Abstract
Chronic itch can be extremely devastating and, in many cases, difficult to treat. One challenge in treating itch disorders is the limited understanding of the multitude of chemical players involved in the communication of itch sensation from the peripheral to central nervous system. Neuropeptides are intercellular signaling molecules that are known to be involved in the transmission of itch signals from primary afferent neurons, which detect itch in the skin, to higher-order circuits in the spinal cord and brain. To investigate the role neuropeptides play in transmitting itch signals, we generated two mouse models of chronic itch—Acetone-Ether-Water (AEW, dry skin) and calcipotriol (MC903, atopic dermatitis). For peptide identification and quantitation, we analyzed the peptide content of dorsal root ganglia (DRG) and dorsal horn (DH) tissues from chronically itchy mice using liquid chromatography coupled to tandem mass spectrometry. De novo-assisted database searching facilitated the identification and quantitation of 335 peptides for DH MC903, 318 for DH AEW, 266 for DRG MC903, and 271 for DRG AEW. Of these quantifiable peptides, we detected 30 that were differentially regulated in the tested models, after accounting for multiple testing correction (q≤0.1). These include several peptide candidates derived from neuropeptide precursors, such as proSAAS, protachykinin-1, proenkephalin and calcitonin gene-related peptide, some of them previously linked to itch. The peptides identified in this study may help elucidate our understanding about these debilitating disorders. Data are available via ProteomeXchange with identifier PXD015949.
Keywords: Label-free quantitation, Liquid chromatography, Mass spectrometry, Skyline, Peptidomics
Graphical Abstract
INTRODUCTION
Pruritus, also known as itch, has been estimated to afflict approximately 16% of the population1 and can be a significant symptom of many disorders, including chronic conditions like psoriasis, atopic dermatitis, and cholestasis. The symptoms of pruritus are typically characterized as an irritating sensation that causes the victim to scratch the affected area and, when chronic, can cause significant distress to the point of impairing the affected individual’s quality of life. Unfortunately, chronic pruritis is an important health issue for which we do not have reliable treatments, partly because we do not yet understand the wide range of molecular players involved in the transmission of itch sensation from the peripheral to central nervous system (CNS).
Itch can be classified into four categories: neurogenic, psychogenic, neuropathic, and pruritoceptive, with pruritoceptive being the most common type.2 A diverse class of molecules have been implicated in the transmission of itch from the primary sensory neurons to higher-order spinal and brain structures, including amines, interleukins, neuropeptides, cannabinoids, and eicosanoids.3 The itch-producing stimulus is first detected in the skin by primary sensory neurons, which then transmit this information to central neurons in the spinal or medullary dorsal horn (DH) neurons. These afferent fibers that innervate the skin can be classified as either Aβ, Aδ, or C fibers, based on their size, transmission speed, and myelination status.4 Most well-characterized pruritoceptive primary afferents are unmyelinated, small diameter C-fiber neurons.5–6
Primary sensory neurons within spinal nerves have a single axon that bifurcates and projects from the peripheral nerve endings to the DH of the spinal cord, transmitting important sensory information to the CNS.4,6 These neurons have their cell bodies located within the dorsal root ganglia (DRG), which are clusters of neurons that lie in the intervertebral foramen of the spinal cord.7–9 The sensory information from the external stimuli is transmitted to the CNS through the release of a variety of signaling molecules, including neurotransmitters, neuropeptides, and neuromodulators.
Given the large diversity of neuropeptides, and their ability to serve as neurotransmitters, neurohormones, and neuromodulators, the current study focuses on the identification and quantitation of those that play a role in the transmission of itch. Neuropeptides are a class of signaling molecules, typically about 3–40 amino acid residues in length, that are synthesized and released by neurons to modulate various physiological processes in the body. These processes include itch, learning, and reproduction; cardiovascular, gastrointestinal, and respiratory control; food and water intake; and analgesia and pain.10 Neuropeptides are typically produced from the cleavage of larger precursor proteins, which is sometimes followed by enzymatic post- translational modifications (PTMs) such as C-terminal amidation,11–12 acetylation,13 phosphorylation,14–15 sulfation,16–17 and pyroglutamination.18 Previous studies have shown that several neuropeptides, such as substance P (SP) (TAC1[58–68]; P41539), bradykinin (KNG1[380–388]; O08677), calcitonin gene-related peptide (CGRP) (CALCA[83–119]; Q99JA0), neuromedin B (NMB[47–56]; Q9CR53), gastrin-releasing peptide (GRP[24–52]; Q8R1I2), and natriuretic polypeptide B (ANFB[103–134]; P16860) are linked to pruritus.19–21 However, there has yet to be an untargeted study of the peptide repertoire in the DH and DRG regions to identify the dynamics of neuropeptide changes in chronic itch.
Here liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS) was used for the identification and quantification of neuropeptides. LC–MS/MS is a valuable technique for performing peptidomic analyses; complex samples can be simplified by separation and elution on a chromatographic column, followed by subsequent analysis and fragmentation on a mass spectrometer.22–24 The resulting fragmentation spectra of the eluted peptides can then be searched against a protein database of the studied organism for identification of the peptides.25
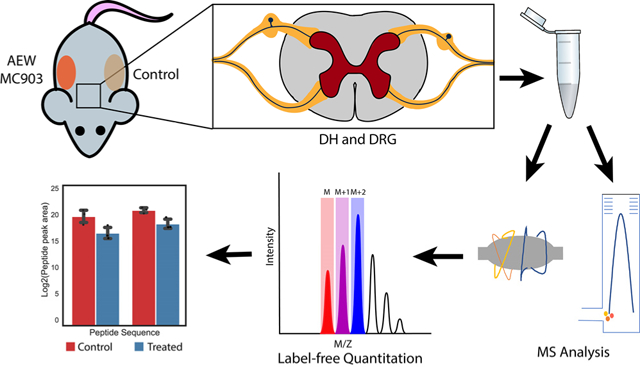
In this study, we generated two well-described chronic itch models: an atopic dermatitis- like model, using calcipotriol (MC903), and a dry skin model using Acetone-Ether and Water (AEW). Atopic dermatitis, a skin disorder in which eczematous lesions appear on the skin, is associated with intense itch.26 MC903 is a synthetic analogue of vitamin D3 that has been shown to induce atopic dermatitis-like inflammation and itch when applied topically to the skin.27–29 Chronic itch has also been associated with dry skin; mice that have had a mixture of acetone and diethyl ether, followed by water, applied topically to the skin show an increase in scratching bouts at the affected area.30–31 The peptidomic analysis was performed on both DRG and DH tissues from mice that were subjected to MC903 treatment to induce atopic dermatitis-like symptoms, or AEW treatment to induce scratching at the affected areas; a label-free quantification was performed to determine the relative changes in the abundance of peptides between treated and control mice (Figure 1).
Figure 1.
Workflow for label-free relative quantitation of peptides in itch models of mice.
EXPERIMENTAL PROCEDURES
Additional details about the experimental design are presented in Table S1.
Chemicals
Reagents and solvents were obtained from either MilliporeSigma (St. Louis, MO) or Thermo Fisher Scientific (Waltham, MA), unless stated otherwise.
Animals
Twenty, 8-week-old, male B6J WT mice (Jackson Laboratories, https://www.jax.org/; stock #000664) were housed on a 12 h light/dark cycle and fed ad libitum. All experimental procedures, including euthanasia, were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Washington University School of Medicine, and in full compliance with both federal and ARRIVE guidelines for the humane care and treatment of animals.
AEW (Dry Skin) Model
Ten mice were shaved on both their left and right thoracic flanks; 5 mice were treated on their shaved right flank and the other 5 mice were treated on their shaved left flank. A 1:1 mixture of acetone:diethyl ether was topically applied to the treated side in an approximately 1 cm × 2 cm area for 15 s followed by milliQ water for 30 s. The treatment was applied daily for 7 d; the untreated side served as the control.32
MC903 (Atopic Dermatitis) Model
Ten mice were shaved on either their left or right thoracic flank; 5 mice were treated on their shaved right flank and the other 5 mice were treated on their shaved left flank. MC903 (40 μl of 0.1 mM in ethanol; Tocris, Minneapolis, MN) was topically applied in an approximately 1 cm × 2 cm area under anesthesia (3% isoflurane) to the treated side for 7 consecutive days to induce atopic dermatitis-like disease;33 the untreated side served as the control.
Behavioral Assessment
Animals were video recorded in their cages for 50 min before their first treatment to serve as a baseline measurement, and again after the final day of treatment to confirm development of spontaneous itch at the treated sites. Scratch bouts directed at the treated sites were quantified from the recorded videos for 30 min, starting after a 20-min habituation period.
Tissue Collection
On the seventh day, approximately 170–180 h after the first treatment, 24 (MC903) or 17 (AEW) h after the final treatment, and immediately after the last behavioral recording, the mice were sacrificed and DH and DRG from approximately the T4–T10 spinal levels were dissected from both the treated and control sides. Immediately following dissection, the DH the DRG were stabilized using the Stabilizor T1 system (Denator AB, Uppsala, Sweden) to minimize peptide degradation by peptidases.34–35 The stabilized tissues were then stored at −80 °C until prepared for peptide extraction.
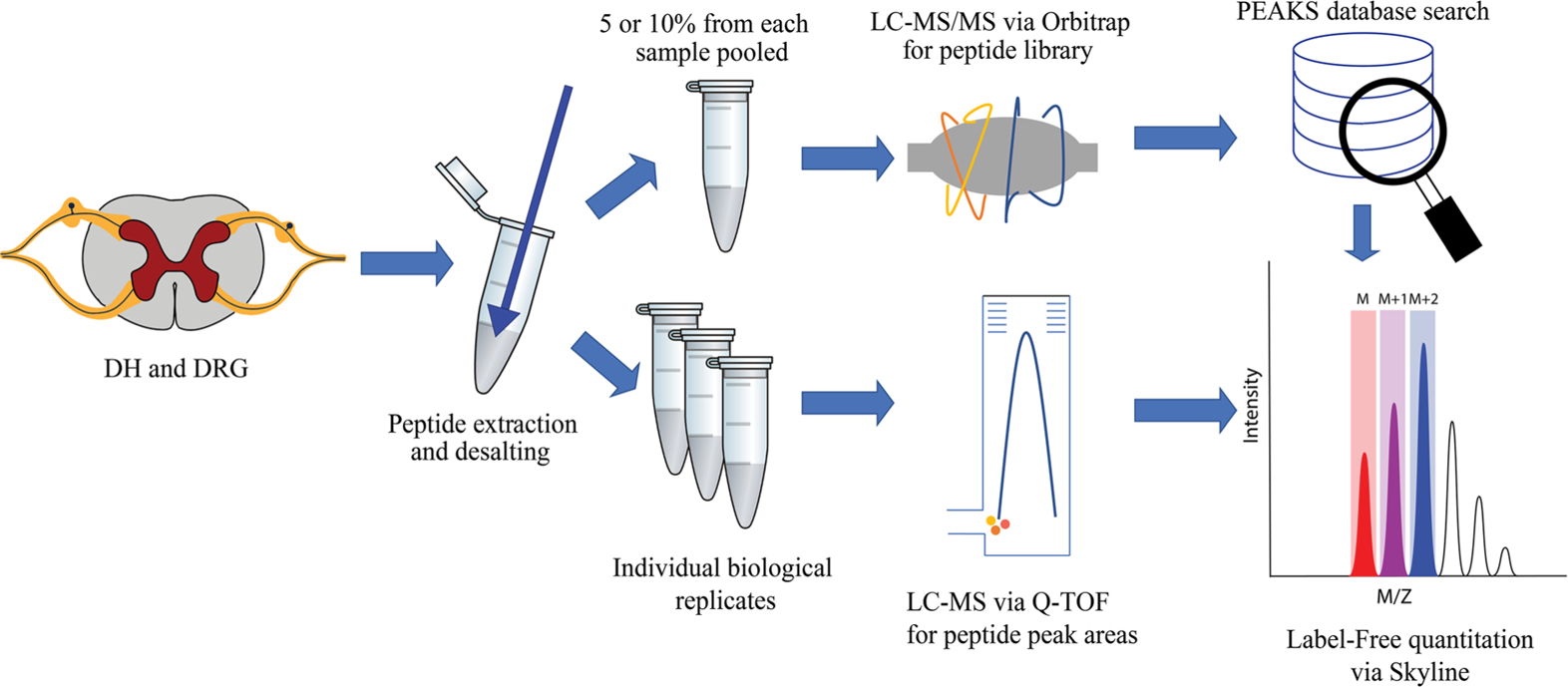
Peptide Extraction
The sampling and peptide extraction approaches were modeled after our prior studies.17,35–36 The treated and the control samples were pooled such that two tissue samples corresponding to either treated or control were combined. Ice-cold LC–MS grade water (400 μL) was then added to each of the pooled tissues and homogenized using a pellet pestle cordless motor. The homogenized samples were left to incubate on ice for 40 min. Following incubation, the samples were centrifuged for 20 min at 16000 × g at 4 °C. Following centrifugation, the supernatant was transferred to a new tube, and the pellet was re-suspended in 400 μL of 40:5:5 methanol:formic acid (FA):water and left to incubate on ice for 40 min. The sample was then centrifuged for 20 min at 16000 × g at 4 °C, and afterwards, this supernatant was combined with the supernatant from the previous step. The combined supernatants were then evaporated on a SpeedVac evaporator (Genevac, Ipswich, Suffolk, UK). While the supernatants were evaporating, the pellets were re-suspended in 400 μL of 0.25% FA in LC–MS grade water and incubated on ice until the combined supernatants were dry (~60 min). The reconstituted pellets were then centrifuged for 20 min at 16000 × g at 4 °C. The pellets were discarded, and the supernatants added to the vial containing the dried extracts from the previous incubations.
Peptide Cleanup and De-Salting
The samples were loaded onto an equilibrated Pierce C18 spin column (Thermo Fisher Scientific) and washed twice with 200 μL of 95:5:0.1:0.01 water:acetonitrile (ACN):FA:trifluoroacetic acid (TFA); the peptides were then eluted twice with 50 μL of 50:50:0.1:0.01 water:ACN:FA:TFA and twice with 50 μL of 20:80:0.1:0.01 water:ACN:FA:TFA. After eluting the peptides, the samples were evaporated until dry on the SpeedVac evaporator. The dried samples were stored at −80 °C until analysis by LC–MS/MS.
Nano-LC for Peptide Identification and Quantitation
For the peptide library construction, the pooled samples were reconstituted in 10 μL of 95:5:0.1 water:ACN:FA and loaded onto an Acclaim PepMap100 C18 trap column at 15 μL/min. After 3 min, the trap column was then connected in-line with the analytical column (Acclaim PepMap 2 Å, 75 μm × 150 mm, Thermo Fisher Scientific) using a Thermo Scientific Ultimate 3000 RSLC system. The solvents used were water with 0.1% FA, and ACN with 0.1% FA, as solvents A and B, respectively, at a flow rate of 300 nL/min. A 120-min gradient elution was used with the following parameters: 0–3 min, 1–1% B, 3–6 min, 1–10% B, 6–90 min, 10–70% B, 90–100 min, 70–99% B, 100–110 min, 99–1% B, 110–120 min, 1–1% B. For the peptide quantitation, the dried samples were reconstituted in 95:5:0.1 water:ACN:FA and then centrifuged for 20 min at 16000 × g. An 8-μL sample was then transferred to an autosampler vial, with 7 μL being injected for analysis on the Ultimate 3000 RSLC coupled to an Impact Q-TOF mass spectrometer (Bruker, Billerica, MA).
Orbitrap Parameters
Top speed data-dependent precursor selection was used on a Thermo Scientific Quadrupole-Ion Trap-Orbitrap Fusion mass spectrometer with a cycle time of 3 s. Parent ions were scanned with an Orbitrap resolution of 120K with an AGC target of 200,000. Dynamic exclusion was used with the following settings: exclusion time = 60 s, mass tolerance = ± 10 ppm, repeat count = 3. For the Orbitrap detection, the parent ions were scanned in the range of 200–1400 m/z, the fragment ions were scanned with the ion trap detector, a maximum injection time of 35 ms, and an AGC target of 10,000. Precursor ions with a charge ranging from +1 to +7 were considered and a normalized collision energy of 35% was used for the CID fragmentation.
Q-TOF Parameters
The samples used for the peptide quantitation were analyzed using a Bruker Impact HD QqTOF mass spectrometer equipped with a CaptiveSpray nanosource. Data was acquired with MS1, with a mass range of 290–3000 m/z, a cycle time of 3 s, and a scan rate of 1 Hz. An absolute intensity threshold of 694 counts was used for spectra collection.
Peptide Library Construction
The .RAW files obtained from the Thermo Orbitrap Fusion were imported into the PEAKS 8 software (Bioinformatics Solutions Inc., Canada); the spectra were searched against a mouse proteome database with 81,515 proteins from UniProt.37 For the database search, the following parameters were used: precursor mass tolerance = 10 ppm; fragment mass tolerance = 0.1 Da; no enzymatic cleavage; variable PTMs selected, which included acetylation, amidation, phosphorylation, half-disulfide bond, pyroglutamination, and methionine oxidation; and a maximum number of variable PTMs. A false discovery rate (FDR) threshold of 1% peptide spectrum match was used to filter out the identified peptide sequences. The peptide libraries for the DH and DRG are provided in Table S2.
Peptide Quantitation via Skyline
Relative quantitation of the identified peptides was performed using Skyline (version 4.2.0) software.38 A peptide library was constructed that consisted of individual peptide details, including amino acid sequence, PTMs, m/z, and retention time. After the library construction, the individual LC–MS data files corresponding to each of the studied regions were imported into the Skyline project. An extracted ion chromatogram (XIC) for each of the peptides present in the library was then generated by Skyline from the LC–MS files. The integrated peak areas of the XIC for each peptide were then exported to a CSV file for further statistical analysis.
Statistical Analysis
Following peptide quantitation, the summed peak areas were subjected to a locally weighted regression analysis using the Normalyzer tool.39 The purpose of performing this normalization was to account for inter-run variability brought about by sample preparation inconsistencies and instrument variability. The normalization was performed with the basic assumption that the sum of all the quantifiable peptide areas that were matched to the peptide library should be equal / similar across the biological replicates of a given treatment. The data was transformed into the standard M (log ratio) and A (mean average) scales within a specific treatment group. Next a plot of M vs. A values was constructed and the individual peptide peak areas for each sample were corrected based on a locally weighted regression algorithm as described by Chawade et al.39 The plots corresponding to the peptide peak areas are included in the Supporting Information (Figure S1). To determine if there was a significant change in peptide levels between the treated and the control samples, a student’s t-test was performed; p values that were less than 0.05 were considered significantly different. A permutation-based FDR correction was then applied to account for false-positive values using the Perseus computational platform (Version 1.6.2.3),40 in which the values that had a q-value of less than or equal to 0.1 were considered. The peptide quantitation workflow presented here has been implemented and validated in our prior work.41
Principal component analysis (PCA) was performed using the statistical package of R.42 Briefly, the dataset was first scanned for missing values. Since PCA cannot handle missing values, the missing values for peptide peak areas were imputed based on random draw from a truncated normal distribution-based approach, which is known to perform well in case of left-censored missing values; i.e., values that are missing due to low intensity / peak area.43 Following the imputation, the data is centered to ensure that the first principal component (PC1) does indeed describe the direction of maximum variance. PCA was then performed on this centered data. Once the PCA was performed, a clustering analysis was performed using the first 3 PCs to identify the pairs that capture the maximum variance and separate the two treatment groups. A loadings plot was further constructed with the scores corresponding to these PCs and a cutoff of where columns represent the number of quantified peptides. Given the number of peptides quantified in both of the regions, this cutoff is between 0.05–0.06.
RESULTS
Behavioral Model Validation
Animals were video recorded from above in their cages for at least 1 h at baseline and after 7 d of treatment for the AEW mice and 5 d of treatment for the MC903 mice. Upon review of the videos, the scratch bouts per side for each animal were counted and recorded. When compared with baseline or with the untreated flank, all animals showed significantly increased spontaneous scratching directed at the treatment site; confirming that the AEW and MC903 treatments successfully produced chronic itch in our model animals (Figure 2A, B).
Figure 2.
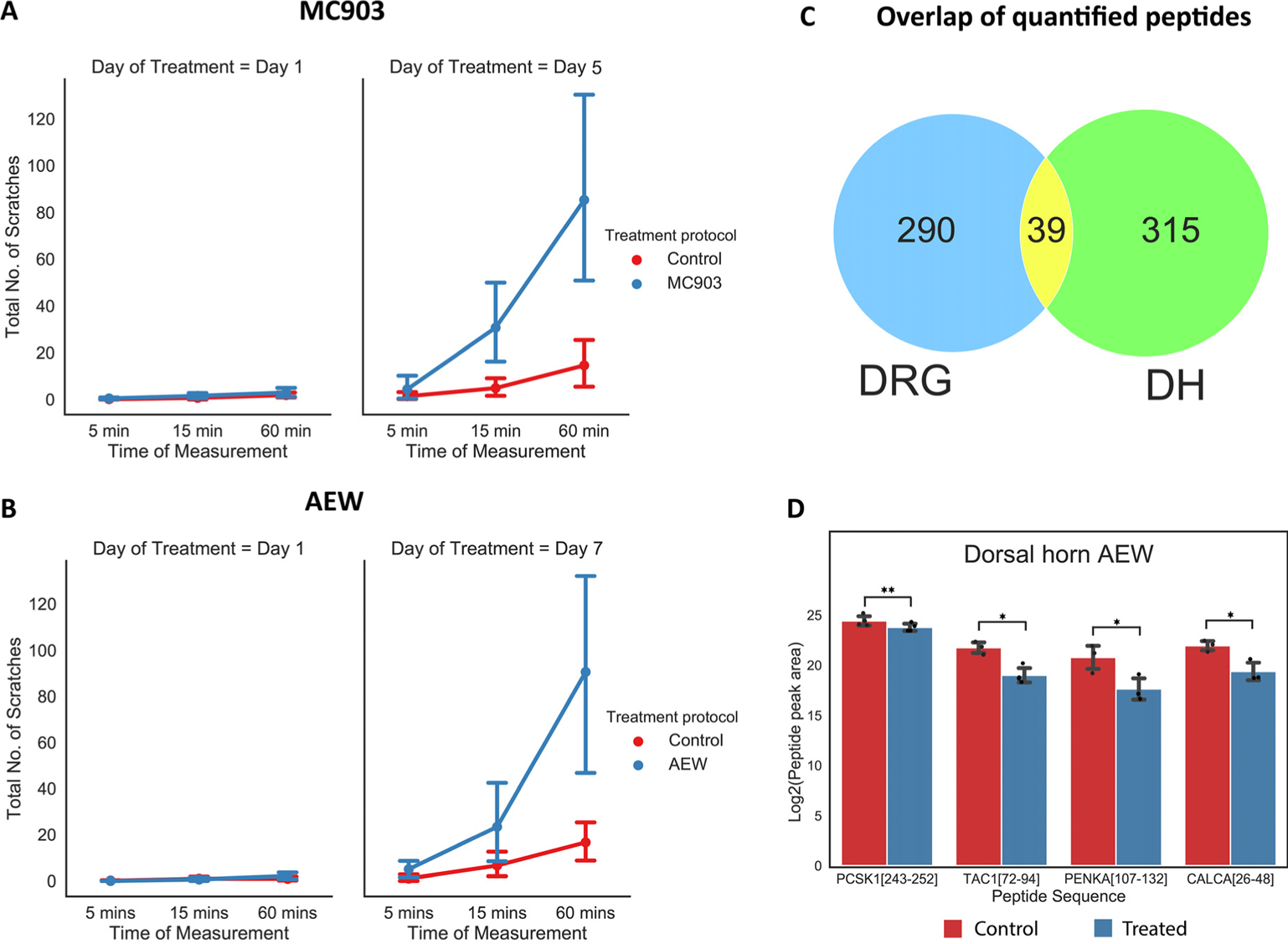
Number of scratches on the control and treated sides for the (A) MC903 and (B) AEW models. (C) Overlap of all the quantified peptides between the DH and DRG regions (in both models combined). (D) Select peptides that were significantly different between the treated and control DH AEW. *p<0.05, **p<0.01
Itch-Related Peptides Detected by LC–MS/MS Analysis
This study was performed in two different cohorts. The initial cohort consisted of N=3 replicates per model (AEW and MC903) per condition (treated and control) with tissue from N=1 animal being used per replicate. Though we did find some encouraging results with peptides such as somatostatin-28 (SST[89–116]; P60041), dynorphin B29 (PDYN[221–248], O35417), β-preprotachykinin C-terminal flanking peptide (TAC1[111–126], P41539), SP (TAC1[58–68], P41539), nocistatin (PNOC[98–138], Q64387), and PEN (PCSK1[219–240], Q9QXV0), which were differentially regulated in treated vs. control samples (p<0.05), none of the peptides crossed the set threshold (q≤0.1) for multiple testing correction by FDR estimation. Therefore, we performed a second independent cohort experiment with a higher number of replicates (N=4 per condition per treatment) and also a higher number of animals per replicate (tissues of N=2 animals pooled into one). De novo-assisted database searching followed by quantitation facilitated the quantitation of 329 and 354 peptides from the DRG and DH, respectively (Figure 2C). Of these total quantified peptides, 266 are from MC903 DRG and 271 from DRG AEW, and 335 peptides are from DH MC903 and 318 from DH AEW. Moreover, increasing the sample size led to an increase in the power (the probability of rejecting the null hypothesis when, in fact, it is false) of the hypothesis test used to investigate the differences between the treated and control mice; hence, only the results from the second cohort were considered (Table S3).
Differentially Regulated Peptides in Itch Models
Among the quantified peptides (Figure 2C) in the DRG and DH (cohort 2, AEW and MC903 models), 30 peptides were differentially regulated after accounting for the multiple testing correction (q≤0.1) (Table 1). All 30 peptides are from the DH of the AEW model and include several derived from prohormones such as proSAAS (PCSK1[243–252], Q9QXV0), protachykinin-1 (TAC1[72–94], P41539), proenkephalin (PENKA[107–132], P22005), and CGRP (CALCA[26–48]; Q99JA0) (Figure 2D). Additionally, we noticed several other prohormone-derived peptides had a p-value<0.05. Though these peptides did not cross the multiple testing correction threshold (q≤0.1) a further careful analysis may reveal their potential role in mediating itch. These include:: secretogranin1 (SCG1, P16014), cerebellin-1 (CBLN1, Q9R171), and tachykinin-3(TKNK; P55099) in DH MC903; proSAAS (PCSK1, Q9QXV0) in DRG AEW; and CGRP (CALCA, Q99JA0) and proSAAS in DRG MC903.
Table 1.
Peptides and the corresponding precursor proteins from DH AEW after accounting for multiple testing correction (q≤0.1).
| Precursor Protein with Accession Number | Peptide Sequence | Log2 Fold Change |
|---|---|---|
| Beta-hexosaminidase subunit beta (P20060) | ARLQPALWPFPRSVQMF* | 0.728 |
| Calcitonin gene-related peptide 1 (Q99JA0) | VPLRSILESSPGMATLSEEEVRL* | 1.306 |
| Calmodulin-1 (P62204) | A(+42.01)DQLTEEQIAEFKEAFSLFD* | 1.875 |
| Cathepsin B (P10605) | IDLPETFDAREQWSN* | 2.432 |
| Cathepsin D (P18242) | PVFDNLMQQKLVDKNIF* | 0.987 |
| Macrophage migration inhibitory factor (P34884) | DMNAANVGWNGSTFA** | 1.363 |
| PMFIVNTNV* | 2.135 | |
| PMFIVNTNVPRASVPEGFLSEL** | 1.416 | |
| PMFIVNTNVPRASVPEGFLSELTQQL* | 1.466 | |
| Neuroendocrine convertase 2 (P21661) | Q(−17.03)ELEEELDEAVERSLQSILRKN* | 0.784 |
| Neuroendocrine protein 7B2 (P12961) | YSPRTPDRVSETDIQRLLHGVMEQL* | 1.113 |
| Neurosecretory protein VGF (Q0VGU4) | Q(−17.03)AEATRQAAAQEERLADLASDLLLQYLLQGGARQ* | 0.885 |
| Peptidyl-prolyl cis-trans isomerase A (P17742) | EDENFILKHTGPGILSM** | 3.838 |
| V(+42.01)NPTVFFDIT*** | 1.488 | |
| Pituitary Adenylate cyclase activating enzyme (O70176) | Q(−17.03)MAVKKYLAAVL(−0.98)** | 0.366 |
| Phosphatidylethanolamine-binding protein 1 (P70296) | A(+42.01)ADISQW** | 1.030 |
| AGVTVDELGKVLTPTQV* | 2.268 | |
| MNRPSSISWDGLDPGKLYTL* | 1.585 | |
| PSSISWDGLDPGKLYTL* | 2.293 | |
| QAEWDDYVPKLYEQLSGK* | 0.857 | |
| Proenkephalin-A (P22005) | FAESLPSDEEGENYS(+79.97)KEVPEIE* | 2.303 |
| YGGFMKKMDELYPMEPEEEANGGEIL* | −0.329 | |
| ProSAAS (Q9QXV0) | AGDETPDVDPELLRYLLGRILTGSSEPEAAPAPRRL* | 0.073 |
| LENPSPQAPA** | 0.677 | |
| Protachykinin-1 (P41539) | DADSS(+79.97)VEKQVALLKALYGHGQIS* | 3.012 |
| Protein AF1q (P97783) | PIASIHSVDLDLL** | 1.022 |
| Protein virilizer homolog (A2AIV2) | EAFLRST** | 1.470 |
| Secretogranin-1 (P16014) | SFARAPQLDL* | 0.545 |
| Thioredoxin (P10639) | VKLIESKEAFQEALAAAGDKLVVVDF* | 0.956 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 (Q9R0P9) | MQLKPMEINPEMLNKVLAKLGVAGQWRFADVL* | 0.858 |
p<0.001;
p<0.01;
p<0.05.
Clustering Analysis of the Quantified Peptides
Changes in the levels of individual peptides / proteins usually do not capture the entire picture of the dynamic changes that they undergo under a specific physiological condition. Oftentimes a group of peptides or proteins have correlated levels of change or directions of change. Here, we performed additional clustering analysis to identify if a particular peptide / group of peptides are responsible for differentiation between the treated and control groups. Moreover, clustering analysis also identifies if a certain group of peptides / proteins have a similar pattern of regulation. First, by PCA (Figure 3), a widely used technique for dimensionality reduction and cluster visualization,44–46 we were able to distinguish between the treated and control group of samples for DH AEW, DRG AEW, and DRG MC903. A further loadings plot analysis was performed to gain more insights into the peptides that contributed to this separation (based on the cutoff described earlier) (Table S4). From this analysis, several prohormone-derived and signaling pathway-related peptides contribute towards differentiating the control and treated groups: PENK-A (P22005), TAC1 (P41539), CGRP (Q99JA0), pituitary adenylate cyclase-activating polypeptide (O70176), and proSAAS (Q9QXV0) for DH AEW; CGRP (Q99JA0), neurofilament light polypeptide (NFL-P08551), and mast cell protease 4 (Q3UN88) for DRG AEW; CGRP (Q99JA0), SCG (Q03517), mast cell protease 4 (Q3UN88) and cathepsinB (CATB-P10605) for DRG MC903.
Figure 3.
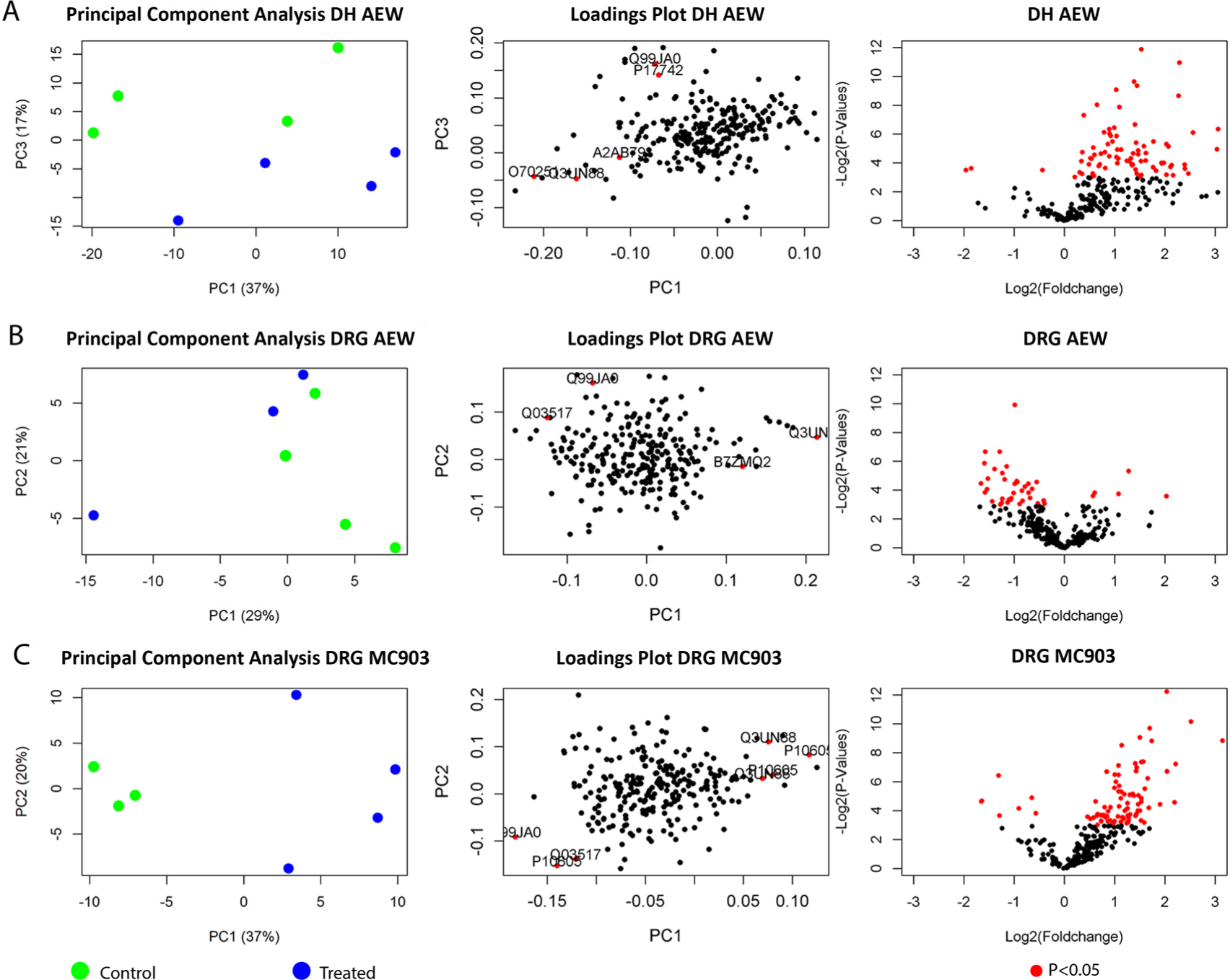
Volcano plot, PCA, and loadings plots of the (A) DH AEW, (B) DRG AEW, and (C) DRG MC903 models.
Functional Classification of Precursor Proteins Involved in Itch
Precursor proteins corresponding to the significantly changed peptides (p<0.05, after accounting for multiple testing correction (q≤0.1), only from DH AEW) were further grouped according to their protein class. These proteins were subjected to a gene ontology (GO) classification using PANTHER.47 Though all of the identified peptides fall into 11 different classes of precursor proteins, the 30 significantly changed peptides from DH AEW were mapped to three classes: signaling molecules, hydrolase, and enzyme modulators (Figure 4).
Figure 4.
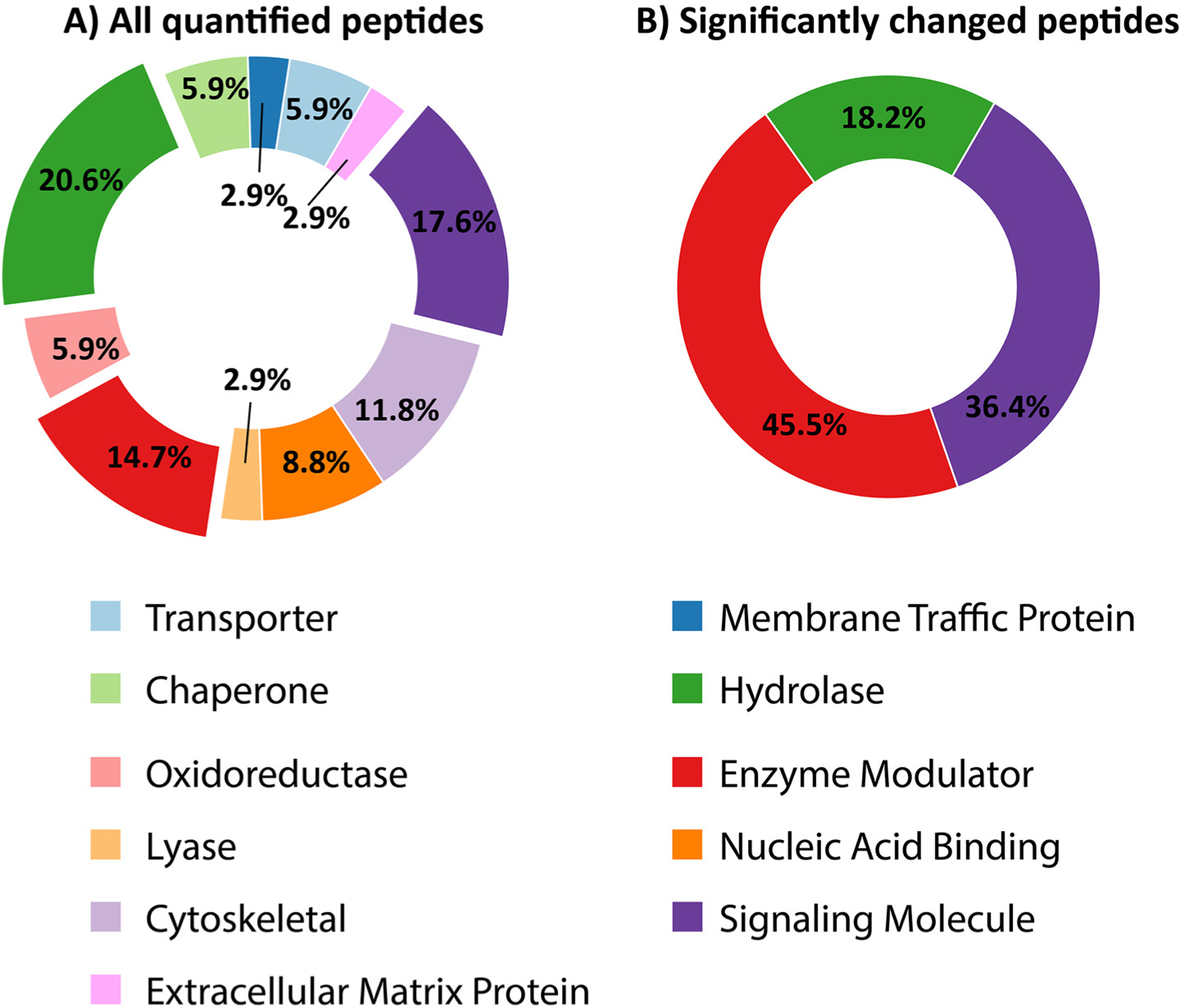
Gene ontology analysis of the precursor proteins corresponding to (A) All the quantified peptides and (B) peptides that were significantly different (p<0.05, q≤0.1) in DW AEW. The hydrolase, enzyme modulators, and signaling molecule classes of enzymes are depicted with an exploded view.
Validation of Detected Neuropeptide Localization in the Nervous System
To verify that the differentially detected peptides associated with itch are present in the analyzed regions of the nervous system, we checked the gene expression profiles of the DH in the Allen Brain Atlas (https://portal.brain-map.org/).48 From this analysis, we found that all significantly changed peptides have their precursor protein gene expressed in the DH (except for Protein Virilizer homolog (A2AIV2) and neuroendocrine protein 7B2 (P12961), for which the expression information was not available). This comparative analysis indicates that genes or transcripts corresponding to the identified candidates are indeed present in the mouse DH (Table S5).
DISCUSSION
To the best of our knowledge, this is the first MS-based peptidomics analysis of models of chronic itch. We performed a comprehensive peptidomic analysis on DRG and DH samples from control and treated regions of mouse models for atopic dermatitis and dry skin. The quantitation results were analyzed to investigate peptide level changes between the control and treated sides of subject animals and statistically significant differences were found after accounting for multiple testing correction. Additionally, an unsupervised clustering based on reduced dimensions (via PCA) revealed that the change in peptide levels indeed contributed towards differentiating the treated vs. control groups in three of the four tested conditions (Figure 3 and Table S4). A further analysis via loadings plot demonstrated that several known signaling molecules, as well as important signaling regulators, do make a major contribution towards this differentiability.
Importantly, many of the peptides detected in our study have already been reported to be involved in itch. For example, SP acts as a neurotransmitter in the spinal cord and induces scratching responses when injected intrathecally into mice.49 CGRP serves as a neuromodulator and potentiates glutamatergic itch signals from TRPV1+ primary afferents.50 Additionally, these neuropeptides are also released from the peripheral terminals of DRG neurons in the skin. There, CGRP and SP have specifically been shown to recruit and activate mast cells, respectively, and potentiate chronic itch.51
Within the list of peptides that have significantly changed in the DH AEW model after accounting for multiple testing correction are two known endogenous neuropeptides and significant portions of two other known full-length neuropeptides. These are PCSK1[243–252] (little LEN), PENK[238–259] (P22005), the first 7 residues of hippocampal cholinergic neurostimulatory peptide (PEBP1[2–9]; P70286), and the first 22 residues of neuropeptide K (TAC1[72–94]; P41539). Additionally, along with PENK[238–259], another PENK-derived peptide; PENK[107–132], which contains the sequence for met-enkephalin (PENK[107–111]), was significantly changed.
Little LEN, first identified by Fricker et al.,52 is derived from the prohormone proSAAS. Not much is known about the function of this peptide, except that it may have a neuroendocrine function. While it has been found in the mouse hypothalamus and pituitary, little LEN does not localize to neuropeptide Y-expressing cells or influence feeding behavior like other proSAAS-derived peptides do.53–54 One recent study reported that the expression of the proSAAS encoding gene, PCSK1N, is enriched in Mrgpra3+ expression in DRG neurons, which are selective neurons for detecting itch.55
PENK[238–259] and met-enkephalin are peptides derived from proenkephalin-A, a member of the endogenous opiate enkephalin family. In general, opiates decrease the sensitivity of neurons to pain by inhibiting their firing rates and decreasing neurotransmitter release.10,56 Particularly, the enkephalins play a role in pain regulation at the spinal level through the functioning of met-enkephalin interneurons.10,57 However, it has been noted that while opiates provide relief from pain processes, they cause an increase in itch sensation.58 This differential sensory response to opiate stimuli has in fact been used to support the idea that there are separate sensory pathways for pain and itch sensation. Interestingly, PENK expression has been found to be increased within the skin in various skin diseases, including psoriasis,59 which indicates a role for PENK in the diseases we were modeling in this study.
Hippocampal cholinergic neurostimulatory peptide is derived from phosphatidylethanolamine-binding protein 1 and has been shown to modulate acetylcholine synthesis through either stimulation of or suppression of acetylcholine transferase, depending on location and length of exposure.51,60 Acetylcholine is a monoamine neurotransmitter that plays multiple roles in the nervous system, one of which involves communication between neurons and mast cells, which are involved in inflammatory response.51,61 Within this context, acetylcholine has been shown to induce itch in patients with atopic disease compared with pain that is induced in controls under the same circumstances,62 suggesting a possible role for an acetylcholine modulatory peptide within atopic disease states.
Finally, the particular shortened form of neuropeptide K detected within our experiments has been previously detected and tentatively identified in a peptidomics study comparing levels of peptides in the brains of Cpefat/fat mice to wild type controls performed by the Fricker group.63 Neuropeptide K is derived from protachykinin, which is also the precursor of the well-known pain peptide SP. While TAC1[72–94] (P41539) was not detected in enough samples in the aforementioned Fricker study for the difference to be statistically significant, a similar peptide with one additional amino acid on its C terminus, TAC1[72–95] (P41539), was significant. An alternative but equally interesting lead related to this result is that neurokinin A, another peptide derived from protachykinin, expressed in sensory neurons and distributed similarly to SP in the spinal cord,64 is formed by cleavage at a nearby dibasic site to our detected peptide. Therefore, the detection of this peptide may be a result of the processing of the prohormone to make neurokinin A.
Within the GO analysis of the precursor proteins, the protein levels that changed the most between treatment and control samples were those that were characterized as signaling molecules, enzyme modulators, and hydrolases. Neuropeptides are typically produced from the cleavage of larger precursor proteins through a sequence of processing steps that use a variety of enzymes, including prohormone convertases, endopeptidase, carboxypeptidase, and aminopeptidase. Therefore, in addition to the observation that the levels of signaling molecules are changing between the treatment and control groups, changes in the proteins used by the cell to synthesize and process specific neuropeptides is a promising result.
Finally, it might seem paradoxical that the peptide levels are decreasing in the treatment groups compared to the control groups when we hypothesize that these peptides are involved in the transmission of sensory information, including itch. Two aspects of this study make these observations more reasonable. First, MS measures the amount of peptide in the tissue at the time of sample collection. If a peptide undergoes activity-dependent release, its amount within the tissue may decrease. A longer-term use of a peptide within the system can result in more synthesis and accumulation. However, it could also result in decreased synthesis as the organism tries to modulate its sensitivity to the stimulus over time. Thus, we expect the amount of a peptide involved in sensory transmission within our itch model to change, but the direction of change depends on peptide release / synthesis dynamics and is difficult to predict. Additionally, neuropeptides involved in signal transmission may originate from either the DRG or spinal neurons, making their detection complex.
Neuropeptide signaling is a dynamic system in which location, timing, and sampling methods all influence which peptides are present, as well as if we can detect that change. Our study began the process of identifying peptides to target for follow-up studies within itch model systems. Overall, we are hopeful that the increased insight into this complex system provided by this work helps to identify important peptide candidates for further investigation into the mechanistic processes leading to itch and other peripheral nervous system-related disorders in the future.
Supplementary Material
Table S1: Details of the experimental design.
Table S3: List of all the quantified peptides from both regions (DH and DRG) and both models (AEW and MC903).
Table S2: List of all the identified peptides via PEAKS search from the DH and DRG.
Table S4: Loading scores for the first 3 principal components from both regions (DH and DRG) and both models (AEW and MC903)
Table S5: Gene expression validation from the Allen Brain Atlas (spinal cord). (MS Excel files)
Figure S1. Fold-change (M) vs. average (A) plots for the quantified peptides and the box plots corresponding to all quantified peptides before and after normalization. Figure S2. Plot of qvalues for all the quantified peptides from the DH AEW model. (PDF)
ACKNOWLEDGMENTS
This project was supported by the National Institutes of Health under awards from the National Institute on Drug Abuse, P30DA018310 (to J.V.S.); the National Eye Institute, R01EY024704, and the National Institute of Allergy and Infectious Disease, R01AI125743 (to Q.L.); and a Pew Scholar Award (to Q.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the awarding agencies.
Footnotes
Data Availability
The mass spectrometry peptidomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015949 and DOI:10.6019/PXD015949. Reviewer account details: Username: reviewer38452@ebi.ac.uk, Password: 6F699oue
The authors declare no competing interests.
REFERENCES
- (1).Matterne U; Apfelbacher CJ; Vogelgsang L; Loerbroks A; Weisshaar E, Incidence and determinants of chronic pruritus: a population-based cohort study. Acta Derm. Venereol 2013, 93, 532–537. [DOI] [PubMed] [Google Scholar]
- (2).Garibyan L; Rheingold CG; Lerner EA, Understanding the pathophysiology of itch. Dermatol. Ther 2013, 26, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Song J; Xian D; Yang L; Xiong X; Lai R; Zhong J, Pruritus: Progress toward Pathogenesis and Treatment. Biomed. Res. Int 2018, 2018, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).McGlone F; Reilly D, The cutaneous sensory system. In Neurosci. Biobehav. Rev, 2010; Vol. 34, pp 148–159. [DOI] [PubMed] [Google Scholar]
- (5).Brown AG, The dorsal horn of the spinal cord. Q. J. Exp. Physiol 1982, 67, 193–212. [DOI] [PubMed] [Google Scholar]
- (6).Potenzieri C; Undem BJ, Basic mechanisms of itch. Clin. Exp. Allergy 2012, 42, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Devor M, Unexplained peculiarities of the dorsal root ganglion. Pain 1999, Suppl 6, S27–35. [DOI] [PubMed] [Google Scholar]
- (8).Hogan QH, Labat lecture: the primary sensory neuron: where it is, what it does, and why it matters. Reg. Anesth. Pain Med 2010, 35, 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ikoma A; Rukwied R; Stander S; Steinhoff M; Miyachi Y; Schmelz M, Neurophysiology of pruritus: interaction of itch and pain. Arch. Dermatol 2003, 139, 1475–1478. [DOI] [PubMed] [Google Scholar]
- (10).Strand F, Neuropeptides: Regulators of Physiological Processes. MIT Press: Cambridge, MA, 1999; p 480. [Google Scholar]
- (11).Eipper BA; Stoffers DA; Mains RE, The biosynthesis of neuropeptides: peptide alpha-amidation. Annu. Rev. Neurosci 1992, 15, 57–85. [DOI] [PubMed] [Google Scholar]
- (12).Saria A, The Role of Substance P and Other Neuropeptides in Transmission of Pain In Pain, Brihaye J; Loew F; Pia HW, Eds. Springer Vienna: Vienna, 1987; pp 33–35. [DOI] [PubMed] [Google Scholar]
- (13).Tsujii S; Bray GA, Acetylation alters the feeding response to MSH and beta-endorphin. Brain Res. Bull 1989, 23, 165–169. [DOI] [PubMed] [Google Scholar]
- (14).Baldwin GS; Knesel J; Monckton JM, Phosphorylation of gastrin-17 by epidermal growth factor-stimulated tyrosine kinase. Nature 1983, 301, 435–437. [DOI] [PubMed] [Google Scholar]
- (15).Lietz CB; Toneff T; Mosier C; Podvin S; O’Donoghue AJ; Hook V, Phosphopeptidomics Reveals Differential Phosphorylation States and Novel SxE Phosphosite Motifs of Neuropeptides in Dense Core Secretory Vesicles. J. Am. Soc. Mass Spectrom 2018, 29, 935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Huttner WB, Tyrosine sulfation and the secretory pathway. Annu. Rev. Physiol 1988, 50, 363–376. [DOI] [PubMed] [Google Scholar]
- (17).Yang N; Anapindi KDB; Rubakhin SS; Wei P; Yu Q; Li L; Kenny PJ; Sweedler JV, Neuropeptidomics of the Rat Habenular Nuclei. J. Proteome Res 2018, 17, 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Garden RW; Moroz TP; Gleeson JM; Floyd PD; Li L; Rubakhin SS; Sweedler JV, Formation of N-pyroglutamyl peptides from N-Glu and N-Gln precursors in Aplysia neurons. J. Neurochem 1999, 72, 676–681. [DOI] [PubMed] [Google Scholar]
- (19).Pitake S; Ralph PC; DeBrecht J; Mishra SK, Atopic Dermatitis Linked Cytokine Interleukin-31 Induced Itch Mediated via a Neuropeptide Natriuretic Polypeptide B. Acta Derm. Venereol 2018, 98, 795–796. [DOI] [PubMed] [Google Scholar]
- (20).Lee H; Ko M-C, Distinct functions of opioid-related peptides and gastrin-releasing peptide in regulating itch and pain in the spinal cord of primates. Sci. Rep 2015, 5, 11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wan L; Jin H; Liu XY; Jeffry J; Barry DM; Shen KF; Peng JH; Liu XT; Jin JH; Sun Y; Kim R; Meng QT; Mo P; Yin J; Tao A; Bardoni R; Chen ZF, Distinct roles of NMB and GRP in itch transmission. Sci. Rep 2017, 7, 15466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Dallas DC; Guerrero A; Parker EA; Robinson RC; Gan J; German JB; Barile D; Lebrilla CB, Current peptidomics: applications, purification, identification, quantification, and functional analysis. Proteomics 2015, 15, 1026–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Romanova EV; Sweedler JV, Peptidomics for the discovery and characterization of neuropeptides and hormones. Trends Pharmacol. Sci 2015, 36, 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sandor K; Krishnan S; Agalave NM; Krock E; Salcido JV; Fernandez-Zafra T; Khoonsari PE; Svensson CI; Kultima K, Spinal injection of newly identified cerebellin-1 and cerebellin-2 peptides induce mechanical hypersensitivity in mice. Neuropeptides 2018, 69, 53–59. [DOI] [PubMed] [Google Scholar]
- (25).Yates JR 3rd; Eng JK; McCormack AL; Schieltz D, Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem 1995, 67, 1426–1436. [DOI] [PubMed] [Google Scholar]
- (26).Bieber T, Atopic Dermatitis. N. Engl. J. Med 2008, 358, 1483–1494. [DOI] [PubMed] [Google Scholar]
- (27).Choi J; Kim JR; Kim H; Kim YA; Lee HJ; Kim J; Lee KW, The atopic dermatitis-like symptoms induced by MC903 were alleviated in JNK1 knockout mice. Toxicol. Sci 2013, 136, 443–449. [DOI] [PubMed] [Google Scholar]
- (28).Moosbrugger-Martinz V; Schmuth M; Dubrac S, A Mouse Model for Atopic Dermatitis Using Topical Application of Vitamin D3 or of Its Analog MC903. Methods Mol Biol 2017, 1559, 91–106. [DOI] [PubMed] [Google Scholar]
- (29).Li M; Hener P; Zhang Z; Kato S; Metzger D; Chambon P, Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 11736–11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Akiyama T; Carstens MI; Carstens E, Enhanced scratching evoked by PAR-2 agonist and 5-HT but not histamine in a mouse model of chronic dry skin itch. Pain 2010, 151, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Miyamoto T; Nojima H; Shinkado T; Nakahashi T; Kuraishi Y, Itch-associated response induced by experimental dry skin in mice. Jpn. J. Pharmacol 2002, 88, 285–292. [DOI] [PubMed] [Google Scholar]
- (32).Han L; Ma C; Liu Q; Weng HJ; Cui Y; Tang Z; Kim Y; Nie H; Qu L; Patel KN; Li Z; McNeil B; He S; Guan Y; Xiao B; Lamotte RH; Dong X, A subpopulation of nociceptors specifically linked to itch. Nat. Neurosci 2013, 16, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Oetjen LK; Mack MR; Feng J; Whelan TM; Niu H; Guo CJ; Chen S; Trier AM; Xu AZ; Tripathi SV; Luo J; Gao X; Yang L; Hamilton SL; Wang PL; Brestoff JR; Council ML; Brasington R; Schaffer A; Brombacher F; Hsieh CS; Gereau R. W. t.; Miller MJ; Chen ZF; Hu H; Davidson S; Liu Q; Kim BS, Sensory Neurons Co-opt Classical Immune Signaling Pathways to Mediate Chronic Itch. Cell 2017, 171, 217–228 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Roush JK; McLaughlin RM; Radlinsky MAG, Understanding the pathophysiology of osteoarthritis. Vet. Med 2002, 97, 108–112. [Google Scholar]
- (35).Yang N; Anapindi KDB; Romanova EV; Rubakhin SS; Sweedler JV, Improved identification and quantitation of mature endogenous peptides in the rodent hypothalamus using a rapid conductive sample heating system. Analyst 2017, 142, 4476–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bora A; Annangudi SP; Millet LJ; Rubakhin SS; Forbes AJ; Kelleher NL; Gillette MU; Sweedler JV, Neuropeptidomics of the supraoptic rat nucleus. J. Proteome Res. 2008, 7, 4992–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).UniProt Consortium, UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).MacLean B; Tomazela DM; Shulman N; Chambers M; Finney GL; Frewen B; Kern R; Tabb DL; Liebler DC; MacCoss MJ, Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Chawade A; Alexandersson E; Levander F, Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 2014, 13, 3114–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T; Mann M; Cox J, The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [DOI] [PubMed] [Google Scholar]
- (41).Anapindi KDB; Yang N; Romanova EV; Rubakhin SS; Tipton A; Dripps I; Sheets Z; Sweedler JV; Pradhan AA, PACAP and other neuropeptides link chronic migraine and opioid-induced hyperalgesia in mouse models. Mol. Cell. Proteomics 2019, mcp.RA119.001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).R Core Team R: A language and environment for statistical computing. https://www.R-project.org/.
- (43).Webb-Robertson BJ; Wiberg HK; Matzke MM; Brown JN; Wang J; McDermott JE; Smith RD; Rodland KD; Metz TO; Pounds JG; Waters KM, Review, evaluation, and discussion of the challenges of missing value imputation for mass spectrometry-based label-free global proteomics. J. Proteome Res 2015, 14, 1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).David CC; Jacobs DJ, Principal component analysis: a method for determining the essential dynamics of proteins. Methods Mol. Biol 2014, 1084, 193–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lambers TT; Gloerich J; van Hoffen E; Alkema W; Hondmann DH; van Tol EA, Clustering analyses in peptidomics revealed that peptide profiles of infant formulae are descriptive. Food Sci. Nutr 2015, 3, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Romanova EV; Rubakhin SS; Ossyra JR; Zombeck JA; Nosek MR; Sweedler JV; Rhodes JS, Differential peptidomics assessment of strain and age differences in mice in response to acute cocaine administration. J. Neurochem 2015, 135, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Thomas PD; Campbell MJ; Kejariwal A; Mi H; Karlak B; Daverman R; Diemer K; Muruganujan A; Narechania A, PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003, 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Hawrylycz MJ; Lein ES; Guillozet-Bongaarts AL; Shen EH; Ng L; Miller JA; van de Lagemaat LN; Smith KA; Ebbert A; Riley ZL; Abajian C; Beckmann CF; Bernard A; Bertagnolli D; Boe AF; Cartagena PM; Chakravarty MM; Chapin M; Chong J; Dalley RA; David Daly B; Dang C; Datta S; Dee N; Dolbeare TA; Faber V; Feng D; Fowler DR; Goldy J; Gregor BW; Haradon Z; Haynor DR; Hohmann JG; Horvath S; Howard RE; Jeromin A; Jochim JM; Kinnunen M; Lau C; Lazarz ET; Lee C; Lemon TA; Li L; Li Y; Morris JA; Overly CC; Parker PD; Parry SE; Reding M; Royall JJ; Schulkin J; Sequeira PA; Slaughterbeck CR; Smith SC; Sodt AJ; Sunkin SM; Swanson BE; Vawter MP; Williams D; Wohnoutka P; Zielke HR; Geschwind DH; Hof PR; Smith SM; Koch C; Grant SGN; Jones AR, An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012, 489, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kuraishi Y; Nagasawa T; Hayashi K; Satoh M, Scratching behavior induced by pruritogenic but not algesiogenic agents in mice. Eur. J. Pharmacol 1995, 275, 229–233. [DOI] [PubMed] [Google Scholar]
- (50).Rogoz K; Andersen HH; Lagerstrom MC; Kullander K, Multimodal use of calcitonin gene-related peptide and substance P in itch and acute pain uncovered by the elimination of vesicular glutamate transporter 2 from transient receptor potential cation channel subfamily V member 1 neurons. J. Neurosci 2014, 34, 14055–14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kleij HP; Bienenstock J, Significance of Conversation between Mast Cells and Nerves. Allergy Asthma Clin. Immunol 2005, 1, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Fricker LD; McKinzie AA; Sun J; Curran E; Qian Y; Yan L; Patterson SD; Courchesne PL; Richards B; Levin N; Mzhavia N; Devi LA; Douglass J, Identification and Characterization of proSAAS, a Granin-Like Neuroendocrine Peptide Precursor that Inhibits Prohormone Processing. J. Neurosci 2000, 20, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wardman JH; Fricker LD, ProSAAS-derived peptides are differentially processed and sorted in mouse brain and AtT-20 cells. PLoS One 2014, 9, e104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Ye H; Wang J; Tian Z; Ma F; Dowell J; Bremer Q; Lu G; Baldo B; Li L, Quantitative Mass Spectrometry Reveals Food Intake-Induced Neuropeptide Level Changes in Rat Brain: Functional Assessment of Selected Neuropeptides as Feeding Regulators. Mol. Cell. Proteomics 2017, 16, 1922–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Xing Y; Chen J; Hilley H; Steele H; Yang J; Han L, Molecular signature of pruriceptive MrgprA3 + neurons. bioRxiv 2019, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Priestley JV, Neuropeptides: Sensory Systems. In Encyclopedia of Neuroscience, 2010; pp 935–943. [Google Scholar]
- (57).Mudge AW; Leeman SE; Fischbach GD, Enkephalin inhibits release of substance P from sensory neurons in culture and decreases action potential duration. Proc. Natl. Acad. Sci. U. S. A 1979, 76, 526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Akiyama T; Carstens E, Neural processing of itch. Neuroscience 2013, 250, 697–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Slominski AT; Zmijewski MA; Zbytek B; Brozyna AA; Granese J; Pisarchik A; Szczesniewski A; Tobin DJ, Regulated proenkephalin expression in human skin and cultured skin cells. J. Invest. Dermatol 2011, 131, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ojika K; Mitake S; Tohdoh N; Appel SH; Otsuka Y; Katada E; Matsukawa N, Hippocampal cholinergic neurostimulating peptides (HCNP). Prog. Neurobiol 2000, 60, 37–83. [DOI] [PubMed] [Google Scholar]
- (61).Hirschmann JV; Lawlor F; English JS; Louback JB; Winkelmann RK; Greaves MW, Cholinergic urticaria. A clinical and histologic study. Arch. Dermatol 1987, 123, 462–467. [DOI] [PubMed] [Google Scholar]
- (62).Heyer GR; Hornstein OP, Recent studies of cutaneous nociception in atopic and non-atopic subjects. J. Dermatol 1999, 26, 77–86. [DOI] [PubMed] [Google Scholar]
- (63).Zhang X; Che FY; Berezniuk I; Sonmez K; Toll L; Fricker LD, Peptidomics of Cpe(fat/fat) mouse brain regions: implications for neuropeptide processing. J. Neurochem 2008, 107, 1596–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Swain CJ, 2 - Neurokinin Receptor Antagonists In Prog. Med. Chem, Ellis GP; Luscombe DK; Oxford AW, Eds. Elsevier: 1998; Vol. 35, pp 57–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Details of the experimental design.
Table S3: List of all the quantified peptides from both regions (DH and DRG) and both models (AEW and MC903).
Table S2: List of all the identified peptides via PEAKS search from the DH and DRG.
Table S4: Loading scores for the first 3 principal components from both regions (DH and DRG) and both models (AEW and MC903)
Table S5: Gene expression validation from the Allen Brain Atlas (spinal cord). (MS Excel files)
Figure S1. Fold-change (M) vs. average (A) plots for the quantified peptides and the box plots corresponding to all quantified peptides before and after normalization. Figure S2. Plot of qvalues for all the quantified peptides from the DH AEW model. (PDF)


