Significance
Most protein degradation in eukaryotic cells is catalyzed by the 26S proteasome, which digests proteins marked for destruction by a chain of ubiquitin molecules. Typically, 26S proteasomes are present in cells as inactive particles that can become active when a ubiquitylated substrate binds. Proteasomes are usually viewed as containing a characteristic set of subunits, but, in cells, many important proteins associate transiently with proteasomes to deliver substrates or provide additional activities. Here, we show that a major class of proteasome-binding proteins that contain a ubiquitin-like (Ubl) domain can also enhance the 26S proteasome’s multiple enzymatic activities that are critical for protein degradation. Thus Ubl-containing proteins, in addition to their other functions, have this unexpected regulatory role.
Keywords: Rad23, Parkin, Usp7, Bag6, Ddi2
Abstract
During protein degradation by the ubiquitin–proteasome pathway, latent 26S proteasomes in the cytosol must assume an active form. Proteasomes are activated when ubiquitylated substrates bind to them and interact with the proteasome-bound deubiquitylase Usp14/Ubp6. The resulting increase in the proteasome’s degradative activity was recently shown to be mediated by Usp14’s ubiquitin-like (Ubl) domain, which, by itself, can trigger proteasome activation. Many other proteins with diverse cellular functions also contain Ubl domains and can associate with 26S proteasomes. We therefore tested if various Ubl-containing proteins that have important roles in protein homeostasis or disease also activate 26S proteasomes. All seven Ubl-containing proteins tested—the shuttling factors Rad23A, Rad23B, and Ddi2; the deubiquitylase Usp7, the ubiquitin ligase Parkin, the cochaperone Bag6, and the protein phosphatase UBLCP1—stimulated peptide hydrolysis two- to fivefold. Rather than enhancing already active proteasomes, Rad23B and its Ubl domain activated previously latent 26S particles. Also, Ubl-containing proteins (if present with an unfolded protein) increased proteasomal adenosine 5′-triphosphate (ATP) hydrolysis, the step which commits substrates to degradation. Surprisingly, some of these proteins also could stimulate peptide hydrolysis even when their Ubl domains were deleted. However, their Ubl domains were required for the increased ATPase activity. Thus, upon binding to proteasomes, Ubl-containing proteins not only deliver substrates (e.g., the shuttling factors) or provide additional enzymatic activities (e.g., Parkin) to proteasomes, but also increase their capacity for proteolysis.
Degradation of intracellular proteins through the ubiquitin proteasome pathway is controlled through substrate ubiquitylation and by adjusting the activity of 26S proteasomes (1, 2). Unlike the control of ubiquitylation, the regulation of proteasome activity and amounts has received little attention even though they also influence overall rates of protein breakdown under normal conditions and in disease. Rates of degradation may be altered by the transient association of 26S proteasomes with proteins that deliver ubiquitylated substrates, provide additional enzymatic activities, or enhance its degradative capacity. For example, although many ubiquitylated proteins directly bind to proteasomes and are degraded, some proteasome-associated proteins (e.g., Rad23) function as “shuttling factors” that ferry ubiquitylated substrates to proteasomes (3–8). Other proteasome-associated proteins are enzymes that modify the ubiquitin chains on substrates either by adding and extending chains (e.g., UBE3C/Hul5, PRKN/Parkin, or the BAG6 complex) (9–11) or by catalyzing the deubiquitylation (e.g., USP14, UCHL5/Uch37, and USP7/HAUSP) (9, 12). In addition to modifying the ubiquitylated substrate, which influences its likelihood of degradation, some 26S-associated proteins can also alter the proteasomes’ degradative capacity, such as the phosphatase UBLCP1 (13) or the ubiquitin ligase UBE3C (14, 15).
Many proteins that transiently bind to 26S proteasomes contain a ubiquitin-like (Ubl) domain. This domain, which is present in over 60 human proteins, is defined by its structural similarity to ubiquitin. Like ubiquitin and its homologs (e.g., SUMO and Nedd8), the Ubl domain contains five β-sheets surrounding an α-helix in what is called a β-grasp fold (16). Unlike ubiquitin-family members, Ubl-containing proteins cannot be conjugated to other proteins. Instead, the Ubl domain promotes noncovalent binding to other proteins, especially the 26S proteasome (17). In yeast, the fusion of any yeast Ubl domain to a loosely folded protein will target it to proteasomes for degradation without ubiquitylation (18).
Recent tomographic electron microscopy of cultured neurons indicated that about two-thirds of proteasomes appear not to be engaged in conjugate degradation (19). This finding supports the model that most 26S particles are in a latent form and must be activated. The proteasomes’ degradative capacity can be stimulated by phosphorylation (20, 21) and by specific proteins that associate with it, such as PSME4/PA200/Blm10 (22, 23) or ZFAND5 (24), and by the binding of ubiquitylated proteins (25–27). When bound to ubiquitylated proteins, the proteasome’s 19S regulatory complex assumes a more active form in which the substrate channel through the six ATPases widens and aligns with the gated entry channel into the 20S particle, which also opens (28–30). Consequently, peptide entry and hydrolysis by the core particle’s chymotrypsin-like, trypsin-like, and caspase-like sites increase simultaneously (31). In contrast to small peptides, proteins are efficiently degraded by 26S proteasomes only if they contain both a ubiquitin chain and a loosely folded region (32, 33). If a ubiquitylated protein fulfilling these criteria binds proteasomes, proteasomal the adenosine 5′-triphosphate (ATP) hydrolysis rate also increases (34), driving the unfolding and translocation of the substrate into the 20S core particle (2). Although the increased peptide hydrolysis and the associated changes in 26S structure upon substrate binding are easily reversed (35), the increased ATP hydrolysis leads to a tighter association of the ubiquitylated substrate with the proteasome and induces further structural changes that favor translocation that commits the protein to degradation (2, 35). Thus, the rate of ATP consumption determines the rate of degradation (36, 37).
The initial activation of proteasomes by substrates is triggered by the association of the ubiquitin chains with the catalytic domains of the deubiquitylases (DUBs) Usp14 (Ubp6 in Saccharomyces cerevisiae) or Uch37 (34). In the absence of ubiquitin chains, Usp14/Ubp6 allosterically inhibits multiple proteasome activities (38–41). This inhibition is reversed by the binding of ubiquitin chains or ubiquitin aldehyde (a transition-state inhibitor of Usp14’s active site). Additionally, Kim and Goldberg (42) recently showed that the Ubl domain of Usp14 by itself can induce the same changes in activity induced by binding a ubiquitin chain: 1) increased hydrolysis of small peptides, 2) increased disassembly of ubiquitin chains by Rpn11, 3) increased ATPase activity, provided that a loosely folded protein (e.g., casein) is also present to mimic a protein substrate, and, consequently, 4) increased rates of degradation of both ubiquitylated and nonubiquitylated proteins. These findings imply that proteasome activation by the binding of ubiquitin chains to Usp14 is mediated by Usp14’s Ubl domain interacting with Rpn1. However, Usp14 only binds a minority of cellular proteasomes (43). Moreover, the Ubl domains from the shuttling factors Rad23B and Ubiquilin/PLIC could also stimulate proteasome activity, and simply expressing the Ubl domain of Rad23B in cells increases overall protein degradation by the ubiquitin–proteasome pathway (42). These observations led to the proposal that shuttling factors with Ubl-containing proteins may also regulate proteasome function (42) although this activation had not been tested with full-length proteins.
We therefore investigated whether various full-length proteins that contain Ubl domains and bind proteasomes may also stimulate its several activities. In addition to the shuttling factor Rad23B, we used a variety of recombinant Ubl-containing proteins with diverse cellular functions: the shuttling factors Rad23A and Ddi2, the ubiquitin ligase Parkin, the DUB Usp7, the phosphatase UBLCP1, and Ubl4A (part of the chaperone Bag6 complex). We studied the abilities of these various proteins to enhance 26S peptidase activity and ATP hydrolysis, which is directly linked to degradation and more tightly regulated than peptidase activity because stimulating ATPases requires both a ubiquitin chain and an unfolded polypeptide (34, 44). These studies demonstrate an unexpected role for these and presumably other Ubl-containing proteins in regulating proteasomal activity by converting latent proteasomes into active species.
Results
The Shuttling Factors Rad23A and Rad23B Stimulate 26S Peptidase Activity through Rpn1.
The peptidase activity of purified 26S proteasomes is stimulated upon binding a linear ubiquitin chain, a ubiquitin conjugate, or ubiquitin-aldehyde, all of which can activate by binding to Usp14’s catalytic site (25). Ligand-bound Usp14 appears to use its Ubl domain to activate proteasomes as this domain, without the rest of Usp14, causes a similar increase in peptide hydrolysis (42). We confirmed that the Rad23B Ubl domain could also enhance peptide entry and hydrolysis by 26S proteasomes purified from rabbit muscles. The maximal stimulation was a four- to fivefold increase over proteasomes alone, and the EC50 (effective concentration, 50%) for activation was 200 nM (Fig. 1A). By contrast, free ubiquitin, despite its similar amino acid sequence (64%) and remarkably similar structure to Rad23B’s Ubl, did not stimulate peptidase activity (25) (Fig. 1A). Also, neither phosphorylated ubiquitin (phospho-Ser-65), which is produced by the PINK1 kinase when mitochondria are depolarized (45, 46) (Fig. 1A), nor the ubiquitin family member, SUMO2/3, could increase peptide hydrolysis (Fig. 1B).
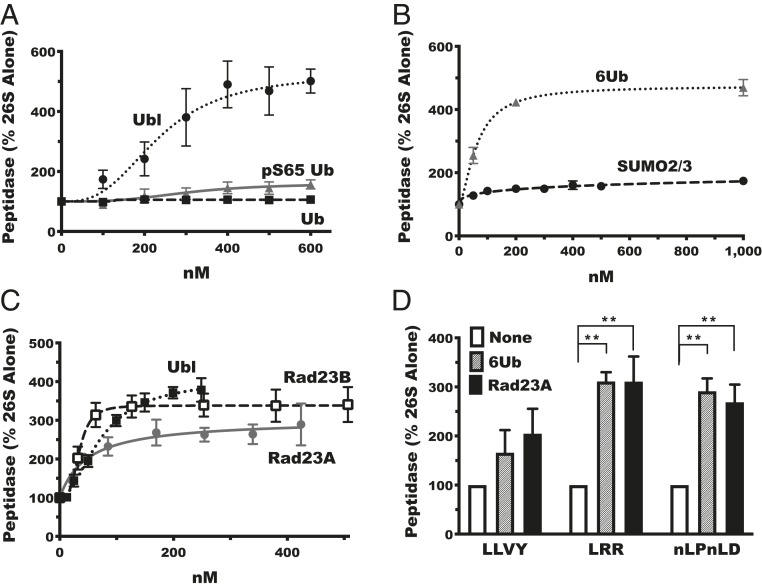
Fig. 1.
The shuttling factors Rad23A and Rad23B stimulate 26S proteasome peptidase activity. (A) The Rad23B Ubl domain (Ubl), but not monomeric ubiquitin (Ub) or phosphorylated monomeric ubiquitin (pS65 Ub), stimulates peptidase activity of purified rabbit muscle proteasomes measured by the hydrolysis of ac-Nle-Pro-Nle-Asp-amc. (B) SUMO2/3, unlike linear hexa-ubiquitin chains (6Ub), does not stimulate peptidase activity. (C) Full-length Rad23A and Rad23B and the Ubl domain of Rad23b (Ubl) stimulate peptidase activity similarly. (D) Linear chains of six ubiquitin (6Ub) and Rad23A stimulate the activity of all three peptidase activities of rabbit proteasomes: chymotrypsin-like activity measured with suc-Leu-Leu-Val-Tyr-amc (LLVY), trypsin-like activity measured with boc-Leu-Arg-Arg-amc (LRR), and caspase-like activity measured with ac-Nle-Pro-Nle-Asp-amc (nLPnLD). Error bars represent SEM (n = 3). **P < 0.01.
Importantly, this activation is not a peculiar feature of this isolated Ubl domain because addition of full-length human Rad23B to the purified 26S proteasomes stimulated its peptidase activity to a similar extent (three- to fourfold), but with higher affinity (EC50 30 nM) than its isolated Ubl domain (Fig. 1C). Likewise, its close homolog, Rad23A, stimulated the proteasomes’ peptidase activity with a similar EC50 although the increase was consistently smaller than with Rad23B (Fig. 1C). Ubiquitin chains activate peptide hydrolysis by inducing large structural rearrangements in the proteasome—widening and aligning the ATPase central channel and opening the gate in the 20S particle’s outer ring (30, 47). Consequently, the entry of peptide substrates increases, and more substrates are cleaved. Accordingly, full-length Rad23A, like linear ubiquitin chains, enhanced hydrolysis of peptides specific for the chymotrypsin-like, trypsin-like, and caspase-like sites (Fig. 1D), presumably by facilitating substrate entry. Interestingly, the relative increases in the caspase-like activities by both Rad23B and Rad23A were consistently larger than the changes in chymotrypsin-like activity, as had been found with linear ubiquitin chains and other activators of gate opening (42). Therefore, to monitor the activation of peptide hydrolysis in subsequent experiments, we followed the cleavage of the fluorogenic peptide ac-Nle-Pro-Nle-Asp-amc by the caspase site.
Activating peptide hydrolysis by ubiquitin chains requires their binding to Usp14, which is present on only 20 to 30% of proteasomes (43). We therefore compared the abilities of Rad23A to enhance peptidase activity of proteasomes isolated from mouse embryonic fibroblasts (MEFs) lacking Usp14 (knockout [KO]) and from wild-type (WT) MEF cells. Rad23A stimulated peptide hydrolysis very similarly in proteasomes from Usp14 KO and wild-type cells (Fig. 2A). Thus, Rad23A stimulates proteasome activity independently of Usp14 and does not act by displacing Usp14, which, in the absence of a substrate, inhibits proteolysis allosterically (39, 40). Moreover, the similar stimulation in Usp14KO and WT cells implies that these Ubl-containing proteins activate primarily by associating with those particles lacking Usp14.
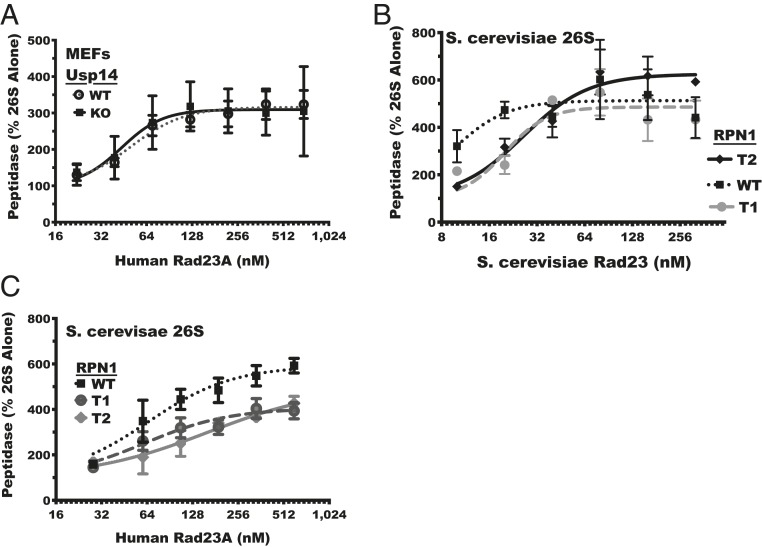
Fig. 2.
Rad23 can interact with multiple binding sites to stimulate peptidase activity. (A) Hydrolysis rates of ac-Nle-Pro-Nle-Asp-amc with Rad23 present relative to those without Rad23A by 26S proteasomes purified from Usp14+/+ (WT) or from Usp14−/−(KO) mouse embryonic fibroblasts (MEFs). Error bars represent SEM (n = 3). (B) Yeast Rad23 activates ac-Nle-Pro-Nle-Asp-amc hydrolysis in proteasomes purified from yeast bearing a deletion of the Rpn10 UIM and an inactivating mutation in the Rpn13 PRU domain but expressing either wild-type Rpn1 (WT) or a T1 mutant Rpn1 (T1), T2 mutant Rpn1 (T2). Error bars represent SEM (n = 3). (C) Human Rad23A activates ac-Nle-Pro-Nle-Asp-amc hydrolysis in proteasomes purified from yeast bearing a deletion of the Rpn10 UIM and an inactivating mutation in the Rpn13 Pru domain but expressing either wild-type Rpn1 (WT) or a T1 mutant Rpn1 (T1), T2 mutant Rpn1 (T2). Error bars represent SEM (n = 3).
The proteasome’s 19S regulatory complex contains at least five different sites that seem capable of binding ubiquitin or Ubl domains. Both PSMD4/Rpn10’s UIM and ADRM1/Rpn13’s PRU domains can bind Rad23 (48, 49), and, in yeast, Rad23’s Ubl domain binds to Rpn1 at a region called T1 (50, 51). However, the Ubl of Ubp6, the yeast homolog of Usp14, binds preferentially to Rpn1 at the T2 site (50), and Usp14’s Ubl seems to activate yeast proteasomes exclusively through this site (42). We tested which of these two Rpn1 regions is important for activation by full-length Rad23 using mutant yeast strains with Rpn10 UIM deleted and Rpn13 PRU domain inactivated. Loss of the Rpn10 UIM and disruption of the Rpn13 PRU domain decreases proteasome binding to GST-Rad23A by about one-third (SI Appendix, Fig. S1). When yeast Rad23 was added to these purified mutant yeast proteasomes, which contain a wild-type Rpn1, Rad23 still activated peptide hydrolysis (Fig. 2B). Thus, yeast, like mammalian, proteasomes are activated by Ubl domain-containing proteins, and the Rpn10 UIM and Rpn13 PRU domains are not required for this activation. We further tested if mutations in yeast Rpn1 that should disrupt the binding of Rad23’s Ubl (T1) or Ubp6’s Ubl binding (T2) reduce the ability of Rad23 to activate peptide hydrolysis. These proteasomes are not generally defective for peptidase activity and hydrolyze nLPnLPD-amc at least as well as, if not better, than the wild-type Rpn1 proteasomes (SI Appendix, Fig. S2). Surprisingly, with mutations of either the T1 or T2 site, the maximal increase in peptide hydrolysis was similar to that in wild-type proteasomes, but higher yeast Rad23 concentrations were necessary to activate peptide hydrolysis (Fig. 2B). Human Rad23A also stimulated yeast proteasomes, but, unlike yeast Rad23, mutation of either T1 or T2 sites in Rpn1 decreased the magnitude of stimulation (Fig. 2C). Thus, both sites on Rpn1 seem to contribute to proteasome activation by yeast Rad23 and human Rad23A.
Various Ubl-Containing Proteins Enhance Proteasomal Peptidase Activity.
Because full-length Rad23A and Rad23B, as well as the Ubl domains of both Usp14 and Rad23B, could activate the purified proteasomes (42), we tested if other Ubl-containing proteins reported to associate with proteasomes could also stimulate its peptidase activity. Parkin is a ubiquitin ligase which, when mutated, can lead to Parkinson’s disease (52, 53). Upon depolarization of the mitochondrial membrane, PINK1 phosphorylates Parkin on its Ubl domain, leading to Parkin’s association with 26S proteasomes (10, 54). Parkin binding has been reported to enhance proteasomal peptidase activity (55) although the concentrations of Parkin needed were not analyzed. Adding full-length Parkin to purified rabbit muscle proteasomes caused a two- to threefold increase in peptide hydrolysis (Fig. 3A) at roughly similar concentrations (EC50 of 70 nM) as Rad23A and Rad23B. Furthermore, deletion of Parkin’s Ubl domain prevented this activation of the peptidase activity (Fig. 3A) while addition of the isolated Parkin Ubl domain stimulated this activity (Fig. 3B). Thus, this effect clearly depends on Parkin’s Ubl domain. Moreover, phosphorylation of the Ubl domain at S65 did not alter its ability to increase peptidase activity (Fig. 3B). However, Parkin, with an R42P mutation in its Ubl domain, which causes early onset Parkinson’s disease (56), induced a much greater increase in peptide hydrolysis than did wild-type Parkin (Fig. 3A).
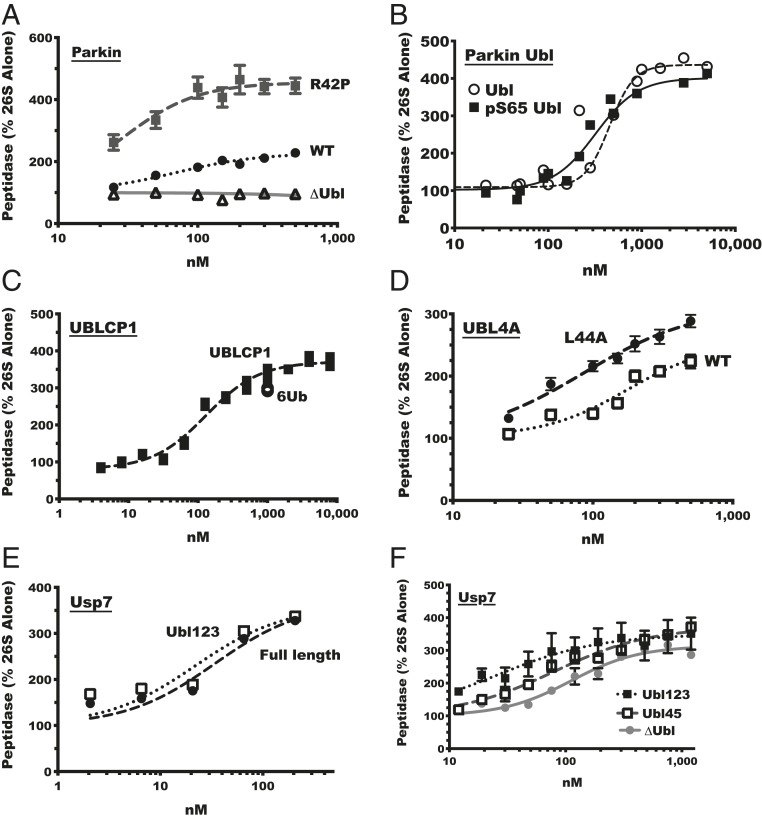
Fig. 3.
Various mammalian Ubl-containing proteins enhance proteasomal peptidase activity. (A) The wild-type ubiquitin ligase Parkin and R42P mutant Parkin, but not ∆Ubl Parkin, stimulate ac-Nle-Pro-Nle-Asp-amc hydrolysis by rabbit muscle 26S proteasomes (n = 3). (B) The Parkin Ubl domain and phosphor-S65 Parkin Ubl stimulate peptidase activity, assayed as in A (n = 2; individual points are shown). (C) The protein phosphatase UBLCP1, like linear hexa-ubiquitin chains (6Ub), stimulates peptidase activity of proteasomes purified from mouse embryonic fibroblast (n = 3; individual points are shown). (D) Wild-type UBL4A (WT) and L44A UBL4A stimulate peptidase activity analyzed as in A (n = 3). (E) The full-length deubiquitylating enzyme Usp7 (WT) and its catalytic domain plus the first three of its five Ubl domains (Ubl123) stimulated peptidase activity as in A. (F) The catalytic domain of Usp7 plus its first three of the five Ubl domains (Ubl123), the last two of its five Ubl domains (UBL45), or no Ubl domains (∆Ubl) stimulated peptidase activity analyzed as in A (n = 3). Error bars represent SEM.
The nuclear protein phosphatase UBLCP1 binds to the proteasome subunit Rpn1 through its Ubl domain (13, 57). Incubation of 26S proteasomes with UBLCP1 was reported to decrease protein degradation and peptidase activity by dephosphorylating proteasomes (and perhaps substrates) (13, 58). However, the possible stimulatory effects of its binding to proteasomes were not analyzed. We therefore tested if wild-type UBLCP1 could activate peptide hydrolysis. We found that UBLCP1, like linear hexa-ubiquitin chains (6Ub), stimulated 26S peptidase activity up to three- to fourfold with an EC50 of 130 nM (Fig. 3C). This direct activation by UBLCP1 binding appears to oppose the reported inhibitory effects of its phosphatase activity (13, 58).
The Bag6 chaperone complex serves important functions in protein quality control by processing of misfolded and misdirected tail-anchored membrane proteins (59). Two components of the Bag6 complex contain Ubl domains, UBL4A and BAG6, and both copurify with proteasomes (11, 60). When UBL4A was added to purified proteasomes, it stimulated peptide entry and hydrolysis twofold (Fig. 3D). Many ubiquitin and Ubl interactions with other proteins involve their hydrophobic surface containing key residues I/L44 and G47. To test if this surface is also important in the activation of proteasomes, we used the L44A mutation in UBL4A. This mutant stimulated 26S peptidase activity as well as, if not better than, wild-type UBL4A (Fig. 3D).
Usp7/HAUSP is an important DUB that regulates the degradation of many cell proteins (e.g., p53) (61) and is one of several DUBs that copurifies with proteasomes (9). Usp7 has the unusual feature of containing five C-terminal Ubl domains (62). Full-length Usp7 increased peptidase activity fourfold with a high affinity (EC50 of 30 nM). A fragment of Usp7 with the catalytic domain (CD) plus the first three of the five Ubl domains (Usp7CD-Ubl123) also stimulated peptide hydrolysis with high affinity (EC50 of 20 nM) (Fig. 3E). This finding suggested that the final two Ubl domains may be superfluous. We therefore compared the stimulatory ability of Usp7CD-Ubl123 and a similar fragment of Usp7’s catalytic domain attached to only the final two Ubl domains (Usp7CD-UBL45). Although Usp7CD-UBL45 had a weaker affinity for proteasomes (EC50 of 80 nM), it still increased the peptidase activity three- to fourfold (Fig. 3F). Surprisingly, even without the Ubl domain, Usp7CD could activate peptide hydrolysis (discussed below).
Rad23B Increases the Number of Proteasomes That Are Active.
Proteasomal peptidase activity is also enhanced by the nonhydrolyzable ATP analog ATPγS. Although this nucleotide is believed to open the gate maximally and to align the ATPases (63, 64), the Ubl domain of Usp14 increased peptidase activity in the presence of ATPγS (42). Similarly, while ATPγS by itself raised peptidase activity at least 10-fold over that seen with adenosine 5′-diphosphate (ADP) or ATP, full-length Rad23B stimulated peptide hydrolysis three- to fourfold regardless of whether ADP, ATP, or ATPγS was present (Fig. 4A). Consequently, Rad23B caused a larger net increase in peptide hydrolysis with ATPγS present.
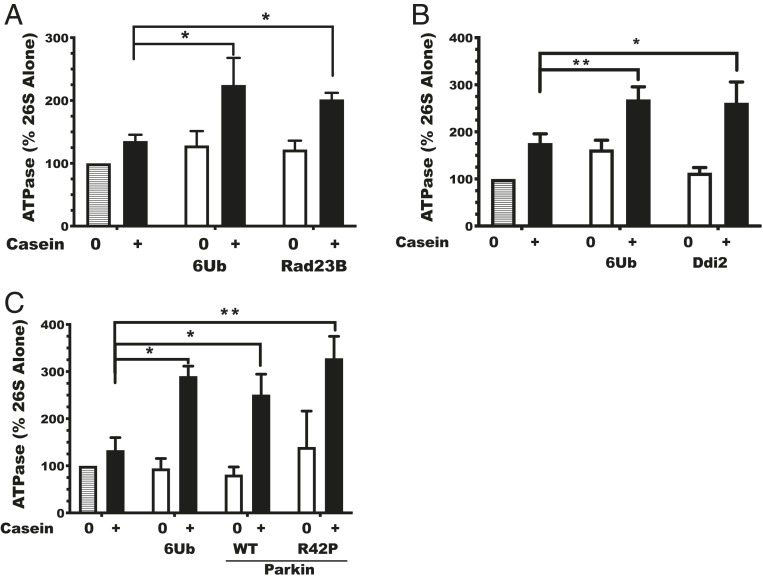
Fig. 4.
Rad23B increases the number of proteasomes activated in peptide hydrolysis. (A) The Rad23B Ubl domain and full-length Rad23B stimulate hydrolysis of ac-Nle-Pro-Nle-Asp-amc purified rabbit muscle proteasomes in the presence of ADP (1 mM), ATP (1 mM), or ATPγS (0.1 mM) (n = 3). *P < 0.05, ***P < 0.001. (B) Representative image of a sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis (PAGE) gel of rabbit muscle proteasomes labeled at PSMB5/β5 with the MVB127 activity probe (66) in a reaction with ATP with and without the Rad23B Ubl domain (n = 2). (C) Quantification of bands signal from each gel represented in B reported as arbitrary fluorescent units (AFUs). Individual points plotted from two independent experiments are shown. (D) Representative image of an SDS/PAGE gel of rabbit muscle proteasomes labeled by the MVB127 activity probe in a reaction with ATPγS with and without the Rad23B Ubl domain (n = 3). (E) Quantification of bands signal from each gel represented in D reported as AFUs. Individual data points shown from three independent experiments. (F) Quantification of MVB127-labeled bands of 26S proteasomes from MEF with Rad23B, Rad23B Ubl domain, or 26S only (n = 3). Error bars represent SEM.
Two mechanisms may explain this remarkable ability of Ubl domains from Rad23 and Usp14 to stimulate peptide hydrolysis beyond what was widely assumed to be the maximal increase of the substrate entry channel (63): 1) The Ubl domain could open the channel through the ATPases and 20S gate wider than has been observed with ATPγS alone (65), or 2) the Ubl domain with ATPγS might activate a population of proteasomes not activated by nucleotide binding alone. In fact, in recent cryo-electron microscopy (cryo-EM) images of purified 26S proteasomes saturated with ATPγS, only a subpopulation of the particles had opened gates (64). To distinguish between these explanations, we used a fluorescent, irreversible inhibitor of the proteasome’s chymotryptic-like site, MVB127 (66). This inhibitor accesses active sites only when the substrate channel and the 20S gate are open. Because MVB127 binds irreversibly, this fluorescent probe essentially allows single turnover analysis. If the Rad23B Ubl domain opens the substrate entry channel wider (mechanism 1), then labeling proteasomes with MVB127 should result in faster initial labeling, without increasing the number of proteasomes ultimately labeled. Alternatively, if the Rad23B Ubl increases the number of activated proteasomes (mechanism 2), then the total number of proteasomes modified by MVB127 should increase.
Total proteasome labeling was low in the presence of ATP (Fig. 4 B and C), but the addition of the Ubl domain increased the number of 26S proteasomes labeled severalfold. With ATPγS, labeling was already high (Fig. 4 D and E), yet the Ubl domain of Rad23B further increased (twofold) maximal labeling. In neither case did the Ubl domain increase the initial rate of labeling. Likewise, when full-length Rad23B was added to 26S proteasomes in the presence of ATPγS, it increased the number of proteasomes labeled by MVB127 (Fig. 4F). Thus, both Rad23B and its Ubl domain activate a latent population of proteasomes not already active in the presence of ATPγS alone. This conclusion does not require hypothesizing a totally new structure for the ATPases’ central channel and the 20S gate, and it suggests that the reason why many proteasomes are not found to be active in cells (19) or after purification is that they lack a bound Ubl-containing protein.
Like Ubiquitin Chains, Ubl-Containing Proteins Together with an Unfolded Protein Activate 26S ATPases.
The stimulation of ATPase activity by a ubiquitylated protein triggers the tighter binding of the ubiquitylated substrate to the proteasome and commits it to degradation (35). Unlike the activation of peptide hydrolysis, which only needs a ubiquitin chain, increasing ATP hydrolysis requires both a ubiquitin chain and a loosely folded polypeptide although the ubiquitin chain and polypeptide need not be covalently linked as they are in most 26S substrates (34). The Ubl domain of Usp14 stimulates ATP hydrolysis by proteasomes when present with casein (42). Therefore, we tested if full-length Ubl-containing proteins could also activate the proteasomal ATPases, with or without a loosely folded protein. Rad23B by itself did not stimulate ATP hydrolysis. Likewise, casein produced only a slight increase in ATPase activity (Fig. 5A), but, when Rad23B and casein were added together to proteasomes, they doubled the rate of ATP hydrolysis (Fig. 5A). This activation by Rad23B thus resembles the activation observed with casein plus a ubiquitin chain (6Ub) (Fig. 5A), ubiquitin aldehyde (34), or the Ubl domain of Usp14 (42).
Fig. 5.
Ubl-containing proteins together with the loosely folded protein casein activate proteasomal ATPases and thus replace the requirement for ubiquitin chains. (A) Rates of ATP hydrolysis as percent activity of 26S proteasomes alone (gray shaded bar). Rates of ATP hydrolysis upon addition of linear hexa-ubiquitin chains (6Ub) or Rad23B with (black bars) or without (white bars) casein. (B) Rates of ATP hydrolysis as percent of the activity of 26S proteasomes alone (gray shaded bar). Rates of ATP hydrolysis upon addition of 6Ub or addition of Ddi2 with (black bars) or without (white bars) casein. (C) Rates of ATP hydrolysis as percent activity of 26S proteasomes alone (gray shaded bar). Rates of ATP hydrolysis upon addition of 6Ub, wild-type Parkin (WT), or Parkin with the R42P mutation (R42P) with (black bars) or without (white bars) casein. Error bars are SEM (n = 3). *P < 0.05; **P < 0.01.
We also tested the effects of another Ubl-containing shuttling factor, Ddi2, on ATP hydrolysis (67). If casein was present, Ddi2 activated the 26S ATPases two- to threefold (Fig. 5B). Similarly, addition of Parkin to proteasomes increased ATP hydrolysis two- to threefold, but only in the presence of casein (Fig. 5C). Furthermore, the R42P mutation in Parkin, which enhanced peptide hydrolysis more potently than wild-type Parkin (Fig. 3A), also seemed to activate ATP hydrolysis to a greater extent than wild-type Parkin, and this effect also required casein (Fig. 5C). This ability of Parkin R42P to activate the proteasomes’ ATPase and peptidase activities was surprising because the isolated Ubl domain with the R42P mutation was reported to be unstructured by NMR (68). However, the large activation of both peptidase and ATPase activities suggests that, within the full-length protein, the R42P Ubl domain maintains some key structural features and behaves similarly (or even better) than other Ubl domain proteins.
Some Proteins Activate Peptide Hydrolysis Even after Deletion of Their Ubl Domain, but ATPase Activation Requires Their Ubl Domain.
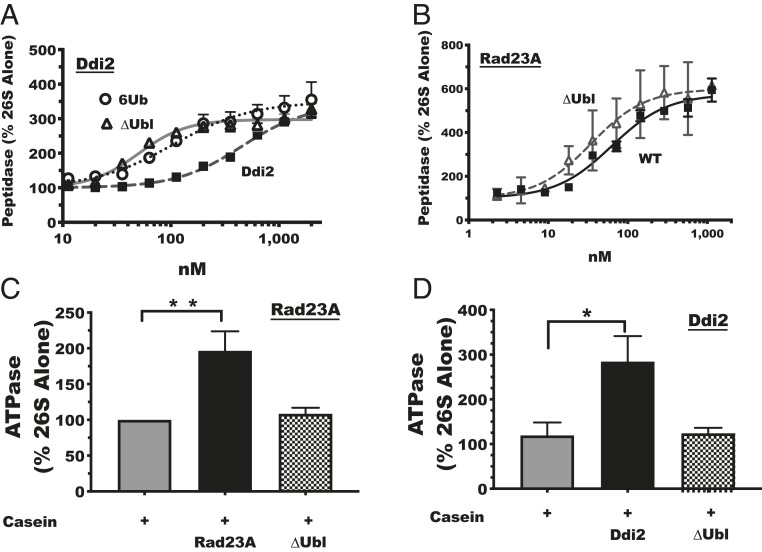
Because Ubl domains by themselves can activate peptide hydrolysis (Fig. 1A) (42) and the Ubl domain of Parkin is essential for its stimulatory effects (Fig. 3A), we were surprised that a fragment of Usp7CD without any Ubl domains, but containing only a short C-terminal peptide (Usp7CD-CTP), could also increase peptide hydrolysis nearly as potently as the other Usp7 fragments (about threefold) (Fig. 3F) and with only slightly less affinity (EC50 of 110 nM). Thus, Usp7’s Ubl domains are clearly not essential to enhance peptide hydrolysis. Therefore, we tested if the Ubl domain was required for the stimulation of peptidase activity by another proteasome-interacting protein, Ddi2. When Ddi2 was incubated with proteasomes, it enhanced peptidase activity three- to fourfold (EC50 of 400 nM) (Fig. 6A). However, a version of Ddi2 lacking its Ubl domain (∆Ubl) still stimulated proteasomes’ peptidase activity threefold, yet with an even greater affinity than wild-type Ddi2 (EC50 of 50 nM) (Fig. 6A).
Fig. 6.
Activation of the 26S ATPases, unlike peptidases, absolutely requires Ubl domains. (A) Full-length Ddi2 (WT), as well as Ddi2 lacking its Ubl domain (∆Ubl), stimulated peptidase activity, as performed in Fig. 3A. 6Ub denotes linear hexa-ubiquitin chains. Error bars are SEM (n = 3). (B) Full-length Rad23A (WT), as well as Rad23A lacking its Ubl domain (∆Ubl), stimulated peptidase activity as in A. Error bars represent the range from two independent experiments. (C) Rates of ATP hydrolysis as the percent of the activity of 26S proteasomes with casein present. Rad23A (black bar) but not Rad23A ∆Ubl (checkered bar) stimulated ATP hydrolysis. Casein without Rad23A (gray bar) did not stimulate ATP hydrolysis. (D) Rates of ATP hydrolysis as percent of 26S proteasomes activity alone. Ddi2 with casein (black bar) but not Ddi2 ∆Ubl with casein (checkered bar) stimulated ATP hydrolysis. Casein without Ddi2 (gray bar) did not stimulate ATP hydrolysis. n = 3. *P < 0.05, **P < 0.01.
Thus, unlike Parkin, which required its Ubl domain to activate peptide hydrolysis (Fig. 3A), Usp7 and Ddi2 must contain an additional region that induces peptidase activity (Figs. 4F and 6A). Because much of our study focused on Rad23’s Ubl domain, we tested if Rad23A required its Ubl domain to stimulate peptide entry and hydrolysis. Surprisingly, Rad23A ∆Ubl resembled wild-type Rad23A in its ability to increase peptidase activity (Fig. 6B). This finding was quite unexpected because Rad23’s Ubl domain is believed to be necessary to bind the proteasome (69–74). Thus, although the Ubl domain is sufficient to enhance peptidase activity, some proteasome-interacting proteins (for example Rad23A, Usp7, and Ddi2) contain a mechanism to activate peptide hydrolysis.
Because of the surprising finding that the Ubl domains of Rad23A and Ddi2 were sufficient but not necessary to enhance proteasomes’ peptidase activity, we tested if the Ubl domains of these proteins are essential for the activation of ATP hydrolysis, which is critical in driving substrate degradation. Even in the presence of casein, the Rad23A ∆Ubl (Fig. 6C) and Ddi2 ∆Ubl (Fig. 6D) proteins did not enhance ATPase activity. Thus, although both shuttling factors enhanced peptide hydrolysis without their Ubl domains, the stimulation of ATP hydrolysis occurred only if their Ubl domains were present. These different structural requirements further indicate that peptidase and ATPase activation are two distinct steps in proteasome activation. Together, these various findings revealed an important feature of many Ubl-containing proteins: their ability to enhance the proteasome’s capacity for proteolysis and increase the likelihood of substrate degradation.
Discussion
This study developed from our earlier discovery that the proteasome’s degradative capacity increases when ubiquitylated proteins bind to the proteasome-associated DUB Usp14 (25, 34) and from our recent finding that Usp14’s Ubl domain by itself can stimulate multiple proteasome activities in a similar fashion to ubiquitin chains (42). Those observations suggested that the Ubl domain of Usp14 mediates the activation of proteasomes triggered by ubiquitin conjugates, and the finding that the Ubl domains of Rad23 and ubiquilin could also stimulate peptidase activity led to the proposal that these shuttling factors and perhaps other proteasome-interacting Ubl-containing proteins might also enhance proteasome activity (42). The human genome encodes over 60 Ubl-containing proteins. Only about 15 Ubl-containing proteins are known to bind to proteasomes, perhaps because the capacity of most of these proteins to associate with proteasomes has not been studied. In yeast, the fusion of any of the Ubl domains in the S. cerevisiae genome can target a protein to proteasomes for degradation (18). The different Ubl domains tested not only represent proteins with a diverse set of functions, but also reflect the structural diversity within Ubl domains (SI Appendix, Fig. S3). Surprisingly, multiple sequence alignments of these domains did not reveal a clear separation of those domains that activate proteasomes from ubiquitin or SUMO2, which do not increase peptidase activity (SI Appendix, Fig. S3). Nevertheless, structural features of the Ubl must be important because a small change in Parkin’s Ubl domain (R42P) produced a large increase in the activation of proteasomes (Figs. 3A and 5C). (Incidentally, unlike the isolated R42P Ubl domain, the full-length Parkin R42P protein must contain the critical structural features allowing activation.) Because all seven proteasome-interacting Ubl-containing proteins studied stimulated peptide hydrolysis two- to fivefold and all of the Ubl-containing proteins we tested also enhanced ATP hydrolysis, the ability to enhance proteasome activity is likely to be a feature of many, perhaps most, mammalian Ubl-containing proteins. However, several surprising features of this activation were uncovered.
Another potent activator of the 26S proteasomes peptidase activity is ATPγS (63, 75, 76), which maintains the ATPase entry channel and the 20S gate in open conformations (26, 63, 77). In fact, ATPγS is believed to maximally open the channel through the ATPases and the gate into the 20S (64, 65). ATPγS and ubiquitin conjugates produce similar structural changes in the 26S, leading to increased peptide hydrolysis (1, 78). Therefore, we were surprised that the largest Ubl-mediated increases in peptidase activity were observed even when proteasomes were already activated by ATPγS. It seems unlikely that a two- to fivefold increase in peptide hydrolysis can be explained by further enlargement of the ATPases’ central channel or the opening in the 20S particle beyond what is evident in recent cryo-EM structures of proteasomes with ATPγS or ubiquitin conjugates bound (28–30, 64). Our experiments with the covalent active-site probe MVB127 resolved this issue by demonstrating that the Ubl domain does not stimulate the proteasomes already activated by ATPγS but instead increases the total number of active particles by somehow inducing activity in particles not activated by ATPγS alone. This conclusion complements recent reports that, even after ATPγS addition, most 26S proteasomes are in inactive conformations (64), as was also reported for proteasomes in cultured neurons (19). Accordingly, in related studies, we also found that 26S proteasomes are present in mammalian cells in at least fourfold excess over what is needed to support normal rates of protein degradation. Presumably, these inactive particles are the ones that are activated upon binding a Ubl-containing protein. These observations emphasize that 26S proteasomes, within a cell or even after purification, are heterogeneous, differing in composition, functional state, and modes of activation.
Multiple sites within the proteasome can bind ubiquitin and Ubl domains (48–51, 79, 80), but the activation of proteolysis depends on the association of the Ubl domains with Rpn1, to which the Ubl domains from Usp14 and Rad23A associate (50). However, these two Ubl domains seem to activate through distinct interactions in yeast and mammalian proteasomes. In yeast proteasomes, activation by the Ubl domain of Usp14 requires only the T2 binding site on Rpn1 (42) while mutating either the T1 or T2 sites on Rpn1 in mammalian particles reduced the Ka of Rad23A by half. The basis for this difference is puzzling because both Rad23A and Usp14 Ubl domains increase the same enzymatic activities in both types of proteasomes.
Unlike the activation of peptide entry, the stimulation of proteasomal ATP hydrolysis by ubiquitylated substrates is tightly linked to proteolysis and has a key additional requirement (36). Like degradation of ubiquitylated proteins (32, 33), activation of the ATPases requires a ubiquitin chain and an unfolded region in the protein (34). Although the ubiquitin chain is normally covalently linked to the protein, free ubiquitin chains, ubiquitin aldehyde, or, as shown here, a Ubl-containing protein can activate ATP hydrolysis if an unlinked unfolded protein is also present. Full-length Rad23A and Ddi2 could substitute for a ubiquitin chain to activate ATP hydrolysis, and this effect required their Ubl domains (Fig. 6). Activating the ATPases triggers the tighter binding of ubiquitylated proteins to proteasomes and commits them to degradation. The ability of these Ubl-containing proteins, together with a loosely folded substrate, to stimulate ATP hydrolysis implies that these proteins can promote the breakdown of ubiquitin conjugates. Accordingly, expression of Rad23B’s Ubl domain in HEK293 cells enhances overall protein breakdown by stimulating degradation of ubiquitin conjugates (42).
These findings fit with various biochemical studies showing that ATP binding stimulates peptide entry into the mammalian 26S and into the archaeal PAN-20S complex (63, 81). However, recent cryo-EM studies of proteasomes engaged in conjugate degradation (28, 29) depicted structural changes in the ATPases before the 20S gate opened. Although those observations suggest that ATP hydrolysis increases before peptidase activity rises, another recent study (30) reported that free ubiquitin chains induce similarly large changes in the ATPases and open the 20S gate, which should suffice to promote entry of small peptides (82). Presumably, the structural changes in the 26S particle leading to its activation upon binding a ubiquitin chain resemble the changes induced by the binding of Ubl-containing proteins. Although the basal level of ATP hydrolysis, when coupled with binding of a ubiquitin chain or a Ubl domain, may allow peptide entry, increased ATP hydrolysis must occur for efficient protein degradation, especially for highly structured proteins whose unfolding is rate limiting (37). Structural changes responsible for the increased rates of ATP hydrolysis have not been reported and may be missed by the large changes that permit substrate translocation. Moreover, recent cryo-EM studies of 26S proteasomes focused on elucidating the structural steps in ubiquitin chain removal and protein translocation occurring after the initial binding of a ubiquitin conjugate. Protein degradation may involve a different reaction sequence when the ubiquitin conjugate is delivered by a shuttling factor or when a proteasome is activated by a Ubl-containing protein.
We were quite surprised to find that, although Ubl domains alone can activate proteasomal peptidases, other regions in Ddi2, Usp7, and Rad23A could also do so. This result was particularly unexpected because of the frequent claims based on immunoprecipitation that Rad23’s Ubl domain is essential for Rad23 binding to proteasomes (69–74). However, peptidase activation may depend on interactions that are more transient than those required for coprecipitation. In addition, other proteasome-interacting proteins that lack a Ubl domain may also activate proteolysis, as was evident upon deletion of the Ubl domains of Rad23A, Usp7, and Ddi2. In fact, ZFAND5/ZNF216, a zinc finger protein, which is not a Ubl-containing protein, binds to the 26S proteasome and stimulates its multiple activities (24).
Although ubiquitin conjugates and Ubl-containing proteins stimulate the several proteasome activities similarly, important differences probably exist between their biochemical and cellular consequences. Ubiquitylated proteins activate proteasomes through interactions with the 26S-associated DUBs, Usp14 or Uch37, which are not stoichiometric components of proteasomes. Usp14, in particular, is present, at most, on a third of proteasomes (43). Most likely, the binding of other Ubl-containing proteins can activate the remaining two-thirds of proteasomes. Ubl-containing proteins possibly promote the degradation of some ubiquitin conjugates that would otherwise not be hydrolyzed if they bound directly to Rpn1, Rpn10, or Rpn13 where their loosely folded regions may not be able to interact with the ATPases and activate proteolysis. These distinct modes of activation are likely to be important in the degradation of different cell proteins. For example, following mitochondrial membrane depolarization, Parkin binds to the 26S and ubiquitylates mitochondrial surface proteins (54) while ubiquitin conjugates generated during ERAD are delivered to the 26S by Rad23 (6, 70, 83). Because these Ubl proteins transiently bind to proteasomes only under specific conditions [e.g., Rad23 and Usp14 when ubiquitin conjugates are abundant (43) or Parkin when it is phosphorylated by PINK1 (54)], activation of proteasomes in cells by Ubl-containing proteins is linked to specific physiological stimuli. Consequently, most 26S complexes in cells remain in the latent state.
In delivering conjugates to proteasomes, Ubl-containing proteins may also make degradation more efficient; Ubl-containing proteins not only activate the particles, but also inhibit the actions of DUBs (84). On the proteasome, there is a kinetic competition between deubiquitylation, leading to the release of the bound conjugate, and substrate capture by the ATPases (1, 85). Well-folded, ubiquitylated substrates may escape proteolysis if they are deubiquitylated and dissociate from proteasomes before the activation of ATPases, tight binding, and degradation occur (2, 39). If a Ubl-containing protein activates ATP hydrolysis and increases the capture of a loosely folded region by the ATPases, it should reduce the chance of substrate escape. Thus, the critical kinetic competition on the proteasome, which determines whether a polypeptide is degraded or continues to exist, is altered by the presence of Ubl-containing proteins, which increase the efficiency of the proteasome and the likelihood of substrate degradation.
Materials and Methods
Purification of Rad23A and Rad23B.
BL21 Star (DE3) Escherichia coli (C601003; Thermo Scientific) with pGEX-6P-Rad23A or pGEX-6P-Rad23B (86) induced with 0.5 mM isopropyl ß-d-1-thiogalactopyranoside (IPTG) at optical density at 600 nm (OD600) of 0.6 to 0.8 for 3 h at 37 °C. Bacteria were lysed through an emulsiflex homogenizer (Avestin) at 10,000 psi in 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 5 mM dithiothreitol (DTT) supplemented with 1 µg/mL DNase I and 1 mM phenylmethylsulfonyl fluoride (PMSF) and clarified by centrifugation (10,000 × g, 15 min). Clarified lysate was mixed with GSH-Sepharose (17-0756-01; GE Healthcare) at 4 °C for 1 h. The resin was washed with lysis buffer, and protein was eluted by cleavage with PreScission protease. Protein was dialyzed against 25 mM Hepes-KOH (pH 7.5), 40 mM KCl, 1 mM DTT, and 10% glycerol.
Purification of Rad23B Ubl.
BL21-AI E. coli (C607003; Thermo Scientific) with pDEST15-Ubl-hHR23B (9) induced with 0.1% l-arabinose at OD600 0.6 for 3 h at 37 °C. Bacteria were lysed through an emulsiflex homogenizer (Avestin) at 10,000 psi in PBS supplemented with 10 mM MgCl2 and 1 mM DTT, and clarified by centrifugation (100,000 × g, 1 h). Clarified lysate was mixed with GSH-Sepharose (17-0756-01; GE Healthcare) at 4 °C for 90 min. The resin was washed with lysis buffer, and protein was eluted by cleavage with thrombin (27-0846-01; GE Healthcare), which was inactivated and removed with Benzamidine-Sepharose 6B (17-0568-01; GE Healthcare). Protein was dialyzed against 25 mM Hepes-KOH (pH 7.4), 40 mM KCl, 5 mM MgCl2, 10% glycerol, and 1 mM DTT.
Purification of Rad23A ∆Ubl and Yeast Rad23.
BL21 Star (DE3) E. coli (C601003; Thermo Scientific) with pGEX-2TK HHR23A ∆Ubl (87) or pGEX-4T Rad23 (88) induced with 0.5 mM IPTG at OD600 0.6 to 0.8 for 3 h at 37 °C. Bacteria were lysed through an emulsiflex homogenizer (Avestin) at 10,000 psi in 25 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 5 mM DTT supplemented with 1 µg/mL DNase I and 1 mM PMSF and clarified by centrifugation (10,000 × g, 15 min). Clarified lysate was mixed with GSH-Sepharose (17-0756-01; GE Healthcare) at 4 °C for 1 h. The resin was washed with lysis buffer, and protein was eluted by cleavage with thrombin (27-0846-01; GE Healthcare), which was inactivated and removed with Benzamidine-Sepharose 6B (17-0568-01; GE Healthcare). Protein was dialyzed against 25 mM Hepes-KOH (pH 7.4), 40 mM KCl, 5 mM MgCl2, 10% glycerol, and 1 mM DTT.
Purification of UBLCP1.
BL21 Codon Plus (DE3) RPx E. coli (Agilent 230275) with pSJ2-His-UBLCP1 (13) induced with 0.5 mM IPTG at OD600 0.6 to 0.8 for 20 h at 16 °C. Bacteria were lysed through an emulsiflex homogenizer (Avestin) at 10,000 psi in 50 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 5 mM MgCl2, 5 mM 2-mercaptoethanol, and 20 mM imidazole supplemented with 1 mM PMSF. The lysate was incubated on ice with RNase A and DNase I and then clarified by centrifugation (10,000 × g, 30 min). Clarified lysate was mixed with Ni-NTA agarose (H-350; Gold Biotechnology) at 4 °C for 1 h. The resin was washed with lysis buffer, and protein was eluted with 50 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 5 mM MgCl2, 5 mM 2-mercaptoethanol, 200 mM imidazole and concentrated in a 10 molecular weight cut off (MWCO) spin filter before exchanging buffer using a PD-10 column (17-0851-01; GE Healthcare) to 50 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 30% glycerol.
Purification of USP7-CD-Ubl123.
BL21 Star (DE3) with pGEX4T-USP7CD-Ubl123 (62) induced with 0.5 mM IPTG at OD600 0.6 to 0.8 for 20 h at 16 °C. Bacteria were lysed through an emulsiflex homogenizer (Avestin) at 10,000 psi in 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 15% glycerol, 0.1% Triton X-100, 1 mM DTT, and complete protease inhibitor and clarified by centrifugation (186,000 × g, 1 h). Clarified lysate was mixed with GSH-Sepharose (17-0756-01; GE Healthcare) at 4 °C for 1 h. The resin was washed with 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 15% glycerol, 1 mM DTT, and protein was eluted by cleavage with thrombin (27-0846-01; GE Healthcare), which was inactivated and removed with Benzamidine-Sepharose 6B (17-0568-01; GE Healthcare). Protein was concentrated through a 30 MWCO spin filter further purified by size exclusion chromatography using a Superdex 200 16/60 column (17-1088-01; GE Healthcare) in 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 15% glycerol, and 1 mM DTT.
Purification of USP7-CD-Ubl45 and USP7-CD-CTP.
BL21 Star (DE3) with pET-Usp7CD-UBL45 and pET-Usp7CD-CTP (62) induced with 0.5 mM IPTG at OD600 0.6 to 0.8 for 20 h at 16 °C. Bacteria were lysed through an emulsiflex homogenizer (Avestin) at 10,000 psi in 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 15% glycerol, 0.1% Triton X-100, 1 mM DTT, and complete protease inhibitor and clarified by centrifugation (186,000 × g, 1 h). Clarified lysate was mixed with Ni-NTA agarose (H-350; Gold Biotechnology) at 4 °C for 1 h. The resin was washed with 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 30 mM imidazole, 15% glycerol, and 0.5 mM DTT and eluted with 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 300 mM imidazole, 15% glycerol, and 0.5 mM DTT. The eluate was treated with tobacco etch virus (TEV) protease (T4555; Sigma) and concentrated through a 30 MWCO spin filter before being further purified using a Superdex 200 16/60 column (17-1088-01; GE Healthcare) in 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 15% glycerol, and 1 mM DTT.
Purification of DDI2.
BL21 RIL (DE3) Codon Plus E. coli (230245; Agilent) with pET28b Ddi2 or pET28b Ddi2-RVP-UIM (89) induced with 0.5 mM IPTG at OD600 0.6 to 0.8 for 20 h at 16 °C. Bacteria were lysed by sonication in 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole (pH 8.0) supplemented with 1 mM PMSF and clarified by centrifugation (10,000 × g, 20 min). Clarified lysate was mixed with Ni-NTA agarose (H-350; Gold Biotechnology) at 4 °C for 90 min. The resin was washed with 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole (pH 8.0) and eluted with 50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole (pH 8.0). Protein was concentrated by centrifugation in Amicon Ultra spin filters (30 MWCO for Ddi2 and 10 MWCO for Ddi2 RVP-UIM) and purified through a Superdex 200 16/60 column (17-1088-01; GE Healthcare) in 25 mM Hepes (pH 8.0), 40 mM potassium acetate, 10% glycerol, and 1 mM DTT.
Mammalian 26S Proteasome Purification.
The 26S proteasomes were isolated from frozen rabbit muscle tissue or mouse embryonic fibroblasts as indicated as described previously (9, 31). Rabbit muscle frozen muscles were from albino rabbits, 4.75 to 5.75 lbs., ∼8 to 12 wk old of mixed gender (41225-2; Pel-Freez). Mouse embryonic fibroblasts (90) were cultured in DMEM plus 10% FBS at 37 °C, 5% CO2. Tissues or cells were lysed in 25 mM Hepes-KOH, pH 7.5, 5 mM MgCl2, 10% glycerol, 1 mM ATP, and 1 mM DTT and clarified by centrifugation at 100,000 × g, 30 min. Clarified lysate with GST-Ubl (RAD23B) and glutathione-Sepharose (17-0765-01; GE Healthcare) were combined at 4 °C for 90 min. The resin was washed twice with lysis buffer and eluted with His10-UIM2(S5a) in lysis buffer. His10-UIM2(S5a) was removed by incubation with Ni-NTA agarose (H-350; Gold Biotechnology) for 20 min and separated through a 500-µL, 0.45-µm spin filter.
Yeast 26S Proteasome Purification.
The 26S proteasomes were purified from yeast [SY1214 (WT), SY1210 (T1), and SY1724 (T2)] (50) as described by Leggett et al. (91). One liter of yeast culture at OD600 6 to 8 collected by centrifugation (3,000 × g, 10 min) was lysed in two-pellet volumes of 50 mM Tris⋅HCl (pH 8.0), 5 mM MgCl2, 1 mM (ethylenedinitrilo)tetraacetic acid (EDTA), and 1 mM ATP by passing through an emulsiflex at 10,000 psi. Lysates were clarified by centrifugation (20, 000 × g, 30 min) and filtered through cheesecloth. Clarified lysate was bound to 500 µL of IgG resin at 4 °C for 1 h. The resin was washed twice with 25 mL of 50 mM Tris⋅HCl (pH 7.5), 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, and 100 mM NaCl and once with 25 mL 50 mM Tris⋅HCl (pH 7.5), 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, and 1 mM DTT. Proteasomes were eluted with 1 mL of TEV protease (T4455; Sigma) in 50 mM Tris⋅HCl (pH 7.5), 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, and 1 mM DTT at 30 °C for 1 h. Eluate was collected onto 100 µL of Ni-NTA agarose (H-350; Gold Biotechnology) with a wash of the IgG resin of 2.5 mL of 50 mM Tris⋅HCl (pH 7.5), 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, and 1 mM DTT. Proteasomes with the Ni-NTA agarose were incubated at 4 °C for 10 min to remove TEV protease. This mix was centrifuged (100 × g, 5 min), and the supernatant was transferred to 30 MWCO spin filters (UFC903096; Millipore) and concentrated to about 500 µL. Glycerol was added to a final concentration of 10% by volume.
Peptidase Assay.
Typically, peptidase assays contained 1 nM purified proteasomes and were incubated with indicated concentrations of linear hexa-ubiquitin chains or Ubl-containing proteins in 50 mM Tris⋅HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.1 mM ATPγS, and 10 µM ac-Nle-Pro-Nle-Asp-amc. Reactions were incubated at 37 °C, and the released fluorescent amc was monitored once per minute for 20 to 40 min at fluorescence excitation wavelength (λex) 380 nm and emission wavelength (λem) 460 nm.
In Fig. 1D, this protocol was modified to measure multiple proteolytic sites by exchanging 10 µM ac-Nle-Pro-Nle-Asp-amc with 10 µM suc-Leu-Leu-Val-Try-amc or 10 µM boc-Leu-Arg-Arg-amc.
In Fig. 2 B and C, the protocol was modified to accommodate yeast proteasomes by using 30 °C instead of 37 °C. In Fig. 4A, the protocol was modified to measure nucleotide effects by using 1 mM ADP, 1mM ATP, or 0.1 mM ATPγS, respectively.
Activity Probe Labeling.
The 2 nM 26S proteasomes (purified from MEFs by Ubl Affinity) were incubated in 50 mM Tris⋅HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.1 mM ATPγS (or 1 mM ATP), 1 mM DTT, and 500 nM MVB127 activity probe at 37 °C. Rad23B Ubl or Rad23B was added at 500 nM. Reactions were stopped at the indicated time points by adding LDS sample buffer and heating at 70 °C for 10 min. Samples were run on Bolt 4 to 12% Bis-Tris Plus 1-mm acrylamide gels (Thermo Scientific) in 1× MES Running Buffer and scanned with an AI600 RGB camera with λex 520 nm and λem 593 nm. Quantification of the bands was done with the IQTL software package from GE Healthcare Sciences.
ATPase Assay.
ATP hydrolysis was assayed by a malachite green reaction as previously described (31). Five time points from 0 to 40 min were set with 10 nM 26S, 1 µM casein, and 500 nM Ubl-containing protein or linear hexa-ubiquitin in 25 mM Hepes-KOH (pH 8.0), 2.5 mM MgCl2, 125 mM potassium acetate, 0.025% Triton X-100, 0.5 mM DTT, 1 mM ATP, and 0.1 g/L BSA at 37 °C until the end of the time course, when they were briefly centrifuged and transferred to ice. During the time course, a solution of 3:1 0.045% malachite green HCl to 4.2% ammonium molybdate (4 M HCl) was prepared and filtered. To 10 µL of the samples, 170 µL of 0.0337% malachite green and 0.84% ammonium molybdate was added followed with 20 µL of 34% (wt/vol) citric acid. The absorbance at 650 nm was measured. Rates of ATP hydrolysis were calculated as a linear slope through the five time points. The background signal of casein and Ubl-containing protein without 26S was subtracted, and the rates were normalized to the control sample of 26S without casein and without Ubl-containing protein.
Quantification and Statistical Analysis.
Statistical analysis was performed with Prism 8 for macOS (version 8.1.2.) software. Unless indicated otherwise, errors represent the standard error of the mean (SEM) from three independent experiments. Peptidase activation curves were modeled with the equation Y = Bottom + (X^Hillslope) × (Top-Bottom)/(X^HillSlope + EC50^HillSlope) to estimate activation. Binding curves were modeled with the equation Y = Bmax × X/(Kd + X). Comparison between groups used one-way ANOVA with post hoc Dunnett’s t test and P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Data Availability.
Requests for reagents or further information should be directed to A.L.G. (alfred_goldberg@hms.harvard.edu). The following items are restricted by material transfer agreements: for Ddi2, Ddi2 Ubl, and Ddi2 RVP UIM. For plasmids, please contact Klara Saskova (saskova@uochb.cas.cz), and, for Usp7 constructs, please contact Genentech.
Supplementary Material
Acknowledgments
We thank Hyoung Tae Kim for initial assay development and valuable discussions; Beyza Erbil for purifying and characterizing Ddi2; Megan LaChance for assistance in preparing this manuscript; and several colleagues for kindly providing valuable reagents: Parkin from Jennifer Johnston (AN2H), Parkin Ubl and phospho-Parkin Ubl from Wade Harper (Harvard Medical School), UBL4A from Susan Shao (Harvard Medical School), and the MVB127 activity probe from Herman Overkleeft (Universiteit Leiden). The plasmid for UBLCP1 was a gift from Jack Dixon (University of California, San Diego); for Usp7CD, Usp7CD-Ubl123, Usp7CD-45, and Usp7CD-CTP from Genentech; Ddi2 from Klára Šašková (Charles University, Prague); and Rad23A ∆Ubl from Martin Scheffner (Universität Konstanz). This project was supported by grants to A.L.G. from NIH-National Institute of General Medical Sciences (R01 GM51923) and Cure Alzheimer’s Disease.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915534117/-/DCSupplemental.
References
- 1.Finley D., Prado M. A., The proteasome and its network: Engineering for adaptability. Cold Spring Harb. Perspect. Biol., 12, a033985 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins G. A., Goldberg A. L., The logic of the 26S proteasome. Cell 169, 792–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L., Madura K., Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22, 4902–4913 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleijnen M. F., et al. , The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 6, 409–419 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Saeki Y., Saitoh A., Toh-e A., Yokosawa H., Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 293, 986–992 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Richly H., et al. , A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Tsuchiya H., et al. , In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol. Cell 66, 488–502.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Itakura E., et al. , Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol. Cell 63, 21–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besche H. C., Haas W., Gygi S. P., Goldberg A. L., Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48, 2538–2549 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakata E., et al. , Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 4, 301–306 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., et al. , The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 292, 16310–16320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Poot S. A. H., Tian G., Finley D., Meddling with fate: The proteasomal deubiquitinating enzymes. J. Mol. Biol. 429, 3525–3545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X., et al. , UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc. Natl. Acad. Sci. U.S.A. 108, 18649–18654 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besche H. C., et al. , Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 33, 1159–1176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosas B., et al. , Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Hochstrasser M., Origin and function of ubiquitin-like proteins. Nature 458, 422–429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H., Matouschek A., Recognition of client proteins by the proteasome. Annu. Rev. Biophys. 46, 149–173 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Yu H., Kago G., Yellman C. M., Matouschek A., Ubiquitin-like domains can target to the proteasome but proteolysis requires a disordered region. EMBO J. 35, 1522–1536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asano S., et al. , Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Guo X., Huang X., Chen M. J., Reversible phosphorylation of the 26S proteasome. Protein Cell 8, 255–272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VerPlank J. J. S., Goldberg A. L., Regulating protein breakdown through proteasome phosphorylation. Biochem. J. 474, 3355–3371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt M., et al. , The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 12, 294–303 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Ortega J., et al. , The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 346, 1221–1227 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Lee D., Takayama S., Goldberg A. L., ZFAND5/ZNF216 is an activator of the 26S proteasome that stimulates overall protein degradation. Proc. Natl. Acad. Sci. U.S.A. 115, E9550–E9559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peth A., Besche H. C., Goldberg A. L., Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Demartino G. N., Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem. J. 421, 397–404 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bech-Otschir D., et al. , Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat. Struct. Mol. Biol. 16, 219–225 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Dong Y., et al. , Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565, 49–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Peña A. H., Goodall E. A., Gates S. N., Lander G. C., Martin A., Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362, eaav0725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Z., et al. , Structural snapshots of 26S proteasome reveal tetraubiquitin-induced conformations. Mol. Cell 73, 1150–1161.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Kim H. T., Collins G. A., Goldberg A. L., Measurement of the multiple activities of 26S proteasomes. Methods Mol. Biol. 1844, 289–308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inobe T., Fishbain S., Prakash S., Matouschek A., Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 7, 161–167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishbain S., et al. , Sequence composition of disordered regions fine-tunes protein half-life. Nat. Struct. Mol. Biol. 22, 214–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peth A., Kukushkin N., Bossé M., Goldberg A. L., Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J. Biol. Chem. 288, 7781–7790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peth A., Uchiki T., Goldberg A. L., ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell 40, 671–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peth A., Nathan J. A., Goldberg A. L., The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J. Biol. Chem. 288, 29215–29222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bard J. A. M., Bashore C., Dong K. C., Martin A., The 26S proteasome utilizes a kinetic gateway to prioritize substrate degradation. Cell 177, 286–298.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bashore C., et al. , Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat. Struct. Mol. Biol. 22, 712–719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanna J., et al. , Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Kim H. T., Goldberg A. L., The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis. J. Biol. Chem. 292, 9830–9839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aufderheide A., et al. , Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 112, 8626–8631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H. T., Goldberg A. L., UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proc. Natl. Acad. Sci. U.S.A. 115, E11642–E11650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo C. L., Goldberg A. L., Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc. Natl. Acad. Sci. U.S.A. 114, E3404–E3413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A., An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Ordureau A., et al. , Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koyano F., et al. , Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Matyskiela M. E., Lander G. C., Martin A., Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deveraux Q., Ustrell V., Pickart C., Rechsteiner M., A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269, 7059–7061 (1994). [PubMed] [Google Scholar]
- 49.Husnjak K., et al. , Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y., et al. , Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351, aad9421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X., et al. , Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 reveal distinct binding mechanisms between substrate receptors and shuttle factors of the proteasome. Structure 24, 1257–1270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitada T., et al. , Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Riley B. E., et al. , Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarraf S. A., et al. , Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Um J. W., et al. , Parkin directly modulates 26S proteasome activity. J. Neurosci. 30, 11805–11814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terreni L., Calabrese E., Calella A. M., Forloni G., Mariani C., New mutation (R42P) of the parkin gene in the ubiquitinlike domain associated with parkinsonism. Neurology 56, 463–466 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Wang X., et al. , Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell. Proteomics 16, 840–854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun S., et al. , Phosphatase UBLCP1 controls proteasome assembly. Open Biol. 7, 170042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chio U. S., Cho H., Shan S. O., Mechanisms of tail-anchored membrane protein targeting and insertion. Annu. Rev. Cell Dev. Biol. 33, 417–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lehner B., et al. , Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics 83, 153–167 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Eletr Z. M., Wilkinson K. D., Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 1843, 114–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rougé L., et al. , Molecular understanding of USP7 substrate recognition and C-terminal activation. Structure 24, 1335–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Smith D. M., et al. , ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell 20, 687–698 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Eisele M. R., et al. , Expanded coverage of the 26S proteasome conformational landscape reveals mechanisms of peptidase gating. Cell Rep. 24, 1301–1315.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Śledź P., et al. , Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 7264–7269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li N., et al. , Relative quantification of proteasome activity by activity-based protein profiling and LC-MS/MS. Nat. Protoc. 8, 1155–1168 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Auesukaree C., Fuchigami I., Homma T., Kaneko Y., Harashima S., Ddi1p and Rad23p play a cooperative role as negative regulators in the PHO pathway in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 365, 821–825 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Safadi S. S., Shaw G. S., A disease state mutation unfolds the parkin ubiquitin-like domain. Biochemistry 46, 14162–14169 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Goh A. M., et al. , Components of the ubiquitin-proteasome pathway compete for surfaces on Rad23 family proteins. BMC Biochem. 9, 4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim I., Mi K., Rao H., Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol. Biol. Cell 15, 3357–3365 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambertson D., Chen L., Madura K., Investigating the importance of proteasome-interaction for Rad23 function. Curr. Genet. 42, 199–208 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Saeki Y., Sone T., Toh-e A., Yokosawa H., Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 296, 813–819 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Wilkinson C. R., et al. , Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3, 939–943 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Schauber C., et al. , Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391, 715–718 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Liu C. W., et al. , ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol. Cell 24, 39–50 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D., Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17, 4909–4919 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horwitz A. A., et al. , ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J. Biol. Chem. 282, 22921–22929 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Bard J. A. M., et al. , Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walters K. J., Kleijnen M. F., Goh A. M., Wagner G., Howley P. M., Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41, 1767–1777 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Hiyama H., et al. , Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J. Biol. Chem. 274, 28019–28025 (1999). [DOI] [PubMed] [Google Scholar]
- 81.Smith D. M., Fraga H., Reis C., Kafri G., Goldberg A. L., ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144, 526–538 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bajorek M., Finley D., Glickman M. H., Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 13, 1140–1144 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Medicherla B., Kostova Z., Schaefer A., Wolf D. H., A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 5, 692–697 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raasi S., Pickart C. M., Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 278, 8951–8959 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Lee B. H., et al. , USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532, 398–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nathan J. A., Kim H. T., Ting L., Gygi S. P., Goldberg A. L., Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glockzin S., Ogi F. X., Hengstermann A., Scheffner M., Blattner C., Involvement of the DNA repair protein hHR23 in p53 degradation. Mol. Cell. Biol. 23, 8960–8969 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D., Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279, 26817–26822 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Sivá M., et al. , Human DNA-damage-inducible 2 protein is structurally and functionally distinct from its yeast ortholog. Sci. Rep. 6, 30443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee B. H., et al. , Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leggett D. S., et al. , Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for reagents or further information should be directed to A.L.G. (alfred_goldberg@hms.harvard.edu). The following items are restricted by material transfer agreements: for Ddi2, Ddi2 Ubl, and Ddi2 RVP UIM. For plasmids, please contact Klara Saskova (saskova@uochb.cas.cz), and, for Usp7 constructs, please contact Genentech.