Significance
Reliably detecting the presence of aneuploidy in clinical samples has implications for a broad range of diagnostic applications, including noninvasive prenatal testing, preimplantation genetic diagnosis, the evaluation of congenital abnormalities, and cancer diagnostics. Next generation sequencing protocols, such as whole-genome sequencing, are typically used to detect aneuploidy, but amplicon-based protocols achieve high coverage depth at relatively low cost and can be used when only tiny amounts of DNA are available. In this paper, we describe a simple PCR-based approach to detect the presence of aneuploidy in liquid biopsies, even when only small amounts of blood are available for assay. This approach detected cancers in 49% of 883 nonmetastatic patients with cancer but in less than 1% of 812 healthy controls.
Keywords: liquid biopsy, aneuploidy, early cancer detection, circulating tumor DNA
Abstract
We report a sensitive PCR-based assay called Repetitive Element AneupLoidy Sequencing System (RealSeqS) that can detect aneuploidy in samples containing as little as 3 pg of DNA. Using a single primer pair, we amplified ∼350,000 amplicons distributed throughout the genome. Aneuploidy was detected in 49% of liquid biopsies from a total of 883 nonmetastatic, clinically detected cancers of the colorectum, esophagus, liver, lung, ovary, pancreas, breast, or stomach. Combining aneuploidy with somatic mutation detection and eight standard protein biomarkers yielded a median sensitivity of 80% in these eight cancer types, while only 1% of 812 healthy controls scored positive.
As a result of drastic reductions in costs, whole-genome sequencing (WGS) is now commonly used to detect chromosome copy number variations, also known as aneuploidy (1). Identifying the presence of aneuploidy has a broad range of diagnostic applications, including noninvasive prenatal testing (NIPT) (2), preimplantation genetic diagnosis (3), evaluation of congenital abnormalities (4), and cancer diagnostics (5).
Shallow (0.1 to 1×) WGS is used for aneuploidy detection in a large number of commercially available tests (6). WGS is typically used in NIPT, where a relatively high fraction (5 to 25%) of the total DNA is derived from the fetus (7). A companion diagnostic is frequently used to estimate the fetal fraction, and NIPT is often not performed when the fraction of fetal DNA is less than 4% (8, 9). Sequencing depth becomes a major issue for the assessment of aneuploidy in cell-free DNA (cfDNA) from patients with cancer, where the fraction of DNA derived from cancer cells is often much less than 1% of the total input DNA (10).
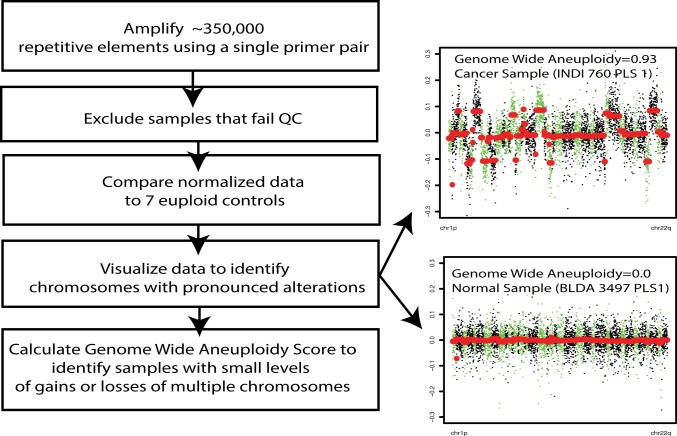
Amplicon-based methods using sequence-specific primers have been proposed as an alternative to WGS for the assessment of aneuploidy (11–13). Amplicon-based protocols offer many advantages over WGS (or exome sequencing), including a simpler workflow that does not require library construction, a reduced requirement for input DNA, and a simplified computational analysis. Here, we report a substantially improved amplicon-based approach to detect the presence of aneuploidy named the Repetitive Element AneupLoidy Sequencing System (RealSeqS). Using a single PCR primer pair, RealSeqS amplifies ∼350,000 genomic loci with an average size of 43 bp spread throughout the genome (Fig. 1).
Fig. 1.
Overview of RealSeqS.
Results and Discussion
Primer Development.
The FAST-SeqS approach described in Kinde et al. (11) was the first aneuploidy detection method to use a single primer pair to amplify numerous repetitive long interspersed nucleotide elements spread throughout the genome. However, due to the low genomic density of these amplicons (a total of 38,000 across the entire genome), its power to detect focal amplifications and deletions (<5 Mb) was limited. Additionally, FAST-SeqS amplicons ranged in size from 120 to 145 bp; this was suboptimal for assessing cfDNA, which has an average size of ∼140 bp. Accordingly, FAST-SeqS was only able to detect aneuploidy in 22% of liquid biopsy samples containing more than 1% of tumor-derived DNA (14).
Based on the limitations described above, we attempted to identify a single primer pair that could amplify far more than 38,000 amplicons of a size far less than 120 to 145 bp. To generate a list of candidate primers, we first calculated the frequency of all possible 6-mers (46 = 4,096) within the RepeatMasker track of hg19. Next, we calculated the frequency of all possible 4-mers (44 = 256) within 75 bp upstream or downstream of the 6-mers. Joining the 6-mers with the 4-mers (SI Appendix, Fig. S1) generated 2,097,152 candidate pairs. We narrowed these pairs based on the number of unique genomic loci expected from their PCR-mediated amplification, the average size between the 6-mer and its corresponding 4-mers, and the distribution of these sizes, aiming for a unimodal distribution. These filtering criteria generated seven potential k-mer pairs, leading to the design of seven primer pairs that incorporated these k-mer pairs at their 3 ends (SI Appendix, Table S1). Two of these primer pairs (REAL1 and REAL2) outperformed the remaining five primers when experimentally assessed by the number of unique loci that were amplified and the size distribution of the amplicons. After further experimental testing of REAL1 and REAL2 on 100 euploid peripheral blood samples, the REAL1a primer pair was chosen for the experiments reported herein (SI Appendix, Fig. S1). REAL1a amplifies up to 745,154 unique amplicons residing within various repetitive elements defined by the RepeatMasker track (Dataset S1). Off-target amplicons outside the predefined regions were not analyzed. The average amplicon size of REAL1 was 43 bp, not including the length of the forward and reverse primers (SI Appendix, Fig. S2). The average number of amplicons observed in cfDNA was ∼350,000 (SI Appendix, Fig. S3). Details of the primer selection methods, experimental procedures, and analytic techniques are described in SI Appendix, SI Materials and Methods and Figs. S3–S5.
Comparison with Other Next Generation Sequencing Technologies.
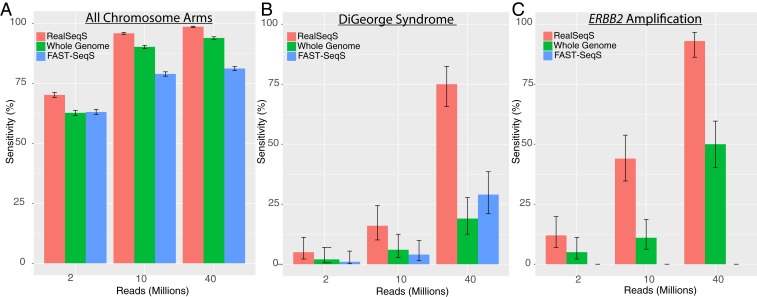
In the most common form of NIPT, detection of a gain or loss of a chromosome (e.g., chromosome 21 in Down syndrome) is the goal. We used WGS (SI Appendix, Table S2), FAST-SeqS (SI Appendix, Table S3), and RealSeqS (SI Appendix, Table S4) to assess performance on samples for DNA admixtures typically encountered in NIPT (i.e., when the fraction of fetal DNA was 5%). For this purpose, we used actual data obtained with the three methods and then added a defined number of reads from various chromosome regions from the same samples to simulate what would happen if there was aneuploidy in these regions. The pseudocode used to generate these in silico-simulated samples is described in SI Appendix, Figs. S6 and S7. We calculated performance using a frequently used z score that compares the observed fraction of reads on a particular chromosome arm with the average fraction of reads from a normal panel divided by the SD in the normal panel (SI Appendix, SI Materials and Methods). We reported results in total reads needed for all three approaches assuming single-end 100-bp reads and accounting for differences in alignment rates and filtering criteria typically used (SI Appendix, Tables S2–S4). While this approach relies on numerous assumptions, RealSeqS consistently achieved higher sensitivity at lower amounts of sequencing. For example, RealSeqS had 99% sensitivity (at 99% specificity) for monosomies and trisomies at a 5% cell fraction, while WGS and FAST-SeqS had 94 and 81% sensitivities, respectively (Fig. 2A).
Fig. 2.
Detection of aneuploidy using next generation sequencing technologies. Sensitivities were calculated at 99% specificity. Error bars represent 95% CIs. (A) Comparison of sensitivity for monosomies and trisomies across all 39 nonacrocentric chromosome arms at 5% cell fraction. (B) Comparison of sensitivity for the 1.5-Mb DiGeorge deletion on 22q at 5% cell fraction. (C) Comparison of sensitivity for a 20-copy ERBB2 focal amplification at 1% cell fraction.
Another important aspect of assays for copy number variation is the detection of relatively small regions which are deleted or amplified. For example, DiGeorge syndrome deletions are often as small as 1.5 Mb (15). For data simulating a 5% deletion-containing cell fraction, RealSeqS had 75.0% sensitivity for the 1.5-Mb DiGeorge deletion (at 99% specificity), while WGS and FAST-SeqS had 19.0 and 29.0% sensitivity, respectively (Fig. 2B; example in Fig. 3 A and B).
Fig. 3.
Examples of plasma samples with focal deletions or amplifications. (A) RealSeqS data on a plasma sample from a normal individual with an ∼3-Mb deletion of chromosome 22, characteristic of DiGeorge syndrome. Note that many patients with microdeletions at this locus have mild signs and symptoms and are clinically undetected. (B) RealSeqS data on a typical plasma sample from a normal individual, showing no deletion at the DiGeorge locus. (C) RealSeqS data on a plasma sample from a patient with cancer showing a 2.5-Mb focal amplification that includes the ERBB2 locus on chromosome 17q. (D) RealSeqS data on a typical plasma sample from a normal individual, showing no amplification at the ERBB2 locus.
The detection of amplifications, such as those on ERBB2 in breast cancer, is critical for deciding whether patients should be treated with trastuzumab or other targeted therapies. Following the same strategy described above, we generated in silico-simulated samples with focal amplifications of the ∼42-kb ERBB2 gene (20 copies) for WGS, FAST-SeqS, and RealSeqS. RealSeqS could detect such amplifications in the in silico-simulated samples with significantly less sequencing than could WGS or Fast-SeqS. For a 1% cell fraction, RealSeqS had a 91.0% sensitivity, while WGS had 50.0% (Fig. 2C; example in Fig. 3 C and D). FAST-SeqS did not have enough spatial coverage in this genomic region to detect ERBB2 amplifications.
Reduced Input DNA.
Reliably detecting aneuploidy in only a few picograms of DNA is necessary for preimplantation diagnostics as well as forensic applications. In preimplantation diagnosis, a few cells picked from a blastocyst are used to assess copy number variations, such as those responsible for Down syndrome. To test the limit of detection of RealSeqS with respect to input DNA, we analyzed genomic DNA (gDNA) from 10 trisomy 21 samples at input DNA concentrations ranging from 3 to 34 pg (Dataset S2) and 6 euploid controls. Trisomy 21 was detected in all of these samples, even those from 3 pg of DNA, representing half of a diploid cell. No chromosome arms other than chromosome 21 were found to be aneuploid in the trisomy 21 samples. Additionally, no chromosome arms, including chromosome 21, were found to be aneuploid in the euploid controls used in these experiments. The reduced requirement for input DNA also enables retrospective testing of samples from biobanks for either aneuploidy or identification purposes (using single-nucleotide polymorphisms within the amplified repeated sequences) (SI Appendix, SI Materials and Methods).
Detection of an Admixture of Cellular DNA with cfDNA.
DNA that has leaked out of leukocytes, either during phlebotomy or preparation of plasma DNA, can “contaminate” cfDNA. Plasma “cell-free” DNA is relatively short, with the vast majority less than 200 bp in size (16, 17). In contrast, lysed “cellular DNA” from leukocytes is much longer than 200 bp and can complicate the interpretation of any cfDNA analysis, including the analysis of aneuploidy. For example, it can mask aneuploidy, particularly when the contribution of DNA from the tumor is minor compared with the fraction derived from other sources of cfDNA. RealSeqS enables the detection of leukocyte DNA contamination by virtue of the differently sized amplicons generated with REAL1 primers. This led to two simple methods to detect contaminating leukocyte DNA. We first calculated the fraction of reads within amplicons >50 bp in size. If this fraction was >8.15%, we considered it contaminated with leukocyte DNA (SI Appendix, SI Materials and Methods and Fig. S4). For a more sensitive analysis of gDNA contamination, we identified 1,241 amplicons typically present in gDNA but not in cfDNA (Dataset S3). Reads at these amplicons thereby indicated leukocyte contamination in plasma samples (Dataset S3). Through experimental mixing of leukocyte DNA with cell-free plasma DNA from the same individual, we were able to demonstrate that samples containing >4% of leukocyte DNA could be detected by this metric (SI Appendix, Table S5). For validation of this result, we also evaluated an independent set of 457 peripheral blood samples (leukocyte DNA) and 2,181 plasma samples (cfDNA). All 457 leukocyte DNA samples had higher coverages of the 1,241 amplicons than the 2,181 cfDNA samples.
Detection of Aneuploidy in Liquid Biopsies.
DNA from cancer cells is shed into the bloodstream, fostering the analysis of cfDNA in plasma (“liquid biopsies”) to detect the presence of cancers. Several features of cancer DNA, including point mutations, aberrant DNA methylation, and aneuploidy, have been used to assess liquid biopsies (10, 18–20). Because aneuploidy is a feature of virtually every cancer type (>90%), it is well suited for this purpose (14, 21).
In preliminary experiments with plasma or peripheral white blood cells (WBC) DNA from normal individuals prior to the evaluation of any samples reported in this paper, we noticed that certain chromosome arms were heavily enriched for the type of amplicons that were most variable among patients. The proportional representation of these chromosome arms was highly correlated (SI Appendix, Table S7). We therefore designed a quality control (QC) metric that could be used to identify outlier samples (explicitly represented in SI Appendix, Fig. S5). We then applied this prespecified QC metric (Fig. S5) as well as the prespecified length metric (Fig. S4) to the 2,319 plasma samples analyzed in this work. This resulted in exclusion of 88 samples (45 samples or 3.2% of the 1,393 samples from normal controls and 43 samples or 4.6% of 926 samples from patients harboring surgically resected cancers). Aneuploidy was assessed in the remaining 2,231 samples.
RealSeqS was used to detect aneuploidy in cell-free plasma DNA from the 883 patients harboring cancers of eight different cancer types: ovary, colorectum, esophagus, liver, lung, pancreas, stomach, and breast (Table 1). Each plasma sample was given a RealSeqS score based on a machine learning-based algorithm described in SI Appendix, SI Materials and Methods. Machine learning scores were generated using 10-fold cross-validation (SI Appendix, SI Materials and Methods) so that all 2,231 samples had a corresponding genome-wide aneuploidy score. Aneuploidy was scored in the samples from cancer patients at a threshold of 99% specificity derived from the analysis of the 1,348 plasma samples from healthy individuals (Datasets S4 and S5). The plasma samples from cancer patients had previously been analyzed for somatic point mutations and small insertions or deletions using a sensitive mutation detection technique based on 61 genomic regions that are frequently altered in cancer (10). Mutations in the plasma samples were also scored at a threshold of 99% specificity.
Table 1.
Description of the cancer samples evaluated with RealSeqS
| Breast | Colorectum | Esophagus | Liver | Lung | Ovary | Pancreas | Stomach | |
| Stage I | 27 | 71 | 5 | 5 | 41 | 9 | 4 | 18 |
| Stage II | 96 | 174 | 27 | 15 | 26 | 3 | 72 | 26 |
| Stage III | 51 | 101 | 9 | 18 | 27 | 36 | 6 | 16 |
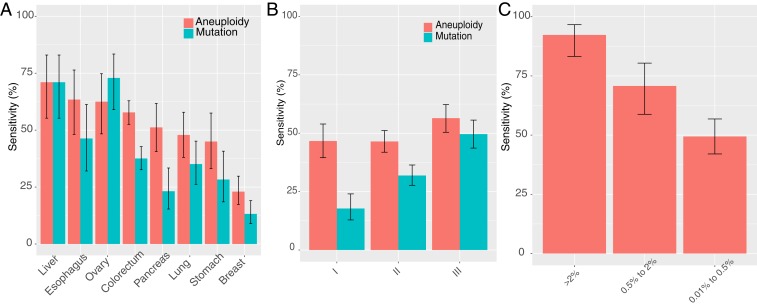
Overall, we found that aneuploidy was detected more commonly than mutations in plasma samples from cancer patients (49 and 34% of 883 samples, respectively; P < 10−20, one-sided binomial test) (Fig. 4A). With respect to tissue type, aneuploidy was detected more commonly than mutations in samples from patients with cancers of the esophagus, colorectum, pancreas, lung, stomach, and breast (all P values < 0.01); less commonly in ovary (P = 0.048); and equally commonly in liver cancer (Fig. 4A). With respect to stage, aneuploidy was detected more commonly than mutations in all stages, especially stages I and II (Fig. 4B) (P values < 10−9).
Fig. 4.
Detection of cancer in liquid biopsies from samples with nonmetastatic cancers of eight different types. Sensitivities were calculated at 99% specificity during cross-validation. Error bars represent 95% CIs. (A) Comparison of aneuploidy status as assessed by RealSeqS with somatic mutations status with respect to tumor type. (B) Comparison of aneuploidy status as calculated by RealSeqS with somatic mutations status with respect to cancer stage. (C) RealSeqS sensitivity for plasma samples containing various amounts of tumor-derived DNA. The amount of tumor-derived DNA was estimated by the mutant allele frequency of driver gene mutations present in the plasma sample and indicated on the x axis (10).
We then assessed whether the sensitivity for detecting aneuploidy was higher in samples that had a higher concentration of tumor-derived DNA. We considered this as an important “sanity check” as any type of liquid biopsy metric should reflect the amount of tumor-derived DNA in the plasma. There were 302 samples in which the mutant allele fraction had been determined by the analysis of mutations that were present in plasma (10). Aneuploidy was detected in 92% of 65 samples that had mutant allele fraction ≥2%, 71% of 65 samples with mutant allele fractions of 0.5 to 2%, and 49% of 172 samples with mutant allele frequencies ranging from 0.01 to 0.5% (Fig. 4C). The differences in aneuploidy among these three classes of samples were significant (P < 10−3, one-sided binomial test). The expectation that aneuploidy should be related to an orthogonal measure of circulating tumor DNA was thus confirmed.
Mutations in the plasma originating from clonal hematopoiesis of indeterminant potential (CHIP), rather than from cancer cells, have confounded previous analyses of mutations in cfDNA. This confounder was mitigated with aneuploidy detection; 0 of 17 samples that harbored CHIP mutations in both plasma and leukocytes scored positively for aneuploidy via RealSeqS. We also tested leukocyte DNA from 18 patients whose plasma samples scored positive for aneuploidy with RealSeqS. Only one of these leukocyte samples was aneuploid as assessed by RealSeqS.
We then scored the set of 88 samples that failed quality control. The rate of aneuploidy was much higher in this cohort for both the normal controls as well as the patients with cancers. In this cohort, 8 of 45 normal controls and 31 of 43 patients with cancers were called aneuploid using the 99% specific threshold defined above (Dataset S5).
Combining Tests.
Aneuploidy was detected in 242 (42%) of the plasma samples in which no mutations were detected in the study of Cohen et al. (10). Conversely, mutations were detected in 112 (25%) of the plasma samples in which aneuploidy was not detected. In combination, either aneuploidy or mutations could be detected in 525 (61%) of the 883 plasma samples while still maintaining 99% specificity in the normal samples (Fig. 5A). Further increases in sensitivity could be achieved by combining aneuploidy with elevated levels of protein biomarkers. For this purpose, eight standard protein biomarkers were evaluated in the 883 cancer samples and 812 normal samples as described in ref. 10. We scored all plasma samples using 10 iterations of 10-fold cross-validation using the logistic regression model. Due to multiple iterations of cross-validation, we maintained an aggregate specificity of 99% (i.e., no more than 1% of the 8,120 normal samples [812 normal samples repeated in 10 different iterations] were scored positive in the combination assay) (Datasets S6–S8 and SI Appendix, Table S6). In the plasma samples of patients harboring cancers of seven tissue types (liver, ovary, pancreas, esophagus, stomach, colorectal, and lung), the sensitivity ranged from 77 to 97%, while in breast cancers, it was lower (38%) (Fig. 5).
Fig. 5.
Multianalyte tests including RealSeqS. (A) The sensitivity of a test incorporating aneuploidy, mutations, and abnormally high levels of eight proteins was compared with that of a test comparing only aneuploidy + proteins or only mutations and proteins. All sensitivities were calculated at an aggregate of 99% specificity (i.e., only 1% of the plasma samples were positive for aneuploidy, mutations, or proteins in the test incorporating aneuploidy, mutations, and proteins using 10 iterations of 10-fold cross-validation). (B) Receiver operating characteristic (ROC) curves for the various single-analyte and multianalyte tests.
Screening tests do not exist for five of the eight cancer types shown in Fig. 5. Our results show that more than 75% of these cancer cases could be detected using a combination test incorporating aneuploidy, mutations, and protein biomarkers. This performance is likely an underestimate of the maximum possible sensitivity: more mutations might have been detected if more amplicons were sequenced, and additional aneuploidy might have been identified at a greater sequencing depth. However, in practice there must be a balance between sensitivity and cost, thus limiting the amount of sequencing that can be performed in a screening setting.
Several limitations of our study should be acknowledged. The study does not compare RealSeqS against WGS and FAST-SeqS on the identical samples. Comparison of each of these technologies is challenging due to differences in input DNA requirements, sequence coverage requirements, statistical methods used to evaluate the sequencing data, and algorithms used to integrate chromosomal aneuploidies into a single predictive score. Due to the large number of samples in this study, the cost required to analyze all 2,231 samples with the three next generation sequencing (NGS) technologies was prohibitive. However, we performed RealSeqS on 21 highly aneuploid plasma samples from cancer patients and 173 plasma samples from normal individuals who had previously been analyzed by FAST-SeqS. RealSeqS and FAST-SeqS had high concordance among these 193 samples (Dataset S9). RealSeqS scored all 21 cancer samples as aneuploid and 171 of 173 normal samples as euploid. FAST-SeqS scored all 21 cancer samples as aneuploid and 172 of 173 normal samples as euploid. Because the amplicons evaluated by RealSeqS were nearly completely distinct from those amplified by FASTSeqS, this concordance provides an orthogonal measure of reliability of both assays.
To further confirm that the chromosome arms scored as aneuploid by RealSeqS were indeed aneuploid, we compared chromosomal gains or losses in the plasma with those observed in primary tumors from the same patients. If RealSeqS data indicating aneuploidy were “real,” one would expect that those chromosome arms exhibiting gains in plasma would also exhibit gains in the corresponding primary tumors. Similarly, one would expect that those chromosome arms exhibiting losses in the plasma would also exhibit losses in the corresponding primary tumors. We were able to perform this analysis in 243 instances (214 patients) in which chromosome arm losses or gains were significant (z scores: z > 4 or z < −4) in plasma DNA (SI Appendix, SI Materials and Methods). Of these 243, 188 (77%) were found to be concordant in their respective tumors (SI Appendix and Dataset S10). Note that concordance was directional; if a gain of a chromosome arm was found in the plasma, a gain (rather than a loss) had to be identified in the primary tumor and vice versa.
Even though no patients had metastatic disease on study entry, most individuals were diagnosed on the basis of symptoms. In a true screening setting, patients would likely have less advanced disease resulting in reduced sensitivity. Our healthy controls were not age or gender matched. When moving to an age-matched screening setting, a small number of individuals without cancer might have inflammatory or other diseases that could decrease the reported specificity. Though cross-validation is frequently used to assess robust performance, cross-validation is not as reliable as a completely independent validation set; we did not use a completely independent validation set. Additionally, the RealSeqS inclusion criteria for quality control were based on preliminary pilot experiments from a small number of plasma and WBC samples. Future studies may determine that our criteria are too restrictive and could be relaxed to analyze samples with gDNA contamination. Last, the proportion of cancers of each type in our cohort was purposefully not representative of those in the United States as a whole so as to be able to assess a reasonable number of each of the eight cancer types given the resources that we had available. To actually establish the clinical utility of RealSeqS and to demonstrate that it can save lives, prospective studies of all incident cancer types in a large population will be required.
In summary, RealSeqS is exceedingly simple to perform, requires only a single primer pair, and is relatively sensitive and cost effective (∼$100 per assay). We anticipate that it will be used to assess aneuploidy in a variety of clinical contexts.
Materials and Methods
Detailed materials and methods are available in SI Appendix, SI Materials and Methods. Plasma was purified from healthy individuals and patients with cancer using Qiagen kit catalog #937556 (QIAsymphony DSP Circulating DNA Kit) or Biochain kit catalog #K5011625MA. All individuals participating in the study provided written informed consent after approval by the institutional review board (IRB) at the patients’ participating institutions. Their demographic information is included in Dataset S6. The full study protocol was approved by the Johns Hopkins IRB.
Data Availability.
Summaries of the sequencing data are provided in Datasets S4 and S5. All code used in the manuscript is available in a Zenodo repository, https://doi.org/10.5281/zenodo.3656943.
Supplementary Material
Acknowledgments
This work was supported by the Lustgarten Foundation for Pancreatic Cancer Research; The Marcus Foundation; Richard W. TeLinde Endowment; The Virginia and D. K. Ludwig Fund for Cancer Research; The Commonwealth Fund; a Burroughs Wellcome Career Award for Medical Scientists; The Honorable Tina Brozman Foundation; the Gray Foundation; the Conrad N. Hilton Foundation; the Rolf Foundation; The John Templeton Foundation; NIH Grant T32-GM007309; and National Cancer Institute Grants U01CA230691-01, P50CA228991, U01CA200469, U01 CA152753, R37 CA230400-01, CA62924, CA210170, and CA208723. All sequencing was performed in the The Sol Goldman Sequencing Facility at Johns Hopkins.
Footnotes
Competing interest statement: K.W.K., N.P., and B.V. are founders of, hold equity in, and are consultants to Thrive and Personal Genome Diagnostics. K.W.K. and N.P. are on the Board of Directors of Thrive. K.W.K. and B.V. are consultants to Sysmex, Eisai, and CAGE Pharma. B.V. is also a consultant to Nexus, and K.W.K., N.P., and B.V. are consultants to Neophore. C.B. is a consultant to Depuy-Synthes. C.T. is a paid consultant to Thrive and Bayer. C.D. is a consultant to Thrive. The companies named above as well as other companies have licensed previously described technologies related to the work described in this paper from Johns Hopkins University. C.D., J.D.C., R.K., C.B., K.W.K., N.P., and B.V. are inventors on some of these technologies. Licenses to these technologies are or will be associated with equity or royalty payments to the inventors as well as to Johns Hopkins University. Additional patent applications on the work described in this paper may be filed by Johns Hopkins University. The terms of all of these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. N.R. is a cofounder and Chief Scientific Officer of Inivata.
Data deposition: Summaries of the sequencing data are provided in Datasets S4 and S5. All code used in the manuscript is available in a Zenodo repository, https://doi.org/10.5281/zenodo.3656943.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910041117/-/DCSupplemental.
References
- 1.Raman L., Dheedene A., De Smet M., Van Dorpe J., Menten B., WisecondorX: Improved copy number detection for routine shallow whole-genome sequencing. Nucleic Acids Res. 47, 1605–1614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dheedene A., et al. , Implementation of non-invasive prenatal testing by semiconductor sequencing in a genetic laboratory. Prenat. Diagn. 36, 699–707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deleye L., et al. , Shallow whole genome sequencing is well suited for the detection of chromosomal aberrations in human blastocysts. Fertil. Steril. 104, 1276–1285.e1 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Liang D., et al. , Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J. Mol. Diagn. 16, 519–526 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Leary R. J., et al. , Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 4, 162ra154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho E.-H., Whole genome sequencing based noninvasive prenatal test. J. Genet. Med. 12, 61–65 (2015). [Google Scholar]
- 7.Suzumori N., et al. ; Japan NIPT Consortium , Fetal cell-free DNA fraction in maternal plasma is affected by fetal trisomy. J. Hum. Genet. 61, 647–652 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Grömminger S., et al. , Fetal aneuploidy detection by cell-free DNA sequencing for multiple pregnancies and quality issues with vanishing twins. J. Clin. Med. 3, 679–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolaides K. H., Syngelaki A., Ashoor G., Birdir C., Touzet G., Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am. J. Obstet. Gynecol. 207, 374.e1–6 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Cohen J. D., et al. , Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinde I., Papadopoulos N., Kinzler K. W., Vogelstein B., FAST-SeqS: A simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLoS One 7, e41162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasso C., et al. , Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J. Mol. Diagn. 17, 53–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan C., et al. , A multiplex droplet digital PCR assay for non-invasive prenatal testing of fetal aneuploidies. Analyst (Lond.) 144, 2239–2247 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Douville C., et al. , Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs). Proc. Natl. Acad. Sci. U.S.A. 115, 1871–1876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Packham E. A., Brook J. D., T-box genes in human disorders. Hum. Mol. Genet. 12, R37–R44 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Yu S. C., et al. , Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc. Natl. Acad. Sci. U.S.A. 111, 8583–8588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Underhill H. R., et al. , Fragment length of circulating tumor DNA. PLoS Genet. 12, e1006162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder M. W., Kircher M., Hill A. J., Daza R. M., Shendure J., Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan J. C. M., et al. , Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Sun K., et al. , Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–E5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knouse K. A., Davoli T., Elledge S. J., Amon A., Aneuploidy in cancer: Seq-ing answers to old questions. Annu. Rev. Cancer Biol. 1, 335–354 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summaries of the sequencing data are provided in Datasets S4 and S5. All code used in the manuscript is available in a Zenodo repository, https://doi.org/10.5281/zenodo.3656943.