Significance
Flowering plants exhibit exceptional diversity of form and stature. Their ability to achieve great heights is supported in part by their water-conducting specialized cells, the vessel elements, which have gradually evolved and resulted in increased hydraulic efficiency. Here we report the discovery, functional, and evolutionary analysis of a previously uncharacterized gene, ENLARGED VESSEL ELEMENT (EVE), that contributes to vessel element dimensions in the perennial woody species Populus. EVE’s appearance among the streptophyte algae and ultimate expansion in flowering plants may represent an important addition to the genetic toolkit required for plant vascular development. Surprisingly, EVE homologs are also detected in algae-infecting prasinoviruses, suggesting that it has been horizontally transferred.
Keywords: vessel, xylem, EVE, vessel dimension, phycodnavirus
Abstract
The radiation of angiosperms led to the emergence of the vast majority of today’s plant species and all our major food crops. Their extraordinary diversification occurred in conjunction with the evolution of a more efficient vascular system for the transport of water, composed of vessel elements. The physical dimensions of these water-conducting specialized cells have played a critical role in angiosperm evolution; they determine resistance to water flow, influence photosynthesis rate, and contribute to plant stature. However, the genetic factors that determine their dimensions are unclear. Here we show that a previously uncharacterized gene, ENLARGED VESSEL ELEMENT (EVE), contributes to the dimensions of vessel elements in Populus, impacting hydraulic conductivity. Our data suggest that EVE is localized in the plasma membrane and is involved in potassium uptake of differentiating xylem cells during vessel development. In plants, EVE first emerged in streptophyte algae, but expanded dramatically among vessel-containing angiosperms. The phylogeny, structure and composition of EVE indicates that it may have been involved in an ancient horizontal gene-transfer event.
The development of a vascular system for the efficient transport of water and nutrients contributed to the successful adaptation of vascular plants to a diverse range of terrestrial environments. In angiosperms, water transport occurs in highly specialized, water-conducting cells in the xylem: The vessel elements. Several vessel elements join vertically to form vessels, the multicellular conduit of water. The physical characteristics of these specialized cells have continuously influenced plant growth and evolution, gradually evolving to increase hydraulic efficiency (1). Wider water-transporting tracheary elements of angiosperms provide lower resistance to water flow and support higher rates of photosynthesis than those in other living land plants (2, 3). This allowed the development of larger plant bodies, while maintaining an optimal balance between hydraulic conductivity and vulnerability to cavitation (4, 5).
Genes that contribute to the development of vessel elements have been identified, including NAC domain transcription factors. The NAC acronym is derived from three genes that were initially discovered to contain a particular domain: NAM, ATAF1-2, and CUC2 (6, 7). In vascular plants, genes in the VASCULAR-RELATED NAC-DOMAIN (VND)/NAC secondary wall thickening promoting factor/secondary wall-associated NAC domain protein (SND) clade act as key regulators of xylem differentiation (8). The initial differentiation of meristematic cells into vessel elements is followed by cell proliferation, expansion, programmed cell death, and patterned secondary wall deposition (9). It is during this well-coordinated process that the dimension of vessel elements is determined. Vessel diameter is universally associated positively with stem length in angiosperms. Thus, the developmental stage that determines vessel dimension is critical to support the size that a given plant can achieve.
Potassium (K+) plays a critical active role in plant cell turgor regulation, including cell expansion during wood development in trees, contributing to the size of newly formed vessel elements (10). Moreover, K+ seasonal variation has been observed in poplar cambium, the site of xylogenesis, a high K+ content occurs during spring and summer and a significant reduction during fall and winter (11, 12). Although there is evidence for a role of K+ in wood formation and vessel size in the developing xylem, the cellular mechanism and the transporters relevant to K+ transport and homeostasis, as well as the genetic factors that regulate vessel element dimensions, have remained largely unknown.
Here we show that a previously uncharacterized gene, ENLARGED VESSEL ELEMENT (EVE), contributes to the dimension of vessel elements in poplar trees, impacting hydraulic conductivity, photosynthesis, and growth. Our data suggest that EVE is localized in the plasma membrane and is involved in potassium uptake in differentiating xylem cells during vessel development. In plants, EVE appears to have emerged initially in streptophyte algae, and expanded dramatically among vessel-containing angiosperm. The presence of EVE homologs in algae-infecting phycodnaviruses suggests that this gene has also been involved in extensive horizontal movement in the tree of life.
Results
EVE Is Involved in the Development of Vessel Elements.
To identify genes that contribute to vessel element dimensions, we measured vessel diameter in a mapping population of Populus trichocarpa × Populus deltoids hybrid (13, 14), and identified the most significant quantitative trait locus (QTL) between genetic markers at positions 28.5 to 36.9 Mb of chromosome 1 (SI Appendix, Fig. S1). Of the 827 genes within the QTL interval, we previously reported 45 (SI Appendix, Fig. S2 and Table S1) as expressed primarily in wood forming tissues (15), where differentiation of vessel elements occurs. Variation in xylem mRNA levels of these genes was analyzed as a quantitative phenotype (14). Four cis-regulated genes were identified (SI Appendix, Fig. S2 and Table S1), of which Potri.001G329000 was the only one functionally uncharacterized and, therefore, selected for further analysis.
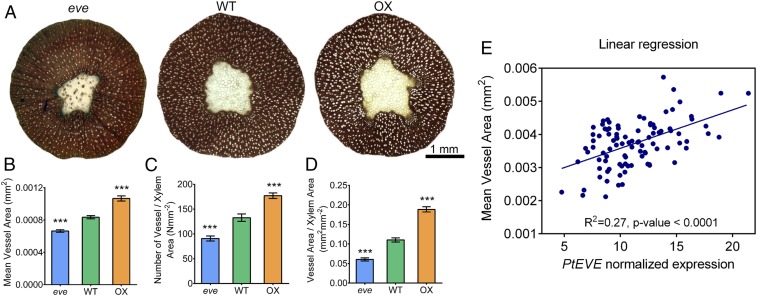
To evaluate the effect of Potri.001G329000 in vessel element dimensions, we overexpressed it under the control of the 35S promoter in hybrid poplar (Populus tremula × alba). We observed that xylem mean vessel area and vessel count were significantly higher (70% and 35%, respectively) in the three transgenic lines evaluated, compared to wild-type (Fig. 1 A–C and SI Appendix, Figs. S3 and S4). The longitudinal dimension of individual vessel elements extracted from transgenic lines were also significantly longer (SI Appendix, Fig. S5). We also generated CRISPR/Cas9-mediated homozygous mutants (SI Appendix, Fig. S6). Phenotypic results from three mutant transgenic lines showed the opposite pattern relative to plants that overexpressed Potri.001G329000 (Fig. 1 A–C and SI Appendix, Fig. S7), with a significant reduction in mean vessel area (18%) and vessel count (31%). Overall, the area occupied by vessels was 129% higher in overexpressing and 43% lower in mutant lines, compared to wild-type trees (Fig. 1D and SI Appendix, Figs. S4 and S7). In contrast to vessel properties, the mean fiber area from transgenic mutant and overexpressing lines was not significantly different from wild-type plants (SI Appendix, Fig. S8).
Fig. 1.
Vessel number and dimensions are modified in transgenic EVE plants. (A) Xylem cross-section of 2-mo-old, greenhouse-grown P. tremula × alba wild-type and transgenic (CRISPR/Cas9 mutant: eve; overexpression: OX) EVE plants. Cell walls are stained with phloroglucinol and vessels are white. (B) Mean vessel area, (C) vessel count per area, and (D) overall vessel area per unit total xylem area in wild-type and transgenic lines of EVE. Data are presented as means ± SE (n = three biological replicates per genotype, with at least three cross-sections measured per replicate). Asterisks indicate significantly lower measures in the mutant eve line, and higher in the transgenic OX line, compared to wild-type, at P < 0.001 (one-way ANOVA followed by Dunnett’s test). Results from three independent OX and eve transgenic lines are presented in SI Appendix, Figs. S4 and S6. (E) Correlation between mean individual vessel area and expression of EVE in a genetically unstructured population of P. trichocarpa. Area of individual vessel elements were measured in partial stem sections of 96 unrelated P. trichocarpa trees grown in common garden for 4 y. Transcript abundance of EVE was quantified in each individual tree by RNA-sequencing, and the normalized expression was determined by the number of fragments per kilobase of transcript, per million mapped reads. Linear models fitting vessel phenotypes (number and area) and PotriEVE transcript abundance were generated in Prism 6.01 software. The slope of this model was significant nonzero (P < 0.0001). ***P < 0.001 (one-way ANOVA followed by Dunnett’s test).
Vessel dimension is a plastic phenotype influenced by environmental conditions, such as water or nutrient availability (10, 16, 17). Thus, changes of molecular or cellular mechanisms that impact water or nutrient uptake (e.g., photosynthesis, root system architecture) could potentially impact vessel size indirectly. To verify that the phenotypes observed in the transgenic plants were due to the direct effects of the expression of Potri.001G329000 within vessel-forming cells, we quantified vessel element diameter in in vitro root culture. Roots were generated by Agrobacterium rhizogenes transformation using leaf explants of wild-type, overexpressing, and mutant plants. After 4 wk of growth in the dark, we observed similar phenotypes in hairy roots as those observed in whole plants. A significant increase (39%) and decrease (23%) in vessel diameter was detected in the overexpressing and mutant lines, respectively, compared to wild-type (SI Appendix, Fig. S9).
To further verify the role of Potri.001G329000 in vessel development, we also examined the correlation of its expression with individual vessel area variation in a natural, genetically unstructured population of P. trichocarpa (18). A highly significant, positive correlation (r2 = 0.27, P < 0.0001), confirmed that the expression of Potri.001G329000 impacts vessel dimension (Fig. 1E).
These molecular and quantitative genetic approaches led us to conclude that Potri.001G329000 contributes to the differentiation and expansion of cambial derivatives into vessel elements in Populus, affecting the number and dimension of these cells. Consequently, we renamed it EVE.
EVE Is a Membrane Protein That Forms a Pentameric Complex.
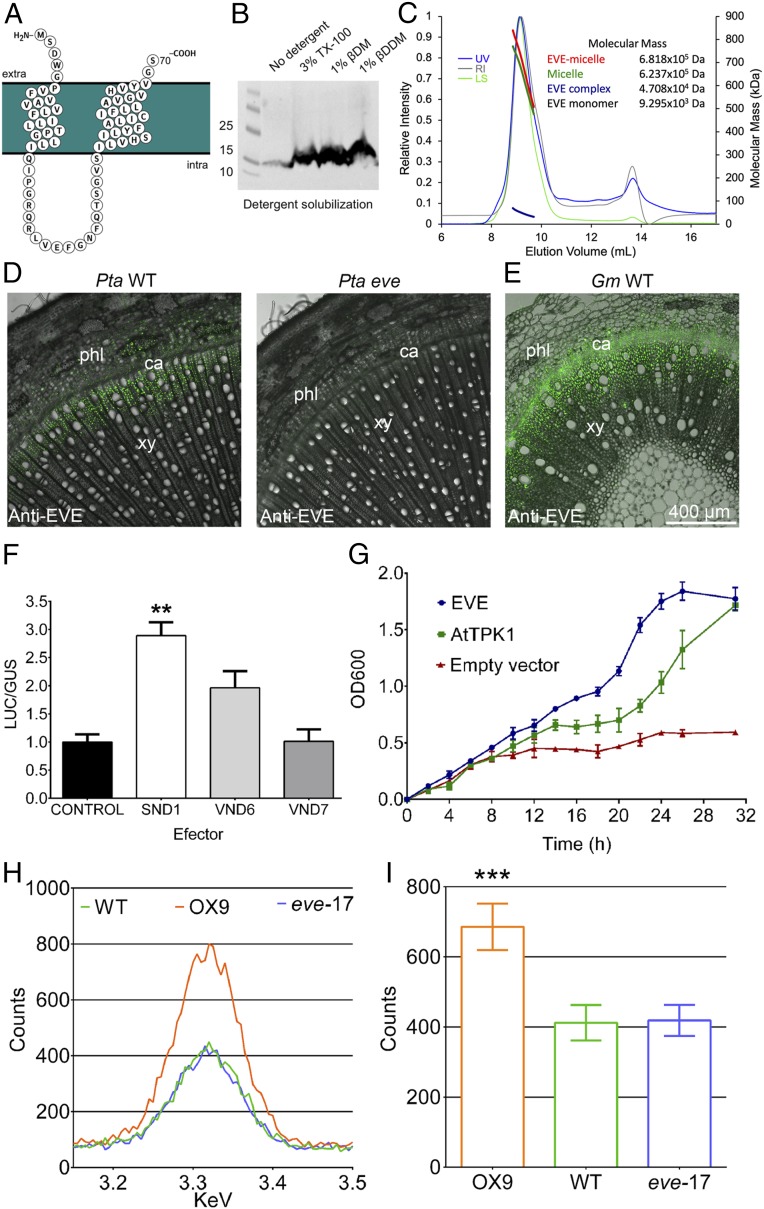
The P. trichocarpa EVE (PotriEVE) coding sequence translates into a 70-amino acid protein, consisting almost entirely of the 68 residues of the DUF3339 domain (Fig. 2A), with a theoretical mass of 9.295 kDa. PotriEVE is predicted to contain two transmembrane domains of 20 and 23 residues but no cleavable signal peptide (Fig. 2A and SI Appendix, Fig. S10). To confirm that EVE is localized in the membrane, we overexpressed the EVE homolog of Physcomitrella patens (PpatEVE) (Fig. 2B) in Pichia pastoris in a heterologous expression system. Western blot analysis showed that detergent solubilization of membrane proteins from the disrupted Pichia lysate is required for visualization of EVE, indicating its presence in the Pichia membrane (Fig. 2B). For further protein characterization, PpatEVE overexpressed in Pichia was purified to yield highly pure and monodispersed protein. PpatEVE protein was then analyzed using size-exclusion chromatography in combination with multiangle laser light scattering (SEC-MALLS) (Fig. 2C). This allowed us to determine the molecular masses of the EVE protein complex (4.708 ± 0.1 kDa). Overexpression of PpatEVE in Pichia and exclusion chromatography suggest that EVE exists as a membrane conjugate likely consisting of five molecules.
Fig. 2.
EVE predicted protein structure, cellular localization, regulation, and activity. (A) The PotriEVE is composed almost entirely of the domain DUF3339 with two predicted transmembrane domains. (B) Western blot of P. patens EVE in P. pastoris cells. Detergents are needed to solubilize the membrane prior the visualization of EVE. (C) Chromatographs from SEC-MALLS analysis of PpatEVE show UV (blue), refractive index (RI, gray), and light scattering (LS, green) detector readings normalized to the peak maxima (left axis). The thick lines indicate the calculated molecular masses (right axis; kDa) of the complete protein/detergent complex (red, 681.8 ± 15.9 kDa), as well as the contributions of the detergent (green, 623.7 ± 22.2 kDa) and EVE protein complex (blue, 4.708 ± 0.1 kDa) throughout the elution peaks. The theoretical molecular mass of the PpatEVE monomer is 9.295 kDa, suggesting that the EVE protein complex is composed of five units (47.08/9.295 ∼ 5). (D) EVE immunolocalizations in 2-mo-old plants of P. tremula × alba stem cross-sections show the protein in the differentiating xylem in wild-type plants, while the signal is absent in eve trees. (E) EVE localization in 1-mo-old plants of G. max cross-sections also reveals EVE in the differentiating xylem. (F) Effect of PotriSND1, PotriVND6 and PotriVND7 on the regulation of PotriEVE. 35S::GUS::tNOS and pEVE::LUC::tNOS were used as controls. LUC and GUS enzymatic activity was expressed as LUC/GUS relative fluorescence units in each biological replicate, normalized to the wild-type control. Data are presented as means ± SE (n = five biological replicates for each effector/reporter combination). Statistical differences between effector and control were determined by one-way ANOVA followed by Dunnett’s test. **P ≤ 0.01. (G) A complementation assay using the E. coli strain LB2003 showed growth complementation in cells growing in low K+ media (3 mM K+) by expression of EVE and AtTPK19. The plotted values are the means ± SD of three biological replicates. (H) Carbon-coated stem cross-sections of EVE OX, CAS9, and wild-type lines were examined in a TESCAN Mira 3 electron microscope fitted with an EDAX Octane Pro X-ray analyzer. For each vessel, the characteristic peak of the X-ray counts of potassium was recorded. In the graph, data from an individual vessel for OX, wild-type, and Cas9 lines are plotted. (I) The average of the counts peak for potassium recorded in three biological replicates of OX, wild-type, and Cas9 lines showed significantly higher potassium content in EVE OX lines, compared to wild-type trees (one-way ANOVA followed by Dunnett’s test). Data are presented as means ± SE. ***P ≤0.001. Results from two independent OX and Cas9 transgenic lines are presented in SI Appendix, Fig. S14.
To verify the subcellular localization of EVE in planta, we carried out a Western blot analysis of soluble and membrane fractions from tobacco plants transiently expressing the EVE:FLAG fusion protein, which indicated the presence of EVE in the membrane fraction (SI Appendix, Fig. S11A). To resolve the membrane location of EVE, we performed a sucrose density-gradient fractionation, and identified fractions associated with each specific membrane using known markers. The fractions containing EVE overlapped largely with those associated with the plasma membrane, suggesting localization in that compartment (SI Appendix, Fig. S11B). Finally, we analyzed tobacco epidermis cells transiently expressing the fusion of EVE and tdTOMATO fluorescence protein (EVE:tdTOMATO:EVE) and detected similar subcellular localization as the full-length fusion of the aquaporin PIP2A to mCherry, which occurs in the plasma membrane (SI Appendix, Fig. S11 C–F).
EVE Is Expressed Primarily in Differentiating Xylem and Is Activated by SND1.
By qRT-PCR analyses we observed that, among all homologs identified in the P. trichocarpa genome, EVE is the most highly expressed in differentiating xylem (SI Appendix, Fig. S12). EVE is also the most consistently, highly expressed gene in this family, in the cell layers that extend until the region of vessel cell expansion (19). EVE protein localization was verified by immunolocalization, showing its presence in the vascular cambium and initial layer of dividing and expanding cells of xylem, where vessel formation occurs (Fig. 2D). Since the Populus antibody also recognizes the epitope in the soybean (Glycine max) homolog of EVE, it was also used for immunolocalization in this herbaceous species where it was present in xylem cells differentiating into vessels (Fig. 2E). High-magnification images show the presence of EVE in fibers and developing vessels, before they are fully formed (SI Appendix, Fig. S13).
In Arabidopsis and Populus, the differentiation of the vascular cambium into xylem is regulated by secondary wall-associated NAC domain transcription factors (8, 20–22). Transcriptomics studies in Arabidopsis demonstrated that EVE homologs are direct targets of the SND1 (20), a key transcriptional switch that activates differentiation of xylem cells. We evaluated if PotriEVE is regulated by this and other secondary wall-associated NAC domain transcription factors by transactivation assays in vivo. We used Nicotiana benthamiana leaves to evaluate the transcriptional induction of the LUCIFERASE enzyme, driven by the PotriEVE promoter, in the presence and the absence of Populus SND1 homologs, and two other transcriptional regulators involved in vessel development (VND6 and VND7). Of all of the transcriptional regulators evaluated, only SND1 showed significantly higher activation of the LUCIFERASE, compared to the control (Fig. 2F). While in Arabidopsis these transcription factors exhibit fiber- (SND1) or vessel- (VND6, VND7) specific expression, in the woody P. trichocarpa SND1 orthologs (PtrWND1A&B, PtrWND2A&B) are expressed in the vessels, xylary fibers, and ray parenchyma cells in developing xylem (23). Expectedly, we found that the PotriEVE promoter contains a palindromic 19-bp consensus sequence secondary wall NAC binding element to which SND1 binds (24).
EVE Contributes to K+ Uptake.
Potassium (K)+ concentration is distinctly higher in vessels compared to fibers (10). Furthermore, K+ contributes to vessel expansion by regulating it osmotically (11). Because high concentration of K+ occurs where EVE is localized (cambial and differentiating xylem zones), and EVE’s impact on vessel dimensions, we hypothesized that it is involved in K+ uptake. To evaluate this hypothesis, a complementation assay was performed using the Escherichia coli strain LB2003, a K+-uptake–deficient triple mutant. Cells grown in low K+ media showed growth complementation by expression of EVE (Fig. 2G) that parallels the growth observed with the Arabidopsis two-pore potassium channel AtTPK1 (25), used as a positive control.
To further verify that EVE is implicated in K+ uptake, we quantified the levels of potassium in vessels of EVE-overexpressing lines, CRISPR/Cas9 mutants and wild-type plants grown in hydroponic liquid nutrient solution. Initial levels of potassium (3.75 mM) were decreased to 0.375 mM after 2 wk. Three weeks later, potassium was measured using energy-dispersive X-ray spectroscopy (EDS). X-ray counts of K+ were significantly higher in the overexpressing transgenic lines, compared to the CRISPR/Cas9 mutants and wild-type plants (Fig. 2 H and I), suggesting that EVE contributes to K+ uptake. While EVE appears to be involved in the uptake of K+, its specific functional role in that process remains unknown.
Plants Overexpressing EVE Have Higher Height Growth and Net Photosynthesis.
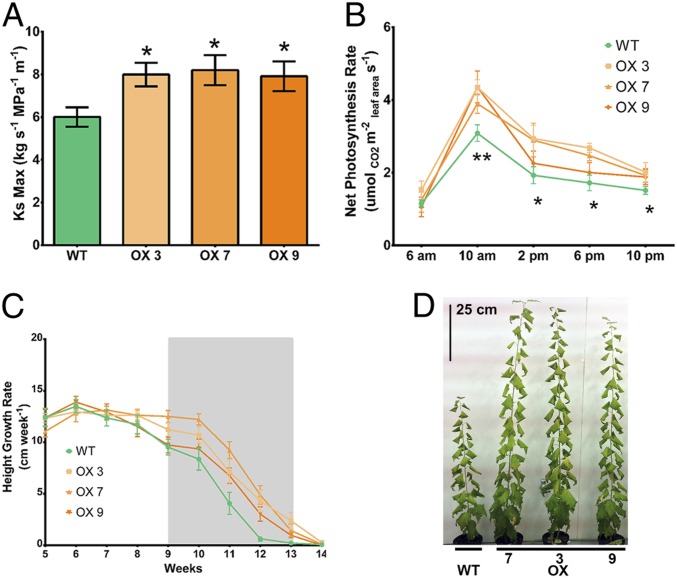
Wider vessels are expected to provide lower resistance to water flow in the plant vascular system. To evaluate this hypothesis, we measured hydraulic conductivity in wild-type and transgenic poplar overexpressing PotriEVE and verified a significant increase in hydraulic conductivity (∼28%) (Fig. 3A). Wider vessels also support increased transpiration rates associated with higher stomatal conductance and photosynthetic rates, and are highly positively correlated with stem length (height) in angiosperms (26). Thus, we evaluated whether larger vessel area contributed to stem height growth and net photosynthesis in three overexpressing and wild-type lines, under conditions of high evaporative demand (diurnal temperature range of 29 to 35 °C). Height growth and net photosynthesis were significantly higher in plants overexpressing EVE compared to wild-type (Fig. 3 B and C). This was not due to differences in biochemical photosynthetic capacity between the lines, as measured by plots of net CO2 assimilation rate versus calculated substomatal CO2 concentration, but strictly to stomatal closure limitations in the wild-type trees (SI Appendix, Table S2). In contrast, lower evaporative demand conditions (diurnal temperature of 25 °C) resulted in no significant phenotypic differences in photosynthesis or growth between transgenic and wild-type plants.
Fig. 3.
Transgenic plants overexpressing EVE have higher hydraulic conductivity and support higher growth and photosynthesis under high evaporative demand. (A) Maximum hydraulic conductivity (means ± SE) is significantly higher in overexpressing transgenic EVE, compared to wild-type trees, when grown at 29 to 35 °C. (B) Net photosynthesis rate (means ± SE) is significantly higher in all overexpressing transgenic EVE lines compared to wild-type trees at 10:00 AM. At 2:00 PM, 6:00 PM, and 10:00 PM, net photosynthesis rate is significantly higher in one or more overexpressing transgenic lines compared to wild-type. Net photosynthesis rate was measured repeatedly on the same plants during the experiment, on six trees per genotype. For (A and B), asterisks indicate statistically significant difference between each transgenic line and wild-type, at **P < 0.01 and *P < 0.05 by two-tailed Student’s t test. (C) Overexpressing transgenic EVE lines maintain active growth and higher height growth rates of each transgenic line compared to wild-type. The plotted values are the means ± SE. The gray box indicates the growth phase where the overexpressing transgenic lines outperformed wild-type trees and showed a significantly higher growth rate at P = 0.05 (two-tailed Student’s t-test). Measurements were taken repeatedly on the same plants during the experiment, on six trees per genotype. (D) Wild-type and overexpressing transgenic EVE lines final growth after 14 wk in growth chamber.
EVE Appeared Prior to the Colonization of Land and May Have Originated From a Horizontal Gene Transfer.
The development of an improved vascular system had a significant role in the evolution of plants. Thus, we evaluated the occurrence of EVE-like genes across plant evolution and detected putative homologs in nearly all land plant genomes sequenced to date (SI Appendix, Table S3), with notable absences in the ferns Azolla and Salvinia. We supplemented our database with transcriptomes from hornworts and green algae (OneKP.org) (27). This allowed us to identify putative homologs in all three bryophyte lineages (one to three copies) and all seed plants. Compared to other land plants, a large expansion and diversification of EVE-like genes occurred among most flowering plants (monocots have 5 to 19 copies, eudicots 2 to 20 copies).
To investigate the origin of EVE, we expanded our search for possible ancestors in taxa that precedes the land plants evolutionarily. We detected several EVE-like genes in streptophyte algae (28) (SI Appendix, Table S3). Streptophyte algae are a paraphyletic grade of algae, one lineage of which represents the closest relatives to land plants. This observation points to the appearance of EVE among green plants prior to colonization of land. The search in chlorophyte genomes, a clade of green algae distantly related to land plants, as well as Bacterial or Archaea domains, showed no evidence of EVE. Surprisingly, EVE-like genes were identified in numerous prasinoviruses of the Phycodnaviridae family and in one species of amoebae (Acanthamoeba castellanii). Prasinoviruses, also known as “giant viruses,” are lytic and lysogenic viruses with a genome ranging from 160 to 560 kb (29).
To further evaluate homology, we searched and confirmed that EVE-like genes in Phycodnaviridae contain the characteristic features of the DUF3339 domain present in land plants (http://pfam.xfam.org/family/PF11820). A phylogeny of EVE-like genes across viruses, algae, and land plants was constructed. The tree revealed a close phylogenetic relationship between viral and streptophyte copies (SI Appendix, Figs. S15 and S16).
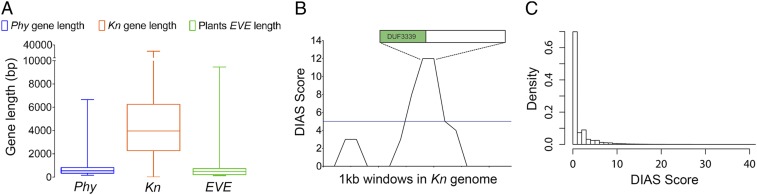
The absence of putative EVE homologs in chlorophyte algae, together with the closer phylogenetic relation between viral and streptophyte algae, is suggestive of a horizontal gene transfer (HGT) event between prasinoviruses and an ancestral streptophyte. The length of putative EVE homologs is significantly shorter than the average gene size in plants, but similar to that of prasinoviruses (Fig. 4A). To further evaluate the HGT origin of EVE, we investigated nucleotide base composition in the genome of Klebsormidium nitens (28), which shares the most ancient common ancestor of EVE putative homologs in plants (Fig. 4B). We identified a highly significant genomic islands (discrete interval accumulative score [DIAS] = 12) possibly originated by HGT, containing the algae EVE homolog (Fig. 4 B and C). To discard the possibility that the presence of an EVE-like gene in the K. nitens genome is due to contamination with prasinoviruses, we confirmed the presence of the EVE genomic island within the K. nitens genome by PCR in independent algae cultures (SI Appendix, Fig. S17).
Fig. 4.
HGT origin of EVE. (A) Box-plot describing the length of all genes in Phycodnaviruses (Phy), in the earliest plant species known to contain a putative EVE homolog (K. nitens, Kn), and the length of EVE-like genes across plant species (EVE), showing that EVE structure in plants resemble other viral genes. (B) SigHunt analysis of the K. nitens genome identifies a highly significant genomic island associated with a HGT event, at the location of the putative EVE homolog. The 1-kb window containing EVE was scored based on the number of the 4-mer frequencies that deviate from the credibility interval of their local genomic density, using a DIAS. (C) Density of DIAS in the K. nitens genome shows that high scores, such as that observed in the location of the putative EVE homolog, are rare.
Discussion
We performed QTL and xylem transcriptome analysis of a mapping population of P. trichocarpa × P. deltoides and identified a previously uncharacterized gene, EVE, involved in vessel development. Analysis of vessel dimensions in transgenic poplar trees overexpressing EVE and CRISPR/Cas9-mediated homozygous mutants, as well as transcript variation of EVE in a natural population of P. trichocarpa, showed significant changes of vessel area that are compatible with a role in the regulation of vessel dimensions. Interestingly, CRISPR/Cas9-mediated homozygous mutants didn’t abolish entirely the occurrence of structurally normal vessel elements. This may have occurred because of functional redundancy from EVE-like genes expressed in differentiating xylem, or the compensatory contribution from other genes with a similar function.
The observation that EVE complements the growth phenotype of the E. coli strain LB2003, a K+-uptake–deficient triple mutant, suggests a functional role in cell K+ uptake. K+ is the most abundant cation in higher plants, and is the major inorganic, osmotically active substance in differentiating cambium cells (30, 31). K+ has a critical role in numerous aspects of xylogenesis, such as cell expansion through an increase of symplastic K+ content and xylem development (11). In plants growing under nonlimiting K+ supply, the developing xylem zone and the cambial K+ levels are significantly higher than in plants growing under limiting K+ (10). An increase in K+ supply also results in an increase of vessel size; and treatment with tetraethylammonium, a K+ channel blocker, significantly reduces the size of newly formed vessels (10). To further verify the role of EVE in K+ uptake, we performed an EDS assay, where plants were grown under low K+ conditions (0.375 mM KCl) (12, 32). Under these conditions, overexpressing lines showed a significant increase in K+ concentration in the vessels, compared to wild-type plants. In contrast, no difference was detected between CRISPR/Cas9 mutants and wild-type. This suggests that, under these growth conditions, the absence of a functional copy of EVE may be compensated by other potassium transporters or EVE-like family members. The functional role of EVE under different levels of K+ availability, and the implications to vessel dimensions, should be evaluated in the future.
Results of the localization experiments showed that EVE is not expressed exclusively in cambium cells differentiating into vessels. Instead, expression was also detected in fiber cells of the differentiating xylem. However, we did not observe any difference in fiber structure in the transgenic plants, compared to wild-type. The absence of differences between fibers of transgenic and wild-type plants suggests that EVE may promote K+ uptake in both developing fibers and vessels, but the presence of an outward K+ channel expressed only in fibers could compensate for this uptake. PTORK, which is absent in vessels but expressed in new fibers, could play such a role by mediating K+ efflux (33). In this scenario, potassium released from fibers could further contribute to its accumulation in differentiating vessels.
To further uncover the cellular role of EVE, we performed heterologous overexpression in yeast and observed its localization in the membrane. Exclusion chromatography suggests that it exists as a conjugate of five copies. Experiments designed to evaluate the localization of EVE in planta pointed to its presence in the plasma membrane. Finally, we showed that EVE expression is directly activated by the poplar SND1 homolog, which is a regulator of vascular development. The combination of structural, spatial, and functional data suggests that EVE influences vessel dimensions by physical uptake of K+ during the process of cell expansion in the differentiating xylem.
Our measurement of water conductivity showed, as expected, that the larger vessels of EVE-overexpressing lines lead to more efficient water transport. Interestingly, we observed that under high temperature, plants overexpressing EVE exhibited significantly greater height growth and net photosynthesis. Thus, the high evaporative demand triggered by warmer temperatures was compensated by the improved hydraulic conductivity in plants overexpressing EVE, possibly by allowing stomata to remain open under increased evaporative demand. These results are consistent with the previously reported linear relationship between stem hydraulic conductivity and growth (34–42). Our results indicate that, in the absence of water deficit-mediated cavitation, wider vessels lead to higher growth rates under high evaporative conditions, in agreement with the hypothesis that vessel widening contributed to the expansion of land plants with larger body size (4, 5).
In the search for the evolutionary origin of EVE, we showed that putative homologs are present in most land species and in the streptophyte algae, which are the closest aquatic relatives to land plants. This result points that the origin of EVE precedes land colonization by plants. However, the functional role of EVE in algae and land plant species that lack a vascular system remains unknown. An evaluation of the number of copies showed that a large expansion and diversification of EVE-like genes occurred among most flowering plants, which coincides with the expansion of the vessels across land plants (43). Thus, the functional role of EVE in vessel development is derived from its function in streptophyte algae and nonvascular land plants. The increased number of EVE-like genes in conifers suggests that the expansion is coincident in evolution with the establishment of the secondary growth mediated by the vascular cambium in land plants (43). In the future, the functional role of EVE in the water-conducting cells of conifers, the tracheids, should be evaluated.
Surprisingly, the search for EVE putative homologs resulted in its detection in Prasinovirus, a genus of the Phycodnaviridae family, which infects eukaryotic algae from both fresh and marine waters, and often integrates part of their genome into that of their hosts (44, 45). Phycodnaviruses belong to the monophyletic clade of nucleocytoplasmic large DNA viruses (NCLDVs), characterized by a large double-stranded DNA genome (46, 47). Recent studies showed that certain eukaryotes contain fragments of NCLDV DNA integrated to their genome, when surprisingly many of these organisms were not previously shown to be infected by NCLDVs (48). The eukaryotic groups most impacted with the presence of NCLDV viral homolog genes are brown algae (Phaeophyceae), Amoebozoa, and green algae (Chlorophyta and Streptophyta). Of 11 genomes available for amoebas (amoebadb.org), one contains a DUF3339 copy, A. castellanii. In fact, the A. castallanii genome sequencing revealed multiple fragments of prasinoviruses DNA integrated in its genome (49). The Acanthamoeba genus is one of the most efficient laboratory hosts to isolate giant viruses from aquatic samples (50–53). Taking these data together, we show that the presence of EVE copies in green algae and amoeba suggests that multiple HGT events occurred between an ancestor of the prasinovirus lineage and an ancestral streptophyte and amoeba, likely facilitated by sharing the same aquatic habitats.
Materials and Methods
Additional information can be found in SI Appendix, Material and Methods.
QTL Analysis of Vessel Development.
A hybrid Populus backcross population of 397 individuals was genotyped as described previously (54). Cross-sections were obtained from the stem harvested from 100 individuals representing the most informative recombinations. Vessels were measured by visualizing the sections in a microscope and analyzing the images obtained. Individual vessel areas were converted to diameters (d) and counted (n), and mean hydraulic diameter (Dh, μm, [(Σd4)n-1]1/4) was calculated (2). QTL analysis of vessel mean hydraulic diameter (Dh) was carried out using composite interval mapping implemented in QTL Cartographer v2.5 (55).
Generation and Phenotyping of Transgenic Plants Overexpressing EVE and CRISPR/Cas9 EVE Mutant Plants.
Overexpressing transgenic lines were generated with EVE under control of the 35S cauliflower promoter. For generation of CRISPR/Cas9 EVE mutant plants, a guide RNA (gRNA) targeting EVE was selected from http://aspendb.uga.edu/ (56), and cloned as previously described (57). Constructs were transferred into Agrobacterium and transformation of the hybrid (P. tremula × alba) poplar genotype 717-1B4 was performed (58). Vessel properties were measured as described above. Measurement of vessel element longitudinal dimensions and stem fibers was achieved by isolating them (59), visualizing them in a microscope and quantifying their properties. Vessel elements of transgenic poplar hairy roots were characterized using a similar approach.
Measurement of Hydraulic Conductivity.
Conductivity was measured in stem segments connected to a low-pressure flow apparatus (60). Flow of a perfusion solution through the stems was performed at low pressure (4 kPa), and weight of the receiving container was recorded. Based on these data, the initial “native” conductivity (Knative) was calculated according to Darcy’s law (2). After removal of all embolisms in the vascular system, the flow was recorded again and converted to maximum conductivity (Ks max, kg s−1 MPa−1 m−1).
Measurement of Vessel Properties and EVE Expression in a P. trichocarpa Association Population.
Vessel phenotypes and PotriEVE gene expression were measured in 96 unrelated, 4-y-old black cottonwood (P. trichocarpa) genotypes grown in a common garden, as described previously (18). Cross-sections were obtained and vessel properties were quantified as described above. RNA from xylem scrapings collected from the stem of each tree was purified and quantified prior to RNA-sequencing (RNA-seq) library preparation and sequencing in an Illumina HiSeq. 2000. The raw RNA-seq data were analyzed as described previously (61).
Measurement of Growth and Photosynthesis under Normal and High Evaporative Demand in Transgenic Poplar Plants.
Two temperature experiments were conducted in a growth chamber, with plants grown under soil moisture saturation and long day lengths. Following acclimation at 25 °C for 6 wk, plants were grown either under normal conditions (20 °C to 25 °C) or under high evaporative demand (29 °C to 35 °C). The diurnal net photosynthesis rate and curves of net photosynthesis relative to leaf internal CO2 concentration (A-Ci curves) were measured on the first fully expanded leaves. The maximum rate of carboxylation of Rubisco (Vcmax) and maximum rate of electron transport (Jmax) were derived from A-Ci curves, as described previously (62).
EVE Purification and Structure Analysis.
The EVE gene from P. patens (PpatEVE) was cloned into the pPICZc vector and expressed using P. pastoris prior to purification by nickel affinity chromatography and SEC. The structure of PpatEVE was analyzed using SEC-MALLS.
RT‐PCR Expression Analysis of EVE and Its Homologs in Differentiating Xylem of P. trichocarpa.
Differentiating xylem was collected from Populus wild-type plants and RNA was extracted (63). qRT-PCR analyses were performed and gene expression was quantified as described previously (64).
EVE Antibody Synthesis and Immunohistochemical Analysis.
EVE antibodies were made by Genscript. For immunohistochemical analysis, basal segments of stems of Populus wild-type, EVE homozygous CRISPR/Cas9 mutant lines and soybean (G. max) wild-type were formaldehyde-fixed and preserved, as previously described (65). Segments were then sectioned and permeabilized before incubation with anti-EVE antibody and the corresponding secondary antibody, prior to washes and visualization under a confocal microscope.
Subcellular Localization of EVE.
Golden Gate technology was used to assemble p35S::EVE:FLAG::t35S transcriptional unit and clone it into the destination vector pAGM4673 (66). A set of binary vectors containing organelle-targeting proteins fused with mCherry were acquired from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH). To verify the subcellular localization of EVE, we expressed transiently the constructs described above in N. benthamiana leaves through agroinfiltration, and performed a sucrose density-gradient fractionation, as previously described (67). Subcellular localization of EVE was also determined using microscopy. The Golden Gate system was employed to assemble the p35S::EVE:tdTOMATO:EVE::t35S transcriptional unit. After agroinfiltration, leaves were sectioned and placed under the confocal microscope for visualization of the fluorescence emitted by tdTOMATO and mCherry.
Growth Complementation Assay.
Competent LB2003 cells were transformed with a plasmid carrying the coding sequence of EVE, a plasmid carrying the coding sequence of an Arabidopsis two-pore potassium channel (AtTPK1), and an empty vector. Cells were grown overnight, harvested, and washed to remove any residual potassium. Expression of EVE and AtTPK1 was induced by isfopropyl-β-d-thiogalactopyranoside (IPTG) and OD600 values were obtained for each genotype (EVE, AtTPK1, and empty vector) every 2 h for a period of 32 h.
EDS Assay.
Plants from EVE CRISPR/Cas9 mutant lines, overexpressing transgenic lines and wild-type were grown in aerated Hocking’s Complete Solution (Hocking, 1971) containing 3.75 mM KCl for 2 wk. Next, plants were transferred to a solution containing 0.375 mM of KCl. After 3 wk, internode cross sections were obtained, freeze-dried, coated with carbon, and examined in a TESCAN Mira 3 scanning electron microscope fitted with a EDAX Octane Pro X‐ray analyzer. The potassium content of vessels was measured as the potassium counts peak (minus background) recorded after 50 s.
Transactivation Assay.
PtaSND1 (Potri.011G153300), PtaVND6 (Potri.012G126500), and PtaVND7 (Potri.013G113100) were amplified from P. tremula × alba cDNA samples and a 2,000-bp fragment of the PtaEVE promoter was amplified from P. tremula × alba gDNA. Fragments were purified and cloned into the pUPD vector. Golden Braid technology (68) was employed to place the 35S::PtaSND1, PtaVND6 or PtaVND7::tNOS (effector) together with 35S::GUS::tNOS (reporter) and pEVE::LUC::tNOS in the 2omega1 plasmid. Transient N. benthamiana agroinfiltration experiments were completed as previously described (69). N. benthamiana leaves were collected 3 d postinfiltration for protein extraction and GUS or LUC enzymatic activity measurements.
Evolution of EVE.
We searched for EVE in previously sequenced plant genomes and transcriptome data from the One Thousand Plant Transcriptome Project (1KP, onekp.com). From the final set of detected sequences with similarity to EVE, copy number in each lineage was tabulated. Maximum-likelihood analysis of 56 plant DUF3339 proteins from representative species was conducted with RAxML v. 8.2.12 (70).
Identification of Genomic Islands due to HGT.
SigHunt (71) was employed to identify genomic islands originated from HGT events in the K. nitens genome (28). We estimated 4-mer density and its variance in sliding windows of chromosome DNA sequence, as described previously (71). The windows were then scored using a DIAS with the improved variant of the method (71).
Data Availability.
Sequences of EVE homologs identified can be accessed from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Sequences containing the DUF3339 domain are available in PFAM (http://pfam.xfam.org/family/PF11820).
Supplementary Material
Acknowledgments
We thank Pieter Bass, Ron Sederoff, Pamela Soltis, and Douglas Soltis for critical reading of the manuscript; and Thomas Wood for the gift of the LB2003 Escherichia coli. This work was supported by the US Department of Agriculture Plant Feedstock Genomics for Bioenergy Program (Grant 2009-65504-05697) and the Department of Energy Office of Science Biological and Environmental Research (Grant DE-SC0003893).
Footnotes
Competing interest statement: The use of the gene described in the paper to improve plant growth and productivity is the subject of a patent (US# 9,650,646) and is currently licensed.
This article is a PNAS Direct Submission. S.M.B. is a guest editor invited by the Editorial Board.
Data deposition: Sequences of EVE homologs identified can be accessed from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Sequences containing the DUF3339 domain are available in PFAM (http://pfam.xfam.org/family/PF11820).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912434117/-/DCSupplemental.
References
- 1.Sperry J. S., Hacke U. G., Feild T. S., Sano Y., Sikkema E. H., Hydraulic consequences of vessel evolution in angiosperms. Int. J. Plant Sci. 168, 1127–1139 (2007). [Google Scholar]
- 2.Tyree M. T., Zimmermann M. H., Zimmermann M. H., Xylem Structure and the Ascent of Sap (Springer, 2002). [Google Scholar]
- 3.Lucas W. J., et al. , The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 55, 294–388 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Olson M. E., Rosell J. A., Vessel diameter-stem diameter scaling across woody angiosperms and the ecological causes of xylem vessel diameter variation. New Phytol. 197, 1204–1213 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Rosell J. A., Olson M. E., Anfodillo T., Scaling of xylem vessel diameter with plant size: Causes, predictions, and outstanding questions. Curr. For. Rep. 3, 46–59 (2017). [Google Scholar]
- 6.Souer E., van Houwelingen A., Kloos D., Mol J., Koes R., The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Aida M., Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohtani M., et al. , Evolution of plant conducting cells: Perspectives from key regulators of vascular cell differentiation. J. Exp. Bot. 68, 17–26 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Bollhöner B., Prestele J., Tuominen H., Xylem cell death: Emerging understanding of regulation and function. J. Exp. Bot. 63, 1081–1094 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Langer K., et al. , Poplar potassium transporters capable of controlling K+ homeostasis and K+-dependent xylogenesis. Plant J. 32, 997–1009 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Fromm J., Wood formation of trees in relation to potassium and calcium nutrition. Tree Physiol. 30, 1140–1147 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Wind C., Arend M., Fromm J., Potassium-dependent cambial growth in poplar. Plant Biol. (Stuttg) 6, 30–37 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Novaes E., et al. , Quantitative genetic analysis of biomass and wood chemistry of Populus under different nitrogen levels. New Phytol. 182, 878–890 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Drost D. R., et al. , Diversification in the genetic architecture of gene expression and transcriptional networks in organ differentiation of Populus. Proc. Natl. Acad. Sci. U.S.A. 107, 8492–8497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quesada T., et al. , Comparative analysis of the transcriptomes of Populus trichocarpa and Arabidopsis thaliana suggests extensive evolution of gene expression regulation in angiosperms. New Phytol. 180, 408–420 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Lovisolo C., Schubert A., Effects of water stress on vessel size and xylem hydraulic conductivity in Vitis vinifera L. J. Exp. Bot. 49, 693–700 (1998). [Google Scholar]
- 17.Schreiber S. G., Hacke U. G., Hamann A., Variation of xylem vessel diameters across a climate gradient: Insight from a reciprocal transplant experiment with a widespread boreal tree. Funct. Ecol. 29, 1392–1401 (2015). [Google Scholar]
- 18.McKown A. D., et al. , Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 201, 1263–1276 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Sundell D., et al. , Aspwood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29, 1585–1604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong R., Lee C., Ye Z.-H., Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 3, 1087–1103 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Ohashi-Ito K., Oda Y., Fukuda H., Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi M., et al. , VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 66, 579–590 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Zhong R., Lee C., Ye Z. H., Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy R. L., Zhong R., Ye Z. H., Secondary wall NAC binding element (SNBE), a key cis-acting element required for target gene activation by secondary wall NAC master switches. Plant Signal. Behav. 6, 1282–1285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czempinski K., Zimmermann S., Ehrhardt T., Müller-Röber B., New structure and function in plant K+ channels: KCO1, an outward rectifier with a steep Ca2+ dependency. EMBO J. 16, 2565–2575 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson M. E., et al. , Universal hydraulics of the flowering plants: Vessel diameter scales with stem length across angiosperm lineages, habits and climates. Ecol. Lett. 17, 988–997 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Wickett N. J., et al. , Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111, E4859–E4868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hori K., et al. , Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5, 3978 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Etten J. L., Graves M. V., Müller D. G., Boland W., Delaroque N., Phycodnaviridae—Large DNA algal viruses. Arch. Virol. 147, 1479–1516 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Gierth M., Mäser P., Potassium transporters in plants—Involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 581, 2348–2356 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Barrelet T., Ulrich A., Rennenberg H., Krähenbühl U., Seasonal profiles of sulphur, phosphorus, and potassium in Norway spruce wood. Plant Biol. (Stuttg) 8, 462–469 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Fromm J., “Xylem development in trees: From cambial divisions to mature wood cells” in Cellular Aspects of Wood Formation, Fromm J., Ed. (Plant Cell Monographs, Springer, Berlin, Heidelberg, 2013), vol. 20, pp. 3–39. [Google Scholar]
- 33.Arend M., et al. , Polar-localised poplar K+ channel capable of controlling electrical properties of wood-forming cells. Planta 223, 140–148 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Tyree M. T., Ewers F. W., The hydraulic architecture of trees and other woody plants. New Phytol. 119, 345–360 (1991). [Google Scholar]
- 35.Machado J. L., Tyree M. T., Patterns of hydraulic architecture and water relations of two tropical canopy trees with contrasting leaf phenologies: Ochroma pyramidale and Pseudobombax septenatum. Tree Physiol. 14, 219–240 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Brodribb T. J., Feild T. S., Stem hydraulic supply is linked to leaf photosynthetic capacity: Evidence from New Caledonian and Tasmanian rainforests. Plant Cell Environ. 23, 1381–1388 (2000). [Google Scholar]
- 37.Brodribb T. J., Holbrook N. M., Gutiérrez M. V., Hydraulic and photosynthetic co-ordination in seasonally dry tropical forest trees. Plant Cell Environ. 25, 1435–1444 (2002). [Google Scholar]
- 38.Atkinson C. J., Else M. A., Taylor L., Dover C. J., Root and stem hydraulic conductivity as determinants of growth potential in grafted trees of apple (Malus pumila Mill.). J. Exp. Bot. 54, 1221–1229 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Lovelock C. E., et al. , Linking physiological processes with mangrove forest structure: Phosphorus deficiency limits canopy development, hydraulic conductivity and photosynthetic carbon gain in dwarf Rhizophora mangle. Plant Cell Environ. 29, 793–802 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Petit G., Anfodillo T., Carraro V., Grani F., Carrer M., Hydraulic constraints limit height growth in trees at high altitude. New Phytol. 189, 241–252 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Fan Z. X., Zhang S. B., Hao G. Y., Ferry Slik J. W., Cao K. F., Hydraulic conductivity traits predict growth rates and adult stature of 40 Asian tropical tree species better than wood density. J. Ecol. 100, 732–741 (2012). [Google Scholar]
- 42.Hajek P., Leuschner C., Hertel D., Delzon S., Schuldt B., Trade-offs between xylem hydraulic properties, wood anatomy and yield in Populus. Tree Physiol. 34, 744–756 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Spicer R., Groover A., Evolution of development of vascular cambia and secondary growth. New Phytol. 186, 577–592 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Read B. A., et al. ; Emiliania huxleyi Annotation Consortium , Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Blanc G., et al. , The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22, 2943–2955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iyer L. M., Balaji S., Koonin E. V., Aravind L., Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117, 156–184 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Koonin E. V., Yutin N., Nucleo-cytoplasmic large DNA viruses (NCLDV) of Eukaryotes. eLS, 10.1002/9780470015902.a0023268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallot-Lavallée L., Blanc G., A glimpse of nucleo-cytoplasmic large DNA virus biodiversity through the eukaryotic genomics window. Viruses 9, E17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke M., et al. , Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14, R11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arslan D., Legendre M., Seltzer V., Abergel C., Claverie J.-M., Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. U.S.A. 108, 17486–17491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raoult D., Boyer M., Amoebae as genitors and reservoirs of giant viruses. Intervirology 53, 321–329 (2010). [DOI] [PubMed] [Google Scholar]
- 52.La Scola B., et al. , A giant virus in amoebae. Science 299, 2033–2033 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Thomas V., et al. , Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 13, 1454–1466 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Drost D. R., et al. , A microarray-based genotyping and genetic mapping approach for highly heterozygous outcrossing species enables localization of a large fraction of the unassembled Populus trichocarpa genome sequence. Plant J. 58, 1054–1067 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Wang S., Basten C. J., Zeng Z.-B., Windows QTL Cartographer 2.5 (Department of Statistics, North Carolina State University, Raleigh, NC, 2012).
- 56.Zhou X., Jacobs T. B., Xue L.-J., Harding S. A., Tsai C.-J., Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol. 208, 298–301 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Jacobs T. B., Martin G. B., High-throughput CRISPR vector construction and characterization of DNA modifications by generation of tomato hairy roots. J. Vis. Exp., e53843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gallardo F., et al. , Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta 210, 19–26 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Ruzin S. E., Plant Microtechnique and Microscopy (Oxford University Press, 1999). [Google Scholar]
- 60.Sperry J. S., Donnelly J. R., Tyree M. T., A method for measuring hydraulic conductivity and embolism in xylem. Plant Cell Environ. 11, 35–40 (1988). [Google Scholar]
- 61.Hefer C. A., Mizrachi E., Myburg A. A., Douglas C. J., Mansfield S. D., Comparative interrogation of the developing xylem transcriptomes of two wood-forming species: Populus trichocarpa and Eucalyptus grandis. New Phytol. 206, 1391–1405 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Dubois J.-J. B., Fiscus E. L., Booker F. L., Flowers M. D., Reid C. D., Optimizing the statistical estimation of the parameters of the Farquhar-von Caemmerer-Berry model of photosynthesis. New Phytol. 176, 402–414 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Chang S., Puryear J., Cairney J., A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 11, 113–116 (1993). [Google Scholar]
- 64.Ibañez C., et al. , Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS One 3, e3567 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conde D., González-Melendi P., Allona I., Poplar stems show opposite epigenetic patterns during winter dormancy and vegetative growth. Trees (Berl.) 27, 311–320 (2013). [Google Scholar]
- 66.Engler C., et al. , A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Yang H., Murphy A., Membrane preparation, sucrose density gradients and two-phase separation fractionation from five-day-old Arabidopsis seedlings. Bio Protoc. 3, e1014 (2013). [Google Scholar]
- 68.Sarrion-Perdigones A., et al. , GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS One 6, e21622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramos-Sánchez J. M., et al. , Real-time monitoring of PtaHMGB activity in poplar transactivation assays. Plant Methods 13, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaron K. S., Moravec J. C., Martínková N., SigHunt: Horizontal gene transfer finder optimized for eukaryotic genomes. Bioinformatics 30, 1081–1086 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences of EVE homologs identified can be accessed from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Sequences containing the DUF3339 domain are available in PFAM (http://pfam.xfam.org/family/PF11820).