Abstract
Forces generated by molecular motors and the cytoskeleton move the nucleus and genome during many cellular processes, including cell migration and division. How these forces impact the genome, and whether cells regulate cytoskeletal forces to preserve genome integrity is unclear. We recently demonstrated that, in budding yeast, mutants that stabilize the microtubule cytoskeleton cause excessive movement of the mitotic spindle and nucleus. We found that increased nuclear movement results in DNA damage and increased time to repair the damage through homology-directed repair. Our results indicate nuclear movement impairs DNA repair through increased tension on chromosomes and nuclear deformation. However, previous studies have shown genome mobility, driven by cytoskeleton-based forces, aids in homology-directed DNA repair. This sets up an apparent paradox where genome mobility may prevent or promote DNA repair. Hence this review explores how the genome is affected by nuclear movement and how genome mobility could aid or hinder homology-directed repair.
Keywords: DNA damage, HDR, Dynein, Microtubule, Cytoskeleton, Nucleus
The maintenance of a stable genome is necessary for cellular homeostasis. Cells are routinely confronted with challenges that can alter the genome, including DNA damage. Pathways that repair DNA damage are critical for maintaining genome stability, and disruption of these pathways therefore represents a common mechanism of genomic instability (Loeb et al., 1974; Nowell, 1976; Tubbs 2017). This review explores how forces that move the genome during the routine processes of cell division and migration impact the efficiency of DNA damage repair, and thus are a key determinant of genome stability.
The cytoskeleton plays essential roles in positioning the genome within the cell. During cell division, the position of the genome is controlled by the mitotic spindle, which provides two key functions: segregating sister chromosomes to opposite spindle poles and guiding the spindle poles to regions of the cytoplasm that will become the daughter cells. While the former is necessary to prevent aneuploidy, the latter plays a determining role in cell differentiation and contributes to tissue architecture and morphogenesis. In animal cells, the position of the spindle determines the site of division or cytokinesis (Canman et al. 2003; Green, Paluch, and Oegema 2012). Symmetric spindle placement results in daughter cells of equal size. Asymmetric cell divisions, such as those that give rise to distinct daughter cells, require careful spindle positioning to set the plane of cytokinesis and direct the inheritance of different regions of cytoplasm (Canman et al. 2003). In contrast, budding yeast contain a pre-determined site of cytokinesis, called the bud neck, and must move the spindle to that site for every cell division. Importantly, all of these cases require precise genome positioning before cytokinesis.
Spindle positioning is an active process involving motor proteins and the cytoskeleton. In this mini-review, we will focus on two major players that drive spindle movement: microtubules and dynein. Microtubules are dynamic protein polymers that form trafficking networks and the mitotic spindle within cells. The mitotic spindle includes two spindle poles nucleating microtubules that either extend inward to attach to chromosomes and stabilize the spindle, or astral microtubules that extend outward into the cytoplasm where they interact with the motor protein dynein to coordinate the position of the spindle in the dividing cell (Carminati and Stearns 1997; Raspelli and Fraschini 2019). Dynein is responsible for the majority of minus end-directed movement along microtubules, and is needed for vesicle transport, mitotic spindle assembly, and spindle positioning. A wealth of knowledge on dynein and its regulatory pathway has come from budding yeast where dynein has one role: positioning the mitotic spindle at the bud neck. To move the spindle to the right place and at the right time, dynein is highly regulated through interactions with adaptor proteins and the microtubule substrate. Whereas null mutations in dynein and its regulators disrupt spindle positioning and increase the frequency of failed mitoses that produce bi-nucleate and anucleate daughter cells (Carminati and Stearns, 1997; Yeh, 1995; Li et al., 1993; Eshel et al., 1993; Muhua et al., 1994), partial loss of function mutations impair the accuracy and efficiency of spindle positioning. For example, dynein requires the activating complex dynactin to pull the nucleus into the bud neck. Dynactin includes a Nip100/p150 protein subunit with its own microtubule-binding activity, and removing the microtubule-binding CAP-Gly domain from Nip100/p150 impairs dynein activity, reducing the frequency and duration of spindle movements (Moore et al., 2009). Another important point of dynein regulation is the microtubule substrate. Mutations that enhance the stability of microtubules increase dynein-dependent movement of the spindle, resulting in increased displacement of the spindle from the bud neck (Estrem, Fees, and Moore 2017). Thus, spindle positioning uses precisely regulated microtubule forces to deliver a genome to an incipient daughter cell.
While we know how dynein and microtubules move the mitotic spindle to a destination within a cell, we understand less about how these forces might impact the organization of the nucleus and the integrity of the genome within. We have recently addressed this possibility by focusing on the interplay between forces on astral microtubules, nuclear shape and genome stability. We demonstrated that, in budding yeast, mutants that stabilize microtubules and hyperactivate dynein cause excessive movement of the mitotic spindle and concomitantly increase the frequency of double stranded DNA breaks. In this review, we present the major findings of our study and discuss how they may shed light on our understanding of how cytoskeletal forces can alter DNA and DNA repair. We propose that cytoskeletal forces on the nucleus must be coordinated with DNA replication and repair to minimize genome damage.
1.). How do mechanical forces in the cytoplasm affect the nucleus and genome?
Cells are naturally exposed to many mechanical forces -- from outside the cell, during tissue organization and migration, to inside the cell, during replication and division. The cytoskeleton and nuclear envelope are two crucial components that allow a cell to withstand changes in mechanical forces. The cytoskeleton includes networks of protein polymers, motor proteins and other binding proteins that can both generate mechanical forces from within the cell and mitigate external forces applied to the cell. Actin filaments provide structure to the plasma membrane and rearrange to form stress fibers when cells are exposed to mechanical force (Hornberger et al. 2005; Miller et al. 2004). Microtubules act as scaffolds for the organization of intracellular organelles such as the nucleus and mitochondria (Schroer 2000). Microtubules can bear compressive forces and are the most rigid component of the cytoskeleton (Brangwynne et al. 2006; Fletcher and Mullins 2010). The nucleus is surrounded by an additional protective element - the nuclear envelope. The mammalian nuclear envelope is composed of inner and outer nuclear membranes that form a double bilayer, nuclear pore complexes that regulate transit across the envelope, and lamina that provide rigidity to the envelope and help organize the genome (Hetzer 2010). Disruption of the nuclear lamina impairs nuclear stiffness, size, and shape (Dahl et al. 2004; Liu et al. 2000). Together, the cytoskeleton and nuclear envelope protect the genome against damage from changes in mechanical forces.
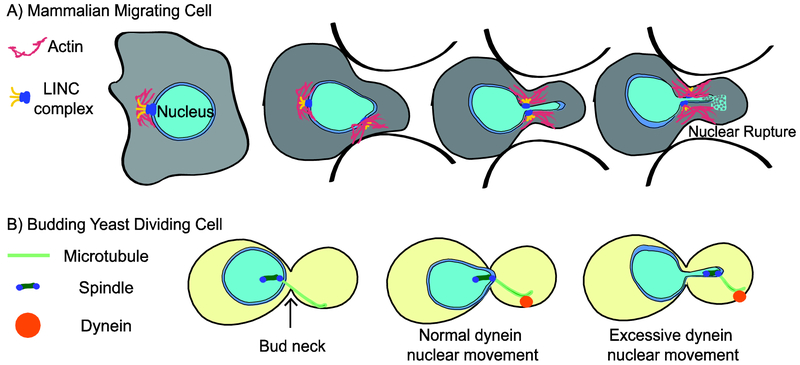
In interphase cells, defects in nuclear lamina structure and/or the application of mechanical forces can deform the nucleus and can even lead to rupture of the nuclear envelope (Chow, Factor, and Ullman 2012; Denais et al. 2016; Ho and Lammerding 2012; Irianto et al. 2017; Raab et al. 2016; De vos et al. 2011; Xia et al. 2019). Recent studies have shown that under mechanical strain, chromatin can protrude through gaps in the lamina, expand, and rupture the nuclear envelope; resulting in the exchange between the nuclear and cytoplasmic contents (Denais et al. 2016; Raab et al. 2016; Xia et al. 2019). For example, forcing mammalian cells to migrate through a pore as small as 3.5 μm compresses the nucleus and causes envelope rupture (Le Berre, Aubertin, and Piel 2012; Broers, Hutchison, and Ramaekers 2004)(Figure 1A). Repeatedly forcing cells through several rounds of nuclear compression results in a greater increase in DNA mutation and chromosome copy number changes, but is not always accompanied by envelope rupture (Irianto et al. 2017). These studies clearly show that mechanical force on the nucleus can lead to genome damage, but the mechanism of how forces cause damage is not understood. Although one model posits that DNA damage results from exposure to cytoplasmic nucleases at sites nearby envelope rupture (Denais et al. 2016; Raab et al. 2016), the location and extent of damage varies and is not always associated with envelope rupture (for a review of these DNA damage mechanisms see Shah, Wolf, and Lammerding 2017).
Figure 1. Nuclear migration.
A) Schematic depicting a mammalian cell migrating through a narrow pore. The actin cytoskeleton exerts force on the nucleus causing deformation, disruption of nuclear lamina, and nuclear envelope rupture.
B)Schematic depicting movement of the spindle and nucleus during budding yeast cell division. Dynein exerts force on the astral microtubule connected to the spindle pole body. This force pulls the spindle and intact nucleus into the bud neck between the mother and bud. Normal dynein movement shows the minimal deformation of the nucleus, while excessive dynein movement shows a highly deformed nucleus and aberrant spindle positioning.
While many possibilities have been proposed for the mechanism of DNA damage during interphase, several lines of evidence clearly implicate forces from the cytoskeleton in causing nuclear envelope rupture and/or genome damage. Hatch and Hetzer found that actin fibers constrict the nucleus (Figure 1A), and can cause nuclear envelope rupture in cells with weakened nuclear lamina; but this could be prevented by disruption of the actin cytoskeleton with destabilizing drugs (cytochalasin D or latrunculin A) or inhibition of myosin motors (Hatch and Hetzer 2016). In addition, disruption of the linker of nucleoskeleton and cytoskeleton (LINC) complex, which physically connects the nuclear envelope to the cytoskeleton, also reduces the frequency of nuclear rupture and changes perinuclear actin organization (Hatch and Hetzer 2016). Taken together, these results show that the actin cytoskeleton that surrounds the nucleus can promote nuclear rupture and subsequent DNA damage. The LINC complex also attaches the nucleus to other crucial components of the cytoskeleton -- microtubules and dynein. This raises the question of whether movement of the nucleus by microtubules and dynein could damage the genome?
The possibility of dynein-driven movement causing nuclear envelope rupture in animal cells is supported by recent work on C. elegans zygotes. Here, increased mechanical forces generated by dynein and applied to microtubules during pronuclear movement resulted in envelope rupture (Penfield et al. 2018). Penfield and colleagues found that in zygotes with defective lamina, dynein forces increased the severity of nuclear envelope rupture; but limiting dynein activity decreased the frequency of ruptures, and when a rupture did occur, the rate of repair of the nuclear envelope was faster (Penfield et al. 2018). This study is important because it reveals that forces from microtubules and dynein can rupture the nuclear envelope during interphase.
In our recent paper, we explore how forces from dynein are transmitted through microtubules and the mitotic spindle to cause nuclear deformation and disrupt genome integrity in Saccharomyces cerevisiae. Because budding yeast undergo a closed mitosis, dynein is necessary to position the mitotic spindle and the surrounding nucleus at the bud neck, between the mother and bud (Figure 1B)(Byers and Goetsch 1975). We asked whether forces from dynein on astral microtubules are transmitted through the mitotic spindle. Normally, dynein pulls on astral microtubules from the cortex of the bud and the entire spindle moves in the direction of dynein pulling force (Figure 1B). Even though dynein is exerting force on only one side of the spindle, the spindle length normally does not change due to the activity of proteins that crosslink microtubules from the two spindle poles. However, when we disrupted crosslinks inside the spindle, we found that the spindle elongates when dynein pulls on astral microtubules, with the proximal spindle pole moving in the direction of dynein pulling while the distal pole lagged behind (Estrem and Moore 2019). These results indicate that forces on astral microtubules are transmitted through the mitotic spindle, raising the possibility that these forces could also be experienced by the genome through microtubule-kinetochore interactions. To test this, we used a series of mutants to stabilize microtubules and increase the frequency of dynein-dependent movement of the spindle and nucleus. We found increased movement by dynein led to increased tension between sister chromatids and increased the frequency of DNA double strand breaks (DSBs), as indicated by the focal accumulation of the repair factor, Rad52 (Estrem and Moore 2019). Importantly, in our experiments increased movement through the bud neck deformed the nucleus, but did not cause nuclear rupture. These results demonstrate that forces from microtubules in the cytoplasm can promote DNA damage without rupturing the nuclear envelope.
Our findings raise the question of whether increasing microtubule stability might also increase DNA damage in other cellular contexts. Several cancer drugs target microtubules and increase their stability, including the prevalent chemotherapeutic, paclitaxel or taxol (Fanale et al. 2015; Tangutur et al. 2017). Many labs have shown that these drugs impair mitotic spindle function, arrest the cell cycle, and lead to cell death (Abal, Andreu, and Barasoain 2003; Bhalla 2003; Jablonski, Liu, and Yen 2003; Snyder and Mullins 1993; Sorger et al. 1997; Wang, Wang, and Soong 2000). However, far less is known about the effect these drugs have on the genome. Rankin and Wordeman found that increasing astral microtubule stability with taxol or by depleting a microtubule depolymerizing protein dramatically increases spindle movement in mammalian cells (Rankin and Wordeman 2010). Whether these conditions also promote DNA damage, similar to our studies from budding yeast, has not been addressed; but could represent an additional mechanism of action.
2.). Dynamics within the nucleus: Does DNA mobility hinder or help homology-directed DNA repair?
DNA can be damaged in many ways, but DSBs are among the most deleterious. Cells have evolved a highly conserved mechanism to correct these by homology-directed repair (HDR) (Jasin and Berg 1988; Orr-Weaver, Szostak, and Rothstein 1981; Szostak et al. 1983). During HDR, the cell repairs DSBs by finding a homologous region of DNA and using it as a donor template strand (reviewed in Bordelet and Dubrana 2019). The first proteins recruited to the break site in HDR are the MRN complex, including MRE11, RAD50, and NBS1 (Ivanov et al. 1994; Mimitou and Symington 2010; Moriel-Carretero, Pasero, and Pardo 2019). This complex then recruits and activates Ataxia Telangiectasia Mutated (ATM) protein kinase to phosphorylate histone H2A around the break site (Rogakou et al. 1998). The phosphorylation of H2A causes chromatin reorganization and opens around two megabases of chromatin to allow for repair (Soria, Polo, and Almouzni 2012). Thus, HDR involves a complex reorganization of the genome to prepare the damaged site for repair and recruit the homologous region.
In addition to histones, many other proteins bind DNA to organize and, ultimately, protect it. This is particularly important during mitosis, when there are ample opportunities for damage from replication stress and from forces generated by the mitotic spindle. Inner nuclear membrane proteins tether telomeres to the nuclear envelope (Bupp et al. 2007; Hediger et al. 2002; Schober et al. 2009), Kinetochores tether centromeres to microtubules, and cohesion tethers sister chromatids together (Verdaasdonk et al. 2013). For each of these DNA tethers, the cell uses mechanisms to release chromatin in response to DNA damage, to allow for efficient repair. For example, upon DNA damage by multiple, random breaks or a single, site-specific DSB, telomeres are released from the nuclear membrane, increasing the motion of DNA proximal to break regions (Chung et al. 2015; Lawrimore et al. 2017). Centromeres respond to DNA damage by loosening their attachments to kinetochores and declustering with respect to other centromeres (Lawrimore et al. 2017). Cohesins become phosphorylated in response to DSBs, thereby relaxing the connections between sister chromatids (Strecker et al. 2016; Wu and Yu 2012). Collectively, these findings indicate that releasing DNA from various protein tethers is an important component of DSB repair, and may aid in the search for the homologous region of the genome.
Interestingly, recent work has shown that HDR employs mobility throughout the genome, not just near sites of repair. Studies have shown that upon DNA damage, the entire genome experiences an increase in mobility that cannot be attributed to the release from specific chromatin tethers or remodeling factors (Lawrimore et al. 2017; Miné-Hattab and Rothstein 2012; Zimmer and Fabre 2019). The utility of global genome mobility during DNA repair is still unclear. One proposed model is that genome mobility aids the pairing of the damage site with the homologous donor region (Lawrimore et al. 2017; Lee et al. 2016; Strecker et al. 2016). Lee et al. confirm that the search for the homologous region is the rate-limiting step in DSB repair, and failure to repair DNA is most likely due to a failed search (Lee et al., 2016). Mine-Hattab et al. showed that genome mobility is not stimulated by the DNA break alone but requires the recruitment of the repair factors (Miné-Hattab and Rothstein 2012). Other groups have shown microtubules and motor proteins inside and outside of the nucleus contribute to chromosomal movement upon DNA damage (Lawrimore et al. 2017; Lottersberger et al. 2015; Oshidari et al. 2018; Snijders Blok et al. 2015). Lawrimore and colleagues found that the largest contributing factor to global chromosomal movement after DNA damage was microtubules and motor proteins outside of the nucleus (Lawrimore et al. 2017). While these studies demonstrate the importance of genome mobility and cytoskeletal forces in facilitating DSB repair, how forces are regulated to promote efficient repair is unknown.
Another example in which genome movement is crucial for finding homologous regions of DNA is in meiosis. The pairing of homologous chromosomes in meiosis is a crucial step in development. Inhibiting the movement of chromosomes results in mispairing and diminished recombination, indicating that movement aids in the homology search (Ding et al. 2004; Labrador et al. 2013; Parvinen and Soderstorm 1976; Phillips et al. 2009; Sato et al. 2009; Scherthan et al. 2007; Woglar and Jantsch 2014; Wynne et al. 2012; Yamamoto et al. 1999). In the fission yeast, Schizosaccharomyces pombe, the homology search is driven by nuclear oscillations during prophase, a process called horsetail nuclear movements. Here, the nucleus oscillates back and forth along the long axis of the cell for two hours in prophase I (Chikashige et al. 1994). This drastic and prolonged movement of the nucleus is dependent upon astral microtubules and dynein (Wynne et al. 2012; Yamamoto et al. 1999). In addition to aiding the finding of homologous chromosomes, nuclear oscillations prevent unwanted non-homologous connections, suggesting that movement selects for higher affinity binding between regions of homology over the lower affinity binding of mispaired regions (Conrad et al. 2008; Koszul and Kleckner 2009). While this nuclear movement by dynein in meiosis is beneficial, one can imagine that too much movement or movement at the wrong time might be detrimental.
How might too much and/or ill-timed microtubule force be detrimental to repair? Our experiments in budding yeast indicate that excessive dynein-dependent nuclear migration inhibits HDR. Under these conditions, cells attempt HDR while the entire nucleus, spindle, and genome are continually pulled back-and-forth through the bud neck, and we found that HDR took on average 2.5 times longer to repair DSBs (Estrem and Moore 2019). In this case, genome mobility inhibited, rather than promoted, HDR. Our data shows that when forces from dynein in the cytoplasm are transmitted to the spindle, tension on the pericentromeric regions between sister chromatids is increased. Furthermore, mutants that increase tension across sister chromatids are sufficient to promote high levels of DSBs, even when nuclear movement is normal (Estrem and Moore 2019). In contrast, decreasing tension around centromeres by disrupting the recruitment of cohesin to the pericentromeric chromatin (Fees et al. 2016; Goshima and Yanagida 2000; Ng et al. 2009) decreases the frequency of DSBs and restores the efficiency of HDR (Estrem and Moore 2019). These data suggest that applying too much force to the genome – from hyperactive dynein, to the microtubule-kinetochore interface, and through sister chromatid cohesion – inhibits efficient HDR. By analogy to the meiosis example in fission yeast, perhaps too much energy from microtubule-based movement may be sufficient to disrupt even the high affinity pairing between homologues. Testing this model will require further study.
3.). How is spindle movement inhibited during DNA repair?
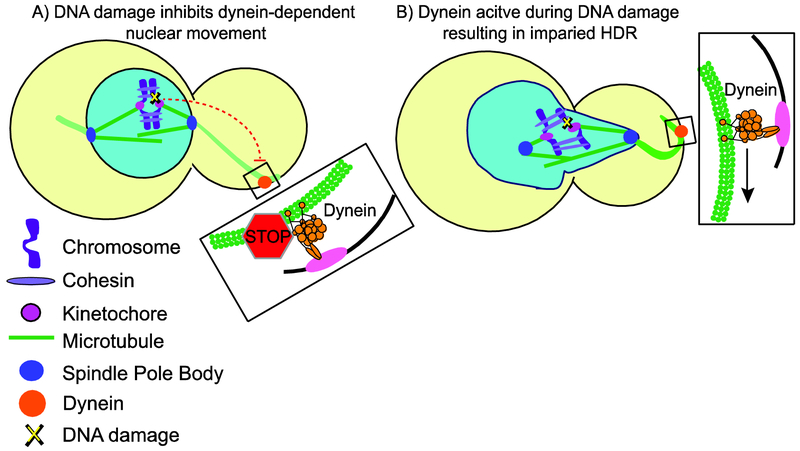
If DNA is damaged before mitosis, the cell cycle is paused through a checkpoint-dependent mechanism until the damage is repaired. In a normal budding yeast cell division, dynein pulls the spindle and nucleus into the bud neck and keeps it there through anaphase. Dotiwala et al. showed that dynein-dependent movement of the spindle and nucleus is diminished during DNA repair (Figure 2A); but ablating either of the DNA damage checkpoint genes RAD53 or CHK1 restores movement and tension on pericentromeric DNA (Dotiwala et al. 2007). This hints at a mechanism to inhibit dynein from moving the spindle and nucleus during HDR. We speculate that dynein-dependent movement of the nucleus is inhibited to relieve tension on chromosomes within the spindle and allow for relaxation of the genome and efficient HDR. Consistent with this model, our results show that increasing dynein-dependent nuclear movement increases tension on pericentromeric DNA and inhibits HDR (Figure 2B). A remaining question is how DNA damage checkpoint signaling inhibits dynein.
Figure 2. Hyperactive Dynein disrupts HDR.
A)Schematic depicting normal DNA damage signaling, leading to inhibition of dynein in the cytoplasm and halting nuclear movement.
B) Schematic depicting hyperactive dynein pulling the nucleus far into the bud during DNA damage, and thereby disrupting HDR.
One possible mechanism involves a phosphorylation cascade starting with Chk1 kinase and ending with inhibition of dynein activity. However, the mechanism of how Chk1 could be interacting with dynein is unknown. Astral microtubules are also attractive candidates for a putative mechanism to regulate dynein activity. Astral microtubules must be long enough to bring dynein to the cortex yet dynamic so that dynein pulling forces are properly terminated by microtubule depolymerization (Estrem, Fees, and Moore 2017). Therefore, one possible mechanism of dynein inhibition could involve destabilizing microtubules. In this scenario, activation of a microtubule destabilizer would cause astral microtubules to fall away from dynein and remain too short to reach the cell cortex, thereby preventing movement of the nucleus.
A second potential mechanism involves the yeast protein phosphatase 1 (PP1) complex Bud14p-Glc7p, which localizes to the cell cortex where it regulates dynein activity (Knaus et al. 2005). The catalytic subunit, Glc7p, is known to also regulate the microtubule binding activity of kinetochores inside the nucleus (Sassoon et al. 1999). Upon DNA damage, PP1/Glc7 could be exported out of the nucleus to bind to Bud14, which is anchored at the cortex, leading to inhibition of dynein motor by either modifying the motor or destabilizing astral microtubules.
Another possible mechanism of dynein inhibition could be through its activating partner dynactin. Only when dynactin binds to dynein can the motor attach to the cell cortex and generate pulling forces on astral microtubules. A regulator of dynactin, called She1, prevents dynactin from localizing to the microtubule until G2 of the cell cycle and directly inhibits dynein motor activity (Ecklund et al. 2017; Woodruff, Drubin, and Barnes 2009). It is possible that DNA damage factors signal to She1, which is localized inside and outside of the nucleus, to inhibit dynein. The signaling mechanisms that regulate She1 are currently undefined.
Beyond budding yeast, carefully regulated dynein-dependent movement of the nucleus occurs in many other contexts, including neuronal migration. Neuroepithelial cells undergo interkinetic nuclear migration, coordinating the cell cycle with dynein recruitment to the nucleus through the LINC complex (Crisp et al., 2006; Yu et al., 2011; Zhang et al., 2009; Baffet et al., 2015). The nucleus moves to the basal region of the cell for G1 and S phase. Then during G2, the nucleus rapidly moves back to the apical surface for M phase (Willardsen and Link 2011). The G2 to M cyclin-dependent kinase 1 (Cdk1) phosphorylates RanBP2, a nucleoporin, which recruits dynein adaptor protein BicD2, which subsequently recruits dynein to the nucleus (Baffet, Hu, and Vallee 2015). Future studies could reveal whether the cell cycle recruitment of dynein is important to regulate time-dependent activity and/or to limit forces on the nucleus and preserve genome integrity.
The studies discussed here identify an important need to regulate dynein to achieve a proper level of genome mobility during nuclear positioning and during DNA repair. Dynein is a highly regulated motor protein, and recent studies have revealed a multitude of adapter proteins that are required for proper activity and function (Olenick and Holzbaur 2019; Reck-Peterson et al. 2018). Due to the high number of proteins that interact with dynein, it is possible that a mutation in any of these regulators could alter dynein’s activity resulting in DNA damage or a hinderance of repair. Accordingly, it will be important to determine whether DNA damage or inhibited HDR are involved in diseases where dynein and its regulators are implicated.
This review provides evidence that nuclear movement is a highly regulated process not only for proper chromosome segregation but also for limiting DNA damage and aiding in HDR. In budding yeast, we found increased microtubule-based movement delays HDR. We propose two possible models for how increased nuclear movement could inhibit HDR. In the first model, nuclear movement restricts the search for the homologous strand of DNA by increasing tension on microtubule connections to chromosomes, thereby decreasing Rad51 accessibility to search the genome. In the second model, which is not mutually exclusive, the increased energy from microtubule-based forces disrupts the homologous pairing required for HDR. Future work is needed to test these models and determine the effects of increased microtubule-based movement on disrupting HDR.
Acknowledgements:
This work was supported by National Institutes of Health Grant no. R01GM-112893 (to J.K.M.).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Abal M, Andreu JM, and Barasoain I. 2003. “Taxanes: Microtubule and Centrosome Targets, and Cell Cycle Dependent Mechanisms of Action.” Current cancer drug targets 3(3): 193–203. http://www.ncbi.nlm.nih.gov/pubmed/12769688 [DOI] [PubMed] [Google Scholar]
- Baffet Alexandre D., Hu Daniel J., and Vallee Richard B.. 2015. “Cdk1 Activates Pre-Mitotic Nuclear Envelope Dynein Recruitment and Apical Nuclear Migration in Neural Stem Cells.” Developmental Cell 33(6): 703–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berre Le, Maël Johannes Aubertin, and Piel Matthieu. 2012. “Fine Control of Nuclear Confinement Identifies a Threshold Deformation Leading to Lamina Rupture and Induction of Specific Genes.” Integrative Biology 4(11): 1406 http://xlink.rsc.org/?DOI=c2ib20056b [DOI] [PubMed] [Google Scholar]
- Bhalla Kapil N. 2003. “Microtubule-Targeted Anticancer Agents and Apoptosis.” Oncogene 22(56): 9075–86. http://www.nature.com/articles/1207233 [DOI] [PubMed] [Google Scholar]
- Bordelet Hélène, and Dubrana Karine. 2019. “Keep Moving and Stay in a Good Shape to Find Your Homologous Recombination Partner.” Current genetics 65(1): 29–39. http://link.springer.com/10.1007/s00294-018-0873-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne Clifford P. et al. 2006. “Microtubules Can Bear Enhanced Compressive Loads in Living Cells Because of Lateral Reinforcement.” The Journal of Cell Biology 173(5): 733–41. http://www.ncbi.nlm.nih.gov/pubmed/16754957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers Jos L. V., Hutchison Chris J., and Ramaekers Frans C.S.. 2004. “Laminopathies.” Journal of Pathology 204(4): 478–88. www.interscience.wiley.com [DOI] [PubMed] [Google Scholar]
- Bupp Jennifer M., Martin Adriana E., Stensrud Elizabeth S., and Jaspersen Sue L.. 2007. “Telomere Anchoring at the Nuclear Periphery Requires the Budding Yeast Sad1-UNC-84 Domain Protein Mps3.” The Journal of Cell Biology 179(5): 845–54. http://www.ncbi.nlm.nih.gov/pubmed/18039933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, and Goetsch L. 1975. 124 Journal of Bacteriology Behavior of Spindles and Spindle Plaques in the Cell Cycle and Conjugation of Saccharomyces Cerevisiae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman Julie C. et al. 2003. “Determining the Position of the Cell Division Plane.” Nature 424(6952): 1074–78. [DOI] [PubMed] [Google Scholar]
- Carminati Janet L., and Stearns Tim. 1997. “Microtubules Orient the Mitotic Spindle in Yeast through Dynein-Dependent Interactions with the Cell Cortex.” Journal of Cell Biology 138(3): 629–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y et al. 1994. “Telomere-Led Premeiotic Chromosome Movement in Fission Yeast.” Science (New York, N.Y.) 264(5156): 270–73. http://www.sciencemag.org/cgi/doi/10.1126/science.8146661 [DOI] [PubMed] [Google Scholar]
- Chow Kin Hoe, Factor Rachel E., and Ullman Katharine S.. 2012. “The Nuclear Envelope Environment and Its Cancer Connections.” Nature Reviews Cancer 12(3): 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Daniel K C et al. 2015. “Perinuclear Tethers License Telomeric DSBs for a Broad Kinesin- and NPC-Dependent DNA Repair Process.” Nature communications 6(1): 7742 http://www.nature.com/articles/ncomms8742 [DOI] [PubMed] [Google Scholar]
- Conrad Michael N. et al. 2008. “Rapid Telomere Movement in Meiotic Prophase Is Promoted By NDJ1, MPS3, and CSM4 and Is Modulated by Recombination.” Cell 133(7): 1175–87. http://www.ncbi.nlm.nih.gov/pubmed/18585352 [DOI] [PubMed] [Google Scholar]
- Dahl Kris Noel, Kahn Samuel M, Wilson Katherine L, and Discher Dennis E. 2004. “The Nuclear Envelope Lamina Network Has Elasticity and a Compressibility Limit Suggestive of a Molecular Shock Absorber.” Journal of cell science 117 (Pt 20): 4779–86. http://www.ncbi.nlm.nih.gov/pubmed/8832400 [DOI] [PubMed] [Google Scholar]
- Denais Celine M. et al. 2016. “Nuclear Envelope Rupture and Repair during Cancer Cell Migration.” Science 352(6283): 353–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Da-Qiao, Yamamoto Ayumu, Haraguchi Tokuko, and Hiraoka Yasushi. 2004. “Dynamics of Homologous Chromosome Pairing during Meiotic Prophase in Fission Yeast.” Developmental cell 6(3): 329–41. http://www.ncbi.nlm.nih.gov/pubmed/15030757 [DOI] [PubMed] [Google Scholar]
- Dotiwala Farokh et al. 2007. 104 Proceedings of the National Academy of Sciences of the United States of America The Yeast DNA Damage Checkpoint Proteins Control a Cytoplasmic Response to DNA Damage. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1896138&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecklund Kari H. et al. 2017. “She1 Affects Dynein through Direct Interactions with the Microtubule and the Dynein Microtubule-Binding Domain.” Nature Communications 8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem Cassi, Fees Colby P., and Moore Jeffrey K.. 2017. “Dynein Is Regulated by the Stability of Its Microtubule Track.” Journal of Cell Biology 216(7): 2047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem Cassi, and Moore Jeffrey K.. 2019. “Astral Microtubule Forces Alter Nuclear Organization and Inhibit DNA Repair in Budding Yeast” ed. Bloom Kerry. Molecular Biology of the Cell: mbc.E18-12-0808. http://www.ncbi.nlm.nih.gov/pubmed/31067146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanale Daniele et al. 2015. “Stabilizing versus Destabilizing the Microtubules: A Double-Edge Sword for an Effective Cancer Treatment Option?” Analytical Cellular Pathology 2015: 1–19. http://www.ncbi.nlm.nih.gov/pubmed/26484003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fees Colby P. et al. 2016. “The Negatively Charged Carboxy-Terminal Tail of β-Tubulin Promotes Proper Chromosome Segregation.” Molecular Biology of the Cell 27(11): 1786–96. http://www.molbiolcell.org/lookup/doi/10.1091/mbc.E15-05-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher Daniel A, and Mullins R Dyche. 2010. “Cell Mechanics and the Cytoskeleton.” Nature 463(7280): 485–92. http://www.ncbi.nlm.nih.gov/pubmed/20110992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima Gohta, and Yanagida Mitsuhiro. 2000. “Establishing Biorientation Occurs with Precocious Separation of the Sister Kinetochores, but Not the Arms, in the Early Spindle of Budding Yeast.” Cell 100(6): 619–33. [DOI] [PubMed] [Google Scholar]
- Green Rebecca A, Ewa Paluch, and Oegema Karen. 2012. “Cytokinesis in Animal Cells.” Annu. Rev. Cell Dev. Biol 28: 29–58. http://www.annualreviews. [DOI] [PubMed] [Google Scholar]
- Hatch Emily M., and Hetzer Martin W.. 2016. “Nuclear Envelope Rupture Is Induced by Actin-Based Nucleus Confinement.” Journal of Cell Biology 215(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger Florence et al. 2002. “Live Imaging of Telomeres: YKu and Sir Proteins Define Redundant Telomere-Anchoring Pathways in Yeast.” Current biology : CB 12(24): 2076–89. http://www.ncbi.nlm.nih.gov/pubmed/12498682 [DOI] [PubMed] [Google Scholar]
- Hetzer Martin W. 2010. “The Nuclear Envelope.” Cold Spring Harbor perspectives in biology 2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, and Lammerding J. 2012. “Lamins at a Glance.” Journal of Cell Science 125(9): 2087–93. http://jcs.biologists.org/cgi/doi/10.1242/jcs.087288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger Troy A. et al. 2005. “Intracellular Signaling Specificity in Response to Uniaxial vs. Multiaxial Stretch: Implications for Mechanotransduction. ” American Journal of Physiology-Cell Physiology 288(1): C185–94. http://www.physiology.org/doi/10.1152/ajpcell.00207.2004 [DOI] [PubMed] [Google Scholar]
- Irianto Jerome et al. 2017. “DNA Damage Follows Repair Factor Depletion and Portends Genome Variation in Cancer Cells after Pore Migration.” Current Biology 27(2): 210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL et al. 1994. “Mutations in XRS2 and RAD50 Delay but Do Not Prevent Mating-Type Switching in Saccharomyces Cerevisiae.” Molecular and cellular biology 14(5): 3414–25. http://www.ncbi.nlm.nih.gov/pubmed/8164689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski SA, Liu ST, and Yen TJ. 2003. “Targeting the Kinetochore for Mitosis-Specific Inhibitors.” Cancer-Biology & Therapy 2(3): 236–41. http://www.ncbi.nlm.nih.gov/pubmed/12878855 [DOI] [PubMed] [Google Scholar]
- Jasin M, and Berg P. 1988. “Homologous Integration in Mammalian Cells without Target Gene Selection.” Genes & development 2(11): 1353–63. http://www.ncbi.nlm.nih.gov/pubmed/2850258 [DOI] [PubMed] [Google Scholar]
- Knaus Michèle et al. 2005. “The Bud14p-Glc7p Complex Functions as a Cortical Regulator of Dynein in Budding Yeast.” EMBO Journal 24(17): 3000–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul Romain, and Kleckner Nancy. 2009. “Dynamic Chromosome Movements during Meiosis: A Way to Eliminate Unwanted Connections?” Trends in Cell Biology 19(12): 716–24. http://www.ncbi.nlm.nih.gov/pubmed/19854056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador Leticia et al. 2013. “Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Caenorhabditis Elegans Meiosis” ed. Colaiácovo Mónica P.. PLoS Genetics 9(5): e1003497 http://www.ncbi.nlm.nih.gov/pubmed/23671424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore Josh et al. 2017. “Microtubule Dynamics Drive Enhanced Chromatin Motion and Mobilize Telomeres in Response to DNA Damage.” Molecular Biology of the Cell 28(12): 1701–11. http://www.molbiolcell.org/lookup/doi/10.1091/mbc.E16-12-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Cheng-Sheng et al. 2016. “Chromosome Position Determines the Success of Double-Strand Break Repair.” Proceedings of the National Academy of Sciences 113(2): E146–54. http://www.ncbi.nlm.nih.gov/pubmed/26715752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Jun et al. 2000. “Essential Roles for Caenorhabditis Elegans Lamin Gene in Nuclear Organization, Cell Cycle Progression, and Spatial Organization of Nuclear Pore Complexes” ed. Joseph Gall. Molecular Biology of the Cell 11(11): 3937–47. http://www.molbiolcell.org/doi/10.1091/mbc.11.11.3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottersberger Francisca, Karssemeijer Roos Anna, Nadya Dimitrova, and de Lange Titia. 2015. “53BP1 and the LINC Complex Promote Microtubule-Dependent DSB Mobility and DNA Repair.” Cell 163(4): 880–93. http://www.ncbi.nlm.nih.gov/pubmed/26544937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Henk Granzier, Elisabeth Ehler, and Gregorio Carol C. 2004. “The Sensitive Giant: The Role of Titin-Based Stretch Sensing Complexes in the Heart.” Trends in Cell Biology 14(3): 119–26. http://www.ncbi.nlm.nih.gov/pubmed/15003620 [DOI] [PubMed] [Google Scholar]
- Mimitou Eleni P, and Symington Lorraine S. 2010. “Ku Prevents Exo1 and Sgs1-Dependent Resection of DNA Ends in the Absence of a Functional MRX Complex or Sae2.” The EMBO Journal 29: 3358–69www.embojournal.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miné-Hattab Judith, and Rothstein Rodney. 2012. “Increased Chromosome Mobility Facilitates Homology Search during Recombination.” Nature Cell Biology 14(5): 510–17. [DOI] [PubMed] [Google Scholar]
- Moore JK, Sept D, and Cooper JA. 2009. 106 Proceedings of the National Academy of Sciences Neurodegeneration Mutations in Dynactin Impair Dynein-Dependent Nuclear Migration. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2664072&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel-Carretero María, Pasero Philippe, and Pardo Benjamin. 2019. “DDR Inc., One Business, Two Associates.” Current Genetics 65(2): 445–51. http://www.ncbi.nlm.nih.gov/pubmed/30467717 [DOI] [PubMed] [Google Scholar]
- Ng Tessie M, Waples William G, Lavoie Brigitte D, and Sue Biggins. 2009. “Pericentromeric Sister Chromatid Cohesion Promotes Kinetochore Biorientation.” Molecular Biology of the Cell 20: 3818–27. http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenick Mara A, and Holzbaur Erika L F. 2019. “Dynein Activators and Adaptors at a Glance.” Journal of cell science 132(6): jcs227132 http://jcs.biologists.org/lookup/doi/10.1242/jcs.227132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, and Rothstein RJ. 1981. “Yeast Transformation: A Model System for the Study of Recombination.” Proceedings of the National Academy of Sciences 78(10): 6354–58. http://www.pnas.org/cgi/doi/10.1073/pnas.78.10.6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshidari Roxanne et al. 2018. “Nuclear Microtubule Filaments Mediate Non-Linear Directional Motion of Chromatin and Promote DNA Repair.” Nature communications 9(1): 2567 http://www.nature.com/articles/s41467-018-05009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvinen Martti, and Soderstrom Karl-ove. 1976. “Chromosome Rotation and Formation of Synapsis.” Nature 260(5551): 534–35. http://www.ncbi.nlm.nih.gov/pubmed/1264213 [DOI] [PubMed] [Google Scholar]
- Penfield Lauren et al. 2018. “Dynein-Pulling Forces Counteract Lamin-Mediated Nuclear Stability during Nuclear Envelope Repair.” Molecular Biology of the Cell: mbc.E17-06-0374. http://www.molbiolcell.org/lookup/doi/10.1091/mbc.E17-06-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips Carolyn M. et al. 2009. “Identification of Chromosome Sequence Motifs That Mediate Meiotic Pairing and Synapsis in C. Elegans.” Nature Cell Biology 11(8): 934–42. http://www.ncbi.nlm.nih.gov/pubmed/19620970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M et al. 2016. “ESCRT III Repairs Nuclear Envelope Ruptures during Cell Migration to Limit DNA Damage and Cell Death.” Science 352(6283): 359–62. http://www.ncbi.nlm.nih.gov/pubmed/27013426 [DOI] [PubMed] [Google Scholar]
- Rankin Kathleen E, and Linda Wordeman. 2010. “Long Astral Microtubules Uncouple Mitotic Spindles from the Cytokinetic Furrow.” The Journal of cell biology 190(1): 35–43. http://www.ncbi.nlm.nih.gov/pubmed/20603328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspelli Erica, and Fraschini Roberta. 2019. “Spindle Pole Power in Health and Disease.” Current Genetics 65(4): 851–55. http://www.ncbi.nlm.nih.gov/pubmed/30788566 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson Samara L., Redwine William B., Vale Ronald D., and Carter Andrew P.. 2018. “The Cytoplasmic Dynein Transport Machinery and Its Many Cargoes.” Nature Reviews Molecular Cell Biology 19(6): 382–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou Emmy P. et al. 1998. “DNA Double-Stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139.” Journal of Biological Chemistry 273(10): 5858–68. http://www.ncbi.nlm.nih.gov/pubmed/9488723 [DOI] [PubMed] [Google Scholar]
- Sassoon Ingrid et al. 1999. “Regulation of Saccharomyces Cerevisiae Kinetochores by the Type 1 Phosphatase Glc7p.” Genes and Development 13(5): 545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Aya et al. 2009. “Cytoskeletal Forces Span the Nuclear Envelope to Coordinate Meiotic Chromosome Pairing and Synapsis.” Cell 139(5): 907–19. http://www.ncbi.nlm.nih.gov/pubmed/19913287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H et al. 2007. “Chromosome Mobility during Meiotic Prophase in Saccharomyces Cerevisiae.” Proceedings of the National Academy of Sciences 104(43): 16934–39. http://www.ncbi.nlm.nih.gov/pubmed/17939997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H et al. 2009. “Yeast Telomerase and the SUN Domain Protein Mps3 Anchor Telomeres and Repress Subtelomeric Recombination.” Genes & Development 23(8): 928–38. http://www.ncbi.nlm.nih.gov/pubmed/19390087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer Trina A. 2000. “Motors, Clutches and Brakes for Membrane Traffic: A Commemorative Review in Honor of Thomas Kreis.” Traffic 1(1): 3–10. http://doi.wiley.com/10.1034/j.1600-0854.2000.010102.x [DOI] [PubMed] [Google Scholar]
- Shah Pragya, Wolf Katarina, and Lammerding Jan. 2017. “Bursting the Bubble – Nuclear Envelope Rupture as a Path to Genomic Instability?” Trends in Cell Biology 27(8): 546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders Blok Lot, et al. 2015. “Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling.” The American Journal of Human Genetics 97(2): 343–52. http://www.ncbi.nlm.nih.gov/pubmed/26235985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder J, and Mullins JM. 1993. “Analysis of Spindle Microtubule Organization in Untreated and Taxol-Treated PtK1 Cells.” Cell Biology International 17(12): 1075–84. http://www.ncbi.nlm.nih.gov/pubmed/7906984 [DOI] [PubMed] [Google Scholar]
- Sorger PK, Dobles M, Tournebize R, and Hyman AA. 1997. “Coupling Cell Division and Cell Death to Microtubule Dynamics.” Current opinion in cell biology 9(6): 807–14. http://www.ncbi.nlm.nih.gov/pubmed/9425345 [DOI] [PubMed] [Google Scholar]
- Soria Gaston, Polo Sophie E, and Geneviève Almouzni. 2012. “Prime, Repair, Restore: The Active Role of Chromatin in the DNA Damage Response.” Molecular cell 46(6): 722–34. https://linkinghub.elsevier.com/retrieve/pii/S1097276512004911 [DOI] [PubMed] [Google Scholar]
- Strecker Jonathan et al. 2016. “DNA Damage Signalling Targets the Kinetochore to Promote Chromatin Mobility.” Nature Cell Biology 18(3): 281–90. [DOI] [PubMed] [Google Scholar]
- Szostak Jack W., Terry L Orr-Weaver, Rothstein Rodney J., and Stahl Franklin W.. 1983. “The Double-Strand-Break Repair Model for Recombination.” Cell 33(1): 25–35. [DOI] [PubMed] [Google Scholar]
- Tangutur Anjana Devi, Kumar Dinesh, Kommalapati Vamsi Krishna, and Kantevari Srinivas. 2017. “Microtubule Targeting Agents as Cancer Chemotherapeutics: An Overview of Molecular Hybrids as Stabilizing and Destabilizing Agents.” Current Topics in Medicinal Chemistry 17(22): 2523–37. http://www.ncbi.nlm.nih.gov/pubmed/28056738 [DOI] [PubMed] [Google Scholar]
- Verdaasdonk Jolien Suzanne et al. 2013. “Centromere Tethering Confines Chromosome Domains.” Molecular Cell 52(6): 819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De vos Winnok H. et al. 2011. “Repetitive Disruptions of the Nuclear Envelope Invoke Temporary Loss of Cellular Compartmentalization in Laminopathies.” Human Molecular Genetics 20(21): 4175–86. [DOI] [PubMed] [Google Scholar]
- Wang TH Wang HS, and Soong YK. 2000. “Paclitaxel-Induced Cell Death: Where the Cell Cycle and Apoptosis Come Together.” Cancer 88(11): 2619–28. http://www.ncbi.nlm.nih.gov/pubmed/10861441 [DOI] [PubMed] [Google Scholar]
- Willardsen Minde I., and Link Brian A.. 2011. “Cell Biological Regulation of Division Fate in Vertebrate Neuroepithelial Cells.” Developmental Dynamics 240(8): 1865–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar Alexander, and Jantsch Verena. 2014. “Chromosome Movement in Meiosis I Prophase of Caenorhabditis Elegans.” Chromosoma 123(1–2): 15–24. http://www.ncbi.nlm.nih.gov/pubmed/24036686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff Jeffrey B, Drubin David G, and Georjana Barnes. 2009. “Dynein-Driven Mitotic Spindle Positioning Restricted to Anaphase by She1p Inhibition of Dynactin Recruitment Dynein Is a Minus-End-Directed Microtubule Motor Important for Mitotic Spindle Positioning.” Molecular Biology of the Cell 20: 3003–11. http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Nan, and Yu Hongtao. 2012. “The Smc Complexes in DNA Damage Response.” Cell & Bioscience 2(1): 5 http://cellandbioscience.biomedcentral.com/articles/10.1186/2045-3701-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne David J., Rog Ofer, Carlton Peter M., and Dernburg Abby F.. 2012. “Dynein-Dependent Processive Chromosome Motions Promote Homologous Pairing in C. Elegans Meiosis.” The Journal of Cell Biology 196(1): 47–64. http://www.ncbi.nlm.nih.gov/pubmed/22232701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Yuntao et al. 2019. “Rescue of DNA Damage after Constricted Migration Reveals a Mechano-Regulated Threshold for Cell Cycle.” The Journal of cell biology: jcb.201811100 http://www.jcb.org/lookup/doi/10.1083/jcb.201811100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Ayumu, West Robert R., Richard McIntosh J, and Yasushi Hiraoka. 1999. “A Cytoplasmic Dynein Heavy Chain Is Required for Oscillatory Nuclear Movement of Meiotic Prophase and Efficient Meiotic Recombination in Fission Yeast.” Journal of Cell Biology 145(6): 1233–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E 1995. “Spindle Dynamics and Cell Cycle Regulation of Dynein in the Budding Yeast, Saccharomyces Cerevisiae.” The Journal of Cell Biology 130(3): 687–700. http://www.jcb.Org/cgi/doi/10.1083/jcb.130.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Juehua et al. 2011. “KASH Protein Syne-2/Nesprin-2 and SUN Proteins SUN1/2 Mediate Nuclear Migration during Mammalian Retinal Development.” Human Molecular Genetics 20(6): 1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Xiaochang et al. 2009. “SUN1/2 and Syne/Nesprin-1/2 Complexes Connect Centrosome to the Nucleus during Neurogenesis and Neuronal Migration in Mice.” Neuron 64(2): 173–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer Christophe, and Fabre Emmanuelle. 2019. “Chromatin Mobility upon DNA Damage: State of the Art and Remaining Questions.” Current Genetics 65(1): 1–9. http://www.ncbi.nlm.nih.gov/pubmed/29947969 [DOI] [PubMed] [Google Scholar]