Significance
In this study, we find that ARID2 expression is negatively correlated with HCC metastasis and positively associated with prognosis of HCC patients. We demonstrate that ARID2 inhibits metastasis of HCC cells by recruiting DNMT1 to the promoter of Snail, which elevates DNA methylation and suppresses Snail transcription. Notably, we discover a C2H2 truncated mutation (3817C > T) in metastatic HCC tissue. We further reveal that C2H2 domain is required for ARID2–DNMT1 interaction and find loss-of-function ARID2 mutants with disrupted C2H2 domain are positively associated with HCC metastasis and poor survival of HCC patients. Taken together, our study not only expands comprehensive understanding of ARID2 and its mutations in HCC, but also provides indication for treatment of ARID2-deficient HCC.
Keywords: hepatocellular carcinoma, metastases, epigenetic modification, methylation, mutations
Abstract
Recurrence and metastasis remain the major obstacles to successful treatment of hepatocellular carcinoma (HCC). Chromatin remodeling factor ARID2 is commonly mutated in HCC, indicating its important role in cancer development. However, its role in HCC metastasis is largely elusive. In this study, we find that ARID2 expression is significantly decreased in metastatic HCC tissues, showing negative correlation with pathological grade, organ metastasis and positive association with survival of HCC patients. ARID2 inhibits migration and invasion of HCC cells in vitro and metastasis in vivo. Moreover, ARID2 knockout promotes pulmonary metastasis in different HCC mouse models. Mechanistic study reveals that ARID2 represses epithelial–mesenchymal transition (EMT) of HCC cells by recruiting DNMT1 to Snail promoter, which increases promoter methylation and inhibits Snail transcription. In addition, we discover that ARID2 mutants with disrupted C2H2 domain lose the metastasis suppressor function, exhibiting a positive association with HCC metastasis and poor prognosis. In conclusion, our study reveals the metastasis suppressor role as well as the underlying mechanism of ARID2 in HCC and provides a potential therapeutic target for ARID2-deficient HCC.
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide (1, 2). Frequent intrahepatic and extrahepatic metastases of HCC are responsible for poor clinical prognosis of HCC patients (3). Epithelial–mesenchymal transition (EMT), orchestrated by a restricted number of transcription factors—mainly the three Snail, Twist, and Zeb families (4)—confers metastatic properties upon cancer cells by enhancing mobility, invasion, and resistance. Therefore, identification of EMT suppressors and clarification of underlying mechanisms will provide therapeutic benefit for HCC.
ARID2 is one subunit of chromatin remodeling complex and is involved in various biological processes, including transcriptional regulation (5), cell cycle modulation (6, 7), embryonic development (8), and DNA damage repair (9). ARID2 mutations can be detected in most cancers, especially in HCC and often lead to partial or complete inactivation of ARID2 protein (10–12). Chromatin remodeling complex affects epigenetic modification (13), and ARID2 is supposed to modulate epigenetic modifications on DNA or histone. Although it is indicated that ARID2 may be a tumor suppressor by regulating epigenetic modification, this hypothesis has not yet been proved, and the role of ARID2 in metastasis of HCC remains largely elusive.
DNMT1 plays a dominant role in genome-wide DNA methylation maintenance and gene silencing (14, 15). Previous research indicated that DNMT1 was involved in cell metastasis, and down-regulation or inhibition of DNMT1 could facilitate the metastasis of cancer cells (16). However, target genes of DNMT1 involved in metastasis are poorly understood.
In this study, we found that ARID2 expression was significantly reduced in metastatic HCC tissues. Decreased ARID2 expression was positively correlated with organ metastasis and associated with poor prognosis of HCC patients. Moreover, ARID2 inhibited migration and invasion of HCC cells in vitro and decreased the intrahepatic metastases and distant seeding of HCC cells in vivo. Arid2 knockout in primary HCC mouse models promoted the pulmonary metastases of cancer cells. Further study revealed that ARID2 suppressed HCC metastasis by repressing EMT of HCC cells. ARID2 recruited DNMT1 to Snail promoter, which enhanced DNA methylation and suppressed Snail transcription. Moreover, we discovered that the C2H2 mutants of ARID2 could not interact with DNMT1, thus losing the metastasis suppressor function. It is interesting to note that these C2H2 truncated mutations were associated with HCC metastasis, and HCC patients with such mutations had significantly shorter survival times compared to those with intact C2H2 domain. Therefore, our study not only clarified that ARID2 acted as a metastasis suppressor in HCC, but also revealed the mechanism underlying the loss-of-function mutation of ARID2, which provided a potential therapeutic target for the treatment of ARID2-deficient HCC.
Results
Expression of ARID2 Was Significantly Decreased in Metastatic HCC and Positively Correlated with Good Prognosis of HCC Patients.
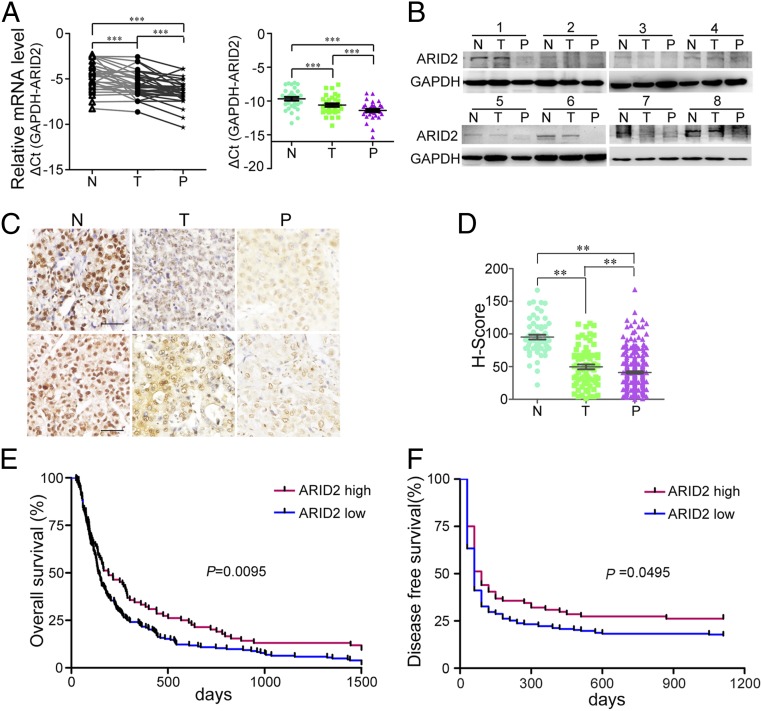
Study indicated that ARID2 was down-regulated in primary HCC tissues and suppressed proliferation of HCC cells (6), but the role of ARID2 in HCC metastasis was unclear. Therefore, we examined mRNA level expression of ARID2 in 34 paired primary HCC tissues (T), metastatic HCC tissues/portal vein tumor thrombus (PVTT) tissues (P), and the matched noncancerous adjacent normal tissues (N). As shown in Fig. 1A, ARID2 mRNA was dramatically reduced in primary HCC tissues, and PVTT tissues showed a lower ARID2 mRNA level than primary HCC tissues (Fig. 1A). Moreover, the gradual decrease in ARID2 expression from noncancerous normal tissues, primary HCC tissues, to PVTT tissues was confirmed by Western blot (Fig. 1B) and immunohistochemistry (Fig. 1C). To further evaluate the clinical significance of ARID2 in HCC metastasis, HCC tissue microarray was stained with ARID2 antibody, and the gradual decline of ARID2 expression was also detected (Fig. 1D). In addition, HCC cells with high metastatic potential displayed low expression of ARID2 (SI Appendix, Fig. S1). Taken together, these results indicated that ARID2 expression was inversely correlated with HCC metastasis.
Fig. 1.
ARID2 expression was down-regulated in metastatic HCC and positively correlated with good prognosis of patients. (A and B) Expression levels of ARID2 mRNA and protein in clinical HCC tissues, metastatic HCC tissues (hepatic portal vein carcinoma thrombus), and their adjacent noncarcinoma normal tissues were investigated by RT-qPCR (n = 34) and Western blot, respectively. (C) Immunohistochemistry was employed to identify the expression of ARID2 protein in N, T, and P tissues. (Scale bar, 250 μm.) (D) H-scores of ARID2 protein in N (n = 60), T (n = 73), and P (n = 214) HCC tissues. (E and F) The overall survival (ARID2 high, n = 84; ARID2 low, n = 203; P = 0.0095) and disease-free survival (ARID2 high, n = 62; ARID2 low, n = 150; P = 0.0495) of patients with ARID2 low and high expression. Data were analyzed using Student’s t test. All **P < 0.01, ***P < 0.001.
Based on the score of ARID2 staining intensity in tissue microarray, the patients were divided into ARID2 low (H score < 50) and high (H score ≥ 50) expression groups, and the correlation between ARID2 expression and clinical pathology features was assessed. A low level of ARID2 was positively correlated with pathological grade, tumor size, and organ metastasis (SI Appendix, Table S1). Meanwhile, decreased ARID2 expression was associated with poor overall survival (OS) (Fig. 1E) and disease-free survival (DFS) (Fig. 1F) of HCC patients. Together, these data further confirmed the negative correlation between ARID2 expression and HCC metastasis, and indicated that low ARID2 expression was associated with a poor prognosis for HCC patients.
ARID2 Suppressed Migration and Invasion of HCC Cells In Vitro and Metastasis In Vivo.
The above observations prompted us to investigate the function of ARID2 in HCC metastasis. We constructed ARID2-overexpressing and ARID2 knockdown cells using lentiviruses (Fig. 2A), and effects of ARID2 on migration and invasion of HCC cells were assessed by Boyden chamber assay and transwell assay, respectively. We found that forced expression of ARID2 notably inhibited the migration and invasion of MHCC97-H and PVTT cells (Fig. 2B and SI Appendix, Fig. S2A). In contrast, knockdown of ARID2 significantly promoted migration and invasion of PLC/PRF/5 and YY-8103 cells (Fig. 2C and SI Appendix, Fig. S2B). Therefore, these results suggested that ARID2 suppressed migration and invasion of HCC cells in vitro.
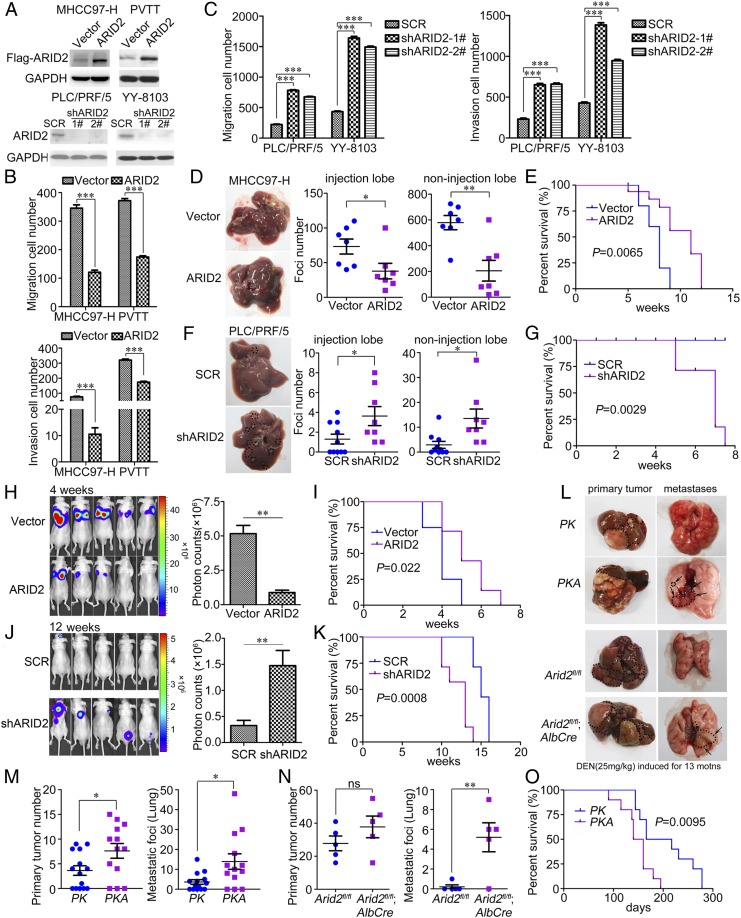
Fig. 2.
ARID2 suppressed the migration and invasion of HCC cells in vitro and inhibited the metastasis of HCC cells in vivo. (A) The protein levels of ARID2 in stably transfected HCC cells were examined by Western blot. (B and C) The effects of ARID2 on migration and invasion of HCC cells were investigated by Boyden chamber and transwell assay. (D and F) Gross morphology of livers injected with stably transfected HCC cells or control cells in left lobe and statistical analysis of foci number in injection lobe and noninjection lobes. Metastases on surface of injection lobe and noninjection lobes were counted under microscope. n(MHCC97-H-Vector) and n(MHCC97-H-ARID2)=7; n(PLC/PRF/5-SCR)=10, n(PLC/PRF/5-shARID2)=8. (E and G) Survival of the mice in intrahepatic metastasis mouse model injected with stably transfected HCC cells or control cells. MHCC97-H–ARID2 and control cells, n = 10, P = 0.0065; PLC/PRF/5-shARID2 and control cells, n = 7, P = 0.0029. (H and J) Fluorescence of metastases generated in distant seeding mouse model tail-vein-injected with stably transfected HCC cells or control cells was monitored, n = 5. (I and K) Survival of mice in the distant-seeding mouse model. MHCC97-H-ARID2 and control cells, n = 8, P = 0.022; PLC/PRF/5-shARID2 and control cells, n = 7, P = 0.0008. (L) Primary tumors and metastatic lesions of HCC mouse models. Mice of P53/RasG12D driven and DEN (25 mg/kg) induced HCC mouse models were euthanized at 4 and 13 months old, respectively. (M and N) Number of primary tumors (liver) and metastatic lesions (lung) of HCC mouse models. PK, n = 14; PKA, n = 13; Arid2fl/fl and Arid2fl/fl; AlbCre, n = 5. (O) Survival of PK and PKA mice. n = 10, P = 0.0095. Data were analyzed using Student’s t test. PK: P53fl/fl; LSL-RasG12D; AlbCre. PKA: P53fl/fl; LSL-RasG12D; Arid2fl/fl; AlbCre. All *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant.
An intrahepatic metastasis mouse model was used to verify the suppressive effect of ARID2 in HCC metastasis. As shown in Fig. 2D, fewer foci were detected on injected and noninjected lobes of the nude mice injected with MHCC97-H–ARID2 cells (Fig. 2D and SI Appendix, Fig. S3A), compared with those injected with control cells. The mice with MHCC97-H–ARID2 cells had longer survival than control mice (Fig. 2E). In contrast, ARID2 knockdown remarkably increased foci formation on both injected and noninjected liver lobes (Fig. 2F and SI Appendix, Fig. S3A). Accordingly, mice in shARID2 group exhibited shorter overall survival than those in scramble shRNA (SCR) group (Fig. 2G). These results suggested that ARID2 extended the survival time of tumor-bearing mice by repressing metastasis of HCC cells.
We also investigated the effect of ARID2 on distant seeding of HCC cells using a tail vein injection model. Four weeks after injection, lung seeding was notably weaker in mice injected with MHCC97-H–ARID2 cells compared to the control group, according to the fluorescence intensity (Fig. 2H), associated with prolonged survival (Fig. 2I). On the contrary, PLC/PRF/5-shARID2 cells exhibited an increase in metastatic seeding compared with SCR cells (Fig. 2J), and the mice in PLC/PRF/5-shARID2 group had poorer survival than those in SCR group (Fig. 2K). Consistent with the in vitro study, these results demonstrated that ARID2 repressed metastasis of HCC cells in vivo.
To further verify the suppressive effect of ARID2 on HCC metastasis, a primary HCC mouse model P53fl/fl; LSL-RasG12D was employed, and hepatic ARID2 was specifically deleted by Alb-Cre. In 4-month-old Alb-Cre; P53fl/fl; LSL-RasG12D; Arid2fl/fl (PKA) mice, more pulmonary metastases were detected compared with control (PK) mice (Fig. 2 L and M and SI Appendix, Fig. S3B). Moreover, hepatic deletion of Arid2 significantly increased pulmonary metastases in N-nitrosodiethylamine (DEN; 25 mg/kg)–induced HCC mouse model (Fig. 2 L and N and SI Appendix, Fig. S3B). Consistently, PKA mice had shorter survival than the control mice (Fig. 2O). Therefore, liver-specific deletion of ARID2 promoted HCC metastasis in different mouse models. Taken together, we demonstrated that ARID2 acted as a metastasis suppressor in HCC.
ARID2 Repressed EMT of HCC Cells.
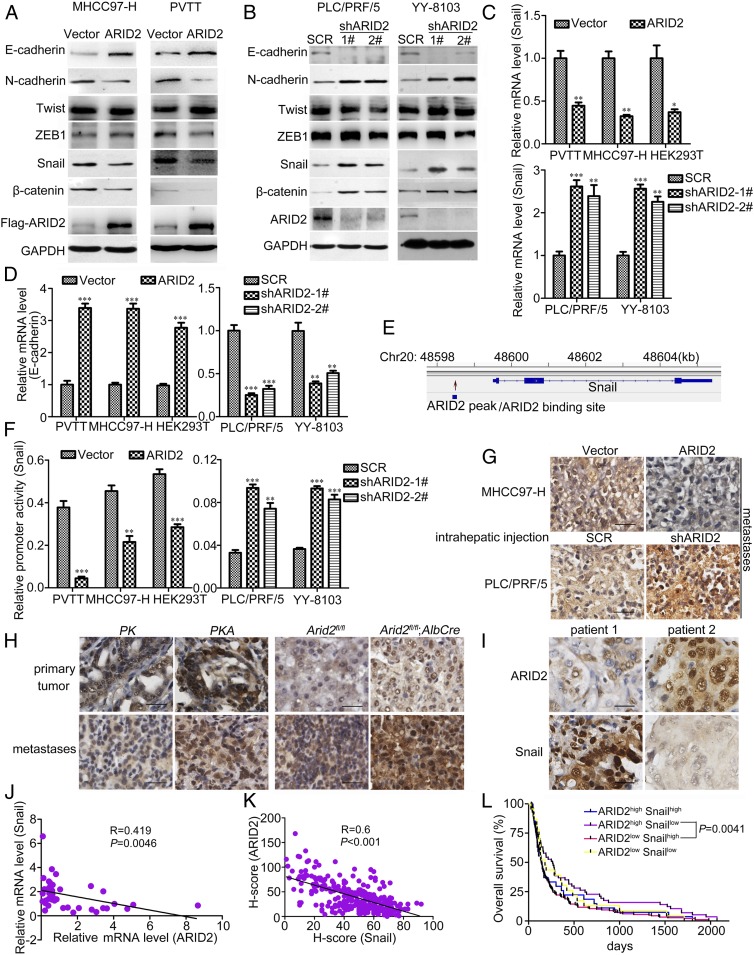
During the EMT program, genes involved in cell adhesion, migration, and invasion were transcriptionally altered (4). Therefore, we hypothesized that ARID2 might suppress metastasis of HCC by inhibiting EMT of HCC cells. To identify this hypothesis, we examined the expression of EMT genes and found that ARID2 overexpression repressed the expression of mesenchymal marker N-cadherin, Snail, and β-catenin and increased the expression of epithelial marker E-cadherin (Fig. 3A). Down-regulation of ARID2 increased the expression of N-cadherin, Snail, and β-catenin, while attenuating the expression of E-cadherin in HCC cells (Fig. 3B). Furthermore, a consistent expression pattern of EMT genes was observed in primary hepatocytes from ARID2-knockout mice (SI Appendix, Fig. S4A). Therefore, our data suggested that ARID2 inhibited metastasis of HCC by suppressing EMT.
Fig. 3.
ARID2 suppressed EMT and Snail transcription of HCC cells. (A and B) The expression of EMT associated factors in ARID2 knockdown and overexpressing HCC cells were examined by Western blot. (C and D) mRNA levels of Snail and E-cadherin in stably transfected HCC cell lines were examined by RT-qPCR. (E) Based on previous ChIP-seq data from HepG2 cells, ARID2 potentially bound at the promoter region of Snail gene. (F) Effect of ARID2 on activity of Snail promoter in stably transfected HCC cells was assessed by reporter assay. (G and H) Snail expression in metastatic lesions of intrahepatic metastasis mouse model and HCC mouse models was evaluated by immunohistochemistry. (Scale bar, 25 μm.) (I) The converse ARID2 and Snail expression pattern in clinical HCC tissues were assessed by immunohistochemistry. (J), (K) The correlation of expression between ARID2 and Snail at mRNA (n = 34, P = 0.0046) and protein (n = 287, P < 0.001) levels were analyzed. (L) A survival time comparison between HCC patients with ARID2highSnaillow expression with patients with ARID2lowSnailhigh expression was performed, P = 0.0041. According to the immunostaining intensity of ARID2 and Snail (high, H-score ≥ 38; low, H-score < 38) protein, patients (shown in Fig. 1 E and F) were divided into ARID2highSnailhigh (n = 25), ARID2highSnaillow (n = 59), ARID2lowSnailhigh (n = 181) and ARID2lowSnaillow (n = 22) group. Data were analyzed using Student’s t test. All *P < 0.05, **P < 0.01, ***P < 0.001.
ARID2 Inhibited Snail Transcription in HCC Cells.
Molecular reprogramming occurring during EMT is triggered and orchestrated by EMT-activating transcriptional factors, including Snail family, Twist family, and the zinc finger E-box binding homeobox proteins (17–19). Among them, we found that only the expression of Snail was significantly correlated with ARID2 (Fig. 3 A and B). To further elucidate their relationship, RT-qPCR was used to examine the expression of Snail in cells with modulated ARID2 expression. Forced expression of ARID2 decreased Snail transcription while increasing the transcription of its target gene E-cadherin in MHCC97-H and PVTT cells, whereas ARID2 knockdown in HCC cells and Arid2 knockout in primary hepatocytes increased Snail transcription and decreased E-cadherin transcription (Fig. 3 C and D and SI Appendix, Fig. S4B). In addition, we found that ARID2 potentially binds to the promoter of Snail (Fig. 3E), based on the chromatin immunoprecipitation sequencing (ChIP-seq) data (GSE69568) in HepG2 cells (20). We speculated that ARID2 might regulate the transcription of Snail. Luciferase reporter assay revealed that the activity of Snail promoter was inhibited by ARID2 overexpression and activated in ARID2 knockdown HCC cells (Fig. 3F). Furthermore, metastatic lesions generated by MHCC97-H–ARID2 cells in an intrahepatic metastasis mouse model displayed lower expression of Snail and higher expression of E-cadherin compared with lesions generated by control cells. In contrast, metastatic lesions generated by PLC/PRF/5-shARID2 cells demonstrated a converse Snail and E-cadherin expression pattern (Fig. 3G and SI Appendix, Fig. S4C). Besides, expression of Snail protein was increased while E-cadherin was decreased in pulmonary metastatic lesions of PKA mice and DEN-treated ARID2-knockout mice when compared to control mice (Fig. 3H and SI Appendix, Fig. S4D). In addition, HCC tissues with higher level of ARID2 expression showed lower level of Snail expression, and vice versa (Fig. 3I). Statistical analysis revealed that ARID2 and Snail were inversely correlated at the level of mRNA and protein in clinical samples (Fig. 3 J and K). Finally, HCC patients with ARID2low Snailhigh expression had shortest overall survival time, while the patients with ARID2high Snaillow expression had the longest survival time (Fig. 3L). Therefore, these data disclosed that ARID2 suppressed the expression of EMT-regulator Snail by inhibiting its promoter activity. We also revealed the relationship and the clinical significance of ARID2 and Snail in HCC. To investigate whether ARID2 suppressed HCC metastasis mainly through the regulation of Snail, Snail was overexpressed in ARID2-overexpressing HCC cells and was knocked down with shRNA in ARID2 knockdown HCC cells (SI Appendix, Fig. S5A). Migration and invasion of these HCC cells were assessed by Boyden chamber assay and transwell assay, respectively. We found that the migration and invasion capability of ARID2-overexpressing HCC cells were rescued by forced expression of Snail, while down-regulation of Snail expression effectively inhibited the migration and invasion of ARID2 knockdown HCC cells (SI Appendix, Fig. S5 B and C). Accordingly, overexpression of Snail restored the metastatic capacity of MHCC97-H–ARID2 cells and significantly increased the number of metastatic foci in both injection lobes and noninjection lobes. Down-regulation of Snail expression inhibited the intrahepatic metastasis of PLC/PRF/5-shARID2 cells and effectively reduced the number of foci in injection lobes and noninjection lobes (SI Appendix, Fig. S5D). To further confirm the contribution of Snail to inhibition of HCC metastasis by ARID2, we investigated the distant seeding of HCC cells using tail vein injection model. Forced expression of Snail notably promoted the lung seeding of MHCC97-H–ARID2 cells, while Snail knockdown remarkably reduced the lung seeding of PLC/PRF/5-shARID2 cells (SI Appendix, Fig. S5E). These results suggested that ARID2 inhibited the migration, invasion, and metastasis of HCC cells mainly through the regulation of Snail.
ARID2 Recruited DNMT1 to Snail Promoter to Enhance DNA Methylation.
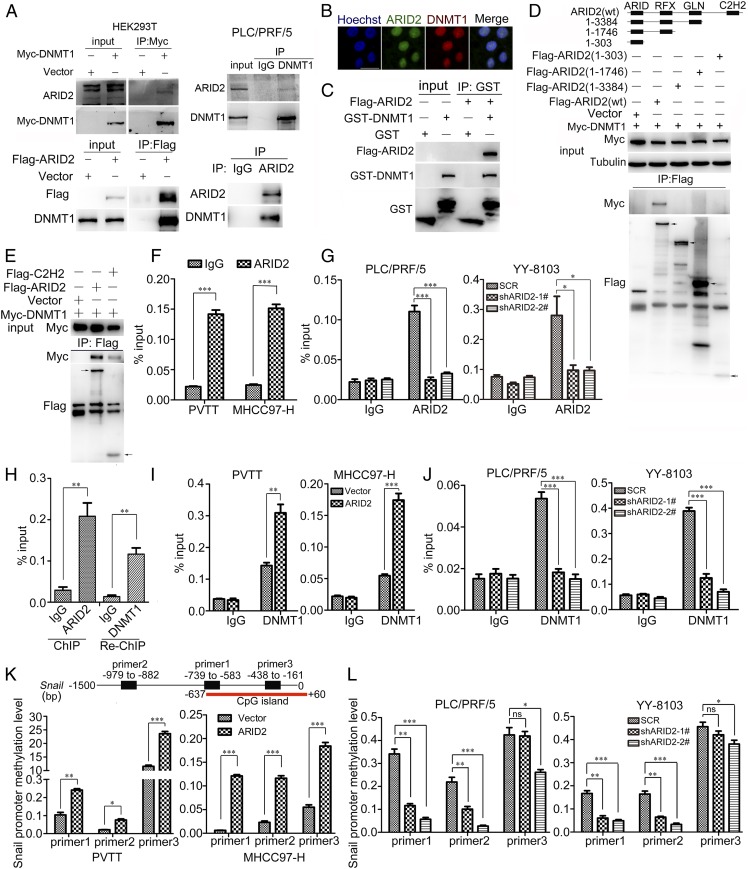
Next, the mechanism underlying transcriptional suppression of Snail by ARID2 was explored. As a subunit of SWI/SNF complex, ARID2 potentially interacted with epigenetic regulators modulating gene transcription (21). Therefore, we examined the interaction between ARID2 and several candidate epigenetic modifiers. We found that ARID2 interacted with DNA methyltransferase DNMT1, either exogenously or endogenously, which was further confirmed by their colocalization in the nucleus (Fig. 4 A and B). Glutathione-S-transferase (GST) pull-down assay further confirmed the direct interaction between ARID2 and DNMT1 (Fig. 4C). Moreover, various truncated mutants of ARID2 were constructed, and truncation assay demonstrated that C2H2 domain was indispensable for ARID2–DNMT1 interaction (Fig. 4D). Furthermore, we detected interaction between DNMT1 and C2H2 domain (Fig. 4E). These findings suggested that the interaction between ARID2 and DNMT1 was mediated via C2H2 domain.
Fig. 4.
ARID2 up-regulated the methylation of Snail promoter by recruiting DNMT1. (A) Endogenous ARID2 interacted with exogenous and endogenous DNMT1. (B) Localization of ARID2 and DMNT1 protein in HEK293T was examined by immunofluorescence assay. (Scale bar, 25 μm.) (C) GST pull-down assay was performed to verify the direct interaction between ARID2 and DNMT1. (D) Coimmunoprecipitation was used to examine the interaction between exogenous DNMT1 and truncations of ARID2 in HEK293T cells. Truncations of ARID2 were constructed as shown in graphic. (E) The interaction between C2H2 domain and DNMT1 was examined by coimmunoprecipitation in HEK293T cells. (F and G) The binding of ARID2 on Snail promoter in ARID2-overexpressing and knockdown HCC cells was examined by ChIP assay. (H) ChIP-Re-ChIP assay was used to verify that the ARID2–DNMT1 interaction occurred at Snail promoter. (I and J) The binding of DNMT1 on Snail promoter in ARID2-overexpressing and knockdown HCC cells was examined by ChIP assay. (K and L) MeDIP-PCR was performed to assess the methylation level of Snail promoter in ARID2-overexpressing and knockdown HCC cells. CpG island (−637∼+60 bp) was shown in diagram. Data were analyzed using Student’s t test. All *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant.
The previous ChIP-seq data suggested direct binding of ARID2 to the promoter region of Snail (20), and chromatin immunoprecipitation (ChIP) assay confirmed this possibility. An enrichment of ARID2 protein was observed at −740 to −615 bp upstream of the transcription start site of Snail gene in MHCC97-H and PVTT cells, while ARID2 occupancy at this region was reduced in PLC/PRF/5 and YY-8103 ARID2 knockdown cells (Fig. 4 F and G). Furthermore, ChIP-Re-ChIP assay suggested that DNMT1 co-occupied at the same promoter region of Snail with ARID2 (−740 to −615 bp) (Fig. 4H), which overlapped with CpG island (−637 to +60 bp). In addition, we observed increased binding of DNMT1 to the Snail promoter in ARID2-overexpressing HCC cells while decreased binding in ARID2 knockdown cells, suggesting that ARID2 recruited DNMT1 to Snail promoter (Fig. 4 I and J). DNA methylation of promoter is commonly associated with gene silencing (15). We found that DNA methylation approximate to the binding site of ARID2 and DNMT1 in Snail promoter was up-regulated in ARID2-overexpressing HCC cells and down-regulated in ARID2-knockdown HCC cells, which was coincident with DNMT1 binding (Fig. 4 K and L). Therefore, ARID2 suppressed Snail transcription by recruiting DNMT1 to Snail promoter, which resulted in elevated promoter methylation in HCC cells.
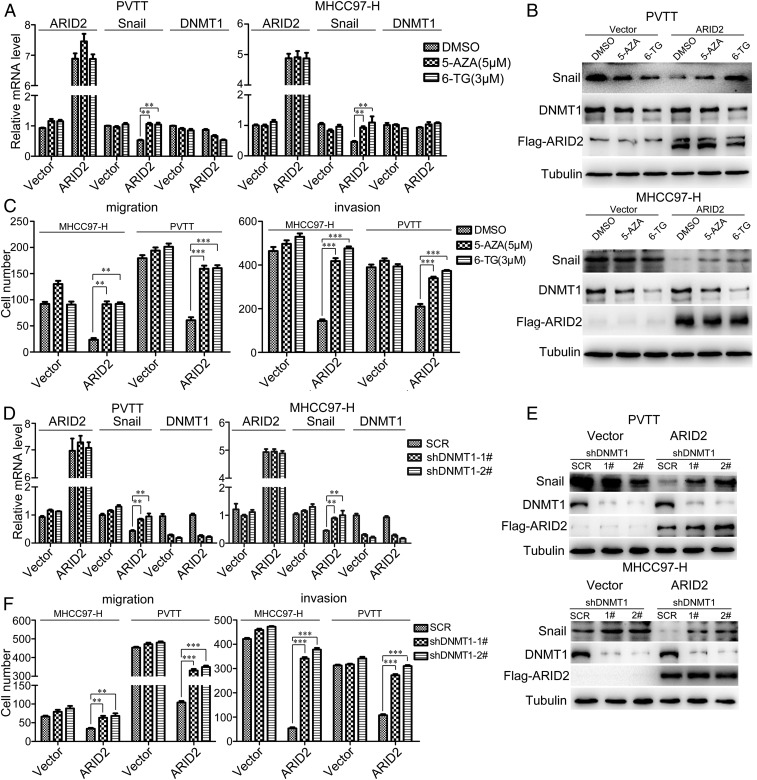
Inhibition of DNMT1 Rescued the Suppression of Migration and Invasion by ARID2 in HCC Cells.
To further evaluate the involvement of DNMT1 in the metastasis suppressor role of ARID2, we examined whether loss of DNMT1 could restore the phenotype and Snail expression in ARID2-overerxpressing HCC cells. Two inhibitors of DNMT1, 5-azacitidine (5-AZA) and 6-thioguanine (6-TG), decreased the expression of DNMT1 protein and increased the mRNA and protein level of Snail in ARID2-overexpressing MHCC97-H and PVTT cells (Fig. 5 A and B). Moreover, the inhibition of migration and invasion by ARID2 in HCC cells were relieved upon inhibitor treatment (Fig. 5C and SI Appendix, Fig. S6A). To further confirm these observations, we knocked down DNMT1 in HCC cells which overexpressed with ARID2 and also found that mRNA and protein level of Snail were notably increased (Fig. 5 D and E). In addition, DNMT1 knockdown rescued the suppressed migration and invasion in ARID2-overexpressing HCC cells (Fig. 5F and SI Appendix, Fig. S6B). Our data suggested that either inhibition of DNMT1 activity or down-regulation of DNMT1 expression could restore the inhibition of migration and invasion by ARID2 in HCC cells via restoring Snail expression.
Fig. 5.
Inhibition of DNMT1 rescued the suppression of migration and invasion by ARID2 in HCC cells. (A and D) RT-qPCR was performed to investigate the mRNA levels of ARID2, DNMT1 and Snail of ARID2 overexpressing HCC cells and control cells treated with shRNA targeted DNMT1 or DNMT1 inhibitors 5-AZA (5 μM) and 6-TG (3 μM) for 48 h. (B and E) The alterations of Snail and DNMT1 protein level were examined by Western blot in ARID2-overexpressing HCC cells and control cells treated with shRNA targeted DNMT1 or DNMT1 inhibitors for 48 h. (C and F) Boyden chamber and transwell assay were employed to assess the migration and invasion of ARID2-overexpressing MHCC97-H and PVTT cells which DNMT1 was inhibited by shRNA or treated with DNMT1 inhibitors for 48 h. Data were analyzed using Student’s t test. All **P < 0.01, ***P < 0.001.
ARID2 Mutation Disrupting C2H2 Domain Was Positively Associated with HCC Metastasis.
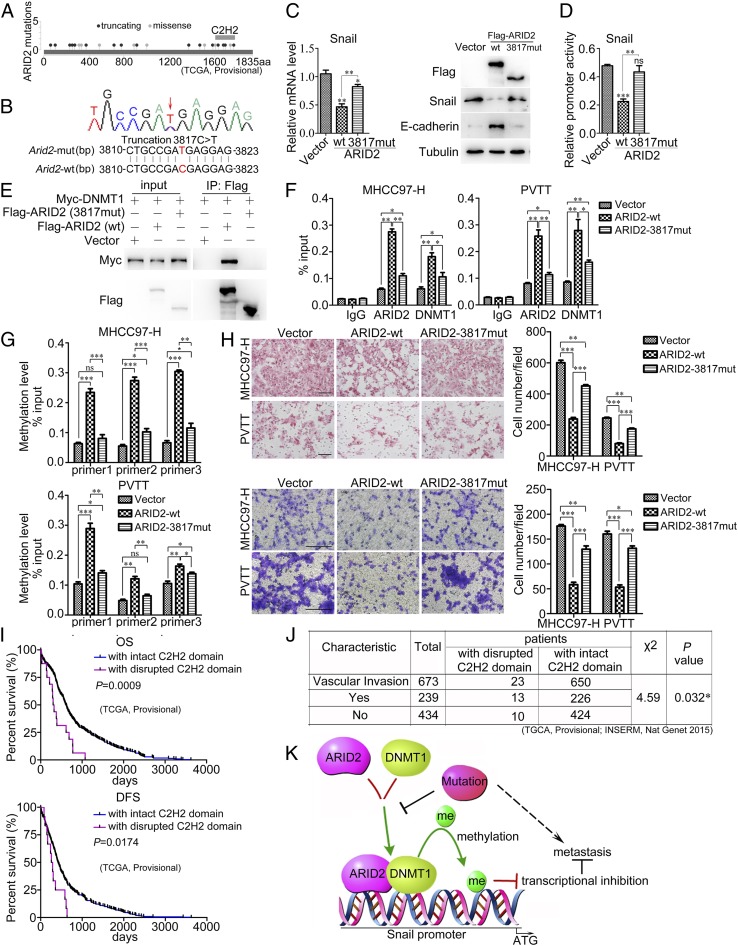
ARID2 shows high mutation frequency in HCC. Based on the data from TCGA database (TGCA; Provisional), we found that the mutations were distributed randomly throughout ARID2 gene. However, approximate 40% (6/16) truncated mutations were enriched at the zinc finger domain of ARID2 (C2H2 domain) and over 60% (16/24) mutations led to disruption of C2H2 domain, suggesting that C2H2 domain was crucial for ARID2 (Fig. 6A). We discovered a truncated mutation of ARID2 (3817C > T or R1273*) in a clinical PVTT tissue-derived cell by exon sequencing, which was also reported by TCGA data (TCGA-G3-A5SL-01) (Fig. 6B). We constructed this truncation mutation and examined the distinct features of this mutant from wild-type ARID2. RT-qPCR and Western blot disclosed that 3817-truncated mutant showed little influence on Snail expression (Fig. 6C). Accordingly, transcriptional activity of Snail promoter was unaffected by 3817-truncated mutant compared with vector control (Fig. 6D). Moreover, no interaction was detected between 3817-truncated mutant and DNMT1 by Co-IP; 3817-truncated mutation disturbed the interaction between ARID2 and DNMT1 (Fig. 6E). Furthermore, DNMT1 occupancy at Snail promoter was remarkably weakened by 3817-truncated mutant compared with wild-type ARID2 in HCC cells, this indicated that 3817-truncated mutation impeded the recruitment of DNMT1 to Snail promoter mediated by ARID2 (Fig. 6F). Simultaneously, DNMT1-mediated promoter methylation of Snail was also decreased in HCC cells overexpressing 3817-truncated mutant compared with those overexpressing wild-type ARID2 (Fig. 6G). Furthermore, 3817-truncated mutant only slightly decreased the migration and invasion of HCC cells, and the suppressive effect was much weaker than wild-type ARID2 (Fig. 6H). In addition, we analyzed the TCGA data and found that ARID2 mutations disrupting C2H2 domain were positively associated with vascular metastasis of cancer cells (TGCA, Provisional; INSERM, Nat Genet 2015), poor overall survival, and disease-free survival of HCC patients (TCGA, Provisional) (Fig. 6 I and J). Taken together, these data revealed that C2H2 mutations abolished the metastasis suppressor function of ARID2 by losing the ability to recruit DNMT1 to Snail promoter.
Fig. 6.
ARID2 mutation disrupting C2H2 domain was positively associated with HCC metastasis. (A) ARID2 mutations shown in TCGA data (Provisional); (B) 3817C > T mutation was detected in a clinical PVTT tissue-derived cell by exon sequencing. (C) mRNA and protein levels of Snail were investigated in HEK293T cells transfected with wild-type ARID2, ARID2 3817-mutant, or vector. (D) Activity of Snail promoter in HEK293T cells transfected with wild-type ARID2, ARID2 3817-mutant or vector. (E) Coimmunoprecipitation was employed to examine the interaction of ARID2 3817-mutant and DNMT1. (F and G) Effects of the binding of DNMT1 and methylation on Snail promoter in MHCC97-H and PVTT cells mediated by ARID2 3817-mutant were examined by ChIP and MeDIP-PCR assay, respectively. (H) The migration and invasion of HCC cells forced expression of ARID2 3817-mutant were assessed by Boyden chamber and transwell assay. (Scale bar, 100 μm.) (I) According to database (TGCA; Provisional), both OS (P = 0.0009) and DFS (P = 0.0174) survival of HCC patients with disrupted C2H2 domain (OS, n = 16; DFS, n = 12) and with intact C2H2 domain (OS, n = 362; DFS, n = 315) were analyzed. (J) Based on database (TGCA, Provisional and INSERM, Nat Genet 2015), statistical analysis was used to verify the correlation between ARID2 mutations disrupted the C2H2 domain and HCC metastasis, P = 0.032. (K) Schematic representation of the molecular mechanism of wild-type ARID2 and ARID2 loss-of-function mutants in HCC metastasis. Data were analyzed using Student’s t test. All *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Mutation ratio of SWI/SNF complex ranked third in cancers (22). As a component of SWI/SNF complex, ARID2 had a high mutation frequency in HCC, and these mutations often resulted in partial or complete loss of physiological function (23). Therefore, ARID2 is supposed to play a vital role in carcinogenesis and cancer progression. As previously reported, ARID2 modulated DNA damage and cell cycle in HCC, which contributed its suppressor function. However, the role of ARID2 in HCC metastasis remains largely unknown. Our study showed that ARID2 expression was further reduced in metastatic HCC tissues when compared to primary HCC tissues, and its expression was negatively correlated with extrahepatic metastasis of HCC, while positively associated with survival of HCC patients. These findings indicated an important role of ARID2 in HCC metastasis. This speculation was confirmed by the suppression on migration and invasion of HCC cells in vitro, and repression of intrahepatic metastasis and pulmonary seeding of HCC cells in vivo by ARID2. We also demonstrated the metastasis suppressor role of ARID2 in different primary HCC mouse models. Therefore, we disclosed a metastasis suppressor function of ARID2 in HCC in this work.
EMT process was positively correlated with the metastasis of cancer cells and altered the expression of factors involved in maintaining the epithelial or mesenchymal characteristics (18). Recently, it was indicated that deletion of ARID2 elevated the expression of genes involved in EMT in mouse melanoma cells (24). Consistently, we clarified that ARID2 repressed the EMT of HCC cells. After screening the major EMT-activating transcription factors (ATFs), including members of Snail, Zeb, and Twist family, and analyzing the previous ChIP-seq data (GSE69568) in HepG2 cells, we found that only Snail exhibited mutually exclusive expression pattern with ARID2 in HCC cells with modulated ARID2 expression. We also observed the similar expression pattern of Snail and ARID2 in metastatic lesions in mouse models and clinical samples. Interestingly, ARID2highSnaillow HCC patients showed the longest survival time, while the shortest survival time was observed in ARID2lowSnailhigh patients. All these evidence uncovered the inverse correlation between Snail and ARID2 and revealed the prognostic significance of these two molecules.
Chromatin remodeling molecules often regulate gene expression cooperating with epigenetic modulators. E2F4–RBL2–HDAC1–BRM complex suppressed PRAP1 transcription (25), mSin3A/HDAC2 and PRMT5-containing BRG1 complex was involved in transcriptional repression of the Myc target gene cad (26). To find the epigenetic regulator involved in the regulation of Snail transcription by ARID2, we examined the interaction between ARID2 and several candidates. Finally, we identified DNMT1 as the epigenetic modulator assisting ARID2. DNMT1 mediated DNA methylation maintaining in CG dinucleotide-rich regions for transcriptional repression (27). Recent studies reported that reduced expression of DNMT1 and DNA methylation induced EMT in prostate cancer (16, 28), but the downstream gene and regulation manner of DNMT1 in EMT were unclear. We confirmed the interaction between ARID2 and DNMT1. Furthermore, we verified the co-occupancy of ARID2 and DNMT1 at the same region of Snail promoter. In addition, the binding of DNMT1 to Snail promoter enhanced DNA methylation and was regulated by ARID2 expression. These suggested that DNMT1 was recruited to Snail promoter by ARID2. To verify the involvement of DNMT1 in the metastasis suppressor role of ARID2, we used DNMT1 inhibitors and modulated the expression of DNMT1. Both of them demonstrated that inhibition of DNMT1 rescued the inhibition of migration and invasion by ARID2. Taken together, our work disclosed a mechanism underlying EMT suppressor function of ARID2: ARID2 recruited DNMT1 to Snail promoter, elevated the DNA methylation, and suppressed Snail transcription, leading to subsequent repression of EMT.
A previous study reported that Snail promoter CpG island was hypermethylated during the early stage of tumorigenesis and was demethylated when cancer cells progressed from epithelial phenotype to a highly metastatic spindle morphology at the latter stage of tumorigenesis (29). In our study, we revealed that ARID2 repressed the metastasis of HCC cells by recruiting DNMT1 to Snail promoter, which increased promoter methylation and inhibited Snail transcription. ARID2 shows high mutation frequency in HCC, and the mutations disrupting C2H2 domain could not recruit DNMT1 to Snail promoter, leading to decreased methylation of Snail promoter, which was associated with vascular metastasis and poor prognosis. Meanwhile, inhibition of DNMT1 increased Snail expression and relieved the inhibition of migration and invasion by ARID2. Based on these findings, we proposed a hypothesis: ARID2 loss or ARID2 mutation with disrupted C2H2 domain occurred during HCC progression, which impaired recruitment of DNMT1 to Snail promoter, leading to decreased methylation of Snail promoter CpG island and increased Snail expression. Therefore, our observations were consistent with ref. 29, and provided a potential explanation for DNA methylation alterations in promoter regions of Snail when cancer cells obtained the metastasis capacity at later stage of tumor progression.
Although ARID2 was highly mutated in HCC, the function of its mutations remains largely unknown. Domain mapping disclosed that the C2H2 domain was required for ARID2–DNMT1 interaction. ARID2 mutation was randomly localized throughout the gene; however, we found that about 40% truncated mutation occurred approximately to the C2H2 domain and over 60% mutations disrupted the C2H2 domain. Interestingly, mutations disrupting the C2H2 domain were positively associated with HCC metastasis and poor prognosis of HCC patients. Coincidently, we found that one of these mutations, 3817C > T, lost the ability to interact with DNMT1. Subsequently, this mutation showed little effect on Snail transcription as well as migration and invasion of HCC cells. It seemed that the metastasis suppressor function of ARID2 was abolished by disruption of C2H2 domain, which was consistent with the correlation between the mutations and survival of HCC patients.
In conclusion, we identified the metastasis suppressor role of ARID2 in HCC and disclosed the underlying mechanism. ARID2 recruited DNMT1 to the promoter of Snail, suppressed Snail transcription by enhancing DNA methylation, leading to repression of EMT. In addition, we clarified the loss-of-function mechanism for ARID2 mutants and the correlation between ARID2 mutations disrupted the C2H2 domain and HCC metastasis (Fig. 6K). About 60% mutations of ARID2 were C2H2 truncation, thus our findings may provide far-reaching implications to therapeutic targets for ARID2-mutant HCC.
Materials and Methods
Cell Lines and Tumor Samples.
PVTT cells was established by our laboratory, the details of establishing PVTT cell line and clinical information about the patient who provided the tumor tissue has been described previously (30). HEK293T and other human HCC cells were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai Institute of Cell Biology, Chinese Academy of Sciences. Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco) supplemented with 1% antibiotics and 10% fetal bovine serum (FBS) were used to culture the cells, the culture incubator was sterilized at 37 °C with 5% CO2.
After obtaining written informed consent, all HCC and paired adjacent tissues were collected from Eastern Hepatobiliary Surgery Hospital, Second Military Medical University (Shanghai, China) between 2013 and 2015. To evaluate the expression of ARID2 and Snail, we examined the tissues array with different cohort of HCC patients and analyzed the correlation between ARID2 and clinical features, Snail expression. These experiments were approved by the Ethical Committee of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) following the declaration of Helsinki ethical guidelines.
Statistical Analysis.
The correlations between the clinicopathological features and ARID2 staining scores were analyzed using the chi-square (χ2) test. Survival curves were plotted by the Kaplan–Meier method and analyzed by the log-rank test. Statistical analyses were performed by GraphPad Prism 5 and SPSS17.0 software. The results are representative of at least three independent experiments performed in triplicate and are expressed as the means ±SD. The data were analyzed using Student’s t test.
Data Availability.
Correlation between ARID2 expression and clinicopathologic features of HCC is available in SI Appendix, Table S1. Detail of plasmid generation is described in SI Appendix, Methods and Materials. All data used for the study are within the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFC1604404 and 2018YFC1603002); the “Personalized Medicines-Molecular Signature-based Drug Discovery and Development,” Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDA12010316); National Natural Science Foundation of China (31520103907, 81730083, 31771538, and 81972757); Youth Innovation Promotion Association of Chinese Academy of Sciences fund and Sanofi-SIBS 2018 Young Faculty Award to J.-J.L.; and Postdoctoral Science Foundation of China (2017M622677).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914937117/-/DCSupplemental.
References
- 1.Bray F., et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Nieto M. A., Huang R. Y., Jackson R. A., Thiery J. P., Emt: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Aldrighetti L., et al. , Liver resection with portal vein thrombectomy for hepatocellular carcinoma with vascular invasion. Ann. Surg. Oncol. 16, 1254 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Yang J., Weinberg R. A., Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Zhang X., Azhar G., Zhong Y., Wei J. Y., Zipzap/p200 is a novel zinc finger protein contributing to cardiac gene regulation. Biochem. Biophys. Res. Commun. 346, 794–801 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Duan Y., et al. , Chromatin remodeling gene ARID2 targets cyclin D1 and cyclin E1 to suppress hepatoma cell progression. Oncotarget 7, 45863–45875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., et al. , MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int. J. Oncol. 48, 2425–2434 (2016). [DOI] [PubMed] [Google Scholar]
- 8.He L., et al. , BAF200 is required for heart morphogenesis and coronary artery development. PLoS One 9, e109493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oba A., et al. , ARID2 modulates DNA damage response in human hepatocellular carcinoma cells. J. Hepatol. 66, 942–951 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Cajuso T., et al. , Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int. J. Cancer 135, 611–623 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Li M., et al. , Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodis E., et al. , A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hargreaves D. C., Crabtree G. R., ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 21, 396–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bigey P., Ramchandani S., Theberge J., Araujo F. D., Szyf M., Transcriptional regulation of the human DNA Methyltransferase (dnmt1) gene. Gene 242, 407–418 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Szyf M., Detich N., Regulation of the DNA methylation machinery and its role in cellular transformation. Prog. Nucleic Acid Res. Mol. Biol. 69, 47–79 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Lee E., et al. , DNMT1 regulates epithelial-mesenchymal transition and cancer stem cells, which promotes prostate cancer metastasis. Neoplasia 18, 553–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z. F., Behringer R. R., Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9, 686–699 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Peinado H., Olmeda D., Cano A., Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Spaderna S., et al. , The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 68, 537–544 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Raab J. R., Resnick S., Magnuson T., Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet. 11, e1005748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson B. G., Roberts C. W., SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Guichard C., et al. , Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44, 694–698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao H., et al. , ARID2: A new tumor suppressor gene in hepatocellular carcinoma. Oncotarget 2, 886–891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan D., et al. , A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiśnik E., Płoszaj T., Robaszkiewicz A., Downregulation of PARP1 transcription by promoter-associated E2F4-RBL2-HDAC1-BRM complex contributes to repression of pluripotency stem cell factors in human monocytes. Sci. Rep. 7, 9483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pal S., et al. , mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23, 7475–7487 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharif J., et al. , The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Ateeq B., Unterberger A., Szyf M., Rabbani S. A., Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia 10, 266–278 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraga M. F., et al. , A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 64, 5527–5534 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Wang T., et al. , Characterisation of a novel cell line (CSQT-2) with high metastatic activity derived from portal vein tumour thrombus of hepatocellular carcinoma. Br. J. Cancer 102, 1618–1626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Correlation between ARID2 expression and clinicopathologic features of HCC is available in SI Appendix, Table S1. Detail of plasmid generation is described in SI Appendix, Methods and Materials. All data used for the study are within the manuscript and SI Appendix.