Abstract
Background
Ding Chuan Tang (DCT), a traditional Chinese herbal formula, has been consistently prescribed for the therapeutic management of wheezing and asthma-related indications since the Song Dynasty (960–1279 AD). This study aimed to identify molecular network pharmacology connections to understand the biological asthma-linked mechanisms of action of DCT and potentially identify novel avenues for asthma drug development.
Methods
Employing molecular docking (AutoDock Vina) and computational analysis (Cytoscape 3.6.0) strategies for DCT compounds permitted examination of docking connections for proteins that were targets of DCT compounds and asthma genes. These identified protein targets were further analyzed to establish and interpret network connections associated with asthma disease pathways.
Results
A total of 396 DCT compounds and 234 asthma genes were identified through database search. Computational molecular docking of DCT compounds identified five proteins (ESR1, KDR, LTA4H, PDE4D and PPARG) mutually targeted by asthma genes and DCT compounds and 155 docking connections associated with cellular pathways involved in the biological mechanisms of asthma.
Conclusions
DCT compounds directly target biological pathways connected with the pathogenesis of asthma including inflammatory and metabolic signaling pathways.
Keywords: Herbal medicine, Complementary medicine, Natural product, Asthma, Respiratory disease, Network pharmacology, Computational analysis
Introduction
Asthma is defined and classified by its major physical characteristics including airflow obstruction and airway inflammation. Currently the specific cause(s) of asthma remain unknown and curative drugs are not available (Global Initiative for Asthma, 2018).
Historical Chinese medical texts have documented the use of the traditional Chinese herbal formula Ding Chuan Tang (DCT), also known as the Arrest Wheezing Decoction, for the use of asthma-like symptoms such as wheeze and cough since the Song dynasty (960–1279 AD) and the formulation in contemporary use, has been prescribed continually since the Ming Dynasty (1368–1644) (Hoizey & Hoizey, 1993). Contemporary DCT continues to be indicated for wheeze and cough. Modern clinical and experimental evidence suggests its herbal ingredients and their constituent chemical compounds can moderate and manage the symptoms of asthma, including reduction of airway inflammation, hyper-responsiveness and respiratory mucous production, as well as ameliorate damage to the airway epithelium caused by chronic inflammation (Bensky & Barolet, 2009; Chan et al., 2006; Kao et al., 2004). The contemporary DCT formula contains nine herbs: Ginkgo biloba L. (Bai Guo), Ephedra sinica Stapf (Ma Huang), Perilla frutescens (L.) Britton (Su Zi), Glycyrrhiza uralensis Fisch (Gan Cao), Tussilago farfara L. (Kuan Dong Hua), Prunus armeniaca L. (Xing Ren), Morus alba L. (Sang Bai Pi), Scutellaria baicalensis Georgi (Huang Qin) and Pinellia ternata (Thunb.) Makino (Ban Xia) (Bensky & Barolet, 2009; The Plant List, 2013).
As DCT is a multi-herbal formula, identification and analysis of the molecular properties of the chemical compounds could theoretically disclose the biological mechanisms and pathways targeted associated with asthma (Chillistone & Hardman, 2014; Lipinski et al., 2001). Additionally, using a network pharmacology approach through molecular docking potentially identifies DCT-compound target proteins and signaling pathways and how these relate to asthma pathology.
Network pharmacology focuses on the biological networks between ligands (such as DCT-compounds) and disease pathways including target proteins, protein–protein interactions, cellular signaling and biochemical cascades (Shi et al., 2014; Ursa et al., 2011). Computational software was applied to identify potential target proteins for DCT-compounds through the use of pharmacophore models and molecular docking. Molecular docking assists in three-dimensional computer-assisted drug design and within this study, will identify and predict the possible binding sites of DCT-compounds with proteins targeted by asthma genes; thus, identifying the pathological processes in asthma that are directly associated with DCT (Li & Zhang, 2013; Yu & MacKerell, 2017).
Materials and Methods
The molecular modeling software used included: Discovery Studio 4.5 system from Accelrys Inc. (DS 4.5; San Diego, CA, USA) (BIOVIA, 2015), AutoDock Vina software (Trott & Olson, 2010) and Cytoscape 3.6.0 software (Shannon et al., 2003).
DCT-compounds identification
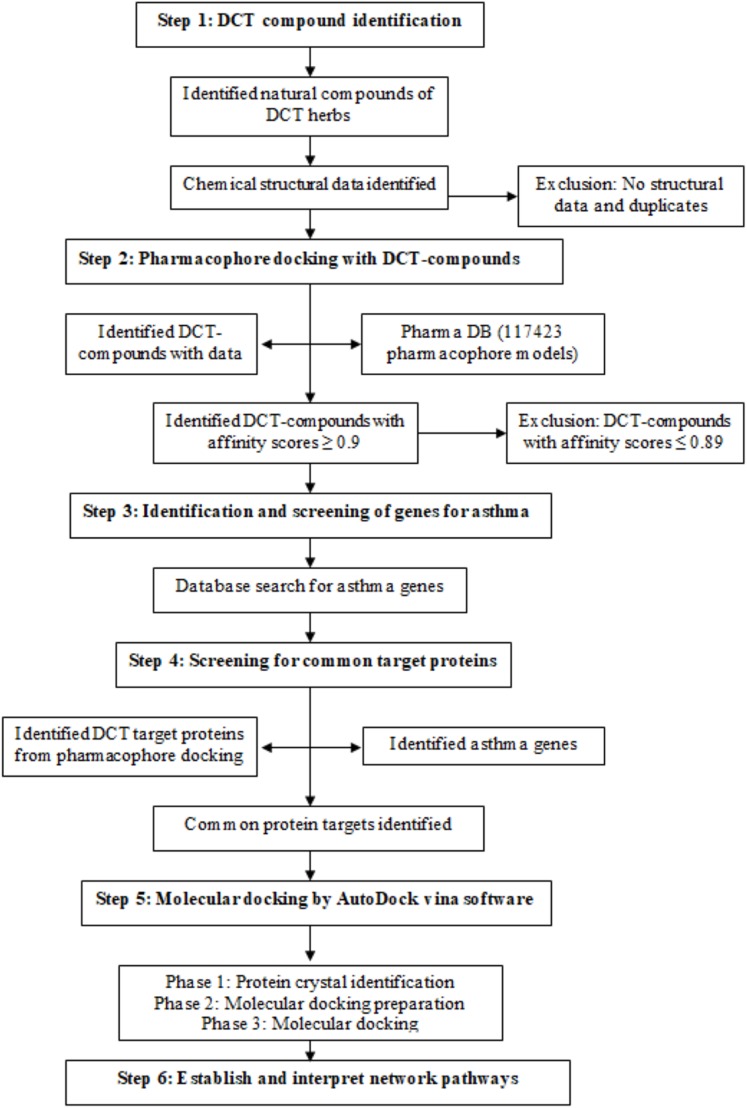
DCT-compounds were identified by searching the keywords DCT and each of the nine DCT herb names (Chinese, English (common), pharmaceutical and Latin) within five English databases (PubMed, Embase, Science direct, HIT database and CINAHL) and two reference books (Traditional Chinese medicines: Molecular structures, natural sources and applications (Zhou, Yan & Xie, 2003), Chinese herbal medicine: Materia medica (Bensky, Clavey & Stoger, 2004)). The three-dimensional chemical structures of the DCT-compounds were downloaded from PubChem, and DS 4.5 was used for further optimization (step 1 in Fig. 1).
Figure 1. Method flowchart for computational analysis for Ding Chuan Tang.
Pharmacophore screening with DCT-compounds
Pharmacophore models are constructed from multiple pharmacological features and targets and are used in computational drug discovery to recognize the molecular features of one or more compounds (ligand) required for a lock and key fit with macromolecules (proteins) with the same or similar biological activity (Thangapandian et al., 2011; Wieder et al., 2017). The DS 4.5 software contains a Ligand pharmacophore profiler (LPP) protocol, allowing for virtual screening of multiple chemical compounds against a pharmacophore database (PharmaDB) containing 117,423 pharmacophore models and constructed from 7,028 protein databank (PDB) protein-ligand X-ray crystal structures. This protocol investigates the potential biological actions for the ligands using the pharmacophore models, with the results for the docking within the LPP calculated in fit-values, as a high fit-value indicates the likely comparability of small molecules and target protein binding (Ming, Yi & Yang, 2018). The identified 396 DCT-compounds were virtually screened with the PharmaDB database and the inclusion criteria was set as fit-value 0.9 and above (1.0 fit-value being perfect prediction), to identify the target proteins with the high comparability score (step 2 in Fig. 1).
Identification and screening of genes for asthma
Genes associated with the asthma disease pathways were identified using “asthma” as a keyword to search the following four databases: Disease gene search engine with evidence (Kim et al., 2013), Genetic Association Database (Becker et al., 2004), Pubmed_Gene (Benson et al., 2013) and GeneCards (Stelzer et al., 2016) (step 3 in Fig. 1). The identified asthma genes were further screened with the pharmacophore and DCT-compound docking results to identify mutual target proteins (step 4 in Fig. 1).
Molecular docking of DCT-compounds and potential target proteins
DCT-compounds that resulted with fit-value scores ≥0.9 were docked using AutoDock Vina software (Trott & Olson, 2010) with the target proteins identified in the asthma gene and pharmacophore screening (step 5 in Fig. 1). Molecular docking included three phases: (1) crystal complex identification, (2) crystal complex preparation, and (3) molecular docking.
Phase 1: crystal complex identification
Docking the DCT-compounds with protein-ligand co-crystallization complexes allows a more specific prediction of binding site locations, docking pose prediction and the likelihood of binding affinity. Phase one of molecular docking required the identification of three-dimensional crystal complex structures of all identified mutual target proteins from RCSB-Protein Data Bank database. For the crystal complexes to be included they must be a human protein, result in an affinity binding score of ≥−9.0 kcal/mol with the identified mutual target proteins, be in the form of a protein-molecule crystal and have a resolution of less than 3 Å.
Phase 2: crystal complex preparation
Phase two prepared the identified crystal complexes for docking, with each crystal defined by a configuration (config) file to determine the actions. These config files determined the following for each target protein: the center of the active site (centre_x, centre_y, centre_z), the size of the active site (size_x, size_y, size_z), the maximum number of binding modes to output (nummodes) (set to 10) and exhaustiveness (set to 50). During the docking process, a PDBQT file was used in addition to the config file, as the PDBQT file determined the information of potential ligands and receptors required for the target protein docking process.
Phase 3: molecular docking
Phase three consisted of AutoDock Vina molecular docking; using Lamarckian genetic algorithm to establish results. The results output as .pdbqt* and log*.txt files and were scored according to virtual docking binding free energy to establish the affinity binding score between the target protein receptors and ligands. For this investigation, affinity binding results ≥−9.0 kcal/mol were selected for the establishment of networks between DCT-compounds and target proteins, as these values suggest strong binding affinity between the DCT-compounds and proteins (Hopkins & Paolini, 2007).
Establishment of networks
Cytoscape 3.6.0 software (Shannon et al., 2003) was used to establish the networks between docked DCT-compounds and the identified target proteins (step 6 in Fig. 1). The DCT-compounds and target proteins were defined as the nodes of the network, while the affinity binding energy value was the edge of the network. The Network analysis function in Cytoscape 3.6.0 analyzed the features of the network structure through the yfiles plug-in module to draw the network image. This further optimized the layout of the network and identified the most suitable protein targets for DCT-compounds. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa et al., 2017) includes an advanced mapping tool (Search and Color pathway) linked to Cytoscape 3.6.0 software and accessing the affinity binding results from DCT-compounds and protein docking, identified the signaling pathways targeted by DCT.
Data provided from the Protein Data Bank database (PDB) aided in the establishment of network connections between the DCT-compounds and the target proteins post the molecular docking process. Furthermore, the KEGG database (Kanehisa et al., 2017), UniProt database (UniProt Consortium, 2018), Wikipathways (Slenter et al., 2018) and Cell signaling technology pathways (Cell Signaling Technology, 2018) were searched to define and interpret the identified network pathways for the potential mechanisms of action and relevance to the management of asthma.
Results
DCT-compound identification
Following the literature search strategies, the original search identified 644 DCT-compounds; however, 109 duplicates and one toxic compound (cyanide) were removed and a further 138 were excluded due to lack of compound identification data such as the molecular structure, molecular formula, simplified molecular-input line-entry system data and physicochemical property data. This resulted in 396 DCT-compounds included for analysis, with 40 from Ginkgo biloba L. (Bai Guo), 40 from Ephedra sinica Stapf (Ma Huang), 30 from Perilla frutescens (L.) Britton (Su Zi), 100 from Glycyrrhiza uralensis Fisch (Gan Cao), 15 from Tussilago farfara L. (Kuan Dong Hua), 17 from Prunus armeniaca L. (Xing Ren), 43 from Morus alba L. (Sang Bai Pi), 33 from Scutellaria baicalensis Georgi (Huang Qin) and 78 from Pinellia ternata (Thunb.) Makino (Ban Xia).
Pharmacophore screening with DCT-compounds
The virtual screening of the 396 DCT-compounds with the 117,423 pharmacophore models within the PharmaDB resulted in 159 pharmacophore models (potential target proteins). Following this initial hit list, this step resulted in two further sets of outcomes. The first outcome was the reduction of the original hit list of 159 potential target proteins down to 53 due to the inclusion parameter set at fit-value 0.9 and above (Table S1). The second outcome was the reduction of 396 DCT-compounds to 243 DCT-compounds, also due to the 0.9 and above fit-value inclusion parameter (Table S2). This second outcome narrowed down the number of compounds compared during the molecular docking phase to only those DCT-compounds with high affinity binding; as a high fit-value indicates high comparability of small molecules and target protein binding (Ming, Yi & Yang, 2018).
Identification of genes for asthma
Asthma is classified as a “complex” heritable disease with multiple genes implicated and contributing to its development (Global Initiative for Asthma, 2018). A total of 234 asthma genes were identified, with the results listed in Table S3.
Screening for common target proteins for DCT target proteins and asthma genes
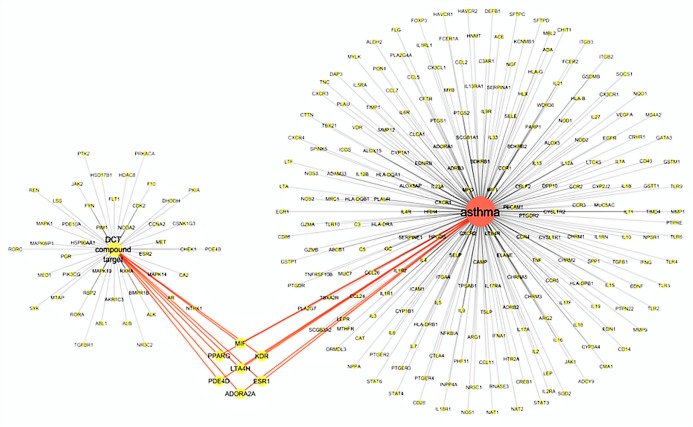
The virtual screening of 53 DCT potential target proteins identified from pharmacophore docking and the 234 identified asthma genes, resulted in seven mutual targets as centrally displayed in Fig. 2, with the list of all proteins provided in Tables S1 and S3. These mutual protein targets included Adenosine A2A receptor (ADORA2A), Estrogen receptors alpha (ESR1), Kinase insert domain receptor (KDR), Leukotriene A4 hydrolase (LTA4H), Macrophage migration inhibitory factor (MIF), cAMP-specific 3′,5′-cyclic phosphodiesterase 4D (PDE4D) and Peroxisome proliferator-activated receptor gamma (PPARG).
Figure 2. Virtual screening results for Ding Chuan Tang and asthma genes target proteins.
In Fig. 2, the group on the left-hand side (Group 1) is the 53 DCT protein targets resulting from pharmacophore virtual screening. Group 2: On the right are all the identified genes associated with asthma. In the center highlighted are the seven proteins both groups target: ADORA2A, ESR1, KDR, LTA4H, MIF, PDE4D and PPARG. At the time of this investigation, the active binding sites, required for Autodock Vina, could not be identified for ADORA2A and MIF; therefore, they were not included in further examination. Five target proteins (ESR1, KDR, LTA4H, PDE4D and PPARG) were included in the next step: Molecular docking with DCT-compounds.
Molecular docking of active compounds and potential target proteins
Phase 1 results: crystal complex identification
Post molecular docking of the five target proteins and the 243 DCT-compounds, a total of 18 human protein-molecule crystal complexes with a resolution of less than 3 Å were identified with affinity scores of ≥−9.0 kcal/mol including three for ESR1 (3erd, 2yja, 2jfa), four for KDR (3vo3, 3vhe, 3cjg, 3cjf), two for LTA4H (4dpr, 3fts), four for PDE4D (1xom, 1tbb, 1tb7, 1zkn), and five for PPARG (5lsg(1), 4jaz, 3sz1_MYR, 3sz1_LU, 3adx) (PDB). The descriptions of the protein-ligand complex and the classifications of these protein crystal complexes as an agonist (promotor or activator) or antagonist (inhibitor or blocker) are defined in Table S4.
Phase 2 results: crystal complex preparation
To prepare for molecular docking, the config files for the three-dimensional crystal structures of the five identified target proteins (ESR1, KDR, LTA4H, PDE4D and PPARG) were determined and the parameters for each target protein and corresponding crystals were defined (Table S4).
Phase 3 results: molecular docking
The molecular docking results for the specific numbers of compounds from each DCT herb that targeted and docked with affinity scores equal to and over −9.0 kcal/mol to each protein target crystal complex are outlined in Table 1; with many DCT-compounds docking with multiple crystal complexes of the same protein. The three ESR1 crystal complexes resulted in eight docked DCT-compounds from two DCT herbs, G. uralensis Fisch (Gan Cao) and M. alba L. (Sang Bai Pi) (Table 1). The four KDR crystal complexes resulted in 119 docked DCT-compounds from eight DCT herbs, excluding P. armeniaca L. (Xing Ren) (Table 1). The two LTA4H crystal complexes resulted in 139 docked DCT-compounds from all nine DCT herbs, while the four PDE4D crystal complexes resulted in 103 docked DCT-compounds from all nine DCT herbs (Table 1). The five PPARG crystal complexes resulted in 109 docked DCT-compounds from eight DCT herbs, excluding P. armeniaca L. (Xing Ren) (Table 1).
Table 1. Molecular docking results for Ding Chuan Tang-compounds and target proteins with affinity scores ≥−9.0 kcal/mol.
| Target protein Crystal complex (n = total compounds docked) |
Ding chuan tang herbal ingredient | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ginkgo biloba L. (Bai Guo) | Ephedra sinica Stapf (Ma Huang) | Perilla frutescens (L.) Britton (Su Zi) |
Glycyrrhiza uralensis Fisch (Gan Cao) |
Tussilago farfara L. (Kuan Dong Hua) |
Prunus armeniaca L. (Xing Ren) |
Morus alba L. (Sang Bai Pi) |
Scutellaria baicalensis Georgi (Huang Qin) | Pinellia ternata (Thunb.) Makino (Ban Xia) | |
| ESR1_3erd (n = 1) |
1 | ||||||||
| ESR1_2yja (n = 2) |
1 | 1 | |||||||
| ESR1_2jfa (n = 6) |
4 | 2 | |||||||
| KDR_3vo3 (n = 88) |
3 | 3 | 8 | 44 | 2 | 20 | 7 | 1 | |
| KDR_3vhe (n = 78) |
5 | 1 | 7 | 39 | 1 | 14 | 9 | 2 | |
| KDR_3cjg (n = 52) |
3 | 1 | 3 | 26 | 2 | 14 | 2 | 1 | |
| KDR_3cjf (n = 18) |
2 | 1 | 6 | 1 | 7 | 1 | |||
| LTA4H_4dpr (n = 122) | 10 | 4 | 7 | 59 | 3 | 1 | 21 | 13 | 4 |
| LTA4H_3fts (n = 54) |
7 | 1 | 8 | 22 | 9 | 2 | 5 | ||
| PDE4D_1xom (n = 82) | 2 | 3 | 48 | 3 | 1 | 17 | 5 | 3 | |
| PDE4D_1tbb (n = 63) | 3 | 3 | 29 | 2 | 1 | 19 | 3 | 3 | |
| PDE4D_1tb7 (n = 59) | 2 | 1 | 3 | 25 | 1 | 1 | 17 | 7 | 2 |
| PDE4D_1zkn (n = 78) | 2 | 2 | 45 | 2 | 1 | 18 | 5 | 3 | |
| PPARG_5slg(1) (n = 36) | 2 | 2 | 12 | 15 | 4 | 1 | |||
| PPARG_4jaz (n = 71) | 4 | 1 | 6 | 34 | 1 | 17 | 5 | 3 | |
| PPARG_3sz1_MYR (n = 70) | 2 | 1 | 5 | 32 | 13 | 16 | 1 | ||
| PPARG_3sz1_LU (n = 69) | 2 | 1 | 5 | 32 | 13 | 15 | 1 | ||
| PPARG_3adx (n = 66) | 3 | 5 | 37 | 1 | 14 | 5 | 1 | ||
Note:
ESR1, Estrogen receptors alpha; KDR, Kinase insert domain receptor; LTA4H, Leukotriene A4 hydrolase; PDE4D, cAMP-specific 3’,5’-cyclic phosphodiesterase 4D; PPARG, Peroxisome proliferator-activated receptor gamma.
Affinity binding scores ≥−9.0 kcal/mol resulted with a range between −9 and −13.4 kcal/mol. The range for each target protein is as follows: ESR1 (−9 to −9.9 kcal/mol), KDR (−9 to −11.9 kcal/mol), LTA4H (−9 to −13.4 kcal/mol), PDE4D (−9 to −12 kcal/mol) and PPARG (−9 to −11.8 kcal/mol).
Establish network connections
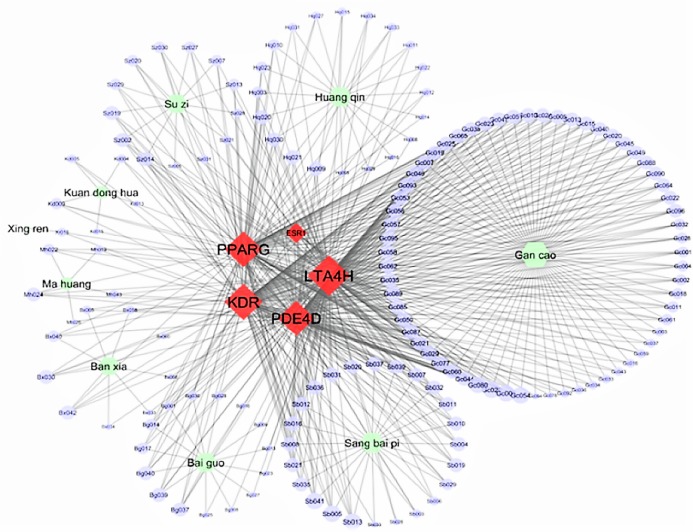
Figure 3 displays the Cytoscape graphic network connections from the molecular docking results between the 243 DCT-compounds and the five target proteins. The Cytoscape 3.6.0 yfiles plug-in module was used to draw and optimize the layout of the network image (Shannon et al., 2003). All five target proteins resulted in 155 network connections with compounds from the nine DCT herbs including: Ginkgo biloba L. (Bai Guo) (n = 15), Ephedra sinica Stapf (Ma Huang) (n = 5), Perilla frutescens (L.) Britton (Su Zi) (n = 13), Glycyrrhiza uralensis Fisch (Gan Cao) (n = 63), Tussilago farfara L. (Kuan Dong Hua) (n = 5), Prunus armeniaca L. (Xing Ren) (n = 1), Morus alba L. (Sang Bai Pi) (n = 24), Scutellaria baicalensis Georgi (Huang Qin) (n = 20) and Pinellia ternata (Thunb.) Makino (Ban Xia) (n = 9) (Fig. 2). Table 2 contains the compound names for each connection node code seen in Fig. 3.
Figure 3. Network connections between Ding Chuan Tang compounds and five target proteins.
ESR1, Estrogen receptors alpha; KDR, Kinase insert domain receptor; LTA4H, Leukotriene A4 hydrolase; PDE4D, cAMP-specific 3′,5′-cyclic phosphodiesterase 4D; PPARG, Peroxisome proliferator-activated receptor gamma.
Table 2. 155 Target protein connections identified from Ding Chuan Tang compound molecular docking.
| Ding Chuan Tang herb | Compound code | Herb chemical compound | Target protein | ||||
|---|---|---|---|---|---|---|---|
| ESR1 | KDR | LTA4H | PDE4D | PPARG | |||
|
Ginkgo biloba L. (Bai Guo) |
Bg017 | catechin | √ | √ | √ | ||
| Bg014 | campherol | √ | √ | √ | |||
| Bg040 | β-sitosterol | √ | √ | √ | √ | ||
| Bg039 | stigmasterol | √ | √ | √ | √ | ||
| Bg037 | sciadopitysin | √ | √ | √ | √ | ||
| Bg036 | rutin | √ | √ | ||||
| Bg028 | isorhamnetine | √ | √ | ||||
| Bg001 | (−)-epicatechin | √ | √ | ||||
| Bg027 | hydroginkgolinic acid | √ | |||||
| Bg025 | ginkgolide C | √ | |||||
| Bg023 | ginkgolid B | √ | |||||
| Bg016 | cardanol | √ | |||||
| Bg013 | bilobil | √ | |||||
| Bg009 | anacardic acid B | √ | |||||
| Bg008 | anacardic acid A | √ | |||||
|
Ephedra sinica Stapf (Ma Huang) |
Mh026 | leucodelphinidin | √ | ||||
| Mh019 | dobutamine | √ | √ | ||||
| Mh022 | herbacetin | √ | √ | √ | |||
| Mh024 | kaempferol rhamnoside | √ | √ | √ | √ | ||
| Mh043 | tricin | √ | √ | ||||
| Perilla frutescens (L.) Britton (Su Zi) | Sz020 | rosmarinic acid | √ | √ | √ | ||
| Sz013 | luteolin | √ | √ | √ | |||
| Sz007 | chrysoeriol | √ | √ | √ | |||
| Sz019 | phytosterols | √ | √ | √ | √ | ||
| Sz014 | luteolin-7-O-glucoside | √ | √ | √ | √ | ||
| Sz002 | apigenin-7-O-glucoside | √ | √ | √ | √ | ||
| Sz031 | δ-tocopherol | √ | √ | √ | |||
| Sz030 | γ-tocopherol | √ | √ | √ | |||
| Sz029 | β-tocopherol | √ | √ | √ | |||
| Sz027 | α-tocopherol | √ | √ | ||||
| Sz005 | caffeic acid-3-O-glucoside | √ | |||||
| Sz028 | β-amyrin | √ | √ | ||||
| Sz021 | rosmarinic acid methyl ester | √ | √ | ||||
| Glycyrrhiza uralensis Fisch (Gan Cao) | Gc001 | 11-deoxoglycyrrhetinic | √ | √ | √ | ||
| Gc002 | 18β-glycyrrhetinic acid | √ | √ | √ | |||
| Gc003 | 2,4,4’-trihydroxychalcone | √ | √ | ||||
| Gc004 | 3,3’-dimethylquercetin | √ | √ | √ | |||
| Gc005 | 3’-methoxyglabridin | √ | √ | √ | √ | ||
| Gc006 | 3-hydroxyglabrol | √ | √ | √ | √ | √ | |
| Gc007 | 4′-O-methylglabridin | √ | √ | √ | √ | ||
| Gc010 | 5-O-methyl licoricidin | √ | √ | √ | √ | ||
| Gc011 | 7-O-methylluteone | √ | √ | √ | |||
| Gc013 | apioside | √ | √ | √ | √ | ||
| Gc015 | dehydroglyasperin C | √ | √ | √ | √ | ||
| Gc016 | dehydroglyasperin D | √ | √ | ||||
| Gc018 | formononetin | √ | √ | √ | |||
| Gc019 | gancaonin A | √ | √ | √ | √ | ||
| Gc020 | gancaonin B | √ | √ | √ | √ | ||
| Gc021 | gancaonin C | √ | √ | √ | √ | ||
| Gc022 | gancaonin D | √ | √ | √ | √ | ||
| Gc023 | gancaonin E | √ | √ | √ | √ | ||
| Gc024 | gancaonin F | √ | √ | √ | √ | ||
| Gc025 | gancaonin p-3′-methylether | √ | √ | √ | √ | ||
| Gc026 | glabranin | √ | √ | √ | √ | √ | |
| Gc028 | glabrol | √ | √ | √ | |||
| Gc029 | glisoflavanone | √ | √ | √ | √ | ||
| Gc032 | glycocoumarin | √ | √ | √ | |||
| Gc033 | glycybenzofuran | √ | √ | ||||
| Gc034 | glycyrin | √ | √ | ||||
| Gc035 | glycyrol | √ | √ | √ | √ | ||
| Gc036 | glycyrrhetic acid acetate | √ | √ | ||||
| Gc037 | glycyrrhetol | √ | √ | ||||
| Gc038 | glycyrrhisoflavanone | √ | √ | √ | √ | ||
| Gc040 | glycyrrhiza-flavonol A | √ | √ | √ | √ | ||
| Gc043 | glyuranolide | √ | √ | ||||
| Gc044 | glyzaglabrin | √ | √ | √ | √ | ||
| Gc045 | hemileiocarpin | √ | √ | √ | √ | ||
| Gc046 | hispaglabridin A | √ | √ | √ | √ | ||
| Gc047 | hispaglabridin B | √ | √ | √ | √ | ||
| Gc049 | isoglycyrol | √ | √ | √ | √ | ||
| Gc050 | isolicoflavonol | √ | √ | √ | √ | ||
| Gc051 | isoliquiritin | √ | √ | √ | √ | ||
| Gc053 | isoquercitrin | √ | √ | √ | √ | ||
| Gc054 | isotrifoliol | √ | √ | √ | √ | √ | |
| Gc056 | kanzonol F | √ | √ | √ | √ | ||
| Gc057 | kanzonol h | √ | √ | √ | √ | ||
| Gc058 | licocoumarone | √ | √ | √ | √ | ||
| Gc059 | licochalcone A | √ | √ | ||||
| Gc060 | licocoumarone | √ | √ | √ | √ | ||
| Gc061 | licofuranocoumarin | √ | √ | √ | |||
| Gc062 | licoisoflavone | √ | √ | √ | √ | ||
| Gc064 | licoleafol | √ | √ | √ | √ | ||
| Gc065 | licopyranocoumarin | √ | √ | √ | √ | ||
| Gc077 | licoricidin | √ | √ | √ | √ | ||
| Gc078 | licoricone | √ | |||||
| Gc080 | liquiritin | √ | √ | √ | √ | √ | |
| Gc085 | neoliquiritin | √ | √ | √ | √ | ||
| Gc087 | ononin | √ | √ | √ | √ | ||
| Gc088 | paratocarpin B | √ | √ | √ | √ | ||
| Gc089 | phaseollinisoflavan | √ | √ | √ | √ | ||
| Gc090 | semilicoisoflavone B | √ | √ | √ | √ | ||
| Gc092 | uralene | √ | √ | ||||
| Gc093 | uralenin | √ | √ | √ | √ | ||
| Gc094 | uralenneoside | √ | |||||
| Gc095 | uralenol | √ | √ | √ | √ | ||
| Gc096 | uralenol-3-methylether | √ | √ | √ | √ | ||
|
Tussilago farfara L. (Kuan Dong Hua) |
Kd004 | bauerenol | √ | √ | |||
| Kd005 | faradiol | √ | √ | ||||
| Kd009 | kaempferol-3-O-glucoside | √ | √ | √ | |||
| Kd013 | senecionine | √ | |||||
| Kd015 | tussilagonone | √ | |||||
| Prunus armeniaca L. (Xing Ren) | Xr016 | prunasine | √ | √ | |||
|
Morus alba L. (Sang Bai Pi) |
Sb003 | α-acetyl-amyrin | √ | √ | |||
| Sb004 | cyclomorusin | √ | √ | √ | √ | ||
| Sb005 | cyclomulberrin | √ | √ | √ | √ | √ | |
| Sb006 | ecdysterone | √ | √ | ||||
| Sb007 | eudraflavone β hydroperoxide | √ | √ | √ | √ | ||
| Sb008 | kuwanon C | √ | √ | √ | √ | ||
| Sb010 | kuwanon L | √ | √ | √ | √ | ||
| Sb011 | kuwanon S | √ | √ | √ | √ | ||
| Sb012 | kuwanon T | √ | √ | √ | √ | ||
| Sb013 | leachianone G | √ | √ | √ | √ | √ | |
| Sb016 | moracin E | √ | √ | √ | √ | ||
| Sb019 | morocin P | √ | √ | √ | √ | ||
| Sb020 | morusin | √ | √ | √ | √ | ||
| Sb021 | mulberrofuran B | √ | √ | √ | √ | ||
| Sb026 | mulberrofuran N | √ | |||||
| Sb029 | mulberrofuran Q | √ | √ | √ | |||
| Sb031 | mulberroside C | √ | √ | √ | √ | ||
| Sb032 | oxydihydromorusin | √ | √ | √ | √ | ||
| Sb033 | oxyresveratrol | √ | |||||
| Sb035 | sanggenol A | √ | √ | √ | √ | ||
| Sb036 | sanggenol L | √ | √ | √ | √ | ||
| Sb037 | sanggenol O | √ | √ | √ | √ | ||
| Sb039 | sanggenon F | √ | √ | √ | √ | √ | |
| Sb041 | sanggenon N | √ | √ | √ | √ | √ | |
| Scutellaria baicalensis Georgi (Huang Qin) | Hq003 | 2′,3′,5,7-tetrahydroxyflavone | √ | √ | √ | ||
| Hq005 | 3,5,7,2′,6′-pentahydroxy flavanon | √ | |||||
| Hq008 | 5,2′-dihydroxy-6,7,8-trimethoxyflav | √ | |||||
| Hq009 | 5,6-dihydroxy-7-O-glucoside-flavone | √ | √ | √ | √ | ||
| Hq010 | 5,7,2′,3′-tetrahydroxyflavone | √ | √ | √ | |||
| Hq011 | 5,7,2′,6′-tetrahydroxyflavone | √ | √ | ||||
| Hq012 | 5,7,2′-trihydroxyflavon | √ | √ | ||||
| Hq014 | 5,8,2′-trihydroxy-7- methoxyflavone | √ | |||||
| Hq015 | 5,8-dihydroxy-6,7-dimethoxyflavone | √ | √ | √ | |||
| Hq016 | 5-hydroxy-7,8-dimethoxyflavone | √ | |||||
| Hq020 | chrysin-7-glucuronide | √ | √ | √ | √ | ||
| Hq021 | dihydrobaicalin | √ | √ | √ | √ | ||
| Hq022 | dihydrooroxylin A | √ | √ | ||||
| Hq023 | eriodictyol | √ | √ | √ | |||
| Hq027 | oroxylin A | √ | √ | ||||
| Hq029 | panicolin | √ | |||||
| Hq030 | scutellarin | √ | √ | √ | √ | ||
| Hq031 | scutevulin | √ | √ | ||||
| Hq033 | wogonin | √ | √ | ||||
| Hq034 | wogonoside | √ | √ | ||||
|
Pinellia
ternata (Thunb.) Makino (Ban Xia) |
Bx030 | baicalin | √ | √ | √ | √ | |
| Bx033 | bis-(4-hydroxybenzyl) ether | √ | |||||
| Bx034 | bisabolene | √ | |||||
| Bx040 | cycloartenol | √ | √ | √ | |||
| Bx042 | daucosterol | √ | √ | √ | √ | ||
| Bx058 | methyl phenanthrene | √ | √ | ||||
| Bx060 | octadecane | √ | |||||
| Bx065 | sachaliside 1 | √ | √ | ||||
| Bx066 | shogaol | √ | |||||
Note:
Bg, Bai Guo; Bx, Ban Xia; ESR1, Estrogen receptors alpha; Gc, Gan Cao; Hq, Huang Qin; KDR, Kinase insert domain receptor; Kd, Kuan Dong Hua; LTA4H, Leukotriene A4 hydrolase; Mh, Ma Huang; PDE4D, cAMP-specific 3’,5’-cyclic phosphodiesterase 4D; PPARG, Peroxisome proliferator-activated receptor gamma; Sb, Sang Bai Pi; Sz, Su Zi; Xr, Xing Ren.
Discussion
Asthma is a complex disease, with multi-cellular interactions, networks and complexes associated and recognized as the foundation of the pathology (KEGG). The five identified target proteins, ESR1, KDR, LTA4H, PDE4D and PPARG, show evidence of involvement in the pathological expression of asthma, with three classified as receptors (ESR1, KDR, PPARG), and two categorized as enzymes (LTA4H, PDE4D) (KEGG, UniProt). These results are significant for the investigation of DCT for asthma, as 155 DCT-compounds resulted in high affinity bonding with these five proteins. Inferences from the molecular docking of DCT-compounds to the target proteins based on the KEGG asthma pathway (Fig. 4) indicate the potential of DCT-compounds to function as agonists (promoting) or antagonists (inhibiting); thus affecting the signaling pathways involved in asthma pathology.
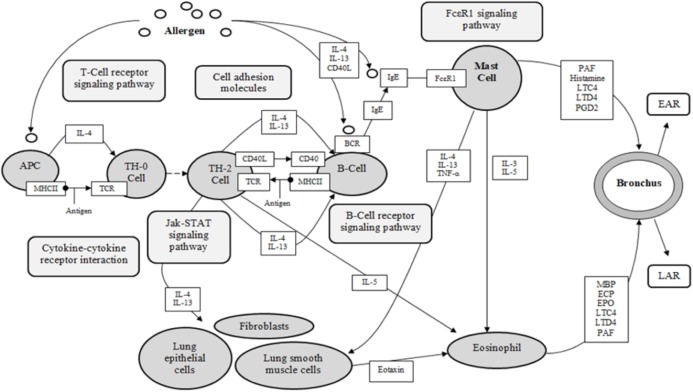
Figure 4. KEGG asthma pathway (adapted from “Asthma pathway- Homo sapiens” on KEGG database (Kanehisa et al., 2017)).
APC, antigen presenting cell; BCR, B-cell receptor; CD40, cluster of differentiation 40; CD40L, cluster of differentiation 40 L receptor; EAR, early asthmatic response; ECP, eosinophil cationic protein; EPO, eosinophil peroxidase; FcεR1, high affinity immunoglobulin E receptor; IgE, immunoglobulin E; IL-#, Interleukin-#; LAR, late asthmatic response; LTC4, leukotriene C4; LTD4, leukotriene D4; MBP, major basic protein; MHCII, major histocompatibility complexes II; PAF, platelet activating factor; PGD2, Prostaglandin D2; TCR, T-Cell surface receptor; TH-2, T-helper 2 cells; TH-0, naïve T-cells; TNF-α, tumor necrosis factor-α.
Determining target protein network connections of all identified DCT-compounds, as opposed to only highlighting the top binding compounds, is important for herbal medicine research, as DCT is prescribed as a multi-herbal formula. Cytoscape 3.6.0 includes applications to associate network connections between the 155 DCT-compounds and target proteins to biological pathways involved in functions such as biological signaling and metabolism through the KEGG database (Shannon et al., 2003). Identifying the biological KEGG pathways targeted by each target protein, as seen in Fig. 5, is the initial step towards understanding the biological activities of the DCT-compounds and how these relate to the disease pathways of asthma. Although some disease pathways are not usually directly associated with asthma, these results suggest there are possible future novel therapeutic approaches for the treatment of asthma to be investigated, as the disease pathways identified all involve the inflammatory process which is the primary pathogenesis of asthma (KEGG; Global Initiative for Asthma, 2018).
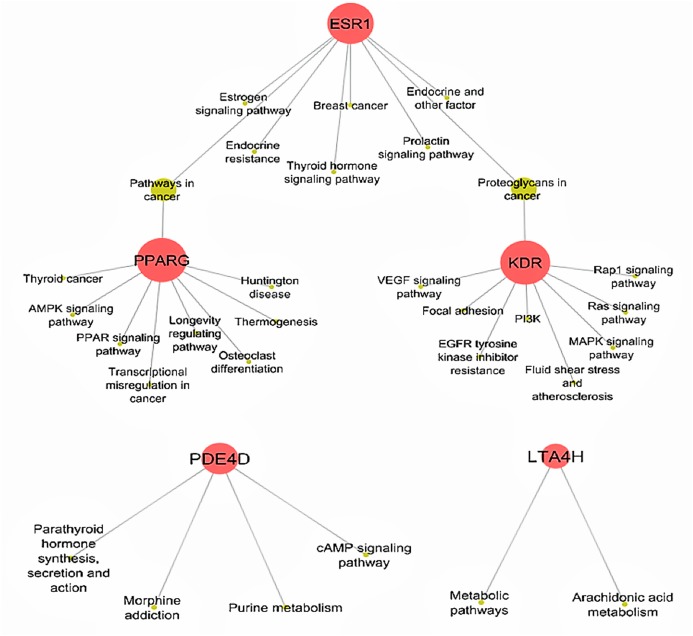
Figure 5. Biological KEGG pathways involved in ESR1, KDR, LTA4H, PDE4D and PPARG.
ESR1 (Estrogen receptors alpha) targets eight pathways and shares the Proteoglycans in cancer pathway with KDR (kinase insert domain receptor) and pathways in cancer with PPARG (peroxisome proliferator-activated receptor gamma). All Ding Chuan Tang compounds that target these three proteins potentially increase the synergistic therapeutic outcomes specifically to those pathways. AMPK, 5′ AMP-activated protein kinase; cAMP, cyclic-3′,5′-adenosine monophosphate; EGFR, epidermal growth factor receptor; LTA4H, leukotriene A4 hydrolase; MAPK, mitogen-activated protein kinase; PDE4D, cAMP-specific 3′,5′-cyclic phosphodiesterase 4D; PI3K, phosphatidylinositol 3″-kinase; PPAR, peroxisome proliferator-activated receptor; Rap1, Ras-related protein 1; VEGF, vascular endothelial growth factor.
Interpreting network connections
The KEGG database resource is based on current experimental knowledge for “understanding high-level functions and utilities of the biological system” (Kanehisa et al., 2017). The KEGG asthma pathway (Fig. 4) displays the known molecular/cellular interactions and signaling pathways involved in chronic inflammation leading to asthma symptoms. As seen in Fig. 4, there are four signaling pathways involved in the KEGG asthma pathways: (1) T-Cell receptor signaling pathway; (2) B-Cell receptor signaling pathway; (3) the JAK/STAT signaling pathway and (4) the FcεRI signaling pathway. Within this discussion, these pathways will be referred to as the primary pathways. Following on, the pathways further involved within the four primary signaling pathways are referred to as the secondary pathways.
Each of the five identified target proteins, ESR1, KDR, LTA4H, PDE4D and PPARG, have established potential to play significant roles involving both the primary and secondary asthma pathways through many signaling cascade pathways.
ESR1
Nuclear hormone receptor, ESR1, in respect to the pathology of asthma, is involved in cell proliferation in tissues within the respiratory system, increasing and maintaining the number of cells in the target tissues through an increase in cell division and altering the process of cell death (UniProt). ESR1 participates in two primary KEGG asthma pathways: the B-Cell signaling pathway and the JAK-STAT signaling pathway, through the Prolactin signaling pathway (KEGG). ESR1 acts downstream of both the B-Cell signaling pathway and the JAK-STAT signaling pathway and modulates the immune response through regulation of macrophages, T and B lymphocytes and fibroblasts (KEGG). The Prolactin signaling pathway, targeted by ESR1, activates pathways involved in the inflammatory response including the KEGG secondary MAPK pathway and PI3K pathway and is also involved in the secondary KEGG asthma NF-κB signaling pathway; with ESR1 again acting downstream (KEGG). A protein–protein interaction with ESR1 and NF-κB inhibits NF-κB signaling, resulting in a decrease in the inflammatory response through a reduction of pro-inflammatory cytokines and a dampening of inflammatory gene expression in smooth muscle cells (Kovats, 2015). ESR1 also acts downstream within the Thyroid hormone signaling pathway, involved in the secondary Calcium signaling pathways and the MAPK signaling pathway through moderation of gene transcription and cell proliferation (KEGG). ESR1 participates and acts upstream in the Estrogen signaling pathway involved in the secondary KEGG asthma PI3K-Akt signaling pathway, Calcium signaling pathway and MAPK signaling pathway; targeting immune genes, regulating cell cycles and cell adhesion molecules (KEGG). As ESR1 acts upstream of the Estrogen signaling pathway, there is potential that targeting this gene may inhibit or reduce the effects of those three secondary asthma pathways.
Docking results for ESR1 included two crystal complexes classified as antagonists of ESR1 (2fja and 3erd) and one agonist of ESR1 (2yja) (Table 1). Cyclomulberrin is the single compound binding with multiple ESR1 crystals, suggesting both agonist and antagonist functions. The selective estrogen receptor modulator drug tamoxifen is a known ESR1 agonist and antagonist of ESR1, supporting the findings of cyclomulberrin (Gallo & Kaufman, 1997). The research community recognizes the disparity of inflammatory and anti-inflammatory effects of ESR1, hypothesizing the effects are dependent on the available levels of circulating estrogens or the tissue the ESR1 is present (Yung, Fuseini & Newcomb, 2018; Keselman & Heller, 2015; Dijkstra et al., 2006). Inhibiting ESR1 has been found to (1) reduce airway hyperresponsiveness and thymic development, (2) regulate and decrease pro-inflammatory mediators and (3) hinder the development and advancement of airway changes and remodeling, while promoting ESR1 has been found to impede pro-inflammatory responses in both human and animal studies (Yung, Fuseini & Newcomb, 2018; Keselman & Heller, 2015; Dijkstra et al., 2006).
KDR
KDR is classified as a type III receptor tyrosine kinase, with alternative names including vascular endothelial growth factor receptor 2 and Fetal Liver Kinase 1. KDR acts as a cell surface receptor for vascular endothelial growth factors A, C and D, all involved in the migration, growth and survival of endothelial cells (Duffy, Bouchier-Hayes & Harmey, 2013; UniProt). KDR is expressed throughout human tissues, acting upstream to the primary KEGG asthma pathway T-cell receptor signaling pathway through the Ras signaling pathway and effects cell growth, survival, cell cycle progression and gene expression (Pate et al. 2010; KEGG). KDR is involved in three of the secondary KEGG asthma pathways including PI3K-Akt signaling pathway, Calcium signaling pathway and MAPK signaling pathway; with the Ras signaling pathway additionally involved in these (KEGG). Two cascade pathways include KDR acting upstream of these three secondary asthma pathways potentially inhibiting the inflammatory outcomes; the VEGF signaling pathway- resulting in cell survival, migration and proliferation, and the RAP1 signaling pathway- involving cell adhesion, migration, proliferation, survival and gene activation (KEGG). KDR acts upstream from MAPK pathway in an additional pathway associated with focal adhesion, with effects including inflammatory cell proliferation and survival (KEGG).
All four KDR crystal complexes are classified as antagonists of KDR functions (KEGG); therefore, the DCT-compounds that docked with all four crystals provide strong evidence as KDR antagonists (Table 1). Evidence supporting the inhibition of KDR as beneficial for asthma involves a variety of pathological pathways including a dampening of the inflammatory asthma response through a reduction in recruitment of pro-inflammatory cells, a decrease circulating cytokines and cell adhesion molecules and the reduction of airway remodeling through smooth muscle hyperplasia (PDB; Uniprot).
LTA4H
The protein that the human gene LTA4H encodes is an enzyme containing both: (a) hydrolase, with the function of converting leukotriene A4 (LTA4) into leukotriene B4 (LTB4), a proinflammatory mediator and; (b) aminopeptidase, recognized as a biological characteristic of chronic obstructive pulmonary disease (Uniprot; PubMed_Gene). LTB4 promotes the inflammatory response as a mediator of leukocytes including monocytes, eosinophils, macrophages and dendritic cells, along with specific chemotactic function for neutrophils and regulating differentiation of T-cells (Di Gennaro & Haeggstrom, 2014; Liu & Yokomizo, 2015). The primary KEGG asthma FcεRI signaling pathway includes LTA4H through its function in AA metabolism (KEGG). The Eicosanoids pathway instigates both the immune and the inflammatory response through utilizing LTA4H downstream for both the FcεRI signaling pathway and the secondary AA metabolism pathway (KEGG). Pro-inflammatory cells including leukocytes, platelets and fibroblasts, in addition to target tissue of the respiratory system, widely express LTA4H (Uniprot).
Both LTA4H crystal complexes are classified as antagonists of LTA4H, therefore the 38 DCT-compounds that docked with both crystals of LTA4H (Table 1) potentially reduce asthma symptoms such as increased mucous and persistent airway inflammation, through blocking the conversion of LTA4 to LTB4 (PDB; Uniprot).
PDE4D
Nine isoforms have been identified for the PDE4D gene, each encoding hydrolyzing proteins that break down the second messenger Cyclic adenosine monophosphate (cAMP) (UniProt). The immune effects of cAMP regulate both the innate and adaptive immune responses, with many cells within the respiratory system including leukocytes, macrophages, B-cells and T-cells affected and modulated (Billington et al., 2013; Raker, Becker & Steinbrink, 2016). Increased levels of intracellular cAMP inhibit asthma pathways leading to broncho-constriction through the contraction of respiratory smooth muscle, while as a drug target, it amplifies the anti-inflammatory response and simultaneously inhibits pro-inflammatory immune and cellular factors (Billington et al., 2013; Raker, Becker & Steinbrink, 2016). The secondary KEGG asthma disease pathway PI3K-Akt signaling pathway, involves PDE4D through the cAMP signaling pathway, leading to cell growth, survival and proliferation (KEGG). PDE4D acts upstream of the PI3K-Akt signaling pathway; effectively inhibiting the outcomes and thus reducing inflammatory mediated symptoms (KEGG; Billington et al., 2013).
The four investigated PDE4D crystal complexes have all been shown to be antagonists of PDE4D functions, thus the DCT-compounds docked with them (Table 1) present the potential to reduce bronchoconstriction and reduce the asthma inflammatory cellular mediated symptoms (Billington et al., 2013; Raker, Becker & Steinbrink, 2016; UniProt). PDE4D inhibitors have been used successfully as drug targets for asthma, such as theophylline since the 1930s (Spina, 2008) and Ibudilast, an oral asthma drug approved in Asia (Rolan et al., 2008). These anti-asthma drugs present strong evidence for DCT-compounds as potential future drug candidates through computational demonstration to target PDE4D.
PPARG
The type II nuclear receptor PPARG is known primarily for the function of regulating the fatty acid pathways and glucose metabolism (UniProt). Asthma development implicates PPARG in regard to the reduction of respiratory inflammation and the permanent remodeling that occurs due to the chronic asthma response (Oh et al., 2009). The activation of PPARG moderates the initiation of pro-inflammatory Th2 cytokines, eosinophils and macrophages, while allowing the expression of anti-inflammatory cellular factors within the lung (Huang & Glass, 2010; Oh et al., 2009). PPARG is involved and acts downstream from the secondary KEGG asthma disease pathway PI3K-Akt signaling pathway through the AMP-activated protein kinase (AMPK) signaling pathway, to inhibit cell growth, cell survival and protein synthesis (KEGG).
All five PPARG crystal complexes are classified as PPARG agonists, suggesting all 108 docked DCT-compounds (Table 1) promote the reduction of pro-inflammatory Th2 cytokines, eosinophils and macrophages, along with lessening asthma chronicity (PDB; Uniprot). Rosiglitazone, a strong PPARG agonist, has shown to significantly improve lung function and decrease IL-5 and IgE in a murine model (El-Naa et al., 2015). Furthermore, a current review of rosiglitazone defines the PPARG agonist as a key regulator of airway inflammation, with strong evidence as an asthma therapeutic target (Banno et al., 2018).
ADORA2A and MIF
Although these two target proteins were excluded, they are still worth discussion due to virtual screening results. The excluded target protein ADORA2A is a receptor for adenosine, a signaling transductor and second messenger (Uniprot). Adenosine mediates G proteins and in turn activates adenylyl cyclase (an enzyme that synthesizes cAMP), associated with immune function and inflammatory pathways (Uniprot). Adenosine has long been associated with the inhibition of the inflammatory response through the effects on neutrophil and macrophage regulation (Hasko & Cronstein, 2013). The biological processes associated with ADORA2A demonstrate involvement in cell–cell signaling, the cellular defense response and the inflammatory response (Uniprot). Multiple experimental studies have shown agonist ligands of ADORA2A reduce respiratory inflammation (Van Waarde et al., 2018). This research suggests the prospect of the DCT-compounds successfully docked with ADORA2A to display similar agonistic biological effects for asthma.
MIF is a pro-inflammatory cytokine with a regulatory function of the innate immune response (Uniprot). The identified molecular functions of MIF identified and associated to asthma are (1) macrophage mediator and regulator, (2) chemokine attraction and (3) signaling receptor binding (Uniprot). The biological processes of MIF include (1) the inflammatory response, (2) cell proliferation, (3) cell surface receptor signaling pathways, (4) macrophage regulation, (5) leukocyte regulation and (6) B-cell maturation regulation and (7) the innate immune response (Uniprot). Through the direct activation of immune cells and the contribution to airway remodeling in asthma experimental studies (Lan et al., 2018; Hoi, Iskander & Morand, 2007), inhibitors of MIF prove to be a strategic therapeutic target for asthma; thus indicating the potential favorable benefits of the docked DCT-compounds.
Pharmacological contributions to asthma management
Single pharmaceutical compounds, such as the majority of asthma medications, fundamentally target single molecular mechanisms and as a result may be less efficient at treating complex diseases that have multi-cellular/tissue pathology (Kumar & Zhang, 2018; Aggarwal et al., 2007). It has been proposed that pharmaceuticals targeting molecular signaling pathways are the future for drug discovery as they target multiple protein targets and pathways (Kumar & Zhang, 2018; Aggarwal et al., 2007). The network connection results of this investigation are in line with this theory, as the pharmacological and mechanistic predictions provide evidence, albeit computational, of 155 strong affinity binding connections (≥−9.0 kcal/mol) between DCT-compounds and five proteins associated with multiple asthma KEGG disease pathways (Fig. 3).
Specific DCT-compound insights include the compounds that targeted all crystal complexes of KDR, LTA4H, PDE4D or PPARG (Table 3), as each of the crystal complexes for these proteins are in the same classification; agonist or antagonist. These results suggest that these compounds may promote or inhibit the biological actions associated with each protein through accumulation and synergism. Furthermore, the identified DCT-compounds that bound with all investigated crystal complexes of PDE4D and PPARG are significant justifications for the historical clinical evidence of DCT reducing the symptoms of bronchoconstriction and airway inflammation, as we know these are the outcomes of registered asthma pharmaceutical compounds theophylline (PDE4D antagonist), ibudilast (PDE4D antagonist) and rosiglitazone (PPARG agonist). Within these DCT-compounds there are 12 that bound to all PDE4D crystal complexes and all PPARG crystal complexes and are especially highlighted for further investigation as PDE4D antagonists or PPARG agonists. These included four from G. uralensis Fisch (Gan Cao) (apioside-Gc013, gancaonin F-Gc024, paratocarpin B-Gc088 and semilicoisoflavone B-Gc090), five from M. alba L. (Sang Bai Pi) (cyclomulberrin-Sb005, kuwanon S-Sb011, moracin E-Sb016, morusin-Sb020 and sanggenon N-Sb041) and three from S. baicalensis Georgi (Huang Qin) (chrysin-7-glucuronide-Hq020, dihydrobaicalin-Hq021 and scutellarin-Hq030).
Table 3. Ding Chuan Tang-compounds that docked (≥−9.0 kcal/mol) with all investigated crystal complexes.
| Target proteins (investigated crystal complexes) | Classification of investigated crystal complexes | DCT-compounds that docked (≥−9.0 kcal/mol) with all investigated crystal complexes |
|---|---|---|
| KDR (3vo3; 3vhe; 3cjg; 3cjf) |
KDR antagonists | Sciadopitysin-Bg037; gancaonin F-Gc024; hemileiocarpin-Gc045; hispaglabridin B-Gc04; isoglycyrol- Gc049; semilicoisoflavone B-Gc090; uralenin-Gc093; bauerenol- Kd004; cyclomorusin-Sb004; sanggenon N-Sb041; sanggenol O-Sb037 |
| LTA4H (4dpr; 3fts) |
LTA4H antagonists | Catechin-Bg017; rutin-Bg036; apigenin-7-O-glucoside-Sz002; rosmarinic acid-Sz020; rosmarinic acid methyl ester-Sz021; dehydroglyasperin C-Gc015; gancaonin B-Gc020; gancaonin C-Gc021; gancaonin D-Gc022; gancaonin F-Gc024; glabranin-Gc026; glisoflavanone-Gc029; glycyrol-Gc035; glycyrrhisoflavanone-Gc038; glycyrrhiza-flavonol A-Gc040; glyzaglabrin-Gc044; hemileiocarpin-Gc04; hispaglabridin A-Gc046; isoglycyrol-Gc049; isoliquiritin-Gc051; isotrifoliol-Gc054; licoleafol-Gc064; ononin-Gc087; paratocarpin B-Gc088; phaseollinisoflavan-Gc089; semilicoisoflavone B-Gc090; kuwanon S-Sb011; moracin E-Sb016; morusin-Sb020; mulberrofuran B-Sb021; mulberroside C-Sb031; sanggenol L-Sb036; sanggenol O-Sb037; sanggenon F-Sb039; sanggenon N-Sb041; scutellarin-Hq030; 5,6-Dihydroxy-7-O-glucoside-flavone-Hq009; baicalin-Bx030 |
| PDE4D (1xom; 1tbb; 1tb7; 1zkn) |
PDE4D antagonists | Sciadopitysin-Bg037; stigmasterol-Bg039; luteolin-7-O-glucoside-Sz014; apioside-Gc013; gancaonin E-Gc023; gancaonin F-Gc024; glycyrol-Gc035; glycyrrhisoflavanone-Gc038; glycyrrhiza-flavonol A-Gc040; hemileiocarpin-Gc045; hispaglabridin B-Gc047; hydroxyglabrol-Gc006; isolicoflavonol-Gc050; licocoumarone-Gc058; licoleafol-Gc064; liquiritin-Gc080; paratocarpin B-Gc088; semilicoisoflavone B-Gc090; uralenin-Gc093; 3’-Methoxyglabridin-Gc005; 3-uralene-Gc092; eudraflavone β hydroperoxide-Sb007; cyclomorusin-Sb004; cyclomulberrin-Sb005; kuwanon L-Sb010; kuwanon S-Sb011; moracin E-Sb016; morusin-Sb020; mulberrofuran Q-Sb029; mulberroside C-Sb031; sanggenol L-Sb036; sanggenol O-Sb037; sanggenon N-Sb041; prunasine-Xr016; chrysin-7-glucuronide-Hq020; dihydrobaicalin-Hq021; scutellarin-Hq030 |
| PPARG (5slg(1); 4jaz; 3sz1_MYR; 3sz1_LU; 3adx) |
PPARG agonists | Apigenin-7-O-glucoside-Sz002; apioside-Gc013; gancaonin F-Gc024; neoliquiritin-Gc085; paratocarpin B-Gc088,semilicoisoflavone B-Gc090; cyclomulberrin-Sb005; kuwanon S-Sb011; kuwanon T-Sb012; moracin E-Sb016; morusin-Sb020; oxydihydromorusin-Sb032; sanggenol A-Sb035; sanggenon N-Sb041; chrysin-7-glucuronide-Hq020; dihydrobaicalin-Hq021; scutellarin-Hq030; 5,6-Dihydroxy-7-O-glucoside-flavone-Hq009; baicalin-Bx030 |
Note:
Bg, Bai Guo; Bx, Ban Xia; ESR1, Estrogen receptors alpha; Gc, Gan Cao; Hq, Huang Qin; KDR, Kinase insert domain receptor; LTA4H, Leukotriene A4 hydrolase; PDE4D, cAMP-specific 3′,5′-cyclic phosphodiesterase 4D; PPARG, Peroxisome proliferator-activated receptor gamma; Sb, Sang Bai Pi; Sz, Su Zi; Xr, Xing Ren.
Conclusions
Network pharmacology has facilitated a fundamental, evidence-based platform to determine the biological mechanisms of action for DCT, demonstrated through the multi-asthma protein target capabilities of DCT-compounds. The 155 network docking connections that DCT-compounds formed with proteins also targeted by asthma genes suggest the potential for DCT-compounds to implement therapeutic effects, through inhibition or promotion, to proteins associated with asthma disease pathways and biological networks.
Supplemental Information
Funding Statement
Allison Clyne was supported by an Australian Commonwealth Government Research Training Program Scholarship for her PhD study. The authors received no other funding for this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Allison Clyne performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Liping Yang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Ming Yang performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Brian May analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Angela Wei Hong Yang conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.
References
- Aggarwal et al. (2007).Aggarwal BB, Sethi G, Baladandayuthapani V, Krishnan S, Shishodia S. Targeting cell signaling pathways for drug discovery: an old lock needs a new key. Journal of Cellular Biochemistry. 2007;102(3):580–592. doi: 10.1002/jcb.21500. [DOI] [PubMed] [Google Scholar]
- Banno et al. (2018).Banno A, Reddy AT, Lakshmi SP, Reddy RC. PPARs: key regulators of airway inflammation and potential therapeutic targets in asthma. Nuclear Receptor Research. 2018;5(3):622. doi: 10.11131/2018/101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker et al. (2004).Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nature Genetics. 2004;36(5):431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- Bensky & Barolet (2009).Bensky D, Barolet R. Chinese herbal medicine formulas and strategies. Seattle: Eastland Press; 2009. [Google Scholar]
- Bensky, Clavey & Stoger (2004).Bensky D, Clavey S, Stoger E. Chinese herbal medicine: materia medica. Seattle: Eastland Press; 2004. [Google Scholar]
- Benson et al. (2013).Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Research. 2013;41(D1):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington et al. (2013).Billington CK, Ojo OO, Penn RB, Ito S. cAMP regulation of airway smooth muscle function. Pulmonary Pharmacology & Therapeutics. 2013;26(1):112–120. doi: 10.1016/j.pupt.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIOVIA (2015).BIOVIA . Discovery studio. San Diego: Dassault Systèmes; 2015. [Google Scholar]
- Cell Signaling Technology (2018).Cell Signaling Technology . CST pathways. Arundel: Genesearch PTY. Ltd; 2018. [Google Scholar]
- Chan et al. (2006).Chan C-K, Kuo M-L, Shen J-J, See L-C, Chang H-H, Huang J-L. Ding Chuan Tang, a Chinese herb decoction, could improve airway hyper-responsiveness in stabilized asthmatic children: a randomized, double-blind clinical trial. Pediatric Allergy and Immunology. 2006;17(5):316–322. doi: 10.1111/j.1399-3038.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- Chillistone & Hardman (2014).Chillistone S, Hardman JG. Factors affecting drug absorption and distribution. Anaesthesia & Intensive Care Medicine. 2014;15(7):309–313. doi: 10.1016/j.mpaic.2014.04.004. [DOI] [Google Scholar]
- Di Gennaro & Haeggstrom (2014).Di Gennaro A, Haeggstrom JZ. Targeting leukotriene B4 in inflammation. Expert Opinion on Therapeutic Targets. 2014;18(1):79–93. doi: 10.1517/14728222.2013.843671. [DOI] [PubMed] [Google Scholar]
- Dijkstra et al. (2006).Dijkstra A, Howard TD, Vonk JM, Ampleford EJ, Lange LA, Bleecker ER, Meyers DA, Postma DS. Estrogen receptor 1 polymorphisms are associated with airway hyperresponsiveness and lung function decline, particularly in female subjects with asthma. Journal of Allergy and Clinical Immunology. 2006;117(3):604–611. doi: 10.1016/j.jaci.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Duffy, Bouchier-Hayes & Harmey (2013).Duffy A, Bouchier-Hayes D, Harmey J. Vascular endothelial growth factor (VEGF) and its role in non-endothelial cells: autocrine signalling by VEGF. Austin: Landes Bioscience; 2013. [Google Scholar]
- El-Naa et al. (2015).El-Naa MM, El-Refaei MF, Nasif WA, Abduljawad SH, El-Brairy AI, El-Readi MZ. In-vivo antioxidant and anti-inflammatory activity of rosiglitazone, a peroxisome proliferator-activated receptor-gamma (PPAR-gamma) agonists in animal model of bronchial asthma. Journal of Pharmacy and Pharmacology. 2015;67(10):1421–1430. doi: 10.1111/jphp.12445. [DOI] [PubMed] [Google Scholar]
- Gallo & Kaufman (1997).Gallo MA, Kaufman D. Antagonistic and agonistic effects of tamoxifen: significance in human cancer. Seminars in Oncology. 1997;24(Suppl. 1):S1-71–S1-80. [PubMed] [Google Scholar]
- Global Initiative for Asthma (2018).Global Initiative for Asthma Global strategy for asthma management and prevention. 2018. www.ginasthma.org. [20 June 2019]. www.ginasthma.org
- Hasko & Cronstein (2013).Hasko G, Cronstein B. Regulation of inflammation by adenosine. Frontiers in Immunology. 2013;4:85. doi: 10.3389/fimmu.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi, Iskander & Morand (2007).Hoi AY, Iskander MN, Morand EF. Macrophage migration inhibitory factor: a therapeutic target across inflammatory diseases. Inflammation & Allergy Drug Targets. 2007;6(3):183–190. doi: 10.2174/187152807781696455. [DOI] [PubMed] [Google Scholar]
- Hoizey & Hoizey (1993).Hoizey D, Hoizey MJ. A history of Chinese medicine. Edinburgh: Edinburgh University Press; 1993. [Google Scholar]
- Hopkins & Paolini (2007).Hopkins AL, Paolini GV. Chemogenomics in drug discovery—the druggable genome and target class properties. In: Taylor JB, Triggle DJ, editors. Comprehensive Medicinal Chemistry II. Oxford: Elsevier; 2007. pp. 421–433. [Google Scholar]
- Huang & Glass (2010).Huang W, Glass CK. Nuclear receptors and inflammation control: molecular mechanisms and pathophysiological relevance. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010;30(8):1542–1549. doi: 10.1161/ATVBAHA.109.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa et al. (2017).Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao et al. (2004).Kao S-T, Chang C-H, Chen Y-S, Chiang S-Y, Lin J-G. Effects of ding-chuan-tang on bronchoconstriction and airway leucocyte infiltration in sensitized guinea pigs. Immunopharmacology and Immunotoxicology. 2004;26(1):113–124. doi: 10.1081/IPH-120029949. [DOI] [PubMed] [Google Scholar]
- Keselman & Heller (2015).Keselman A, Heller N. Estrogen signaling modulates allergic inflammation and contributes to sex differences in asthma. Frontiers in Immunology. 2015;6(813–816):568. doi: 10.3389/fimmu.2015.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim et al. (2013).Kim J, So S, Lee H-J, Park JC, Kim J-J, Lee H. DigSee: disease gene search engine with evidence sentences (version cancer) Nucleic Acids Research. 2013;41(W1):W510–W517. doi: 10.1093/nar/gkt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovats (2015).Kovats S. Estrogen receptors regulate innate immune cells and signaling pathways. Celullular Immunology. 2015;294(2):63–69. doi: 10.1016/j.cellimm.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar & Zhang (2018).Kumar A, Zhang KYJ. Advances in the development of shape similarity methods and their application in drug discovery. Frontiers in Chemistry. 2018;6:315. doi: 10.3389/fchem.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan et al. (2018).Lan H, Wang N, Chen Y, Wang X, Gong Y, Qi X, Luo Y, Yao F. Macrophage migration inhibitory factor (MIF) promotes rat airway muscle cell proliferation and migration mediated by ERK1/2 and FAK signaling. Cell Biology International. 2018;42(1):75–83. doi: 10.1002/cbin.10863. [DOI] [PubMed] [Google Scholar]
- Li & Zhang (2013).Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese Journal of Natural Medicines. 2013;11(2):110–120. doi: 10.1016/S1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- Lipinski et al. (2001).Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 2001;46(1–3):3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Liu & Yokomizo (2015).Liu M, Yokomizo T. The role of leukotrienes in allergic diseases. Allergology International. 2015;64(1):17–26. doi: 10.1016/j.alit.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Ming, Yi & Yang (2018).Ming Y, Yi J, Yang L-P. Screening for cyclooxygenase 2 inhibitors from natural compounds of Radix Glycyrrhizae using computer simulation. Traditional Medicine Research. 2018;3:115–130. doi: 10.12032/tmr201811070. [DOI] [Google Scholar]
- Oh et al. (2009).Oh S-H, Park S-M, Lee YH, Cha JY, Lee J-Y, Shin EK, Park J-S, Park BL, Shin HD, Park CS. Association of peroxisome proliferator-activated receptor-gamma gene polymorphisms with the development of asthma. Respiratory Medicine. 2009;103(7):1020–1024. doi: 10.1016/j.rmed.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Pate et al. (2010).Pate M, Damarla V, Chi DS, Negi S, Krishnaswamy G. Endothelial cell biology: role in the inflammatory response. Advandces in Clinical Chemistry. 2010;52:109–130. doi: 10.1016/S0065-2423(10)52004-3. [DOI] [PubMed] [Google Scholar]
- Raker, Becker & Steinbrink (2016).Raker VK, Becker C, Steinbrink K. The cAMP pathway as therapeutic target in autoimmune and inflammatory diseases. Frontiers in Immunology. 2016;7(Pt. 1):123. doi: 10.3389/fimmu.2016.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolan et al. (2008).Rolan P, Gibbons JA, He L, Chang E, Jones D, Gross MI, Davidson JB, Sanftner LM, Johnson KW. Ibudilast in healthy volunteers: safety, tolerability and pharmacokinetics with single and multiple doses. British Journal of Clinical Pharmacology. 2008;66(6):792–801. doi: 10.1111/j.1365-2125.2008.03270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon et al. (2003).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2014).Shi S-H, Cai Y-P, Cai X-J, Zheng X-Y, Cao D-S, Ye F-Q, Xiang Z. A network pharmacology approach to understanding the mechanisms of action of traditional medicine: bushenhuoxue formula for treatment of chronic kidney disease. PLOS ONE. 2014;9(3):e89123. doi: 10.1371/journal.pone.0089123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slenter et al. (2018).Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, Melius J, Cirillo E, Coort SL, Digles D, Ehrhart F, Giesbertz P, Kalafati M, Martens M, Miller R, Nishida K, Rieswijk L, Waagmeester A, Eijssen LMT, Evelo CT, Pico AR, Willighagen EL. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Research. 2018;46(D1):D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina (2008).Spina D. PDE4 inhibitors: current status. British Journal of Pharmacology. 2008;155(3):308–315. doi: 10.1038/bjp.2008.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer et al. (2016).Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The genecards suite: from gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics. 2016;54(1):1 30 31–31 30 33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- Thangapandian et al. (2011).Thangapandian S, John S, Sakkiah S, Lee KW. Potential virtual lead identification in the discovery of renin inhibitors: application of ligand and structure-based pharmacophore modeling approaches. European Journal of Medicinal Chemistry. 2011;46(6):2469–2476. doi: 10.1016/j.ejmech.2011.03.035. [DOI] [PubMed] [Google Scholar]
- The Plant List (2013).The Plant List The Plant List. Version 1.1www.theplantlist.org/ [20 June 2019];2013
- Trott & Olson (2010).Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2018).UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Research. 2018;46(5):2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursa et al. (2011).Ursa O, Rayan A, Goldblum A, Oprea TI. Understanding drug-likeness. Computational Molecular Science. 2011;1(5):760–781. doi: 10.1002/wcms.52. [DOI] [Google Scholar]
- Van Waarde et al. (2018).Van Waarde A, Dierckx R, Zhou X, Khanapur S, Tsukada H, Ishiwata K, Luurtsema G, De Vries EFJ, Elsinga PH. Potential therapeutic applications of adenosine A2A receptor ligands and opportunities for A2A receptor imaging. Medicinal Research Reviews. 2018;38(1):5–56. doi: 10.1002/med.21432. [DOI] [PubMed] [Google Scholar]
- Wieder et al. (2017).Wieder M, Garon A, Perricone U, Boresch S, Seidel T, Almerico AM, Langer T. Common hits approach: combining pharmacophore modeling and molecular dynamics simulations. Journal of Chemical Information and Modeling. 2017;57(2):365–385. doi: 10.1021/acs.jcim.6b00674. [DOI] [PubMed] [Google Scholar]
- Yu & MacKerell (2017).Yu W, MacKerell AD., Jr Computer-aided drug design methods. Methods in Molecular Biology. 2017;1520:85–106. doi: 10.1007/978-1-4939-6634-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung, Fuseini & Newcomb (2018).Yung JA, Fuseini H, Newcomb DC. Hormones, sex, and asthma. Annals of allergy, asthma & immunology: official publication of the American College of Allergy. Annals of Allergy, Asthma & Immunology. 2018;120(5):488–494. doi: 10.1016/j.anai.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Yan & Xie (2003).Zhou J, Yan X, Xie G. Traditional Chinese medicines: molecular structures, natural sources and applications. Burlington: Ashgate; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are available in the Supplemental Files.