Significance
Turner syndrome (TS) is caused by having only one X chromosome (X0), and Klinefelter syndrome (KS) by having two X chromosomes and one Y chromosome (XXY). In this study we carried out a direct comparison analysis of the effect these chromosome copy number aberrations have on gene expression networks, analyzing genes located on the X chromosome or anywhere else in the genome, in primary samples from KS and TS patients. In both KS and TS, we found gene expression level changes not only in genes on the X chromosome, but also in many genes on all the other chromosomes, revealing a genomewide ripple effect of the chromosome X copy number aberrations.
Keywords: sex chromosome aneuploidies, Turner syndrome, Klinefelter syndrome, transcriptome, methylation
Abstract
In both Turner syndrome (TS) and Klinefelter syndrome (KS) copy number aberrations of the X chromosome lead to various developmental symptoms. We report a comparative analysis of TS vs. KS regarding differences at the genomic network level measured in primary samples by analyzing gene expression, DNA methylation, and chromatin conformation. X-chromosome inactivation (XCI) silences transcription from one X chromosome in female mammals, on which most genes are inactive, and some genes escape from XCI. In TS, almost all differentially expressed escape genes are down-regulated but most differentially expressed inactive genes are up-regulated. In KS, differentially expressed escape genes are up-regulated while the majority of inactive genes appear unchanged. Interestingly, 94 differentially expressed genes (DEGs) overlapped between TS and female and KS and male comparisons; and these almost uniformly display expression changes into opposite directions. DEGs on the X chromosome and the autosomes are coexpressed in both syndromes, indicating that there are molecular ripple effects of the changes in X chromosome dosage. Six potential candidate genes (RPS4X, SEPT6, NKRF, CX0rf57, NAA10, and FLNA) for KS are identified on Xq, as well as candidate central genes on Xp for TS. Only promoters of inactive genes are differentially methylated in both syndromes while escape gene promoters remain unchanged. The intrachromosomal contact map of the X chromosome in TS exhibits the structure of an active X chromosome. The discovery of shared DEGs indicates the existence of common molecular mechanisms for gene regulation in TS and KS that transmit the gene dosage changes to the transcriptome.
Turner syndrome (TS) (45X or X0) and Klinefelter syndrome (KS) (47XXY or XXY) are common sex chromosome aneuploidies (SCAs) in humans with an approximate occurrence of 1 in 2,000 female and 1 in 600 male livebirths, respectively (1). TS is due to the partial or complete absence of an X chromosome in females. Clinical features of Turner syndrome may include short stature, structural cardiac abnormalities, autoimmune disease, infertility, and learning disorders (2). KS is characterized by the presence of an additional X chromosome in males with 47XXY being the most prevalent type. Characteristics associated with Klinefelter syndrome may include tall stature, small testicles, hypogonadism, and language-based learning disorders (3). Importantly, a subset of phenotypic characteristics of TS and KS appear to follow a linear dose-dependent relationship across sex chromosome number, including stature and performance in cognitive subdomains of language and visuospatial ability (2).
Surprisingly for diseases that are as common as these, our knowledge of genotype–phenotype relationships is rather limited, with very few specific candidate genes linked to clinical features. The only well-established association is that of the SHOX gene with short stature in TS (4) and tall stature in KS (5). X-chromosome inactivation (XCI) transcriptionally silences one X chromosome in female mammals; as such, most genes on the inactivated X chromosome are silenced and inactive (hereinafter referred to as inactive genes). However, some genes escape XCI (hereinafter referred to as escape genes) and have also been hypothesized to contribute to the phenotypes of SCAs (6, 7). Several studies have analyzed gene expression profiles, but almost all in either TS or KS only. For TS, gene expression patterns have been documented in peripheral blood mononuclear cells (PBMCs) (8), leukocytes (9), amniotic fluid (10), fibroblast cells (11, 12), and in a single induced pluripotent cell line (13). For KS, gene expression profiles have been measured in testis (14, 15), whole blood (16, 17), leukocytes (18), and in a single brain sample (19). Only one very recent study included multiple SCAs and also KS and TS in an extensive comparative analysis on the level of gene expression patterns, using microarrays and studying Epstein–Barr virus (EBV)-transformed B lymphoblastoid cell lines (LCLs) (20). Low-resolution methylation profiles generated by microarrays have been described in leukocytes (9), peripheral blood (21, 22), and fibroblasts (12, 23) for TS, and in leukocytes (18), peripheral blood (22, 24), and one brain sample (19) for KS.
Our knowledge about molecular network effects caused by SCA in TS and KS is still limited, particularly in regard to epigenomic levels of control and regulation of gene activity. To close this gap, we report on a direct comparative analysis of TS versus KS and integrative analysis over three levels of genomic and functional genomic activity, in primary PBMCs derived from individuals with SCAs and typically developing controls to examine levels of gene expression and DNA methylation, and in LCLs for chromosome folding patterns. We analyzed gene expression patterns with RNA-Seq and DNA methylation patterns with genomewide targeted-capture bisulfite sequencing from clinically well-characterized TS and KS cohorts together with sex-matched controls in PBMCs. Using in situ Hi-C, we also investigated the three-dimensional (3D) chromatin structure of individuals with TS and KS relative to euploid controls in LCLs. As the dataset includes both monosomic and trisomic conditions, it allows for integrated multiomics analysis over a linear distribution of sex chromosome copy numbers. We carried out direct comparative analysis on the molecular level in TS vs. KS and discovered evidence for the existence of shared molecular mechanisms of control that in both TS and KS appear to be transmitting the gene dosage changes to the transcriptome.
Results
Genotyping Data Verified the Karyotypes of TS and KS Patients and Resolved the Occurrence of the Nondisjunction Events during Meiosis in KS Patients.
To validate the karyotypes of the subjects, 55 individuals—14 females with TS (X0), 14 males with KS (XXY), and 13 male (XY) and 14 female (XX) typically developing controls—were genotyped on high-density oligonucleotide arrays. All but 1 of the girls with TS carried only one X chromosome and all males with KS carried two (SI Appendix, Fig. S1). No mosaicism was observed in the patients. The one individual misdiagnosed with TS, carrying the normal two X chromosomes, was excluded from further analyses.
The second X chromosome in KS arises through a nondisjunction event either during paternal or maternal meiosis I where homologous X chromosomes fail to separate, which leads to two distinct X chromosomes (one paternal and one maternal or both maternal), or maternal meiosis II where sister chromatids fail to separate, which results in two identical maternal X chromosomes. Of the 14 males with KS, 12 have two distinct X chromosomes and 2 have identical ones (SI Appendix, Fig. S1).
Differentially Expressed Escape Genes Are Almost Unanimously Down-Regulated but Most Differentially Expressed Inactive Genes Are Up-Regulated in TS.
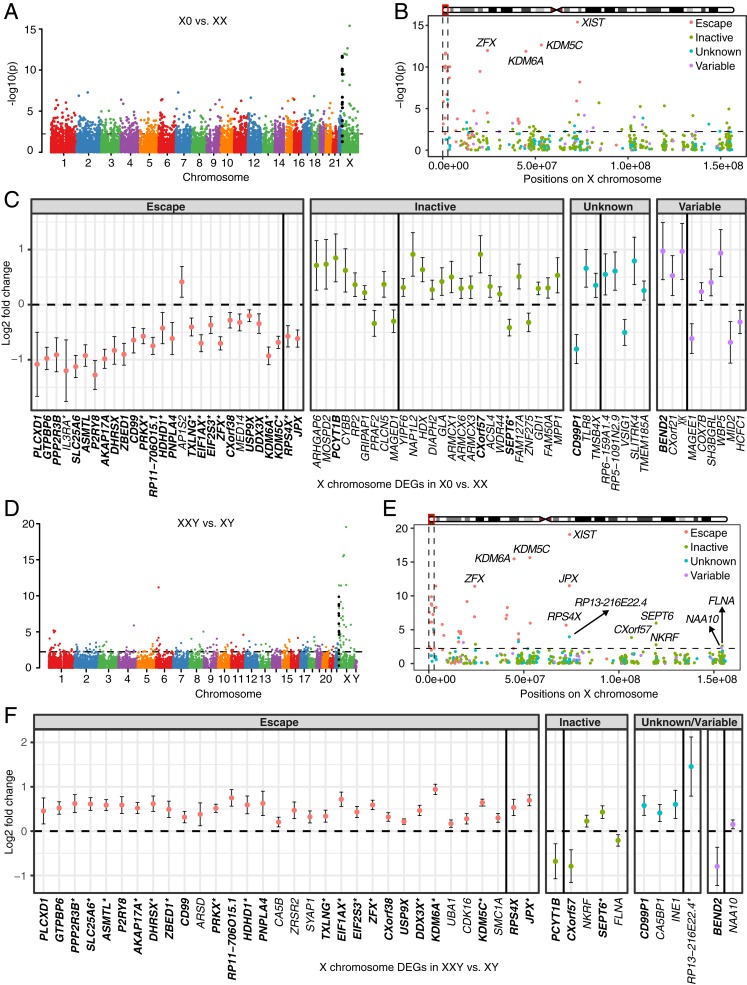
To find the TS-associated genes, differential expression analysis was performed between X0 vs. XX. Among the 14,314 expressed genes, there were 1,142 differentially expressed genes (DEGs) (513 down-regulated genes and 629 up-regulated genes in X0) (Dataset S1). The most significant signals were situated on the X chromosome, especially on Xp, the short arm of the X chromosome (Fig. 1A). The 14 most significantly differentially expressed genes (all down-regulated in X0) were all on Xp except JPX and XIST, which are long noncoding RNAs (lncRNAs) in the X-inactivation center (XIC) (Fig. 1B). Intriguingly, previous studies have reported that females with TS with a total or partial deletion of Xp or a 46,X,i(Xq) karyotype (i.e., deletion of Xp but duplication of Xq on one X chromosome) cannot be differentiated phenotypically from 45,X TS individuals (25, 26), whereas females with a deletion of Xq (i.e., the long arm of the X chromosome) do not show, or show a significantly attenuated phenotype characteristic of TS (27, 28), indicating that the causative genes of TS are mainly on Xp. Our results provided further evidence of this finding on the molecular level.
Fig. 1.
Differential expression analysis between TS patients and female controls (A–C), and between KS patients and male controls (D–F). −log10(P values) across the genome are shown in A for X0 vs. XX and in D for XXY vs. XY. Genes in PAR1 are colored in black. −log10(P values) across the X chromosome are shown in B for X0 vs. XX and in E for XXY vs. XY. Genomewide significance is based on false discovery rate (FDR) <0.05 indicated by the horizontal lines. PAR1 region is represented by the vertical black lines and genes are shown in four colors based on their XCI status in B and E. Log2 fold change and 95% confidence interval of DEGs on the X chromosome are shown in C for X0 vs. XX and in F for XXY vs. XY. DEGs are shown in four categories based on their XCI status. DEGs labeled by asterisk in C and F were also reported in Trolle et al. (9) and Skakkebæk et al. (18), respectively. Gene XIST (log2 fold change −11.4, 95% confidence interval [−12.9, −10] for X0 vs. XX; log2 fold change 12.9, 95% confidence interval [11.7, 14.0] for XXY vs. XY) is omitted for viewing purpose. Genes on the Xp and the Xq are separated by the black vertical line within each category. DEGs shared between TS and KS are highlighted in bold.
A previous study identified 20 X chromosome DEGs between TS and female controls (9). We detected expression of 18 of them in our study, among which 11 were differentially expressed. Among the 72 X chromosome DEGs identified in our study, 29 were known escape genes (9 more were variable escape genes, Fig. 1C) (29). The enrichment of X chromosome DEGs for escape genes was significant (Fisher’s exact test P value = 2.09E-06) compared with 49 escape genes out of the 456 expressed genes on the X chromosome. As only one copy of the X chromosome is present in X0, we expected lower expression of the DEGs on the X chromosome in X0. However, of the 72 DEGs on the X chromosome, only 37 showed lower expression in X0 but the other 35 showed higher expression (Fig. 1C). Interestingly, almost all of the escape DEGs (28 of 29) showed lower expression in X0 (Fig. 1C). Of the other 9 genes with lower expression in X0, 3 were variable escape genes, 2 were of unknown XCI status, and only 4 were subject to X inactivation. By contrast, 22 of the 35 DEGs with higher expression were subject to XCI, 6 were of unknown XCI status, only 6 were variable escape genes, and 1 was an escape gene (Fig. 1C). Taken together, almost all of the escape DEGs showed lower expression but most of the inactive DEGs showed higher expression in X0, indicating that different mechanisms might underlie the diverging expression of escape and inactive genes in TS.
While most significant differential expression occurred on the X chromosome, the majority of DEGs (1,070 of 1,142) were observed on the autosomes. These autosomal DEGs were enriched in genes with sex-biased expression (145 of 1,070, Fisher’s exact test P value = 0.007) identified by a recent study (30). Pathway enrichment analysis of all of the DEGs showed that one of the most significant pathways was immune response (P value = 1.52E-10) (SI Appendix, Fig. S2A). Other pathways enriched with DEGs included cell adhesion, regulation of cell death, cell–cell signaling, and neurological system process (SI Appendix, Fig. S2A).
Differentially Expressed Escape Genes Were Up-Regulated While Inactive Genes Appeared Unchanged in KS.
Differential expression analysis between XXY vs. XY identified 241 DEGs (111 down-regulated and 130 up-regulated in XXY, Dataset S2) genomewide (Fig. 1D). The number of DEGs relative to same-sex controls is far less in XXY than in X0. Interestingly, KS patients have a significantly less pronounced phenotype relative to individuals with TS (2), which was in line with our observation on the molecular level that only comparatively mild global expression changes occurred in KS. Pathway analysis found that only biological adhesion and cell adhesion were enriched with DEGs (SI Appendix, Fig. S2B).
Similar to TS, the most significant expression changes were on the X chromosome, especially in pseudoautosomal region 1 (PAR1) (Fig. 1E). A previous study identified 27 X chromosome DEGs between KS and male controls (18). We observed expression of 19 of them in our study and all of them were differentially expressed. However, in contrast to TS, the vast majority of the DEGs on the X chromosome (40 of 44) were up-regulated in XXY relative to XY (Fig. 1F), which was to be expected because an extra copy of X chromosome genes is present in XXY. Most of these DEGs were driven by escape genes—of the 40 DEGs with higher expression in XXY, 33 were escape genes, 4 were of unknown XCI status, 1 was a variable escape gene, and only 2 were inactive (Fig. 1F). Similar to X0, escape genes were significantly enriched in the DEGs between XXY vs. XY (Fisher’s exact test P value = 1.24E-11). The only 4 genes with lower expression were BEND2, FLNA, PCYT1B, and CX0rf57. BEND2 has variable XCI escaping status and the other 3 are subject to XCI (29). Taken together, most of the DEGs on the X chromosome were escape genes that were up-regulated in KS.
Although most of the DEGs on the X chromosome were on Xp (35 of 44), previous case reports found that individuals with 47,X,i(Xq),Y karyotypes exhibit typical clinical features of KS, excluding tall stature (31, 32). This suggests that genes located on the long arm of the X chromosome are responsible for the clinical features in KS. We identified 9 DEGs on Xq in analysis of XXY vs. XY (Fig. 1 E and F). Three of them—XIST, JPX, and RP13-216E22.4—are lncRNAs in the XIC. Among the other 6 genes, RPS4X escapes X inactivation and has a homologous gene—RPS4Y—on the Y chromosome, NAA10 is a variable escape gene, whereas SEPT6, NKRF, FLNA, and CX0rf57 are subject to XCI. No genes on the Y chromosome were differentially expressed. These results open further perspectives for finding causative genes for KS.
XCI Occurs in XXY but Not X0.
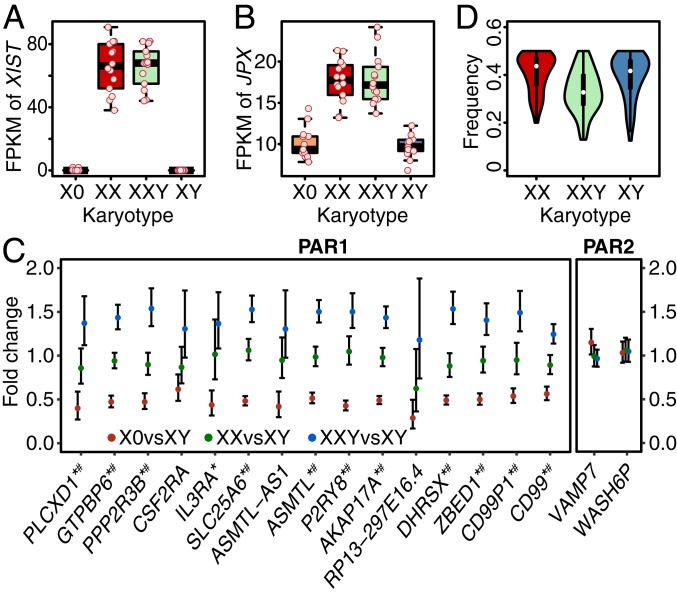
XCI is achieved through the XIC on the X chromosome, which is dominated by lncRNAs such as XIST and JPX. XIST triggers XCI while JPX is an RNA-based activator of XIST (33, 34). We observed significantly differential expression of both lncRNAs in comparison of both XXY vs. XY (P value = 2.86E-20 for XIST, P value = 3.13E-12 for JPX) and X0 vs. XX (P value = 4.07E-16 and 6.51E-09). XIST displayed high expression in XXY and XX but almost no expression in X0 and XY (Fig. 2A). Consistently, JPX showed higher expression in XXY than XY (log2 fold change = 0.69) and lower expression in X0 than XX (log2 fold change = −0.62) (Fig. 2B). Both XIST and JPX showed similar levels of expression in XXY and XX, indicating that XCI occurs similarly in XXY and XX controls. The lack of expression of XIST and lower expression of JPX in X0 and XY indicates the absence of XCI.
Fig. 2.
Expression levels of XIST (A) and JPX (B), and expression of genes in PARs (C and D). FPKM values are shown as points for all individuals for XIST (A) and JPX (B). Fold change of average expression levels of all expressed genes (FPKM > 1) in PAR1 and PAR2 are shown in C for X0 vs. XY, XX vs. XY, and XXY vs. XY. Expressions of these genes in males are the aggregated expressions of both X and Y chromosomes as sequencing reads cannot be distinguished in PARs due to homology. Genes labeled by asterisk are DEGs in X0 vs. XX and those labeled by number sign are DEGs in XXY vs. XY. Frequencies of the number of the reads mapped to the allele with lower expression are shown in D for exonic SNPs in PAR1 genes genotyped by array.
X Chromosome Dosage Compensation Remains Intact in TS and KS.
Previous research has demonstrated compensation of sex chromosome gene expression between males and females, to balance X chromosome expression, given the unequal genetic material between X and Y chromosomes. We found that expression levels for the X chromosome were relatively consistent across euploid samples and those with aneuploidies, regardless of minimum expression levels (one-way ANOVA P value >0.53, SI Appendix, Fig. S3A), indicating global equivalence of X chromosome gene expression.
While XCI allows male–female equivalence in X chromosome gene expression, dosage compensation between sex chromosomes and autosomes has been debated, with some studies finding that sex chromosome expression is decreased relative to autosomes, with the ratio of X chromosome gene expression relative to autosomal gene expression (X:A) being ∼0.5 (35), while others have reported an X:A ratio closer to 1, indicating up-regulation from the sex chromosomes to match autosomal output (36). More recent studies have proposed a partial dosage compensation mechanism where only the expressions of dosage-sensitive genes on the X chromosome such as components of large protein complexes are doubled to balance their dosage with autosomes (7, 37), resulting in an X:A gene expression ratio between 0.5 and 1. We did not observe evidence of different X:A ratios across the four cohorts (SI Appendix, Fig. S3B), suggesting that the dosage compensation mechanisms maintaining expression balance between the X chromosome and autosomes are intact across different X chromosome numbers. Furthermore, the X:A ratios across all cohorts were between 0.5 and 1, which was in line with partial compensation for the dosage of the X chromosome relative to autosomes as reported in earlier literature (7).
PAR1 Genes Are Dosage Sensitive to Sex Chromosome Number but Not PAR2.
PARs are homologous domains at the distal ends of the X and Y chromosomes and thus the copy number of genes in these regions are consistent with the sex chromosome number. Given that the majority of these genes escape XCI (29), but demonstrate homology across the X and Y chromosomes, we predicted expression from these regions to follow a dosage stoichiometry based on sex chromosome number, which is 1:2:2:3 for X0:XX:XY:XXY. Expression profiles followed this pattern in PAR1 (distal Xp) but not in PAR2 (distal Xq) (Fig. 2C). Expression from each PAR1 gene is the lowest in monosomic TS (X0), roughly double in XX and XY, and, as expected given the presence of three PAR sets in trisomic KS (XXY), expression levels were approximately triple the TS output (Fig. 2C).
To further investigate if PAR1 genes on both the X and Y chromosomes are expressed in KS, we performed allele-specific expression (ASE) analysis on the heterozygous exonic SNPs genotyped by the array. The frequency of the number of sequencing reads from the lower-expressed allele was calculated for each SNP. For XX and XY, both alleles are expressed, as the frequency is around 0.5 (Fig. 2D). For XXY, the frequency is around 0.33 as there are three copies and two of them should have the same allele (Fig. 2D). This indicates both X and Y chromosomes express PAR1 genes in XY and XXY, and both X chromosomes express PAR1 genes in XX.
The only two genes—VAMP7 and WASH6P—with detectable expression in PAR2 did not follow the pattern of 1:2:2:3. They showed comparable expression across aneuploid and control cohorts. A previous study has shown that VAMP7 undergoes XCI and is also inactive on the Y chromosome (38), indicating that in all cohorts only one copy of VAMP7 is active, which was in line with our observation. Although WASH6P has been reported to escape XCI and to be expressed from the Y chromosome (39), its XCI-escape status is not well established. Our results showed that WASH6P is subject to XCI and inactive on the Y chromosome.
Shared DEGs between X0 vs. XX and XXY vs. XY Exhibited Divergent Expression Changes.
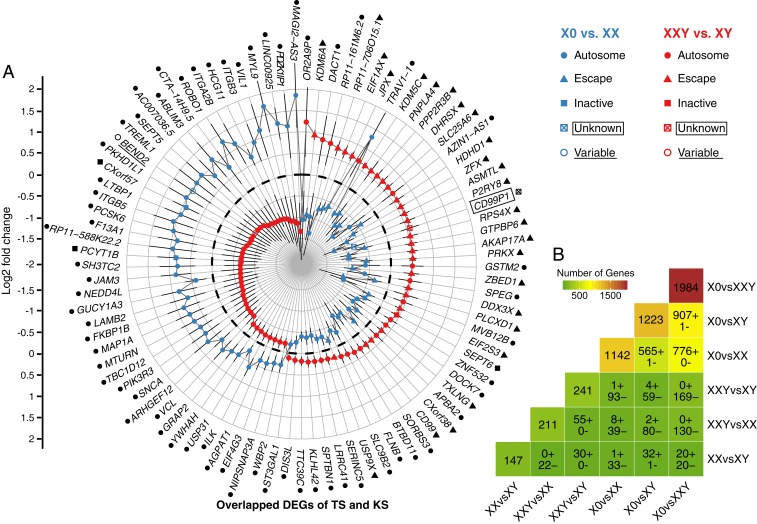
Some clinical features are shared between TS and KS, including gross impairments in executive functioning, motor skills, and higher-order social cognitive ability (2). On the other hand, other clinical features exhibit divergent patterns when comparing TS and KS, such as height and performance with language, an observation which points to an apparent dose effect driven by the number of sex chromosomes. This prompted us to investigate the overlapping changes between TS and KS relative to their same-sex controls on the molecular level. Interestingly, of the 241 DEGs in XXY vs. XY and the 1,142 DEGs in X0 vs. XX, 94 DEGs were overlapping between the two comparisons, i.e., these 94 genes were differentially expressed in both TS and KS (Fig. 3A). Of these 94 overlapping or shared DEGs, only 31 are located on the X chromosome, while the other 63 are autosomal genes. The 31 shared DEGs on the X chromosome are enriched with genes that have been reported by previous studies to escape XCI (26 of 31, Fisher’s exact test P value = 1.67E-10) (6, 40). All of these 26 escapee genes were down-regulated in X0 but up-regulated in XXY, in line with the expected dosage effect of the number of sex chromosomes moderated by this dosage compensation mechanism. Our results indicate that the overlapping dosage effect on the X chromosome in TS and KS mostly impacts escape genes but not inactive genes.
Fig. 3.
Shared DEGs between comparisons. (A) Shared DEGs in TS and KS. XIST is omitted for viewing purpose. DEGs on the X chromosome are shown in four categories based on XCI status: escape, variable escape, inactive and unknown status. Dashed black circle represents log2 fold change = 0. Autosomal genes and X chromosome genes with different XCI statuses are also labeled with black symbols after the gene names. (B) Number of DEGs in each comparison and shared DEGs between different comparisons. The number of DEGs for each comparison is shown in the diagonal cells. The number of shared DEGs with expression changes in the same direction (denoted by “+”) or opposite direction (denoted by “−”) between comparisons is shown in other cells. For the shared DEGs between X0vsXXY and XXvsXY, 15 of the 20 DEGs with the same direction of expression changes are on the Y chromosome, which is expected as the Y chromosome genes only express in XXY and XY.
All but 1 of the 94 shared DEGs displayed expression changes in opposite directions in X0 and XXY compared with their sex-matched controls. While we expected this pattern of gene expression on the X chromosome, evidence of dosage effects extending to genes on the autosomes was unexpected (Fig. 3A). Additionally, DEGs on the X chromosome and a proportion of autosomal DEGs, demonstrated an expected dose–response correlating to sex chromosome number, with 49 shared DEGs (28 on the X chromosome and 21 on the autosomes) down-regulated in X0 relative to XX, and up-regulated in XXY relative to XY. However, we found that a substantial portion of autosomal DEGs demonstrated an inverse relationship between expression and sex chromosome number, with other 44 shared DEGs (3 on the X chromosome and 41 on the autosomes) up-regulated in X0 but down-regulated in XXY. This finding suggests that the dosage effect of the number of sex chromosomes on the gene expression spectrum, while strongly evident on the X chromosome, also extends to the autosomes. Furthermore, although the dosage effect of the number of sex chromosomes was positively correlated with expression changes for the majority of shared DEGs on the X chromosome, expression level changes occurred in both directions for autosomal DEGs. This discovery of shared DEGs implicates the existence of common molecular mechanisms for the regulation of gene expression levels that function in a linear fashion when transmitting the X chromosome dosage changes in TS and KS to the transcriptome.
Dosage Effects of Sex Chromosome Copy Number Are Conserved across Sex Chromosome Karyotypes.
We next examined the DEGs discovered when comparing other groupings of the samples. Among the 6 comparisons that were carried out, X0 vs. XXY showed the most differential expression spectrum with 1,984 DEGs (Fig. 3B), which was in line with the larger difference in sex chromosome number between these two cohorts than between any other two cohorts, and is also in line with the additive effects of the number of X chromosomes on escape gene expression and Y chromosomal gene expression effects. The number of DEGs when comparing X0 vs. XY was 1,223, similar to X0 vs. XX (1,142 DEGs), and both of these comparisons demonstrated much higher DEG counts relative to other comparisons, indicating that X monosomy resulted in the most pronounced expression differences, regardless of whether the second missing chromosome is X or Y. This finding indicates that the predominant impact of sex chromosome number is driven by changes in X-Y homologous PAR regions. The relative difference of 81 DEGs between the X0 vs. XX and X0 vs. XY comparisons may be consistent with an admixture of effects related to XCI escape or gene expression related to the Y chromosome.
To further evaluate the impact of Y chromosome expression, we also assessed DEG comparisons for XXY vs. XX and X0 vs. XY. We observed a similar pattern of dose-related differences nonuniformly weighted toward X monosomy. We identified 1,223 DEGs in X0 vs. XY but only 211 DEGs in XXY vs. XX (SI Appendix, Fig. S4). The difference in DEG count is particularly worthy of consideration given the same disparity of one Y chromosome in both comparisons. It appears that an additional Y chromosome can have different effects subject to the occurrence of the X chromosome inactivation. On the X chromosome, there were 58 DEGs in X0 vs. XY and 20 DEGs in XXY vs. XX (SI Appendix, Fig. S4). All but 2 of the 82 shared DEGs between the two comparisons exhibited expression differences in opposite directions (SI Appendix, Fig. S5). Our finding highlights the preservation of X-Y dosage compensation mechanisms, given similarities between XX and XY DEGs number in comparison to X monosomy, while also raising the potential effect of additional Y chromosome-specific effects.
We then examined the degree of overlap between these two cohort comparisons—given the divergent expression changes of shared disease-associated genes in TS and KS, we hypothesized that overlapping DEGs between any two comparisons would follow the same pattern due to dosage effects of differential sex chromosome number. As expected, almost all of the overlapping DEGs demonstrate similar directions of differential expression, depending on the order of comparison (Fig. 3B). The number of overlapping DEGs was the largest for the comparisons X0 vs. XXY and X0 vs. XY, followed by X0 vs. XXY and X0 vs. XX. Taken together, dosage effects of differential sex chromosome numbers were conserved across comparisons of any two cohorts.
We also identified DEGs for the comparison of XX vs. XY karyotypes, which are equivalent in sex chromosome number, but divergent in X-Y chromosome effects. When examining overlapping DEGs between this comparison with X0 vs. XX and XX vs. XXY comparisons, we identified a subset of genes with shared differential expression patterns (SI Appendix, Table S1), which is consistent with XCI escape effects across the X chromosome and autosomes. It is noteworthy that this subset of 17 overlapping DEGs are not specific to aneuploidy, but present even in typically developing cohorts.
Expressions of DEGs on the X Chromosome and Autosomes Are Correlated in TS and KS.
Considering that X chromosome dosage-sensitive DEGs were distributed across the transcriptome, we performed weighted correlation network analysis (WGCNA) (41) of gene expressions to identify the relationship between these genes. Analysis of all individuals of X0 and XX together identified 25 coexpression modules. Among the 8 modules significantly associated with X monosomy (SI Appendix, Fig. S6 A and B), the yellow module was enriched with immune function-related pathways (SI Appendix, Fig. S6C), which was in line with the finding by pathway enrichment analysis of DEGs. Interestingly, the “regulation of body fluid levels” pathway was enriched in the cyan module (SI Appendix, Fig. S6D), a biological effect that might be associated with one of the phenotypic traits commonly identified in TS—lymphedema of the hands and feet in early development. WGCNA of all individuals of XXY and XY together identified 33 coexpression modules; however, no module was significantly associated with KS after multiple testing correction (SI Appendix, Fig. S7 A and B). The most related module was the light-yellow module, which was also enriched with pathways (SI Appendix, Fig. S7C) grossly overlapping with the cyan module from the analysis of X0 and XX.
To investigate if the DEGs on the X chromosome and on the autosomes coexpress, we calculated the number of these genes within each module. For X0 vs. XX, all of the 72 X chromosome DEGs were assigned to 12 modules (SI Appendix, Table S2). Of the 1,070 autosomal DEGs, 975 were also assigned to these modules, which was an extremely significant enrichment (Fisher’s exact test P value <2.2E-16, SI Appendix, Table S2). For XXY vs. XY, 40 of the 44 DEGs on the X chromosome were assigned to 11 modules, which contained 129 of 197 autosome DEGs (SI Appendix, Table S3). This also resulted in significant enrichment of autosomal DEGs within the same modules as the X chromosome DEGs (Fisher’s exact test P value = 5.99E-04). Taken together, the X chromosome DEGs and autosomal DEGs tend to be assigned to the same modules and coexpress both in X0 vs. XX and in XXY vs. XY comparison, which implies that expression changes of autosomal genes are ripple effects of the X chromosome genes propagated through regulation of expression networks.
WGCNA on all four cohorts together identified 28 modules. Remarkably, 53 of the 63 shared autosomal DEGs in TS and KS were located within the same 3 coexpression modules with the shared X chromosome DEGs (SI Appendix, Table S4 and Dataset S3, Fisher’s exact test P value <2.20E-16). Expression ratios of the X chromosome DEGs between the four groups were conserved on the autosomal DEGs, indicating the existence of overlapping ripple effects of the X chromosome genes on autosomal genes between TS and KS. Intriguingly, the eigengenes of 3 modules were significantly correlated with the number of sex chromosomes (SI Appendix, Fig. S8 A and B): positive correlation for the purple module (P value = 4.73E-11, SI Appendix, Fig. S8C) and negative correlation for the light-yellow (P value = 6.67E-06, SI Appendix, Fig. S8D) and blue module (P value = 1.29E-04, SI Appendix, Fig. S8E). Among the 26 shared differentially expressed escape genes between TS and KS, 14 were clustered in the purple module and 8 were clustered in the turquoise module (Dataset S3).
ZFX May Play a Key Role in Ripple Network Effects of X Chromosome Dosage Change on Global Transcriptome in TS.
The enrichment of the X chromosome dosage-sensitive genes within the same coexpression modules implicates shared transcriptional activity mediated by a regulatory network. We proposed that this could take the form of DEGs encoding transcription factors (TFs) located on the X chromosome, that subsequently regulate functional targets on autosomes as well as on the X chromosome itself. To test this hypothesis, we performed an analysis identifying enrichment of TF binding motifs in the promoter and enhancer regions for all of the DEGs identified in TS and KS comparisons. Enhancer–target interactions were derived from existing data characterizing the transcriptional regulatory network in primary T cells and B cells, which was constructed using the paired expression and chromatin accessibility (PECA) model (42). We observed that the binding motif of ZFX was significantly enriched in the enhancers of down-regulated DEGs in TS (P value = 1.00E-6) but not in KS. Of note, ZFX is an escape gene on the X chromosome and was significantly down-regulated in TS, but up-regulated in KS. Our results identified ZFX as one of the potential hub genes mediating the regulatory network changes in TS in primary cells, after having been implicated in such a role in EBV-transformed cell lines (20).
Interestingly, eight DEGs on the X chromosome characterized as undergoing XCI, are targets for regulation by ZFX directly, or by differentially expressed TFs encoded on the autosomes. Some of these autosomal TFs are also targets for regulation by ZFX. Moreover, the up-regulated expression for six of these eight inactive genes in X0 is consistent with the annotation (activation/repression) by the PECA model (SI Appendix, Fig. S9), indicating that ZFX can impact inactive genes on the X chromosome via transcription network.
The X Chromosome Is Hypomethylated in TS but Hypermethylated in KS.
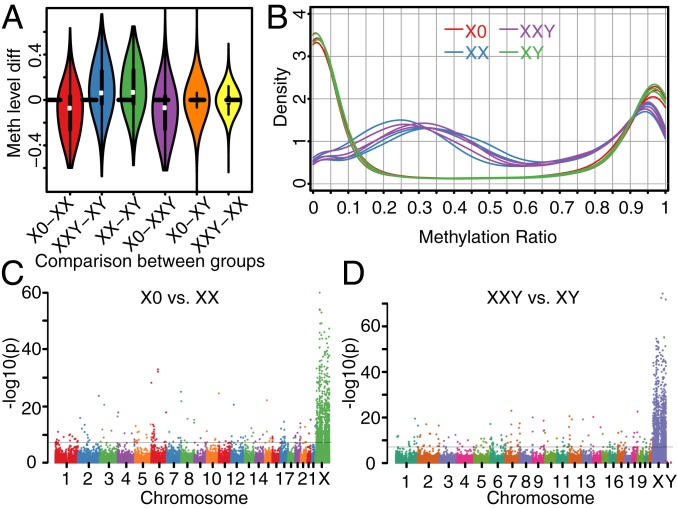
To analyze the genomewide methylation profiles associated with TS and KS, we performed genomewide targeted-capture bisulfite sequencing on the genomic DNA from primary cells from 12 individuals (3 X0, 3 XX, 3 XY, and 3 XXY). The inactive X chromosome in XX karyotypes has been reported to show increased methylation levels relative to the active X chromosome at the majority of CpGs but also decreased methylation level for 7% of CpGs (21). As XX and XXY similarly carry both an active and inactive X chromosome, the methylation levels measured represent a combination of methylation patterns for two X chromosomes. We observed that the X chromosome methylation patterns in XX and XXY were similar and showed pronounced differences compared to X0 and XY (SI Appendix, Fig. S10). Specifically, the methylation levels of the X chromosome were 10.1% lower in X0 compared to XX and 9.5% higher in XXY compared to XY (Fig. 4A), demonstrating that the methylation profile was determined by the X chromosome number (Fig. 4A).
Fig. 4.
Comparison of methylation levels across groups. (A) Methylation level difference of the CpGs on the X chromosome for comparisons of different groups. (B) Distribution of methylation levels of CpGs on the X chromosome for individuals. (C) Differential methylation analysis between TS patients and female controls. (D) Differential methylation analysis between KS patients and male controls. −log10 (P values) across the genome are shown and genomewide significance is based on FDR <0.05 indicated by the horizontal black lines in C and D.
Further analysis identified a decrease in CpGs with methylation levels of 0 to 10% and >90% but a concomitant increase in CpGs between 10% and 90% methylation in XX and XXY compared to X0 and XY (Fig. 4B). While the methylation profile shift from low overall methylation (∼5%) to medium levels (∼35%) in XX and XXY was consistent with the generally increased methylation of the inactive X chromosome, the reductions in methylation level of >90% were a reflection of the CpGs with decreased methylation.
Inactive X Chromosome Genes with Differentially Methylated Promoters Were Hypomethylated in TS but Hypermethylated in KS.
We performed differential methylation analysis between patients and the corresponding euploid controls to identify differentially methylated regions (DMRs) in TS and KS. Genomewide, we detected 559 DMRs in X0 vs. XX (Dataset S4) and 677 DMRs in XXY vs. XY (Dataset S5). As expected, the majority of these DMRs (495 for TS and 613 for KS) were located on the X chromosome in both TS and KS (Fig. 4 C and D). Interestingly, we also identified DMRs on autosomes; namely, we identified 39 hypomethylated DMRs and 25 hypermethylated DMRs in X0 compared to XX, and 30 hypomethylated DMRs and 34 hypermethylated DMRs in XXY compared to XY.
Only 7 of the 64 autosomal DMRs from XXY vs. XY comparison in our study overlapped with the differentially methylated positions reported by a recent study (18), which performed DNA methylation profiling of KS patients in leukocytes using the human 450K-Illumina Infinium assay. This may relate to the different approaches to profiling methylation patterns. The human 450K-Illumina Infinium assay is a microarray platform containing ∼480,000 CpG sites, while the SeqCap Epi CpGiant used in our study is a capture system followed by sequencing to interrogate far more CpGs (>5.5 million).
Unsurprisingly, these DMRs on the X chromosome were significantly enriched within promoters of genes categorized as undergoing XCI (Fisher’s exact test P value = 2.10E-05 for TS and 3.34E-05 for KS). For X0 vs. XX, among the 197 known inactive genes (29) with DMRs in their promoters, all were hypomethylated in X0. For XXY vs. XY, all but 1 of the 229 known inactive genes (29) with DMRs in their promoters were hypermethylated in XXY. The majority of these genes (186 genes) were shared between the X0 vs. XX and XXY vs. XY comparisons (SI Appendix, Fig. S11).
In contrast to genes subject to XCI, only six escape genes in X0 and seven in XXY displayed differentially methylated promoters; specifically they were all hypomethylated in X0 and all hypermethylated in XXY. All six of the hypomethylated escape genes in X0 overlapped with the hypermethylated escape genes in XXY.
Methylation and Expression Changes Are Complementary Rather than Overlapping in both TS and KS.
In X0 vs. XX, among the 369 genes with DMRs in their promoters, 220 were expressed. Consistent with our hypothesis that the majority of these DMRs correlate to genes undergoing XCI which would compensate for dose imbalance across X0 and XX karyotypes, we identified only 30 DMRs in promoters for genes that were also identified as DEGs. Notably, 26 of them were hypomethylated and showed up-regulated gene expression in X0.
Among the 423 genes with DMRs in their promoters in the XXY vs. XY comparison, 242 were expressed and only 4 genes—CX0rf57, NKRF, FLNA, and HCG11—were DEGs. The promoters of these 4 genes were hypermethylated. CX0rf57, FLNA, and HCG11 were down-regulated in XXY while NKRF was up-regulated. Our results suggest that the methylation changes in TS and KS are not necessarily reflected in the gene expression level in PBMCs.
Chromatin Conformations of the X Chromosome in TS Exhibit Features of an Active X Chromosome.
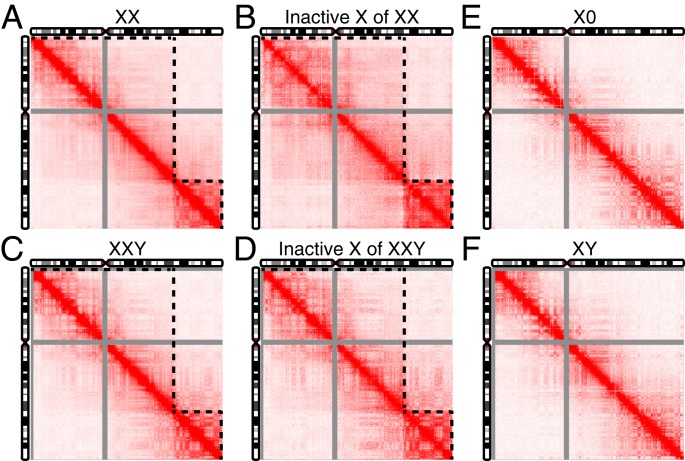
To investigate the 3D architecture of the X chromosome in TS and KS, we constructed in situ Hi-C chromosome contacts in LCLs derived from individuals with TS and KS and corresponding euploid controls. Previous studies have reported that the Hi-C map of the inactive X chromosome in females is partitioned into two superdomains but the active X chromosome is not (43, 44). Consistent with this, the two superdomains were observed in both the diploid contact map of two X chromosomes combined (Fig. 5A) and the haploid contact map of only the inactive X chromosome (Fig. 5B) in the 46XX control. The same structure was seen in the male with KS (Fig. 5 C and D). However, the two superdomains were not present in the haploid contact map of the X chromosome in the individual with TS (Fig. 5E), nor in the male control (Fig. 5F), consistent with single X chromosome status for both these karyotypes. Furthermore, the A/B compartments of the X chromosome in the individual with TS and the male control showed the same pattern (SI Appendix, Fig. S12). Taken together, the X chromosome in TS exhibits chromatin conformations characteristic of an active X chromosome in a 46XX karyotype, while the 3D contacts of the two X chromosomes in KS resemble the diploid 46XX karyotype.
Fig. 5.
Chromatin conformations of the X chromosome in TS and KS patients and their euploid control. (A) Diploid contact map of the X chromosome of female control. (B) Haploid contact map of the inactive X chromosome of female control. (C) Diploid contact map of the X chromosome of KS patient. (D) Haploid contact map of the inactive X chromosome of KS patient. (E) Haploid contact map of the X chromosome of TS patient. (F) Haploid contact map of the X chromosome of male control. The contact map in A and C is a combination of contacts of the active and the inactive X chromosome for female control and KS patient, respectively. Contact matrix in B was generated by subtracting the contacts of the X chromosome in the TS patient from the combined contacts of two X chromosomes of the female control. Contact matrix in D was generated by subtracting the contacts of the X chromosome in the male control from the combined contacts of two X chromosomes of the KS patient. All of the contact maps were normalized using the Knight–Ruiz matrix balancing algorithm. Intensity of contacts is represented by a scale from 0 (white) to 250 (deep red). Contact maps are shown at 200-kbp resolution. Contacts of PAR1 and PAR2 are not included for the KS patient and male control due to homology of the X and Y chromosome. Dashed black lines indicate the two superdomains.
Discussion
Here we carried out comparative and integrative network analysis of transcriptomes, DNA methylation profiles, and chromatin conformations of two SCAs—TS and KS. This study utilizes RNA-Seq, bisulfite sequencing, and in situ Hi-C to study both monosomic and trisomic SCAs in direct comparison and in primary patient cells (and in patient-derived LCLs for the Hi-C analyses).
As expected, we found that genes in PAR1 were uniformly down-regulated in X0 and up-regulated in XXY relative to euploid karyotypes, indicating that these genes are sensitive to sex chromosome dosage. However, we did not observe the same pattern for genes in PAR2. Similar to PAR1 genes, escape genes which were differentially expressed between SCAs and euploids were also down-regulated in X0 and up-regulated in XXY, and thus shown to be specifically sensitive to X chromosome dosage. Interestingly, the vast majority of these differentially expressed escape genes are shared between TS and KS, suggesting that the same set of escape genes may play a substantial role in the development of phenotypic traits associated with SCAs. In particular, the genes in PAR1 and escape genes were differentially expressed in opposite directions in TS and KS relative to sex-matched controls, providing a compelling premise for phenotypic observations in specific traits that appear to also demonstrate inverse correlation across these syndromes.
We found that some inactive genes on the X chromosome are also dosage sensitive, which is in line with a previous study (20). Comparison of TS with female controls demonstrated overexpression of 35 genes that were mostly subject to XCI, which is the opposite of what would be expected. Among the 22 up-regulated inactive DEGs in X0, 18 of them were reported in a previous study on LCLs (20) and most of them (13 of 18) were similarly up-regulated in X0 vs. XX. The up-regulation of these genes may represent an indirect effect of X chromosome loss in TS, where the loss of the X chromosome primarily results in down-regulation of PAR1 genes and escape genes, causing downstream expression changes in autosomal genes through the regulatory network, which in turn exerts a counterintuitive up-regulation of inactive genes on the X chromosome. In contrast, only four DEGs on the X chromosome were found to be down-regulated in XXY relative to XY, with the remainder following an expected up-regulated expression pattern, suggesting genes subject to XCI in the second X chromosome in KS, have a much less pronounced impact on the KS phenotype compared to TS, which is consistent with observations of clinical phenotype.
We also found X chromosome dosage sensitivity extending beyond the sex chromosomes into autosomal regions in both TS and KS. Our WGCNA results show that DEGs on the X chromosome and autosomes are coexpressed across the four groups, indicating that autosomal DEGs are consequences of X chromosome dosage changes via transcription network regulation. Interestingly, we observed two TF genes—ZFX and ZBED1—to be differentially expressed in both TS and KS patients. Target genes of ZFX are significantly enriched in the down-regulated DEGs in TS, suggesting ZFX might be a key mediator in the regulatory network. Moreover, the transcriptome changes in SCAs appear not to be caused by ASE or expression quantitative trait loci (eQTLs) as we did not observe evidence of different patterns of ASE and eQTLs between X0 vs. XX or between XXY vs. XY, suggesting the dosage effect of the X chromosome as being the major contributing factor of transcription network changes in SCAs.
Although DEGs on the X chromosome are preferentially concentrated on Xp in both TS and KS, which raises questions for future research about soluble and nonsoluble factors involved in access to chromatin and chromatin conformations of short and long arms of the X chromosome, our findings support a central role of Xp genes underlying TS, whereas an elevated dose of these genes does not play a major role in KS. Previous studies focused on phenotypes indicated that the causative genes of TS are mainly on Xp (25–28) while the causative genes of KS are primarily located on Xq (32). This also demonstrates that overexpression of Xq plays a much more important role in the development of KS than dosage insufficiency of Xq in TS. Additionally, most females with Xp duplication appear phenotypically normal (45), while males with Xq duplication are more severely affected (46). In summary, the consequences of dosage insufficiency of Xp genes in females are more severe than overexpression of Xp genes in males, whereas the overexpression of Xq genes in males exerts larger impacts than both dosage insufficiency and overexpression of Xq genes in females.
While insufficiency and overdose of genes on the X chromosome impact phenotypes to varying extents, the same phenomenon was also observed for autosomal genes. For instance, genes in the pathway “regulation of body fluid levels” were enriched within coexpression modules associated with both TS and KS as identified by WGCNA. The seven autosomal DEGs (F13A1, ILK, ITGB3, TREML1, MYL9, ITGA2B, and VCL) that are shared between TS and KS in this pathway were all up-regulated in TS and down-regulated in KS. However, only TS has relevant symptoms such as lymphedema of the hands and feet in early development, whereas KS does not, indicating that up-regulation of the pathway might lead to disease while down-regulation not, which is reminiscent of the importance of the directions of expression changes together with the aforementioned varying effects of insufficiency and overexpression of Xp and Xq.
We identified 41 DEGs on Xp associated with TS, among which 26 were escape genes whose expression changes were presumably direct effects of X chromosome dosage change. A previous study has predicted a list of candidate genes for X aneuploidy syndromes based on dosage-sensitive genes involved in large complexes (7). We observed differential expression of 5 of them—EIF1AX, USP9X, MED14, HCFC1, and MAGEE1—in TS. Additionally, escape genes with a Y homolog experience the strongest purifying selection during sex-chromosome evolution and thus the persistence of a Y homolog suggests the importance of dosage balance for these genes (47), indicating that disruption of the strong constraints on dosage of these genes may results in disease. In our PBMC samples we detected expressions of 11 of the 19 genes which have X and Y homologs (48). Nine of the 10 differentially expressed X–Y pair genes (EIF1AX and USP9X included) between TS and female controls were on Xp (SI Appendix, Table S5). While the genotype–phenotype relationships for these genes warrant further study, our results provide top candidate genes for the TS phenotype. As an example, one of the most significant pathways identified from TS DEGs was immune response (SI Appendix, Fig. S2A). Consistently, TS has been associated with a number of autoimmune manifestations such as autoimmune thyroiditis and inflammatory bowel disease (49). Two X chromosome genes—TLR8 and CYBB—in this pathway were differentially expressed in X0 vs. XX. As lymphocytes are major components of the immune response, these two genes may contribute to the predisposition for autoimmune disease in TS.
We were also able to identify six potential candidate genes—RPS4X, SEPT6, NKRF, CX0rf57, NAA10, and FLNA—for KS on Xq. RPS4X exhibited the highest expression in XXY among all of the DEGs on the X chromosome between XXY vs. XY (sixth highest among all expressed genes on the X chromosome). Higher expression was observed in XXY (fragments per kilobase of transcript per million mapped reads [FPKM] 860.8) than XY (FPKM 597.8) due to escape from XCI. RPS4X encodes ribosomal protein S4, a component of cytoplasmic ribosomes. Ribosomal protein S4 can also be encoded by RPS4Y in males, whose isoforms are not identical, but are functionally equivalent to RPS4X. However, no differential expression was observed for RPS4Y between XXY vs. XY (FPKM 204.8 vs. 209.4), indicating that the significant increase in expression of ribosomal protein S4 results exclusively from higher expression of RPS4X in XXY. RPS4X is highly expressed in sex organs such as breast in both females and males (also ovary and uterus in females and prostate in males) (50). One common clinical feature of KS patients is gynecomastia (i.e., breast enlargement) and KS patients have been reported to have an increased risk of developing breast cancer (3), and RPSX4 may be a candidate gene for contributing to these phenotypic characteristics. Regarding SEPT6, it has been reported to be subject to X inactivation and displays female-biased expression (29). We observed higher expression of SEPT6 in XXY vs. XY (FPKM 85.4 vs. 66.0). SEPT6 is a member of the septin gene family, which are small GTPase proteins required for proper functioning of actin and the microtubule cytoskeleton (51). Together with other septins, SEPT6 is an essential structural component of the human sperm annulus and required for sperm motility during postmeiotic differentiation (52). Testis is one of the few tissues with high expression of SEPT6 in males. Interestingly, small testes and azoospermia are common features observed in KS patients. Lastly, we also observed higher expression of NKRF and lower expression of CX0rf57 in XXY relative to XY. Although both genes exhibited low expression in our PBMC samples, they are almost exclusively highly expressed in brain tissues (e.g., frontal cortex, cerebellar hemisphere, and cerebellum for NKRF; pituitary, hypothalamus, and nucleus accumbens for CX0rf57) (50). Previous studies have reported aberrant brain structure in the prefrontal cortex, cerebellum, and lateral ventricles of KS patients (53, 54). Furthermore, impairments in motor function are common features of KS patients, which might be due to neuroanatomical and functional changes in the associated brain regions. Our results pinpoint two candidate genes for the genetic basis of cognitive and neurological features of KS patients. Higher expression of NAA10 and lower expression of FLNA were observed in XXY. Both genes are important for development.
Genomewide, we identified 1,142 DEGs in TS and 241 DEGs in KS. Interestingly, there is a “core-group” of 94 DEGs that are present in both TS and KS, which are located on both the X chromosome (31 DEGs) and autosomes (63 DEGs). Strikingly, all but 1 of these 94 core-group DEGs change their levels of expression into opposite directions in TS vs. KS relative to respective controls (XX and XY). Further investigation of these genes in the dataset of a previous study, that used LCLs from TS and KS patients (20), showed that most of them (47 of 67 genes in both datasets) also exhibited the same pattern in LCLs. One explanation for this pattern could be that the copy number changes of the X chromosome in X0 and XXY result in the divergent expression pattern of the shared X chromosome DEGs (mainly escape genes, which are X chromosome dosage sensitive). The effect of the divergent expression of the shared X chromosome DEGs are then transmitted to the shared autosomal DEGs through gene expression network regulation, as these genes are coexpressed. Both the shared autosomal and X chromosome DEGs might underlie the fact that a subset of phenotypic characteristics of TS and KS appear to follow a linear dose-dependent relationship.
Broad hypomethylation of the X chromosome is observed in TS, whereas hypermethylation of chromosome X is present in KS. The methylation profile of the X chromosome in TS resembles that of male controls while the profile in KS resembles that of female controls. Inactive genes with DMRs in promoters are hypomethylated in TS but hypermethylated in KS while the methylation levels of escape genes mostly remain unchanged. Both hypomethylated and hypermethylated regions are present on autosomes in both TS and KS. A previous study has reported that methylation and expression changes are not overlapping but complementary in TS (9). Our results confirmed this finding in TS and also extended the finding to KS by showing that genes with methylation changes and expression changes tend not to overlap. However, the few genes exhibiting both methylation and expression changes are of particular interest and warrant further investigation, especially CX0rf57 and NKRF in KS.
Our in situ Hi-C results showed that the chromatin conformations of the one X chromosome in a TS patient resembled the active X chromosome in male and female control. The diploid contact map of X chromosomes in the KS patient exhibited the same pattern as in the female control, suggesting that the Y chromosome might have little impact on the 3D architecture of the X chromosome.
One of the strengths of our study, the use of primary tissue from both TS and KS in a direct comparative functional genomics analysis, also constitutes one of its limitations, since the primary cells in question were PBMCs, which are only one of the relevant tissues for TS and KS. However, the analysis presented here provides a foundation for further progress into understanding causative mechanisms of the phenotypes seen in SCAs that are likely far reaching and warrants further similar investigation in other tissue types. Additional questions that are of immediate interest in follow-up studies are the effects of Y chromosome dosage and those of complex epigenetic factors such as chromatin conformation.
Given that patients with TS and KS are prone to autoimmune diseases and one of the most significant pathways identified was immune response, altered cell composition might be speculated in the blood of the patients. However, we did not observe strong evidence of distinct fractions of any immune cell types between patients and their sex-matched controls by deconvolving our RNA-Seq data (SI Appendix, Fig. S13) and methylation sequencing data (SI Appendix, Fig. S14). Another question open for interrogation is whether heterogeneity of epigenomic changes exists among different immune cell types in patients with TS and KS. Future studies using single-cell technologies will help to determine the molecular basis of these epigenomic changes at higher resolution.
Methods
The local institutional review board of the Stanford University School of Medicine approved this study and informed written consent was obtained from a legal guardian for all participants, as well as written assent for participants greater than 7 y of age. Details of sample cohorts, experimental procedures, and data analysis are included in SI Appendix.
Data Availability.
RNA-Seq, DNA methylation, and in situ Hi-C data from this study have been deposited to GEO under accession no. GSE126712.
Supplementary Material
Acknowledgments
This research was supported by external funding to A.E.U. from the National Human Genome Research Institute grant HG007735-01 (Principal Investigator (PI) Howard Chang to A.E.U.), as well as funds from Stanford Child Health Research Institute (Tashia and John Morgridge Faculty Scholar) and departmental funds from the Stanford Department of Psychiatry and Behavioral Sciences. We thank Dr. Armin Raznahan (National Institute of Mental Health) for sharing the results of differential expression analysis of X0, XX, XXY, and XY in LCLs.
Footnotes
Reviewers: C.J.B., University of British Columbia; and J.F.C., Emory University School of Medicine.
The authors declare no competing interest.
Data deposition: RNA-Seq, DNA methylation, and in situ Hi-C data from this study have been deposited to GEO under accession no. GSE126712.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910003117/-/DCSupplemental.
References
- 1.Nielsen J., Wohlert M., Chromosome abnormalities found among 34,910 newborn children: Results from a 13-year incidence study in Arhus, Denmark. Hum. Genet. 87, 81–83 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Hong D. S., Reiss A. L., Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol. 13, 306–318 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Bojesen A., Gravholt C. H., Klinefelter syndrome in clinical practice. Nat. Clin. Pract. Urol. 4, 192–204 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Rao E., et al. , Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat. Genet. 16, 54–63 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Ottesen A. M., et al. , Increased number of sex chromosomes affects height in a nonlinear fashion: A study of 305 patients with sex chromosome aneuploidy. Am. J. Med. Genet. A. 152A, 1206–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrel L., Willard H. F., X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Pessia E., Makino T., Bailly-Bechet M., McLysaght A., Marais G. A., Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc. Natl. Acad. Sci. U.S.A. 109, 5346–5351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakalov V. K., Cheng C., Zhou J., Bondy C. A., X-chromosome gene dosage and the risk of diabetes in Turner syndrome. J. Clin. Endocrinol. Metab. 94, 3289–3296 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trolle C., et al. , Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Sci. Rep. 6, 34220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massingham L. J., et al. , Amniotic fluid RNA gene expression profiling provides insights into the phenotype of Turner syndrome. Hum. Genet. 133, 1075–1082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajpathak S. N., et al. , Human 45,X fibroblast transcriptome reveals distinct differentially expressed genes including long noncoding RNAs potentially associated with the pathophysiology of Turner syndrome. PLoS One 9, e100076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajpathak S. N., Deobagkar D. D., Micro RNAs and DNA methylation are regulatory players in human cells with altered X chromosome to autosome balance. Sci. Rep. 7, 43235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R., et al. , Gene expression analysis of induced pluripotent stem cells from aneuploid chromosomal syndromes. BMC Genom. 14 (suppl. 5), S8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Aurora M., et al. , Deregulation of sertoli and leydig cells function in patients with Klinefelter syndrome as evidenced by testis transcriptome analysis. BMC Genom. 16, 156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winge S. B., et al. , Transcriptome profiling of fetal Klinefelter testis tissue reveals a possible involvement of long non-coding RNAs in gonocyte maturation. Hum. Mol. Genet. 27, 430–439 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Huang J., et al. , Global transcriptome analysis of peripheral blood identifies the most significantly down-regulated genes associated with metabolism regulation in Klinefelter syndrome. Mol. Reprod. Dev. 82, 17–25 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Zitzmann M., et al. , Gene expression patterns in relation to the clinical phenotype in Klinefelter syndrome. J. Clin. Endocrinol. Metab. 100, E518–E523 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Skakkebæk A., et al. , DNA hypermethylation and differential gene expression associated with Klinefelter syndrome. Sci. Rep. 8, 13740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viana J., et al. , Epigenomic and transcriptomic signatures of a Klinefelter syndrome (47,XXY) karyotype in the brain. Epigenetics 9, 587–599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raznahan A., et al. , Sex-chromosome dosage effects on gene expression in humans. Proc. Natl. Acad. Sci. U.S.A. 115, 7398–7403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp A. J., et al. , DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 21, 1592–1600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma A., et al. , DNA methylation signature in peripheral blood reveals distinct characteristics of human X chromosome numerical aberrations. Clin. Epigenetics 7, 76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajpathak S. N., Deobagkar D. D., Evidence for epigenetic alterations in Turner syndrome opens up feasibility of new pharmaceutical interventions. Curr. Pharm. Des. 20, 1778–1785 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Wan E. S., et al. , Genome-wide site-specific differential methylation in the blood of individuals with Klinefelter syndrome. Mol. Reprod. Dev. 82, 377–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geerkens C., Just W., Held K. R., Vogel W., Ullrich-Turner syndrome is not caused by haploinsufficiency of RPS4X. Hum. Genet. 97, 39–44 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Lachlan K. L., Youings S., Costa T., Jacobs P. A., Thomas N. S., A clinical and molecular study of 26 females with Xp deletions with special emphasis on inherited deletions. Hum. Genet. 118, 640–651 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Kaiser P., Harprecht W., Steuernagel P., Daume E., Long arm deletions of the X chromosome and their symptoms: A new case (bp q24) and a short review of the literature. Clin. Genet. 26, 433–439 (1984). [DOI] [PubMed] [Google Scholar]
- 28.Geerkens C., Just W., Vogel W., Deletions of Xq and growth deficit: A review. Am. J. Med. Genet. 50, 105–113 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Tukiainen T., et al. ; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz , Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naqvi S., et al. , Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 365, eaaw7317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Höckner M., et al. , Unravelling the parental origin and mechanism of formation of the 47,XY,i(X)(q10) Klinefelter karyotype variant. Fertil. Steril. 90, 2009.e13-7 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Song S. H., et al. , A case of the rare variant of Klinefelter syndrome 47,XY,i(X)(q10). Clin. Exp. Reprod. Med. 40, 174–176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian D., Sun S., Lee J. T., The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Augui S., Nora E. P., Heard E., Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429–442 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Xiong Y., et al. , RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 42, 1043–1047 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Nguyen D. K., Disteche C. M., Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38, 47–53 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Lin F., Xing K., Zhang J., He X., Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc. Natl. Acad. Sci. U.S.A. 109, 11752–11757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Esposito M., et al. , A synaptobrevin-like gene in the Xq28 pseudoautosomal region undergoes X inactivation. Nat. Genet. 13, 227–229 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Ciccodicola A., et al. , Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum. Mol. Genet. 9, 395–401 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Cotton A. M., et al. , Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 14, R122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duren Z., Chen X., Jiang R., Wang Y., Wong W. H., Modeling gene regulation from paired expression and chromatin accessibility data. Proc. Natl. Acad. Sci. U.S.A. 114, E4914–E4923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darrow E. M., et al. , Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. U.S.A. 113, E4504–E4512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armstrong L., McGowan-Jordan J., Brierley K., Allanson J. E., De novo dup(X)(q22.3q26) in a girl with evidence that functional disomy of X material is the cause of her abnormal phenotype. Am. J. Med. Genet. A. 116A, 71–76 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Cheng S. F., Rauen K. A., Pinkel D., Albertson D. G., Cotter P. D., Xq chromosome duplication in males: Clinical, cytogenetic and array CGH characterization of a new case and review. Am. J. Med. Genet. A. 135, 308–313 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Park C., Carrel L., Makova K. D., Strong purifying selection at genes escaping X chromosome inactivation. Mol. Biol. Evol. 27, 2446–2450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bellott D. W., et al. , Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 (2014). Erratum in: Nature514, 126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lleo A., Moroni L., Caliari L., Invernizzi P., Autoimmunity and Turner’s syndrome. Autoimmun. Rev. 11, A538–A543 (2012). [DOI] [PubMed] [Google Scholar]
- 50.GTEx Consortium , The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weirich C. S., Erzberger J. P., Barral Y., The septin family of GTPases: Architecture and dynamics. Nat. Rev. Mol. Cell Biol. 9, 478–489 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Kuo Y. C., et al. , SEPT12 orchestrates the formation of mammalian sperm annulus by organizing core octameric complexes with other SEPT proteins. J. Cell Sci. 128, 923–934 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Itti E., et al. , The structural brain correlates of cognitive deficits in adults with Klinefelter’s syndrome. J. Clin. Endocrinol. Metab. 91, 1423–1427 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Lentini E., Kasahara M., Arver S., Savic I., Sex differences in the human brain and the impact of sex chromosomes and sex hormones. Cereb. Cortex 23, 2322–2336 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq, DNA methylation, and in situ Hi-C data from this study have been deposited to GEO under accession no. GSE126712.