Significance
The application of heterosis (hybrid vigor) in hybrid rice since the 1970s has tremendously improved rice productivity worldwide. But how breeders construct hybrid parents to obtain hybrid rice heterosis remains unclear. Here, by genome analysis, we found that breeders introduced different introgressed exogenous genomes of other rice subpopulations to construct male and female parents. The differentiated introgression in parents shaped heterotic loci in the hybrid rice. Genetic origin analysis revealed that heterotic loci existed in wild rice and were divergently selected among rice subpopulations. Our results traced the origin of heterotic loci of hybrid rice and uncovered genetic change of heterotic loci across rice evolution and breeding stages, which could facilitate the future breeding of more superior hybrid rice varieties.
Keywords: hybrid rice, heterosis, divergent selection, genetic introgression
Abstract
The successful application of heterosis in hybrid rice has dramatically improved rice productivity, but the genetic mechanism for heterosis in the hybrid rice remains unclear. In this study, we generated two populations of rice F1 hybrids with present-day commercial hybrid parents, genotyped the parents with 50k SNP chip and genome resequencing, and recorded the phenotype of ∼2,000 hybrids at three field trials. By integrating these data with the collected genotypes of ∼4,200 rice landraces and improved varieties that were reported previously, we found that the male and female parents have different levels of genome introgressions from other rice subpopulations, including indica, aus, and japonica, therefore shaping heterotic loci in the hybrids. Among the introgressed exogenous genome, we found that heterotic loci, including Ghd8/DTH8, Gn1a, and IPA1 existed in wild rice, but were significantly divergently selected among the rice subpopulations, suggesting these loci were subject to environmental adaptation. During modern rice hybrid breeding, heterotic loci were further selected by removing loci with negative effect and fixing loci with positive effect and pyramid breeding. Our results provide insight into the genetic basis underlying the heterosis of elite hybrid rice varieties, which could facilitate a better understanding of heterosis and rice hybrid breeding.
The phenomenon that hybrid progenies often perform better (e.g., higher growth rate or biomass) than their homozygous parents, which is known as heterosis (or hybrid vigor), is widespread in crop plants like rice and maize (1–4). The successful application of heterosis in hybrid crop breeding has improved food production during the past several decades. Today, hybrid seeds are used in about half of all rice crops in China and nearly all maize crops in the United States (5, 6). Historical efforts to genetically improve hybrid parental lines have led to significant increases in commercial hybrid rice production in China, from ∼3.8 t/ha in the 1970s to ∼6.8 t/ha in recent years (7). Recent studies have mapped hundreds of agronomical traits heterotic quantitative trait loci (QTLs) whose performance of heterozygous genotypes differed from the mean performance of two homozygous genotypes in modern rice hybrids (1, 2, 8–10). Many QTLs contribute to heterosis by dominant or overdominant effects, with some exhibiting strong heterotic effects and including candidate genes implicated in important agronomical traits such as grain yield and flowering time (8–10). But the mechanisms that explain how past breeding efforts led to heterotic QTLs in modern hybrids and how these heterotic QTLs improved hybrid performance remain unclear.
Rice genome studies in recent years have detected a large scale of genetic variations in Asian cultivated rice germplasm (11–14). By population structure analysis, the Asian cultivated rice germplasm could be divided into six canonical subpopulations (11, 13, 14): three indica subpopulations (South China origin, International Rice Research Institute [IRRI]-bred lines, and South Asia origin/Southeast Asia origin indica subpopulations), two japonica subpopulations (tropical and temperate japonica), and aus, most of which associated with their geographic origin, indicating rich diversity. Based on the pedigree records, the indica subpopulation from south China and the one which comprises IRRI-bred lines, were frequently employed during modern rice breeding. An investigation of genomewide breeding signatures at these two subpopulations found numerous selected genomic regions which encompassed large numbers of important functional genes and QTLs and may be the breeding targets during modern rice genetic improvement (13). These studies enhanced our understanding toward the population structure and genetic diversity of rice germplasm, but our knowledge about how breeders utilized the genetic diversity to breed the hybrid parents, and what genomic regions/genes had been selected during the parental breeding, is still limited.
In this study, we constructed two rice hybrid populations comprising ∼1,000 hybrids each using current commercial hybrid parents. We then genotyped the hybrid parents using a 50k SNP chip (RiceSNP50) (15) and genome resequencing, and conducted phenotyping of ∼2,000 hybrids based on nine important agronomical traits in three experimental trials. Our genetic analysis indicates that divergent selection among the subpopulations shaped different heterotic alleles in different subpopulations. The male and female parents of hybrid rice had genetic variations, when females displayed a much higher level of genome introgression from other subpopulations, which gave rise to allele differences at heterotic loci between male and female parents and shaped heterotic loci in the hybrids. Heterotic loci were then selected based on hybrid performance, which fixed heterotic QTLs but removed QTLs that caused hybrid depression. Our findings reveal the mechanisms underlying how heterotic loci were developed in hybrid rice and how hybrid performance was improved with intensive breeding efforts. These results yield genetic insights into hybrid rice heterosis and may lead to improved breeding of parental lines that produce superior hybrids.
Results
Heterosis in Hybrid Rice Was Contributed by Multiple Loci with Positive Dominant/Overdominant Effects.
To study the genetic basis of heterosis in hybrid rice, we employed present-day commercial male and female parental lines and constructed two populations of rice F1 hybrids (designated Pop I and II, SI Appendix, Table S1 and Dataset S1), which included ∼1,000 F1 hybrids each. Pop I is comprised of 53 three-line hybrids and 947 two-line hybrids, and Pop II is comprised of two-line hybrids only. The female parents of two-line hybrids in Pop I shared one of the derived parents with the female parents of Pop II, but the male parents of Pop I are totally difference from that of Pop II. We performed phenotyping for 10 important agronomical traits in the hybrids from three experimental field trials (Pop I was phenotyped at ChangSha City, China in the year 2014 [denoted as 2014CS], Pop II was phenotyped at ChangSha City, China in the year 2015 [denoted as 2015CS], and HeFei City in the year 2015 [denoted as 2015HF]) over a period of 2 y. Then all 171 male and 104 female parents (including 11 three-line and 93 two-line male sterility lines, Dataset S1) were genotyped using a 50k SNP array, and the hybrid genotypes were obtained by combining the haploid genotypes of both parents (SI Appendix).
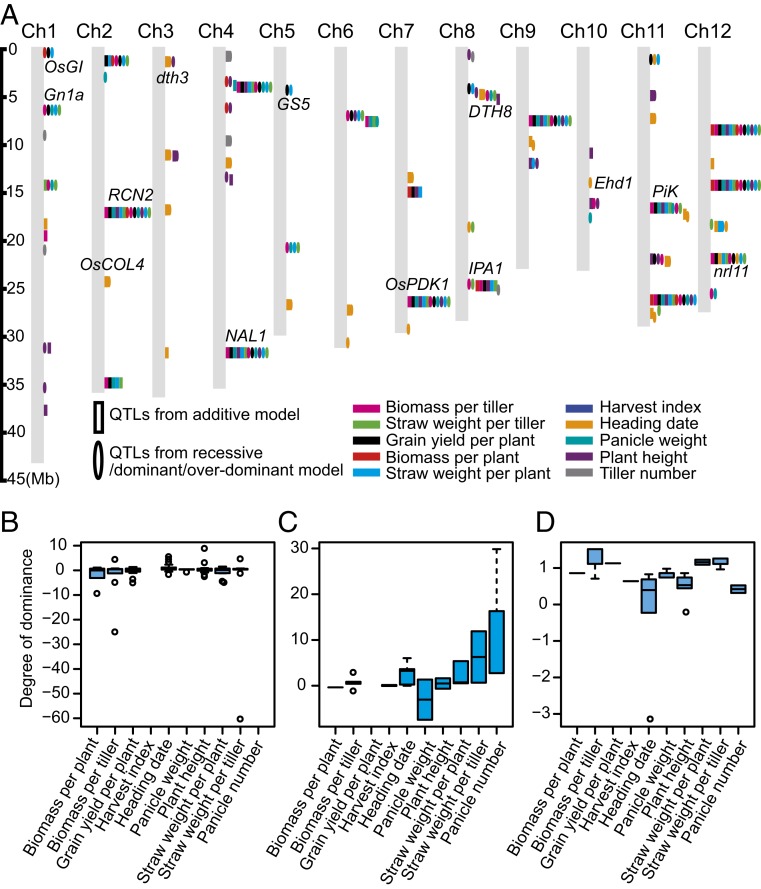
To identify heterotic loci whose trait value of heterozygous genotype differed from the mean performance of homozygous genotypes in the hybrids, we performed additive, dominant, and overdominant model genomewide association studies (GWAS) using EMMAX (16), which corrected cryptic genetic relatedness. The GWAS analyses revealed 143 significant loci including 50 pleiotropic QTL clusters (genomewide significance cutoff: P value ≤10−5; false discovery rate <0.05; 300 repetitions of permutation test; Fig. 1A and SI Appendix, Fig. S1). These loci overlapped a wide range of canonical genes previously implicated in important agronomical traits, including Ghd8/DTH8 (17, 18), NAL1 (19), IPA1 (20), Gn1a (21), and RCN2 (22), and some of these loci were also detected at previous studies in indica hybrid populations (9, 10). An evaluation of the dominant/overdominant effects across loci with three genotypes (two parental homozygous genotypes and one heterozygous genotype) across all traits revealed that the number of positive dominant/overdominant loci was higher than negative dominant/overdominant loci (Fig. 1B). This supports the notion that heterosis was the overall net effect of multiple loci (8).
Fig. 1.
A genomewide association study was used to identify potential heterotic loci in the hybrids. (A) Chromosomal distribution of heterotic loci identified in the 2014CS trial. Ellipses indicate the heterotic loci detected using the dominant/overdominant model, while rectangles represent those detected using the additive model. Different colors represent different traits. (B–D) Distribution of the degree of dominance of heterotic loci in the 2014CS (B), 2015CS (C), and 2015HF (D) trials. Only loci representing all three genotypes (one heterozygous and two homozygous genotypes) in the hybrids were investigated.
Male and Female Parents of Hybrids Have Different Levels of Genome Introgression from Rice Subpopulations Which Relate to Heterosis in Hybrids.
We inferred the population structure of the parents of the hybrids based on previously reported resequencing data for landrace strains and improved varieties (4,214 lines in total) (11–14) by using the ADMIXTURE tool (23). In accordance with recent studies (13, 24), the landraces and improved varieties exhibited six canonical distinct groups (SI Appendix): three indica groups (designated Ind I, Ind II, and Ind III), two canonical japonica groups (Tropical Japonica and Temperate Japonica, designated TroJ and TemJ, respectively), and aus (SI Appendix, Fig. S2A). Ind I predominantly comprised the landraces and conventional rice from South China, Ind II predominantly comprised IRRI-bred germplasm including elite varieties like IR8 and IR24, and Ind III predominantly comprised landraces from South Asia and Southeast Asia.
Based on the population structure of landraces and improved varieties, both the male and female parents of the hybrids displayed high proportion of the Ind II genome (an average 94.26% in male parents and 77.43% in female parents, Fig. 2A). However, the parents displayed significantly different levels of population admixture, with the male parent showing a low level of genome introgression from other subpopulations (average 5.74% of the genome, range from 0 to 37.21%) and the female parent showing a high level (average 22.57% of the genome, range from 2.86 to 46.40%). In the male parent, aus contributed an average of 2.34% exogenous genome introgression (range from 0 to 8.09%), Ind I contributed 1.46% (range from 0 to 17.36%), Ind III contributed 0.85% (range from 0 to 11.23%), TroJ contributed 0.60% (range from 0 to 10.78%), and TemJ contributed 0.50% (range from 0 to 9.81%). For the female parent, aus contributed 4.88% (range from 1.90 to 9.25%), Ind I contributed 14.49% (range from 0 to 34.26%), Ind III contributed 0.31% (range from 0 to 8.00%), TroJ contributed 1.71% (range from 0 to 9.30%), and TemJ contributed 0.50% (range from 0 to 7.52%). Overall, hybrid male and female parents displayed different levels of exogenous genome introgression, which were introduced by breeders from Asian cultivated rice.
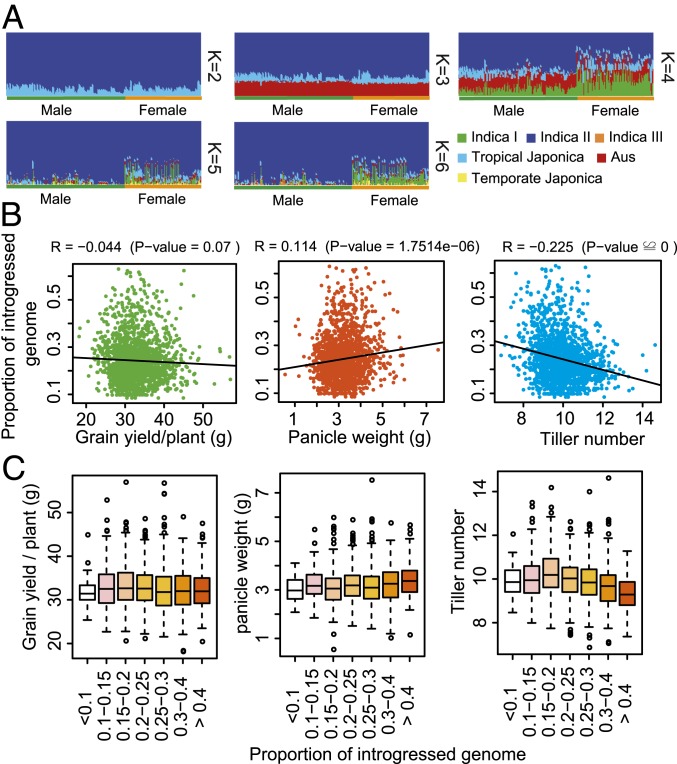
Fig. 2.
The heterotic level of hybrids increased with increasing exogenous genome introgression. (A) Population structure difference between male and female parents under different numbers of ancestor populations (K represents the prior number of ancestor populations). All 171 male and 104 female parents were integrated with ∼4,200 previously reported resequenced landraces and improved varieties to conduct the population structure analysis. The reasonable number of ancestor populations was determined using five-fold crossvalidation. Only SNPs on the 50k SNP chip were used to conduct the population structure analysis. Aus represents the south Asia origin aus rice subpopulation. (B) Correlation between degree of genome introgression and grain yield per plant (Left), panicle weight (Middle), and tiller number (Right) of hybrids. The effects of year, location, and population were regressed out using a general linear model for each trait, and the trait values for all hybrids in the study were used to conduct the analysis. (C) Comparison of hybrid yield traits with level of exogenous genome introgression. The yield traits included grain yield per plant (Left), panicle weight (Middle), and tiller number (Right). Hybrids were grouped by their cumulative levels of exogenous genome introgression (male parent + female parent). The bar in the middle of each box plot indicates the 50th quantile of the trait value for the group.
As the three-line and two-line male sterility lines are two different hybrid systems during hybrid breeding, the comparison of their population structure showed that the three-line male sterility lines (38.13% of exogenous introgressed genome and 29.02% of Ind I genome on average) usually have higher proportion of exogenous introgressed genome and Ind I genome than these of two-line male sterility lines (20.91% of exogenous genome and 13.30% of Ind I genome on average). However, the male parents of both types have no significant difference in the proportion of exogenous introgressed genome.
To evaluate the impact of exogenous genome introgression on hybrids, we investigated the correlation between the degree of genome introgression (proportion of nonInd II genome) and yield traits in the hybrids. We found that the degree of genome introgression significantly positively correlated with panicle weight (PW) and significantly negatively correlated with tiller number (TN) (Fig. 2B), suggesting that a medium level of introgression may exert a maximum increasing effect on grain yield. To test this hypothesis, we divided the hybrids into groups based on their levels of genome introgression: ≤10%, 10–15%, 15–20%, 20–25%, 25–30%, 30–40%, and ≥40% of the genome. Hybrids with 15–20% introgressed genome had the highest grain yield per plant (GYPP) and TN, and the lowest PW (Fig. 2C). In addition, because the general combining ability (GCA) of parents is an indicator of the heterotic potential of their hybrids, we also investigated the relationship between the GCA of male and female parents and their degree of genome introgression. We found that the degree of genome introgression of both parents correlated positively with the GCA of PW and correlated negatively with the GCA of TN (SI Appendix, Fig. S2 B–D and Tables S1 and S2). Hybrid parents with a medium level of introgressed genome had the highest grain yield GCA (SI Appendix, Fig. S2 D and F). Thus, we suggested that the level of grain yield heterosis in hybrids and the grain yield GCA of the parents could be improved by increasing the level of exogenous genome introgression in parents up to a certain point. Introducing exogenous genomes into parents of hybrids and considering the tradeoff between PW and TN heterosis is crucial for increasing the yield GCA of parents and the yield heterosis of hybrids.
Genome Introgression from Rice Subpopulations Shaped Heterotic Loci in Hybrids.
To detect introgressed regions in parent genomes, we resequenced 36 core female parental lines and 79 male lines (Dataset S1), removed the parental lines of three-line hybrids and screened introgressed regions using the four-taxon fd statistic, which calculates excessively shared derived variants between two taxa (25). We calculated the fd statistic with a window size of 25 kb and a step of 10 kb, removed windows with fewer than three informative SNPs or with a meaningless result (fd > 1, fd < 0 or with Patterson’s D statistic <0) (25), and used the population admixture result (see SI Appendix) to determine the fd cutoff for detecting introgressed regions. In total, we detected 349 (16.80 Mb) and 664 (37.42 Mb) introgressed regions in male and female parents, respectively (Fig. 3A and Datasets S3 and S4). Male and female parents shared 5.38 Mb (10.25%) of introgressed regions, but only 23.42% of these shared regions (1.26 Mb, 2.40% of all introgressed regions) derived from the same subpopulation, suggesting that 97.60% of introgressed regions differ between male and female parents (Fig. 3A and Datasets S3 and S4). We found that the parental introgressed regions included many important genes controlling grain yield, heading date, and biotic resistance (Fig. 3A), such as, Gn1a (21), GW2 (26), IPA1 (20), DTH8 (17, 18), Bph14, and Xa27 (27). Male and female parents had different alleles for these genes (SI Appendix, Fig. S3A) because they derived from different subpopulations, that shaped heterozygous genotypes at these regions in the hybrids.
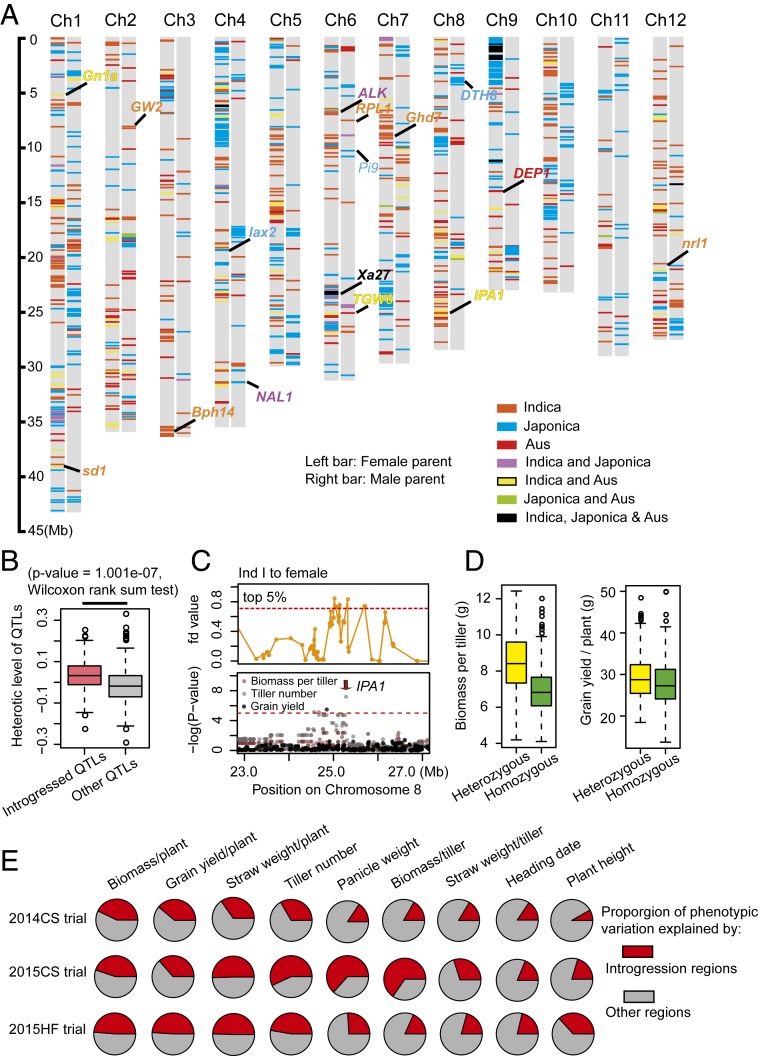
Fig. 3.
Exogenous genome introgression differences between male and female parents shape heterotic loci in the hybrids. (A) Distribution of introgressed genome regions in female (Left bar) and male (Right bar) parents. Introgressed regions were screened using the four-taxon fd statistic, which calculates excessively shared derived variants between two taxa. The cutoff to define introgressed regions from each subpopulation was determined based on the results of the population structure analysis. Different colors represent the origin of introgressed genomic segments. Aus represents the south Asia origin aus rice subpopulation. (B) Heterotic loci in the introgressed regions had greater heterotic effects than loci in other regions. The heterotic effect of each locus was defined as the proportion of the trait value in the heterozygous genotype that was different from the trait value in the homozygous genotype. (C) IPA1 was involved in genetic introgression from Ind I to female parents (Top) and was significantly associated with the variation in GYPP, TN, and biomass per plant (Bottom). (Top) Distribution of the fd value at the IPA1 locus. (Bottom) GWAS Manhattan plot of GYPP, TN, and biomass per tiller for the 2014CS trial at the IPA1 locus. (D) IPA1 locus has heterotic effect at biomass per tiller (Left) and grain yield per plant (Right). (E) Estimated heritability of introgressed regions and other regions across traits and trials. Genetic relatedness matrixes among individuals were calculated using variants within and outside of the introgressed regions. Heritability of the introgressed regions and other regions was then estimated from their genetic relatedness matrixes using the genome-based best linear unbiased prediction (G-BLUP) approach.
To investigate whether heterotic loci of hybrids are related to introgression differences between male and female parents, we compared heterotic loci to the introgressed regions and found 48 out of 143 heterotic loci (33.57%) overlapped with the introgressed regions (Dataset S5). Of these loci, 24 of which were contributed by indica subpopulations, 28 were contributed by japonica, and 11 by aus. We defined the heterotic effect of the QTLs as the proportion of the trait value of the heterozygous genotype that was different from that of the homozygous genotype. We observed that the heterotic QTLs in the introgressed regions had an average 4.50% greater heterotic effect (P value = 1.001e-07, Wilcoxon rank sum test) than those in other genomic regions (Fig. 3B). Meanwhile, we observed many large-effect heterotic QTL clusters that affected multiple traits located in the introgressed regions. For example, the heterotic locus on chromosome 8 that overlapped with heterotic gene IPA1 (20), a gene that encodes a squamosa promoter-binding-like transcription factor and a semidominant regulator of panicle branching and plant architecture, had a strong heterotic effect on the biomass per tiller trait and was involved in genetic introgression from Ind I and aus to the female parent (Fig. 3 C and D). Other heterotic loci, such as the ones located on chromosome 11 and chromosome 12, both of which had strong heterotic effects for multiple traits, including biomass, panicle weight, and grain yield (SI Appendix, Fig. S3 B–E), were involved in genetic introgression from Ind I and TroJ to the female parent, respectively. By heritability partitioning, we found that the introgressed regions explained an average of 41.44% grain yield heritability, 45.46% biomass heritability, 45.86% tiller number heritability, and 35.02% panicle weight heritability in the hybrids. Given that the introgressed regions comprised only 13.90% of the rice genome (Fig. 3E), which suggested that many heterotic loci in the hybrids were shaped by genome introgression from different subpopulations.
The introgressed regions detected above did not cover all heterotic loci due to false-positive controls or the lack of informative variants located near the loci. Therefore, to further investigate whether other heterotic loci were due to differences in parental genetic introgression, we constructed polygenetic trees for all heterotic loci with their 25-kb flanking variants using FastTree2 (28) and analyzed the tree topology to detect potential introgression events (SI Appendix, Fig. S4). By this method, we uncovered the origins of male and female alleles for 141 of heterotic loci (98.60% of all loci, SI Appendix, Table S6). We detected 125 of the heterotic loci that were involved in parental genetic introgression. Among them, 87 of the heterotic loci were in introgression from the indica subpopulations, 61 from the japonica subpopulation, and 29 from other subpopulations. These results demonstrate that large proportions of heterotic loci in the hybrids were potentially affected by parental genetic introgression.
Heterotic Loci Were Potentially Divergently Selected Among Rice Subpopulations.
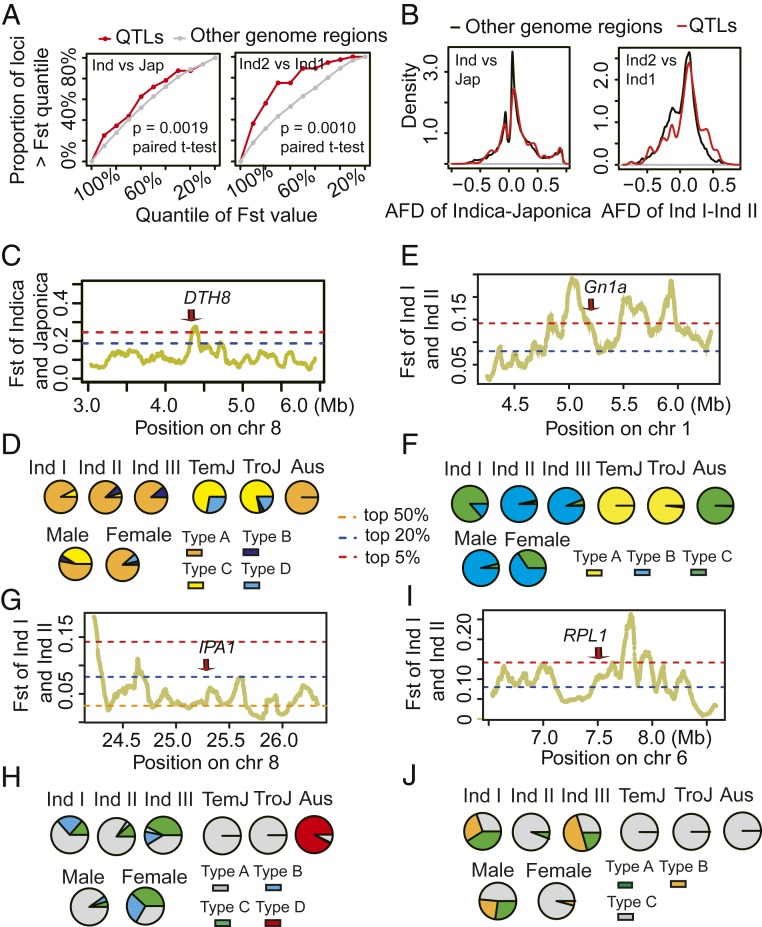
Our polygenetic analysis of heterotic loci indicates that lines were clustered by subpopulation in a large proportion of trees (SI Appendix, Fig. S4). This suggests that the two heterotic alleles (the introgressed allele and the local allele) might have been divergently selected among different subpopulations during evolution. Then we investigated genomewide Fst values (29) for introgression donor and recipient populations using a window size of 100 kb and a step size of 10 kb. At heterotic loci involved in introgression from indica or japonica subpopulations, we consistently observed that heterotic loci were enriched in regions with high Fst values (Fig. 4A). We also investigated the allele frequency difference (AFD) between the introgression donor and recipient subpopulations and observed that the AFDs of heterotic loci were higher than those of the genome background (P value = 1.05e-252, Wilcoxon rank sum test, Fig. 4B). For example, the pleiotropic heterotic locus on chromosome 8 overlaps with flowering time, plant height, and the grain yield regulator gene Ghd8/DTH8 (17, 18), which encodes a CCAAT box-binding transcription factor associated with yield heterosis in modern hybrid rice (9, 10). This locus involves in the genetic introgression from japonica to male parent (SI Appendix, Fig. S5A). The Fst value between indica and japonica was higher than the genomewide 95th quantile (Fig. 4C), indicating a high level of divergence. The haplotype network and haplotype frequency of Ghd8/DTH8, which reveal the relationship among haplotypes from different subpopulations, clearly show that the gene was divergently selected between indica and japonica (Fig. 4D and SI Appendix, Fig. S5A). As another example, the heterotic locus at chromosome 1 overlaps with Gn1a (21), a gene that encodes cytokinin oxidase/dehydrogenase and regulates grain number per panicle. This locus involves in the genetic introgression from Ind I to the female parent (SI Appendix, Fig. S5B). The Fst value between Ind I and Ind II was higher than the genomewide 95th quantile (Fig. 4E), indicating a high level of divergence. The haplotype network and haplotype frequency show that the gene was divergently selected between Ind I and other indica subpopulations (Fig. 4F and SI Appendix, Fig. S5B). Furthermore, we also observed that IPA1 (20) was involved in genetic introgression from Ind I to the female parent (SI Appendix, Fig. S5C). The Fst value between Ind I and Ind II was higher than the genomewide 60th quantile (Fig. 4G), indicating a medium level of divergence. The haplotype network and haplotype frequency show that the gene was divergently selected between Ind II and the other indica subpopulations (Fig. 4H and SI Appendix, Fig. S5C).
Fig. 4.
Alleles of heterotic loci were divergently selected among their derived subpopulations. (A) Heterotic loci originating from both the indica–japonica origin and the Ind I–Ind II origin were enriched in divergent regions between their derived subpopulations. The origins of heterotic loci alleles were analyzed using a polygenetic tree profile constructed from the 25-kb flanking variants of the loci. Alleles from indica and japonica were classified as indica–japonica-origin heterotic loci. Alleles from Ind I and Ind II were classified as Ind I–Ind II-origin heterotic loci. The highest Fst values located within 5 kb of the heterotic loci were used to analyze divergence within the loci. We conducted 100 repeats to generate random loci, with each repeat QTL sampling equal to the number of loci, and calculated the Fst of these loci as the genomewide negative control. (B) Heterotic loci exhibited greater AFDs of derived subpopulations than that of genomewide background. The AFD of the variants in the 25-kb regions flanking the heterotic loci were selected to investigate divergence at these loci. The distribution of AFD of variants at other genome regions was investigated to serve as genomewide negative control. (C and D) Fst (C) and haplotype frequency (D) indicate divergent selection between indica and japonica at the Ghd8/DTH8 locus. (E and F) Fst (E) and haplotype frequency (F) indicate divergent selection between Ind I and Ind II at the Gn1a locus. (G and H) Fst (G) and haplotype frequency (H) indicate divergent selection between Ind I and Ind II at the IPA1 locus. (I and J) Fst (I) and haplotype frequency (J) indicate divergent selection between Ind I and Ind II at the RPL1 locus. Haplotype networks for these genes were constructed using the genetic variants within 5 kb of the heterotic loci. Haplotypes with frequency <2 were removed before constructing the networks. Aus represents the south Asia origin aus rice subpopulation.
Some other loci are involves in genetic introgression from subpopulation to the male parent. For example, the locus on chromosome 6 had a strong heterotic effect on grain yield and panicle weight and overlaps with RPL1 (SI Appendix, Fig. S5D), a gene that affects epigenetic processes and regulates phenotypic plasticity in different environments (30). The Fst of the locus and haplotype network of the RPL1 gene show that it was divergently selected between Ind I and the genetic background of the male parent Ind II (Fig. 4 I and J and SI Appendix, Fig. S5D). Furthermore, integrating the hybrid parents with the genome of 66 representative accessions of O. sativa and O. rufipogon reported in previous study (31) further confirmed the result of the haplotype network analysis (SI Appendix, Fig. S6). Overall, we observed the heterotic loci alleles were divergently selected among their derived subpopulations during rice evolution.
Rice domestication can cause subpopulation divergence. Three types of variants may be subject to divergent selection: new variants (NVs), arisen after subpopulation divergence; standing variants (SVs), which existed before subpopulation divergence and domestication; and postdomestication standing variants (PSVs), arisen during subpopulation divergence and domestication. To determine which types of variants contributed to the divergence of heterotic loci, we investigated the AFDs of the three types of variants within the 25-kb flanking regions of the heterotic loci. Our results indicate that the SV had a much higher AFD than the PSV (P value ≅ 0, Wilcoxon rank sum test) and the NV (P value ≅ 0, Wilcoxon rank sum test, SI Appendix, Fig. S7). Furthermore, by constructing the haplotype network of heterotic genes, we observed that many alleles involved in the heterosis of these loci existed in wild rice (SI Appendix, Figs. S5 and S6). These results suggest heterotic alleles were shaped by the variants that existed before domestication, but not by the variants that arose during rice domestication or modern breeding (SI Appendix, Fig. S7).
Heterotic Loci Were Further Selected to Improve Heterotic Effect during Rice Hybrid Breeding.
During indica hybrid breeding, introgression regions with potential benefit were introduced from rice subpopulations. But only a few proportions of the introgression were kept in hybrids. To demonstrate how introgression regions were selected through hybrid breeding, we used the cross-population composite likelihood ratio (XP-CLR) approach to screen selective sweeps of the parental genomes using landrace strains as the reference population and male/female parents as the query population (32). Regions with the strongest 5% of the XP-CLR values were considered as the selective sweeps. As expected, we found that tms5 (33), the male-sterile gene which had been widely used to develop hybrid female parents, was located by our selective sweeps (SI Appendix, Fig. S8A). This indicated that our strategy could detect selected genes. In total, we detected 19.96 Mb (5.25% of the genome) and 19.68 Mb (5.18% of the genome) of selective sweeps on the paternal and maternal genomes, respectively (Datasets S7 and S8). We observed that 10.40 Mb of the paternal selective sweeps overlapped with maternal selective sweeps (Datasets S7 and S8), demonstrating that many genomic regions were targeted by breeders in both the male and female parents. We found that 14.90% of the paternal introgressed regions were selected during breeding of the male parent and 20.93% of the maternal introgressed regions were selected during breeding of the female parent. Meanwhile, 27.80% of parental introgressed regions overlapped with paternal or maternal selective sweeps. Of these regions, 15.06% overlapped with the common selective sweeps of male and female parents and 17.25% overlapped with male or female parent-specific selective sweeps.
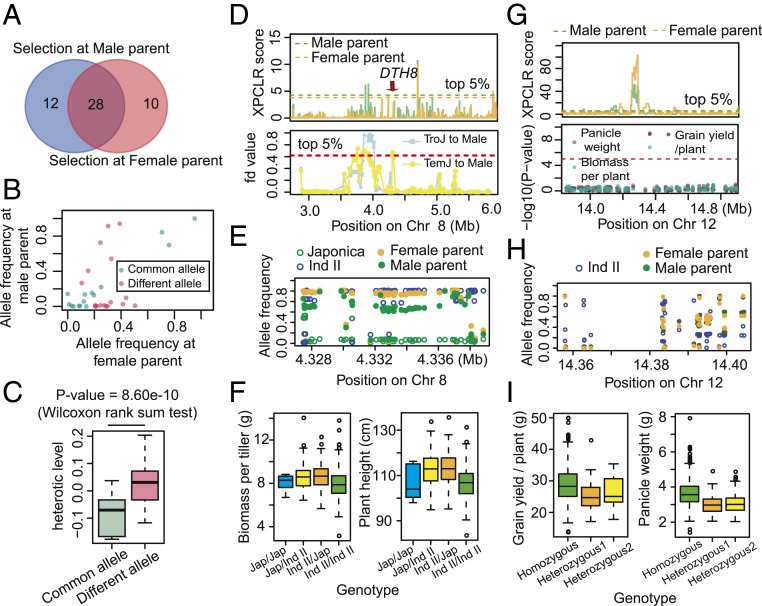
Many important genes in genetic introgression regions were selected during parental breeding. For example, grain yield and flowering gene Ghd7, which encodes a CCT domain protein (34), was involved in the genetic introgression from Ind I to female parent and selected during the breeding of male and female parents (SI Appendix, Fig. S8B). Other genes, including grain yield gene GW2 (26) and biotic resistance gene pi9 (35), were involved in both genetic introgression and selection (SI Appendix, Fig. S8 C and D). We compared the heterotic loci with the parental selective sweeps, and found that 34.96% of the loci (50 out of 143) were selected in the parental genome. Of these, 24.00% (12 out of 50) were specifically selected in the paternal genome, 20.00% (10 out of 50) were specifically selected in the maternal genome, and 56.00% (28 out of 50) were selected in both the paternal and maternal genomes (Fig. 5A and Dataset S9).
Fig. 5.
Heterotic effect-based selection toward heterotic loci improved hybrid performance during breeding. (A) Comparison of heterotic loci selected in the male and female parents. (B) Allele frequency in the male and female parents for the 28 heterotic loci selected in both the male and female parents. Loci with an AFD >0.15 between male and female parents were classified as having different alleles in the male and female parents; loci with an AFD <0.15 were classified as having common alleles in both parents. (C) Comparison of the heterotic effect of heterotic loci with common or different alleles selected in the male and female parents. (D–F) Different alleles were selected at that heterotic locus that overlaps the heterotic gene Ghd8/DTH8. This locus was involved in genetic introgression from japonica to male parents (D, Bottom) and was selected in both male and female parents (D, Top), resulting in different allele frequency spectra between male and female parents (E). This locus was responsible for heterosis of biomass per tiller (F, Left) and plant height (F, Right) in the 2014CS trial. Chr on the x-axis label is the abbreviation for chromosome. (G–I) Common alleles were selected in the hybrid depression locus on chromosome 12. This locus was not involved in any genetic introgression events and was strongly selected in both male and female parents (G, Top), and significantly associated with the variation of grain yield per plant, panicle weight and biomass per plant at 2014CS trial (G, Bottom), resulting in similar allele frequency spectra between male and female parents (H). Heterozygous genotypes had lower trait values than those that were homozygous for grain yield per plant (I, Left) and panicle weight (I, Right).
We defined the parentally differentially selected alleles if their allele frequency differences were higher than 0.15; while other alleles were determined to be commonly selected in both parents (Fig. 5B). The loci with different alleles selected in male or female parents exhibited an average 10.95% higher heterotic effect than the commonly selected alleles across traits and trials (P value = 8.06e-10, Wilcoxon rank sum test). The average heterotic effect of different alleles was 2.61% and the average effect of common alleles was −8.34% (Fig. 5C). These results demonstrate that during hybrid breeding, divergently selected regions among subpopulations were first introgressed into hybrid parents, then they were selected based on their effect. When the loci with positive heterotic effect were selected to improve heterosis, the loci with hybrid depression was also selected to suppress heterozygous genotypes in the hybrids (Fig. 5 D–I).
Discussion
In this study, we generated large-scale genotypic and phenotypic data for two populations of rice hybrids constructed from present day commercial hybrid parents. By combining these data with ∼4,200 resequenced rice landrace strains and conventional varieties (11–14), this comprehensive genetic analysis revealed that those male and female parents have different genetic structure. Female parents have a much higher level of genome introgression from other subpopulations than do male parents (Fig. 2A). These differences in parental population structure shaped heterotic loci in the hybrids and explained a large proportion of variation in biomass, grain yield, and tiller number (Fig. 3). Effect-based selection toward heterotic loci during hybrid breeding reduced the rate of heterozygous depression loci but improved the rate of positive heterotic loci in hybrids, resulting in improved hybrid performance (Fig. 5). Tracing the origin of heterotic alleles suggests that they were divergently selected during evolution of the derived subpopulations (Fig. 4).
Two factors may contribute to the population structure difference of male and female parents. The breeding of hybrid rice employs a male sterility line as female parent and a restorer line which could restore the sterility of female parents as the male parent. The sterility-restorer relationship between female and male parents leads to genetic difference at sterility genes like tms5 (33). As expected, we observed obvious genotypic differences between our two-line male and female parents at the tms5 locus (SI Appendix, Fig. S9A), indicating the sterility-restorer relationship between female and male parents contributes to the genome difference between them. Another factor may be the utilization of dominant/overdominant effect loci during hybrid breeding. Many heterotic loci show dominant or overdominant effect in the hybrids (Fig. 1 B–D). To utilize the dominant and overdominant effect of these loci, breeders may select different alleles in the male and female parents, therefore generating genome difference between male and female parents (SI Appendix, Fig. S9 B and C).
According to the pedigree record (http://www.ricedata.cn/variety), IRRI-bred varieties, like IR8 and IR24, were widely used to develop conventional varieties and parents of hybrids during China’s past hybrid rice breeding efforts because of their outstanding performance. Accordingly, we found that both male and female parents have connections with IRRI-bred varieties. Male parents contained an average of 5.74% introgressed exogenous genomes while the female parents contained 22.57% introgressed exogenous genomes (Fig. 2A). Introgression into the parental genomes explained a large proportion of variation in yield trait phenotypes (Fig. 3E) and was estimated to be involved in 88.60% of heterotic loci in the hybrids. These results demonstrate that differences in parental genetic introgression shape heterotic loci in the hybrids.
With this parental construction strategy, the outstanding performance of elite genetic background varieties, such as high yield and stress resistance, could be directly inherited by the hybrids. However, this strategy was limited by the extent of applied heterotic loci. Previous studies have revealed the importance of accumulating favorable alleles when it comes to improving hybrid performance (8, 10). But the strategy incorporated only a portion of the heterotic loci containing beneficial alleles from the two distinct genomes and thus was unable to exploit the full heterotic potential. Furthermore, Ind I provided the majority of genes introgressed into female parents (64.20% of the introgressed genome, Fig. 2A), indicating that there is room for improving gene introgression from other subpopulations to help shape heterotic loci in the hybrids. Taken together, we conclude that introducing divergently selected regions to parents, improving the proportion of the exogenous genome, then removing hybrid depression loci and fixing heterotic loci would promote the accumulation of additional superior alleles. This strategy could help overcome yield improvement bottlenecks in recent rice hybrid breeding.
The majority of the genome introgressed into female parents was contributed by Ind I, which originated from South China and included varieties that had been intensively used during historical breeding of conventional rice and hybrid parents (Fig. 2A). For example, Aijiaonante from Ind I was the first semidwarf variety released in 1956 in China and offered early semidwarf germplasm for the breeding of semidwarfnese varieties (36). Ind I contributed a large number of important genes associated with yield traits, flowering time, and resistance to abiotic and biotic stresses, including Gn1a (21), GW2 (26), TGW6 (37), Ghd7 (34), IPA1 (20), OsFD1 (38), Xa27 (27), Bph14 (39), and OsPT13 (40) (Fig. 3A). QTL tree topology profiling revealed that Ind I affected 44.06% of heterotic loci in the hybrids, including an average of 34.05% of grain yield loci over all trials (Dataset S6). Other maternal introgressed regions were contributed by Ind III, aus, and japonica. Although japonica introgression contributed only 3.31% of the genome in the parents of the hybrids, it nonetheless contributed alleles associated with important agronomical traits (Fig. 3A), like NAL1 (19), Ghd8/DTH8 (17, 18), and Xa27 (27). Our introgressed regions analysis and polygenetic tree profiling both supported the conclusion that japonica affected an average of ∼37.75% of the grain yield heterotic loci over all trials (Datasets S5 and S6). This indicates that the introduction of japonica germplasm played an important role in indica hybrid breeding. And further improving the proportion of japonica germplasm may facilitate the improvement of indica hybrid rice heterosis. Important genes and heterotic loci in introgressed regions from Ind I and japonica germplasm displayed selective signatures during hybrid breeding (Fig. 5D and SI Appendix, Fig. S8 B–D), suggesting that they were the targets of breeding efforts to improve hybrid performance. Therefore, the introgression of Ind I and japonica germplasm explained a large proportion of heterotic loci in the recent hybrids. These loci would be good targets for future “breeding-by-design” efforts.
In crop breeding, heterosis is associated with the level of divergence between parents, but whether heterotic loci are affected by divergent selection remains unclear. Our results show that heterotic loci enriched in differentiated genome regions of derived subpopulations and important heterotic genes like Ghd8/DTH8 (17, 18), Gn1a (21), and IPA1 (20) were divergently selected during the divergence of their derived subpopulations (Fig. 4 C–H). This suggests that heterotic loci had undergone divergent selection during rice evolution. Genome divergence could potentially be shaped by many factors, including selection arising from regional adaptation, genetic drift, genetic conflict, and chromosomal structure (41). Previous studies on the population structure of landraces and improved varieties found that rice subpopulations were divided by their geographic origin (14). We also observed that divergence at heterotic loci among subpopulations were mainly shaped by predomestication standing variants (SI Appendix, Fig. S6). For specific heterotic loci, the divergence was widespread among subpopulations, rather than limited to two specific subpopulations (Fig. 4 C–J). Some heterotic candidate genes, like Ghd8/DTH8 (17, 18) and RPL1 (30), were involved in the gene–environment interaction. Qualifying the contribution of environmental and geographic factors to genetic differentiation emphasized their critical role in shaping genomewide divergence among subspecies (42). Therefore, we suggest that the divergence at heterotic loci among derived subpopulations arose from adaptation to different environments. Hybrids carried more adaptive alleles than either of their parents, leading the improved adaptability of hybrids to external environments.
Materials and Methods
Hybrid parents were constructed by using restorer lines improved from elite backbone restorer lines or conventional cultivars, and male sterile lines and relative improved lines that were frequently applied during modern breeding. Two hybrid populations (∼1,000 hybrids each) were respectively constructed during the year of 2014 (denote as Pop I) and 2015 (denote as Pop II). Pop I was cultivated in Changsha, China, in the summer of 2014, and Pop II was cultivated in both Changsha and Hefei, China (denoted as 2015CS and 2015HF, respectively), in the summer of 2015. Night important agronomical traits were recorded at all three experimental trials. All hybrid parents were genotyped by RiceSNP50 chip designed by our previous study or by genome resequencing with a depth of ∼3× (Dataset S1). Hybrid genotypes were obtained by combining the haploid genome of their corresponding parents. GWAS was conducted to map heterotic loci across experimental trials. For detail information of population design and mapping of heterotic loci, see SI Appendix. By integrating the hybrid parents with previously reported landraces and improved varieties, the population structure of hybrid parents, the introgressed genome regions, selective sweeps on male and female parental genome, and the potential divergence at heterotic loci were exploited as described at SI Appendix, Materials and Method.
Data Avaliability.
The RiceSNP50 chip genotype of hybrid parents and phenotype data of hybrids are included in Dataset S10. The resequencing data of hybrid parents were deposited to GSA (https://bigd.big.ac.cn/gsa/) with Bioproject ID PRJCA001768.
Supplementary Material
Acknowledgments
We thank Prof. Haiyang Wang for critical reading of the manuscript. This work was supported by National Key Research and Development Program of China Grants 2016YFD0100801 and 2016YFD0101104; National High Technology Research and Development Program of China Grant 2014AA10A602; National Science Foundation of China Grant 31871706; and the Peking-Tsinghua Center for Life Sciences, Peking University.
Footnotes
The authors declare no competing interest.
Data deposition: The resequencing data were deposited to GSA (https://bigd.big.ac.cn/gsa/) with Bioproject ID PRJCA001768.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919086117/-/DCSupplemental.
References
- 1.Xiao J., Li J., Yuan L., Tanksley S. D., Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140, 745–754 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu S. B., et al. , Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. U.S.A. 94, 9226–9231 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moll R. H., Salhuana W., Robinson H., Heterosis and genetic diversity in variety crosses of maize. Crop Sci. 2, 197–198 (1962). [Google Scholar]
- 4.Stupar R. M., Springer N. M., Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173, 2199–2210 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swanson-Wagner R. A., et al. , All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. U.S.A. 103, 6805–6810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng S. H., Zhuang J. Y., Fan Y. Y., Du J. H., Cao L. Y., Progress in research and development on hybrid rice: A super-domesticate in China. Ann. Bot. 100, 959–966 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.YUAN LP , Hybrid rice achievements, development and prospect in China. J. Integr. Agric. 14, 197–205 (2015). [Google Scholar]
- 8.Huang X., et al. , Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 6, 6258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X., et al. , Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Li D., et al. , Integrated analysis of phenome, genome, and transcriptome of hybrid rice uncovered multiple heterosis-related loci for yield increase. Proc. Natl. Acad. Sci. U.S.A. 113, E6026–E6035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X., et al. , Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Huang X., et al. , A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie W., et al. , Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. U.S.A. 112, E5411–E5419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W., et al. , Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H., et al. , A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 7, 541–553 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Kang H. M., et al. , Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X., et al. , DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan W. H., et al. , A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Qi J., et al. , Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao Y., et al. , Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Ashikari M., et al. , Cytokinin oxidase regulates rice grain production. Science 309, 741–745 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa M., Shimamoto K., Kyozuka J., Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H., et al. , RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 43, D1018–D1022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin S. H., Davey J. W., Jiggins C. D., Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X., A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Gu K., et al. , R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Price M. N., Dehal P. S., Arkin A. P., FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weir B. S., Cockerham C. C., Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984). [DOI] [PubMed] [Google Scholar]
- 30.Zhang C. C., Yuan W. Y., Zhang Q. F., RPL1, a gene involved in epigenetic processes regulates phenotypic plasticity in rice. Mol. Plant 5, 482–493 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Zhao Q., et al. , Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50, 278–284 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Chen H., Patterson N., Reich D., Population differentiation as a test for selective sweeps. Genome Res. 20, 393–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H., et al. , RNase Z(S1) processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat. Commun. 5, 4884 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Xue W., et al. , Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Liu G., Lu G., Zeng L., Wang G. L., Two broad-spectrum blast resistance genes, Pi9( t) and Pi2( t), are physically linked on rice chromosome 6. Mol. Genet. Genomics 267, 472–480 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Shen J.-H., “Rice breeding in China” in Rice Improvement in China and Other Asian Countries (International Rice Research Institute, Philippines, 1980), pp. 9–30. [Google Scholar]
- 37.Ishimaru K., et al. , Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Taoka K., et al. , 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Du B., et al. , Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U.S.A. 106, 22163–22168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paszkowski U., Kroken S., Roux C., Briggs S. P., Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 13324–13329 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nosil P., Funk D. J., Ortiz-Barrientos D., Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18, 375–402 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Lee C. R., Mitchell-Olds T., Quantifying effects of environmental and geographical factors on patterns of genetic differentiation. Mol. Ecol. 20, 4631–4642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.