Significance
Plant cells are connected to each other by plasmodesmata (PD), membrane-bound cytosolic channels that cross the cell wall. Diverse molecules, ranging from metabolites to small RNAs and proteins, can traffic between plant cells via PD. During development, PD transport is regulated to control the redistribution of carbohydrates from mature leaves that photosynthesize excess sugars to young leaves that rapidly consume sugars. Here, we show that the ancient eukaryotic TARGET OF RAPAMYCIN (TOR) metabolic signaling network controls PD transport in plants. TOR is a protein kinase that is activated by nutrients, including sugars, and then coordinates growth with nutrient availability. Our study demonstrates that TOR has evolved new roles in plants by regulating cell–cell connectivity via PD.
Keywords: plasmodesmata, cell–cell signaling, rapamycin, Reptin, TARGET OF RAPAMYCIN
Abstract
The coordinated redistribution of sugars from mature “source” leaves to developing “sink” leaves requires tight regulation of sugar transport between cells via plasmodesmata (PD). Although fundamental to plant physiology, the mechanisms that control PD transport and thereby support development of new leaves have remained elusive. From a forward genetic screen for altered PD transport, we discovered that the conserved eukaryotic glucose-TOR (TARGET OF RAPAMYCIN) metabolic signaling network restricts PD transport in leaves. Genetic approaches and chemical or physiological treatments to either promote or disrupt TOR activity demonstrate that glucose-activated TOR decreases PD transport in leaves. We further found that TOR is significantly more active in mature leaves photosynthesizing excess sugars than in young, growing leaves, and that this increase in TOR activity correlates with decreased rates of PD transport. We conclude that leaf cells regulate PD trafficking in response to changing carbohydrate availability monitored by the TOR pathway.
Plant cells are connected by plasmodesmata (PD), nanoscopic membrane-bound channels in the cell wall that permit molecules up to ∼80 kDa to traffic between neighboring cytoplasts (1–3). PD are absolutely essential for plant development and physiology, and their importance is reflected in the repeated, parallel evolution of PD or analogous structures in every lineage of complex multicellular eukaryotes that have cellulosic cell walls (4, 5). The upstream signal transduction pathways that regulate PD transport in plants remain poorly understood, however. We and others discovered that chloroplasts can regulate the rate of PD transport and the biogenesis of PD in pathways that we named “organelle-nucleus-plasmodesmata signaling” (6–11). When plants undergo developmental transitions, such as during lateral root formation, PD transport is often restricted, which is proposed to isolate cells during differentiation and allow signaling molecules (such as phytohormones and transcription factors) to accumulate to high concentrations (3, 12–16). PD transport is also tightly regulated during various stresses, especially upon recognition of infection by a pathogen (5, 17–21).
During plant development, plants redistribute sugars photosynthesized in mature leaves (“sources”) to support the development of young leaves and roots (“sinks”). Tracheophytes traffic sugars via the phloem, specialized vascular tissue that concentrates high levels of sugars for long-distance transport through the plant body. As leaves develop, they switch from importing nutrients out from the phloem (sinks) to exporting sugars into the phloem (sources), a physiological shift that is called the “sink-to-source” transition. Among different plant species, there is considerable variation in how sugars are loaded from the photosynthetic mesophyll into the phloem; here, we will focus on herbaceous plants, which typically use an “apoplastic” loading mechanism. In source leaves of apoplastic loaders, sugars are loaded against a concentration gradient into the phloem companion cells by active transporters in the plasma membranes (22–25). To prevent passive backflow of sugars from the vasculature into the leaf, PD transport is tightly restricted in source leaves (1). Phloem loading has a dual effect: The concentration of solutes in the phloem increases, creating hydrostatic pressure that can promote phloem transport, and perhaps more importantly, the concentration of free sugars in the source leaf is reduced (i.e., export of photo-assimilated carbohydrates from the source to growing sink tissues is maximized) (26). In contrast to source leaves, PD transport is unrestricted in young sink leaves, which allows rapid, passive transport of sugars out from the vasculature into the growing leaf (27–29). Although this change in PD transport as leaves age has been recognized for several decades, little is known about how PD transport between cells is coordinated with leaf development and physiology during the sink-to-source transition.
We conducted a screen to identify mutants defective in regulating PD transport, using Arabidopsis thaliana embryogenesis as a model system (30). Four of these mutants, ise1 through ise4, do not restrict PD transport at the midtorpedo developmental stage of embryogenesis, which is when wild-type embryos become photosynthetic and PD transport rapidly decreases (30). We previously characterized ise1 (At1g12770) and ise2 (At1g70070), which disrupt chloroplast biogenesis and trigger a retrograde signaling pathway that controls intercellular trafficking (6). Here, we characterize ise3 (At2g25570) and ise4 (reptin-1, At5g67630), two mutants that disrupt the TARGET OF RAPAMYCIN (TOR) signaling network.
TOR is an essential, deeply conserved serine/threonine protein kinase that coordinates metabolism with nutrient availability in all eukaryotes (31–33). When nutrients are available, TOR promotes growth and anabolism, but when nutrients are scarce, TOR activity is suppressed and cells become quiescent, using catabolic pathways, such as autophagy, for sustenance (34). TOR is widely studied in biomedical fields, because mutations in TOR and its interacting partners can cause or contribute to a range of diseases, including many cancers (34, 35); much less is known about TOR signaling in plants. TOR is active in complex with at least two other conserved proteins, RAPTOR and LST8, to make the TOR complex 1 (TORC1). In its active form, TORC1 is a dimer; dimerization is promoted by a cochaperone complex called R2TP-TTT (Telo2-Tti1-Tti2) in association with the well-studied protein chaperone, HSP90. R2TP-TTT, named for its yeast components (Pontin/RuvBL1, Reptin/RuvBL2, Spaghetti/Tah1, and Pih1 compose the R2TP subcomplex; Telo2, Telo2-interacting protein 1, and Telo2-interacting protein 2 compose the TTT subcomplex), is proposed to act as an energy sensor upstream from TOR: Pontin and Reptin form an ATPase in R2TP-TTT that drives TORC1 dimerization when ATP is plentiful, but cannot dimerize TORC1 when ATP is scarce (36). When TORC1 is active, it can phosphorylate a number of downstream targets, including a phosphorylation cascade mediated by the TOR substrate S6K (ribosomal protein eS6 Kinase) that can promote cell division, cell growth, and protein translation, among diverse additional metabolic pathways.
Much less is known about TOR signaling in plants. TOR is essential for plant development: tor mutants are lethal at very early stages of embryogenesis (37) and strong inhibition of TOR through chemical inhibition or RNAi causes plant growth to arrest (38, 39). As in other eukaryotes, plant TOR directly phosphorylates S6K (there are two tandem S6K paralogues in A. thaliana, named S6K1 and S6K2) at a conserved serine (pT389 in humans, pT449 in A. thaliana S6K1), and S6K-pT449 levels are therefore a reliable marker for TOR activity in plants (38). A seminal study discovered that photosynthesis-stimulated TOR activity is required for seedling establishment in A. thaliana (40), and then took advantage of this physiological transition to clearly define a set of ∼2,000 genes that are differentially expressed specifically in response to glucose activation of TOR in plants. Interactomics have demonstrated that TOR closely associates with many of the R2TP-TTT subunits (41), and in maize, loss of one of the TTT subunits causes embryo lethality and a drastic reduction in TOR levels, presumably due to protein instability when TOR is not chaperoned by R2TP-TTT to form the TORC1 dimer (42). The biological functions of plant TOR remain to be elucidated, however, and much less is known about the roles of TOR at later developmental stages (such as in mature leaves). Here, we show that TOR has evolved a role in plants to regulate PD transport and the sink-to-source transition in leaves.
Results and Discussion
The R2TP Complex Regulates PD Transport.
ise4 is an embryo-lethal mutant that we identified in a screen for increased PD transport. For these experiments, PD transport is assayed by the movement of a 10-kDa fluorescent dextran tracer that does not readily move between cells at the midtorpedo stage in wild-type, but does move throughout the midtorpedo embryos in ise mutants (Fig. 1A) (30). In the siliques of ise4/+ heterozygous parents, ∼25% of embryos arrest at the midtorpedo stage and exhibit increased PD transport (Fig. 1A). ise4 maps to a single nucleotide missense mutation in At5g67630, which encodes the Arabidopsis ortholog of Reptin (also called RuvBL2/RVB2) (SI Appendix, Fig. S1A and Dataset S1). Genetic complementation confirmed that this At5g67630 allele causes the PD transport phenotype, because transformation with the native promoter and genomic sequence of At5g67630 rescued the ise4 mutants. We therefore renamed ise4 for clarity, and refer to this allele of AtReptin as reptin-1 throughout. An apparent paralogue, previously named AtTIP49b1 (At3g49830) (43), is a pseudogene: It is not expressed in available A. thaliana RNA sequencing (RNA-seq) data (44), we could not amplify AtTIP49b1 transcripts by RT-PCR, and we did not observe any phenotypes in a line carrying a T-DNA insertion in the AtTIP49b1 coding sequence (SALK_036858). Therefore, we focused exclusively on the functional AtReptin gene, At5g67630.
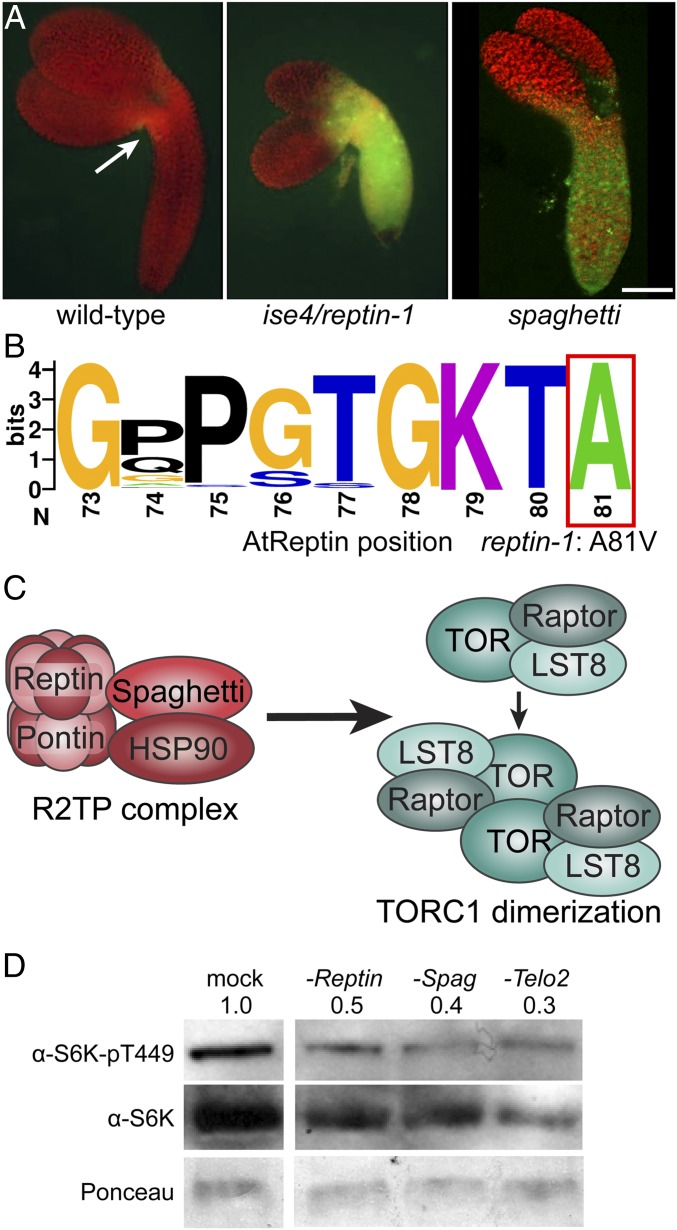
Fig. 1.
R2TP cochaperone complex activity restricts cell–cell transport during embryogenesis. (A) Midtorpedo-stage wild-type, ise4/reptin-1, and spaghetti Arabidopsis embryos were removed from seed coats in a 10 kDa FITC-dextran solution, which loaded into plants at the hypocotyl–cotyledon boundaries (indicated in wild-type with a white arrow). After several washes to remove external FITC-dextran, embryos were observed using fluorescence microscopy (FITC, green; chlorophyll, red). The 10 kDa FITC-dextran rapidly moves through ise4/reptin-1 (Center) and spaghetti (Right), but does not move between cells in their wild-type siblings (Left). (Scale bar, 100 µm.) (B) Consensus Walker A motif across all eukaryotic Reptin orthologs. Amino acid positions follow AtReptin. reptin-1 causes an A81V missense mutation. (C) R2TP is a cochaperone complex composed of Reptin, Pontin, and Spaghetti, which recruits HSP90. R2TP stabilizes the TORC1 (TORC1: dark teal, Raptor; teal, TOR; light teal, LST8) dimer. (D) Knocking down expression of R2TP-TTT cochaperone subunits Reptin, Spaghetti, or Telo2 in Arabidopsis Col-0 with VIGS strikingly decreased levels of the TOR-dependent phosphopeptide S6K-pT449, reflecting significant inhibition of TOR activity. Total S6K levels and Ponceau staining were used for comparison. Note that steady-state levels of total S6K are also somewhat lower in reptin, spaghetti, and telo2 relative to Col-0, as expected in plants with chronic low TOR activity.
Reptin is an AAA+ type ATPase found in all eukaryotes that acts as a chaperone for various multiprotein complexes, and has conserved roles in diverse processes, including chromatin remodeling, cell division, telomerase biogenesis, and R2TP-TTT–mediated phosphatidylinositol 3-kinase-related kinase (PIKK) complex assembly (45, 46). The mutation in reptin-1 is a missense A81V mutation (SI Appendix, Fig. S1 A and D). A81 is a universally conserved residue in eukaryotic Reptin orthologs that is immediately C terminal to the nucleotide-binding Walker A motif (Fig. 1B), hinting that reptin-1 is defective in ATP binding or ATPase activity. For most of its roles, Reptin assembles in a heteromeric complex with a closely related AAA+ ATPase, Pontin (also called RuvBL1/RVB1). As in other eukaryotes and recently confirmed in plants (46), AtReptin-GFP localizes to the nucleus and cytosol, and AtReptin physically associates with AtPontin (encoded by At5g22330) (43, 46) (SI Appendix, Fig. S1 B and E). A homozygous null allele of AtReptin with a T-DNA insertion in the coding sequence, which we named reptin-2, causes female gametophyte lethality (SI Appendix, Fig. S1 A and C), reflecting the essential roles this protein plays in basic cellular functions.
Reptin and Pontin are the only ATPases in R2TP, an HSP90 cochaperone complex (47) that recruits HSP90 via another R2TP subunit, Spaghetti (an ortholog of human RPAP3, a tetratricopeptide-repeat/carboxylate clamp protein with a C-terminal RPAP3-specific domain; for names of other orthologs, see Dataset S1) (Fig. 1C) (47, 48). Although Reptin interacts with several protein complexes, Spaghetti is a component specific to the R2TP complex. Using a null allele of spaghetti in Arabidopsis (encoded by At1g56440) (SI Appendix, Fig. S2) (49), we directly tested whether R2TP specifically regulates PD transport by assaying intercellular trafficking in spaghetti embryos. Strikingly, PD transport is also increased in R2TP-deficient spaghetti mutants at the midtorpedo stage of embryogenesis (Fig. 1A). Thus, we decided to focus further analysis on the roles of R2TP-dependent pathways in regulating PD transport.
TOR Restricts PD Transport.
TOR is a serine/threonine protein kinase that is universally conserved in eukaryotes (31, 32). Nutrients and various hormonal signaling pathways converge to activate TOR, which then promotes growth and anabolism (33). R2TP and the TTT complex (for names of other orthologs, see Dataset S1) are required to stabilize the TOR complex, and R2TP-TTT is proposed to function as a metabolic sensor of intracellular ATP levels to prevent TOR complex assembly when ATP levels are very low (36). A recent report showed that tti2 mutants in maize are embryo-lethal and accumulate drastically lower levels of TOR kinase, reflecting TOR complex instability (42). We directly tested whether R2TP-TTT is required to maintain TOR activity in plants by probing R2TP-TTT knockdowns for differential phosphorylation of the canonical TORC1 substrate, S6 kinase (S6K), which is phosphorylated by TORC1 at T449 in Arabidopsis (38). S6K-pT449 levels are lower in TRV::reptin, TRV::spaghetti, and TRV::telo2 knockdown plants than in mock (TRV::gus) controls (Fig. 1D), demonstrating that R2TP-TTT is required for TOR kinase activity in plants. Furthermore, we used RNA-seq to define the transcriptome of reptin-1 mutants, and found that the reptin-1 and tor transcriptional profiles are significantly similar, as discussed in more detail below (see, for example, Fig. 3 E and G). Given the strong impact of reptin and spaghetti on TOR activity, we next focused on directly testing whether TOR regulates intercellular transport in plants.
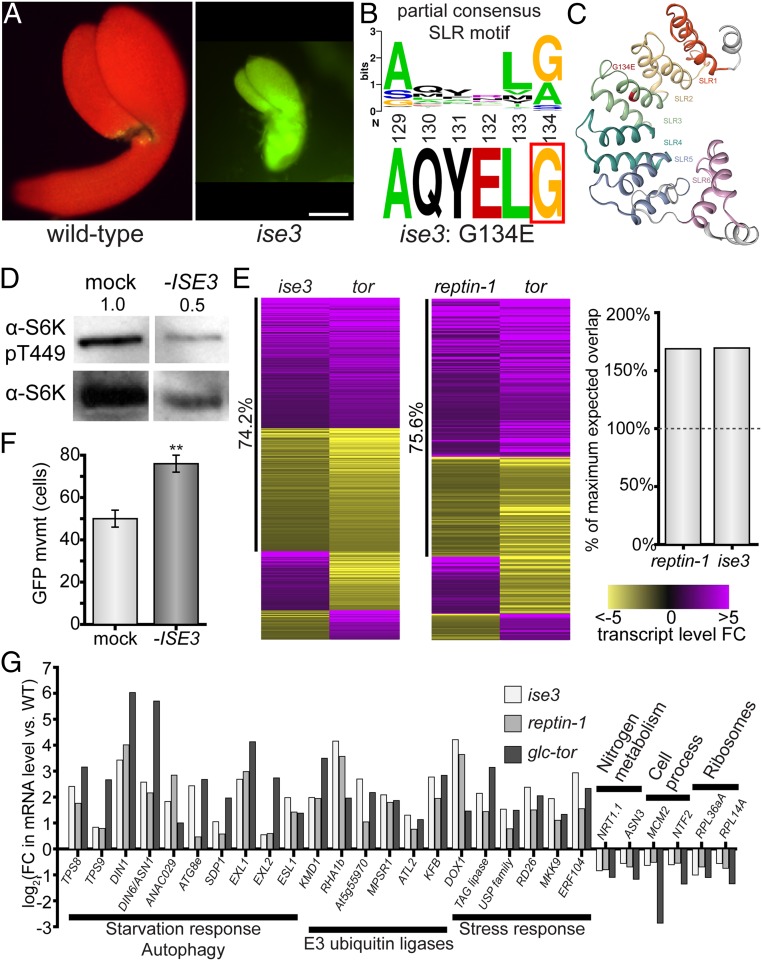
Fig. 3.
Dysfunctional mitochondria inhibit glucose-TOR signaling and increase cell–cell transport. (A) The 10 kDa FITC-dextran rapidly moves through ise3 midtorpedo-stage Arabidopsis embryos (Right) compared to wild-type siblings (Left; FITC, green; chlorophyll, red). (Scale bar, 100 µm.) (B) ise3 is a missense mutation, G134E, at a conserved G/A residue in SLR proteins. (C) Predicted structure of ISE3 protein, after cleavage of the mitochondrial transit peptide. ISE3 is composed of 6 Sel1-like repeats (SLR1–SLR6), of which SLR5 and SLR6 are relatively unstructured. ise3 is a missense G134E mutation at the consensus SLR glycine/alanine residue in SLR3 (indicated in red). (D) Silencing ISE3 with VIGS strongly reduces S6K-pT449 levels in Arabidopsis leaves, demonstrating that ISE3 promotes TOR activity. Total S6K levels are also shown. Numbers indicate S6K-pT449/S6K ratio relative to mock-infected leaves, across three replicates. (E) The glucose-TOR transcriptional program is disrupted in ise3 and reptin-1, reflecting TOR inactivity. Heatmaps show all overlapping significant DEGs between either the ise3 or reptin-1 mutant transcriptome and the tor transcriptome (41), with purple indicating increased transcript abundance and yellow indicating decreased transcript abundance, as indicated by the legend (Right). The number of overlapping genes in each pairwise comparison is significantly more than expected, as shown in the bar chart (Right; “100% maximum expected value” is the number of genes that could overlap with P > 0.05, as determined by a hypergeometric test; specifically P < 10−27 for reptin-1 and P < 10−62 for ise3). reptin-1 and ise3 transcriptomes show strong coregulation with the tor transcriptome (75.6% and 74.2% genes coregulated, respectively) (Datasets S2–S4). (F) Silencing ISE3 expression with VIGS increases PD transport in N. benthamiana leaves (n ≥ 59, **P < 0.001; error bars indicate SEM). (G) Representative DEGs affected in ise3, reptin-1, and tor compared to wild-type. Genes involved in starvation response/autophagy, proteasomal degradation, and stress response are induced in each mutant. Cytosolic ribosomal protein genes, cell process/cell cycle genes, and some nitrogen metabolism genes are repressed (SI Appendix, Fig. S4A and Datasets S2–S4).
TOR forms a complex with two WD40-repeat–containing proteins, RAPTOR and LST8 (Fig. 2A) (37, 50, 51). We cannot directly assay PD transport in null tor and raptor mutants, because they cause complete growth arrest at early stages of embryogenesis under standard growing conditions (37, 50). Arabidopsis lst8 is not lethal under standard growing conditions, however, although it does exhibit pleiotropic mutant phenotypes, including reduced growth and sensitivity to photoperiod (51). We found that lst8 mutants are defective in the restriction of PD transport during embryogenesis (Fig. 2B). Unlike wild-type midtorpedo-stage embryos (Fig. 1A), 10-kDa FITC-dextran rapidly moves via PD throughout lst8 midtorpedo-stage embryos, directly demonstrating that the TOR–RAPTOR–LST8 complex regulates cell–cell trafficking in plants.
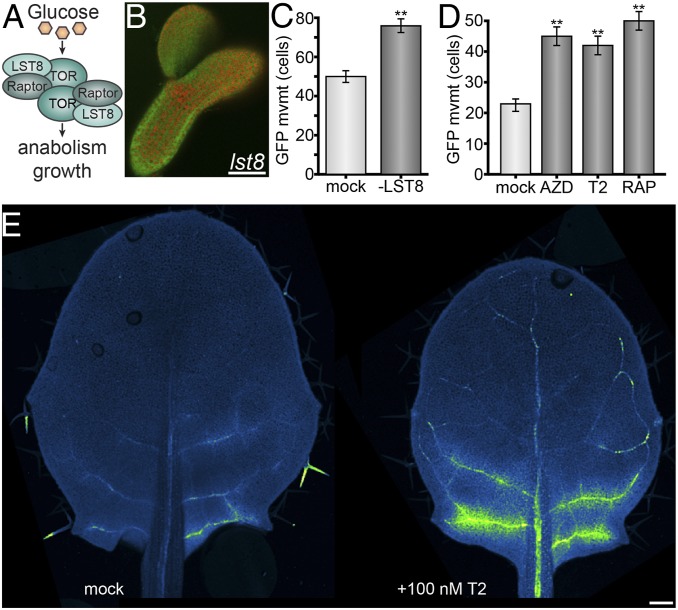
Fig. 2.
TOR restricts cell–cell transport. (A) TORC1 model. TORC1 is a dimer composed of TOR, RAPTOR, and LST8. Glucose stimulates TORC1 to promote anabolism, cell division, and growth, depending on developmental context. (B) The 10 kDa FITC-dextran rapidly moves through lst8 midtorpedo-stage Arabidopsis embryos (for wild-type, see Fig. 1A; FITC, green; chlorophyll, red). (Scale bar, 100 μm.) (C) Silencing LST8 expression with VIGS increases PD transport in N. benthamiana leaves (n ≥ 43, **P < 0.001; error bars indicate SEM). Leaves equivalent to L8 (Fig. 5B) were used for this experiment. (D) Chemically inhibiting TOR activity with ATP-competitive kinase inhibitors [AZD-8055 or Torin 2 (39)] or rapamycin [steric inhibitor through interaction with FKBP12 (38, 75–78)] increases PD transport in N. benthamiana leaves (n ≥ 108, **P < 0.001; error bars indicate SEM) L6 leaves (Fig. 5B) were used for this experiment. (E) Chemically inhibiting TOR activity with Torin 2 in young leaves increased import of GFP from source leaves via the phloem in SUC2PRO:GFP Arabidopsis plants. Mock-treated leaves of the same age and size showed little to no GFP import. GFP fluorescence is shown in green; background chlorophyll autofluorescence is shown in blue; some trichome autofluorescence (not GFP) is also visible and shown in green. (Scale bar, 100 µm.)
Next, we assayed the rate of GFP movement through PD in mature Nicotiana benthamiana leaves after genetically or chemically disrupting TOR activity. Single epidermal cells were transformed by agroinfiltration to express GFP (27 kDa), and GFP movement from the transformed cell into neighboring cells was determined after 48 h using a quantitative assay (52, 53). For all experiments, we visualized GFP movement only in the proximal 25% of the leaf. Results of this movement assay are presented as the number of cells to which GFP spread. Silencing LST8 significantly increased PD transport (Fig. 2C), confirming that LST8 is required to limit intercellular transport in leaves. Treatment with rapamycin or two different ATP-competitive TOR kinase inhibitors, AZD-8055 and Torin 2, also significantly increased PD transport of GFP (Fig. 2D). Collectively, these experiments demonstrate that active TOR restricts trafficking between plant cells.
TOR Promotes the Sink-To-Source Transition in Phloem Transport.
We hypothesized that TOR-mediated restriction of PD transport corresponds to the sink-to-source transition, when tissues switch from phloem importers to phloem exporters. To directly measure phloem transport dynamics after manipulating TOR activity, we took a classic experimental approach using SUC2PRO:GFP transgenic A. thaliana seedlings (28). In this line, GFP is transcribed and translated exclusively in the phloem of source leaves, but the GFP protein then rapidly moves through the phloem and accumulates in cells surrounding the veins in sink leaves. SUC2 transcription has not been found to be regulated by TOR (40, 54). We applied 100 nM of the TOR inhibitor Torin 2 or mock treatment to the shoot apex of SUC2PRO:GFP seedlings (with five emerged leaves), and assayed GFP accumulation in leaves transitioning from sink to source (leaf four [L4], ∼1.5-mm long, which was in direct contact with the applied Torin 2) 1 d later. Across three experiments, we consistently observed GFP unloading from the phloem of leaves after treatment with Torin 2, but little or no GFP unloading from the phloem of leaves that received mock treatment (Fig. 2E). Therefore, inhibiting TOR interferes with the sink-to-source transition in leaves, such that the leaf continues to import resources from the phloem.
TOR Signaling Is Also Disrupted in ise3.
Across eukaryotes, TOR activity is highly sensitive to mitochondrial bioenergetics and the production of ATP by oxidative phosphorylation (OXPHOS) (36, 55). In plants, glucose stimulates TOR through its metabolism via glycolysis and OXPHOS (40). We found that another mutant identified in our genetic screen for increased PD transport is also defective in TOR activity, likely due to disruption of OXPHOS in the glucose-glycolysis-OXPHOS-TOR pathway. ise3 (Fig. 3A) is an embryo-lethal mutant identified in the same genetic screen for altered trafficking of 10 kDa FITC-dextran at the midtorpedo stage of embryogenesis that identified ise1, ise2, and ise4 (30). We mapped ise3 to a missense mutation in At2g25570, which encodes a SEL1-like repeat-containing (SLR) protein that localizes to mitochondria (SI Appendix, Fig. S3 A and D). At2g25570 genomic DNA or At2g25570 coding sequence expression driven by the native At2g25570 promoter complemented the ise3 phenotype, confirming that ise3 is a loss-of-function allele of At2g25570. ISE3 is highly conserved within land plants, but there are no clear orthologs of ISE3 in other eukaryotes (SI Appendix, Fig. S3C). SLR proteins are typically involved in protein–protein interactions; for example, a structurally comparable mitochondrial SLR protein in humans has been implicated in the assembly of complex IV of the OXPHOS chain (56). In Arabidopsis, At2g25570 (ISE3) cofractionates with mitochondrial complex V (ATP synthase), implying that it participates in ATP synthesis (57). The ise3 allele is a G134E missense mutation; G134 is a consensus SLR residue, and G134E is predicted to destabilize the third SLR, presumably disrupting ISE3’s affinity for protein binding partners (Fig. 3 B and C and SI Appendix, Fig. S3 B and D).
We directly tested whether ISE3 is required for TOR activity by silencing ISE3 expression and assaying S6K-T449 phosphorylation, and found that S6K-pT449 levels are significantly lower in TRV::ise3 knockdowns than in mock (TRV::gus) controls (Fig. 3D). Moreover, silencing ISE3 significantly increases PD transport of GFP in leaves (Fig. 3F). Next, we took a comparative transcriptomic approach by comparing significantly differentially expressed genes (DEGs) of ise3, reptin-1, and tor (Datasets S2–S4). The tor transcriptome is the set of DEGs that respond 2 h after glucose activation of TOR in glucose-deprived quiescent Arabidopsis seedlings; these DEGs are specifically sensitive to TOR, because glucose supply has no transcriptomic effect in these seedlings when TOR is genetically or chemically inhibited (40). As mentioned above, the reptin-1 and tor transcriptomes overlapped significantly in DEGs (P < 10−27) (Fig. 3E and Dataset S3), and the vast majority of overlapping DEGs were both induced or both repressed in the two datasets (75.6% coregulated) (Fig. 3E). Similarly, the set of DEGs in ise3 significantly overlaps with the tor transcriptome (P < 10−60, 74.2% coregulated) (Fig. 3E and Dataset S4).
More broadly, both the reptin-1 and ise3 transcriptomes exhibit lowered TOR activity, with high expression of genes related to catabolic recycling and quiescence, as expected for a mutant deficient in TOR activity, instead of gene-expression profiles reflecting the rapid anabolic growth typically associated with embryogenesis (Fig. 3G, SI Appendix, Fig. S4, and Dataset S2). Analysis of significantly enriched categories of DEGs in reptin-1 revealed widespread repression of nuclear-encoded photosynthesis-associated genes and cytosolic ribosomal protein genes, alongside significant induction of genes associated with late embryogenesis and seed maturation (SI Appendix, Fig. S4). Cytosolic ribosomal protein gene expression, in particular, is a well-established target promoted by TOR activity in eukaryotes (58, 59). The ise3 transcriptome shows comparable patterns to reptin-1, with repression of genes encoding ribosomal proteins, histones, cyclin-dependent protein kinases (and related proteins that drive cell cycle progression), components of both photosystems, and Calvin–Benson cycle enzymes (SI Appendix, Fig. S4). Additionally, genes encoding autophagosome components and E3 ubiquitin ligases that target proteins for proteasomal degradation are strongly induced in ise3 (Fig. 3G). Many of these patterns in the ise3 transcriptome are canonical signatures of TOR repression, such as the induction of autophagy-related genes (60–62) and suppression of ribosomal protein genes (58, 59) and cell cycle genes (40, 63). We conclude that the glucose-TOR transcriptional program is inactivated in ise3 and reptin-1, demonstrating their critical roles in promoting TOR activity.
We probed the ise3, reptin-1, and tor transcriptomes for DEGs related to PD, but we did not identify any changes in gene expression that could readily explain the altered PD transport phenotype. For example, PDLP5, which encodes a PD-LOCALIZED PROTEIN that decreases PD transport in leaves (17, 18), is induced 6.7-fold in reptin-1 and 11.9-fold in ise3 (Dataset S2); induction of PDLP5 would not be predicted to increase PD transport in these mutants. If the effect of these mutants on PD transport is not due to transcriptional changes in gene expression, an alternative hypothesis is that TOR could posttranslationally regulate PD transport by phosphorylating proteins that localize to PD or in other ways impact PD trafficking. We expect that ongoing efforts to map the TOR signaling network in plants will illuminate how TOR regulates PD transport at a mechanistic level.
The Glucose-TOR Signaling Axis Restricts PD Transport.
Building on our discovery that mitochondrial ISE3 is critical for maintaining TOR activity, we next interrogated whether the glucose-glycolysis-OXPHOS-TOR signaling axis regulates PD transport in plants using chemical inhibitors (Fig. 4A). First, we treated young leaves (L8) with 15 mM glucose, providing the leaves with a physiologically relevant increase in sugar availability, and assayed GFP movement 48 h later. Glucose treatment significantly decreased PD transport (Fig. 4C). We then inhibited glycolysis in middle-aged leaves (L7) with 15 mM 2-deoxy-d-glucose (2-DG), a glucose analog that cannot undergo glycolysis and competitively inhibits glucose-6-phosphate isomerase, and assayed GFP movement 48 h later. The 2-DG inhibition of glycolysis had the opposite effect of glucose, increasing PD transport (Fig. 4C). Finally, we focused on inhibiting OXPHOS. We previously showed that disrupting mitochondrial energetics with salicylhydroxamic acid increases PD transport (8) (Fig. 4B). Antimycin A, a direct OXPHOS inhibitor that disrupts complex III activity (40), also increases PD transport in leaves (Fig. 4B). Supporting the conclusion that PD transport increases when ATP synthesis is inhibited, we also recently showed that silencing expression of a subunit of the chloroplast ATP synthase, AtpC, increases PD transport in leaves (53). To summarize, stimulating glucose-TOR signaling with glucose decreases PD transport, whereas inhibiting the glucose-glycolysis-OXPHOS signaling axis upstream of TOR increases PD transport.
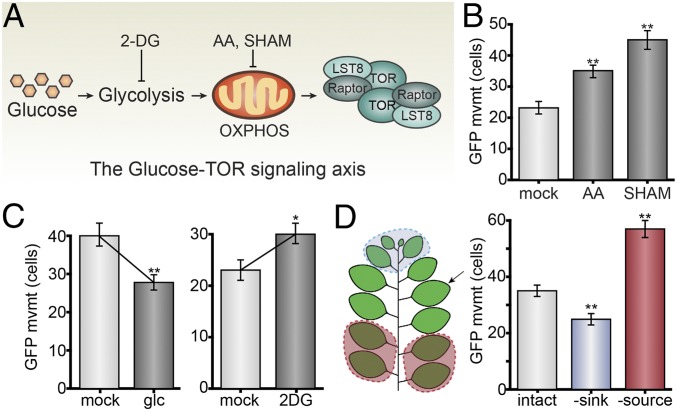
Fig. 4.
Glucose-TOR signaling restricts PD transport in leaves. (A) Summary of the glucose-glycolysis-OXPHOS-TORC1 signaling axis (40) interrogated in this figure, with inhibitors of glycolysis and OXPHOS indicated. (B) Chemically inhibiting OXPHOS with antimycin A (AA) increases PD transport (n ≥ 135, P < 0.001; error bars indicate SEM), similar to the previously reported effect of salicylhydroxamic acid (SHAM) (8). L7 leaves were used for these experiments. (C) 15 mM d-glucose (glc) activates TOR and decreases PD transport in young leaves (n ≥ 95, **P < 0.001; error bars indicate SEM); 15 mM 2-DG inhibits glycolysis and increases PD transport. L7 leaves were used for these experiments. (D) GFP movement was assayed in the proximal 25% of a leaf (L7, indicated with an arrow) that has not yet fully expanded. PD transport increases after limiting sugar availability by source removal (L1 through L4 defoliated for 48 h, red), and PD transport decreases after increasing sugar availability by sink removal (L10 through L13 defoliated for 48 h, blue).
We extended this hypothesis to test whether modulating sink/source relations in shoots could impact PD transport. We defoliated plants to remove either sink or source leaves, followed immediately by agroinfiltrating a middle-aged leaf that had not fully expanded (L7) and assayed PD transport 48 h later. Without source leaves to provide sugars, GFP transport through PD was significantly higher in L7; without sink leaves consuming sugars, GFP transport through PD was significantly lower in L7 (Fig. 4D). These results are consistent with the hypothesis that sugar metabolism and transport dynamically regulate PD transport in leaves.
TOR Acts as a Rheostat to Coordinate Leaf Maturation.
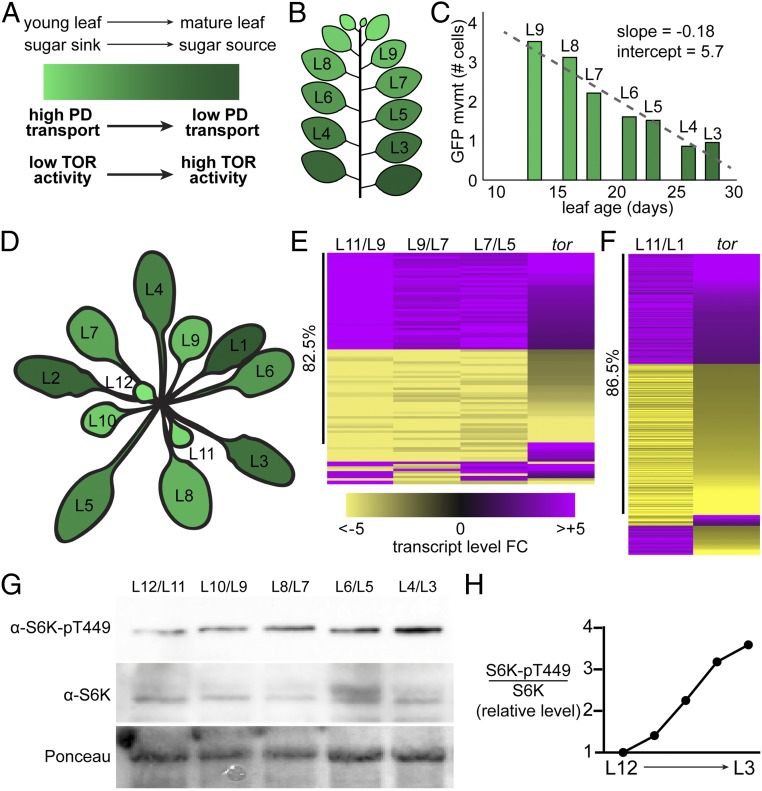
As a working model, we propose that the nutrient sensor TOR regulates PD transport in response to the balance of sugar synthesis and catabolism in leaves. In young leaves that are actively growing, we hypothesize that sugar is metabolically limiting and TOR is relatively inactive, increasing PD transport and phloem import to promote sugar import and limit sugar export (Fig. 5A). In mature leaves, we hypothesize that sugar is synthesized in excess and TOR is strongly active, decreasing PD transport and promoting active sugar export (Fig. 5A). To test this model, we first used the quantitative GFP movement assay (52, 53) to measure rates of PD transport in young to old leaves of mature plants, and found that PD transport steadily declines in a linear relationship with leaf age (Fig. 5 B and C). Next, we directly assayed TOR activity by measuring S6K-pT449 and total S6K levels in each leaf of ∼4-wk-old plants (Fig. 5D), using a transgenic line, 35SPRO:AtS6K1-HA, that drives consistent expression of S6K throughout the plant (38). S6K-pT449 levels gradually and steadily increase as the leaves mature (Fig. 5G), such that the S6K-pT449 to S6K ratio increases ∼3.5-fold (Fig. 5H). These results are further confirmed by analysis of the set of significantly DEGs as a leaf ages. The set of DEGs that gradually change during leaf maturation (L11 to L5) and the set of DEGs between a young sink leaf (L11) and mature source leaf (L1) are significantly coregulated with the glucose-TOR transcriptional signaling network (40) (Fig. 5 D–F and Datasets S7 and S8). Many of these DEGs are canonical TOR targets, including, notably, over half of the cytosolic ribosomal protein genes, which are induced alongside TOR activity in more mature leaves (Dataset S9). Therefore, we propose that TOR acts as a metabolic rheostat (33), coordinating multiple physiological pathways, including the sink-to-source transition, during vegetative development.
Fig. 5.
TOR activity coordinates leaf metabolism with leaf age. (A) As leaves age, they transition from young sinks (light green) that rapidly consume sugars to mature sources (dark green) that photosynthesize excess sugars for export to the rest of the plant. Here, we show that TOR activity steadily increases and PD transport steadily decreases during this transition. (B) Leaf emergence of N. benthamiana plants was tracked for 6 wk, during which time 13 leaves emerged. The first true leaf is labeled leaf 1 (L1), and subsequent leaves are numbered by order of emergence. (C) Six weeks after germination, PD transport in L9 (a relative sink leaf) through L3 (a relative source leaf) was quantified using the GFP movement assay. PD transport was assayed in the proximal 25% of every leaf. PD transport decreases linearly with leaf age (as in ref. 53) in N. benthamiana. (D) Schematic of an Arabidopsis vegetative rosette used for transcriptomes and the Western blot analyses in this figure. (E–H). (E) The sets of genes that are differentially expressed in Arabidopsis leaves of different ages significantly overlap with the tor transcriptome (P < 10−26), and are highly coregulated (82.5%), indicating that the TOR-promoted transcriptional program is activated as leaves mature (Dataset S7). The tor transcriptome is the set of gene-expression changes that respond to glucose only in the presence of TOR (40), and are therefore transcriptional targets of the TOR signaling pathway. Pairwise analyses of DEGs for L11 and L9, L9 and L7, and L7 and L5 are shown, respectively, from left to right. As an example, a gene is considered “induced” in the L11/L9 dataset if it is significantly more highly expressed in L11 than in L9. (F) Over 30% of the glucose-TOR target genes (732 of 2,350, P < 10−138) are significantly differentially expressed between a young sink leaf (L11) and a mature source leaf (L1), and the vast majority of these DEGs (86.5%) are coregulated, suggesting that the glucose-TOR signaling network is more active in source leaves than in sink leaves (Dataset S8). (G) S6K-Thr449 phosphorylation levels, a proxy for TOR activity in plant cells, were assayed by Western blot with a phospho-specific antibody. A 35SPRO:S6K1 transgenic Arabidopsis line was used for this experiment, because it maintains consistent S6K protein levels in leaves of different ages. S6K-Thr449 levels steadily increase with leaf age in an Arabidopsis rosette. Ponceau staining was used to show loading of total protein (Lower). (H) The quantified ratio of S6K-pT449 levels to total S6K levels in leaves of different ages confirms that TOR activity increases with leaf age in a mature Arabidopsis rosette.
We should note that the sink-to-source transition occurs within a leaf during development: For a number of leaves undergoing the transition (in Fig. 5D, approximately L7 through L10) (28), the distal portion of the leaf stops importing nutrients from the phloem while the proximal portion of the leaf continues to grow, undergo cell division, and import nutrients from the phloem. Therefore, while the youngest leaves are exclusively sinks and the more mature leaves are exclusively sources, some leaves represent a composite of sink and source tissue. By conducting all leaf PD transport assays in only the proximal 25% of the leaf, however, we minimized this confounding factor in our analysis. In fact, the results shown here suggest that the reduction in PD transport rates in the leaf epidermis as leaves mature is not a binary switch between “sink” and “source,” but instead a gradual, approximately linear decline with leaf age (Fig. 5C). Similarly, TOR activity gradually increases with leaf age, even after all leaves have completed the transition to phloem exporters (approximately L6 through L1). Thus, rather than simply switching from “low” activity in sink tissues to “high” activity in sources, TOR activity progressively increases as leaves mature and begin to photosynthesize more sugars than they consume. This supports our hypothesis that TOR acts as a metabolic rheostat in leaves, with steady changes in TOR activity causing gradual shifts in PD transport and contributing to the coordination of the discrete sink-to-source transition.
Conclusions.
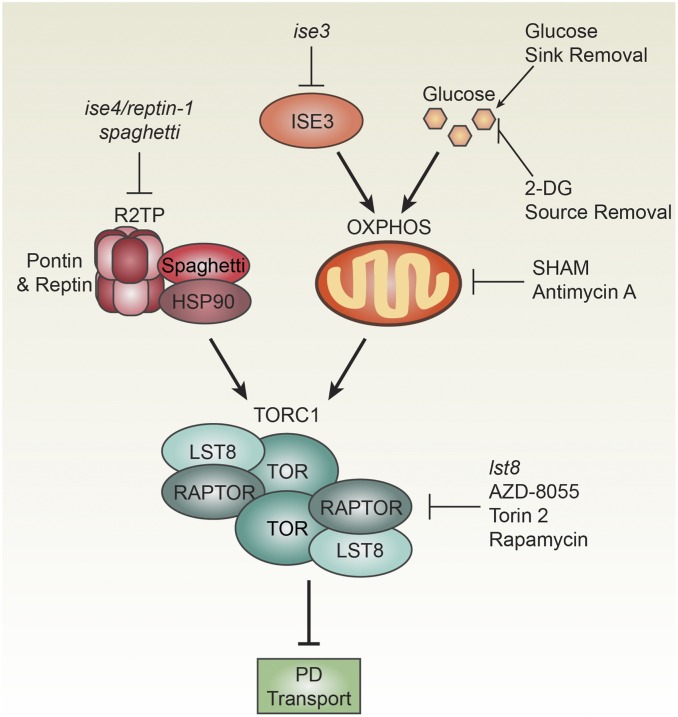
We define a pathway regulating intercellular transport through PD in plants that coordinates growth with sugar availability. Analysis of the reptin-1 mutant led us to postulate that TOR decreases PD transport during plant development, a hypothesis that we then tested directly using multiple genetic, chemical, and physiological approaches (Fig. 6), and elaborated upon this hypothesis by testing how TOR impacts phloem transport. We propose a dynamic, metabolic model of PD and phloem transport, in which relative differences in TOR activity stimulate sugar transport from mature, self-sufficient leaves to younger, rapidly growing leaves. Beyond their roles in nutrient partitioning, PD also traffic a variety of signaling molecules, including transcription factors, cytosolic proteins, and hormones, and are the primary route for viral spread. Dynamic regulation of PD transport by TOR may therefore contribute to a variety of plant biological processes by controlling the mobility of viruses and endogenous micro- and macromolecular signaling molecules. We conclude that TOR, an ancient, conserved mechanism for sensing and responding to metabolic cues, has been coopted in plants to coordinate redistribution of carbohydrates during plant vegetative development.
Fig. 6.
Summary of the glucose-TOR-PD signaling network in plants. A forward genetic screen uncovered two mutants, ise3 and ise4/reptin-1, that decrease TOR activity and increase PD transport. As a component of R2TP, Reptin promotes assembly of the active TORC1 dimer (Fig. 1). spaghetti, another R2TP mutant, also decreases TOR activity and increases PD transport (Fig. 1). OXPHOS promotes TOR activity and is disrupted by loss of ISE3 (Fig. 3) or inhibition of the glucose-glycolysis-OXPHOS-TOR signaling axis (Fig. 4). Direct assays reported here demonstrate that TOR restricts PD transport (Fig. 2), and TOR activity correlates with decreased PD transport during leaf maturation (Fig. 5). Chemical, genetic, and physiological treatments that impact each step in this pathway are indicated; each of these conditions affects PD transport, as described in the text.
Materials and Methods
Plant Materials and Growth Conditions.
Unless otherwise stated, plants were grown under standard conditions with 16-h day/8-h night at ∼120 µmol photons m−2 s−1 of photosynthetically active radiation. ise4/reptin-1 and ise3 were identified in the Landsberg erecta (Ler) background and then mapped in crosses to Col-0. SALK_020547 (spaghetti) (49), GK543F01-021955 (reptin), SALK_036848 (pseudoreptin), and SALK_002459 (lst8-1-1 or lst8a-1) (51) were obtained from the Arabidopsis Biological Resources Center and their genotypes were confirmed by PCR (Dataset S10). Unless otherwise stated, all Arabidopsis experiments used the standard Col-0 ecotype as wild-type. For all N. benthamiana experiments, the reference Nb-1 genotype obtained from the Boyce Thompson Institute was used.
Molecular Cloning.
Virus-induced gene silencing (VIGS) vectors were prepared as previously described (52), using oligonucleotides listed in Dataset S10. RNA was isolated from Nb-1 or Col-0 shoots with the Spectrum Plant Total RNA (Sigma-Aldrich) with on-column DNase I digestion (New England Biolabs). cDNA was synthesized from RNA using random hexamers and SuperScript III reverse transcriptase (Fisher Scientific). Silencing triggers were amplified with Phusion DNA polymerase (New England Biolabs), digested alongside pYL156 with restriction enzymes as listed in Dataset S10 (enzymes from New England Biolabs), and ligated with Promega T4 DNA ligase (Fisher Scientific). Ligations were transformed into house-made chemically competent XL1-Blue Escherichia coli. Kanamycin-resistant bacterial colonies were screened by colony PCR for positive clones. Plasmids were then miniprepped (Bioneer) and Sanger-sequenced to confirm insert sequences. Manufacturer’s protocols were followed throughout. All constructs were then transformed into Agrobacterium GV3101 for plant transformation. For NbLST8, we tested two silencing triggers that had the same effects on NbLST8 expression and PD transport; primers for each silencing trigger are described in Dataset S10.
Next-Generation Mapping of ise3 and ise4.
ise3 and ise4 were identified from a population of embryo-defective ethyl methane sulfonate-mutagenized A. thaliana Ler lines, as previously described (30, 64, 65). F2 mapping populations were constructed by crossing heterozygotes carrying the ise allele with Col-0, and then selfing heterozygous ise/+ F1 plants to generate the F2. Using indel and SNP markers (oligonucleotides for PCR listed in Dataset S10), the mutated ise3 locus mapped to a 112-kb region on chromosome 2 between SNPs SGCSNP606 at At2g25470 and PERL0363471 at At2g25710, and ise4 locus was mapped to a 120-kb region between the SNP PERL1125350 at At5g67310 and the end of chromosome 5. Due to low recombination rates in the ise4 region, we used next-generation mapping to identify the causal mutations for both ise3 and ise4. Eighteen overlapping genomic fragments between 4- and 8-kb long were amplified with iProof high-fidelity DNA polymerase (Bio-Rad Laboratories) from genomic DNA isolated from homozygous ise3 or ise4 mutants. The fragments were purified by gel extraction, and pooled in equal amounts. DNA-seq libraries of bulked DNA were constructed using the Nextera DNA Flex Library Prep Kit (Illumina) following the manufacturer’s protocol, and sequenced on the Illumina HiSEq 2000 platform. Data were mapped to the unmasked Arabidopsis genome and compared using Sequencher (66).
ise3 was complemented by three different constructs (cloned with oligonucleotides in Dataset S10). A full genomic sequence was constructed by cloning from ∼1.8-kb upstream of the At2g25570 start codon through 858 bp downstream from the At2g25570 stop codon. The same genomic ∼1.8-kb promoter was also used to drive expression of the At2g25570 coding sequence or the coding sequence C-terminally tagged with GFP. All constructs rescued ise3 lethality. ise4 was complemented with three different constructs (cloned with oligonucleotides in Dataset S10). A long genomic sequence was composed of ∼2.4-kb upstream of At5g67630, the At5g67630 gene, and ∼1-kb downstream of At5g67630. A gene of unknown function, At5g67640, is within the ∼2.4-kb region upstream of At5g67630; no SNPs were identified in At5g67640 between wild-type and ise4, but to be sure that expression of At5g67640 did not affect the ise4 phenotype, a short genomic sequence lacking the At5g67640 translation start site was also constructed, and this sequence complemented ise4. Finally, the ∼2.4-kb upstream sequence was used to drive expression of the coding sequence of At5g67630 cloned from Arabidopsis cDNA; this construct also complemented ise4. Complementation was performed by transforming ise4/+ heterozygotes with the floral dip method (67), screening for transformants with Basta (0.3% [vol/vol]), and testing for rescue of the embryo-defective phenotype.
PD Movement Assays.
Embryo dye-loading assays were performed as previously reported (30, 68). Briefly, 10 kDa FITC-dextran (Sigma-Aldrich) was prepared at 5 mg/mL in half-strength liquid Murashige and Skoog (MS) (pH = 5.5) media, with any dye less than 10 kDa removed by centrifugal filtration (Amicon Ultra 10K membranes, Millipore Sigma). Embryos were removed from seeds and immediately treated with FITC-dextran for 5 min before several washes with MS media to remove excess dye. Col-0 and ise2 (30) were used as negative and positive controls for dye loading, respectively, alongside each experiment. Experiments were replicated three times. For newly identified ise mutants (including spaghetti and lst8), dye movement throughout the hypocotyl of the embryo was observed in >80% of mutant embryos loaded with 10 kDa FITC-dextran.
GFP movement assays in N. benthamiana were performed as previously reported (52, 53). Briefly, Agrobacterium was grown overnight in LB with antibiotics at 28 °C. Cultures were spun down at 700 × g for 10 min, washed, and resuspended to OD600nm = 1.0 in 10 mM MgCl2, 10 mM 2-(N-morpholino)ethanesulfonic acid (pH = 5.5, adjusted with KOH), and 200 µM acetosyringone from a recently prepared 100-mM stock solution frozen in DMSO. Agrobacteria were left to induce virulence at room temperature for 2 to 4 h with gentle shaking before infiltration. For GFP movement assays, immediately before infiltrating, cells carrying 35SPRO:GFP T-DNA were diluted to OD600nm = 10−4 in the same virulence-inducing media. GFP movement was scored 48 h after infiltration. For VIGS, equal parts of pYL192 (which expressed TRV1) and pYL156 (which expresses TRV2 with silencing triggers) were mixed and infiltrated into leaves. A mock TRV2 vector, pYC1, which is pYL156::GUSfragment (64), was used as a negative control for all VIGS experiments.
Unless otherwise noted, all N. benthamiana movement assays used the fourth-youngest emerged leaf (as counted from the shoot apex basipetally). GFP movement was only visualized in the proximal 25% of the leaf. For chemical treatments and overexpression, 5- to 6-wk-old plants were used. For VIGS experiments, 3-wk-old plants were infiltrated to express TRV, and movement assays were conducted 2 wk later. Heteroscedastic unpaired t tests were used to compare GFP movement between samples throughout.
SUC2PRO:GFP Phloem Transport Assays.
Transgenic A. thaliana SUC2PRO:GFP seeds were grown as described above for 3 wk (at this stage, five leaves had emerged). Five microliters of 100 nM Torin 2 or a mock control was applied by pipette to the shoot apex of seedlings, such that the Torin 2 was in direct contact with L4 and L5. Care was taken not to apply any Torin 2 to the mature source leaves, to prevent any unintended effects of TOR inhibition on translation of GFP in cells with active SUC2 promoters. One day later, young leaves (L4, ∼1.5-mm long) were excised and immediately observed by confocal scanning laser microscopy, collecting z stacks and tiling several fields of view to visualize the entire leaf. At least three mock-treated seedlings and three Torin 2-treated seedlings were observed for each experiment, and the entire experiment was repeated three separate times.
Confirming Gene Silencing with RT-qPCR.
To confirm gene silencing, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed on VIGS tissue in parallel with GFP movement assays, with mock TRV::GUS infected plants as controls. Three biological replicates were included for each experiment. RNA was isolated with the Spectrum Plant Total RNA (Sigma-Aldrich) with on-column DNase I digestion (New England Biolabs). cDNA was synthesized from RNA using oligo (dT)18 and SuperScript III reverse transcriptase (Fisher Scientific). qPCR was performed in parallel for all samples using a validated reference gene (EF1α) and gene-specific oligonucleotides (Dataset S10) for N. benthamiana VIGS experiments (69). Specificity of primers for target genes was validated by resolving RT-qPCR products on 3% agarose gels. VIGS constructs were included in this study only if they reduced gene expression to ≤20% of wild-type levels.
Chemical Inhibitors.
TOR kinase inhibitors used in this report have previously been demonstrated to suppress TOR signaling (38, 39). Stock solutions of rapamycin (10 mM), Torin 2 (10 mM), AZD-8055 (10 mM), cantharidin (100 µM), okadaic acid (100 µM), and antimycin A (5 mM) were prepared within days of use in DMSO (except okadaic acid, prepared in ethanol) and frozen until used. All chemicals were obtained from Cayman Chemicals, except antimycin A (Sigma-Aldrich). Stocks were serially diluted in infiltration media to ensure accurate reporting of inhibitor concentrations. Dexros or 2-DG (Fisher Scientific) was dissolved directly in virulence-inducing infiltration media at 150 mM, and then diluted to 15 mM just prior to infiltration. Concentrations for inhibitor treatments were determined based on previous reports of minimum effective concentrations for rapamycin (38), AZD-8055 (39), Torin 2 (39), antimycin A (40), dextrose (40), and 2-DG (40).
Western Blot Assays.
Leaves from Col-0 or 35SPRO:S6K1-HA plants were snap-frozen in liquid nitrogen. Protein was extracted from leaves in 100 mM Mops (pH 7.6), 100 mM NaCl, 5% SDS, 0.5% β-mercaptoethanol, 10% glycerin, 2 mM PMSF, and 1× PhosSTOP phosphatase inhibitor (Sigma-Aldrich). Specific protein levels were assayed by Western blot protein using primary antibodies against S6K (Santz Cruz Biotechnology, sc-230) and S6K-pThr449 (Abcam no. 207399) along with an HRP-conjugated goat anti-rabbit IgG secondary antibody (Jackson Immuno Research, no. 111-035-003). Total protein was visualized after transfer using Ponceau S red staining. All Western blot experiments were repeated at least three times, with representative results shown in figures.
RNA-seq and Transcriptome Analysis.
Homozygous ise3 or ise4/reptin-1 seeds at the midtorpedo stage of embryogenesis and their wild-type or heterozygous midtorpedo-stage siblings were collected as previously described and stored at −80 °C in RNAlater (6). For each pooled RNA sample, ∼100 to 150 seeds were collected, and four replicates were collected (three replicates were used for RNA-seq, the fourth replicate was stored as back-up). RNA was isolated with the Spectrum Plant Total RNA (Sigma-Aldrich) with on-column DNase I digestion (New England Biolabs). RNA was treated with RiboMinus (Life Technologies) to deplete 16S, 18S, 23S, and 25S rRNA, and then used to prepare Ion Torrent libraries with the Ion Total RNA-Seq Kit v2 (Life Technologies).
Libraries were sequenced with an Ion Proton I Chip. Torrent Suite Software was used to filter polyclonal reads, adapter dimers, reads of very low quality, and to trim 5′ adapter and repetitive sequences. The remaining computational steps were completed using an instance of Galaxy. FASTQ Quality Trimmer was then used to remove low quality 3′ ends (window size 10, step size 1, quality score ≥ 20) from reads, then reads were filtered with Filter FASTQ to remove reads less than 25-bp long.
To determine differences in gene expression between ise4/reptin-1 and wild-type seeds, RNA-seq results were compared using the Tuxedo Suite. Reads were mapped with TopHat2 to the Ler genome and assembled into transcripts with Cufflinks. Cuffmerge was used to assemble the transcriptome with a Ler reference transcriptome. Cuffdiff2 was used to generate a list of differentially expressed genes comparing three replicates of ise3 or ise4/reptin-1 seeds with three replicates of wild-type seeds (P < 0.05, q < 0.05).
The ise3, and ise4/reptin-1 transcriptomes were analyzed using MapMan software, which assigns genes to various categories based on their biological function and uses a Wilcoxon rank sum test, corrected with the Benjamini–Hochberg procedure to reduce the false-positive rate, to identify categories that are significantly altered in gene expression.
DEG lists from different transcriptomes were compared using a hypergeometric test for significant overlap using R software (phyper function). Within the subset of overlapping genes, any genes that were up-regulated or down-regulated (regardless of the measured fold-change in expression) in all transcriptomes were considered “coregulated.” Gene lists were only considered significantly overlapping if the hypergeometric test revealed P < 10−5, and the maximum expected number of overlapping genes was calculated by determining the number of overlapping genes that would yield P = 0.05.
Tables were generated in Excel using TAIR10 and MapMan annotations, including a large table with all transcriptomic data included in this study, and individual tables showing data underlying heatmaps in all figures. In tables, as in the heatmap figures, purple is used to indicate transcript induction, and yellow is used to indicate transcript repression.
Computational Analysis.
ISE3 structure was modeled by Phyre2, which used structures of E. coli EsiB (70) and Pseudomonas fluorescens AlgK (71) as templates. The resulting model was visualized with NGL. SLRs were identified by TPRpred (72) and by comparison to the structure of Helicobacter pylori HcpC (73). ISE3 orthologs were identified using a BLASTp search with the Arabidopsis amino acid sequence as a query against the reference protein sequence database for eukaryotes. BLASTp results were then filtered to remove paralogues within a species and to highlight representative species from different clades. No proteins with worse scores than the closest-matching Homo sapiens protein were included, except for the next-closest match within A. thaliana to root the trees.
Reptin consensus sequences were determined by aligning all Reptin orthologs available in Ensembl databases with MAFFT, trimming the sequence to include only the Walker A motif and surrounding residues, and then generating a consensus sequence logo with WebLogo (74). SLR consensus sequences were determined by aligning the curated seed alignment of SLRs from PFAM, trimming the sequence to remove uncommon insertions sometimes present between residues 134 and 135, and then generating a consensus sequence logo with WebLogo (74).
N. benthamiana orthologs of LST8 and ISE3 were identified by reciprocal BLASTp searches starting with the A. thaliana protein sequences as queries against the Sol Genomics Network N. benthamiana genome v1.0.1 predicted protein database. The resulting aligned protein sequences are available in Dataset S11. There are two homeologues (paralogues derived from an ancestral polyploidization event) of each of these genes in N. benthamiana; therefore, in selecting silencing triggers, we chose regions of the transcripts that are nearly perfectly homologous in nucleic acid sequence between the two homeologues (paralogues), but had no sufficient homology to any other transcripts in N. benthamiana to cause off-target effects. In the case of NbLST8, we made two separate silencing triggers, which had the same effects on NbLST8 expression and PD transport.
Data Availability.
RNA-seq data described in this article have been deposited in National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA600773.
Supplementary Material
Acknowledgments
We thank Kim Gallagher and Ruby O’Lexy for providing SUC2PRO:GFP transgenic seeds; Jen Sheen for providing 35SPRO:S6K1 transgenic seeds; Mary Ahern, Claire Bendix, Michael Busche, Megan Cohn, Nanticha Lutt, Anjuli Matharu, Anne Runkel, Clair Travis, and Jeffrey Tung for providing experimental assistance; and Sarah Hake for constructive comments on experimental design and the manuscript. This project was supported by NIH Grants 5-DP5-OD023072 (to J.O.B.) and 5-R01-GM045244 (to P.Z.). An NSF graduate research fellowship initially supported J.O.B., and S.C. is supported by the Innovative Genomics Institute (IGI) and a National Science Foundation graduate research fellowship. We acknowledge Steve Ruzin and Denise Schichnes at the College of Natural Resources Biological Imaging Facility (University of California, Berkeley, supported by NIH 1-S10-RR026866); and De Wood and Tina Williams at US Department of Agriculture Western Regional Research Center for microscopy support. This work used the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley for Ion Torrent sequencing and Illumina sequencing (supported by NIH S10-RR029668 and S10-RR027303), and the Functional Genomics Laboratory at University of California, Berkeley for microarray analysis.
Footnotes
The authors declare no competing interest.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject PRJNA600773).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919196117/-/DCSupplemental.
References
- 1.Crawford K. M., Zambryski P. C., Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 125, 1802–1812 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paultre D. S. G., Gustin M.-P., Molnar A., Oparka K. J., Lost in transit: Long-distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell 28, 2016–2025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunkard J. O., Runkel A. M., Zambryski P. C., The cytosol must flow: Intercellular transport through plasmodesmata. Curr. Opin. Cell Biol. 35, 13–20 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Raven J., “Evolution of plasmodesmata” in Plasmodesmata, Oparka K. J., Ed. (Blackwell Publishing Ltd, 2005), pp. 33–52. [Google Scholar]
- 5.Brunkard J. O., Zambryski P. C., Plasmodesmata enable multicellularity: New insights into their evolution, biogenesis, and functions in development and immunity. Curr. Opin. Plant Biol. 35, 76–83 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Burch-Smith T. M., Brunkard J. O., Choi Y. G., Zambryski P. C., Organelle-nucleus cross-talk regulates plant intercellular communication via plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 108, E1451–E1460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benitez-Alfonso Y., et al. , Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. U.S.A. 106, 3615–3620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stonebloom S., et al. , Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiol. 158, 190–199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunkard J. O., Runkel A. M., Zambryski P. C., Plasmodesmata dynamics are coordinated by intracellular signaling pathways. Curr. Opin. Plant Biol. 16, 614–620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burch-Smith T. M., Zambryski P. C., Loss of INCREASED SIZE EXCLUSION LIMIT (ISE)1 or ISE2 increases the formation of secondary plasmodesmata. Curr. Biol. 20, 989–993 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burch-Smith T. M., Zambryski P. C., Plasmodesmata paradigm shift: Regulation from without versus within. Annu. Rev. Plant Biol. 63, 239–260 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Gisel A., Hempel F. D., Barella S., Zambryski P., Leaf-to-shoot apex movement of symplastic tracer is restricted coincident with flowering in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 1713–1717 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim I., Kobayashi K., Cho E., Zambryski P. C., Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 11945–11950 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim I., Cho E., Crawford K., Hempel F. D., Zambryski P. C., Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 2227–2231 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benitez-Alfonso Y., et al. , Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell 26, 136–147 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Han X., et al. , Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell 28, 132–146 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Lee J.-Y., et al. , A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23, 3353–3373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X., et al. , Salicylic acid regulates Plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25, 2315–2329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faulkner C., et al. , LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 110, 9166–9170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim G. H., et al. , Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 19, 541–549 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Ganusova E. E., Burch-Smith T. M., Review: Plant-pathogen interactions through the plasmodesma prism. Plant Sci. 279, 70–80 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Riesmeier J. W., Willmitzer L., Frommer W. B., Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer N., Stolz J., SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker’s yeast and identification of the histidine-tagged protein. Plant J. 6, 67–77 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Bürkle L., et al. , The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol. 118, 59–68 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L. Q., et al. , Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Turgeon R., The role of phloem loading reconsidered. Plant Physiol. 152, 1817–1823 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts A. G., et al. , Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9, 1381–1396 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imlau A., Truernit E., Sauer N., Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11, 309–322 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts I. M., et al. , Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218, 31–44 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Kim I., Hempel F. D., Sha K., Pfluger J., Zambryski P. C., Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development 129, 1261–1272 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Chantranupong L., Wolfson R. L., Sabatini D. M., Nutrient-sensing mechanisms across evolution. Cell 161, 67–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi L., Wu Y., Sheen J., TOR signaling in plants: Conservation and innovation. Development 145, dev160887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valvezan A. J., Manning B. D., Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat. Metab. 1, 321–333 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grabiner B. C., et al. , A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 4, 554–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S. G., et al. , Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell 49, 172–185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menand B., et al. , Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U.S.A. 99, 6422–6427 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y., Sheen J., Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287, 2836–2842 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montané M. H., Menand B., ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 64, 4361–4374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y., et al. , Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Leene J., et al. , Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 5, 316–327 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Garcia N., Li Y., Dooner H. K., Messing J., Maize defective kernel mutant generated by insertion of a Ds element in a gene encoding a highly conserved TTI2 cochaperone. Proc. Natl. Acad. Sci. U.S.A. 114, 5165–5170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holt B. F., 3rd, et al. , An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2, 807–817 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Cheng C. Y., et al. , Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Rosenbaum J., et al. , The emergence of the conserved AAA+ ATPases Pontin and Reptin on the signaling landscape. Sci. Signal. 6, mr1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schořová Š., Fajkus J., Záveská Drábková L., Honys D., Schrumpfová P. P., The plant Pontin and Reptin homologues, RuvBL1 and RuvBL2a, colocalize with TERT and TRB proteins in vivo, and participate in telomerase biogenesis. Plant J. 98, 195–212 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Zhao R., et al. , Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120, 715–727 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Henri J., et al. , Deep structural analysis of RPAP3 and PIH1D1, two components of the HSP90 co-chaperone R2TP complex. Structure 26, 1196–1209.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Sotta N., Shantikumar L., Sakamoto T., Matsunaga S., Fujiwara T., TPR5 is involved in directional cell division and is essential for the maintenance of meristem cell organization in Arabidopsis thaliana. J. Exp. Bot. 67, 2401–2411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deprost D., Truong H. N., Robaglia C., Meyer C., An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326, 844–850 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Moreau M., et al. , Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brunkard J. O., Burch-Smith T. M., Runkel A. M., Zambryski P. C., Investigating plasmodesmata genetics with virus-induced gene silencing and an agrobacterium-mediated GFP movement assay Methods Mol. Biol. 1217, 185–198 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Brunkard J. O., Zambryski P. C., Plant cell-cell transport via plasmodesmata is regulated by light and the circadian clock. Plant Physiol. 181, 1459–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong P., et al. , Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 6, 677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D. H., et al. , mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Martinez Lyons A., et al. , COA7 (C1orf163/RESA1) mutations associated with mitochondrial leukoencephalopathy and cytochrome c oxidase deficiency. J. Med. Genet. 53, 846–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senkler J., et al. , The mitochondrial complexome of Arabidopsis thaliana. Plant J. 89, 1079–1092 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J., The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13, 3271–3279 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marion R. M., et al. , Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. U.S.A. 101, 14315–14322 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Settembre C., et al. , A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roczniak-Ferguson A., et al. , The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martina J. A., Chen Y., Gucek M., Puertollano R., MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong Y., Sheen J., Moving beyond translation: Glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle 12, 1989–1990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stonebloom S., et al. , Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 106, 17229–17234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobayashi K., Otegui M. S., Krishnakumar S., Mindrinos M., Zambryski P., INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19, 1885–1897 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H., Ruan J., Durbin R., Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Xu M., Cho E., Burch-Smith T. M., Zambryski P. C., Plasmodesmata formation and cell-to-cell transport are reduced in decreased size exclusion limit 1 during embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 5098–5103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu D., et al. , Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 7, e46451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Urosev D., et al. , Crystal structure of c5321: A protective antigen present in uropathogenic Escherichia coli strains displaying an SLR fold. BMC Struct. Biol. 13, 19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keiski C. L., et al. , AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18, 265–273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karpenahalli M. R., Lupas A. N., Söding J., TPRpred: A tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinf. 8, 2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lüthy L., Grütter M. G., Mittl P. R. E., The crystal structure of Helicobacter cysteine-rich protein C at 2.0 A resolution: Similar peptide-binding sites in TPR and SEL1-like repeat proteins. J. Mol. Biol. 340, 829–841 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cafferkey R., et al. , Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13, 6012–6023 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kunz J., et al. , Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585–596 (1993). [DOI] [PubMed] [Google Scholar]
- 77.Brown E. J., et al. , A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758 (1994). [DOI] [PubMed] [Google Scholar]
- 78.Sabatini D. M., Erdjument-Bromage H., Lui M., Tempst P., Snyder S. H., RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data described in this article have been deposited in National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA600773.