Summary
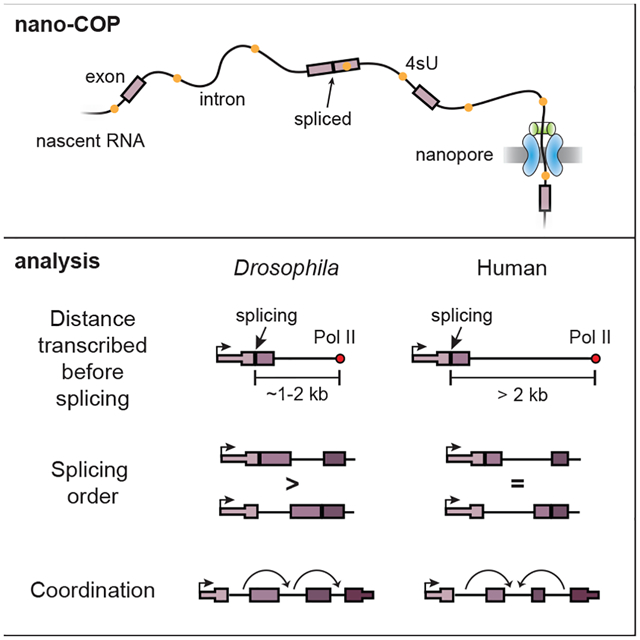
Understanding how splicing events are coordinated across numerous introns in metazoan RNA transcripts requires quantitative analyses of transient RNA processing events in living cells. We developed nanopore analysis of CO-transcriptional Processing (nano-COP), in which nascent RNAs are directly sequenced through nanopores, exposing the dynamics and patterns of RNA splicing without biases introduced by amplification. Long nano-COP reads reveal that in human and Drosophila cells, splicing occurs after RNA polymerase II transcribes several kilobases of pre-mRNA, suggesting that metazoan splicing transpires distally from the transcription machinery. Inhibition of the branch-site recognition complex SF3B rapidly diminished global co-transcriptional splicing. We found that splicing order does not strictly follow the order of transcription and is associated with cis-acting elements, alternative splicing, and RNA-binding factors. Further, neighboring introns in human cells tend to be spliced concurrently, implying that splicing of these introns occurs cooperatively. Thus, nano-COP unveils the organizational complexity of RNA processing.
Graphical Abstract
eTOC blurb
Drexler et al. expose the nascent transcriptome through nanopore analysis of co-transcriptional processing (nano-COP). Long nascent RNAs are directly sequenced through nanopores to provide new insights into the cis- and trans-acting control of splicing order and define the physical proximity between transcription and splicing in metazoans. Furthermore, nano-COP reveals a tendency for proximal introns to have coordinated splicing patterns in human cells.
Introduction
Most metazoan genes contain many long introns with degenerate sequence information at splice sites, requiring sophisticated mechanisms to correctly localize and coordinate the excision of multiple introns within the same nascent transcript. This process requires the multi-megadalton spliceosome complex, which assembles in a stepwise fashion at each intron splice site (SS) within a pre-mRNA (Fica and Nagai, 2017; Wahl et al., 2009). Intron splicing comprises two catalytic transesterification steps, which involve assembly and rearrangements of the spliceosome. First, the branch point adenosine attacks the 5’SS, forming an intron lariat. Second, the free end of the 5’SS attacks the downstream 3’SS, joining the exons and releasing the intron lariat.
Much of our knowledge of splicing mechanisms has been gleaned from in vitro experiments using externally synthesized pre-mRNA (Black et al., 1985; Braun et al., 2018; Hoskins et al., 2011; Krainer et al., 1984). However, because splicing is predominantly co-transcriptional (Beyer and Osheim, 1988; Khodor et al., 2011; Pandya-Jones and Black, 2009; Wuarin and Schibler, 1994), such studies cannot capture a complete view of in vivo splicing. For instance, mutations in the C-terminal domain and trigger loop of RNA polymerase II (Pol II) alter splicing outcomes, suggesting a physical and mechanistic coupling between the transcription and splicing machineries (de la Mata et al., 2003; McCracken et al., 1997). If transcription and splicing are coupled, then the order in which introns are transcribed has been reasoned to influence the order in which they are spliced. In fact, the “first-come, first-served” model of splicing proposes that the first introns transcribed are the first to be committed for splicing (Aebi and Weissman, 1987). However, even though the completion of intron splicing frequently follows a defined order within a gene, the order of splicing is not always concordant with the direction of transcription (Kessler et al., 1993; Kim et al., 2017; Wetterberg et al., 1996). Splicing order has the potential to contribute to alternative splicing mechanisms (Takahara et al., 2002). For example, whether the splicing of each intron is controlled by a defined kinetic window or instead depends on the splicing status of nearby introns has profound consequences for models of splicing regulation. Understanding the control of the order of intron splicing and the coordination of splicing patterns across multi-intron genes is critical to our understanding of alternative splicing.
A variety of approaches have provided estimates of the length of time between the synthesis of an intron and its removal by splicing in living cells. Measurements of intron synthesis, lariat degradation, and the completion of the splicing reaction by quantitative PCR (Singh and Padgett, 2009), metabolic labeling (Pai et al., 2017; Rabani et al., 2014; Wachutka et al., 2019; Windhager et al., 2012), or single-molecule imaging (Coulon et al., 2014; Martin et al., 2013) have revealed that intron removal can take between 30 seconds and an hour in mammalian cells. Although enlightening, these strategies fail to uncover the physical link between transcription and splicing, as well as the patterns of splicing across nascent transcripts. Recent work in the yeast species S. cerevisiae and S. pombe suggests that splicing is completed nearly immediately after an intron is synthesized (Carrillo Oesterreich et al., 2016; Herzel et al., 2018), but it remains to be seen whether this is also the case in organisms with more complex gene structures and abundant alternative splicing, such as Drosophila and humans.
Methods to study the kinetics and regulation of co-transcriptional splicing have been hindered by three main challenges in the quantitative analysis of nascent transcripts. First, nascent RNA is a very small fraction (<0.5%) of cellular RNA (Han and Lillard, 2000; Jackson et al., 1998), presenting a purification challenge that can lead to overestimates of co-transcriptional splicing kinetics if samples are contaminated with spliced mature RNA. Second, length biases that arise during the enzymatic steps of library preparation can systematically distort relative measurements of spliced and unspliced RNAs that differ in size by the length of an intron. Third, traditional short-read sequencing does not provide isoform information across RNA molecules, limiting its ability to detect multi-intron splicing patterns.
Here, we describe nanopore analysis of CO-transcriptional Processing (nano-COP), a technique designed to directly probe the dynamics and regulation of pre-mRNA splicing in vivo. nano-COP uses direct RNA nanopore sequencing to determine the native isoform of long nascent RNA molecules without amplification-associated biases. Application of nano-COP to human and Drosophila cells revealed key features of RNA processing in both species. We observed pronounced differences in splicing kinetics between the two species, discovered global patterns in the order of intron splicing, and identified coordinated patterns of splicing in human cells. In cells treated with the SF3B1 inhibitor pladienolide B, nano-COP detected a near complete abolishment of co-transcriptional splicing. Thus, nano-COP represents an effective strategy for exposing the critical molecular processes that occur during transcription in living metazoan cells.
Results
Unveiling the nascent transcriptome with direct RNA nanopore sequencing
We developed nanopore analysis of CO-transcriptional Processing (nano-COP) in order to observe nascent RNA isoforms as they are transcribed. In this method, stringent purification of nascent RNA followed by long-read nanopore sequencing connects intron splicing to Pol II positions at the 3’ ends of reads, revealing the relationship between transcription and splicing in living cells (Figure 1A). To sequence long RNA molecules without amplification-associated biases, we adopted a direct RNA sequencing approach using Oxford Nanopore Technologies’ MinION and PromethION instruments (Garalde et al., 2018). Since background levels of mature spliced RNA can result in overestimation of co-transcriptional splicing measures, we developed a stringent nascent RNA purification strategy by combining two complementary techniques: cellular fractionation (Pandya-Jones and Black, 2009; Wuarin and Schibler, 1994) (Figure S1A) and 4-thiouridine (4sU) pulse labeling (Dölken et al., 2008; Schwalb et al., 2016).
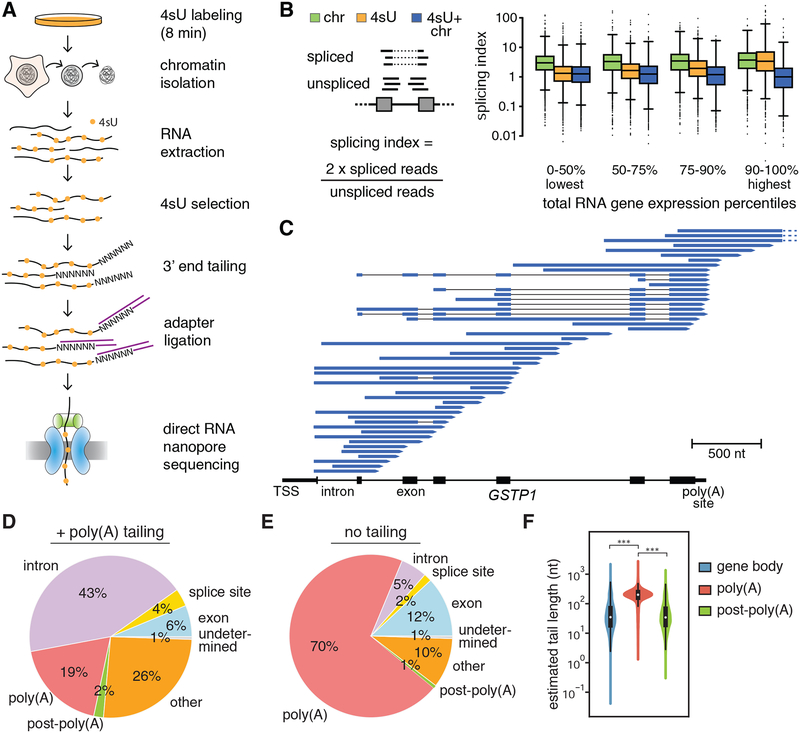
Figure 1. Capturing the nascent transcriptome in human cells with nano-COP.
(A) Schematic of the nano-COP protocol with nascent RNA purification and direct RNA nanopore sequencing. Cells are labeled with 4-thiouridine (4sU); cellular fractionation is performed to isolate chromatin-associated RNA; and 4sU labeled chromatin-associated RNA is selected through biotinylation and affinity purification. Subsequently, a poly(A) or poly(I) tail is added to the purified RNA, represented here as a string of N’s. The sample is subjected to direct RNA library preparation, including 3’ end adapter ligation, followed by nanopore sequencing. (B) Splicing index, which represents the proportion of spliced transcripts in Illumina sequencing datasets, is plotted within different percentiles of total RNA gene expression. The distribution of 4sU labeled chromatin-associated RNA (4sU+chr) differs significantly (t-test p-value < 0.05) between chromatin-associated RNA (chr) and 4sU labeled RNA (4sU) at all gene expression levels. Gene expression percentiles are based on total RNA expression levels out of 9659 genes that have at least 25 reads spanning splice junctions in all datasets. (C) Representative nano-COP reads aligned to the GSTP1 gene in human K562 cells. The gene structure is represented from the transcription start site (TSS) to the poly(A) site, with black boxes representing exons and lines representing introns. Within the reads, blue boxes represent read coverage, black lines represent skipped coverage due to splicing and the start of the read (3’ end of RNA) is represented with an arrow. Dashed lines represent reads that continue beyond the region displayed. (D–E) Distribution of nano-COP 3’ ends by nanopore sequencing in (D) human K562 cells with enzymatic poly(A) tail addition and (E) human K562 cells in the absence of enzymatic poly(A) tail addition. See Methods for descriptions of 3’ end alignment categories. (F) The length of poly(A) tails for sequenced RNAs with enzymatic poly(A) tail addition was estimated using nanopolish-polyA (Loman et al., 2015; Workman et al., 2018). Estimated tail lengths were plotted for RNAs in each sample that have 3’ ends aligning within gene bodies (exon, intron, or splice site), at poly(A) sites, or just downstream of poly(A) sites (*** signifies t-test p-value < 1 × 10−30).
After cells are labeled with 4sU for 8 minutes, they are lysed and subjected to sequential centrifugation steps that yield the chromatin, nucleoplasm and cytoplasm cellular fractions. RNA purified from the chromatin fraction is then subjected to 4sU purification. Even for much longer periods than 8 minutes, 4sU treatment does not show a decrease in splicing levels (Schofield et al., 2018) (Figure S1B–C), supporting the use of this metabolic labeling approach to quantify splicing rates (Pai et al., 2017; Rabani et al., 2014; Wachutka et al., 2019). The combination of cellular fractionation and 4sU purification enriches for RNAs that are both recently transcribed and localized to chromatin. Moreover, the use of both methods in tandem yields higher amounts of unspliced RNA compared to either method alone, resulting in a substantial improvement in the purification of nascent RNA especially at highly expressed genes that have a higher likelihood of mature RNA contamination (Figures 1B and S1D–F). All nascent RNA purification strategies enrich RNA from genes with comparable length distributions (Figure S1G).
During direct RNA nanopore sequencing, each RNA molecule is recruited through the nanopore via an adapter ligated to its polyadenylated [poly(A)] tail, and is consequently read from 3’ to 5’(Garalde et al., 2018). Because we expect most 4sU-labeled chromatin-associated RNAs to be nascent, and therefore not yet polyadenylated, we add poly(A) tails enzymatically using E. coli poly(A) polymerase before ligating the adapter (Figure 1A). After sequencing and aligning reads to the genome, we can distinguish RNAs that have completed transcription with endogenous poly(A) tails from nascent RNAs with artificial poly(A) tails based on where the 3’ end of the RNA aligns. As an alternative method for tailing nascent RNAs, we also utilized a yeast poly(A) polymerase to add poly(I) tails to the ends of nascent RNAs and ligated a custom poly(C) adapter for sequencing (Figure 1A).
We first applied nano-COP to human chronic myeloid leukemia K562 cells using the poly(A) tailing approach. Analysis of datasets from biological triplicates confirmed that nano-COP read coverage was reproducible (Pearson’s R = 0.75–0.91; Figure S2A–B). Read accuracies were similar to those of nanopore datasets acquired using poly(A)-selected mature RNA (Workman et al., 2018), indicating that 4sU does not have a detectable effect on base calling (Figure S2C–D). We obtained a wide range of read lengths (median, 671 nt; longest, 7420 nt; Figure S2E–F) that are shorter than DNA nanopore sequencing read lengths, but comparable to direct RNA read lengths from mRNA (Workman et al., 2018). Over 25% of nascent RNA nanopore reads were greater than 1 kb in length, enabling detection of co-transcriptional RNA processing over long ranges. To assess the variation in processing dynamics between species with diverse gene architectures and splicing regulation, we also applied nano-COP to D. melanogaster S2 cells (Figure S3A).
Each nano-COP sequencing run yielded ~100,000 mapped reads with the MinION instrument and ~500,000 mapped reads with the PromethION instrument (Table S1). In total, over 1.5 million nano-COP reads from human K562 cells aligned to >13,000 genes, with a median of 11 read mappings per gene (Figure S2G–H). For Drosophila S2 nano-COP libraries, reads aligned to >8,500 genes with a median of 4 reads per gene. With current coverage levels, we do not have sufficient depth for the analysis of single genes or introns except for the highest expressed genes. Thus, in this study, we have analyzed aggregated nano-COP data to observe global splicing dynamics that occur on average. nano-COP read coverage correlates with short-read RNA sequencing coverage by the Illumina platform (Pearson’s R = 0.90; Figure S2I), suggesting that the lower sequencing depth of nanopore sequencing does not impact our capacity to capture a wide dynamic range of the transcriptome. Nevertheless, our findings might be limited to the highly expressed genes that are well represented in nano-COP datasets.
Highly expressed genes, such as GSTP1 (Figure 1C), showcase the diversity of nascent transcript isoforms arising from a single gene. The positioning of read 3’ ends within genes, along with the presence of unspliced introns, indicate that the sequenced reads originated from RNAs that were in the midst of being transcribed and processed. In addition to canonical coding genes, nano-COP exposes the transcription and RNA processing of noncoding RNAs. For instance, we observed nano-COP reads that start and end at the boundaries of precursor micro RNAs (pre-miRNA) within unspliced introns (Figure S3B), supporting previous findings that pre-miRNA cleavage can occur before intron splicing (Kim and Kim, 2007). We also detected cases of antisense transcription across long noncoding RNAs (Figure S3C).
Pol II mapping with nano-COP
In native elongating transcript sequencing (NET-seq)-based approaches, sequencing of the 3’ ends of nascent RNAs enables high-resolution mapping of Pol II genome-wide (Churchman and Weissman, 2011; Mayer et al., 2015; Nojima et al., 2015). To determine whether nano-COP also measures Pol II position, we sought to verify that the start of nano-COP reads represents the 3’ ends of nascent RNA. We find that transcript 3’ ends aligned mostly within gene bodies (Figure 1D and S4A, D), consistent with these reads deriving from RNAs in the middle of synthesis. By contrast, libraries constructed from 4sU-labeled chromatin-associated RNA without enzymatic addition of a poly(A) or poly(I) tail aligned predominantly to annotated poly(A) sites (Figure 1E; chi-square p-value < 2.2 × 10−16), and thus represented pre-mRNA that remained associated with chromatin after transcription is complete (Brody et al., 2011). In nano-COP libraries made via poly(A) tailing, the tails at the start of each nanopore read were shorter when aligned within gene bodies and longer when aligned to poly(A) sites, demonstrating that the former were polyadenylated in vitro and therefore distinguishable from the naturally polyadenylated RNAs (Figure 1F and S4B–C; t-test p-value < 1 × 10−30) (Loman et al., 2015; Workman et al., 2018). Nano-COP libraries made using poly(I) tailing resulted in more reads aligning to gene bodies compared with poly(A) tailing libraries (Figure S4D; chi-square p-value < 2.2 × 10−16), providing an increase in the proportion of reads arising from RNAs in the process of transcription for each sequencing run. Importantly, libraries made with poly(A) and poly(I) tailing methods exhibited reproducible read coverage across replicates (Pearson R = 0.935; Figure S4E) and so were combined for all subsequent analyses.
To measure the alignment accuracy of RNA 3’ ends with direct RNA nanopore sequencing, we tailed and sequenced an in vitro transcribed RNA. We found that 85% of sequenced RNA 3’ ends correctly map within 25 nt of the expected transcript end position (Figure S4F). Based on these results, we conclude that the 3’ ends of nascent transcripts with artificial poly(A) or poly(I) tails largely represent active transcription sites within a resolution of 50 nt.
In addition to Pol II position, nano-COP also captures intermediates of the splicing reaction (i.e. free ends of upstream exons after the first catalytic step of splicing), providing insights into the regulation and efficiencies of the catalytic steps of splicing (Burke et al., 2018; Chen et al., 2018) (Figure S4G–H). The alignment positions of nascent RNA 3’ ends allow for the computational removal of reads that have completed transcription (i.e., RNA 3’ ends aligning to poly(A) sites) or have undetermined transcription status (i.e., RNA 3’ ends aligning to splice sites) to focus on those reads arising from RNAs that are being actively transcribed.
nano-COP reveals the relationship between transcription and splicing
Nano-COP simultaneously determines the position of Pol II and the splicing status of each intron in a nascent RNA read. To determine the physical proximity between splicing catalysis and transcription, we tallied the splicing status of introns within each read as a function of Pol II position (i.e., the 3’ end of nascent RNA) (Figure 2A). Focusing initially on constitutively spliced introns, we found that in two human cell lines (myeloid leukemia K562 cells and B lymphoblast BL1184 cells), < 20% of introns are spliced after Pol II has transcribed two kilobases (kb) (Figure 2B and S5A–E). nano-COP analysis in Drosophila S2 cells revealed that the majority of intron splicing occurred more proximally to Pol II compared with human cells (two-way ANOVA p-value < 10−15), such that ~25% of introns were spliced within 1 kb, and more than 50% were spliced within 2 kb from the intron 3’SS (Figure 2B and S5C–D,F). The introns included in these analyses have representative lengths across the two genomes (Figure S5G) and the analysis is consistent across gene expression levels (Figure S5H), suggesting that a small number of overrepresented genes do not dominate the analysis. Since introns at the boundaries of genes, especially first introns, can have different splicing efficiencies (Khodor et al., 2011; Pai et al., 2017; Tilgner et al., 2012), we also repeated the analysis after removing datapoints from first and last introns and found that the results are substantially the same (Figure S5I).
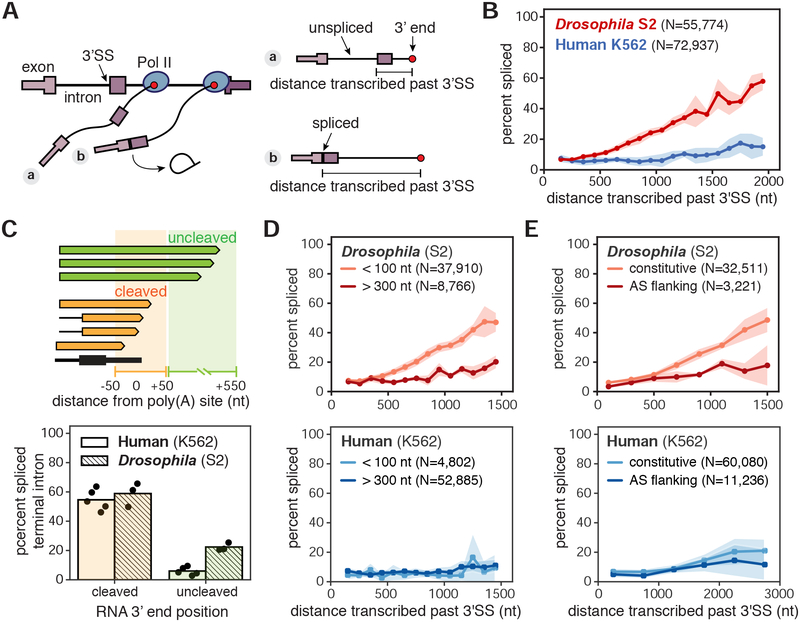
Figure 2. nano-COP measures the physical proximity between transcription and splicing.
(A) Cartoon depicting measurements of distance transcribed past the 3’ splice site (3’SS) and splicing status using nano-COP data from two nascent transcripts. (B) Global analysis of distance transcribed from the 3’SS and the percent of spliced molecules in human K562 (blue) and Drosophila S2 (red) cells (two-way ANOVA p-value < 10−15). (C) Cartoon (top) represents example reads with 3’ ends near gene ends and the regions that differentiate “cleaved” (orange, 50 nt upstream and downstream of poly(A) sites) from “uncleaved” (green, 50–550 nt downstream of poly(A) sites) transcripts. Bar plot (bottom) depicts the proportion of reads with spliced terminal introns in the “cleaved” and “uncleaved” pools of transcripts for human K562 (solid) (N=13,913 cleaved; N=4,293 uncleaved) and Drosophila S2 (hashed) (N=9,534 cleaved; N=6,062 uncleaved) cells (Drosophila S2 chi-square test p-value < 2 × 10−16; human K562 chi-square test p-value < 2 × 10−16). Black points represent results from each biological replicate. (D) Global measurement of the percent spliced as a function of distance transcribed for introns smaller than 100 nt and larger than 300 nt (Drosophila S2 two-way ANOVA p-value < 10−13; human K562 two-way ANOVA p-value = 0.2). (E) Global analysis of transcribed distance from 3’SS and the percent of spliced molecules separated by alternative splicing status of neighboring exons (Drosophila S2 two-way ANOVA p-value < 10−5; human K562 two-way ANOVA p-value = 0.0297). Introns that do not neighbor an alternatively spliced exon are labeled as ‘constitutive’ while introns that do neighbor an alternatively spliced exon are labeled as ‘AS flanking’. Numbers (N) in B, D, and E represent the number of introns within reads that were used to calculate the distance transcribed before splicing plots. Shaded regions (in B, D, and E) represent standard deviation across biological replicates.
A concern in co-transcriptional splicing analyses is the possibility of contamination from fragmented mature RNA. If an RNA 3’ end is formed from fragmentation rather than gene transcription, it does not represent the position of Pol II. If these fragmented RNAs were to be sequenced, it would be depicted as a high level of splicing immediately after the 3’ SS, making it appear as if Pol II has not transcribed as far when splicing occurs for a subpopulation of transcripts. Our data show that only ~5% of reads are spliced within 100 nt of the intron 3’SS and that the proportion rises only when the read end is kilobases away from the intron. Thus, nano-COP data cannot be heavily contaminated by fragmented RNA. Indeed, considering the average human transcription elongation rate of 1–4 kb/min (Danko et al., 2013; Veloso et al., 2014), nano-COP is consistent with previous analyses of mammalian splicing kinetics that describe splicing as generally completing on the order of minutes (Audibert et al., 2002; Coulon et al., 2014; Keohavong et al., 1982; Pandya-Jones et al., 2013; Rabani et al., 2014; Singh and Padgett, 2009; Wachutka et al., 2019).
To assess the extent of post-transcriptional splicing in both species, we focused on the relationship between terminal intron splicing and transcription termination. In human cells, terminal introns were almost exclusively spliced post-transcriptionally (Figure 2C). This finding corroborates established functional links between polyadenylation and terminal intron splicing in vertebrates (Berget, 1995; Niwa and Berget, 1991). In Drosophila cells, ~25% of terminal intron splicing occurred prior to cleavage and polyadenylation (Figure 2C), consistent with the faster co-transcriptional splicing in those cells.
Variations in splicing kinetics between species are likely due to mechanistic differences in splicing regulation. In vitro studies have demonstrated that recruitment and assembly of the spliceosome depends on the predominant gene architecture of an organism. Splicing of short introns (<250 nt) typically occurs through an intron definition model, in which spliceosome factors are initially recruited to the 5’ and 3’ splice sites across an intron to facilitate its splicing. By contrast, long introns (>250 nt) are spliced through an exon definition model, in which splicing factors are recruited to the junctions of an exon at the 5’SS of its downstream intron and the 3’SS of its upstream intron before cross-intron interactions are made to facilitate intron splicing (Berget, 1995; Schneider et al., 2010). Exon definition splicing is presumed to dominate in human cells because most genes contain long introns separated by short exons, whereas in Drosophila melanogaster, which has an abundance of short introns, intron definition splicing is more prevalent (Fox-Walsh et al., 2005; Guo et al., 1993).
In Drosophila cells, we found that short introns (<100 nt) were generally excised faster than long introns (>300 nt) (two-way ANOVA p-value < 10−13; Figure 2D and S5J), and introns neighboring alternative exons were spliced more slowly than those neighboring constitutive exons (two-way ANOVA p-value < 10−5; Figure 2E). However, this difference was partly due to the tendency of long introns to flank alternative exons in Drosophila (Figure S5K) (Fox-Walsh et al., 2005). The abundance of short introns that are rapidly and constitutively spliced in Drosophila cells implies that splicing through intron definition is more efficient than exon definition. These findings are partly in contrast to efficient splicing of long introns (>284 nt) observed using metabolic labeling in Drosophila S2 cells (Pai et al., 2017), which may be due to differences in nascent RNA purification strategies, represented transcripts in each dataset, or calculations of splicing kinetics by time as compared with distance. In human K562 cells, we did not observe the same length dependence for splicing kinetics (two-way ANOVA p-value = 0.2; Figure 2D), and only a slight difference between introns flanking constitutive exons versus alternative exons (two-way ANOVA p-value = 0.0297; Figure 2E). However, as highly expressed genes have shorter introns on average, long introns are less represented in these analyses (Figure S5G), limiting a full analysis of intron size in human cells. A difference at greater distances of Pol II transcription may be found once increased read lengths permit them to be resolved. Nonetheless, these data indicate that the existence of other modes of splicing regulation in humans are likely to be responsible for the differences in splicing kinetics relative to Drosophila.
SF3B activity is required for co-transcriptional splicing
Splicing factors are commonly mutated in certain cancer types (Quesada et al., 2011; Wang et al., 2011; Yoshida et al., 2011), motivating the characterization of splicing inhibitor compounds that exert antitumor activity. For example, mutations in SF3B1, a member of the SF3B complex that performs branch point recognition prior to the first catalytic step of splicing, alter splicing activity and are sufficient to cause cancer phenotypes in mice (Obeng et al., 2016). Several SF3B1 inhibitors, both naturally occurring and synthetically derived, have antitumor properties (Bonnal et al., 2012). However, even though this protein plays a critical role in all splicing reactions, inhibiting SF3B1 by small molecules typically leads to intron retention for a subset of introns as well as other alternative splicing changes in mature mRNA (Sciarrillo et al., 2019; Teng et al., 2017; Vigevani et al., 2017). It remains unclear whether this response is due to differential impacts of SF3B1 on splicing or solely due to variation in RNA turnover or surveillance processes.
To explore this issue, we investigated the impact of the small molecule pladienolide B (PlaB), which competitively binds to SF3B1 (Cretu et al., 2018; Kotake et al., 2007). RT-PCR analysis of cytoplasmic RNA revealed that 1 hour treatment of 100 nM PlaB caused no detectable changes in splice isoforms (Figure 3A). However, an RT-PCR analysis of chromatin-associated RNAs after the same PlaB treatment uncovered an accumulation of unspliced transcripts (Figure 3A). We analyzed published datasets of RNAs that are closer to the site of transcription [nucleoplasm RNA-seq (NP-seq) and mammalian NET-seq (mNET-seq)] and observed only a 2.7% decrease by NP-seq and 10% decrease by mNET-seq in the percent of spliced reads after 4 hours of treatment with 1 μM PlaB (chi-square p-value < 2.2 × 10−16; Figure 3B–C) (Nojima et al., 2015). These results indicate that either PlaB does not have a global influence on splicing or that challenges in analyzing nascent RNA conceal its impact.
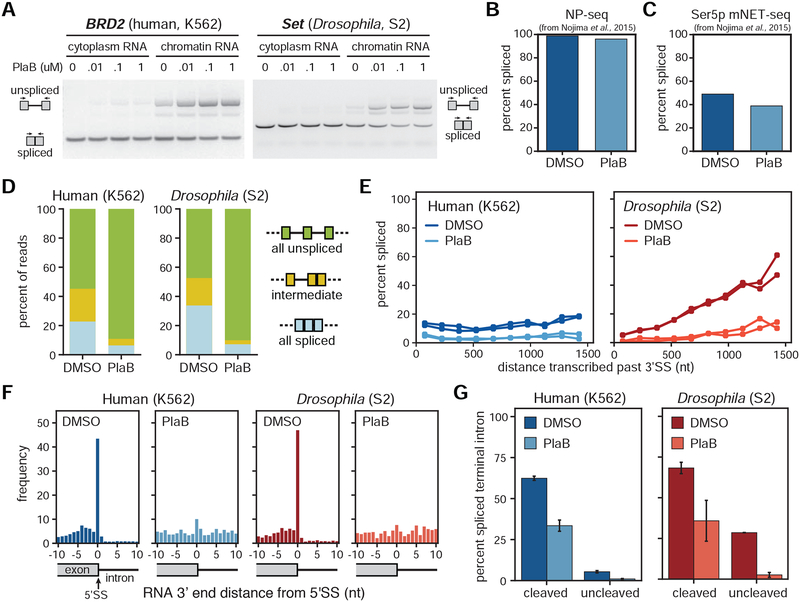
Figure 3. Co-transcriptional splicing is abolished with the splicing inhibitor PlaB.
(A) RT-PCR of example genes in cytoplasmic and chromatin RNA extracted from human K562 cells (left) and Drosophila S2 cells (right) treated with 0.1% DMSO or different concentrations of PlaB (10 nM, 100 nM, and 1 μM). (B) NP-seq and (C) mNET-seq datasets from cells treated with 1 μM PlaB or 0.1% DMSO for 4 hours from (Nojima et al., 2015) were compared for the global percent of spliced reads (spliced reads / total reads aligning to 3’SS junctions). (D) Distribution of splicing patterns found in cells treated with 0.01% DMSO versus 0.1 μM PlaB for 1 hour. In nano-COP reads spanning at least two introns, “all spliced” represents reads where every intron within the read is spliced, “intermediate” represents reads where at least one intron is spliced and one intron is unspliced, and “all unspliced” represents reads where every intron is present and therefore not spliced. (E) Global analysis of distance transcribed past the 3’SS and the percent of spliced molecules in cells treated in biological duplicates with DMSO versus PlaB (human K562 two-way ANOVA p-value < 10−5, Drosophila S2 two-way ANOVA p-value < 10−5). (F) The frequency of RNA ends (the 3’ end of the RNA when aligned) around intronic 5’SS represented as histograms in human and Drosophila cells treated with DMSO or PlaB. High coverage of nascent RNA 3’ ends at 5’SS likely represents free exon ends between the first and second catalytic steps of the splicing reaction. (G) The proportion of reads with spliced terminal introns in the “cleaved” (50 nt upstream and downstream of poly(A) site) and “uncleaved” (50–500 nt downstream of poly(A) site) pools of transcripts from cells treated with DMSO versus PlaB. Bars represent the range from two biological replicates. Chi-square tests were used to compare spliced/unspliced proportions in DMSO vs. PlaB for “cleaved” and “uncleaved” in each species (p-value < 2 × 10−16). In B,C,E-G human K562 samples are represented in blue; Drosophila S2 samples in red; DMSO samples as the darker shade; and PlaB samples as the lighter shade.
To directly investigate how splicing kinetics are affected by interference with SF3B, we performed nano-COP and examined the influence of PlaB on co-transcriptional splicing. When both human K562 and Drosophila S2 cells were treated with 100 nM PlaB for 1 hour, the proportion of spliced nano-COP reads decreased globally by 30% such that less than 10% of reads were spliced in the PlaB datasets (chi-square p-value < 2.2 × 10−16; Figure 3D). Furthermore, co-transcriptional splicing was nearly undetectable in cells treated with the splicing inhibitor by three distinct analyses. First, in PlaB-treated cells from both species, Pol II transcribed more than 1.5 kb before there was any evidence of splicing, which is especially notable in Drosophila which exhibited rapid co-transcriptional splicing under DMSO conditions (Figure 3E). Second, intermediates between the first and second catalytic steps of the splicing reaction, represented by RNAs with 3’ ends at 5’SSs, were abolished in the presence of the splicing inhibitor (Figure 3F). Third, in PlaB-treated cells, terminal introns exhibited no evidence of splicing prior to transcript cleavage and polyadenylation (Figure 3G). Thus, PlaB is a potent and global inhibitor of pre-mRNA splicing. These results also demonstrate the capacity of nano-COP to measure the immediate impact of splicing perturbations.
Intron splicing does not strictly follow the order of transcription
To determine the extent to which transcription impacts splicing order, we analyzed reads spanning pairs of introns in which one intron was spliced and the other was not (Figure S6A, “intermediate”). In contrast to the “first-come, first-served” model of splicing (Aebi and Weissman, 1987), inspection of genes with high coverage frequently revealed examples of downstream introns that were transcribed last but spliced first, such as the third intron of human EIF1 (Figure 4A).
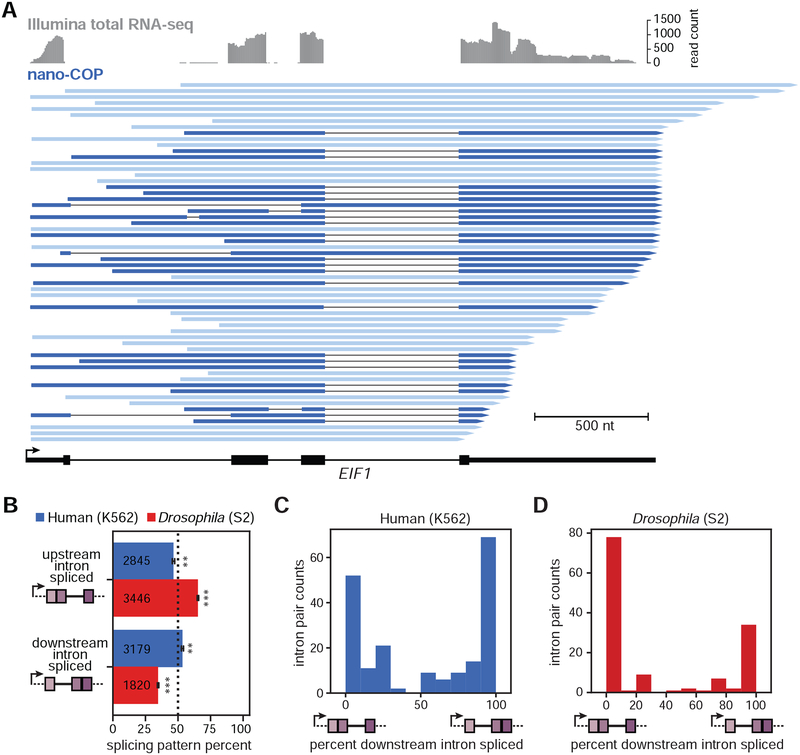
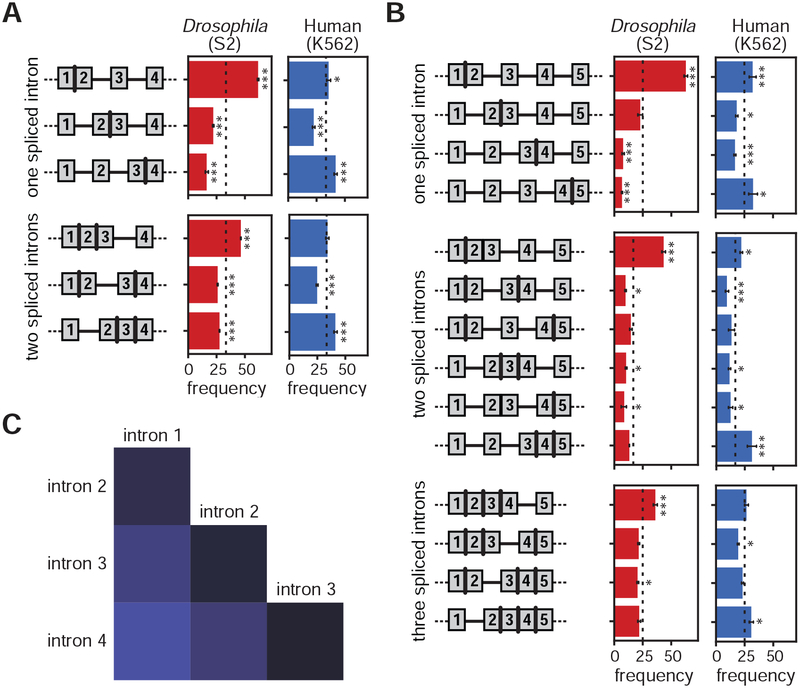
Figure 4. Order of transcription does not strictly dictate splicing order.
(A) Illumina total RNA-seq coverage (top, grey) and nano-COP reads (bottom, blue) that span the 3’SS of all three introns in the EIF1 gene from human K562 cells. Dark blue reads represent RNA transcripts with at least one spliced intron. (B) Measurements of the order of splicing between neighboring intron pairs in both human (blue) and Drosophila (red) cells. For reads that span two or more introns, the top two boxes represent the proportion of reads in which the upstream intron within a pair is spliced first. The bottom two boxes represent the opposite situation, in which the downstream intron within a pair is spliced first. Black bars represent the standard error of the mean across biological replicates. Values within boxes represent the number of datapoints used to calculate splicing order percentages for each subset. A binomial test was used to test whether splicing order percentages differ from random expectations (50%). ** indicates binomial test p-value < 10−4; *** indicates binomial test p-value < 10−100. (C–D) Frequency of spliced patterns within intron pairs that have at least 4 reads aligning to both introns in (C) human K562 (N=193 intron pairs from 127 unique genes) and (D) Drosophila S2 (N=135 intron pairs from 115 unique genes) cells.
To measure splicing order globally, we analyzed all reads that spanned intron pairs and found that, in both K562 and BL1184 cells, splicing does not regularly occur in the order of transcription (Figure 4B and S6B–E), consistent with earlier analyses (Kim et al., 2017). In fact, 54% of the time, the downstream intron within a pair was spliced first in K562 cells, indicating that human introns have a slight bias in favor of splicing from 3’ to 5’, in reverse of the transcription order (binomial test p-value < 2 × 10−5), a trend that does not change across gene expression levels represented in the nano-COP dataset (Figure S6C). By contrast, in Drosophila cells, the upstream intron tended to be spliced first (65% of the time; binomial test p-value < 8 × 10−113) (Figure 4B and S6C–E). This result is consistent with the faster co-transcriptional splicing kinetics and longer exon lengths in this species, as the upstream intron has the possibility of being spliced before the 3’ SS of the downstream intron is transcribed. Because the median nanopore read length is less than 1 kb, the introns examined in this analysis tended to be shorter than the natural intron distributions of the two genomes (Figures S2E–F and S6F). However, analysis of splicing intermediates (in which the 3’ end of the RNA aligns to a 5’SS) is not similarly constrained by intron length (Figure S6G); for reads corresponding to splicing intermediates, the downstream intron is in the process of being spliced regardless of its length. A large proportion of upstream introns within splicing intermediate reads were not spliced (Figure S6H) as observed by previous analyses of splicing intermediates (Nojima et al., 2018), further confirming that splicing does not always follow the order of transcription.
We next asked whether splicing patterns are variable or consistent across the same pair of introns, which would imply that splicing is stochastic or ordered, respectively. For intron pairs that were covered by multiple reads exhibiting intermediate splicing patterns, we measured the frequency of each pattern across the pair. In over 65% of these intron pairs, every read had the same splicing pattern (Figure 4C–D). Thus, although splicing order does not always follow the direction of transcription, especially in human cells, for this set of highly covered intron pairs, it does tend to complete in a preferred order.
Role of cis and trans acting elements in splicing order
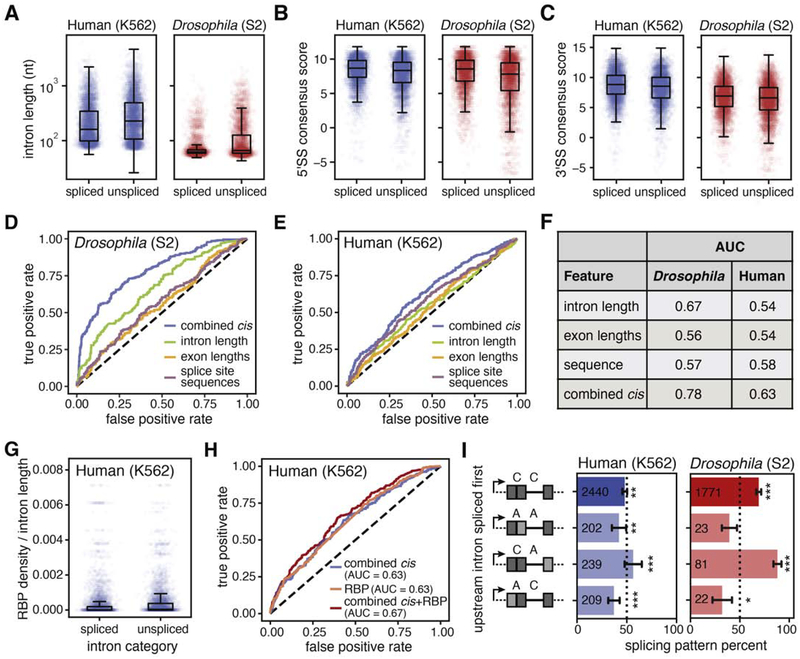
Given that transcription order does not play a dominant role in splicing order for the introns covered in nano-COP datasets, we asked whether cis-acting features are associated with patterns of intron removal. We found that within pairs, spliced introns tended to be shorter (Figures 5A and S6I) and have stronger splice site scores (Figures 5B–C and S6J–K) than the unspliced introns. These trends were especially pronounced in Drosophila S2 cells, but could be observed to a lesser extent in the human K562 dataset.
Figure 5. Cis and trans acting elements influence splicing order.
(A-C) Distributions of (A) intron lengths (human Wilcoxon rank-sum p-value < 10−30; Drosophila Wilcoxon rank-sum p-value < 10−60), (B) 5’ splice site (5’SS) consensus sequence scores (human Wilcoxon rank-sum p-value <10−25; Drosophila Wilcoxon rank-sum p-value < 10−50), and (C) 3’SS consensus sequence scores (human Wilcoxon rank-sum p-value < 10−15; Drosophila Wilcoxon rank-sum p-value < 10−10) in spliced versus unspliced introns within pairs. (D, E) Receiver Operating Characteristic (ROC) curve from a random forest classifier that measures the predictive value of intron features on splicing order in (D) Drosophila S2 cells (3949 training and 1317 testing intron pairs) and (E) human K562 cells (4518 training and 1506 testing intron pairs). (F) Area under the curve (AUC) measurements for ROC curves from random forest models in D and E. (G) Total RNA binding protein (RBP) density across introns was measured using eCLIP peaks from (Van Nostrand et al., 2018). For all reads that span two or more introns where one is spliced and the other is not spliced, total RBP occupancy was determined as the sum of fold change over input for all peaks from all RBPs in the intron, normalized by intron length, and plotted as a boxplot for the spliced and unspliced introns separately (Wilcoxon rank-sum p-value < 10−55). (H) ROC curve from a random forest classifier that measures the predictive value of combined intron features and RBP density on splicing order in human K562 cells (4518 training and 1506 testing intron pairs). (I) The order of intron splicing across gene regions that undergo alternative splicing. For each splicing pattern, the proportion of cases where the upstream intron is spliced first is represented. A binomial test was used to test whether splicing order percentages differ from random expectations (50%). * signifies binomial test p-value < 0.05; ** signifies binomial test p-value < 0.01; *** signifies binomial test p-value < 1 × 10−4. Dark gray boxes represent constitutive exons; light gray boxes represent alternative exons; C, intron that neighbors constitutively spliced exons; A, intron that neighbors an alternatively spliced exon (“AS flanking”). Values within boxes represent the number of datapoints in each category.
To determine the impact of these global trends on splicing order, we tested their predictive power using a random forest model. We found that intron lengths, neighboring exon lengths, and splicing consensus sequences were the strongest predictors of splicing order (Table S2). Combining these three features within the model improved the prediction (Table S2). The combined model for Drosophila splicing performed well (AUC = 0.78) compared to models from each feature alone (AUC = 0.56, 0.57, 0.67 for exon length, SS sequence, intron length respectively) or a random model (AUC = 0.5), suggesting that these cis-acting elements together play critical roles in splicing order (Figure 5D,F). In human K562 cells, the combined model for splicing did not perform as well (AUC = 0.63) (Figure 5E–F), indicating that the cis-acting elements included in the model are not sufficient to explain splicing order for the set of human introns that are well represented in the nano-COP data. As sequencing depth and read lengths of nano-COP increase, it is possible that a broader range of cis-acting elements across human intron pairs will lead to a more predictive model.
To assess the influence of trans-acting factors on splicing order in humans, we analyzed a large eCLIP dataset with occupancy measurements for 120 RNA-binding proteins (RBP) in K562 cells (Van Nostrand et al., 2018). By aggregating signal from all RBPs, we found that total occupancy differed significantly between spliced and unspliced introns within pairs (Wilcoxon rank-sum p-value < 10−55; Figure 5G). Examination of individual proteins revealed that 27 RBPs were significantly more likely to be bound to unspliced introns than spliced introns within pairs, both in terms of the number of introns bound and RBP occupancy levels (Bonferroni-corrected chi-square p-value < 6.6 × 10−4, Bonferroni-corrected Wilcoxon rank-sum p-value < 5 × 10−4). Nine of these RBPs are members of the core spliceosomal machinery, and six are splicing regulators. The rest are uncharacterized RBPs or factors with known roles in other areas of RNA biology (Table S3 and Figure S6L). Combining total RBP occupancy with the above cis-acting features improved the prediction of splicing order with the random forest model (AUC = 0.67; Figure 5H). We were concerned that higher RBP density in unspliced introns could be partly due to the opportunity for slowly-spliced introns to be detected more frequently in eCLIP datasets, even though the RBP density measurements are calculated relative to eCLIP input RNA. However, we do not observe a correlation between RBP occupancy and splicing index in total RNA-seq data, indicating that this is not a major contributing factor (Figure S6M). Interestingly, some of the enriched RBPs in unspliced introns are members of the activated spliceosome (Bact) (Kastner et al., 2019), suggesting that these introns are prepared for splicing but something, perhaps another regulatory factor, is delaying the subsequent reactions.
The order of intron splicing can control alternative splicing decisions (Takahara et al., 2002), so we also investigated the splicing order of intron pairs neighboring alternatively spliced exons. Across intron pairs, introns neighboring constitutively spliced exons show a preference to be spliced before the introns neighboring alternatively spliced exons (binomial test p-value < 3 × 10−5 and < 0.05 for human K562 and Drosophila S2 cells respectively; Figure 5I). Because the splicing of one intron before another could alter the availability of cis-acting elements that influence the splicing regulation in a neighboring exon, this bias has important implications for models of regulated splicing.
Splicing is coordinated across multi-intron genes in humans
In vitro single-molecule studies have demonstrated the capacity for synergistic spliceosomal assembly across human introns (Braun et al., 2018), motivating an analysis of higher-order patterns of splicing in vivo. Long-read sequencing enables analysis of splicing patterns of more than two introns, so we assessed the frequencies of all possible splicing patterns for reads that spanned three or more introns and from RNAs in the process of being spliced (i.e., those containing a mix of spliced and unspliced introns).
In Drosophila cells, the splicing of multiple sequential introns tended to follow the direction of transcription (Figure 6A–B and S7A–B; binomial test p-value < 1 × 10−34), consistent with our analysis of intron pairs (Figure 4B). In human cells, by contrast, intron splicing followed a defined splicing pattern that was not necessarily concordant with the direction of transcription (Figure 6A–B and S7A–B). Furthermore, adjacent introns in human cells were also more likely to have the same splicing status than separated introns (chi-square test p-value < 0.001), supporting a model that proximal introns are coordinately spliced (Figure 6C and S7E–F). These patterns did not change when the first and last introns of all genes were removed from the analysis, confirming that this global pattern is not a result of splicing order differences at the beginnings or ends of genes (Figure S7C–D). Intriguingly, the patterns of intron splicing in human cells showed that the most upstream and downstream introns in each read were more frequently spliced, suggesting that coordinated splicing begins from the ends of transcripts and works inward. Together, these results reveal that higher-order coordinated patterns of splicing are orchestrated across nascent transcripts in human cells.
Figure 6. Splicing is coordinated across neighboring introns in human cells.
(A) Cartoons of each possible intermediate splicing combination across three sequential introns appear to the left of bar plots representing the percentage of each splicing combination among nano-COP reads from Drosophila S2 cells (red, 2,829 reads aligning to 1,209 genes) and human K562 cells (blue, 4116 reads aligning to 1221 genes). The dotted lines at 33% represent the expected percentage if each intron combination were distributed equally. (B) Distribution of splicing patterns in reads that span at least four introns in Drosophila S2 cells (red, 1,214 reads aligning to 565 genes) and human K562 cells (blue, 2268 reads aligning to 710 genes). Dotted lines at 25% for one and three spliced introns and 16.7% for two spliced introns represent the expected percentage if each intron pattern were distributed equally. Error bars represent standard error of the mean across biological replicates. A binomial test was used to test whether percentages differ from random expectations (dotted lines).* signifies binomial test p-value < 0.05; *** signifies binomial test p-value < 1 × 10−5. (C) Heatmap representing the frequency that two introns within a read that spans at least four introns have the same splicing status (both spliced or both not spliced) in human K562 cells. Intron number represents the position of each intron within a read that spans at least four introns such that “intron 1” is the first intron and “intron 4” is the last intron within the set of four introns that a read spans. Results of chi-square tests used to determine statistical significance of differences between pairs are shown in Figure S7E.
Discussion
nano-COP probes the relationship between transcription and splicing in living metazoan cells. This approach works by (1) combining two established approaches for purifying nascent RNA, 4sU labeling and cellular fractionation, thereby substantially improving the enrichment of unspliced pre-mRNA transcripts; and (2) sequencing the stringently purified nascent RNA directly through nanopores, which reveals both the location of transcription and the splice isoform of nascent RNA without amplification based biases. Importantly, treatment with the splicing inhibitor PlaB abolished evidence of co-transcriptional splicing detected by nano-COP, confirming that the approach effectively captures nascent RNA and is sensitive to changes in RNA processing rates.
nano-COP reveals the average physical proximity between nascent RNA ends (or Pol II position) and introns at the moment that splicing completes. We found that the vast majority of human introns were not spliced until after Pol II transcribed 4 kb downstream, whereas in Drosophila S2 cells the majority of splicing occurred within 2 kb of introns. Given that the median transcription rate in both K562 and S2 cells is ~1.25 kb per minute (Ardehali et al., 2009; O’Brien and Lis, 1993; Veloso et al., 2014), our splicing kinetics results are consistent with median splicing half-lives estimated from metabolic labeling data: 2 and 7–14 minutes in Drosophila and mammalian cells, respectively (Pai et al., 2017; Rabani et al., 2014; Wachutka et al., 2019). Our findings suggest that in contrast to S. cerevisiae and S. pombe, in which splicing completes nearly immediately upon synthesis (Carrillo Oesterreich et al., 2016; Herzel et al., 2018), in metazoan cells Pol II is not in close physical proximity to introns when splicing occurs. Since the rate of transcription is proposed to regulate alternative splicing patterns (Dujardin et al., 2014; Fong et al., 2014; Takahara et al., 2002), components of the spliceosome could still assemble on nascent RNA soon after introns are transcribed, in a manner that depends on transcriptional kinetics, even though the transcription machinery is not in close physical proximity when splicing occurs in human cells (Bentley, 2014; Naftelberg et al., 2015).
As nano-COP cannot determine the fate of nascent RNA, it is formally possible that some nano-COP reads originated from RNAs that were destined to be degraded before splicing completion. However, it is unlikely for these species to dominate the sample since most transcribed pre-mRNA splice junctions are successfully spliced (Audibert et al., 2002; Wachutka et al., 2019). We find that introns with differing conversion rates into spliced mRNA junctions (“splicing yield” as measured by RNA metabolic labeling (Wachutka et al., 2019)) exhibit similar splicing dynamics (Figure S7G), albeit with a slight trend for introns with high splicing yields to be spliced more slowly.
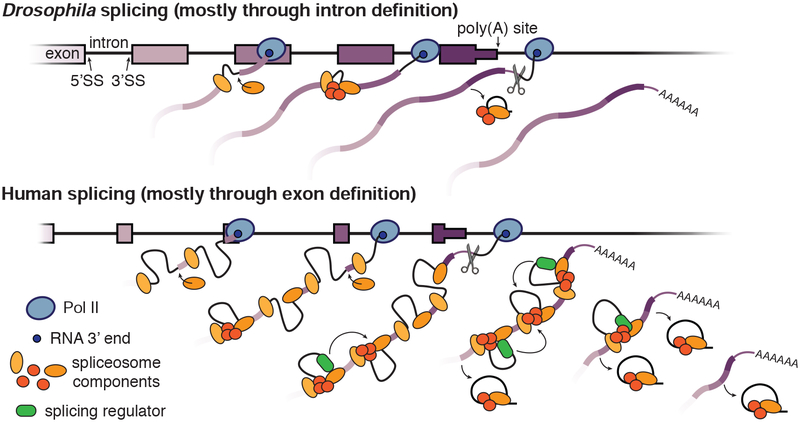
The differences observed by nano-COP between Drosophila and human cells are consistent with previously established models of intron and exon definition splicing, but provide novel insight in both cases (Figure 7). Drosophila has an abundance of short (<100 nt) introns that are assumed to be spliced through intron definition, where the spliceosome assembles at the 5’ and 3’SSs across the intron. Intron definition only requires the coordination of one intron at a time, so the reaction can be completed rapidly in the order of transcription, which we observe. We propose that the prominent role of cis-acting features (e.g. intron length and splice site sequences) in Drosophila splicing regulation is a consequence of intron definition–based splicing in which only one intron is recognized at a time. Human cells, on the other hand, have an abundance of long introns that are predominantly spliced through exon definition, in which the spliceosome initially assembles across exon boundaries before reorganizing to form cross-intron interactions and complete the splicing reaction. The greater distance between Pol II and introns at the time splicing occurs in human cells is in line with exon-definition splicing: the extra step of identifying exon boundaries before intron boundaries presumably increases the time required to splice each individual intron. Furthermore, the extra coordination of multiple introns during exon definition splicing likely relies on accessory factors to direct splicing across multiple introns. We propose that the simultaneous regulation of multiple introns by exon definition drives the coordinated splicing patterns that we observed across groups of three and four introns within human genes. Certainly, some differences between our human and Drosophila observations could be due to cell-type differences rather than species differences, although the human and Drosophila results differed more substantially than our observations across two human cell lines. Nevertheless, nano-COP analysis across many cell types will be critical for determining the variability of splicing dynamics within a single species.
Figure 7. Coupling of transcription and RNA processing during intron definition versus exon definition splicing.
A model depicting the differences in splicing regulation between Drosophila genes (top), which have an abundance of short introns that are spliced through intron definition, and human genes (bottom), which contain mostly long introns that are spliced through exon definition. In Drosophila cells, we observe that splicing typically occurs rapidly, co-transcriptionally, and in the order of transcription. By contrast, in human cells, splicing is slower such that intron splicing does not follow the order of transcription and terminal introns are spliced after cleavage and polyadenylation. Even though splicing catalysis is slower with exon definition, we propose that splicing factors still assemble on the transcribing RNA and likely drive the regulated and coordinated splicing order we observe in human cells.
nano-COP provides a new method to visualize early RNA processing steps in living cells. Our initial nano-COP analyses both corroborated known features of splicing and uncovered intriguing unappreciated aspects of splicing, such as the coordinated splicing across neighboring introns. Future studies will be needed to discover how these splicing patterns are regulated and whether they contribute to alternative splicing decisions. As nanopore sequencing improves in the coming years, nano-COP will only increase in resolution and depth; deeper coverage will allow for analyses of specific introns, longer read lengths will expose patterns across larger transcripts, and greater accuracy will provide allele-specific information.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, L. Stirling Churchman (churchman@genetics.med.harvard.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture.
K562 cells (ATCC, CCL-243) and B lymphoblast BL1184 cells (ATCC, CRL-4959) were maintained at 37°C and 5% CO2 in RPMI 1640 medium (ThermoFisher, 11875119) containing 10% FBS (ThermoFisher, 10437036), 100 U/ml penicillin and 100 ug/ml streptomycin (ThermoFisher, 15140122). L-Glutamine (ThermoFisher, 25030081) was added to the BL1184 medium to a concentration of 2 mM. S2 cells (Expression Systems, 94–005) were maintained at 25°C in Schneider’s Drosophila medium (ThermoFisher, 21720024) containing 10% heat inactivated FBS (ThermoFisher, 16140063), 50 U/ml penicillin and 50 ug/ml streptomycin (ThermoFisher, 15070063). To monitor co-transcriptional splicing in the presence of a splicing inhibitor, cells were incubated with 100 nM Pladienolide B (PlaB, Santa Cruz Biotechnology, sc-391691) or 0.01% DMSO (Sigma, D2438) for 1 hour, unless indicated otherwise.
METHOD DETAILS
4sU labeling.
Cells were labeled in media containing 500 μM 4-thiouridine (4sU, Sigma, T4509) for 8 minutes. We found that 8 minutes was the shortest labeling time to obtain enough nascent RNA for nanopore sequencing (>500 ng) from 100 million human cells or 500 million Drosophila cells. K562 cells were harvested in suspension at 0.8–1 million cells/mL. Drosophila S2 cells were labeled as an adherent layer at 95% confluency and lifted with gentle pipetting before harvesting. Cells were collected by centrifugation at 500 g for 2 minutes and washed once in 1X PBS. Samples for total or 4sU labeled RNA were immediately resuspended in Qiazol lysis reagent (Qiagen, 79306) for RNA extraction. Samples for chromatin-associated RNA purification proceeded immediately to the cellular fractionation protocol.
Cellular fractionation.
The cellular fractionation protocol was performed exactly as described in steps 8–21 of (Mayer and Churchman, 2016). In brief, samples with 10 million K562 cells or 50 million S2 cells were lysed for 2 min with 200 μl cytoplasmic lysis buffer (0.15% (vol/vol) NP-40 (Thermo Fisher Scientific, 28324), 10 mM Tris-HCl (pH 7.0), and 150 mM NaCl), layered over 500 μl of a sucrose cushion (10 mM Tris-HCl (pH 7.0), 150 mM NaCl, 25% (wt/vol) sucrose), and nuclei were collected by centrifugation at 16,000 g for 10 minutes. The nuclei pellet was resuspended in 800 μl wash buffer (0.1% (vol/vol) Triton X-100, 1 mM EDTA, in 1X PBS) and collected by centrifugation at 1,150 g for 1 minute. Washed nuclei were resuspended in 200 μl glycerol buffer (20 mM Tris-HCl (pH 8.0), 75 mM NaCl, 0.5 mM EDTA, 50% (vol/vol) glycerol, 0.85 mM DTT), and mixed with 200 μl nuclei lysis buffer (1% (vol/vol) NP-40, 20 mM HEPES (pH 7.5), 300 mM NaCl, 1 M urea, 0.2 mM EDTA, 1 mM DTT) before pulse vortex and incubation on ice for 2 minutes. The chromatin pellet was collected by centrifugation at 18,500 g for 2 minutes and resuspended in 1X PBS. All steps were performed at 4°C and all buffers were prepared with 25 μM α-amanitin (Sigma, A2263), 0.05U/μl SUPERase. In (ThermoFisher Scientific, AM2694) and protease inhibitor mix (Roche, 11873580001).
Western blot.
Samples for western blot analysis were mixed with 1X PBS to equal volumes for all fractions. The chromatin sample was homogenized by adding 2 μl Benzonase (Sigma, E1014) and mixing with a 1-ml syringe and 22G needle. Western blots were performed using an antibody against Ser2-phosphorylated CTD (transcribing Pol II) (Active Motif, 3E10) or GAPDH (LifeTechnologies, AM4300/6C5) and imaged using a Licor Odyssey.
Splicing inhibitor PlaB RT-PCR.
K562 and S2 cells were incubated with 0.1% DMSO or 10 nM, 100 nM, or 1 μM PlaB splicing inhibitor for 1 hour. The cells were subjected to cellular fractionation to collect cytoplasmic and chromatin-associated RNA. Both RNA samples were extracted using Qiazol lysis reagent (Qiagen, 79306) following the manufacturer’s instructions and DNA was degraded with DNase I (Qiagen, 79254). RNA was reverse transcribed with the SuperScript III First-Strand Synthesis System (Invitrogen, 18080051) using 5 ng/ul random hexamer primers following manufacturer’s instructions. PCR was performed with Phusion High Fidelity polymerase (ThermoFisher, 5F30S), 0.5 μM primers, and 1 μl cDNA from the RT reaction for 1 cycle of 2 min 98°C, 30 cycles of 20 sec 98°C, 30 sec 60°C, and 1.5 min 72°C and 1 cycle of 3 min 72°C. Samples were run on a 1% agarose gel to separate DNA fragments. Primer sequences are shown in Table S4.
Biotinylation and labeled RNA selection.
RNA was first extracted using Qiazol lysis reagent (Qiagen, 79306) following the manufacturer’s instructions. Total RNA (~300 μg per reaction) and chromatin-associated RNA (~50–60 μg per reaction) were subjected to 4sU labeled RNA purification as previously described (Dölken et al., 2008; Schwalb et al., 2016). In brief, labeled RNA (1 μg / 10 μl) was incubated with 10% biotinylation buffer (100mM Tris pH 7.5, 10mM EDTA) and 20% EZ-Link Biotin-HPDP (1 mg/mL resuspended in DMF, Thermo Fisher Scientific, 21341) for 1.5 hours at 800 rpm and 24°C in the dark. RNA was purified by mixing with a 1:1 ratio by volume of chloroform/isoamylacohol (24:1), separating with a phase-lock tube at 16,000 g for 5 min, and performing isopropanol precipitation. Biotinylated RNA separation was performed using the μMACS streptavidin kit (Miltenyi Biotec, 130-074-101). RNA was mixed with μMACS streptavidin beads at a 2:1 ratio by volume at 800 rpm and 24°C for 15 min. RNA-streptavidin beads mix was transferred to the μMACS column and washed with wash buffer (100 mM Tris pH 7.5, 10 mM EDTA, 1 M NaCl, 0.1% Tween 20) at 65°C and room temperature each 3 times. Selected RNA was eluted off the magnet with 0.1M DTT and purified using the miRNeasy micro kit (Qiagen, 217084) with on-column DNase I treatment (Qiagen, 79254).
RT-qPCR splicing analysis.
100 ng RNA was reverse transcribed with the SuperScript III First-Strand Synthesis System (Invitrogen, 18080051) using 5 ng/μl random hexamer primers following the manufacturer’s instructions. qPCR was performed with SsoFast EvaGreen Supermix (Biorad, 1725200) with 1 μM primers using manufacturer’s instructions and a dilution series of cDNA as the standard curve. Samples were denatured at 95°C for 30 seconds followed by 40 cycles of 95°C for 5 seconds and 55°C for 10 seconds. Fluorescence was captured after every step of the 55°C extension. Primers were designed as in the cartoon on the top left of Figure S1D and analysis was performed according to the equation in the right of Figure S1D [percent spliced = 1/(1+2ΔCt(spliced-unspliced))]. Primer sequences are indicated in Table S4.
Illumina sequencing library preparation and sequencing.
Libraries were prepared using the Ovation Universal RNA-seq System (NUGEN, 0343–32) with Universal Human rRNA strand selection reagent (NUGEN, S01859) following the manufacturer’s instructions. All samples were sequenced 2×80 on a NEXTseq 500 sequencer (Illumina, San Diego, CA, USA) in the Biopolymers Facility at Harvard Medical School.
Illumina sequencing data analysis.
Paired-end reads were aligned to ENSEMBLE GRCh38 (release-86) and FlyBase dm6 (r6.19) reference genomes using STAR (v2.5.1a) (Dobin et al., 2013) with default parameters (except for readFilesCommand=cat, limitIObufferSize=200000000, limitBAMsortRAM=64000000000, outReadsUnmapped=Fastx, outSAMtype=BAM SortedByCoordinate, outSAMattributes=All, outFilterMultimapNmax=101, outSJfilterOverhangMin=3 1 1 1, outSJfilterDistToOtherSJmin=0 0 0 0, alignIntronMin=11, alignEndsType=EndToEnd). Total RNA RPKM was measured by counting the number of read pairs within the coding region of each gene divided by the length (in kilobases) of the gene divided by million mapped reads for each sample. Global percent spliced calculations were determined by dividing the number of spliced reads by total (spliced + unspliced) reads that span 3’SS junctions. Splicing index calculations were determined for each gene by counting the number of read pairs that span exon junctions by at least 3 nucleotides and measuring the number of spliced reads divided by unspliced reads; splicing index = 2 × spliced read pairs / (5’SS unspliced + 3’SS unspliced read pairs). NP-seq and mNET-seq datasets from HeLa cells treated with 0.1% DMSO or 1 μM PlaB for 4 hours were retrieved from (Nojima et al., 2015) under the GEO accession number GSE60358. Illumina total RNA sequencing from K562 cells treated with 100 μM 4sU for 4 hours were retrieved from (Schofield et al., 2018) under the GEO accession number GSE95854.
Oxford Nanopore direct RNA library preparation and sequencing.
Ribosomal RNAs were depleted from the 4sU-selected chromatin-associated RNA sample using Ribo-Zero Gold rRNA Removal kit (Illumina, MRZG126) or RiboMinus Eukaryotic Kit v2 (ThermoFisher, A15020) (Table S1). The change in rRNA depletion kits was due to discontinuation of the stand-alone Illumina Ribo-Zero kit between experiments. Poly(A) tails were added to the 3’ ends of the 4sU labeled chromatin-associated RNA with E. coli poly(A) polymerase from New England Biolabs (M0276S) for unperturbed samples or Clontech/Takara (2180) for PlaB/DMSO samples, incubating at 37°C for 1 hour or 7.5 minutes, respectively. Poly(I) tails were added to the 3’ ends of RNA with yeast poly(A) polymerase (ThermoFisher, 74225Z25KU) and Inosine triphosphate (Sigma, I0879) following manufacturer’s instructions and incubating at 37°C for 30 minutes. The “no tailing” sample proceeded to sequencing without the enzymatic addition of a poly(A) or poly(I) tail. The direct RNA sequencing protocol using the SQK-RNA001 kit (Oxford Nanopore Technologies Ltd.) or SQK-RNA002 kit (Oxford Nanopore Technologies Ltd.) (Table S1) was followed exactly as described by the manufacturer with minor modifications described below. In brief, 500 ng of RNA sample was ligated to the provided T10 splinted adapter for poly(A) tailed RNA or custom C10 splinted adapter for poly(I) tailed RNA using T4 DNA ligase (New England Biolabs, 2,000,000 units/mL, M0202M) for 15 minutes at room temperature. The custom C10 splinted adapter was generated by annealing two oligonucleotides (IDT, ONT_oligoA and ONT_oligoB_C10 shown in Table S4) at 1.4 μM in 10 mM Tris-HCl pH 7.5, 50 mM NaCl by heating to 95°C and slowly cooling to room temperature. The ligated RNA sample was reverse transcribed using SuperScript III (Invitrogen, 18080044), as recommended by Oxford Nanopore Technologies. The resulting cDNA does not get sequenced, but instead serves to minimize secondary structures in the RNA that can interfere with RNA threading through the pore, thereby improving sequencing throughput. Samples were purified with Agencourt RNAClean XP beads (Beckman Coulter, A63987) and ligated to the sequencing adapter with preloaded motor protein. After a second purification step with Agencourt RNAClean XP beads, the sample was resuspended in Elution Buffer, mixed with RNA Running Buffer, loaded onto a primed FLO-MIN106 flowcell, and sequenced using MINKNOW software for 48 hours with default settings for direct RNA sequencing. nano-COP datasets from human K562 cells, human BL1184 cells and Drosophila S2 cells were prepared in five, two and three biological replicates, respectively (Table S1). An additional sample from human K562 cells and Drosophila S2 cells was prepared as a technical replicate and was combined with the other technical replicate for downstream analyses (Table S1). nano-COP datasets from each cell type treated with 0.01% DMSO and 100 nM PlaB were prepared in biological duplicates (Table S1).
Direct RNA nanopore sequencing basecalling and alignment.
Raw signal fast5 files from the unperturbed poly(A)-tailed samples were basecalled using Albacore 2.2.7 (Oxford Nanopore Technologies Ltd.) with the following parameters: read_fast5_basecaller.py --flowcell FLO-MIN106 --kit SQK-RNA001 --recursive --output_format fast5,fastq --worker_threads 8 -- save_path ${savePath} --input ${inputPath}. PlaB/DMSO and unperturbed poly(I)-tailed sequences were collected by live basecalling with MinKNOW (release 18.12.6 and 18.12.9). To enable sequence alignment, RNA sequences that pass basecalling thresholds were converted into DNA sequences by substituting U to T bases before mapping. The reference genomes used for K562 and S2 sequence alignments were obtained from ENSEMBLE GRCh38 (release-86) and FlyBase dm6 (r6.19), respectively. Sequences were aligned to the reference genomes using minimap2 (version 2.10-r764-dirty) (Li, 2018) with recommended parameters for Oxford Nanopore Technologies direct RNA sequencing (-ax splice -uf -k14) and GMAP (version 2018-03-25) (Wu and Watanabe, 2005) with default parameters. We observed consistent results between the two aligners and decided to display all analyses using minimap2. All analyses were performed using reads that pass the MINKNOW sequencing threshold (QC>7) and align uniquely to the genome. The direct RNA basecalling step is designed to remove the poly(A) or poly(I) tail when converting between raw signal into nucleotide sequences, but does exhibit higher error at the start of reads. The alignment software minimap2 is designed to manage these errors by soft-clipping the start of nanopore reads until it is confident in the alignment. Many of the analyses in this manuscript used a filtering step to remove reads with large soft-clipping events to prevent inaccuracies in read end alignments.
Read coverage per gene visualization and comparisons.
To display nano-COP reads for figures, split bed files with read information were converted into diagrams using pyGenomeTracks (Ramírez et al., 2018). All sequences from K562 nano-COP replicates were combined when comparing with Illumina sequencing data. Coverage per gene for comparing ONT and Illumina samples was calculated using BEDTools coverage (Dale et al., 2011; Quinlan and Hall, 2010) with parameters s=True and mean=True. Counts per gene for comparing ONT replicates was calculated using BEDTools coverage (Dale et al., 2011; Quinlan and Hall, 2010) with parameters s=True and counts=True. The top 10% of expressed genes for intron distribution plots were determined using nano-COP read counts per gene divided by gene length.
Comparisons between nano-COP and direct RNA sequencing of poly(A)-selected mRNA.
Direct RNA sequencing samples with poly(A) selected RNA from immortalized human B-lymphocyte cells (GM12878) by the Oxford Nanopore RNA Consortium (Hopkins run 1 and UCSC run 1) (Workman et al., 2018) were accessed from https://s3.amazonaws.com/nanopore-human-wgs/rna/fastq/Hopkins_Run1_20170928_DirectRNA.pass.dedup.fastq and https://s3.amazonaws.com/nanopore-human-wgs/rna/fastq/UCSC_Run1_20170907_DirectRNA.pass.dedup.fastq. The reads were aligned to the hg38 reference genome using the same parameters for nano-COP as outlined above. Direct RNA reads from nano-COP and Oxford Nanopore RNA Consortium mRNA were analyzed for ‘match percent’, which represents the number of bases that match the reference genome divided by the total number of bases that align to the reference genome for each read. Samples were also compared for ‘read length’, as determined by the length of the read in the fastq file. To generate confusion matrices from each library to compare sequencing and alignment accuracies, all mapped regions of direct RNA nanopore reads were recorded for their aligned base and reference base. For 100,000 random aligned sequence segments in each sample, the frequency of each base matching the reference was recorded and plotted as a matrix.
Direct RNA 3’ end analysis.
RNA 3’ ends (deriving from the 5’ ends of sequenced reads) were recorded from minimap2 sequence alignments and assigned to gene feature categories. “Intron” and “exon” regions refer to their respective annotated features within protein coding genes from hg38 RefSeq annotations. “Poly(A)” sites are defined as regions within 50 nucleotides of the end coordinate of annotated genes. In K562 datasets, the poly(A) region also includes coordinates within 50 nucleotides of RNA-PET annotations from cytoplasm and chromatin fractions in K562 ENCODE data (ENCODE Project Consortium, 2012). “Post-poly(A)” sites are defined as the region between 50–550 nucleotides after the end of annotated genes. “Splice sites” are defined as 50 nucleotides upstream and 10 nucleotides downstream of annotated 5’ splice sites. “Undetermined” categorizes reads that align to more than one category and “other” represents read ends that do not align in the sense direction of annotated gene features (e.g. antisense transcripts, noncoding RNAs, intergenic transcription, etc.). Significant differences between poly(A) site alignments between samples with and without enzymatic poly(A) tail addition were compared using a chi-square test. Poly(A) tail lengths were estimated using Nanopolish version 0.11.1 (Loman et al., 2015; Workman et al., 2018). Raw signal fast5 files were indexed with nanopolish index; reads were segmented and poly(A) tail lengths were calculated with nanopolish polya using default parameters. Reads with the quality control flag “PASS” and with estimated tail lengths greater than 0 were used to plot tail length distributions for RNA 3’ ends aligning to gene bodies (intron, exon or splice site), poly(A) sites, or post-poly(A) sites based on annotated features from hg38 RefSeq annotations as described above. ERCC-00048 was synthesized as a G-block with a T7 promoter (IDT, see Table S4) and in vitro transcribed using the HiScribe T7 High Yield RNA Synthesis kit (NEB, E2040S). The resulting RNA was purified by gel extraction and tailed using E. coli poly(A) polymerase (Clontech/Takara, 2180) with ATP or yeast poly(A) polymerase (ThermoFisher, 74225Z25KU) with ITP and sequenced with the SQK-RNA002 kit on the MinION instrument (Oxford Nanopore Technologies Ltd.) following the same methods described previously for nano-COP. Live basecalled reads were aligned using the same default parameters of minimap2 (Li, 2018) for nano-COP as outlined above. The 3’ end positions of reads were plotted in relation to the known transcript end site of the ERCC-00048 transcript.
Splicing intermediates analysis.
The prevalence of splicing intermediates, which are reads deriving from the splicing process rather than active transcription, were observed by measuring the coverage of read starts (RNA 3’ ends) at 5’SS’s. Reads with more than 75 nt soft-clipped from the RNA 3’ ends during alignment were discarded from the analysis due to uncertainty in alignment accuracy. RNA 3’ ends that align exactly at 5’SS (or the last base of an exon) are considered to be splicing intermediates because they likely arise from a free RNA end during the splicing reaction rather than active transcription. With the assumption that the downstream intron is actively undergoing splicing when the 3’ end of the read is at the 5’SS of the downstream intron, splicing order was determined for these intermediates by measuring the splicing status of the upstream (first transcribed) intron. Splicing status and order were implemented as described below.
Identifying constitutively spliced introns for splicing analyses.
Constitutively spliced introns were identified using short-read total RNA sequencing data from human K562 cells in this study, human B-lymphoblasts from ENCODE (accession number ENCSR000AEE) and Drosophila S2 cells from (Pai et al., 2017) under the GEO accession number GSE93763. RefSeq annotations of all gene and intron coordinates were extracted from the UCSC table browser both for hg38 and dm6. Constitutively spliced introns were determined to be those that have at least 20 reads that span its 5’ and 3’ splice junctions with at least 4 nt of the read overlapping the junction on both sides (either into the upstream/downstream exon or intron) and more than 80% of the spanning reads are spliced. These constitutively spliced introns were labeled as the “medium stringency” introns and used in all analyses for the distance transcribed before splicing plots. “High stringency” (5 nt overlap; 50 read coverage; 90% spliced), “low stringency” (3 nt overlap; 10 read coverage; 50% spliced), and “no stringency” (3 nt overlap; 0 read coverage, and 0% spliced) intron datasets were also produced to demonstrate the variation in results with different levels of intron retention. Maximum stringency categories for all introns are reported in Tables S5 and S6. Intron “splicing yield” measurements that calculate the ratio of synthesis rates between unspliced intron-exon junctions and the spliced exon-exon junctions were retrieved from (Wachutka et al., 2019).
Determining splicing status of introns in Oxford Nanopore direct RNA sequencing reads.
Due to the high error rate of nanopore sequencing, custom scripts were utilized to determine splicing status of direct RNA nanopore reads. First, aligned reads that overlap annotated intron coordinates (see above section for details on included introns) were identified and characterized using BEDTools intersect (Dale et al., 2011; Quinlan and Hall, 2010). Features of read cigar strings were extracted for the 50 nt around the intron 5’ and 3’ splice sites and the entirety of the intron using the Pysam toolkit (https://github.com/pysam-developers/pysam) (Li et al., 2009). Reads were called as ‘not spliced’ if the alignment file shows no indication of splicing (CIGAR string “N” = 0) within the 50 nt around each splice site, mapped portions of the read (rather than deletions) represent greater than 50% of the 50 nt around each splice site, and at least 75% of the read within the intron is aligned to the reference. Reads were called as ‘spliced’ if the alignment file displays the start or end of a splicing event within the 50 nt around both splice sites and the size of the aligned splicing event is within 90–110% and 100 nt of the intron size. If aligned reads that map to introns do not meet these qualifications, the splicing event is characterized as ‘undetermined’ and not used in subsequent analyses.
Measuring the distance transcribed before splicing.
Reads with transcript 3’ ends that map near poly(A) sites or splice sites were removed from this analysis since they likely correspond to completed RNAs or splicing intermediates, respectively, rather than actively transcribing Pol II. Poly(A) sites were defined as the RefSeq annotated end coordinate of a gene from hg38 and dm6 genome assemblies as well as coordinates of RNA-PET 3’ end signal from cytoplasm and chromatin fractions in K562 ENCODE data (ENCODE Project Consortium, 2012). Reads with 3’ ends within 150 nt upstream or any distance downstream of an annotated poly(A) site were discarded from the analysis. We also discarded reads that end within 50 nt upstream or downstream of a read end alignment in the -poly(A) dataset to avoid signal from unannotated polyadenylation sites. To discard reads that possibly originated from splicing intermediates, RNA sequences with 3’ ends between 50 nt upstream and 10 nt downstream of an annotated intronic 5’ splice site were discarded from the analysis. Reads with more than 150 nt soft-clipped from the RNA 3’ ends during alignment were also discarded from the analysis due to uncertainty in Pol II position. RNA 3’ end mapping was performed with BEDTools intersect (Dale et al., 2011; Quinlan and Hall, 2010). The remaining reads were characterized for intron coverage and splicing status as described previously. In order to avoid read length constraints biasing the distance transcribed before splicing results, we only included cases where the length of the read is greater than the genetic distance from the read end to the intron 3’SS by at least 150 nt. Thus, the intron would be measured regardless of whether or not other introns within the same read are spliced or not spliced. For all reads included in the analysis, the distance between the 3’ end of the read and the 3’SS of the intron(s) it aligns to as well as the splicing status of the intron(s) it aligns to were recorded. Transcribed distances past intron 3’SS’s were binned into 100–500 nt windows depending on the number of datapoints in each bin. For all points within a bin, the percent of spliced introns was measured such that percent spliced = number of spliced molecules / total number of molecules. Statistical significance in differences in the distance transcribed before splicing was tested by two-way ANOVA using the function aov in R, considering percent spliced as the dependent variable and group (e.g. species, intron class, expression level, etc.) and distance bins as the independent variables (percent spliced ~ group + distance).
Characterizing the relationship between transcription termination and last intron splicing.
For this analysis, poly(A) sites were defined as the end coordinate of RefSeq annotated genes from hg38 and dm6. Only the 3’-most poly(A) site for each terminal intron was utilized in this analysis to prevent ambiguity between uncleaved reads and downstream poly(A) sites. Reads with 3’ ends within 50 nt upstream and 50 nt downstream of poly(A) sites are referred to as “cleaved” and likely represent 3’ end processed and polyadenylated transcripts. Reads with 3’ ends between 50 to 500 nt downstream of poly(A) sites are referred to as “uncleaved” and likely represent pre-cleaved nascent transcripts. For reads in these two classes that also span the 3’SS of the terminal intron within the same gene, the splicing status of the intron it spans was determined as described above. The proportion of spliced terminal introns out of total was determined and plotted for each class and species.
Determining introns flanking alternative splicing (“AS flanking”).
Alternative exons were identified from total RNA sequencing data in K562 cells from this study and S2 cells from (Paiet al., 2017) using MISO (Katz et al., 2010). Mean insert length and standard deviation were assessed using CollectInsertSizeMetrics from Picard v.18.15 (http://broadinstitute.github.io/picard). Alternative event annotations (skipped exon, alternative 5’ or 3’ splice sites, and mutually exclusive exons) were obtained from the MISO annotations webpage (https://miso.readthedocs.io/en/fastmiso/annotation.html) for dm3 and hg19 (version 2) and lifted over to dm6 and hg38 using CrossMap (Zhao et al., 2014). Only alternative exons with percent spliced in (PSI) > 0.8 in the corresponding cell type (K562 or S2 cells) were considered for further analyses to use similar criteria for exon inclusion as the “medium stringency” measure used for introns in the distance transcribed before splicing analysis and exclude the confounding effect of partial splicing on kinetics measurements. In addition, annotations of all gene, transcript and exon coordinates were obtained in GTF format from NCBI (RefSeq) for hg38 and from FlyBase for dm6. For S2 cells, introns were considered “AS flanking” when they adjoin exons that are not present in every transcript of an annotated RefSeq gene and are identified as alternative by MISO with PSI > 0.8. For alternative 5’ or 3’ splice sites, only introns in 3’ or in 5’ of the alternative exon, respectively, were considered “AS flanking”. Conversely, introns were considered constitutive when they adjoin exons that are present in every isoform of an annotated RefSeq gene and are not identified as alternative by MISO. Introns that did not meet these criteria were excluded from the analysis. For K562 cells, since most introns neighbor exons that are not present in every isoform of a gene, introns were only considered “AS flanking” when they adjoin exons that are identified as alternative by MISO with PSI > 0.8 and introns were considered constitutive when they adjoin exons that are not identified as alternative by MISO. Introns that did not meet these criteria were excluded from the analysis. For measuring the distance transcribed before splicing of reads spanning “AS flanking” and constitutive introns, we used the same approach described above. Alternative splicing classifications for all introns are reported in Tables S5 and S6.
Determining the order of intron splicing.
Reads that span two or more introns were used to characterize the order of intron splicing by nanopore sequencing. The algorithm requires that the length of the read must be greater than the genetic distance (containing the intron sequences) between the 3’ end of the read and the 5’ splice site of the first intron transcribed in the pair. This step ensures that the size of either intron in the pair does not bias the order of splicing findings. For reads that span two or more introns, the splicing status of introns in each neighboring pair was recorded as described above. For neighboring introns that have different splicing statuses, the frequency at which the upstream intron in the pair (first transcribed) is spliced before the downstream intron in the pair (second transcribed) was recorded for each species. A binomial test was used to assess the statistical significance that splicing order deviates from random (50%). For the splicing order comparisons between intron pairs with multiple spanning reads, we collected all intron pairs with 4 or more reads that span both introns. The splicing order was determined for each read and frequency of the order across all reads spanning the intron pair was plotted. Results were consistent when different coverage thresholds were used and so a coverage of 4 was used to include a reasonable number of intron pairs.
Intron features that influence splicing order and kinetics.
RefSeq annotations of gene and intron coordinates were extracted from the UCSC table browser for hg38 and dm6. Features of all introns in K562 and S2 cells, such as lengths of introns and neighboring exons, were recorded to determine which cis elements influence splicing kinetics and/or order. 5’SS and 3’SS consensus sequence scores were extracted from hg38 and dm6 assemblies using MaxEnt with default parameters (Yeo and Burge, 2004). Intron position within genes (e.g. first, middle, and last) was also determined from the annotation file. Intron features are reported in Tables S5 and S6.
Analysis of eCLIP data in K562 cells.
For assessing RNA-binding protein (RBP) density in K562 cells, the fold change over input for significant peaks in publicly-available enhanced crosslinking and immunoprecipitation (eCLIP) data was obtained for 120 RBPs (Van Nostrand et al., 2018). eCLIP peaks located in introns were identified using BedTools intersect, requiring that at least 50% of the peak overlap with the intron. For each intron, we calculated the sum of the density of all peaks within this intron for all RBPs. RBP density was set to 0 for introns that do not overlap any peaks. Total RBP density per intron is reported in Table S5. To compare RBP density in pairs of spliced and unspliced introns, we used all reads that span two or more introns as described above and normalized by intron length. For each individual RBP, if at least two intron pairs displayed binding in one intron (n=101 RBPs), the difference in binding density between spliced and unspliced introns within pairs was assessed using Wilcoxon rank-sum. In addition, for RBPs binding at least one spliced and one unspliced intron (n=76 RBPs), the number of bound spliced and unspliced introns was compared using a chi-square test. Multiple testing correction was performed using the Bonferroni method.
Random forest model to determine features that influence splicing order in pairs.
The predictive value of intron features for splicing order was determined using a Random Forest model with python scikit learn (Pedregosa et al., 2011). Features for intron length (lengths of upstream and downstream introns), surrounding exon lengths (lengths of upstream, middle, and downstream exons), splice site scores (5’ and 3’ splice sites for both introns), intron position within genes [intron number and position (i.e., first, middle, or last) within gene], and RBP density (for K562 cells only) were compiled for all introns. In total, the compiled dataset included 15 features and compiled + RBP included 16 features. Feature importance scores were generated using a random forest classifier with 75% training and 25% testing sets. The total reads spanning intron pairs in the model are 5266 for S2 cells and 6024 for K562 cells. To avoid the same intron pairs in training and test sets, the intron pairs were sorted by chromosome location with the parameter “shuffle=False” during splitting. The following parameters were used in the random forest classifier, except for random_state = None, n_estimators = 300, max_features = None, max_depth = None. Parameters were determined using a validation set to avoid overfitting. ROC curve and AUC measurements were determined from binary prediction probabilities. Prediction accuracy was determined by measuring the difference between the model’s predictions on a held-out test set and measured values. The baseline score was determined using a “null” parameter that has the same value for every training and testing pair; thus, baseline represents the prediction accuracy with no additional information added to the model.
Splicing coordination across multiple introns.
Reads that span 3 or more introns were collected in the same manner as reads spanning intron pairs (described above) and used to characterize the coordination of intron splicing. In cases where at least one of the three introns in the triplet has a different splicing status than the others, the order of splicing was determined as described for intron pairs. The frequency of triplet and quadruplet splicing patterns was recorded and mapped as a bar plot for all samples in K562 and S2 cells. The heatmaps of intron splicing comparisons in human K562 and BL1184 cells were prepared by compiling all reads that span four introns where at least one of the four introns has a different splicing status than the others. In every read the splicing status of each intron was compared to the splicing status of each other intron within the same read. The frequency at which two distinct introns within a quadruplet have the same splicing status (e.g. both not spliced or both spliced) was plotted as a heatmap. To test statistical significance across pairs in the heatmap, a chi-square test was used to compare each pair of intron pairs and multiple testing correction was performed using the Bonferroni method.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical details for individual experiments can be found in the figure legends, the Results section, or in the STAR Methods. This includes number of observations, number of replicates, statistical tests used, and significance level.
DATA AND CODE AVAILABILITY
All Illumina and nanopore sequencing data generated for this paper are available at Gene Expression Omnibus (GEO) under the accession number GSE123191. Other raw data are published on Mendeley (doi:10.17632/9gfs2kkxbc.2). All scripts and data analyses are available at https://github.com/churchmanlab/nano-COP.
Supplementary Material
Table S1. Key statistics of nano-COP libraries sequenced in this study, Related to STAR methods. Experimental differences and sequencing statistics are described for every nano-COP library that was incorporated into this study. Key experimental points include replicate number, species, cell line, treatment, and tailing approach. Key sequencing statistics include sequencing instrument, total sequenced reads, uniquely mapped reads, and percent uniquely mapped reads.
Table S5. Features of all introns in human K562 cells, Related to STAR methods. The coordinates of the intron are displayed as chromosome, start position, end position, and strand. ‘Gene name’ refers to the RefSeq name for coding genes. ‘Intron pos’ is the intron number in the order of transcription. ‘Intron total’ is the total number of introns within that gene. ‘Intron order’ is the descriptive position of the intron within the gene (first, middle, or last). ‘Distance from start’ and ‘distance from end’ are the lengths in nucleotides from the start and end coordinates of the gene, respectively. ‘Intron length’, ‘upstream exon length’, and ‘downstream exon length’ are the sizes of the intron and surrounding exons in nucleotides. ‘5SS score’ and ‘3SS score’ are the splice site consensus sequence scores at the 5’SS and 3’SS, respectively, as determined by the maximum entropy calculations from (Yeo and Burge, 2004). RPKM refers to the reads per kilobase per million calculations from short read Illumina sequencing. ‘Splicing index’ refers to the splicing index results from short read Illumina sequencing. ‘Alt splicing’ signifies whether the intron neighbors an alternatively spliced exon in this cell line (refer to Materials & Methods). ‘Stringency’ refers to the lowest category of constitutive intron splicing stringency (as defined in the Materials & Methods section). ‘RBP density’ refers to the sum of fold change over input for all RBP peaks in the intron (from (Van Nostrand et al., 2018) and as defined in the Materials & Methods section).
Table S6. Features of all introns in Drosophila S2 cells, Related to STAR methods. The coordinates of the intron are displayed as chromosome, start position, end position, and strand. ‘Gene name’ refers to the RefSeq name for coding genes. ‘Intron pos’ is the intron number in the order of transcription. ‘Intron total’ is the total number of introns within that gene. ‘Intron order’ is the descriptive position of the intron within the gene (first, middle, or last). ‘Distance from start’ and ‘distance from end’ are the lengths in nucleotides from the start and end coordinates of the gene, respectively. ‘Intron length’, ‘upstream exon length’, and ‘downstream exon length’ are the sizes of the intron and surrounding exons in nucleotides. ‘5SS score’ and ‘3SS score’ are the splice site consensus sequence scores at the 5’SS and 3’SS, respectively, as determined by the maximum entropy calculations from (Yeo and Burge, 2004). RPKM refers to the reads per kilobase per million calculations from short read Illumina sequencing from (Pai et al., 2017). ‘Splicing index’ refers to the splicing index results from short read Illumina sequencing. ‘Alt splicing’ signifies whether the intron neighbors an alternatively spliced exon in this cell line (refer to Materials & Methods). ‘Stringency’ refers to the lowest category of constitutive intron splicing stringency (as defined in the Materials & Methods section).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pol II Ser2-phosphorylated CTD | Active Motif | Cat#3E10 |
| GAPDH | Life Technologies | Cat#AM4300/6C5 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Pladienolide B | Santa Cruz Biotechnology | Cat#sc-391691 |
| 4-thiouridine | Sigma | Cat#T4509 |
| EZ-Link Biotin-HPDP | ThermoFisher Scientific | Cat#21341 |
| Inosine triphosphate | Sigma | Cat#I0879 |
| Alpha-Amanitin | Sigma | Cat#A2263 |
| Critical Commercial Assays | ||
| μMACS streptavidin kit | Miltenyi Biotec | Cat#130-074-101 |
| SuperScript III First-Strand Synthesis System | Invitrogen | Cat#18080051 |
| SsoFast EvaGreen Supermix | Biorad | Cat#1725200 |
| Ovation Universal RNA-seq System | NUGEN | Cat#0343-32 |
| Universal Human rRNA strand selection reagent | NUGEN | Cat#S01859 |
| Ribo-Zero Gold rRNA Removal kit | Illumina | Cat#MRZG126 |
| RiboMinus Eukaryotic Kit v2 | ThermoFisher | Cat#A15020 |
| Direct RNA sequencing kit | Oxford Nanopore Technologies | Cat#SQK-RNA001 and SQK-RNA002 |
| T4 DNA ligase (2,000,000 units/mL) | New England Biolabs | Cat#M0202M |
| Deposited Data | ||
| Raw and analyzed data | This study | GEO: GSE123191 |
| Raw experimental data | This study | Mendeley: http://dx.doi.org/10.17632/9gfs2kkxbc.2 |
| Direct nanopore RNA-sequencing of polyA+ RNA | Workman et al., 2018 | https://github.com/nanopore-wgsconsortium/NA12878/blob/master/RNA.md |
| Illumina RNA-sequencing of S2 cells | Pai et al., 2017 | GEO: GSE93763 |
| Illumina RNA-sequencing of K562 cells treated with 4sU | Schofield et al., 2018 | GEO: GSE95854 |
| NP-seq and mNET-seq in HeLa cells | Nojima et al., 2015 | GEO: GSE60358 |
| Experimental Models: Cell Lines | ||
| Human K562 cells | ATCC | Cat#CCL-243 |
| Human B lymphoblasts BL1184 | ATCC | Cat#CRL-4959 |
| Drosophila S2 cells | Expression Systems | Cat#94-005 |
| Oligonucleotides | ||
| See Table S6 for primers and G-blocks used | ||
| Software and Algorithms | ||
| STAR v2.5.1a | Dobin et al.,2013 | N/A |
| Albacore 2.2.7 | Oxford Nanopore Technologies Ltd. | N/A |
| minimap2 version 2.10-r764-dirty | Li, 2018 | N/A |
| GMAP version 2018-03-25 | Wu and Watanabe, 2005 | N/A |
| pyGenomeTracks | Ramirez et al., 2018 | N/A |
| Nanopolish version 0.11.1 | Workman et al., 2018 | N/A |
| MISO | Katz et al., 2010 | N/A |
| Picard v.18.15 | http://broadinstitute.github.io/picard | N/A |
| Pysam | https://github.com/pysam-developers/pysam | N/A |
| Pybedtools | Dale et al., 2011 | N/A |
| MaxEnt | Yeo and Burge, 2004 | N/A |
| Python scikit learn | Pedregosa et al., 2011 | N/A |
| All scripts and data analyses | This study | https://github.com/churchmanlab/nano-COP |
| Other | ||
| E. coli poly(A) polymerase | New England Biolabs | Cat#M0276S |
| E. coli poly(A) polymerase | Clontech/Takara | Cat#2180 |
| Yeast poly(A) polymerase | ThermoFisher | Cat#74225Z25 KU |
| Agencourt RNAClean XP beads | Beckman Coulter | Cat#A63987 |
Highlights.
Direct RNA sequencing exposes splicing dynamics without biases from amplification
Splicing occurs after Pol II transcribes kilobases past introns in metazoans
Introns are frequently removed in a defined order that does not follow transcription
Splicing in humans is coordinated across proximal introns
Acknowledgements
We thank members of the Churchman lab, F. Winston, W. Timp, R. Workman, N. Sadowski, M. Marin, B. Smalec, H. Merens, R. Ietswaart and A. Markham for helpful discussions, advice and assistance; C. Burge, E. McShane, J. Bridgers, R. Ietswaart, K. Tyssowski, H. Merens and C. Patil for critical reading of the manuscript; G. Amador and the DRSC/TRiP facility for guidance culturing Drosophila S2 cells; G. Yeo and E. Wheeler for processed eCLIP data. This work was supported by the NIH (R21-HG009264, R01-HG010538 and R01-GM117333 to L.S.C.; F31-GM122133 to H.L.D.) and the Fonds de Recherche du Québec - Santé (Post-doctoral fellowship award to K.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Aebi M, and Weissman C (1987). Precision and orderliness in splicing. Trends Genet. 3, 102–107. [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, and Lis JT (2009). Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28, 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audibert A, Weil D, and Dautry F (2002). In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol 22, 6706–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2014). Coupling mRNA processing with transcription in time and space. Nat. Rev. Mol. Cell Biol 15, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM (1995). Exon recognition in vertebrate splicing. J. Biol. Chem 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- Beyer AL, and Osheim YN (1988). Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2, 754–765. [DOI] [PubMed] [Google Scholar]
- Black DL, Chabot B, and Steitz JA (1985). U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell 42, 737–750. [DOI] [PubMed] [Google Scholar]
- Bonnal S, Vigevani L, and Valcárcel J (2012). The spliceosome as a target of novel antitumour drugs. Nat. Rev. Drug Discov 11, 847–859. [DOI] [PubMed] [Google Scholar]
- Braun JE, Friedman LJ, Gelles J, and Moore MJ (2018). Synergistic assembly of human pre-spliceosomes across introns and exons. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Böhnlein E-M, Neugebauer KM, Darzacq X, and Shav-Tal Y (2011). The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 9, e1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE, Longhurst AD, Merkurjev D, Sales-Lee J, Rao B, Moresco JJ, Yates JR 3rd, Li JJ, and Madhani HD (2018). Spliceosome Profiling Visualizes Operations of a Dynamic RNP at Nucleotide Resolution. Cell 173, 1014–1030.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Herzel L, Straube K, Hujer K, Howard J, and Neugebauer KM (2016). Splicing of Nascent RNA Coincides with Intron Exit from RNA Polymerase II. Cell 165, 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Moore J, Ozadam H, Shulha HP, Rhind N, Weng Z, and Moore MJ (2018). Transcriptome-wide Interrogation of the Functional Intronome by Spliceosome Profiling. Cell 173, 1031–1044.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, and Weissman JS (2011). Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, and Larson DR (2014). Kinetic competition during the transcription cycle results in stochastic RNA processing. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretu C, Agrawal AA, Cook A, Will CL, Fekkes P, Smith PG, Lührmann R, Larsen N, Buonamici S, and Pena V (2018). Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds. Mol. Cell 70, 265–273.e8. [DOI] [PubMed] [Google Scholar]
- Dale RK, Pedersen BS, and Quinlan AR (2011). Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27, 3423–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, and Kraus WL (2013). Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. (2008). High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14, 1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy EE, Schofield JA, and Simon MD (2019). Gaining insight into transcriptome‐wide RNA population dynamics through the chemistry of 4‐thiouridine. WIREs RNA 10, e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, de la Mata M, Marasco LE, Muñoz MJ, Le Jossic-Corcos C, Corcos L, and Kornblihtt AR (2014). How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54, 683–690. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, and Nagai K (2017). Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol 24, 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu X-D, and Bentley DL (2014). Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 28, 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung S-P, Baldi PF, and Hertel KJ (2005). The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. U. S. A 102, 16176–16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. (2018). Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206. [DOI] [PubMed] [Google Scholar]
- Guo M, Lo PC, and Mount SM (1993). Species-specific signals for the splicing of a short Drosophila intron in vitro. Mol. Cell. Biol 13, 1104–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, and Lillard SJ (2000). In-situ sampling and separation of RNA from individual mammalian cells. Anal. Chem 72, 4073–4079. [DOI] [PubMed] [Google Scholar]
- Herzel L, Straube K, and Neugebauer KM (2018). Long-read sequencing of nascent RNA reveals coupling among RNA processing events. Genome Res. 28, 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, and Moore MJ (2011). Ordered and dynamic assembly of single spliceosomes. Science 331, 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, and Cook PR (1998). Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 9, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Will CL, Stark H, and Lührmann R (2019). Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes. Cold Spring Harb. Perspect. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, and Burge CB (2010). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohavong P, Gattoni R, LeMoullec JM, Jacob M, and Stévenin J (1982). The orderly splicing of the first three leaders of the adenovirus-2 major late transcript. Nucleic Acids Res. 10, 1215–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O, Jiang Y, and Chasin LA (1993). Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol. Cell. Biol 13, 6211–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang C-HA, Marr MT 2nd, and Rosbash M (2011). Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 25, 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-K, and Kim VN (2007). Processing of intronic microRNAs. EMBO J. 26, 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Taggart AJ, Heintzelman C, Cygan KJ, Hull CG, Wang J, Shrestha B, and Fairbrother WG (2017). Widespread intra-dependencies in the removal of introns from human transcripts. Nucleic Acids Res. 45, 9503–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, and Mizui Y (2007). Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol 3, 570–575. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Maniatis T, Ruskin B, and Green MR (1984). Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36, 993–1005. [DOI] [PubMed] [Google Scholar]
- Li H (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, and Simpson JT (2015). A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12, 733–735. [DOI] [PubMed] [Google Scholar]
- Martin RM, Rino J, Carvalho C, Kirchhausen T, and Carmo-Fonseca M (2013). Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. Cell Rep. 4, 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, and Kornblihtt AR (2003). A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532. [DOI] [PubMed] [Google Scholar]
- Mayer A, and Churchman LS (2016). Genome-wide profiling of RNA polymerase transcription at nucleotide resolution in human cells with native elongating transcript sequencing. Nat. Protoc 11, 813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, and Churchman LS (2015). Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, and Bentley DL (1997). The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385, 357–361. [DOI] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G, and Kornblihtt AR (2015). Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem 84, 165–198. [DOI] [PubMed] [Google Scholar]
- Niwa M, and Berget SM (1991). Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 5, 2086–2095. [DOI] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, and Proudfoot NJ (2015). Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell 161, 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Rebelo K, Gomes T, Grosso AR, Proudfoot NJ, and Carmo-Fonseca M (2018). RNA Polymerase II Phosphorylated on CTD Serine 5 Interacts with the Spliceosome during Co-transcriptional Splicing. Molecular Cell 72, 369–379.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, Schneider RK, Lord AM, Wang L, Gambe RG, et al. (2016). Physiologic Expression of Sf3b1K700E Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell 30, 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T, and Lis JT (1993). Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol 13, 3456–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Henriques T, McCue K, Burkholder A, Adelman K, and Burge CB (2017). The kinetics of pre-mRNA splicing in the Drosophila genome and the influence of gene architecture. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, and Black DL (2009). Co-transcriptional splicing of constitutive and alternative exons. RNA 15, 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, Bhatt DM, Lin C-H, Tong A-J, Smale ST, and Black DL (2013). Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA 19, 811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res 12, 2825–2830. [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al. (2011). Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet 44, 47–52. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Raychowdhury R, Jovanovic M, Rooney M, Stumpo DJ, Pauli A, Hacohen N, Schier AF, Blackshear PJ, Friedman N, et al. (2014). High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell 159, 1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, and Manke T (2018). High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun 9, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, and Lührmann R (2010). Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol. Cell 38, 223–235. [DOI] [PubMed] [Google Scholar]
- Schofield JA, Duffy EE, Kiefer L, Sullivan MC, and Simon MD (2018). TimeLapseseq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat. Methods 15, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, and Cramer P (2016). TT-seq maps the human transient transcriptome. Science 352, 1225–1228. [DOI] [PubMed] [Google Scholar]
- Sciarrillo R, Wojtuszkiewicz A, El Hassouni B, Funel N, Gandellini P, Lagerweij T, Buonamici S, Blijlevens M, Zeeuw van der Laan EA, Zaffaroni N, et al. (2019). Splicing modulation as novel therapeutic strategy against diffuse malignant peritoneal mesothelioma. EBioMedicine 39, 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, and Padgett RA (2009). Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol 16, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara K, Schwarze U, Imamura Y, Hoffman GG, Toriello H, Smith LT, Byers PH, and Greenspan DS (2002). Order of intron removal influences multiple splice outcomes, including a two-exon skip, in a COL5A1 acceptor-site mutation that results in abnormal pro-alpha1(V) N-propeptides and Ehlers-Danlos syndrome type I. Am. J. Hum. Genet 71, 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Tsai JHC, Puyang X, Seiler M, Peng S, Prajapati S, Aird D, Buonamici S, Caleb B, Chan B, et al. (2017). Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A–SF3b complex. Nat. Commun 8, 15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, and Guigó R (2012). Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 22, 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Freese P, Pratt GA, Wang X, and Wei X (2018). A large-scale binding and functional map of human RNA binding proteins. bioRxiv doi: 10.1101/179648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, and Ljungman M (2014). Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 24, 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigevani L, Gohr A, Webb T, Irimia M, and Valcárcel J (2017). Molecular basis of differential 3′ splice site sensitivity to anti-tumor drugs targeting U2 snRNP. Nat. Commun 8, 2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachutka L, Caizzi L, Gagneur J, and Cramer P (2019). Global donor and acceptor splicing site kinetics in human cells. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, and Lührmann R (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. [DOI] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. (2011). SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med 365, 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I, Baurén G, and Wieslander L (1996). The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2, 641–651. [PMC free article] [PubMed] [Google Scholar]
- Windhager L, Bonfert T, Burger K, Ruzsics Z, Krebs S, Kaufmann S, Malterer G, L’Hernault A, Schilhabel M, Schreiber S, et al. (2012). Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 22, 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman RE, Tang A, Tang PS, Jain M, Tyson JR, Zuzarte PC, Gilpatrick T, Razaghi R, Quick J, Sadowski N, et al. (2018). Nanopore native RNA sequencing of a human poly(A) transcriptome. bioRxiv doi: 10.1101/459529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, and Watanabe CK (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875. [DOI] [PubMed] [Google Scholar]
- Wuarin J, and Schibler U (1994). Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol 14, 7219–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, and Burge CB (2004). Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol 11, 377–394. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. (2011). Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69. [DOI] [PubMed] [Google Scholar]
- Zhao H, Sun Z, Wang J, Huang H, Kocher J-P, and Wang L (2014). CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Key statistics of nano-COP libraries sequenced in this study, Related to STAR methods. Experimental differences and sequencing statistics are described for every nano-COP library that was incorporated into this study. Key experimental points include replicate number, species, cell line, treatment, and tailing approach. Key sequencing statistics include sequencing instrument, total sequenced reads, uniquely mapped reads, and percent uniquely mapped reads.
Table S5. Features of all introns in human K562 cells, Related to STAR methods. The coordinates of the intron are displayed as chromosome, start position, end position, and strand. ‘Gene name’ refers to the RefSeq name for coding genes. ‘Intron pos’ is the intron number in the order of transcription. ‘Intron total’ is the total number of introns within that gene. ‘Intron order’ is the descriptive position of the intron within the gene (first, middle, or last). ‘Distance from start’ and ‘distance from end’ are the lengths in nucleotides from the start and end coordinates of the gene, respectively. ‘Intron length’, ‘upstream exon length’, and ‘downstream exon length’ are the sizes of the intron and surrounding exons in nucleotides. ‘5SS score’ and ‘3SS score’ are the splice site consensus sequence scores at the 5’SS and 3’SS, respectively, as determined by the maximum entropy calculations from (Yeo and Burge, 2004). RPKM refers to the reads per kilobase per million calculations from short read Illumina sequencing. ‘Splicing index’ refers to the splicing index results from short read Illumina sequencing. ‘Alt splicing’ signifies whether the intron neighbors an alternatively spliced exon in this cell line (refer to Materials & Methods). ‘Stringency’ refers to the lowest category of constitutive intron splicing stringency (as defined in the Materials & Methods section). ‘RBP density’ refers to the sum of fold change over input for all RBP peaks in the intron (from (Van Nostrand et al., 2018) and as defined in the Materials & Methods section).
Table S6. Features of all introns in Drosophila S2 cells, Related to STAR methods. The coordinates of the intron are displayed as chromosome, start position, end position, and strand. ‘Gene name’ refers to the RefSeq name for coding genes. ‘Intron pos’ is the intron number in the order of transcription. ‘Intron total’ is the total number of introns within that gene. ‘Intron order’ is the descriptive position of the intron within the gene (first, middle, or last). ‘Distance from start’ and ‘distance from end’ are the lengths in nucleotides from the start and end coordinates of the gene, respectively. ‘Intron length’, ‘upstream exon length’, and ‘downstream exon length’ are the sizes of the intron and surrounding exons in nucleotides. ‘5SS score’ and ‘3SS score’ are the splice site consensus sequence scores at the 5’SS and 3’SS, respectively, as determined by the maximum entropy calculations from (Yeo and Burge, 2004). RPKM refers to the reads per kilobase per million calculations from short read Illumina sequencing from (Pai et al., 2017). ‘Splicing index’ refers to the splicing index results from short read Illumina sequencing. ‘Alt splicing’ signifies whether the intron neighbors an alternatively spliced exon in this cell line (refer to Materials & Methods). ‘Stringency’ refers to the lowest category of constitutive intron splicing stringency (as defined in the Materials & Methods section).
Data Availability Statement
All Illumina and nanopore sequencing data generated for this paper are available at Gene Expression Omnibus (GEO) under the accession number GSE123191. Other raw data are published on Mendeley (doi:10.17632/9gfs2kkxbc.2). All scripts and data analyses are available at https://github.com/churchmanlab/nano-COP.