Key Laboratory of Tobacco Pest Monitoring, Controlling & Integrated Management, Tobacco Research Institute), (b) NbMLP28 overexpressing transgenic plants (constructed in current study) were grown in a growth chamber with 50–60% humidity and a 16 h/8 h light/dark photoperiod at 25 °C. For inoculation, PVY-GFP (obtained from Key Laboratory of Tobacco Pest Monitoring, Controlling & Integrated Management, Tobacco Research Institute) was used in this study.
Cloning and sequence analysis of NbMLP28
RNA was isolated from N. benthamiana leaves using the TRIzol reagent (Vazyme) and first-strand cDNA synthesis was carried out using 2 μg total RNA and 100 U reverse transcriptase (Vazyme). Gene-specific primers MLP28F and MLP28R (Supplementary Table 1) were designed based on N. benthamiana genome data of Sol Genomics Network and used for PCR amplification; the resulting amplicons were subjected to 1% agarose gel electrophoresis and Sanger-sequenced. The deduced amino acid sequences of MLP28 (designated NbMLP28) were aligned with orthologs in other species in DNAMAN and SWISS-MODEL was employed for structure prediction [37]. The phylogenetic tree was generated using MEGA7 [38]. The potential cis-regulatory elements within NbMLP28 promoter were analyzed using the online program Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Virus-induced gene silencing (VIGS)
TRV vectors were kindly provided by Dr. Yule Liu, Tsinghua University, Beijing, China. Preparation of the pTRV vectors and Agrobacterium tumefaciens for VIGS followed a previously described procedure [39]. For VIGS vector construction, a 200 bp partial coding sequence (CDS) of NbMLP28 was amplified from a cDNA library of N. benthamiana leaf using gene-specific primers MLP28-TRVF and MLP28-TRVR (Supplementary Table 1) and inserted into the pTRV2 vector. For the VIGS assay, pTRV1 or pTRV2 constructs harboring the NbMLP28 fragment were introduced into the Agrobacterium strain LBA4404. Equal amount of Agrobacterium cultures containing pTRV1 and pTRV2 or pTRV2-MLP28 was mixed and used to inoculate the lower leaves of four-leaf stage N. benthamiana plants using a 1-mL needleless syringe. To determine VIGS efficiency, the leaves of tobacco plants 14 days post-inoculation (dpi) were tested by qRT-PCR using primers MLP28 QF and MLP28 QR (Supplementary Table 1), which detected the sequence outside the targeting fragment on the pTRV2-MLP28. Positive silencing plants were selected 14 dpi for analyzing NbMLP28 function. To test the response of NbMLP28 to hormones, we employed the same method described above to silence key hormone signaling genes NPR1, COI1 and EIN2 from the SA, JA and ET signaling pathways, respectively, and compared the expression levels of NbMLP28 and PVY-GFP in N. benthamiana.
Vector construction and agrobacterium-mediated gene transformation
To overexpress NbMLP28, the CDS of NbMLP28 was amplified from N. benthamiana with primers MLP28-35SF and MLP28-35SR, which contain the XbaI and EcoRI restriction sites, respectively (Supplementary Table 1). The resulting PCR fragment was inserted between the restriction sites on the Fu46-RFP entry vector. The target fragment was then inserted into the pEarlyGate100 expression vector that contains the 35S promoter, the resulting construct was introduced into the A. tumefaciens strain LBA4404 using a freeze–thaw method. A. tumefaciens cultures carrying 35S::MLP28::RFP were incubated overnight at 28 °C, harvested the next morning, and resuspended and cultured in an infiltration buffer containing 10 mM MES (pH = 5.6), 10 mM MgCl2, and 150 μM acetosyringone until OD600 reached 0.8. After three-hour incubation at room temperature, the bacterial suspensions were used to infiltrate the lower leaves of N. benthamiana plants using a needleless syringe for transient overexpression experiments.
We also overexpressed NbMLP28 in wild-type N. benthamiana using the same overexpression construct. First, a 5–8 mm disc was taken from a sterile tobacco leaf using a puncher, and the disc was placed on a preculture medium, and cultured at 25 °C for 24 h under light for 18 h. Then, the Agrobacterium of the vector was suspended in a liquid co-cultivation medium, the OD value was adjusted to 0.5–1.0, and the explants were inoculated for 30 min. The explants were placed on the co-culture medium and cultured at 24 °C for 3 days under light for 18 days. After the completion of the co-cultivation, the explants were transferred to a selection medium, cultured at 28 °C, 18 h light, and subcultured once every 2 weeks. In the selection medium, the explants grew longer and the buds grew from the callus. When the bud point grows to a length of 3 mm, it was transferred to the rooting medium. The tobacco plants were moved to the culture soil after about 2 weeks. Positive seedlings were detected with primers E100F/E100R, and the seeds were subcultured. We disinfected the surface of the T3 seeds and placed it on one-half MS medium of 50 mg/L Kan. After 1 week, the seedlings were all green, indicating that homozygous transgenic seeds had been obtained. In addition, tobacco leaves that overexpress NbMLP28 were infected with PVY-GFP after confirming the expression of 35S::MLP28 by PCR and western blotting analysis.
GFP and RFP imaging
The subcellular localization of NbMLP28 was examined using a Leica SP8 confocal microscope (Leica Microsystems, Shanghai) 48 h after the transient expression of NbMLP28 with a RFP tag in N. benthamiana epidermal cells. The plants were grown under a 16 h/8 h light/dark cycle at 25 °C. For the subcellular localization experiment, GFP was excited with a 25 mW, 488 nm argon laser, and emitted light with a wavelength between 495 and 535 nm was captured; RFP was excited with a 25 mW, 552 nm argon laser, and emitted light with a wavelength between 580 and 630 nm was captured. Successive images of 20 μm × 20 μm were scanned sequentially using 488 nm and 552 nm lasers with a 1.0 s scanning interval [40]. For the NbMLP28 silencing and overexpression experiments, in order to visually detect the accumulation of virus in inoculated leaves, we infiltrated the tobacco leaves with PVY-GFP and observed the difference in fluorescence between the treated and the control under a hand-held UV lamp (Ultra-Violet Products, Upland, CA, USA). One inoculated leaf per plant was measured and three biological replicates were analyzed for each line.
Hormone treatment
Four-week-old wild-type N. benthamiana seedlings were grown in a growth chamber under conditions mentioned above. The leaves were sprayed with 0.5 mM SA, 0.1 mM Me-JA, or 0.05 mM Ethephon with 0.02% Tween 20. The control plants were sprayed with water and 0.02% Tween 20. Three biological replicates of the wild-type N. benthamiana were analyzed. The treated leaves were harvested 24 h after the treatments, immediately snap frozen in liquid nitrogen and stored at − 80 °C until use.
Quantitative real-time PCR
Total RNAs isolation and cDNA synthesis followed the same procedures described above. qRT-PCR was performed with the SYBR Premix Ex Taq™ kit (Vazyme) using the Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems, Waltham, MA, USA) following the manufacturers’ instructions. The β-Actin gene was used as the endogenous control. NbMLP28 and β-Actin were amplified using primer pairs MLP28 QF/MLP28 QF and β-Actin QF/β-Actin QR, respectively (Supplementary Table 1). Meanwhile, two PVY primers, PVY-F and PVY-R, were used to detect the changes in virus coat protein expression. The − 2-△△CT method was used to calculate the relative expression level of target gene and three biological replicates were analyzed for each line [41].
Western blotting
For western blotting, protein was isolated from N. benthamiana, and total plant proteins were 1:1 equal volume mixed with 2 × SDS-PAGE buffer. Next, the protein samples were incubated at 95 °C for three min and separated on a 12% SDS-polyacrylamide gel. The separated proteins were then transferred onto nitrocellulose membranes by electroblotting instrument. The PVY CP antibody (SRA20001, Agdia, USA), anti-RFP (ab62341, Abcam, Shanghai) and β-Actin (CW0264M, CWBIO, Beijing) antibody were used for this assay.
Morphological characterization of the transgenic plants
Seeds of the wild-type and NbMLP28::RFP overexpression N. benthamiana lines received in the same batch were surface-sterilized and sown on one-half Murashige & Skoog plates. The plates were stratified at 4 °C for 24 h and let grow vertically at 25 °C with a 16 h/8 h light/dark photoperiod to examine root morphology. Plant root of the plants was measured over a one-week period. These experiments were repeated three times with 100 plants of each of the WT and NbMLP28::RFP lines were used per replicate.
Statistical analysis
Mean values of at least three independent experiments are shown and standard deviations (S.D.) are given. Duncan’s multiple range test analysis of variance (ANOVA), and independent sample t-test were performed in SPSS (v.21, IBM, Armonk, NY, USA). P < 0.05 denotes significant differences between comparisons.
Supplementary information
Additional file 1: Table S1. The primers used in this paper. Figure S1. The efficiency of agrobacterium-mediated virus-induced gene silencing in N. benthamiana. (A) Photographs were taken at 14 days after TRV infiltration. TRV::00 is a negative control, TRV::PDS as a positive control. Experiments were repeated three times with similar results. (B) Silencing efficiency between treatment and control was detected using real-time PCR. Figure S2. Differences in virus expression between overexpressing NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. (A) Fluorescence differences in overexpression of NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. (B) Differences in RNA levels between overexpressing NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. (C) Differences in viral protein between overexpressing NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. Figure S3. The phenotype of 35S::MLP28::RFP transgenic N. benthamiana and wild-type at 2-week old seedlings and 4-week old seedlings. Figure S4. The original western blotting figure of PVY CP differences respective in silencing and transient overexpression NbMLP28. (A) The first four lanes are 35S::00 and 35S::MLP28, and the last four lanes are TRV::MLP28 and TRV::00, Marker (14-120 kDa). (B) The Actin figure of corresponding samples, Marker (14-120 kDa). Figure S5. The original western blotting figure of wild-type and 35S::MLP28::RFP transgenic plants in response to PVY stress. (A) The first and second lanes respective were differences of PCY CP when wild-type and transgenic plants were inoculated with PVY at 7 dpi, Marker (14-120 kDa). (B) The Actin figure of corresponding samples, Marker (14-120 kDa). Figure S6. The full length original images of Gel and blot, presented in Fig. 7. (A) The original map of PCR to detect NbMLP28 highly expressed transgenic plants (We only selected the gel map of the first 5 lanes, showing the differences in expression of NbMLP28 in different transgenic plants, so as to select strong expression of NbMLP28 transgenic plants), Marker (DL2000, Vazyme). (B) The original map of validating the T4 generation stably expressing RFP tags in overexpressed plants, Marker (14-120 kDa). Figure S7. The silencing efficiency of NPR1, COI1 and EIN2 in N. benthamiana.
Acknowledgments
Authors are grateful to teachers and the colleagues who have contributed at any level to this research, with special thanks to Xiaowei Liu and Fangfei Yu for growing plants and maintaining the PVY-GFP source in the growth chamber.
Abbreviations
- 35S::00::RFP
an empty vector carrying an RFP tag
- 35S::MLP28::RFP
an MLP28 overexpression vector carrying an RFP tag
- ABA
Abscisic acid
- CMV
Cucumber mosaic virus
- dpi
day post inoculation
- ET
Ethylene
- HR
Hypersensitive response
- ISR
Induced systemic resistance
- JA
Jasmonic acid
- NbMLP28
MLP-like protein 28 gene in N. benthamiana
- NbMLP28
MLP-like protein 28 in N. benthamiana
- PPV
Plum pox virus
- PVY CP
PVY capsid protein
- PVY
Potato virus Y
- PVY-GFP
PVY with a green fluorescent protein label
- RFP
A red fluorescent protein
- SA
Salicylic acid
- SAR
Systemic acquired resistance
- TMV
Tobacco mosaic virus
- VIGS
Virus-Induced Gene Silencing
Authors’ contributions
YW and JY conceived the project and designed the experiments, LS1, JW, HJ, FW, LS2, AK, YQ, YL1 and KH performed experiments. LS1, JW, HJ and AK analyzed the data. FH, CZ, BL and YL2 interpreted the data. LS1, JW, and HJ drafted the manuscript. The final draft was read and approved by all the authors.
Funding
This research was supported by a grant from State Tobacco Monopoly Bureau (110101601024(LS-04), 110201901041(LS-04)), Jiangxi Tobacco Research Institute (2017.01.002), Sichuan Tobacco Company (SCYC201804), Baoshan Company of Yunnan Tobacco Company (2018530000241014). All the funding was awarded to Jinguang Yang who also conceived the project and designed the experiments. The funders have no role in conducting this research.
Availability of data and materials
Full length sequence of NbMLP28 was submitted in GenBank with the accession number: MK780769. All the raw data is publicly available and included in the manuscript also. Further details can be requested at jiayoulily2009@126.com.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Liyun Song, Jie Wang and Haiyan Jia contributed equally to this work.
Contributor Information
Yuanhua Wu, Email: wuyh09@syau.edu.cn.
Jinguang Yang, Email: yangjinguang@caas.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-020-01725-7.
References
- 1.Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, et al. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011;12:938–954. doi: 10.1111/j.1364-3703.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 2012;13:283. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 3.Wylie SJ, Adams M, Chalam C, Kreuze J, López-Moya JJ, Ohshima K, et al. ICTV virus taxonomy profile: Potyviridae. J Gen Virol. 2017;98:352. doi: 10.1099/jgv.0.000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Liu Y, Donkersley P, Dong Y, Chen X, Zang Y, et al. Preference of the aphid Myzus persicae (Hemiptera: Aphididae) for tobacco plants at specific stages of potato virus Y infection. Arch Virol. 2019;164:1567–1573. doi: 10.1007/s00705-019-04231-y. [DOI] [PubMed] [Google Scholar]
- 5.Quenouille J, Vassilakos N, Moury B. Potato virus Y: a major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Mol Plant Patho. 2013;14:439–452. doi: 10.1111/mpp.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacroix C, Glais L, Kerlan C, Verrier JL, Jacquot E. Biological characterization of French potato virus Y (PVY) isolates collected from PVY-susceptible or-resistant tobacco plants possessing the recessive resistance gene va. Plant Pathol. 2010;59:1133–1143. doi: 10.1111/j.1365-3059.2010.02342.x. [DOI] [Google Scholar]
- 7.Joshi RK, Nayak S. Gene pyramiding-a broad spectrum technique for developing durable stress resistance in crops. Biotechnol Mol Biol Rev. 2010;5:51–60. [Google Scholar]
- 8.Scott JM. Breeding for resistance to viral pathogens. Genet Improv Solanaceous Crops. 2006;2:457–485. doi: 10.1201/b10744-15. [DOI] [Google Scholar]
- 9.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Van Wees SC, De Swart EA, Van Pelt JA, Van Loon LC, Pieterse CM. Enhancement of induced disease resistance by simultaneous activation of salicylate-and jasmonate-dependent defense pathways in Arabidopsis thaliana. P Natl A Sci India B. 2000;97:8711–8716. doi: 10.1073/pnas.130425197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu CM, Murphy JF, Mysore KS, Kloepper JW. Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J. 2004;39:381–392. doi: 10.1111/j.1365-313X.2004.02142.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhu F, Xi DH, Yuan S, Xu F, Zhang DW, Lin HH. Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol Plant Microbe In. 2014;27:567–577. doi: 10.1094/MPMI-11-13-0349-R. [DOI] [PubMed] [Google Scholar]
- 13.Nessler CL, Kurz WG, Pelcher LE. Isolation and analysis of the major latex protein genes of opium poppy. Plant Mol Biol. 1990;15:951–953. doi: 10.1007/BF00039436. [DOI] [PubMed] [Google Scholar]
- 14.Nessler CL, Burnett RJ. Organization of the major latex protein gene family in opium poppy. Plant Mol Biol. 1992;20:749–752. doi: 10.1007/BF00046460. [DOI] [PubMed] [Google Scholar]
- 15.Radauer C, Lackner P, Breiteneder H. The bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggelis A, John I, Karvouni Z, Grierson D. Characterization of two cDNA clones for mRNAs expressed during ripening of melon (Cucumis melo L.) fruits. Plant Mol Biol. 1997;33:313–322. doi: 10.1023/A:1005701730598. [DOI] [PubMed] [Google Scholar]
- 17.Wu FZ, Lu TC, Shen Z, Wang BC, Wang HX. N-terminal acetylation of two major latex proteins from Arabidopsis thaliana using electrospray ionization tandem mass spectrometry. Plant Mol Biol Rep. 2008;26:88–97. doi: 10.1007/s11105-008-0027-6. [DOI] [Google Scholar]
- 18.Nam YW, Tichit L, Leperlier M, Cuerq B, Marty I, Lelièvre JM. Isolation and characterization of mRNAs differentially expressed during ripening of wild strawberry (Fragaria vesca L.) fruits. Plant Mol Biol. 1999;39:629–636. doi: 10.1023/A:1006179928312. [DOI] [PubMed] [Google Scholar]
- 19.Suyama T, Yamada K, Mori H, Takeno K, Yamaki S. Cloning cDNAs for genes preferentially expressed during fruit growth in cucumber. J Am Soc Hortic Sci. 1999;124:136–139. doi: 10.21273/JASHS.124.2.136. [DOI] [Google Scholar]
- 20.Pozueta-Romero J, Klein M, Houlné G, Schantz ML, Meyer B, Schantz R. Characterization of a family of genes encoding a fruit-specific wound-stimulated protein of bell pepper (Capsicum annuum): identification of a new family of transposable elements. Plant Mol Biol. 1995;28:1011–1025. doi: 10.1007/BF00032663. [DOI] [PubMed] [Google Scholar]
- 21.Stanley Kim H, Yu Y, Snesrud EC, Moy LP, Linford LD, Haas BJ, et al. Transcriptional divergence of the duplicated oxidative stress-responsive genes in the Arabidopsis genome. Plant J. 2005;41:212–220. doi: 10.1111/j.1365-313X.2004.02295.x. [DOI] [PubMed] [Google Scholar]
- 22.Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, et al. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. P Natl A Sci. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siemens J, Keller I, Sarx J, Kunz S, Schuller A, Nagel W, et al. Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant Microbe In. 2006;19:480–494. doi: 10.1094/MPMI-19-0480. [DOI] [PubMed] [Google Scholar]
- 24.Malter D, Wolf S. Melon phloem-sap proteome: developmental control and response to viral infection. Protoplasma. 2011;248:217–224. doi: 10.1007/s00709-010-0215-8. [DOI] [PubMed] [Google Scholar]
- 25.Chen JY, Dai XF. Cloning and characterization of the Gossypium hirsutum major latex protein gene and functional analysis in Arabidopsis thaliana. Planta. 2010;231:861–873. doi: 10.1007/s00425-009-1092-2. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 27.Zhu F, Xi DH, Deng XG, Peng XJ, Tang H, Chen YJ, et al. The chilli veinal mottle virus regulates expression of the tobacco mosaic virus resistance gene N and jasmonic acid/ethylene signaling is essential for systemic resistance against chilli veinal mottle virus in tobacco. Plant Mol Biol Rep. 2014;32:382–394. doi: 10.1007/s11105-013-0654-4. [DOI] [Google Scholar]
- 28.Heberle-Bors E, Vicente O. Bet v 1 proteins, the major birch pollen allergens and members of a family of conserved pathogenesis-related proteins, show ribonuclease activity in vitro. Physiol. Plantarum. 1996;96:433–438. doi: 10.1111/j.1399-3054.1996.tb00455.x. [DOI] [Google Scholar]
- 29.Liu JJ, Ekramoddoullah AK. The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant P. 2006;68:3–13. doi: 10.1016/j.pmpp.2006.06.004. [DOI] [Google Scholar]
- 30.Swoboda I, Hoffmann-Sommergruber K, O'Ríordáin G, Scheiner O, Jain S, Kumar A. The pathogenesis related class 10 proteins in plant defense against biotic and abiotic stresses. Adv Plants Agric Res. 2015;3:00077. [Google Scholar]
- 31.Babu M, Griffiths JS, Huang TS, Wang A. Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to plum pox virus infection. BMC Genomics. 2008;9(1):325. doi: 10.1186/1471-2164-9-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu ZL, Wang HY, Xia GX. GhHb1: a nonsymbiotic hemoglobin gene of cotton responsive to infection by Verticillium dahliae. Biochimica et Biophysica Acta (BBA) 2005;1730:103–113. doi: 10.1016/j.bbaexp.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Wang FX, Ma YP, Yang CL, Zhao PM, Yao Y, Jian GL, et al. Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics. 2011;11:4296–4309. doi: 10.1002/pmic.201100062. [DOI] [PubMed] [Google Scholar]
- 34.Zhang WW, Jian GL, Jiang TF, Wang SZ, Qi FJ, Xu SC. Cotton gene expression profiles in resistant Gossypium hirsutum cv. Zhongzhimian KV1 responding to Verticillium dahliae strain V991 infection. Mol Biol Rep. 2012;39:9765–9774. doi: 10.1007/s11033-012-1842-2. [DOI] [PubMed] [Google Scholar]
- 35.Yang CL, Liang S, Wang HY, Han LB, Wang FX, Cheng HQ, et al. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol Plant. 2015;8:399–411. doi: 10.1016/j.molp.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Yang L, Chen X, Ye T, Zhong B, Liu R, et al. Major latex protein-like protein 43 (MLP43) functions as a positive regulator during abscisic acid responses and confers drought tolerance in Arabidopsis thaliana. J Exp Bot. 2015;67:421–434. doi: 10.1093/jxb/erv477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 40.Sun H, Shen L, Qin Y, Liu X, Hao K, Li Y, et al. CLC-Nt1 affects potato virus Y infection via regulation of endoplasmic reticulum luminal Ph. New Phytol. 2018;220:539–552. doi: 10.1111/nph.15310. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 − CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The primers used in this paper. Figure S1. The efficiency of agrobacterium-mediated virus-induced gene silencing in N. benthamiana. (A) Photographs were taken at 14 days after TRV infiltration. TRV::00 is a negative control, TRV::PDS as a positive control. Experiments were repeated three times with similar results. (B) Silencing efficiency between treatment and control was detected using real-time PCR. Figure S2. Differences in virus expression between overexpressing NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. (A) Fluorescence differences in overexpression of NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. (B) Differences in RNA levels between overexpressing NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. (C) Differences in viral protein between overexpressing NbMLP28 transgenic plants and wild type at 7 days of PVY-GFP inoculation. Figure S3. The phenotype of 35S::MLP28::RFP transgenic N. benthamiana and wild-type at 2-week old seedlings and 4-week old seedlings. Figure S4. The original western blotting figure of PVY CP differences respective in silencing and transient overexpression NbMLP28. (A) The first four lanes are 35S::00 and 35S::MLP28, and the last four lanes are TRV::MLP28 and TRV::00, Marker (14-120 kDa). (B) The Actin figure of corresponding samples, Marker (14-120 kDa). Figure S5. The original western blotting figure of wild-type and 35S::MLP28::RFP transgenic plants in response to PVY stress. (A) The first and second lanes respective were differences of PCY CP when wild-type and transgenic plants were inoculated with PVY at 7 dpi, Marker (14-120 kDa). (B) The Actin figure of corresponding samples, Marker (14-120 kDa). Figure S6. The full length original images of Gel and blot, presented in Fig. 7. (A) The original map of PCR to detect NbMLP28 highly expressed transgenic plants (We only selected the gel map of the first 5 lanes, showing the differences in expression of NbMLP28 in different transgenic plants, so as to select strong expression of NbMLP28 transgenic plants), Marker (DL2000, Vazyme). (B) The original map of validating the T4 generation stably expressing RFP tags in overexpressed plants, Marker (14-120 kDa). Figure S7. The silencing efficiency of NPR1, COI1 and EIN2 in N. benthamiana.
Data Availability Statement
Full length sequence of NbMLP28 was submitted in GenBank with the accession number: MK780769. All the raw data is publicly available and included in the manuscript also. Further details can be requested at jiayoulily2009@126.com.