Significance
Terminal uridylyltransferases are writers of poly(U) tails in diverse RNA uridylation pathways. In the let-7 pathway, the LIN28:pre-let-7:TUTase ribonucleoprotein complex regulates the processing and maturation of the let-7 microRNA, and plays an important role in both physiology and disease. The presence of redundant TUTase orthologs and the diverse pathways poses challenges to revealing the molecular mechanisms of TUTases. We developed a precise tool to identify nanobodies that inhibited the LIN28:pre-let-7:TUTase interaction. The nanobody Nb-S2A4 specifically interacted with the LIN28:let-7 interaction fragment in TUT4, which is critical for oligouridylation and monouridylation of let-7. Our data illustrate that the nanobody tool is effective in distinguishing functions among protein orthologs and diverse pathways.
Keywords: nanobody, LIN28, let-7, TUT4, oligouridylation
Abstract
The LIN28:pre-let-7:TUTase ternary complex regulates pluripotency and oncogenesis by controlling processing of the let-7 family of microRNAs. The complex oligouridylates the 3′ ends of pre-let-7 molecules, leading to their degradation via the DIS3L2 exonuclease. Previous studies suggest that components of this complex are potential therapeutic targets in malignancies that aberrantly express LIN28. In this study we developed a functional epitope selection approach to identify nanobody inhibitors of the LIN28:pre-let-7:TUT4 complex. We demonstrate that one of the identified nanobodies, Nb-S2A4, targets the 106-residue LIN28:let-7 interaction (LLI) fragment of TUT4. Nb-S2A4 can effectively inhibit oligouridylation and monouridylation of pre-let-7g in vitro. Expressing Nb-S2A4 allows maturation of the let-7 species in cells expressing LIN28, highlighting the therapeutic potential of targeting the LLI fragment.
The LIN28:pre-let-7:TUTase ribonucleoprotein complex (Fig. 1A) plays a key role in normal development and diseases by modulating the processing and maturation of the let-7 family of microRNAs, which in turn target mRNAs that control differentiation, pluripotency, proliferation, and oncogenesis (1–6). After assembly of the ternary complex, TUTase adds an oligoU tail to pre-let-7 microRNAs and facilitates their recognition and degradation by the exonuclease DIS3L2 (7–10). TUTase may also monouridylate a subset of pre-let-7 microRNAs (group II) independent of LIN28 to promote let-7 maturation (10, 11). Our group has demonstrated that two domains of LIN28, the cold-shock domain and the CCHCx2 zinc knuckle domain (ZKD), bind to the closed loop and the GGAG motif of let-7, respectively (12). We have also shown that the LIN28 ZKD is required and sufficient for TUT4 recruitment (13). Aberrant LIN28 or TUTase expression drives oncogenesis in various organs, including ovarian cancer, colon cancer, Wilms tumor, and liver cancer (14–20). As such, the components of the LIN28:pre-let-7:TUTase complex have been advanced as putative targets for therapy.
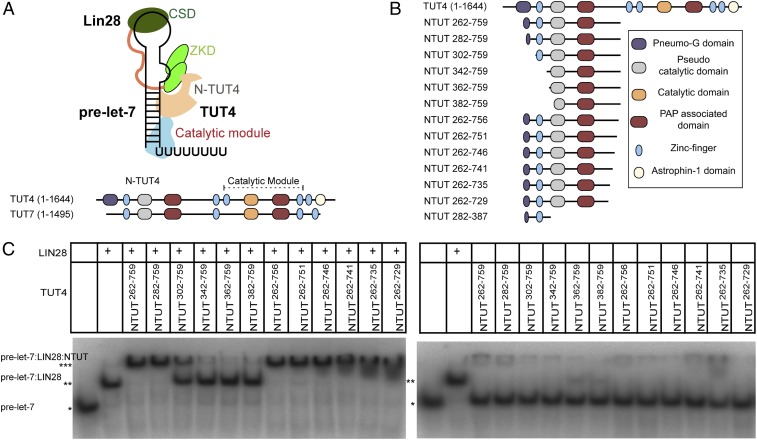
Fig. 1.
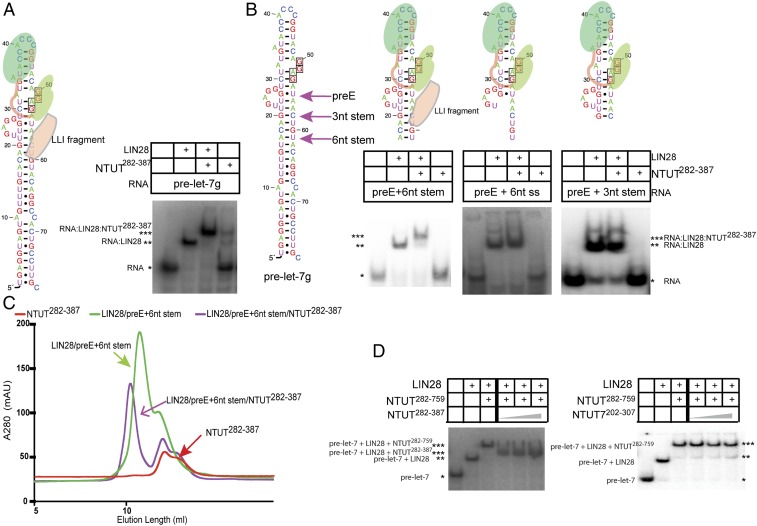
Determination of the functional regions of N-terminal TUT4 that forms a stable LIN28:pre-let-7:TUT4 ternary complex. (A) Schematic representation of LIN28:pre-let-7:TUT4 ternary complex. (B) Schematic representation of constructs of N-terminal TUT4. (C) RNA EMSAs of mouse LIN28 (1.25 µM) and NTUT4 constructs (2 µM) with P32-labeled pre-let-7g. *RNA alone; **RNA:LIN28 binary complex; ***RNA:LIN28:NTUT4 ternary complex.
TUTases are RNA-dependent terminal uridylyl transferases. Two of seven TUTase paralogs in humans, TUT4/Zcchc11 and TUT7/Zcchc6, interact with the LIN28:pre-let-7 complex (11). TUT4 and TUT7 contain two functional modules: The C-terminal catalytic module, which is conserved among all of the TUTase homologs from yeast to mammals, and the N terminus, which is unique to TUT4 and TUT7 (7–11). TUT4 also has two unique domains with unknown functions, an N-terminal Pneumovirinae attachment membrane glycoprotein G-like domain (Pneumo-G domain) and a C-terminal Astrophin-1–like domain (8, 10). The C-terminal catalytic module of TUT4 and TUT7 includes one active catalytic domain, one PAP-associated domain, and two zinc fingers. It is essential in both mono- and oligouridylation of pre-let-7 (11, 21–23). A recent structural analysis has revealed that a C-terminal zinc finger motif, ZK2, engages the oligoU tail of pre-let-7s and is crucial for the oligouridylation activity of TUT7 (21). The N terminus of TUT4/TUT7, which is catalytically inactive, directly interacts with the LIN28:pre-let-7 binary complex, switching the catalytic activity from monouridylation to oligouridylation (13). The atomic mapping of Lin28:pre-let-7:TUTase interactions remains unclear.
Precise molecular tools are needed to dissect the molecular mechanisms of pathogenesis and reveal potential opportunities for the development of novel therapies. Several studies have focused on identifying small-molecule inhibitors of LIN28:let7:TUT4 pathways (24–27). Alternative approaches with biologics, such as antibody fragments, would offer new advantages, including exquisite target selectivity and genetic control of inhibition, neither of which is possible with small molecules. Single domain antibody fragments (VHH) called nanobodies, which are derived from the VH domains of camelid or shark heavy-chain–only antibodies, are ideal candidates for protein inhibitors (28, 29). Scientists have often used nanobodies in protein crystallography and cell biology because of their small size, thermal stability, reversible refolding, high affinity, and target specificity, features that make them useful in clinical diagnosis and therapeutic discovery (30). Nanobodies have also been widely used as bioimaging reagents to detect intracellular signaling molecules and cancer biomarkers (31–33), and a newly developed synthetic nanobody library in yeast is advantageous because it allows rapid selection of targets without immunizing animals (34).
In this study, we devised a rapid functional epitope selection platform, which allowed us to directly isolate nanobodies that bind either to the interaction surface (inhibitory binders) or to the exposed surface (complex binders) of TUT4 in the ternary complex. The inhibitory nanobody Nb-S2A4 blocked TUT4 recruitment to the LIN28:pre-let-7 binary complex, which thereby inhibited the uridylation activities of TUT4 in vitro. Nb-S2A4 recognizes the critical TUT4 sequences, the LIN28:let7 interaction (LLI) fragment, required for assembly of the LIN28:pre-let-7:TUT4 ternary complex. When cells were transfected with Nb-S2A4, they consistently showed increased levels of endogenous let-7 and derepressed let-7 targeted reporters. Nb-S2A4 nanobody can, therefore, effectively rescue let-7 maturation in cells, which demonstrates that targeting assembly of the LIN28:pre-let-7:TUT4 ribonucleoprotein complex with biologics provides a viable venue to control oncogenesis.
Results
Identification of an Optimal Antigen for Nanobody Selection.
To identify an optimal functional unit for LIN28:pre-let-7:TUT4 ternary complex assembly, we generated a series of deletion constructs (Fig. 1B) of the previously identified N-terminal functional construct, NTUT230−759 (13). We compared the ability of each construct in the ternary complex formation using RNA electrophoretic mobility shift assays (EMSA) (Fig. 1 C, Left). N-terminally truncated constructs NTUT262−759 and NTUT282−759 supported full complex formation. In comparison, NTUT302−759 showed more than 50% loss of binding activity, suggesting the importance of the partial Pneumo-G domain or the linker region. More extensive N-terminal deletions (NTUT342−759; NTUT362−759; NTUT382−759) showed complete abrogation of binding activity. C-terminally truncated NTUT262−756 and NTUT262−751 retained full binding activities compared to NTUT262−759, while further C-terminal deletions in NTUT262−759 progressively diminished TUT4 binding to the LIN28:pre-let-7 binary complex. In the absence of LIN28, all TUT4 constructs minimally bound pre-let-7 (Fig. 1 C, Right). Given great stability and high overexpression level, NTUT282−759 was an optimal antigen for nanobody selection.
Library-Based Discovery of Complex and Inhibitory Nanobody Binders.
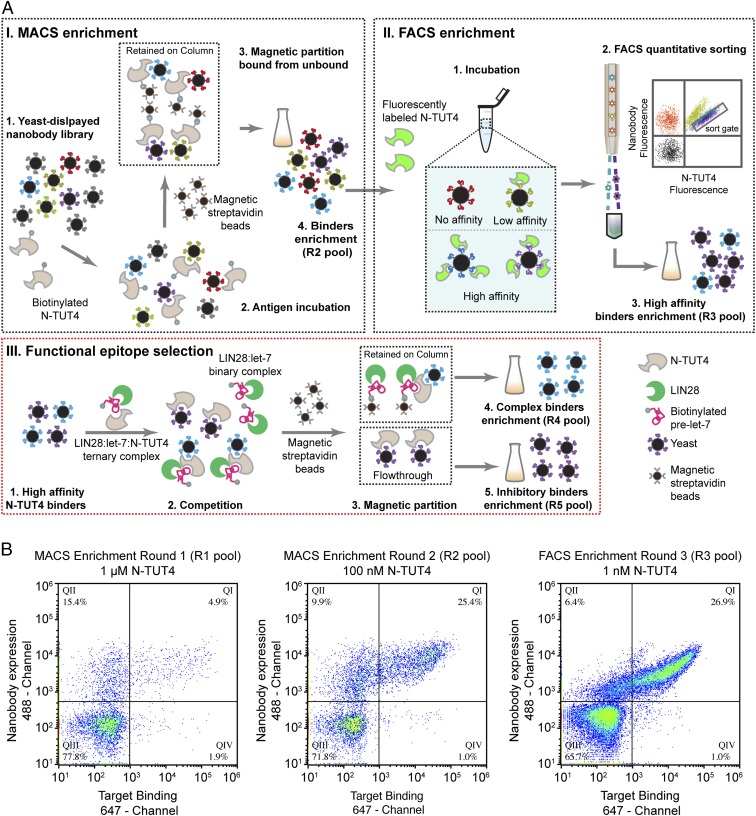
In order to further study the molecular function of the N-terminal TUT4 domain in vitro and in vivo, we developed a rapid functional epitope selection platform to identify nanobody inhibitors for NTUT282−759 (Fig. 2A). NTUT282−759 was lysine-based labeled with biotin or Alexa Fluor 488 by N-hydroxysuccinimide, and we confirmed that the labeling did not influence the binding activity of NTUT282−759 to the LIN28:pre-let-7 binary complex (SI Appendix, Fig. S1A). After two initial rounds of magnetic-activated cell sorting (MACS) (Fig. 2 A, I) enrichment, NTUT282−759 binders represented ∼25% of the total yeast cells in the R2 pool when stained with 100 nM biotinylated NTUT282−759 (Fig. 2B). After a subsequent round of FACS (Fig. 2 A, II) enrichment, ∼27% of the total yeast cells in the R3 pool were positively stained using 1 nM biotinylated NTUT282−759 (Fig. 2B) and their binding affinities were significantly improved compared to those of the R2 pool under staining conditions of 10 pM of biotinylated NTUT282−759 (SI Appendix, Fig. S1B).
Fig. 2.
Discovery of nanobody inhibitors of NTUT4. (A) Schematic flowchart of nanobody discovery for NTUT282−759. (I) MACS enrichment to enrich NTUT282−759 binders. (II) FACS enrichment to sort high-affinity binders. A schematic of flow cytometry data were drawn to illustrate the sort gate. (III) Inhibitor screening by a functional epitope selection process. (B) Flow cytometry of NTUT282−759 nanobody binders in two MACS enrichment pools (round 1 pool and round 2 pool) and FACS enrichment (round 3 pool). Nanobodies expressed on the yeast surface were labeled by Alexa Fluor 488 conjugated anti-HA antibodies. Biotinylated NTUT282−759 was stained with Alexa 647-conjugated streptavidin.
After obtaining the high-affinity R3 pool, we applied a competition assay with the preassembled ternary complex to directly segregate inhibitory nanobody clones, in which the yeast cell displaying nanobodies competed with the biotinylated RNA:LIN28 binary complex to bind to NTUT282−759 (Fig. 2 A, III). Introducing this step eliminated laborious EMSA assays for all candidate nanobodies for individual functional assessment. The nanobodies that recognized the exposed surfaces (R4 pool, labeled as complex binders) were separated from those that recognized the interaction interface (R5 pool, represented as inhibitory binders) of the LIN28:pre-let-7:N-TUT4 ternary complex (Fig. 2 A, III).
We subsequently isolated individual yeast clones from the R3, R4, and R5 pools and, using 96-well autocollection flow cytometry, found that ∼90% of the single clones in these pools displayed positive binding to NTUT282−759. Because NTUT282−759 is a relatively large antigen (54 kDa) and may enrich for a diverse set of nanobody binders, we performed another round of FACS enrichment using the R5 pool stained with 10 pM of Alexa Fluor 488 labeled NTUT282−759 to further enrich high-affinity inhibitory binders. The Nb-S2A4 clone was highly enriched and the complex binder nanobody clones from the R4 pool were diverse, with only 2 of 20 clones identical (named as Nb-CB16). Clones sequenced from each pool showed high diversity in their CDR3 regions (SI Appendix, Fig. S2).
We cloned and purified Nb-S2A4, a nanobody from the inhibitory R5 pool, as well as Nb-CB11 and Nb-CB16, from the complex binder (R4) pool, for further characterization. Two negative control nanobodies were chosen for downstream experiments: Nb.BV025 and Nb-R1A12. Nb.BV025 is a general negative control of the yeast display nanobody library with high expression and solubility but without known binding activity for any antigen (34). Nb-R1A12 is a nanobody displaying no binding activity to NTUT282−759. Through size-exclusion chromatography, the purified Nb-S2A4, Nb-CB11, and Nb-CB16 were able to form stable complexes with NTUT282−759 (SI Appendix, Fig. S3), while the seven purified nanobodies from the R3 pool (Nb-R3A10, Nb-S1C9, Nb-S1B6, Nb-S1A8, Nb-S1B11, Nb-S1G10, and Nb-R3D5) did not, possibly because of lower binding affinity.
Nanobodies Specifically Recognize NTUT282−387 of TUT4.
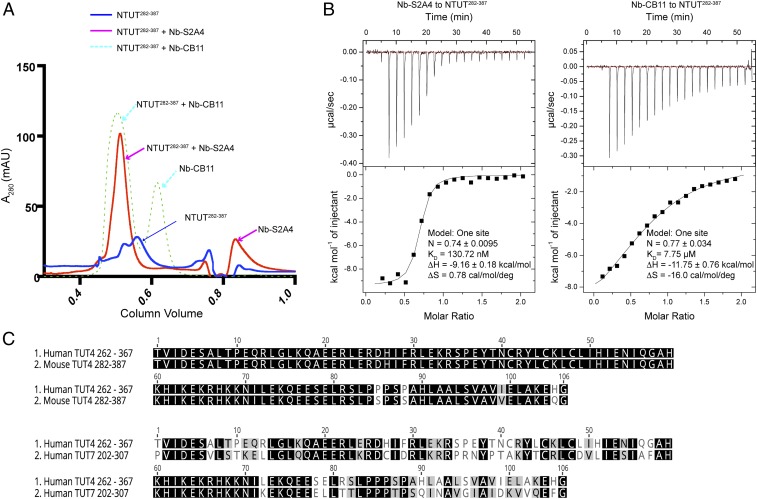
To test the epitopes of TUT4 nanobodies using size-exclusion chromatography, we generated several N-terminal and C-terminal truncation constructs of NTUT282−759. Only NTUT282−387 was overexpressed and purified. Next, 10 µmol of NTUT282−387 and 12 µmol of purified nanobodies were combined and incubated overnight at 4 °C and then applied to a Superdex 75 column. We observed that purified Nb-S2A4 and Nb-CB11 coeluted with NTUT282−387 (Fig. 3A), indicating that residues 282 to 387 contain the binding epitopes of the two nanobodies. This region is highly conserved between mouse and human TUT4, displaying 96.2% identity (Fig. 3C). We next performed isothermal titration calorimetry (ITC) experiments to measure the affinities between the identified nanobodies and NTUT282−387. ITC was carried out by injecting 200 µM nanobody solution into cells containing 20 µM NTUT282−387 and the thermodynamic parameters of the interactions were recorded. As shown in Fig. 3B, binding of Nb-S2A4 and Nb-CB11 to NTUT282−387 were all enthalpically driven, which is consistent with antibody and antigen interactions. The dissociation constants of Nb-S2A4 and Nb-CB11 to NTUT282−387 were 130.7 nM and 7.75 µM, respectively (Fig. 3B and SI Appendix, Fig. S4A). Nb-S2A4 and Nb-CB11 did not cross-react with NTUT7202−307 (54.7% sequence identity to TUT4), as shown in SI Appendix, Fig. S4B. These data indicate that Nb-S2A4 and Nb-CB11 were specific to TUT4, but not TUT7.
Fig. 3.
Nanobodies bind to NTUT282−387 specifically. (A) Gel-filtration binding assay of NTUT282−387 with nanobodies Nb-S2A4 or Nb-CB11. (B) ITC analysis of nanobodies, Nb-S2A4 or Nb-CB11, binding to NTUT282−387. (C) Alignment of the N-terminal sequences of human TUT4, mouse TUT4, and human TUT7. The sequences between mouse NTUT4 (282–387) and human NTUT4 (262–367) are 96.2% identical. The sequences of human NTUT4 (262–367) and human NTUT7 (202–307) are 54.7% identical.
Nb-S2A4 Inhibits LIN28:Prelet-7:TUT4 Ternary Complex Assembly In Vitro.
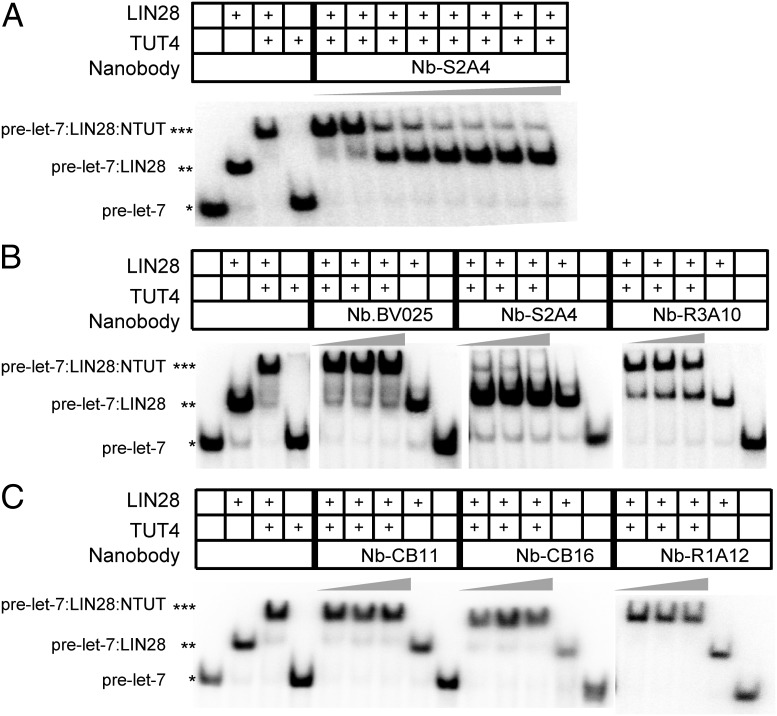
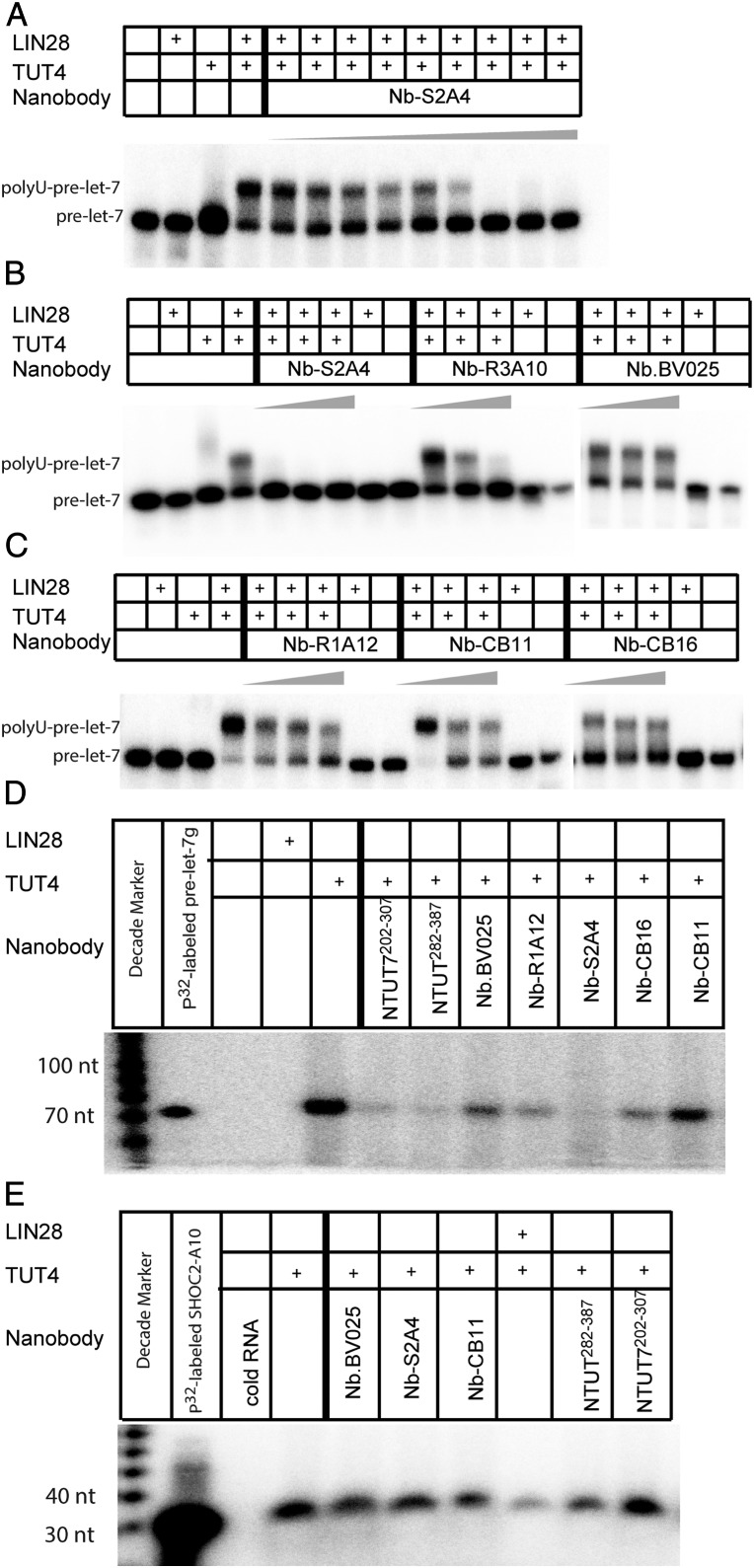
We performed RNA EMSAs to confirm the ability of the nanobodies to interfere with the recruitment of NTUT282−759 to the LIN28:pre-let-7 binary complex, introducing purified nanobodies at 1.5-fold, 3.75-fold, and 7.5-fold molar excess to NTUT282−759. Nb-S2A4 inhibited the formation of the LIN28:pre-let-7:TUT4 ternary complex and outcompeted NTUT282−759 binding at 1.5-fold molar excess of NTUT282−759 (Fig. 4 A and B). Binding of LIN28 to pre-let-7g was not affected by the addition of Nb-S2A4 (Fig. 4 A and B). The Nb-R3A10 nanobody from the R3 pool partially inhibited TUT4 recruitment (Fig. 4B) and, as expected, complex binder nanobodies (Nb-CB11 and NB-CB16) did not block NTUT282−759 recruitment to the LIN28:pre-let-7 binary complex in RNA EMSA (Fig. 4C). As negative controls, Nb.BV025 and Nb-R1A12 did not interfere with the ternary complex formation (Fig. 4 B and C). These data suggest that Nb-S2A4 and Nb-CB11 epitopes are different, even though both antibodies interact with the NTUT282−387 fragment. As expected, the data support binding of Nb-S2A4 to TUT4 on the LIN28:pre-let-7 interface, while Nb-CB11 and Nb-CB16 may bind on the outer surface of the ternary complex. Nanobodies from the R3 pool displayed divergent abilities in interfering with ternary complex formation (SI Appendix, Fig. S5), which validate the efficiency of our functional epitope selection platform.
Fig. 4.
Nanobodies inhibited LIN28:pre-let-7:TUT4 complex assembly in vitro. (A) RNA EMSA of a titration of Nb-S2A4 with LIN28 (1.25 µM), NTUT282−759 (2 µM), and 32P-labeled pre-let-7g. Nb-S2A4 was added at 0.5, 1, 1.5, 2, 2.5, 3, 3.5, and 4 µM, respectively. *RNA alone; **RNA:LIN28 binary complex; ***RNA:LIN28:NTUT4 ternary complex. (B and C) RNA EMSA of LIN28 (1.25 µM), NTUT282−759 (2 µM) with 32P-labeled pre-let-7g, and nanobody inhibitors and complex binders. Nanobodies were added at 1.5-fold, 3.75-fold, and 7.5-fold molar excess to NTUT282−759. Nb.BV025 and Nb-R1A12 were used as negative controls. *RNA alone; **RNA:LIN28 binary complex; ***RNA:LIN28:NTUT4 ternary complex.
NTUT282−387 (LLI Fragment) Is Sufficient to Form a Ternary Complex with LIN28:let-7.
Given that Nb-S2A4 inhibits ternary complex formation, we next asked if NTUT282−387 is sufficient to establish a ternary complex with LIN28:pre-let-7, and confirmed this sufficiency using EMSA binding experiments (Fig. 5A). NTUT282−387, hereafter referred to as the Lin28:let7 interaction (LLI) fragment, contains a C2H2 zinc finger motif and three predicted α-helixes connected by loops. Studies suggest that the C2H2 zinc finger mediates protein–protein interactions between LIN28 and TUT4, and that the mutations (deletion of C2H2 or C326/329A) in this zinc finger resulted in loss of oligouridylation, but not monouridylation, activity of full-length TUT4 (22). When trying to identify the minimal RNA sequence requirement, we observed that binding of the LLI fragment to the LIN28:pre-let-7 binary complex required the presence of the pre-element (preE) and a double-stranded stem with a minimal length of 6 nt (Fig. 5 B, Left). As shown in the Center and Right panels of Fig. 5B, LIN28 with RNA containing either preE + 6-nt single-stranded overhang or the preE + 3-nt double-stranded stem could not recruit NTUT282−387. Because LIN28:pre-let-7 binding does not require double-stranded RNA (12), our data suggest that the LLI fragment not only engages LIN28 but also recognizes the double-stranded RNA in the ribonucleoprotein complex.
Fig. 5.
Determination of NTUT282−387 as the minimal LLI fragment of TUT4. (A) RNA EMSAs of LIN28 (1.25 µM) and the LLI fragment NTUT282−387 (2 µM) with pre-let-7g and the schematic representation of the LIN28:pre-let-7:LLI ternary complex. (B) Schematic representation of pre-let-7 and RNA EMSAs of LIN28 (1.25 µM) and the LLI fragment NTUT282−387 (2 µM) with different pre-let-7 truncation constructs. The sizes of proteins and RNAs drawings are not proportional to their sequences. *RNA alone; **RNA:LIN28 binary complex; ***RNA:LIN28:NTUT4 ternary complex. (C) Gel-filtration binding assay of the LLI fragment (NTUT282−387) with LIN28 and the RNA preE + 6-nt stem construct. (D) RNA competition EMSA of mouse LIN28 (1.25 µM), NTUT282−759 (2 µM), and NTUT282−387 or NTUT7202−307 (6.6 µM, 13.2 µM, and 20 µM) with P32-labeled pre-let-7g. *RNA alone; **RNA:LIN28 binary complex; ***RNA:LIN28:NTUT4 ternary complex.
To further verify the ternary complex containing the LLI fragment, we performed size-exclusion chromatography and demonstrated that NTUT282−387 was able to form a stable, monodisperse ternary complex with preE + 6-nt stem and LIN28 (Fig. 5C). Moreover, the LLI fragment was able to compete out NTUT282−759 in the ternary complex at a threefold molar excess (Fig. 5 D, Left) in the competition EMSA experiments. This result confirmed the critical role of the LLI fragment in the LIN28:pre-let-7:TUT4 ternary complex assembly. In contrast, the paralogous TUT7 fragment, NTUT7202−307, displayed a much weaker competition ability when compared to the LLI fragment of TUT4 (Fig. 5 D, Right). These data suggest that NTUT282−387 is the minimal sequence required for recruitment to the LIN28:pre-let-7 binary complex.
Nb-S2A4 Inhibits Uridylation of Let-7, but Not mRNA Substrates.
Consistent with the EMSA data, Nb-S2A4 inhibited the oligouridylation activity of TUT4 at a concentration of 733 nM (SI Appendix, Fig. S6A and Fig. 6 A and B). Further titration of Nb-S2A4 demonstrated that the inhibitory ability of Nb-S2A4 toward TUT4 oligouridylation activity was concentration-dependent and the IC50 was estimated at ∼200 nM (Fig. 6A). Nb-R3A10 was able to completely inhibit TUT4 oligouridylation activity at 20 µM (Fig. 6B). The complex binders, Nb-CB11 and Nb-CB16, which did not interfere with recruiting TUT4 to the LIN28:pre-let-7 binary complex, did not abolish the catalytic activity of TUT4 at any concentration (Fig. 6 B and C). These data show that Nb-S2A4 effectively inhibits LIN28-dependent oligouridylation activity by TUT4 toward pre-let-7 in vitro. Nb-S2A4 and other nanobodies that we tested did not inhibit TUT7-mediated oligouridylation of pre-let-7 (SI Appendix, Fig. S6B), a result consistent with the specificity of these nanobodies to TUT4. TUT7 showed weaker oligouridylation activity in the LIN28:let-7 pathway compared to TUT4, which is consistent with previous study (11) (SI Appendix, Fig. S6C).
Fig. 6.
Nanobodies inhibited TUT4 uridylation activities in vitro. (A) In vitro TUTase assay with a titration of Nb-S2A4 at 13.3 nM, 33.3 nM, 66.7 nM, 133 nM, 200 nM, 333 nM, 733 nM, 1.33 µM, and 2 µM. (B and C) In vitro TUTase assay with addition of gradient concentrations of both the nanobody inhibitor and complex binders at 3.3 µM, 8.3 µM, and 20 µM. Nb.BV025 and Nb-R1A12 were used as negative controls. (D) In vitro pan-uridylation assay of pre-let-7g carried out by full-length TUT4, 1.6 µM LIN28 with unlabeled pre-let-7g. (E) In vitro pan-uridylation assay of mRNA SHOC2-A10 carried out by full-length TUT4, 1.6 µM LIN28, and unlabeled mRNA SHOC2-A10.
Nb-S2A4, quite interestingly, also completely inhibited TUT4-medidated monouridylation of pre-let-7g (Fig. 6D, lane 10), suggesting that the LLI fragment is also involved in the monouridylation of pre-let-7. Consistent with Nb-S2A4 inhibition of the LLI fragment, both NTUT282−387 and NTUT7202−307 greatly outcompeted the monouridylation conducted by TUT4 (Fig. 6D, lanes 6 and 7). These data suggest a potential direct interaction between the LLI fragment and pre-let-7 in the absence of LIN28, although it is too weak for EMSA detection. Because NTUT282−387 may contain a putative RNA hooking motif, we hypothesized that the monouridylation of pre-let-7 also requires some degree of recognition between N terminus TUT4 and pre-let-7 microRNA, an interaction that might parallel LLI recognition of let-7 in complex with LIN28. The negative control nanobodies, Nb.BV025 and Nb-R1A12, as well as the complex binders, Nb-CB11 and Nb-CB16, did not inhibit the monouridylation activity of TUT4 (Fig. 6D).
We were surprised to find that NTUT282−387, NTUT7202−307, Nb-S2A4, and other nanobodies did not affect TUT4-mediated uridylation of the mRNA samples SHOCK2-A10 and CAML1-A10, which are mRNA substrates of TUT4 and TUT7 identified in a previous study (35) (Fig. 6E and SI Appendix, Fig. S6D). These observations suggest that TUT4 may have a different mRNA recognition mechanism.
Nb-S2A4 Effectively Inhibits TUT4 in Cells.
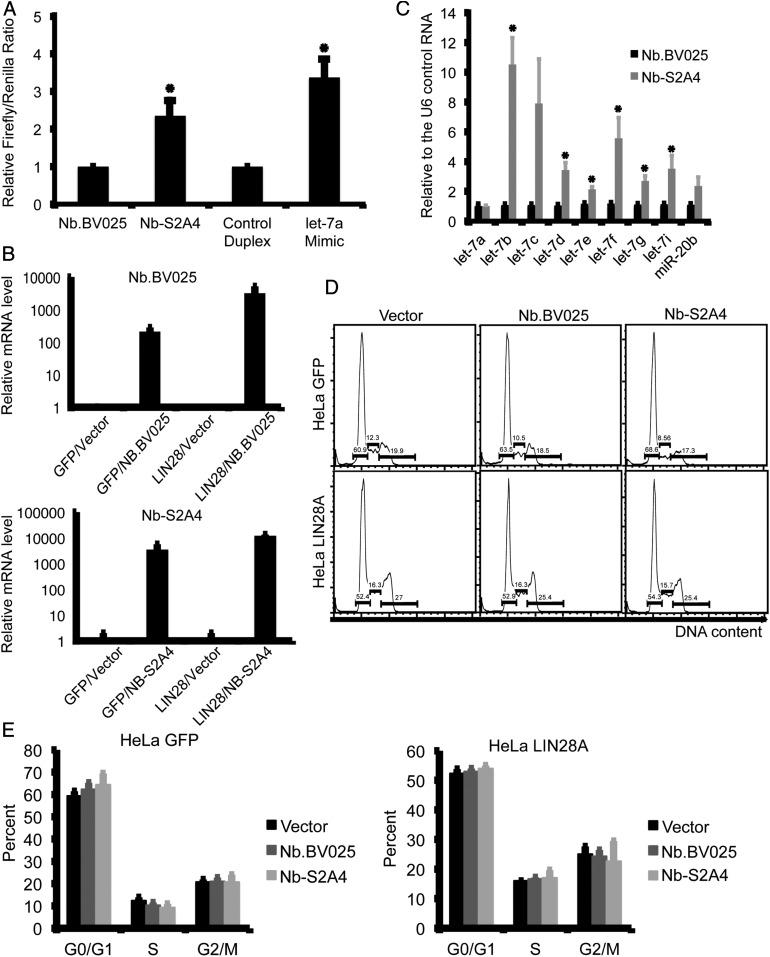
Because mouse NTUT282−387 is 96.2% identical to the homologous region of the human protein, we tested whether these nanobodies recognize TUT4 in human cell lines. We cloned Nb-S2A4 and the negative control Nb.BV025 into a mammalian expression vector and transfected these nanobody vectors into HEK-293T cells, which predominantly express LIN28B (36) along with a let-7–sensitive dual luciferase reporter. This reporter construct contains a constitutively expressed Firefly luciferase as well as a Renilla luciferase with eight tandem let-7 binding sites in the 3′ untranslated region. An increase in mature let-7 would thus be reflected as an increase in the Firefly/Renilla luciferase ratio (SI Appendix, Fig. S7A). We found that overexpression of the LLI fragment in these cells slightly increased the Firefly/Renilla luciferase ratio, suggesting that the LLI fragment outcompeted TUT4 activity (SI Appendix, Fig. S7B). The expression of Nb-S2A4 in these cells was as expected and consistently increased the Firefly/Renilla ratio, in contrast to the negative control nanobody NB.BV025 (Fig. 7A). These data support the importance of the LLI fragment in the LIN28:let-7:TUT4/7 pathway and form the basis of our hypothesis that the nanobody inhibitor Nb-S2A4 to the LLI fragment is able to inhibit TUT4 activity in cells.
Fig. 7.
Nb-S2A4 effectively inhibits TUT4 in cells. (A) HEK-293 cells were transfected with the indicated nanobody constructs, or a control RNA duplex or a let-7a mimic, along with the dual luciferase reporter construct. Forty-eight hours later, Renilla and Firefly luciferase activity were quantified. (B) HeLa cells expressing LIN28 were transfected with the indicated nanobody constructs. After 7 d of selection with G418, levels of the indicated microRNAs were measured relative to the U6 control RNA by quantitative PCR. (C) HeLa cells expressing LIN28 or control GFP were transfected with the indicated nanobody constructs, and 48 h later levels of the nanobody transcripts were measured by quantitative PCR. (D and E) HeLa cells were transfected with the indicated nanobody constructs and after 72-h cell cycle was measured by propidium iodide staining.
To generate stable cell lines expressing either Nb.BV025 or Nb-S2A4, we used a HeLa clone stably expressing exogenous LIN28, because HeLa cells express little, if any, of their own. We found that cells expressing Nb-S2A4 contained higher levels of the mature let-7 microRNA species compared to cells expressing Nb.BV025 (Fig. 7 B and C). Our experiments also confirmed that overexpression of neither Nb-S2A4 nor NB.BV025 affected the cell cycle (Fig. 7 D and E). These data demonstrate that Nb-S2A4 effectively inhibits the LIN28:let7:TUT4 pathway and rescues mature let-7 levels in cells.
Discussion
In this study we developed a rapid and direct functional epitope selection platform and identified a number of nanobodies interacting with components of the LIN28 pathway. The most potent nanobody, Nb-S2A4, inhibits the assembly of the LIN28:pre-let-7:TUT4 ternary complex through direct interaction. We identified a functional region of TUT4, the LLI fragment (residues 282 to 387 of mouse TUT4) that contains a protein–protein interaction motif (C2H2 zinc finger) and an additional sequence, as mediating direct interaction with both the LIN28:pre-let-7 binary complex and Nb-S2A4. Therefore, Nb-S2A4 competes with the LIN28:pre-let-7 binary complex for TUT4 interaction, and inhibits LIN28-mediated oligouridylation of pre-let-7 in vitro. In cells expressing LIN28, the LLI fragment expressed in trans specifically inhibited uridylation of pre-let-7 microRNAs, but not uridylation of SHOCK2-A10 and CAML1-A10 mRNAs, and Nb-S2A4 effectively rescue maturation of the let-7 species. As such, the LLI fragment of TUT4 and the Nb-S2A4 nanobody offer new avenues for the modulation of the LIN28:let-7:TUTase pathway, experimentally and as potential therapeutics.
Our group previously identified a highly conserved 10-residue YRYFACPQKK (724 to 733) motif in the N terminus of TUT4 (NTUT230−759), which is critical for the LIN28:pre-let-7:TUT4 ternary complex formation (13). The binding of this motif to the LIN28:pre-let-7 binary complex requires at least 15 base pairs of the double-stranded stem of pre-let-7 (13). In this study, we demonstrated that the LLI fragment (residues 282 to 387) also contains a motif that interacts with the double-stranded stem of pre-let-7 and requires at least a 6-base pair stem to form the complex. While our understanding of LIN28-TUTase recognition is gradually improving, it is conceivable that additional protein–protein or protein–RNA interactions play critical roles, and a complete understanding of the mechanism of regulation of let-7 maturation will need to await a high-resolution structure of the LIN28:pre-let-7:TUT4 ternary complex.
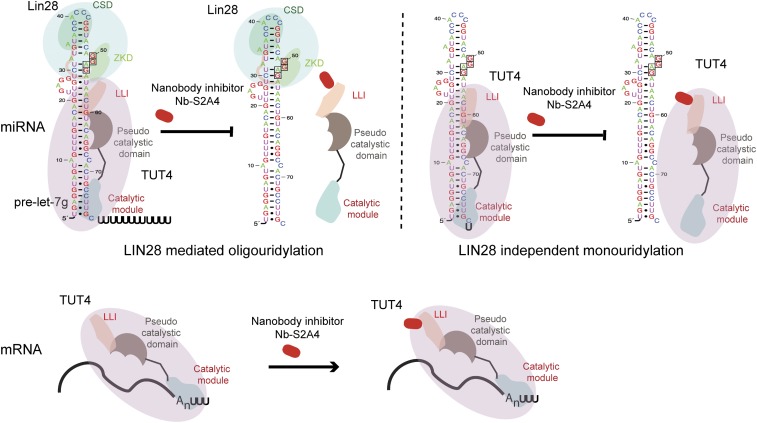
In addition to LIN28-mediated oligouridylation, the LLI fragment is also critical for LIN28-independent monouridylation of the group II pre-let-7s. This finding is unexpected, but consistent with the outcome of the previously reported TUT4 deletion construct that comprised the N-terminal C2H2 zinc finger and the TNF4 domain (22). Our data suggest that an interaction of pre-let-7s with the N-terminal, noncatalytic fragment of TUT4 might be indispensable for the recognition of the microRNA in the absence of LIN28, potentially by locking TUT4 in a conformation that would recognize 1-nt pre-let-7’s overhangs and promote maturation of group II let-7s as the monouridylation pathway described in the previous report (23). We speculate that Nb-S2A4 in LIN28− cell lines may reduce the maturation levels of the group II pre-let-7s. We have also shown that the LLI fragment does not influence TUT4-dependent uridylation of mRNA samples, implying a different molecular mechanism for mRNA recognition by TUT4 (Fig. 8). These results advance our understanding of the molecular mechanisms of TUT4/7 recognition of various RNA species.
Fig. 8.
Schematic of LIN28:let-7:TUT4 pathway with the nanobody inhibitor Nb-S2A4. The sizes of proteins and RNAs in the drawings are not proportional to their sequences.
Many studies have shown that TUT4 and TUT7 paralogs can be functionally redundant in the uridylation of a wide spectrum of RNAs, including rRNA, tRNA, microRNA, and mRNA, acting to regulate RNA degradation as a quality-control mechanism (9, 22, 35, 37–40). However, some substrate preferences have also been identified (41). In the LIN28:let-7:TUTase pathway, TUT4 promotes the oligouridylation of pre-let-7s, while TUT7 was shown to exhibit weak redundancy with TUT4 (11). Correspondingly, inhibition of TUT4 in our system by Nb-S2A4 led to increased levels of mature let-7s. In contrast, in the LIN28-independent monouridylation pathway of pre-let-7s in HeLa cells, TUT7 was shown to promote the modification, while TUT4/2 was less effective (23). TUT7 may therefore compensate the loss of TUT4 in the monouridylation pathway and our specific nanobodies could facilitate analysis of functional overlaps between TUTase paralogs. Nb-S2A4 can also be exploited to investigate the interaction of TUT4 with other binders, such as TIFA (TRAF-interacting protein with a forkhead-associated domain), which was demonstrated to interact with the N-terminal region (residues 1 to 759) of TUTase to regulate Toll-like receptor signaling (42).
Aberrant activation of the LIN28:let-7:TUTase pathway occurs in many malignancies, a finding that has encouraged investigation of the molecular functions of this pathway. Several groups have designed fluorescence resonance energy transfer or fluorescence polarization assays to screen small-molecule inhibitors that block LIN28 and pre-let-7 interactions (24, 25, 27, 43). Small-molecule inhibitors of LIN28 reported to date show minimal efficacy in rescue of let-7 maturation and function in cells (24, 25, 27). A previous study from Lin and Gregory (26) described small molecules that inhibit the catalytic domain of TUT4, but unfortunately these inhibitors have no effect in cells. Our study demonstrates that the LLI fragment of TUTase presents two advantages for targeting by inhibitors. First, the TUT4 LLI has only 54.7% identity with TUT7 LLI, supporting development of the paralog specific inhibitors (identity within the catalytic domain is 70%). The LLI fragment also plays specific roles in let-7 biogenesis, but not in mRNA uridylation pathways, supporting targeting of let-7 rather than mRNA degradation. To illustrate the utility of our approach we have shown that targeting the LLI fragment with the Nb-S2A4 inhibitor, in vitro and in cells, is effective and could inform further investigations of antagonists of the oncogenic LIN28:let-7:TUT4 pathway.
In summary, we developed a rapid and direct functional epitope selection platform and isolated a nanobody inhibitor, Nb-S2A4, which directly and specifically inhibits TUT4 activities. Using this nanobody, we further identified the minimal functional domain in TUT4—the LLI fragment—which is critical for both oligouridylation and monouridylation of pre-let-7 microRNAs, and dispensable for the uridylation of mRNAs. The nanobody inhibitor Nb-S2A4 effectively rescues let-7 biogenesis in cells and validates the LLI fragment as a potential target for inhibition of the oncogenic LIN28:pre-let-7:TUTase pathway.
Materials and Methods
Protein Purification.
Mouse LIN28 protein (residues 16 to 184) (12) and various TUT4 N-terminal constructs were expressed and purified as described previously (12). The human NTUT7 (202 to 307) protein was overexpressed in BL21 (DE3) Rosetta pLysS Escherichia coli and purified using Ni-NTA affinity, ion-exchange (SP column), and size-exclusion chromatography (Superdex 75). Full-length mouse TUT4 was expressed in insect cells and purified using Ni-NTA affinity followed by ion-exchange. Nanobodies were cloned in-frame to a C-terminal 6xHis tag, expressed in E. coli, and purified similarly as for mouse TUT4.
Functional Epitope Nanobody Selection Platform.
Nanobodies were first selected from a synthetic yeast display nanobody library (34) using MACS. The enriched pool was used to further select high-affinity binders by sorting with Alexa Fluor 647-labeled NTUT282−759 using FACS. A functional epitope selection was then performed to enrich nanobodies that bind to the interface (inhibitory binders) of the LIN28:pre-let-7:TUT4 ternary complex. The resultant pool underwent another round of FACS enrichment to further identify inhibitory nanobodies with high binding affinity.
Detailed description of experimental methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Kelly Arnett and the Center for Macromolecular Interactions at Harvard Medical School, and Dr. Jodene K. Moore and the System Biology Flow Cytometry Facility at Harvard Medical School for experimental assistance. This work was supported by a National Cancer Institute Grant R01CA163647 (to P.S.). R.G.R. and G.Q.D. were supported by an Innovation Award from Alex’s Lemonade Stand Foundation. R.G.R. was supported by the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Disorders (Grant K08 DK114527-01). Additional support was provided by a Smith Family Foundation grant (to A.C.K.).
Footnotes
Competing interest statement: G.Q.D. declares that he is a cofounder, consultant, and member of the Scientific Advisory Board of Twentyeightseven Therapeutics, a company that designs therapeutics to proteins that modulate RNA. He also holds a patent related to targeting LIN28 to prevent and treat cancer. P.S. declares that he is a cofounder, consultant, and member of the Scientific Advisory Board of Twentyeightseven Therapeutics. He also holds a patent related to LIN28/let-7 structure, purification, and use in screening for therapeutics. A.C.K., H.W., P.S., and T.U.S. were coauthors on a Nature Communications article in 2016 (PMID:26947396).
Data deposition: The nanobody sequences reported in this paper have been deposited in the GenBank database (accession nos. MN752861 [NB-S2A4], MN752862 [NB-CB11], MN752864 [NB-R1A12], MN752863 [NB-CB16], MN752852 [NB-R3A10], MN752853 [NB-R3D5], MN752854 [NB-R3D12], MN752855 [NB-S1A8], MN752856 [NB-S1B4], MN752857 [NB-S1B11], MN752858 [NB-S1C9], MN752859 [NB-S1G10], and MN752860 [NB-S2B6]).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919409117/-/DCSupplemental.
References
- 1.Büssing I., Slack F. J., Grosshans H., let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 14, 400–409 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Johnson S. M., et al. , RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Mayr C., Hemann M. T., Bartel D. P., Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roush S., Slack F. J., The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Sampson V. B., et al. , MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 67, 9762–9770 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Viswanathan S. R., Daley G. Q., Lin28: A microRNA regulator with a macro role. Cell 140, 445–449 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Chang H. M., Triboulet R., Thornton J. E., Gregory R. I., A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497, 244–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagan J. P., Piskounova E., Gregory R. I., Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16, 1021–1025 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heo I., et al. , Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276–284 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Thornton J. E., et al. , Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res. 42, 11777–11791 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heo I., et al. , TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Nam Y., Chen C., Gregory R. I., Chou J. J., Sliz P., Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080–1091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., et al. , LIN28 zinc knuckle domain is required and sufficient to induce let-7 oligouridylation. Cell Rep. 18, 2664–2675 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Fu X., et al. , miR-26a enhances miRNA biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth and metastasis. Oncogene 33, 4296–4306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King C. E., et al. , LIN28B promotes colon cancer progression and metastasis. Cancer Res. 71, 4260–4268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen L. H., et al. , Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Canc. Cell 26, 248–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng S., Maihle N. J., Huang Y., Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 29, 2153–2159 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Permuth-Wey J., et al. ; Ovarian Cancer Association Consortium , LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 71, 3896–3903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piskounova E., et al. , Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147, 1066–1079 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbach A., et al. , Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 28, 971–982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faehnle C. R., Walleshauser J., Joshua-Tor L., Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis. Nat. Struct. Mol. Biol. 24, 658–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornton J. E., Chang H. M., Piskounova E., Gregory R. I., Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18, 1875–1885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heo I., et al. , Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151, 521–532 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Lightfoot H. L., Miska E. A., Balasubramanian S., Identification of small molecule inhibitors of the Lin28-mediated blockage of pre-let-7g processing. Org. Biomol. Chem. 14, 10208–10216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim D., Byun W. G., Koo J. Y., Park H., Park S. B., Discovery of a small-molecule inhibitor of protein-MicroRNA interaction using binding assay with a site-specifically labeled Lin28. J. Am. Chem. Soc. 138, 13630–13638 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Lin S., Gregory R. I., Identification of small molecule inhibitors of Zcchc11 TUTase activity. RNA Biol. 12, 792–800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roos M., et al. , A small-molecule inhibitor of Lin28. ACS Chem. Biol. 11, 2773–2781 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Feng M., et al. , Construction and next-generation sequencing analysis of a large phage-displayed VNAR single-domain antibody library from six naïve nurse sharks. Antib. Ther. 2, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fridy P. C., et al. , A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., et al. , Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications. Int. J. Nanomedicine 11, 3287–3303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irannejad R., et al. , Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staus D. P., et al. , Regulation of β2-adrenergic receptor function by conformationally selective single-domain intrabodies. Mol. Pharmacol. 85, 472–481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Audenhove I., Gettemans J., Nanobodies as versatile tools to understand, diagnose, visualize and treat cancer. EBioMedicine 8, 40–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon C., et al. , Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 25, 289–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim J., et al. , Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159, 1365–1376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enriquez V. A., et al. , High LIN28A expressing ovarian cancer cells secrete exosomes that induce invasion and migration in HEK293 cells. BioMed Res. Int. 2015, 701390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang H., Lim J., Ha M., Kim V. N., TAIL-seq: Genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53, 1044–1052 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Morgan M., et al. , mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 548, 347–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pirouz M., Du P., Munafò M., Gregory R. I., Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep. 16, 1861–1873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ustianenko D., et al. , TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J. 35, 2179–2191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lackey P. E., Welch J. D., Marzluff W. F., TUT7 catalyzes the uridylation of the 3′ end for rapid degradation of histone mRNA. RNA 22, 1673–1688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minoda Y., et al. , A novel zinc finger protein, ZCCHC11, interacts with TIFA and modulates TLR signaling. Biochem. Biophys. Res. Commun. 344, 1023–1030 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Wang L., et al. , Small-molecule inhibitors disrupt let-7 oligouridylation and release the selective blockade of let-7 processing by LIN28. Cell Rep. 23, 3091–3101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.