Significance
MSTN, a member of the TGF-β family, has been widely shown to suppress muscle growth, leading to an intense effort being directed at targeting MSTN to treat patients with muscle loss. GDF11 is another TGF-β family member closely related to MSTN, but its postnatal function is less clear. Using conditional knockout techniques, we show that GDF11 enhances bone mass in contrast to MSTN, emphasizing that MSTN/GDF11 inhibitors, such as FST, can induce adverse effects on bone through GDF11 inhibition. Because most MSTN inhibitors also inhibit GDF11 due to the high sequence similarity between MSTN and GDF11, our findings suggest that their opposing roles must be carefully considered when developing MSTN inhibitors for clinical applications.
Keywords: GDF11, myostatin, follistatin, osteogenesis, BMP
Abstract
Growth and differentiation factor 11 (GDF11) and myostatin (MSTN) are closely related transforming growth factor β (TGF-β) family members, but their biological functions are quite distinct. While MSTN has been widely shown to inhibit muscle growth, GDF11 regulates skeletal patterning and organ development during embryogenesis. Postnatal functions of GDF11, however, remain less clear and controversial. Due to the perinatal lethality of Gdf11 null mice, previous studies used recombinant GDF11 protein to prove its postnatal function. However, recombinant GDF11 and MSTN proteins share nearly identical biochemical properties, and most GDF11-binding molecules have also been shown to bind MSTN, generating the possibility that the effects mediated by recombinant GDF11 protein actually reproduce the endogenous functions of MSTN. To clarify the endogenous functions of GDF11, here, we focus on genetic studies and show that Gdf11 null mice, despite significantly down-regulating Mstn expression, exhibit reduced bone mass through impaired osteoblast (OB) and chondrocyte (CH) maturations and increased osteoclastogenesis, while the opposite is observed in Mstn null mice that display enhanced bone mass. Mechanistically, Mstn deletion up-regulates Gdf11 expression, which activates bone morphogenetic protein (BMP) signaling pathway to enhance osteogenesis. Also, mice overexpressing follistatin (FST), a MSTN/GDF11 inhibitor, exhibit increased muscle mass accompanied by bone fractures, unlike Mstn null mice that display increased muscle mass without fractures, indicating that inhibition of GDF11 impairs bone strength. Together, our findings suggest that GDF11 promotes osteogenesis in contrast to MSTN, and these opposing roles of GDF11 and MSTN must be considered to avoid the detrimental effect of GDF11 inhibition when developing MSTN/GDF11 inhibitors for therapeutic purposes.
GDF11 also known as bone morphogenetic BMP11 and MSTN are closely related TGF-β family members that share high sequence similarity within their mature signaling domain (1, 2). While GDF11 and MSTN have been reported to share similar receptors, binding molecules, and signaling pathways (3–5), they exhibit distinct biological functions (6, 7) due to differences in their tissue/time-specific expression and activation patterns (8). For instance, MSTN is primarily expressed in skeletal muscle and has been widely shown to negatively regulate skeletal muscle growth (1, 9–13). MSTN has also been reported to impair bone development either directly by affecting OB and osteoclast (OC) differentiation (14, 15) or indirectly through regulating muscle mass (16). In contrast, GDF11 is expressed more broadly in diverse tissues and regulates axial skeletal patterning and organ development during embryogenesis (2, 17–21).
Postnatal functions of GDF11 are less clear and controversial. Specifically, a series of studies from Lee and Wagers’ group suggested that GDF11 rejuvenates aged cardiac/skeletal muscle and brain (22–24). However, subsequent conflicting data from Egerman et al. (25) demonstrated that GDF11 and MSTN are essentially identical in suppressing muscle regeneration. Likewise, while Zhang et al. (26) described that GDF11 stimulates bone formation, Lu et al. (27) and Liu et al. (28) later reported the opposite, suggesting that GDF11 inhibits bone formation in a way similar to that of MSTN. Importantly, due to the perinatal lethality of Gdf11 null mice, these previous controversial studies relied mainly on recombinant GDF11 protein to investigate its physiological function. However, because of the high degree of homology between GDF11 and MSTN, their recombinant proteins share almost identical biochemical properties and, therefore, cannot be clearly differentiated, generating the possibility that the effects artificially mediated by recombinant GDF11 actually reproduce the endogenous functions of MSTN. Furthermore, the quality of recombinant GDF11 and MSTN proteins used in earlier studies has been questioned (29, 30), implying that the use of recombinant GDF11 may not be suitable for determining its endogenous physiological function.
Because of the well-established role of MSTN in skeletal muscle, blocking the MSTN signaling pathway has been adopted as a promising therapeutic strategy to prevent or reverse the loss of muscle mass and strength in patients with muscle wasting disorders (31, 32). Among several MSTN binding proteins identified to increase muscle mass, FST has been shown to display the greatest effect when delivered to mice with normal or dystrophic muscle (33). Based on this finding, Fst gene transfer has been applied to patients with various muscular dystrophies in clinical trials, resulting in improved muscle regeneration (34–37). Like most MSTN antagonists, which also bind and inhibit GDF11 due to its homology to MSTN, FST binds and inhibits both MSTN (5) and GDF11 (21). Therefore, if GDF11 and MSTN oppositely regulate the growth and differentiation of musculoskeletal tissues, GDF11 inhibition mediated by FST may lead to undesired side effects.
To overcome the limitation and controversial issues surrounding the use of recombinant GDF11 protein, here, we focused on genetic studies in mice to elucidate the more complex endogenous actions of GDF11, particularly, its role on bone homeostasis. Specifically, we utilize conditional knockout techniques to produce genetically engineered mice in which the Gdf11 gene is deleted in a tissue/time-specific manner and demonstrate that GDF11 systemically enhances osteogenesis in contrast to MSTN. We also show that the enhanced osteogenic effect of Mstn deletion is due to up-regulation of GDF11 and subsequent activation of BMP signaling. Finally, we present data showing that FST increases muscle mass through inhibition of MSTN but impairs bone quality and strength through blocking GDF11. Our findings emphasize that the opposing roles of GDF11 and MSTN in osteogenesis must be carefully considered when developing MSTN inhibitors, which can also bind GDF11, as therapeutic agents for the treatment of musculoskeletal diseases.
Results
Bone Mass Is Reduced in Gdf11−/− and Cdx2-Cre; Gdf11flox/flox Mice but Increased in Mstn−/− Mice.
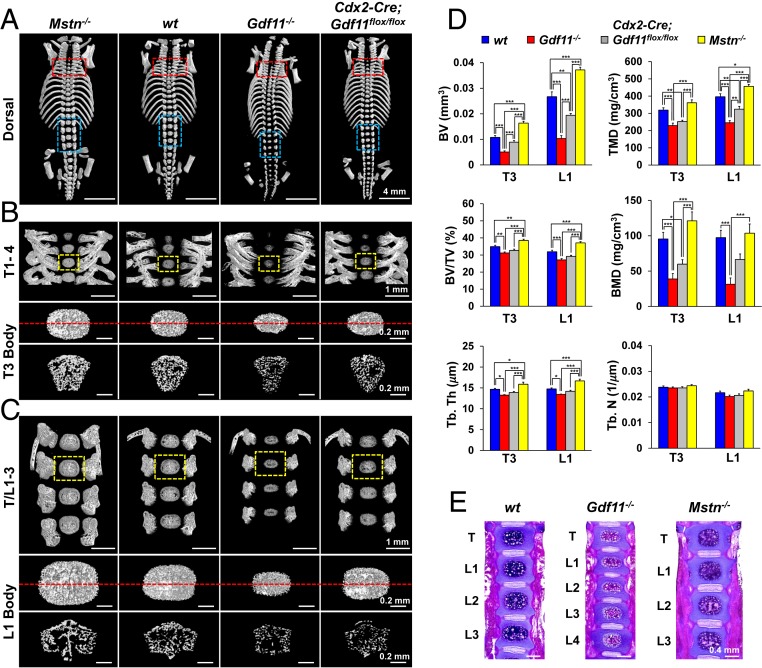
Due to the perinatal lethality of Gdf11−/− mice and the lack of genetic in vivo studies, the reports on the role of GDF11 in bone homeostasis are contradictory, some identifying GDF11 as a negative regulator of bone development (28), while others support the opposite (26). In an attempt to overcome the perinatal lethality of Gdf11−/− mice, we initially used a Cdx2-Cre transgene to generate mosaic mice in which Gdf11 expression is eliminated exclusively in the posterior half tissues (38). However, Cdx2-Cre; Gdf11flox/flox mice also turned out to be perinatally lethal, leading us to first analyze newborn pups. Our microCT analysis of newborn mouse vertebrae, specifically T3 and L1, demonstrated that, in contrast to the enhanced bone mass observed in Mstn−/− mice, bone volume (BV), tissue mineral density (TMD), bone mineral density (BMD), and trabecular thickness (Tb. Th) are significantly decreased in Gdf11−/− mice and mildly reduced in Cdx2-Cre; Gdf11flox/flox mice (Fig. 1 A–D and SI Appendix, Fig. S1A). Tb. separation value was the highest in Mstn−/− mice vertebrae due to their substantially larger volume (SI Appendix, Fig. S1B). Interestingly, there was no differential effect of targeting Gdf11 using the Cdx2-Cre transgene on bone mass between anterior and posterior regions, suggesting that GDF11 systemically promotes osteogenesis. Likewise, hematoxylin and eosin staining of vertebrae sections revealed a decrease in bone area in newborn Gdf11−/− mice and an increase in bone area in Mstn−/− mice (Fig. 1E). Additionally, cranial and sternum ossifications were also affected in a similar pattern: newborn Mstn−/−, Gdf11−/−, and Cdx2-Cre; Gdf11flox/flox mice displayed enhanced, reduced, and mildly reduced bone development, respectively (SI Appendix, Fig. S1C). Delayed limb bone development was also observed in newborn Gdf11−/− mice (SI Appendix, Fig. S1 D and E).
Fig. 1.
Gdf11 deletion reduces bone mass as opposed to Mstn deletion. (A–C) Representative microCT images of newborn mouse vertebrae. Red and blue boxed regions indicate first–fourth thoracic vertebrae (T1–4), last thoracic vertebrae, and first–third lumbar vertebrae (T/L1–3), respectively. Yellow boxed regions in B and C are magnified to display T3 and L1 bodies, respectively. Cross sections of T3 and L1 bodies at positions indicated by the red dashed lines are shown right below them in B and C, respectively. (D) Histomorphometric analysis of T3 and L1 bodies in newborn mice (n = 10 each). BV; TMD; BV/TV, BV/total volume of interest; BMD; Tb. Th; Tb. N, trabecular number. Data represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey’s post hoc test. (E) Representative hematoxylin and eosin staining of frontal vertebrae sections in newborn mice (n = 3 each). All scale bars are displayed with actual size values.
Time-Specific Gdf11 Deletion Reduces Bone Development in Both Embryos and Young Adult Mice.
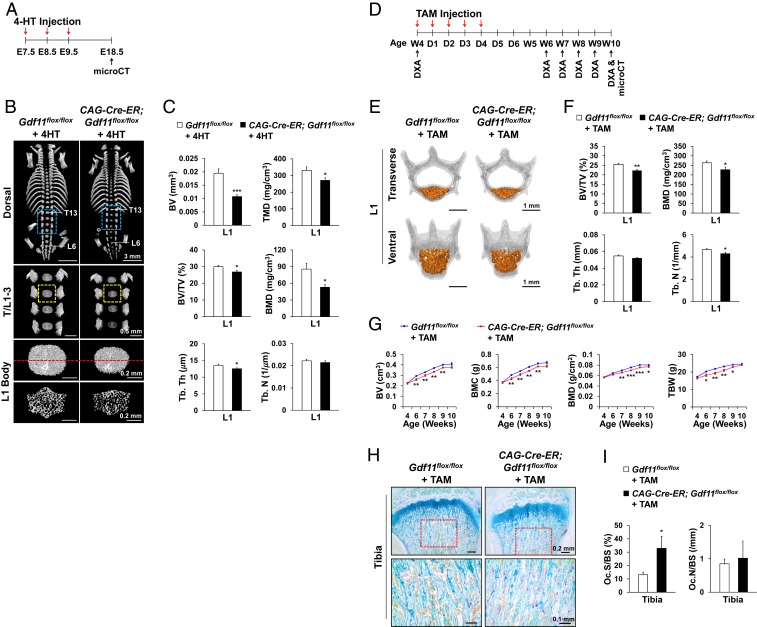
We also examined the effects of Gdf11 deletion on skeletal development by using CAG-Cre-ER to generate inducible ubiquitous conditional knockout mice. Time-specific deletion of Gdf11 in all tissues of CAG-Cre-ER; Gdf11flox/flox embryos induced by daily administration of 4-hydroxytamoxifen (4-HT) from E7.5 to E9.5 (Fig. 2A) resulted in diminished axial bone development at E18.5 as shown by reduced L1 bone mass (Fig. 2 B and C and SI Appendix, Fig. S2 A–F), cranial ossification, and sternum formation (SI Appendix, Fig. S2 B and C). Surprisingly, no homeotic transformations were observed (Fig. 2B and SI Appendix, Fig. S2 C and F), implying that this timing and extent of GDF11 loss was not sufficient to disrupt normal anterior–posterior axial patterning. To elucidate the function of GDF11 in regulating bone development of young adult mice, we also performed daily tamoxifen (TAM) injection for five consecutive days on 4-wk-old CAG-Cre-ER; Gdf11flox/flox mice (Fig. 2D). MicroCT analysis at 10 wk of age revealed a decrease in Tb. bone mass of L1 in TAM-treated CAG-Cre-ER; Gdf11flox/flox mice (Fig. 2 E and F and SI Appendix, Fig. S2 G–I). Similarly, DXA analysis from 4 to 10 wk of age displayed significant declines in BV, BMD, bone mineral content (BMC), total body weight (TBW), and lean mass in TAM-treated CAG-Cre-ER; Gdf11flox/flox mice, although the differences in TBW and lean mass gradually diminished as the mice aged, possibly due to the effect of GDF11 secreted by cells with incomplete deletion of the gene (Fig. 2G and SI Appendix, Fig. S2J). MicroCT analysis at 6 wk of age also revealed a decrease in Tb. bone mass of the humerus in TAM-treated CAG-Cre-ER; Gdf11flox/flox mice (SI Appendix, Fig. S2 K and L). Furthermore, tartrate-resistant acid phosphatase (TRAP) staining of tibias at 6 wk of age showed a significant increase in the OC surface per bone surface (OC.S/BS) in TAM-treated CAG-Cre-ER; Gdf11flox/flox mice, indicating that loss of GDF11 results in increased bone resorption (Fig. 2 H and I). Calcein-alizarin red labeling of L1 at 6 wk also revealed that bone formation is decreased in TAM-treated CAG-Cre-ER; Gdf11flox/flox mice (SI Appendix, Fig. S2 M and N).
Fig. 2.
Time-specific Gdf11 deletion impairs bone development in both E18.5 embryos and young adult mice. (A) 4-HT injection scheme. (B) Representative microCT images of mouse vertebrae at E18.5. Blue boxed regions indicate T/L1–3, and yellow boxed regions are magnified to display L1 bodies. Cross sections of L1 bodies at the position indicated by the red dashed line are shown right below them. (C) Histomorphometric analysis of L1 bodies in E18.5 embryos (n = 10 each). (D) TAM injection scheme. Dual-energy X-ray absorptiometry (DXA). (E) Representative microCT images of 10-wk-old mouse vertebrae (L1). Tb. bones of vertebral bodies are colored in orange. (F) Histomorphometric analysis of L1 Tb. bone in 10-wk-old mice (n = 10 for Gdf11flox/flox mice and n = 8 for CAG-Cre-ER; Gdf11flox/flox mice). (G) DXA analysis of TAM-treated mice from 4 to 10 wk of age (n = 10 for Gdf11flox/flox mice and n = 8 for CAG-Cre-ER; Gdf11flox/flox mice). BMC, Bone mineral content; TBW. (H) Representative TRAP-stained images of 6-wk-old mouse tibia. Magnified images of the red boxed regions are displayed in the Lower panel. (I) Histomorphometric analysis of TRAP-stained images (n = 5 for Gdf11flox/flox mice and n = 3 for CAG-Cre-ER; Gdf11flox/flox mice). OC.S/BS; OC.N/BS, OC.N per BS. All scale bars are displayed with actual size values. All data represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by t test.
Limb Mesenchyme-Specific Deletion of Gdf11 Decreases Bone Mass in Young Adult Mice.
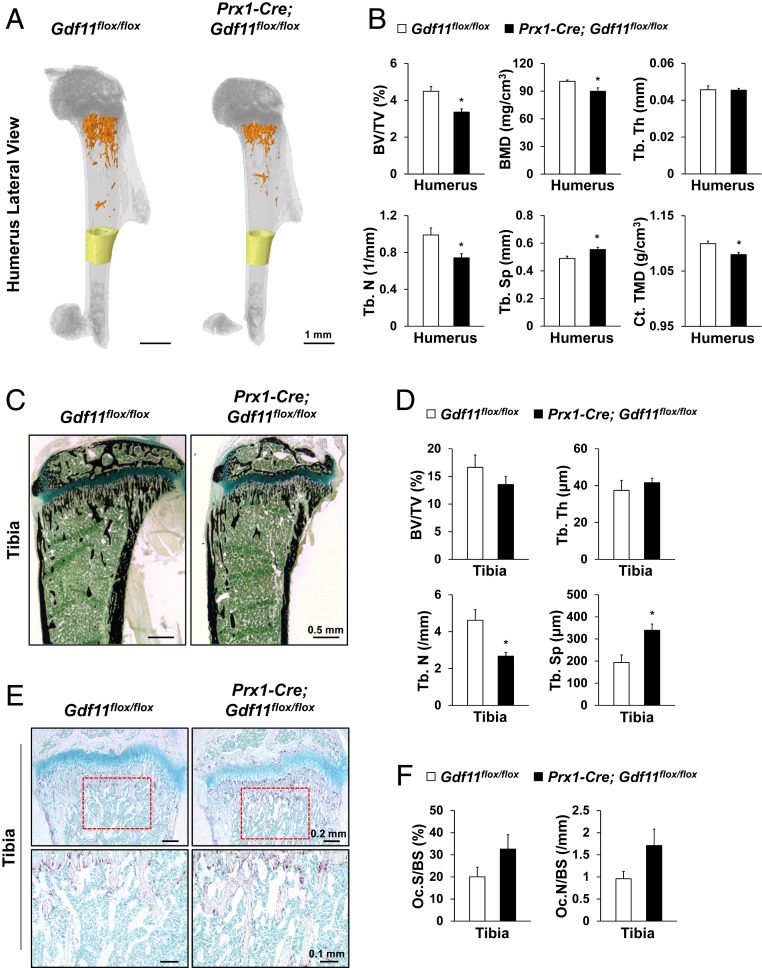
Because Gdf11 has been shown to regulate various organ functions, such as kidney and spleen (17, 19), which can exert indirect effects on skeletal tissues, we examined the outcome of Gdf11 deletion specifically in limb mesenchyme using Prx1-Cre-mediated recombination. Prx1-Cre has been shown to be expressed in both forelimb and hindlimb mesenchyme around E10.5 (39). Fortunately, Prx1-Cre; Gdf11flox/flox mice survived into adulthood, and analysis of these mice at 5 wk of age showed significant reductions in Tb. bone mass, bone density, and cortical (Ct.) bone density of the humerus in Prx1-Cre; Gdf11flox/flox mice (Fig. 3 A and B and SI Appendix, Fig. S3A). Von Kossa staining of tibia sections at 5 wk of age also revealed significantly decreased Tb. number and significantly increased Tb. separation in Prx1-Cre; Gdf11flox/flox mice (Fig. 3 C and D). Furthermore, TRAP staining and calcein-alizarin red labeling of tibia sections at 5 wk of age showed increased OC formation and decreased bone formation, respectively, in Prx1-Cre; Gdf11flox/flox mice (Fig. 3 E and F and SI Appendix, Fig. S3 B and C).
Fig. 3.
Gdf11 deletion in Prx1-Cre-expressing mesenchyme results in decreased bone mass. (A) Representative microCT images of 5-wk-old mouse humerus. Tb. bones of Prx1-Cre; Gdf11flox/flox mice are colored in orange and Ct. bones (1 mm in height) are colored in yellow. (B) Histomorphometric analysis of Tb. and Ct. bones of 5-wk-old mouse humerus (n = 4 each). (C) Representative von Kossa-stained images of 5-wk-old mouse tibia. (D) Histomorphometric analysis of von Kossa-stained sections of 5-wk-old mouse tibia (n = 4 each). (E) Representative TRAP-stained images of 5-wk-old mouse tibia. Magnified images of the red boxed regions are displayed in the Lower panel. (F) Histomorphometric analysis of TRAP-stained images of 5-wk-old mouse tibia (n = 4 each). OC.S/BS; OC.N/BS. All Scale bars are displayed with actual size values. All data represent mean ± SEM. *P < 0.05 by t test.
OB Differentiation and CH Maturation Are Suppressed, and OC Formation Is Enhanced in Gdf11−/− Mice, while the Opposite Is Observed in Mstn−/− Mice.
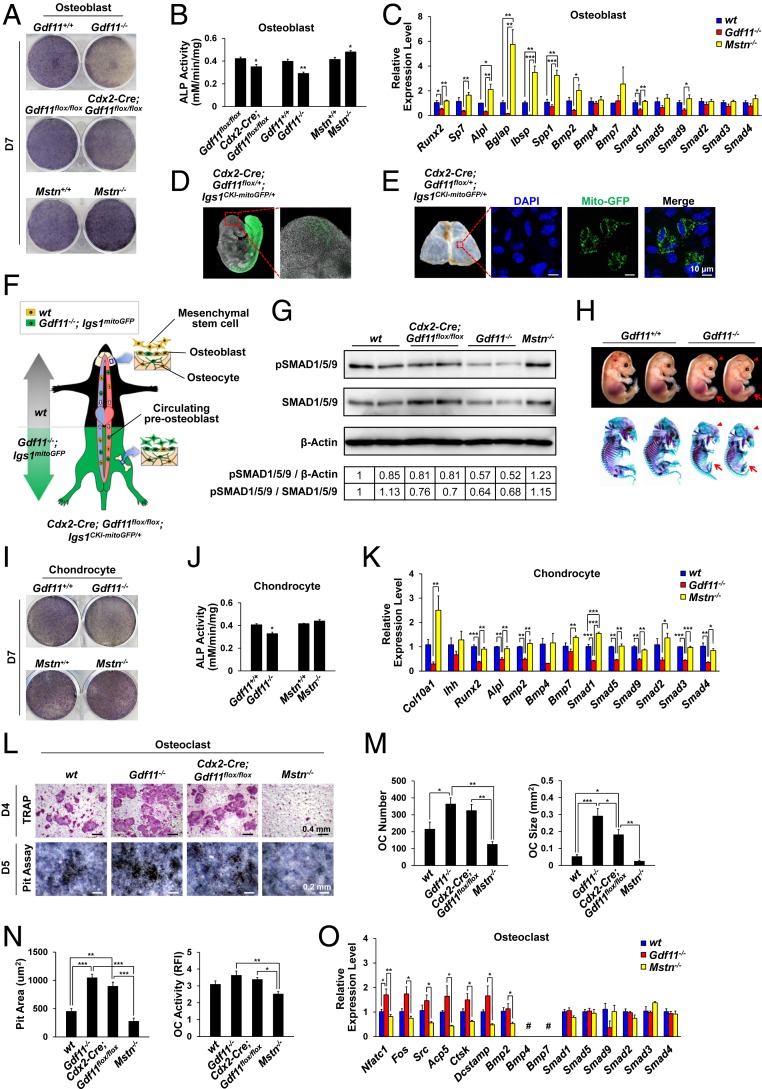
To understand the cellular and molecular mechanisms of the impaired osteogenesis after Gdf11 deletion, we first examined the osteogenic potential of primary OBs isolated from calvaria of live newborn mice. We were able to perform this experiment because some Gdf11−/− mice live up to several hours while others die immediately after birth. Contrary to Mstn−/− OBs that exhibited enhanced osteogenic differentiation, Gdf11−/− and Cdx2-Cre; Gdf11flox/flox OBs showed diminished differentiation activity as demonstrated by alkaline phosphatase (ALP) staining and alizarin red S (ARS) staining (Fig. 4 A and B and SI Appendix, Fig. S4 A and B). We also found that expressions of key osteogenic markers (Runx2, Sp7, Alpl, Bglap, Ibsp, and Spp1), Bmp2, and Smads were reduced in Gdf11−/− OBs but elevated in Mstn−/− OBs (Fig. 4C). Unexpected decline of ALP activity in calvarial OBs of Cdx2-Cre; Gdf11flox/flox mice, which normally express Gdf11 in the upper half tissues, prompted us to investigate whether this effect was due to circulating Gdf11−/− preOBs. Indeed, confocal imaging of Cdx2-Cre; Gdf11flox/+; Igs1CKI-mitoGFP/+ mice, which express GFP exclusively in mitochondria of posterior tissue cells, identified the presence of circulating GFP-positive cells at E9.5 (Fig. 4D) and those embedded in calvarias at week 4 (Fig. 4E), suggesting the function of GDF11 in guiding bone formation is mediated, at least, partially through circulating cells (Fig. 4F). The systemic effect of GDF11 was also confirmed by Western blot analysis of calvarias of newborn mice, which demonstrated decreased BMP signaling in calvarias of Cdx2-Cre; Gdf11flox/flox mice compared to wild type (WT) mice (Fig. 4G). To further explore the role of GDF11 in endochondral ossification during development, we analyzed Gdf11−/− embryos at E15.5 and consistently detected generally underdeveloped embryos with delayed ossification (Fig. 4H and SI Appendix, Fig. S4C). Primary CHs isolated from costal cartilages of live newborn Gdf11−/− mice exhibited decreased hypertrophic maturation in contrast to Mstn−/− CHs as shown by ALP staining (Fig. 4I and SI Appendix, Fig. S4D). ALP activity was significantly reduced in Gdf11−/− CHs compared to WT CHs, and although ALP expression was not significantly increased in Mstn−/− CHs compared to WT CHs, Col10a1 expression, a specific marker of hypertrophic CHs, was dramatically elevated in Mstn−/− CHs (Fig. 4 J and K). Gene expressions of CH maturation markers (Col10a1, Ihh, and Alpl), Runx2, Bmps, and Smads were all down-regulated in Gdf11−/− mice (Fig. 4K). In particular, expressions of Smad1, Smad5, and Smad9 were diminished in Gdf11−/− OBs (Fig. 4C) and significantly down-regulated in Gdf11−/− CHs compared to WT cells (Fig. 4K), suggesting the possible role of GDF11 in activating the BMP signaling pathway, which has been formerly described (40).
Fig. 4.
Gdf11 deletion, in contrast to Mstn deletion, impairs OB differentiation and CH maturation while stimulating OC formation. (A) Representative images of ALP staining of OBs of newborn mice after 7 d of differentiation. (B) ALP activity analysis in OBs after 7 d of differentiation (n = 3 each). (C) Quantitative RT-PCR analysis in OBs after 7 d of differentiation (n = 4 each). (D) Confocal images of a mouse embryo at E9.5 display circulating GFP-positive cells. (E) Confocal images of mouse calvaria at week 4. Note the GFP-positive cells embedded in the calvaria. (F) Western blot analysis of protein extracts from calvaria of newborn mice. Quantification numbers are indicated in the table below the blots. (G) Schematic of circulating preosteoblasts. WT and GFP-positive Gdf11−/− cells are marked in yellow and green, respectively. (H) Alcian blue/alizarin red staining of E15.5 littermate embryos. The arrowheads and arrows indicate underdeveloped skulls and hindlimbs of Gdf11−/− embryos, respectively. (I) Representative images of ALP staining of costal cartilage-derived CHs of newborn mice after 7 d of differentiation. (J) ALP activity analysis in CHs after 7 d of differentiation (n = 3 each). (K) Quantitative RT-PCR analysis in CHs after 7 d of differentiation (n = 4 each). (L) Representative images of TRAP staining of spleen-derived OCs and resorption pit formation after 4 d (for TRAP) or 5 d (for pit assay) of differentiation. (M) OC number and size after TRAP staining (n = 5 each). (N) Resorption pit area and OC activity after pit assay (n = 3 each). RFI, relative fluorescence intensity. (O) Quantitative RT-PCR analysis in OCs after 4 d of differentiation (n = 4 each). #, Expression is too low to evaluate. All scale bars are displayed with actual size values. Data B and J represent mean ± SEM. *P < 0.05, and **P < 0.01 by t test. Data C, K, M, N, and O represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey’s post hoc test.
Previous reports have illustrated that GDF11 is highly associated with anti-inflammatory signaling (41, 42) and inhibits erythroid differentiation (43, 44), while MSTN directly regulates OC formation (15). Therefore, we sought to determine the function of GDF11 in OC differentiation using primary splenocytes isolated from live newborn mutant mice. TRAP staining and resorption pit analysis demonstrated that receptor activator of nuclear factor κ-B ligand (RANKL)-induced OC formation and activity are significantly enhanced in Gdf11−/− splenocytes, moderately increased in Cdx2-Cre; Gdf11flox/flox splenocytes, and depressed in Mstn−/− splenocytes (Fig. 4 L–N and SI Appendix, Fig. S4E). Slight reduction of osteoclastogenesis in Cdx2-Cre; Gdf11flox/flox splenocytes compared to Gdf11−/− splenocytes could be explained by the presence of cells with incomplete Gdf11 deletion, confirmed by the existence of nonrecombined floxed alleles and GFP-negative areas in Cdx2-Cre; Gdf11flox/flox; Igs1CKI-mitoGFP/+ spleen (SI Appendix, Fig. S4F). Moreover, expressions of OC markers, including Nfatc1, Fos, Src, Acp5, Ctsk, and Dcstamp, were elevated in Gdf11−/− OCs, which contrast the expression patterns detected in Mstn−/− OCs (Fig. 4O).
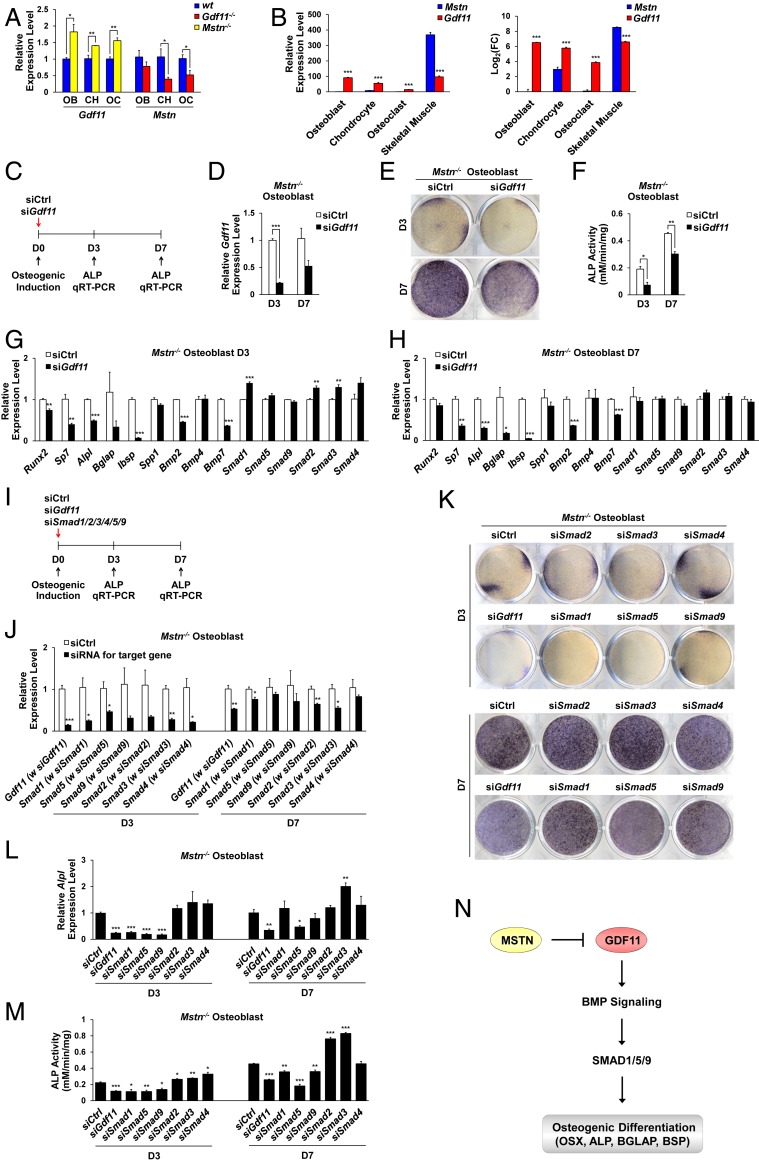
Gdf11 Knockdown Abolishes the Enhanced Osteogenic Effect of Mstn Deletion through Down-Regulation of BMP Signaling.
While Mstn expression was down-regulated in OBs, CHs, and OCs of Gdf11−/− mice with diminished bone mass, Gdf11 expression was significantly up-regulated in those cells of Mstn−/− mice with enlarged bone mass (Fig. 5A), suggesting that increased Gdf11 expression may be responsible for enhanced osteogenic, chondrogenic, and decreased OC activities of Mstn−/− cells. Because Mstn is known to be expressed primarily in skeletal muscle (1), we evaluated the differential expression patterns of Mstn and Gdf11 in OBs, CHs, OCs, and skeletal muscle cells by measuring the relative expression levels of Gdf11 and Mstn in comparison with the Mstn expression level in OBs, which is the lowest. Consistent with a key role of MSTN in skeletal muscle growth, Mstn expression was the greatest in skeletal muscle [data from our previous report (45)] while Gdf11 expression was substantially and significantly greater than that of Mstn in OBs, CHs, and OCs (Fig. 5B), suggesting that GDF11 is the key inducer of osteogenesis, while MSTN more actively regulates myogenesis.
Fig. 5.
Gdf11 knockdown abrogates the enhanced osteogenic effect of Mstn deletion. (A) Quantitative RT-PCR analysis of Gdf11 and Mstn expressions in OBs, CHs, and OCs after 7 d of differentiation for OBs and CHs, and 4 d of differentiation for OCs (n = 4 each). (B) Relative expression levels of Gdf11 and Mstn are evaluated in OBs, CHs, OCs, and skeletal muscle by comparing them with the Mstn expression level in OBs, which is the lowest (n = 4 for OB, CH, OC, and n = 3 for skeletal muscle). Data on gene expression levels in skeletal muscle were taken from our previous report (45). FC, fold change. (C) Gdf11 knockdown scheme. Control (siCtrl) or Gdf11 siRNA (siGdf11) was treated with Mstn−/− OBs at day 0 of differentiation. (D) Quantitative RT-PCR analysis of Gdf11 expression in Mstn−/− OBs 3 and 7 d after Gdf11 knockdown (n = 3 each). (E) Representative images of ALP staining of Mstn−/− OBs 3 and 7 d after Gdf11 knockdown. (F) ALP activity analysis in Mstn−/− OBs 3 and 7 d after Gdf11 knockdown (n = 3 each). (G and H) Quantitative RT-PCR analysis in Mstn−/− OBs 3 d (G) and 7 d (H) after Gdf11 knockdown (n = 3 each). (I) Gdf11 and Smads knockdown scheme. Control, Gdf11, or Smad1–5, or 9 siRNA was individually treated with Mstn−/− OBs at day 0 of differentiation. (J) Quantitative RT-PCR analysis in Mstn−/− OBs 3 and 7 d after knockdown of target genes (n = 3 each). (K) Representative images of ALP staining of Mstn−/− OBs 3 and 7 d after knockdown of target genes. (L) Quantitative RT-PCR analysis of Alpl expression in Mstn−/− OBs 3 and 7 d after knockdown of target genes (n = 3 each). (M) ALP activity analysis in Mstn−/− OBs 3 and 7 d after knockdown of target genes (n = 3 each). (N) A model for regulation of bone formation by GDF11 and MSTN. All data represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by t test.
Using small interfering RNAs (siRNAs), we next examined whether knockdown of increased Gdf11 expression abolishes the enhanced osteogenic ability of Mstn−/− OBs (Fig. 5C). Gdf11 expression was successfully silenced when siGdf11 was treated (Fig. 5D). At both 3 and 7 d after osteogenic induction, ALP activity in Mstn−/− OBs was significantly reduced when Gdf11 expression was down-regulated by siGdf11 (Fig. 5 E and F). Gene expressions of key osteogenic markers (Runx2, Sp7, Alpl, Bglap, Ibsp, and Spp1), Bmp2, and Bmp7 were significantly decreased in Mstn−/− OBs treated with siGdf11 after both 3 and 7 d of differentiation (Fig. 5 G and H), implying that GDF11 stimulates osteogenesis in Mstn−/− OBs by up-regulating the expression of Bmps. To elucidate the downstream signaling pathway activated by GDF11, focusing on SMADs, during osteogenic differentiation, we used siRNAs to individually knockdown Smad1, Smad2, Smad3, Smad4, Smad5, or Smad9 in Mstn−/− OBs and compared the effects with Gdf11 knockdown (Fig. 5I). After confirming successful knockdown of the target genes (Fig. 5J), we observed that both Alpl expression and ALP activity were significantly reduced in Mstn−/− OBs treated with siGdf11, siSmad1, siSmad5, or siSmad9, and increased in those treated with siSmad2, siSmad3, or siSmad4 after 3 d of differentiation (Fig. 5 K–M). ALP activity remained significantly elevated in Mstn−/− OBs treated with siSmad2 or siSmad3 and significantly diminished in those treated with siGdf11, siSmad1, siSmad5, or siSmad9 after 7 d of differentiation (Fig. 5 K and M and SI Appendix, Fig. S5A). Notably, Mstn−/− OBs treated with siSmad5 exhibited the most similar phenotype to those treated with siGdf11 (Fig. 5 K–M and SI Appendix, Fig. S5A), implying that GDF11 positively regulates BMP-SMAD signaling to enhance osteogenesis (Fig. 5N). Also, cell morphological phenotypes of Mstn−/− OBs treated with siGdf11 were similar to those of Mstn−/− OBs treated with siSmad1, siSmad5, or siSmad9, which displayed elongated and less differentiated appearances at 3 d after osteogenic induction (SI Appendix, Fig. S5B). In addition, siRNA-mediated Gdf11 knockdown increased OC formation in Mstn−/− splenocytes, indicating that elevated Gdf11 expression in Mstn−/− splenocytes prevents osteoclastogenesis (SI Appendix, Fig. S5C).
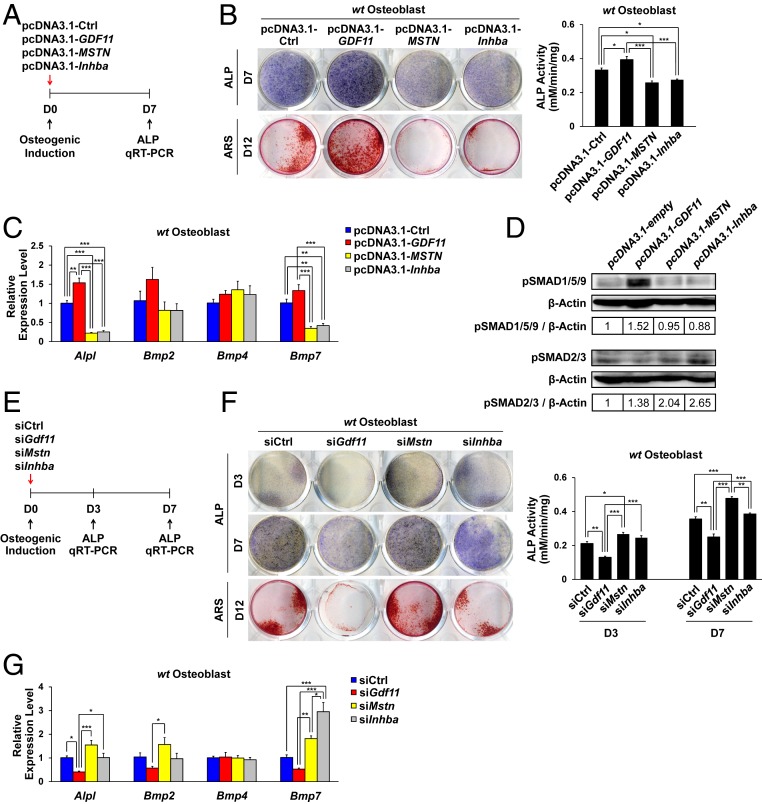
GDF11 Overexpression Stimulates the BMP Signaling Pathway to Promote Osteogenic Differentiation.
Because GDF11 and MSTN recombinant proteins cannot be clearly distinguished in vitro due to their high degree of homology, we overexpressed GDF11, MSTN, and another related TGF-β family member Inhba by transfecting their full-length cDNAs into primary OBs derived from WT newborn mouse calvaria (Fig. 6A and SI Appendix, Fig. S6A). ALP staining and analysis at day 7, and ARS staining at day 12 of osteogenic induction revealed that GDF11 overexpression significantly enhanced OB differentiation and mineralization, while MSTN or Inhba overexpression significantly impaired osteogenic differentiation (Fig. 6B). In addition, GDF11 overexpression up-regulated Bmp2 and Bmp7 expressions (Fig. 6C) and SMAD 1/5/9 phosphorylation at day 7 (Fig. 6D), while the opposite pattern was observed in the MSTN or Inhba overexpression group (Fig. 6 C and D). We also tested the effects of siRNA-mediated knockdown of Gdf11, Mstn, and Inhba on WT primary OBs (Fig. 6E and SI Appendix, Fig. S6B). At both 3 and 7 d after osteogenic induction, Alpl expression and ALP activity were significantly reduced upon siGdf11 treatment, while they were significantly elevated upon siMstn treatment (Fig. 6 F and G and SI Appendix, Fig. S6 C and D). Likewise, ARS staining at day 12 of differentiation revealed the same pattern (Fig. 6F). In addition, gene expression analysis showed that Gdf11 knockdown reduces Bmp2 and Bmp7 expressions, which are up-regulated by Mstn knockdown, further supporting the role of GDF11 in activating the BMP signaling pathway to promote osteogenesis (Fig. 6G). While knockdown of Inhba appeared to slightly increase ALP activity at days 3 and 7, it did not enhance mineralization at day 12 compared to the control group (Fig. 6F), despite a significant increase in Bmp7 expression at day 7 (Fig. 6G).
Fig. 6.
GDF11 overexpression enhances OB differentiation by activating the BMP signaling pathway. (A) GDF11, MSTN, and Inhba overexpression scheme. Plasmids were transfected OBs at day 0 of differentiation. (B) Representative images of ALP staining at day 7, ARS staining at day 12, and ALP activity analysis of WT OBs at day 7 of overexpression (n = 3 each). (C) Quantitative RT-PCR analysis in WT OBs at day 7 of overexpression (n = 4 each). (D) Western blot analysis of WT OBs at day 7 of overexpression. Quantification numbers are indicated in tables below the blots. (E) Gdf11, Mstn, and Inhba knockdown scheme. Control (siCtrl), Gdf11 siRNA (siGdf11), Mstn siRNA (siMstn), or Inhba siRNA (siInhba) was treated with WT OBs at day 0 of differentiation. (F) Representative images of ALP staining at day 3 and 7, ARS staining at day 12, and ALP activity analysis of WT OBs at day 3 and 7 of knockdown of target genes (n = 3 each). (G) Quantitative RT-PCR analysis in WT OBs at day 7 of knockdown of target genes (n = 4 each). Data B and F represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by t test. Data C and G represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey’s post hoc test.
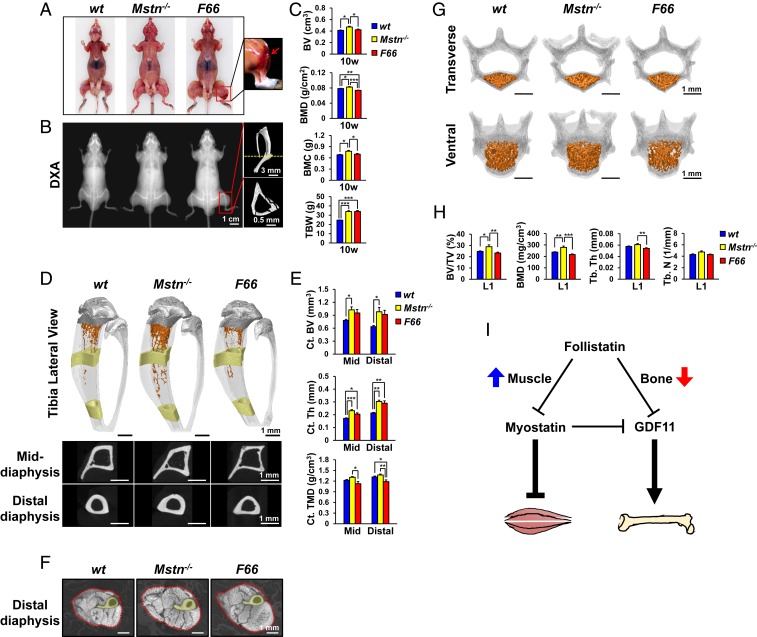
GDF11 Inhibition by FST Decreases BMD and Induces Tibia Fractures.
In previous studies, we showed that transgenic mice overexpressing FST, a MSTN and GDF11 inhibitor (5), from a myosin light chain promoter/enhancer (F66 mice) exhibit about a doubling of skeletal muscle mass comparable to that seen in Mstn−/− mice (46). In further characterizing these mice, we noted the frequent occurrence of fractures, which are not seen in Mstn−/− mice. Specifically, we observed tibial fractures in F66 mice repeatedly at various ages from 10 wk to 1 y (Fig. 7 A and B and SI Appendix, Fig. S7 A and B). This observation guided us to further investigate bone morphometric parameters of F66 mice. DXA analysis at 10 wk revealed that, while TBW and lean mass equally increased in both F66 and Mstn−/− mice, BV, BMD, and BMC are all significantly reduced in F66 mice compared to Mstn−/− mice (Fig. 7C and SI Appendix, Fig. S7C). Notably, overall BMD in F66 mice was significantly decreased even compared to WT mice. Consistent with this, microCT analysis at 10 wk indicated that Ct. TMD is diminished in F66 tibias that also exhibit noticeable Ct. porosity, whereas Ct. BV, Ct. thickness, and Ct. TMD are all significantly elevated in Mstn−/− tibias (Fig. 7 D–F and SI Appendix, Fig. S7D). Furthermore, Tb. bone mass and vertebral BMD of L1 were reduced in 10-wk-old F66 mice compared to WT mice, while they were increased in age-matched Mstn−/− mice compared to WT mice (Fig. 7 G and H, SI Appendix, Fig. S7 E and F, and Movies S1–S3). These results illustrate that while FST promotes skeletal muscle growth by inhibiting MSTN, it can hinder osteogenesis by blocking GDF11 (Fig. 7I).
Fig. 7.
FST overexpression reduces BMD and induces tibia fractures. (A) Representative images of 10-wk-old mice. The boxed region is magnified to display fractured tibia in F66 mice, which is pointed by a red arrow. (B) Representative DXA images of 10-wk-old mice, and microCT images of F66 mouse’s fractured tibia in the boxed region. The cross sectional image at the position indicated by yellow dashed line is shown in the Bottom panel. (C) DXA analysis of 10-wk-old mice (n = 5 for WT mice, n = 5 for Mstn−/− mice, and n = 8 for F66 mice). (D) Representative microCT images of 10-wk-old mouse tibia. Fractured tibias of F66 mice were excluded from microCT analysis. Tb. bones are colored in orange and Ct. bones (1 mm in height) are colored in yellow. Raw cross sectional images of the colored middle and distal diaphysis are displayed in the Bottom panel. (E) Histomorphometric analysis of Ct. bones of 10-wk-old mice (n = 5 each). (F) Representative microCT images of 10-wk-old mouse distal hindlimbs cross sectioned at the level where tibia and fibula start to fuse. The red dotted lines mark muscle boundaries. C. bones are colored in yellow. (G) Representative microCT images of 10-wk-old mouse vertebrae (L1). Tb. bone of the vertebral body is colored in orange. (H) Histomorphometric analysis of L1 Tb. bone in 10-wk-old mice (n = 5 each). (I) A model for regulation of skeletal muscle and bone mass by FST. FST enhances skeletal muscle mass by inhibiting MSTN, while decreasing BMD by blocking GDF11. All scale bars are displayed with actual size values. All data represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA with Tukey’s post hoc test.
Discussion
Both MSTN and its homolog GDF11 have been widely implicated to control developmental and age-related processes. Despite sharing similar structures, binding molecules, and receptors (1–5), GDF11 and MSTN have been shown to exert distinct functions in vivo, likely due to differences in the biological regulation of their tissue/time-specific expression patterns and activity states (8). For instance, while MSTN is expressed primarily in skeletal muscle and acts to limit muscle and bone mass (1, 14–16), GDF11 is more diversely expressed and controls skeletal patterning and organ development during embryogenesis (2, 17–21). Although the functions of MSTN have been well described through Mstn null mice that survive into adulthood, reports on GDF11, especially the ones examining its postnatal functions, are less clear and conflicting. Because Gdf11 null mice die shortly after birth, previous contradictory findings on GDF11, including the ones examining its role in bone development (26–28), have relied primarily on the recombinant GDF11 protein to explain its physiological function. However, due to 89% amino acid sequence identity within the mature signaling domains of GDF11 and MSTN, their recombinant proteins share almost identical biochemical properties and, therefore, are essentially indistinguishable (25), generating the possibility that the effects of recombinant GDF11 reported in previous studies are actually reflecting the endogenous functions of MSTN. Thus, we believed that the use of recombinant GDF11 cannot clearly elucidate the complex biological regulation of GDF11 occurring in vivo, not to mention there can be significant quality differences between recombinant GDF11 proteins (29, 30). To avoid these limitations, here, we focused on genetic studies in mice, demonstrating that GDF11, in contrast to MSTN, promotes osteogenesis.
In this paper, we utilized conditional knockout techniques to produce genetically engineered mice in which the Gdf11 gene is deleted in a tissue/time-specific manner. In detail, we used three different Cre lines, Cdx2-Cre, TAM-inducible CAG-Cre-ER, and Prx1-Cre, to target recombination exclusively in posterior half tissues, in all tissues at specific time points, and specifically in limb mesenchyme, respectively. Our initial analysis revealed that bone mass is significantly diminished in Gdf11 null mice but is slightly recovered in Cdx2-Cre; Gdf11flox/flox mice whose expression of Gdf11 still remains in anterior regions. In contrast to Gdf11 null mice, Mstn null mice exhibited enhanced bone mass. We also observed that time-specific ubiquitous deletion of Gdf11 using CAG-Cre-ER impairs bone development in both embryos and young adult mice. However, because GDF11 plays major roles in the development of various organs, such as kidney and spleen (17, 19), which critically influence bone metabolism (47, 48), it is hard to rule out the possibility that dysfunction in these organs indirectly impeded bone development in CAG-Cre-ER; Gdf11flox/flox mice. Therefore, to avoid possible indirect influence of a dysfunctional organ caused by loss of GDF11 on bone homeostasis, we utilized Prx1-Cre-mediated recombination to examine the direct effects of GDF11 in skeletal tissues. Indeed, Prx1-Cre; Gdf11flox/flox mice at 5 wk of age exhibited reduced bone mass and density in long bones, characterized by decreased bone formation and increased bone resorption. We were also able to isolate primary OBs, CHs, and splenocytes while newborn Gdf11 null mice were still alive and show that OB differentiation and CH maturation were impaired and OC formation was enhanced in Gdf11 null mice, while the opposite was observed in Mstn null mice.
Interestingly, our data from Cdx2-Cre; Gdf11flox/flox mice highlight the important systemic role that GDF11 plays during bone development. In Cdx2-Cre; Gdf11flox/flox mice, although Gdf11 expression is removed exclusively in posterior half tissues, both anterior and posterior vertebrae exhibit similar bone mass, suggesting that GDF11 systemically enhances osteogenesis. However, as circulating GDF11 molar concentration is close to 500 times less than that of circulating MSTN (29), whether circulating GDF11 proteins have physiological relevance has been formerly questioned. Here, we answered this question by identifying the existence of circulating GFP-positive cells and those embedded in calvarial bones in mice designed to express GFP only in posterior tissues (Cdx2-Cre; Gdf11flox/+; Igs1CKI-mitoGFP/+ mice), suggesting that GDF11 exerts its systemic effect on bone formation, at least, partially through circulating cells. We further confirmed the systemic effects of GDF11 through Western blot analysis, which revealed that BMP signaling is down-regulated in calvaria of newborn Cdx2-Cre; Gdf11flox/flox mice compared to that of WT mice.
The precise signaling mechanisms that differentiate GDF11 from MSTN are yet to be fully understood. While both GDF11 and MSTN have been previously shown to signal through activin type II receptors to activate SMAD2 and SMAD3 (3–5), our study revealed that GDF11 stimulates BMP signaling to regulate bone mass. More specifically, Gdf11 null OBs and CHs exhibited a significant decrease in BMP expression, while the opposite was observed in Mstn deficient cells that overexpressed Gdf11. Correspondingly, knockdown of Smad1, Smad5, or Smad9 (especially Smad5), but not Smad2 or Smad3 in Mstn deficient OBs, phenocopied those subjected to Gdf11 knockdown, suggesting that GDF11-mediated activation of OBs involves the BMP signaling pathway, which is inhibited by MSTN through down-regulation of GDF11. Also, transfection of full-length cDNA of GDF11, in contrast to that of MSTN, significantly enhanced OB differentiation and mineralization through SMAD 1/5/9 phosphorylation, while knockdown of Gdf11 in WT OBs significantly reduced osteogenic activity and Bmp expression, in contrast to knockdown of Mstn that consistently produced the opposite results. The activation of the BMP signaling pathway by GDF11 has also been reported in endothelial cells (40), but whether this action of GDF11 occurs through different receptor utilization or through biological regulation of its activity state needs further investigation. Moreover, GDF11 in OCs did not dramatically alter BMP signaling but seemed to involve anti-inflammatory activity through NF-κB inhibition (41), which also requires further examination.
It is noteworthy that while our data described GDF11 as a positive endogenous regulator of bone formation, they do not imply that exogenous administration of GDF11 proteins, which cannot be effectively distinguished from the highly similar MSTN, will exhibit the same effect. In fact, the uncertainty in the effectiveness of GDF11 protein treatment has been well implicated through contradictory results presented by earlier studies using recombinant GDF11. For instance, Zhang et al. (26) reported that recombinant GDF11 delivery improves OB differentiation, but Lu et al. (27) and Liu et al. (28) subsequently reported the opposite, describing the potential risk of exogenous GDF11 treatment in bone development. Based on these and our findings, we suggest that, although care should be taken to avoid inhibiting endogenous GDF11 expression, further research is needed to translate our findings into therapeutic applications involving GDF11 protein delivery.
FST, by antagonizing MSTN, has been shown to effectively promote skeletal muscle hypertrophy in rodents and primates (49), which leads to human trials of Fst gene therapy to treat patients with various forms of muscular dystrophy (34–37). However, our results revealed that, while FST overexpression greatly enhances muscle growth comparable to Mstn deletion in mice, it can cause adverse effects on bone, including decreased BMD and frequent tibia fractures, which is in contrast to significantly increased bone mass and density observed in Mstn null mice. FST has been reported as a secreted protein capable of binding and inhibiting MSTN, GDF11, and activin A (50). Both MSTN and activin A have been shown to strongly suppress bone formation (51), and their inhibition has been shown to enhance bone mass (52), although some early studies report positive effects of activin A on osteogenesis (53, 54). Here, we examined the role of activin A on OB differentiation and observed that overexpression of Inhba significantly impaired ALP activity and mineralization, but knockdown of Inhba also appeared to slightly decrease mineralization at day 12 of differentiation despite a mild increase in ALP activity at day 3 and day 7, implying a possible complex time-dependent regulation of osteogenesis by activin A. This observation needs to be confirmed in further studies including Inhba conditional knockout mice analysis. However, based on our overexpression experiment showing suppressive actions of both MSTN and activin A in mineralization, the decreased skeleton quality by FST overexpression is most likely to be the result of GDF11 inhibition. FST also has been shown to interact with BMPs but with lower affinities (55), still making GDF11 inhibition mainly responsible for the observed fracture phenotype in F66 mice. Here, we demonstrate that FST, a MSTN and GDF11 inhibitor, can induce differential effects on muscle and bone: a dramatic increase in skeletal muscle mass through MSTN inhibition, but a significant decrease in bone quality through GDF11 inhibition.
Currently, numerous MSTN inhibitors, including FST, are being tested in clinical trials to increase skeletal muscle mass by blocking MSTN, a potent negative regulator of muscle growth, for the treatment of muscle wasting and metabolic diseases, including muscular dystrophy, age-related sarcopenia, and cancer cachexia. However, our data suggest that FST also exerts negative effects on bone quality and strength by inhibiting GDF11, a key factor in bone development. Notably, most MSTN inhibitors also bind and inhibit GDF11 due to the high sequence similarity between MSTN and GDF11. Therefore, the crucial role of GDF11 in bone homeostasis must be carefully considered to minimize the potential detrimental effects of GDF11 inhibition on bone when evaluating MSTN-targeting drugs for muscle enhancement.
Materials and Methods
The materials and methods used in this study are described in detail in the SI Appendix. Information includes mice and in vivo experiments, microCT imaging analysis, bone histomorphometry analysis, in vitro cell experiments, and molecular biology experiments. All animal studies were approved by the Institutional Animal Care and Use Committees at Seoul National University.
Data Availability.
All data supporting the findings of this paper are available within the article and SI Appendix.
Supplementary Material
Acknowledgments
We thank Suzanne M. Sebald for assistance with shipping and handling mice; and Je-Yong Choi, Hong-Hee Kim, and Min-Sung Kim for helpful discussions. This work was supported by Creative-Pioneering Researchers Program through Seoul National University (SNU) and Basic Research Laboratory Program (Grant NRF-2017R1A4A1014584) of the National Research Foundation (NRF) funded by the Korean government (to Y.-S.L.).
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916034117/-/DCSupplemental.
References
- 1.McPherron A. C., Lawler A. M., Lee S. J., Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83–90 (1997). [DOI] [PubMed] [Google Scholar]
- 2.McPherron A. C., Lawler A. M., Lee S. J., Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22, 260–264 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Andersson O., Reissmann E., Ibáñez C. F., Growth differentiation factor 11 signals through the transforming growth factor-beta receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep. 7, 831–837 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rebbapragada A., Benchabane H., Wrana J. L., Celeste A. J., Attisano L., Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol. Cell. Biol. 23, 7230–7242 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S. J., McPherron A. C., Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. U.S.A. 98, 9306–9311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker R. G., et al. , Structural basis for potency differences between GDF8 and GDF11. BMC Biol. 15, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPherron A. C., Huynh T. V., Lee S. J., Redundancy of myostatin and growth/differentiation factor 11 function. BMC Dev. Biol. 9, 24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker R. G., et al. , Molecular characterization of latent GDF8 reveals mechanisms of activation. Proc. Natl. Acad. Sci. U.S.A. 115, E866–E875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosher D. S., et al. , A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 3, e79 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clop A., et al. , A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 38, 813–818 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Acosta J., Carpio Y., Borroto I., González O., Estrada M. P., Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J. Biotechnol. 119, 324–331 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Schuelke M., et al. , Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 350, 2682–2688 (2004). [DOI] [PubMed] [Google Scholar]
- 13.McPherron A. C., Lee S. J., Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. U.S.A. 94, 12457–12461 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin Y., et al. , Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: A novel mechanism in muscle-bone communication. J. Biol. Chem. 292, 11021–11033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dankbar B., et al. , Myostatin is a direct regulator of osteoclast differentiation and its inhibition reduces inflammatory joint destruction in mice. Nat. Med. 21, 1085–1090 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Hamrick M. W., et al. , Loss of myostatin (GDF8) function increases osteogenic differentiation of bone marrow-derived mesenchymal stem cells but the osteogenic effect is ablated with unloading. Bone 40, 1544–1553 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmon E. B., et al. , GDF11 modulates NGN3+ islet progenitor cell number and promotes beta-cell differentiation in pancreas development. Development 131, 6163–6174 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Wu H. H., et al. , Autoregulation of neurogenesis by GDF11. Neuron 37, 197–207 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Esquela A. F., Lee S. J., Regulation of metanephric kidney development by growth/differentiation factor 11. Dev. Biol. 257, 356–370 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Nakashima M., Toyono T., Akamine A., Joyner A., Expression of growth/differentiation factor 11, a new member of the BMP/TGFbeta superfamily during mouse embryogenesis. Mech. Dev. 80, 185–189 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Gamer L. W., et al. , A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer in Xenopus embryos. Dev. Biol. 208, 222–232 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Sinha M., et al. , Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 344, 649–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katsimpardi L., et al. , Vascular and neurogenic rejuvenation of the aging mouse brain by young systemic factors. Science 344, 630–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loffredo F. S., et al. , Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153, 828–839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egerman M. A., et al. , GDF11 increases with age and inhibits skeletal muscle regeneration. Cell Metab. 22, 164–174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., et al. , Growth differentiation factor 11 is a protective factor for osteoblastogenesis by targeting PPARgamma. Gene 557, 209–214 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Lu Q., et al. , GDF11 inhibits bone formation by activating smad2/3 in bone marrow mesenchymal stem cells. Calcif. Tissue Int. 99, 500–509 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Liu W., et al. , GDF11 decreases bone mass by stimulating osteoclastogenesis and inhibiting osteoblast differentiation. Nat. Commun. 7, 12794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodgers B. D., Eldridge J. A., Reduced circulating GDF11 is unlikely responsible for age-dependent changes in mouse heart, muscle, and brain. Endocrinology 156, 3885–3888 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Rodgers B. D., et al. , Myostatin stimulates, not inihibits, C2C12 myoblast proliferation. Endocrinology 155, 670–675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lach-Trifilieff E., et al. , An antibody blocking activin type II receptors induces strong skeletal muscle hypertrophy and protects from atrophy. Mol. Cell. Biol. 34, 606–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S. J., et al. , Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 18117–18122 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haidet A. M., et al. , Long-term enhancement of skeletal muscle mass and strength by single gene administration of myostatin inhibitors. Proc. Natl. Acad. Sci. U.S.A. 105, 4318–4322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendell J. R., et al. , Follistatin gene therapy for sporadic inclusion body myositis improves functional outcomes. Mol. Ther. 25, 870–879 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendell J. R., et al. , A phase 1/2a follistatin gene therapy trial for becker muscular dystrophy. Mol. Ther. 23, 192–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al-Zaidy S. A., Sahenk Z., Rodino-Klapac L. R., Kaspar B., Mendell J. R., Follistatin gene therapy improves ambulation in becker muscular dystrophy. J. Neuromuscul. Dis. 2, 185–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodino-Klapac L. R., et al. , Micro-dystrophin and follistatin co-delivery restores muscle function in aged DMD model. Hum. Mol. Genet. 22, 4929–4937 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh J., et al. , Growth differentiation factor 11 locally controls anterior-posterior patterning of the axial skeleton. J. Cell. Physiol. 234, 23360–23368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Logan M., et al. , Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y. H., et al. , GDF11/BMP11 activates both smad1/5/8 and smad2/3 signals but shows no significant effect on proliferation and migration of human umbilical vein endothelial cells. Oncotarget 7, 12063–12074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., et al. , GDF11 antagonizes psoriasis-like skin inflammation via suppression of NF-κB signaling pathway. Inflammation 42, 319–330 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Li W., et al. , GDF11 antagonizes TNF-α-induced inflammation and protects against the development of inflammatory arthritis in mice. FASEB J. 33, 3317–3329 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Dussiot M., et al. , An activin receptor IIA ligand trap corrects ineffective erythropoiesis in β-thalassemia. Nat. Med. 20, 398–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suragani R. N., et al. , Transforming growth factor-β superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat. Med. 20, 408–414 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Lee Y. S., Lee S. J., Regulation of GDF-11 and myostatin activity by GASP-1 and GASP-2. Proc. Natl. Acad. Sci. U.S.A. 110, E3713–E3722 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S. J., Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS One 2, e789 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei K., Yin Z., Xie Y., Roles of the kidney in the formation, remodeling and repair of bone. J. Nephrol. 29, 349–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamichi Y., et al. , Spleen serves as a reservoir of osteoclast precursors through vitamin D-induced IL-34 expression in osteopetrotic op/op mice. Proc. Natl. Acad. Sci. U.S.A. 109, 10006–10011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kota J., et al. , Follistatin gene delivery enhances muscle growth and strength in nonhuman primates. Sci. Transl. Med. 1, 6ra15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneyer A. L., et al. , Differential antagonism of activin, myostatin and growth and differentiation factor 11 by wild-type and mutant follistatin. Endocrinology 149, 4589–4595 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alves R. D., Eijken M., Bezstarosti K., Demmers J. A., van Leeuwen J. P., Activin A suppresses osteoblast mineralization capacity by altering extracellular matrix (ECM) composition and impairing matrix vesicle (MV) production. Mol. Cell. Proteomics 12, 2890–2900 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chantry A. D., et al. , Inhibiting activin-A signaling stimulates bone formation and prevents cancer-induced bone destruction in vivo. J. Bone Miner. Res. 25, 2633–2646 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Gaddy-Kurten D., Coker J. K., Abe E., Jilka R. L., Manolagas S. C., Inhibin suppresses and activin stimulates osteoblastogenesis and osteoclastogenesis in murine bone marrow cultures. Endocrinology 143, 74–83 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Sakai R., Miwa K., Eto Y., Local administration of activin promotes fracture healing in the rat fibula fracture model. Bone 25, 191–196 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Thompson T. B., Lerch T. F., Cook R. W., Woodruff T. K., Jardetzky T. S., The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev. Cell 9, 535–543 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this paper are available within the article and SI Appendix.