Abstract
Unbalanced inflammatory reactions and oxidative stress are inseparably interconnected, and both may play crucial roles in the pathophysiological mechanisms of preeclampsia (PE). In the published previous studies, we have genotyped for SNPs that related to inflammation (rs2227485, rs153109, rs17855750, rs2027432, rs2275913, rs763780, rs4819554, and rs13015714) and oxidative stress (rs1695, rs4680, rs1800566, rs4807542, rs713041, rs7579, rs230813, rs1004467, rs3824755, and rs9932581) to investigate whether these polymorphisms were associated with susceptibility to PE in a Chinese Han population. In this present study, we collected these data of experimental and clinical from above studies for haplotype analysis of inflammation-related SNPs in 631 PE patients and 720 normal pregnancy and oxidative stress-related SNPs in 342 PE patients and 457 normal pregnancies for susceptibility to PE. The data of genotype distribution and allele frequency comparisons after correction for multiple comparisons (P/8 or P/10) showed 2 among the 8 candidate inflammation-related SNPs have significant differences (rs2027432 genotype χ2 = 407.377, p < 0.001, p < 0.00625). Moreover, the minor alleles of rs2027432 T (minor allele χ2 = 450.923, p < 0.001, p < 0.00625; OR = 21.439, 95%CI = 15.181‐30.278) and rs4819554 G (minor allele χ2 = 163.465, p < 0.001, p < 0.00625; OR = 5.814, 95%CI = 4.380‐7.719) were confirmed as risk allele of PE, respectively. Our analysis revealed rs2027432 (TT) of NLRP3 and rs4819554 (GG) of IL-17RA are risk factors for PE. However, no significant difference was found at the oxidative stress-related SNPs. In the candidate loci for oxidative stress, we also identified 3 SNP matches (rs4807542 and rs713041, rs230813 and rs75799, rs1004467 and rs3824755) that had high linkage disequilibrium (LD) with each other and were selected as a block (r2 = 0.98, r2 = 0.97, r2 = 0.97, r2 > 0.9), and the GT and GC haplotypes of rs4807542 and rs713041 in GPX4 showed significant differences between the PE and control groups (χ2 = 5.143, p = 0.0233, p < 0.05; χ2 = 6.373, p = 0.0116, p < 0.05). So, we inferred that polymorphisms of NLRP3 rs2027432 and IL-17RA rs4819554, which are related to inflammation, and the rs713041 variant of GPX4, which is related to oxidative stress, were associated with susceptibility to PE. The GT and GC haplotypes of rs4807542 and rs713041 in GPX4 may increase the risk of PE in the Chinese Han population.
1. Introduction
Preeclampsia (PE) is a serious complication of pregnancy characterized by hypertension and proteinuria after 20 weeks of gestation [1]. It can be accompanied by abnormal changes in the heart, lung, liver, kidney, and other vital organs or the blood system, digestive system, nervous system, and placenta-fetal interface, and thus is a major cause of morbidity and mortality in maternal and fetal medicine [2, 3]. The incidence of PE is estimated to be 3-5% of pregnancies worldwide [4], but 8.1% in developing countries, which the mortality rate for mothers can reach 22.0% [5]. The clinical symptoms of PE are reflected in three aspects; the first involves placental perfusion dysfunction followed by a systemic inflammatory response, the second is vascular endothelial damage, and the last is oxidative stress [1]. These placental factors are released into the maternal body and cause the clinical symptoms of PE [6]. Although studies in recent years have suggested that oxidative stress, inflammatory stimuli, vascular endothelial dysfunction, the immune response, and genetic susceptibility are involved in the development and progression of PE [7, 8], the etiology and mechanism remain elusive.
Th1/Th2 immune status keeps in a steady immune status and plays an important role in normal pregnancy [9]. Th2 cells underlie immune responses mediated by interleukin- (IL-) 4, IL-5, IL-13, and IL-10, whereas Th1 cells are involved in the inflammatory response through interferon-γ (IFN-γ) and IL-2 [10]. Th1 cytokines IL-2, TNF-α, and IFN-γ are significantly increased, while Th2 cytokines IL-4 and IL-10 are significantly reduced PE patients [11]. Obviously, T lymphocytes are inclined toward Th1 cells and produce an increase in Th1 cytokines and a decrease in Th2 cytokines in PE [9, 12]. This unbalanced immunotolerance causes inflammatory cells to be overactive, adheres to the vascular endothelium, and releases inflammatory factors, such as IL family members and the inflammasome, which eventually abnormally remodels the vascular endothelium to cause PE. Overactivation of Th1 cells after combination of IL-33 and IL-1 not only increase the inflammatory response mediated by Th1 but also induce Th1 cells to release IL-12. IL-12 synergizes with IL-27 to induce native CD4 T+ cells to produce increased IFN-γ, which leads to the occurrence of PE [13–15]. In addition, IL-1, bound by the IL-1 receptor family member ST2, initiates NF-κB signaling [16], in which NLRP3, as the core of the inflammatory reaction, plays important roles in the development of PE [17]. Fu et al. indicated that uncontrolled Th17 cells can expand the role of inflammation and tissue damage mediators via IL-17 and IL-22 in PE [18].
Normally, the effect of reactive oxygen species (ROS) can be counteracted by antioxidants, such as glutathione and enzymes, including glutathione S-transferases (GSTs), glutathione peroxidases (GPXs), and cytochrome b-245 alpha chain (CYBA) [19, 20]. Oxidative stress is defined as an imbalance between oxidants and antioxidants in the body in which oxidation is more prone to occur and may be involved in the development of PE [7]. Oxidative stress can also participate in the NF-κB pathway and release inflammatory factors and adhesion molecules, leading to the occurrence of PE [21].
Because genetic factors are involved in the development of PE, in this study, we examined single-nucleotide polymorphisms (SNPs) and haplotypes in inflammation- and oxidative stress-associated candidate genes (inflammation genes IL-22, IL-27, NLRP3, IL-17, and IL-1; oxidative stress genes GSTP1, GPX, COMT, NQO1, SEEP1, CYP17, and CYBA) for susceptibility to PE in a Chinese Han population based on our previous study.
2. Materials and Methods
2.1. Study Population
All PE patients were diagnosed according to guidelines (2015) [22]. The exclusion criteria consisted of chronic hypertension, fetal death, multiple pregnancies, uterine malformation, placental abruption, infection, cancer, in vitro fertilization treatment, gestational diabetes mellitus (GDM), and renal disease or any other potential risk factors for hypertension, including rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE). The control group included women who had no clinical history of PE, with full-term pregnancies and without multiple births, fetal disorder, or any other pathological states. In this present study, a total of 973 patients and 1177 controls were selected from our previous study. That is to say we collected these data of experimental and clinical for the same subjects based on our previous studies for genetic analysis of inflammation-related SNPs in 631 PE patients and 720 normal pregnancy and oxidative stress-related SNPs in 342 PE patients and 457 normal pregnancy for susceptibility to PE in Chinese Han women. The research project was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University.
2.2. PCR Amplification/Genotyping
DNA was extracted from peripheral venous blood samples and stored at -20°C. Genotyping of 8 and 10 candidate SNPs related to inflammatory and oxidative stress, respectively, (Tables 1 and 2) was performed using predesigned TaqMan allelic discrimination real-time PCR followed by partial validation by Sanger sequencing. All women who were genotyped were retrospectively confirmed from our previous studies [23–34].
Table 1.
Genotype distribution and allele frequencies of the human inflammation related gene in cases and controls.
| Genotype frequency of cases | Genotype frequency of controls | Genotype frequency | Minor allele | Minor allele frequencies | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID (allele1/allele2) | Position | 1/1 | 1/2 | 2/2 | 1/1 | 1/2 | 2/2 | χ 2 | p | Case | Control | χ 2 | p | OR | 95% CI | |
| rs2227485 (C/T) | 68253933 | 107 | 188 | 47 | 152 | 234 | 71 | 1.187 | 0.552 | T | 0.412 | 0.411 | 0.001 | 0.971 | 0.996 | 0.815-1.218 |
| rs153109 (C/T) | 28507775 | 60 | 154 | 128 | 62 | 217 | 178 | 2.399 | 0.301 | C | 0.401 | 0.373 | 1.250 | 0.264 | 1.123 | 0.916-1.376 |
| rs17855750 (G/T) | 28503907 | 8 | 64 | 270 | 11 | 104 | 342 | 1.957 | 0.376 | G | 0.117 | 0.138 | 1.521 | 0.217 | 0.828 | 0.614-1.118 |
| rs2027432 (C/T) | 247415139 | 304 | 36 | 2 | 79 | 234 | 144 | 407.377 | ≤0.001 | T | 0.058 | 0.571 | 450.923 | ≤0.001 | 21.439 | 15.181-30.278 |
| rs2275913 (A/G) | 52186235 | 65 | 155 | 122 | 82 | 215 | 160 | 0.270 | 0.874 | A | 0.417 | 0.415 | 0.006 | 0.936 | 1.008 | 0.825-1.233 |
| rs763780 (C/T) | 52236941 | 2 | 59 | 281 | 5 | 68 | 384 | 1.353 | 0.508 | C | 0.092 | 0.085 | 0.223 | 0.637 | 1.087 | 0.768-1.540 |
| rs4819554 (A/G) | 17084145 | 99 | 181 | 62 | 146 | 230 | 81 | 0.848 | 0.654 | G | 0.446 | 0.429 | 163.465 | ≤0.001 | 5.814 | 4.380-7.719 |
| rs13015714 (G/T) | 102355405 | 103 | 156 | 83 | 125 | 232 | 100 | 2.080 | 0.353 | T | 0.471 | 0.473 | 0.006 | 0.940 | 1.008 | 0.826-1.229 |
Table 1 shows the results of inflammation-related genotype distribution and allele frequencies in PE and control groups. After correction for multiple comparisons (P/8), 2 among 8 groups of the candidate inflammation-related SNPs have significant differences (rs2027432 genotype χ2 = 407.377, p < 0.001, p < 0.00625). Moreover, the minor allele of rs2027432 T (minor allele χ2 = 450.923, p < 0.001, p < 0.00625; OR = 21.439, 95%CI = 15.181‐30.278) and rs4819554 G (minor allele χ2 = 163.465, p < 0.001, p < 0.00625; OR = 5.814, 95%CI = 4.380‐7.719) were confirmed as risk allele of PE, respectively. According to Table 1, there were no significant differences in the remaining candidate SNP loci (rs2227485: for genotype, p = 0.552, for allele, χ2 = 0.001, p = 0.971, OR = 0.996, 95%CI = 0.815‐1.218; rs153109: for genotype, p = 0.301, for allele, χ2 = 1.250, p = 0.264, OR = 1.123, 95%CI = 0.916‐1.376; rs17855750: for genotype, p = 0.376, for allele, χ2 = 1.521, p = 0.217, OR = 0.828, 95%CI = 0.614‐1.118; rs2275913: for genotype, p = 0.874, for allele, χ2 = 0.006, p = 0.936, OR = 1.008, 95%CI = 0.825‐1.233; rs763780: for genotype, p = 0.508, for allele, χ2 = 0.233, p = 0.637, OR = 1.087, 95%CI = 0.768‐1.540; rs4819554: for genotype, p = 0.654; rs13015714: for genotype, p = 0.353, for allele, χ2 = 0.006, p = 0.940, OR = 1.008, 95%CI = 0.826‐1.229).
Table 2.
Genotype distribution and allele frequencies of the human oxidative stress-related gene in cases and controls.
| Genotype frequency of cases | Genotype frequency of controls | Genotype frequency | Minor allele | Minor allele frequencies | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID (allele1/allele2) | Position | 1/1 | 1/2 | 2/2 | 1/1 | 1/2 | 2/2 | χ 2 | p | Case | Control | χ 2 | p | OR | 95% CI | |
| rs1695 (A/G) | 67585218 | 394 | 214 | 23 | 452 | 235 | 33 | 0.885 | 0.642 | G | 0.206 | 0.209 | 0.037 | 0.848 | 1.018 | 0.845-1.227 |
| rs4680 (A/G) | 19963748 | 48 | 227 | 356 | 52 | 273 | 395 | 0.120 | 0.758 | A | 0.256 | 0.262 | 0.120 | 0.729 | 0.970 | 0.816-1.153 |
| rs1800566 (C/T) | 69711242 | 177 | 330 | 124 | 219 | 337 | 164 | 4.239 | 0.120 | C | 0.458 | 0.462 | 0.039 | 0.843 | 1.015 | 0.873-1.182 |
| rs4807542 (A/G) | 1104079 | 9 | 118 | 504 | 7 | 149 | 564 | 1.363 | 0.506 | A | 0.108 | 0.113 | 0.201 | 0.654 | 0.946 | 0.743-1.204 |
| rs713041 (C/T) | 1106616 | 216 | 296 | 119 | 204 | 359 | 157 | 5.796 | 0.055 | T | 0.423 | 0.467 | 5.322 | 0.021 | 1.196 | 1.027-1.393 |
| rs7579 (A/G) | 42800706 | 51 | 249 | 331 | 48 | 302 | 370 | 1.502 | 0.472 | A | 0.278 | 0.276 | 0.010 | 0.920 | 1.009 | 0.852-1.194 |
| rs230813 (C/G) | 42798931 | 105 | 276 | 250 | 104 | 353 | 263 | 3.914 | 0.141 | C | 0.385 | 0.390 | 0.057 | 0.811 | 0.981 | 0.840-1.146 |
| rs1004467 (C/T) | 102834750 | 80 | 279 | 272 | 97 | 326 | 297 | 0.522 | 0.770 | C | 0.348 | 0.361 | 0.516 | 0.473 | 0.944 | 0.806-1.105 |
| rs3824755 (C/G) | 102836092 | 72 | 278 | 281 | 85 | 334 | 301 | 1.029 | 0.598 | C | 0.334 | 0.35 | 0.728 | 0.394 | 0.933 | 0.796-1.094 |
| rs9932581 (C/T) | 88651945 | 107 | 337 | 187 | 159 | 355 | 206 | 5.714 | 0.057 | C | 0.437 | 0.467 | 2.567 | 0.109 | 0.883 | 0.759-1.028 |
Table 2 shows the results of oxidative stress-related genotype distribution and allele frequencies in PE and control groups. After correction for multiple comparisons (P/10), there was no significant difference at the oxidative stress-related SNPs among 10 groups, although the SNP rs713041 with a C/T polymorphism, the T allele looks like a risk allele for predisposition to PE (minor allele χ2 = 5.322, p = 0.021, p < 0.05; OR = 1.196, 95%CI = 1.027‐1.393). Similarly, for all other SNPs in Table 2, there were no significant differences in the remaining SNP loci between PE and control groups (rs1695: for genotype, p = 0.642, for allele, χ2 = 0.037, p = 0.848, OR = 1.018, 95%CI = 0.845‐1.227; rs4680: for genotype, p = 0.758, for allele, χ2 = 0.120, p = 0.729, OR = 0.970, 95%CI = 0.816‐1.153; rs1800566: for genotype, p = 0.120, for allele, χ2 = 0.039, p = 0.843, OR = 1.015, 95%CI = 0.873‐1.182; rs4807542: for genotype, p = 0.506, for allele, χ2 = 0.201, p = 0.654, OR = 0.946, 95%CI = 0.743‐1.204; rs713041: for genotype, p = 0.055; rs7579: for genotype, p = 0.472, for allele, χ2 = 0.010, p = 0.920, OR = 1.009, 95%CI = 0.852‐1.194; rs230813: for genotype, p = 0.141, for allele, χ2 = 0.057, p = 0.811, OR = 0.981, 95%CI = 0.840‐1.146; rs1004467: for genotype, p = 0.770, for allele, χ2 = 0.516, p = 0.473, OR = 0.944, 95%CI = 0.806‐1.105; rs3824755: for genotype, p = 0.598, for allele, χ2 = 0.728, p = 0.394, OR = 0.933, 95%CI = 0.796‐1.094; rs9932581: for genotype, p = 0.057, for allele, χ2 = 2.567, p = 0.109, OR = 0.883, 95%CI = 0.759‐1.028).
2.3. Haplotype Analysis
Haplotype analysis was predicted from genotype data by the computer program Haploview 4.2. Only women with all SNPs successfully genotyped were included in the haplotype analysis (ninflammation = 799; ncase = 342, ncontrols = 457; noxidative stress = 1351; ncase = 631, ncontrols = 720).
2.4. Statistical Analysis
All statistical analyses were performed with the statistical software package SPSS 20 (IBM SPSS Statistics 20). The chi-square test was used to calculate genotypic and allelic frequencies and evaluate the Hardy-Weinberg equilibrium (HWE) in the control group to confirm genetic equilibrium. The relative risk is indicated by the ORs and 95% CIs. In Tables 1 and 2 genotype distribution and allele frequency comparisons, the statistical significance after correction for multiple comparisons (P/8 or P/10) is set at p < 0.00625 or p < 0.005. Other than that, statistical significance was set at p < 0.05. Additionally, linkage disequilibrium blocks and haplotype association risk analyses were conducted using the Haploview 4.2 program.
3. Results
3.1. Demographic and Clinical Characteristics of the Study Population
The demographic and clinical characteristics of the PE cohort and normal pregnant women in the inflammation and oxidative stress groups are shown in Table 3. No difference was observed in maternal age, gravidity, or number of abortions among the PE and control groups (all p > 0.05). The PE group had a higher prevalence of preterm birth, and the gestational age at delivery was lower than that of the control group (p < 0.001). The birth weight of newborns in the PE group was lower than that of the control group (p < 0.001). The systolic and diastolic blood pressure values of the PE group were significantly higher than those of the control group (p < 0.001). As shown in Table 3, the number of white blood cells in the PE group was significantly higher than that in the control group (p < 0.001). In the inflammatory group, the PE group had higher neutrophil counts than the control group (p = 0.015); however, no significant difference was found in the neutrophil counts for the oxidative stress group (p = 0.130).
Table 3.
Demographic and clinical characteristics of the inflammation/oxidative stress (1/2) groups.
| Characteristics | PE (342/631) | Controls (457/720) | T | p value |
|---|---|---|---|---|
| 1/2 | 1/2 | 1 vs. 1/2 vs. 2 | 1 vs. 1/2 vs. 2 | |
| Maternal age (years) | 30.45 ± 5.82/30.23 ± 5.59 | 30.80 ± 4.52/30.15 ± 3.92 | -0.930/0.295 | 0.353/0.768 |
| Gravidity (times) | 2.25 ± 1.21/2.25 ± 1.27 | 2, 38 ± 1.24/2.19 ± 1.08 | -1.422/1.001 | 0.156/0.317 |
| Abortion (times) | 0.67 ± 0.91/0.62 ± 0.93 | 0.75 ± 0.94/0.62 ± 0.80 | -1.290/0.016 | 0.197/0.987 |
| Gestational age (weeks) | 34.59 ± 3.81/35.00 ± 3.67 | 38.71 ± 1.60/39.15 ± 1.43 | -18.812/-26.697 | <0.001/<0.001 |
| Gestational age at delivery (weeks) | 35.27 ± 3.10/35.56 ± 3.32 | 39.01 ± 1.32/39.45 ± 1.11 | -20.928/-28.067 | <0.001/<0.001 |
| Birth weight (kg) | 2.38 ± 0.90/2.48 ± 0.92 | 3.38 ± 0.35/3.43 ± 0.34 | -19.467/-24.676 | <0.001/<0.001 |
| Systolic blood pressure (mmHg) | 161.49 ± 19.48/160.74 ± 19.22 | 113.78 ± 10.29/114.24 ± 9.36 | 41.197/55.286 | <0.001/<0.001 |
| Diastolic blood pressure (mmHg) | 105.06 ± 14.32/104.97 ± 14.05 | 74.19 ± 7.87/73.29 ± 7.84 | 36.004/50.194 | <0.001/<0.001 |
| White blood cells (×109/L) | 9.80 ± 2.95/9.79 ± 3.86 | 8.87 ± 2.49/7.33 ± 3.78 | 4.733/4.421 | <0.001/<0.001 |
| Neutrophil (×109/L) | 7.34 ± 2.69/8.99 ± 2.51 | 6.76 ± 3.71/7.03 ± 3.29 | 2.433/1.515 | 0.015/0.130 |
3.2. The Distribution of Genotypes and Allele Frequency
The collected control samples in the study conformed to HWE (rs2227485: χ2 = 1.500, p = 0.221; rs153109 : χ2 = 0.104, p = 0.747; rs17855750 : χ2 = 0.830, p = 0.362; rs2027432 : χ2 = 0.934, p = 0.334; rs2275913 : χ2 = 0.435, p = 0.510; rs763780 : χ2 = 1.004, p = 0.316; rs4819554 : χ2 = 0.342, p = 0.559; rs13015714 : χ2 = 0.154, p = 0.695; rs1695 : χ2 = 0.121, p = 0.728; rs4680 : χ2 = 0.261, p = 0.609; rs1800566 : χ2 = 2.455, p = 0.117; rs4807542 : χ2 = 0.683, p = 0.409; rs713041 : χ2 = 0.002, p = 0.968; rs7579 : χ2 = 1.702, p = 0.192; rs230813 : χ2 = 0.684, p = 0.408; rs1004467 : χ2 = 0.253, p = 0.615; rs3824755 : χ2 = 0.275, p = 0.600; rs9932581 : χ2 = 0.067, p = 0.795). The distributions of the genotypic and allelic frequencies are shown in Tables 1 and 2.
Table 1 shows the inflammation-related genotype distribution and allele frequencies in the PE and control groups. Significant differences were observed for the 2 SNPs among the PE and control groups (rs2027432 genotype χ2 = 407.377, p ≤ 0.00625). Moreover, the minor alleles of rs2027432 T (minor allele χ2 = 450.923, p < 0.001, p < 0.00625; OR = 21.439, 95%CI = 15.181‐30.278) and rs4819554 G (minor allele χ2 = 163.465, p < 0.001, p < 0.00625; OR = 5.814, 95%CI = 4.380‐7.719) were confirmed as risk alleles for PE. According to Table 1, no significant differences were observed in the remaining candidate SNP loci (rs2227485, rs153109, rs17855750, rs2275913, rs763780, and rs13015714).
For the oxidative stress-related genotype distribution and allele frequencies in the PE and control groups, there was no significant difference at the oxidative stress-related SNPs among 10 groups, although the SNP rs713041 with a C/T polymorphism, the T allele looked like a risk allele for predisposition to PE (minor allele χ2 = 5.322, p = 0.021, p < 0.05; OR = 1.196, 95%CI = 1.027‐1.393). Similarly, for all other SNPs in Table 2, there were no significant differences in the remaining SNP loci between PE and control groups.
To further investigate the relationship between the genetic distributions of the PE and control groups, we compared 3 SNPs (rs2027432, rs4819554, rs713041) based on PE classification and staging. First, we divided PE patients into mild and severe PE groups [35]. Table 4 shows the genetic distributions of the mild/severe PE and control groups. The results showed a significant difference in the genetic distribution of rs2027432 in NLRP3 among the mild/severe PE and control groups (mild PE vs. control: for genotype, p < 0.001, for allele, χ2 = 101.849, p < 0.001, OR = 31.959, 95%CI = 11.655‐87.636; severe PE vs. control: for genotype, p < 0.001, for allele, χ2 = 395.685, p < 0.001, OR = 20.270, 95%CI = 14.117‐29.106). However, no significant differences were found in another candidate SNP, rs4819554 in IL17RA (mild PE vs. control: for genotype, p = 0.283, for allele, χ2 = 2.411, p = 0.121, OR = 0.722, 95%CI = 0.477‐1.091; severe PE vs. control: p = 0.808, for allele, χ2 = 0.053, p = 0.818, OR = 0.976, 95%CI = 0.791‐1.203). Table 4 shows a strong association of the genetic distribution of rs713041 in GPX4 between the severe PE and control groups (for allele, χ2 = 5.198, p = 0.023, OR = 1.210, 95%CI = 1.027‐1.425).
Table 4.
The comparison of genetic distributions between mild/severe PE and control groups.
| Group |
N
I/II |
rs2027432 (I) | rs4819554 (I) | rs713041 (II) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | AA | AG | GG | A | G | CC | CT | TT | C | T | ||
| Mild PE | 50/141 | 46 | 4 | 0 | 96 | 4 | 11 | 27 | 12 | 49 | 51 | 47 | 66 | 28 | 160 | 122 |
| Control | 457/720 | 79 | 234 | 144 | 392 | 522 | 146 | 230 | 81 | 522 | 392 | 204 | 359 | 157 | 767 | 673 |
| χ 2 | 135.716 | 101.849 | 2.528 | 2.411 | 1.442 | 1.145 | ||||||||||
| p | <0.001 | <0.001 | 0.283 | 0.121 | 0.486 | 0.285 | ||||||||||
| OR | 31.959 | 0.722 | 1.151 | |||||||||||||
| 95% CI | 11.655-87.636 | 0.477-1.091 | 0.890-1.488 | |||||||||||||
| Severe PE | 292/490 | 258 | 32 | 2 | 548 | 36 | 88 | 154 | 50 | 330 | 254 | 169 | 230 | 91 | 568 | 412 |
| Control | 457/720 | 79 | 234 | 144 | 392 | 522 | 146 | 230 | 81 | 522 | 392 | 204 | 359 | 157 | 767 | 673 |
| χ 2 | 368.100 | 395.685 | 0.426 | 0.053 | 5.584 | 5.198 | ||||||||||
| p | <0.001 | <0.001 | 0.808 | 0.818 | 0.061 | 0.023 | ||||||||||
| OR | 20.270 | 0.976 | 1.210 | |||||||||||||
| 95% CI | 14.117-29.106 | 0.791-1.203 | 1.027-1.425 | |||||||||||||
Table 4 shows the results of genetic distributions between mild/severe PE and control groups. The results show rs2027432 in NLRP3 significant difference between mild/severe PE and control groups in the genetic distributions (mild PE vs. control: for genotype, p < 0.001, for allele, χ2 = 101.849, p < 0.001, OR = 31.959, 95%CI = 11.655‐87.636; severe PE vs. control: for genotype, p < 0.001, for allele, χ2 = 395.685, p < 0.001, OR = 20.270, 95%CI = 14.117‐29.106). While no significant differences were found in another candidate SNP rs4819554 in IL17RA (mild PE vs. control: for genotype, p = 0.283, for allele, χ2 = 2.411, p = 0.121, OR = 0.722, 95%CI = 0.477‐1.091; severe PE vs. control: p = 0.808, for allele, χ2 = 0.053, p = 0.818, OR = 0.976, 95%CI = 0.791‐1.203). In Table 4, it also showed that there was a strong association in the genetic distributions of rs713041 in GPX4 between severe PE and control groups (for allele, χ2 = 5.198, p = 0.023, OR = 1.210, 95%CI = 1.027‐1.425).
Early-onset PE was diagnosed before 34 weeks of gestation, and PE diagnosed at or after 34 weeks of gestation was considered late-onset PE [36]. Table 5 shows a strong association in the genetic distribution of rs2027432 in NLRP3 between the early-onset PE and control groups and between the late-onset PE and control groups (early-onset PE vs. control: for genotype, p < 0.001, for allele, χ2 = 294.95, p < 0.001, OR = 24.881, 95%CI = 15.399‐40.199; late-onset PE vs. control: p < 0.001, for allele, χ2 = 135.044, p < 0.001, OR = 10.335, 95%CI = 6.512‐16.402), whereas no significant differences were found in another candidate SNP, rs4819554 in IL17RA (early-onset PE vs. control: for genotype, p = 0.315, for allele, χ2 = 0.494, p = 0.482, OR = 1.096, 95%CI = 0.849‐1.414; late-onset PE vs. control: p = 0.265, for allele, χ2 = 0.242, p = 0.623, OR = 1.068, 95%CI = 0.822‐1.387). Table 5 also shows a strong association in the genetic distribution of rs713041 in GPX4 between the early-onset PE and control groups (for genotype, p = 0.018, for allele, χ2 = 7.280, p = 0.007, OR = 1.329, 95%CI = 1.081‐1.636).
Table 5.
The comparison of genetic distributions between early/late-onset PE and control groups.
| Group |
N
I/II |
rs2027432(I) | rs4819554(I) | rs713041(II) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | AA | AG | GG | A | G | CC | CT | TT | C | T | ||
| Early-onset PE | 187/249 | 168 | 19 | 0 | 355 | 19 | 52 | 93 | 42 | 197 | 135 | 94 | 112 | 43 | 300 | 198 |
| Control | 457/720 | 79 | 234 | 144 | 392 | 522 | 146 | 230 | 81 | 522 | 392 | 204 | 359 | 157 | 767 | 673 |
| χ 2 | 297.949 | 294.95 | 2.307 | 0.494 | 8.088 | 7.280 | ||||||||||
| p | <0.001 | <0.001 | 0.315 | 0.482 | 0.018 | 0.007 | ||||||||||
| OR | 24.881 | 1.096 | 1.329 | |||||||||||||
| 95% CI | 15.399-40.199 | 0.849-1.414 | 1.081-1.636 | |||||||||||||
| Late-onset PE | 155/382 | 136 | 17 | 2 | 289 | 21 | 47 | 88 | 20 | 182 | 128 | 122 | 184 | 76 | 428 | 336 |
| Control | 457/720 | 79 | 234 | 144 | 392 | 522 | 146 | 230 | 81 | 522 | 392 | 204 | 359 | 157 | 767 | 673 |
| χ 2 | 253.539 | 135.044 | 2.653 | 0.242 | 1.672 | 1.529 | ||||||||||
| p | <0.001 | <0.001 | 0.265 | 0.623 | 0.433 | 0.216 | ||||||||||
| OR | 10.335 | 1.068 | 1.118 | |||||||||||||
| 95% CI | 6.512-16.402 | 0.822-1.387 | 0.937-1.333 | |||||||||||||
Table 5 shows that there existed a strong association in the genetic distributions of rs2027432 in NLRP3 between early-onset PE and control groups, late-onset PE and control groups (early-onset PE vs. control: for genotype, p < 0.001, for allele, χ2 = 294.95, p < 0.001, OR = 24.881, 95%CI = 15.399‐40.199; late-onset PE vs. control: p < 0.001, for allele, χ2 = 135.044, p < 0.001, OR = 10.335, 95%CI = 6.512‐16.402), while no significant differences were found in another candidate SNP rs4819554 in IL17RA (early-onset PE vs. control: for genotype, p = 0.315, for allele, χ2 = 0.494, p = 0.482, OR = 1.096, 95%CI = 0.849‐1.414; late-onset PE vs. control: p = 0.265, for allele, χ2 = 0.242, p = 0.623, OR = 1.068, 95%CI = 0.822‐1.387). In Table 5, it also showed that there was a strong association in the genetic distributions of rs713041 in GPX4 between early-onset PE and control groups (for genotype, p = 0.018, for allele, χ2 = 7.280, p = 0.007, OR = 1.329, 95%CI = 1.081‐1.636).
3.3. LD and Haplotype Analysis
To further examine the association of the candidate SNPs between the PE and control groups, we estimated the LD and haplotype using Haploview 4.2; rs153109 and rs17855750 (IL-27) and rs2275913 and rs763780 (IL-17) were in low LD with each other (r2 = 0.49 and r2 = 0.43, r2 < 0.8, Figure 1. Inflammation-LD plot).
Figure 1.
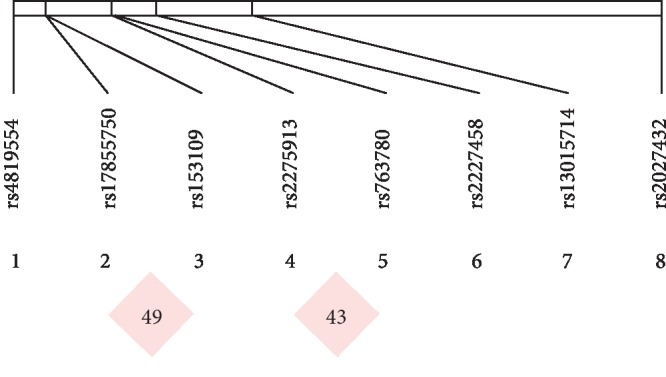
Inflammation-LD plot. We estimated the LD and haplotype using Haploview 4.2; rs153109 and rs17855750 (IL-27) and rs2275913 and rs763780 (IL-17) were in low LD with each other (r2 = 0.49 and r2 = 0.43, r2 < 0.8).
We also estimated the LD and haplotype of the oxidative stress-related candidate SNPs. Three SNPs had strong correlations; rs4807542 and rs713041 (GPX4), rs230813 and rs75799 (SEPP1), and rs1004467 and rs3824755 (CYP17A1) were in high LD with each other and were selected as a block (r2 = 0.98, r2 = 0.97, r2 = 0.97, r2 > 0.9, Figure 2. Oxidative stress-LD plot). rs713041 in GPX4 exhibited high LD (r2 = 0.98) with rs4807542, and both were significantly associated with PE. Table 6 shows the haplotype associations of oxidative stress-related SNPs between the PE and control groups, and two polymorphisms were found (rs4807542/rs713041), indicating that the two primary haplotypes were significantly different. The GT and GC haplotypes from block 1 exhibited the following distribution: 44.6% GT (rs4807542/rs713041), 44.3% GC, and 11% AC. Significant differences in the GT and GC haplotypes were found between the PE and control groups (χ2 = 5.143, p = 0.0233, p < 0.05; χ2 = 6.373, p = 0.0116, p < 0.05), whereas no differences were found for the remaining haplotypes.
Figure 2.
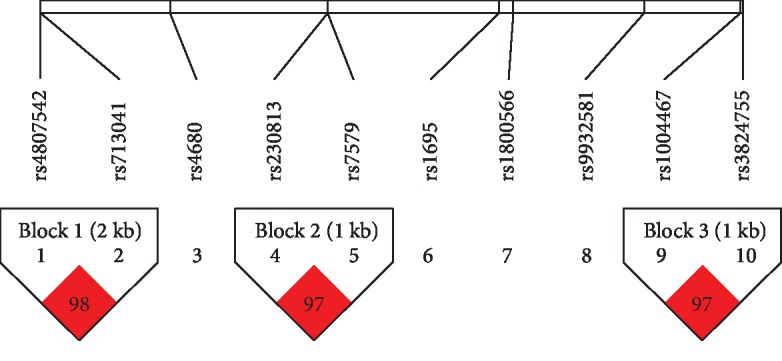
Oxidative stress-LD plot. We estimated the LD and haplotype using Haploview 4.2. Three SNPs had strong correlations; rs4807542 and rs713041(GPX4), rs230813 and rs75799 (SEPP1), and rs1004467 and rs3824755 (CYP17A1) were in high LD with each other and were selected as a block (r2 = 0.98, r2 = 0.97, r2 = 0.97, r2 > 0.9, t). rs713041 in GPX4 exhibited high LD (r2 = 0.98) with rs4807542, and both were significantly associated with PE.
Table 6.
The haplotype associations of oxidative stress-related SNPs between PE and controls.
| Freq. | PE, control ratio counts | PE, control frequencies | Chi-square | p value | |
|---|---|---|---|---|---|
| Block 1 | |||||
| GT | 0.446 | 534.0 : 728.0, 671.0 : 767.0 | 0.423, 0.467 | 5.143 | 0.0233 |
| GC | 0.443 | 592.0 : 670.0, 605.0 : 833.0 | 0.469, 0.421 | 6.373 | 0.0116 |
| AC | 0.11 | 136.0 : 1126.0, 162.0 : 1276.0 | 0.108, 0.113 | 0.164 | 0.6857 |
| Block 2 | |||||
| GG | 0.608 | 771.4 : 490.6, 871.2 : 568.8 | 0.611, 0.605 | 0.109 | 0.7414 |
| CA | 0.273 | 346.4 : 915.6, 390.2 : 1049.8 | 0.274, 0.271 | 0.041 | 0.8397 |
| CG | 0.115 | 139.6 : 1122.4, 170.8 : 1269.2 | 0.111, 0.119 | 0.418 | 0.5177 |
| Block 3 | |||||
| TG | 0.639 | 818.9 : 443.1, 908.9 : 531.1 | 0.649, 0.631 | 0.916 | 0.3385 |
| CC | 0.337 | 417.9 : 844.1, 492.9 : 947.1 | 0.331, 0.342 | 0.373 | 0.5412 |
| CG | 0.018 | 21.1 : 1240.9, 27.1 : 1412.9 | 0.017, 0.019 | 0.171 | 0.6792 |
Apparently, Table 6 indicates the haplotype associations of oxidative stress-related SNPs between PE and controls; there were two polymorphisms (rs4807542/rs713041) stated two primary haplotypes had significant difference, that is the GT and GC haplotypes from block 1, which produced the following distribution: 44.6% GT (rs4807542/rs713041), 44.3% GC, and 11% AC. Significantly statistical differences were identified in haplotype GT and GC between PE and control groups (χ2 = 5.143, p = 0.0233, p < 0.05; χ2 = 6.373, p = 0.0116, p < 0.05), while there were no differences in the rest of haplotypes.
4. Discussion
PE is one of the most common and severe obstetric complications and is characterized by a state of excessive inflammatory and oxidative stress. In the second trimester of pregnancy, Th cells play a significant role in the development of PE [10]. Previous studies have indicated that PE is an excessive inflammatory response and associated with a Th1/Th2 immune imbalance in the maternal body. Normal pregnancy associated with a mild inflammatory Th2-based state favors the maternal and fetal environment. In contrast, PE is a proinflammatory state characterized by a Th1-based state [10, 14]. Many studies also suggested that PE may be due to an increase in Th17 cells and a decrease in Treg cells [10, 37–39]. As the core of the inflammatory response, NLRP3 can be activated by many danger signals to exert an immune response to promote the development of PE [17]. IL-27 regulates the differentiation of T cells in the initial stage and plays a crucial role in promoting T1 differentiation and enhancing the activity of T1 cells [40]. Moreover, IL-27 can inhibit the differentiation of Th17 cells by inhibiting the polarization of naive CD4+ T cells [41]; in this case, IL-27 induces a variety of biological activities, which may cause the inflammatory response to induce the occurrence of PE [18, 42]. IL-27 promotes Th1 cell differentiation and activity by participating in Th initial differentiation [40] and inhibits Th2 and Th17 activation [41, 43]. The imbalance of Th1/Th2 and Th17/Treg may lead to maternal in the PE susceptibility state. The most important role of IL-17 is to amplify the inflammatory reaction of small vascular endothelial cells, which damages vascular endothelial cells, increases the permeability of blood vessels, and results in the release of a large number of oxygen free radicals. IL-22 combined with IL-22R to activate the JAK1 (mobile kinase IL22R1), Tyk2 (mobile kinase IL10R2), and multiple biological pathways, such as AKT, P38, JNK, and ERK1/2, by phosphorylation of serine and tyrosine in STAT 1, 3, and 5, which finally keeps immune homeostasis [44]. A research found the higher level of IL-22 in the PE mother and newborn cord blood compared with controls [45].
Normally, the effect of ROS can be counteracted by antioxidants, including glutathione and enzymes, such as GSTs, GPXs, CYBA, NQO1, SEEP1, and superoxide dismutase [19, 20]. As a selenoprotein, GPX4 exhibits high antioxidant activity in the body to repress the development of oxidative stress, which promotes the development of PE [46]. In addition, polymorphisms of GPX4 may affect the expression and antioxidant activity of GPX4 [47]. COMT plays a crucial role in the degradation of both catecholamines and estrogens [48]. During oxidative stress which is an imbalance between oxidants and antioxidants in the body, oxidation is favored, leading to increased inflammatory infiltration and protease secretion. Additionally, activation of the inflammatory response and oxidative stress in the placenta is closely related to the occurrence of PE.
As PE is a complex multigene hereditary disease that was not only associated with many cytokine candidate genes, such as IL-1 [32, 49], IL-17, IL-22 [39], NLRP1 [50], and vascular-associated genes [51] but also associated with many oxidative stress genes such as GSTs, GPXs, CYBA, NQO1, and SEEP1 [46, 52–58]. In the published previous studies, we have genotyped for SNPs that related to inflammation (rs2227485, rs153109, rs17855750, rs2027432, rs2275913, rs763780, rs4819554, and rs13015714) and oxidative stress (rs1695, rs4680, rs1800566, rs4807542, rs713041, rs7579, rs230813, rs1004467, rs3824755, and rs9932581) to investigate whether these polymorphisms were associated with susceptibility to PE in a Chinese Han population. We found significant difference for the genotype of IL-22 rs2227485 and GPX4 rs713041 associated with the mild, severe, and early-onset PE. Furthermore, the GPX4 rs713041 C allele has the higher risk for pathogenesis of PE [27, 29]. Those that are just single genes without systematic haplotype analysis. Thus, in this present study, we collected these data of experimental and clinical for the same subjects from above studies for genetic contribution and haplotypes of polymorphisms of inflammation-related SNPs in 631 PE patients and 720 normal pregnancy and oxidative stress-related SNPs in 342 PE patients and 457 normal pregnancies for susceptibility to PE in Chinese Han women.
Our study found significant differences in the genetic distributions of rs2027432 in NLRP3, rs4819554 in IL-17RA, and rs713041 in GPX4 between the PE and control groups. We further divided the PE group into mild/severe and early/late-onset subgroups and compared them with the control group. Significant differences were also found for rs2027432 in NLRP3 and rs713041 in GPX4; however, no significant difference was found in the subgroup analysis of rs4819554 in IL-17RA. Our results suggest that the two SNPs, rs2027432 in NLRP3 and rs713041 in GPX4, may be associated with risks for PE in Chinese Han women.
Analysis of LD showed that rs153109 and rs17855750 in IL-27 and rs2275913 and rs763780 in IL-17 were in low LD with each other (r2 < 0.8, Figure 1. Inflammation-LD plot), indicating that no substitution occurs between them. Unfortunately, no haplotype formation was found in the analysis of inflammatory factors, partly due to an imbalance in the HWE or the scatter position distribution of SNPs. Therefore, such contradictory results suggest that our findings should be validated using large samples that include different countries.
However, we identified 3 SNP matches, rs4807542 and rs713041 (GPX4), rs230813 and rs75799 (SEPP1), and rs1004467 and rs3824755 (CYP17A1), among oxidative stress genes that were in high LD, and significant substitutability with each other was observed. The analysis of haplotype correlation showed that the GT and GC haplotypes in block 1 (rs4807542/rs713041) were significantly different, which suggested that pregnant women carrying the GT and/or GC haplotypes were more likely to suffer from PE. To our knowledge, this is the first study of correlations of inflammation and oxidative stress with PE susceptibility in a Chinese Han population with both an LD and haplotype analysis. However, our findings should be confirmed in individuals of different races and geographic locations. Our previous studies demonstrated that the two SNPs (rs2227485 in IL-22 and s713041 in GPX4) are associated with risks for PE. We found that the rs2227485 in IL-22 showed a significant difference in the allele for the early-onset PE group and the genotype of the late-onset PE and control subgroups. The GPX4 rs713041 allele C was associated with an increased risk for PE in a previous study. Additionally, the rs713041 genotype was associated with the mild, severe, and early-onset PE. These genes may play a key role in the pathogenesis of PE.
In conclusion, we found that oxidative stress and the inflammatory response may play an inseparable role in the progression of PE, which provides the basis for revealing the genetic mechanism of PE. As few studies have performed a haplotype analysis of candidate genes related to inflammatory cytokines and oxidative stress in PE, further experiments are needed to verify these findings.
Acknowledgments
We are grateful to all participants who have made contributions of the completion of this study. This work was supported by the Natural Science Foundation of Shandong Province (ZR2019MH127).
Data Availability
All data used to support the findings of this study are included within the article.
Ethical Approval
This study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (2013-01-07).
Conflicts of Interest
All authors claim no conflict of interest.
Authors' Contributions
Aiping Chen and Shiguo Liu conceived and designed the experiments. Huifang Zhao, Jingli Wang, and Ru Zhang analyzed the data. Huifang Zhao, Jingli Wang, Jingjing Liu, Xin Zhao, Congying Li, Xuewen Jia, Xueying Li, Yan Lin, Mingzhen Guo, Sai Li, Chao Liu, and Yuan Li contributed the data. Huifang Zhao wrote the paper. Aiping Chen and Shiguo Liu revised the paper. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work. Aiping Chen and Huifang Zhao are co-first author.
References
- 1.Hansson S. R., Naav A., Erlandsson L. Oxidative stress in preeclampsia and the role of free fetal hemoglobin. Frontiers in Physiology. 2014;5:p. 516. doi: 10.3389/fphys.2014.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell K. H., Savitz D., Werner E. F., et al. Maternal morbidity and risk of death at delivery hospitalization. Obstetrics and Gynecology. 2013;122(3):627–633. doi: 10.1097/AOG.0b013e3182a06f4e. [DOI] [PubMed] [Google Scholar]
- 3.Khan K. S., Wojdyla D., Say L., Gülmezoglu A. M., van Look P. F. A. WHO analysis of causes of maternal death: a systematic review. Lancet. 2006;367(9516):1066–1074. doi: 10.1016/S0140-6736(06)68397-9. [DOI] [PubMed] [Google Scholar]
- 4.Ramos J. G. L., Sass N., Costa S. H. M. Preeclampsia. Rev Bras Ginecol Obstet: revista da Federacao Brasileira das Sociedades de Ginecologia e Obstetricia. 2017;39(9):496–512. doi: 10.1055/s-0037-1604471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordano J. C., Parpinelli M. A., Cecatti J. G., et al. The burden of eclampsia: results from a multicenter study on surveillance of severe maternal morbidity in Brazil. PLoS One. 2014;9(5):p. e97401. doi: 10.1371/journal.pone.0097401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts J. M., Hubel C. A. The two stage model of preeclampsia: variations on the theme. Placenta. 2009;30(Suppl A):32–37. doi: 10.1016/j.placenta.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abad C., Proverbio T., Piñero S., et al. Preeclampsia, placenta, oxidative stress, and PMCA. Hypertension in Pregnancy. 2010;31(4):427–441. doi: 10.3109/10641955.2012.690058. [DOI] [PubMed] [Google Scholar]
- 8.de Sousa Rocha V., Della Rosa F. B., Ruano R., Zugaib M., Colli C. Association between magnesium status, oxidative stress and inflammation in preeclampsia: a case-control study. Clinical Nutrition. 2015;34(6):1166–1171. doi: 10.1016/j.clnu.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Marzi M., Vigano A., Trabattoni D., et al. Characterization of type 1 and type 2 cytokine production profile in physiologic and pathologic human pregnancy. Clinical and Experimental Immunology. 1996;106(1):127–133. doi: 10.1046/j.1365-2249.1996.d01-809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito S., Sakai M. Th1/Th2 balance in preeclampsia. Journal of Reproductive Immunology. 2003;59(2):161–173. doi: 10.1016/s0165-0378(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 11.Darmochwal-Kolarz D., Oleszczuk J. The critical role of Th17 cells, Treg cells and co-stimulatory molecules in the development of pre-eclampsia. Developmental Period Medicine. 2014;18(2):141–147. [PubMed] [Google Scholar]
- 12.Saito S., Umekage H., Sakamoto Y., et al. Increased T-helper-1-type immunity and decreased T-helper-2-type immunity in patients with preeclampsia. American Journal of Reproductive Immunology. 1999;41(5):297–306. doi: 10.1111/j.1600-0897.1999.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 13.Hamzaoui K., Bouali E., Hamzaoui A. Interleukin-33 and Behçet disease: another cytokine among others. Human Immunology. 2015;76(5):301–306. doi: 10.1016/j.humimm.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Laresgoiti-Servitje E., Gomez-Lopez N., Olson D. M. An immunological insight into the origins of pre-eclampsia. Human Reproduction Update. 2010;16(5):510–524. doi: 10.1093/humupd/dmq007. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y., Dutz J. P., MacCalman C. D., Yong P., Tan R., von Dadelszen P. Decidual NK cells alter in vitro first trimester extravillous cytotrophoblast migration: a role for IFN-γ. Journal of Immunology. 2006;177(12):8522–8530. doi: 10.4049/jimmunol.177.12.8522. [DOI] [PubMed] [Google Scholar]
- 16.Kakkar R., Lee R. T. The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nature Reviews Drug Discovery. 2008;7(10):827–840. doi: 10.1038/nrd2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Nardo D., Latz E. NLRP3 inflammasomes link inflammation and metabolic disease. Trends in Immunology. 2011;32(8):373–379. doi: 10.1016/j.it.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu B., Tian Z., Wei H. TH17 cells in human recurrent pregnancy loss and pre-eclampsia. Cellular & Molecular Immunology. 2014;11(6):564–570. doi: 10.1038/cmi.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poranen A. K., Ekblad U., Uotila P., Ahotupa M. Lipid peroxidation and antioxidants in normal and pre-eclamptic pregnancies. Placenta. 1996;17(7):401–405. doi: 10.1016/s0143-4004(96)90021-1. [DOI] [PubMed] [Google Scholar]
- 20.Seidegård J., Ekström G. The role of human glutathione transferases and epoxide hydrolases in the metabolism of xenobiotics. Environmental Health Perspectives. 1997;105(Suppl 4):791–799. doi: 10.1289/ehp.97105s4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaughan J. E., Walsh S. W. Activation of NF-κB in placentas of women with preeclampsia. Hypertension in Pregnancy. 2012;31(2):243–251. doi: 10.3109/10641955.2011.642436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain V. Diagnosis, Evaluation, and Management of the Hypertensive Disorders of Pregnancy: Executive Summary. Journal of Obstetrics and Gynaecology Canada. 2015;37(9):774–775. doi: 10.1016/S1701-2163(15)30145-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X., Liu J., Zhao C., et al. Association between COMT Val158Met polymorphism and preeclampsia in the Chinese Han population. Hypertension in Pregnancy. 2016;35(4):565–572. doi: 10.1080/10641955.2016.1211677. [DOI] [PubMed] [Google Scholar]
- 24.Liu S., Li X., Wang J., et al. The rs9932581 and rs1049255 polymorphisms in CYBA is not associated with preeclampsia in Chinese Han women. Cellular Physiology and Biochemistry : International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2016;39(4):1471–1478. doi: 10.1159/000447850. [DOI] [PubMed] [Google Scholar]
- 25.Gao H., Liu C., Lin P., et al. Effects of GSTP1 and GPX1 polymorphisms on the risk of preeclampsia in Chinese Han women. Cellular Physiology and Biochemistry : International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2016;39(5):2025–2032. doi: 10.1159/000447898. [DOI] [PubMed] [Google Scholar]
- 26.Ren X., Guo M., Liu C., et al. A case-control study indicates that no association exists between polymorphisms of IL-33 and IL-1RL1 and preeclampsia. Cellular Physiology and Biochemistry : International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2016;38(4):1406–1414. doi: 10.1159/000443083. [DOI] [PubMed] [Google Scholar]
- 27.Niu Z., Zhao X., Liu H., et al. Impact of IL-22 and IL-22 receptor alpha 1 polymorphisms on preeclampsia risk in Chinese Han women. Journal of Cellular Biochemistry. 2018;119(6):4656–4663. doi: 10.1002/jcb.26640. [DOI] [PubMed] [Google Scholar]
- 28.Liu B., Li Y., Yao Y., et al. Polymorphisms of the IL27 gene in a Chinese Han population complicated with pre-eclampsia. Scientific Reports. 2016;6(1) doi: 10.1038/srep23029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng X., Lin Y., Li J., et al. Evaluation of glutathione peroxidase 4 role in preeclampsia. Scientific Reports. 2016;6(1) doi: 10.1038/srep33300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H., Jia X., Zhao H., et al. Identification of SEPP1 polymorphisms is not a genetic risk factor for preeclampsia in Chinese Han women: a clinical trial and experimental study. Medicine. 2017;96(28):p. e7249. doi: 10.1097/MD.0000000000007249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao L., Liu J., Tan P., et al. Genetic association of the NQO1 rs1800566 (609C >T) variant with risk of preeclampsia in the Chinese Han population. Pregnancy Hypertension. 2017;10:42–45. doi: 10.1016/j.preghy.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Jiang F., Liang Y., et al. Interleukin-1β-31C/T and -511T/C polymorphisms were associated with preeclampsia in Chinese Han population. PLoS One. 2014;9(9):p. e106919. doi: 10.1371/journal.pone.0106919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Guo M., Liu F., et al. Role of IL-17 variants in preeclampsia in Chinese Han women. PloS One. 2015;10(10):p. e0140118. doi: 10.1371/journal.pone.0140118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Guo M., Li S., et al. The Role of theIL-12polymorphism rs3212227 in preeclampsia in Chinese Han Women. Clinical and Experimental Hypertension. 2016;38(4):388–392. doi: 10.3109/10641963.2015.1131289. [DOI] [PubMed] [Google Scholar]
- 35.Qublan H., Ammarin V., Bataineh O., et al. Lactic dehydrogenase as a biochemical marker of adverse pregnancy outcome in severe pre-eclampsia. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2005;11(8):Cr393–Cr397. [PubMed] [Google Scholar]
- 36.Alvarez-Fernandez I., Prieto B., Rodriguez V., Ruano Y., Escudero A. I., Alvarez F. V. New biomarkers in diagnosis of early onset preeclampsia and imminent delivery prognosis. Clinical Chemistry and Laboratory Medicine. 2014;52(8):1159–1168. doi: 10.1515/cclm-2013-0901. [DOI] [PubMed] [Google Scholar]
- 37.Vargas-Rojas M. I., Solleiro-Villavicencio H., Soto-Vega E. Th1, Th2, Th17 and Treg levels in umbilical cord blood in preeclampsia. The Journal of Maternal-Fetal & Neonatal Medicine. 2015;29(10):1642–1645. doi: 10.3109/14767058.2015.1057811. [DOI] [PubMed] [Google Scholar]
- 38.Darmochwal-Kolarz D., Kludka-Sternik M., Tabarkiewicz J., et al. The predominance of Th17 lymphocytes and decreased number and function of Treg cells in preeclampsia. Journal of Reproductive Immunology. 2012;93(2):75–81. doi: 10.1016/j.jri.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Liu H., Shi Y., et al. Increased circulating Th22 cells correlated with Th17 cells in patients with severe preeclampsia. Hypertension in Pregnancy. 2017;36(1):100–107. doi: 10.1080/10641955.2016.1239737. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida H., Nakaya M., Miyazaki Y. Interleukin 27: a double-edged sword for offense and defense. Journal of Leukocyte Biology. 2009;86(6):1295–1303. doi: 10.1189/jlb.0609445. [DOI] [PubMed] [Google Scholar]
- 41.Diveu C., McGeachy M. J., Boniface K., et al. IL-27 blocks RORc expression to inhibit lineage commitment of Th17 cells. Journal of Immunology. 2009;182(9):5748–5756. doi: 10.4049/jimmunol.0801162. [DOI] [PubMed] [Google Scholar]
- 42.Yin N., Zhang H., Luo X., et al. IL-27 activates human trophoblasts to express IP-10 and IL-6: implications in the immunopathophysiology of preeclampsia. Mediators of Inflammation. 2014;2014:10. doi: 10.1155/2014/926875.926875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimoto T., Yoshimoto T., Yasuda K., Mizuguchi J., Nakanishi K. IL-27 suppresses Th2 cell development and Th2 cytokines production from polarized Th2 cells: a novel therapeutic way for Th2-mediated allergic inflammation. Journal of Immunology. 2007;179(7):4415–4423. doi: 10.4049/jimmunol.179.7.4415. [DOI] [PubMed] [Google Scholar]
- 44.Shabgah A. G., Navashenaq J. G., Shabgah O. G., Mohammadi H., Sahebkar A. Interleukin-22 in human inflammatory diseases and viral infections. Autoimmunity Reviews. 2017;16(12):1209–1218. doi: 10.1016/j.autrev.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Bersani I., de Carolis M. P., Foell D., et al. Interleukin-22: biomarker of maternal and fetal inflammation? Immunologic Research. 2015;61(1-2):4–10. doi: 10.1007/s12026-014-8568-2. [DOI] [PubMed] [Google Scholar]
- 46.Mistry H. D., Kurlak L. O., Williams P. J., Ramsay M. M., Symonds M. E., Broughton Pipkin F. Differential expression and distribution of placental glutathione peroxidases 1, 3 and 4 in normal and preeclamptic pregnancy. Placenta. 2010;31(5):401–408. doi: 10.1016/j.placenta.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 47.Du X. H., Dai X. X., Song R. X., et al. SNP and mRNA expression for glutathione peroxidase 4 in Kashin-Beck disease. The British Journal of Nutrition. 2012;107(2):164–169. doi: 10.1017/S0007114511002704. [DOI] [PubMed] [Google Scholar]
- 48.Roten L. T., Fenstad M. H., Forsmo S., et al. A low COMT activity haplotype is associated with recurrent preeclampsia in a Norwegian population cohort (HUNT2) Molecular Human Reproduction. 2011;17(7):439–446. doi: 10.1093/molehr/gar014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Liu M., Zong J., et al. Genetic variations in IL1A and IL1RN are associated with the risk of preeclampsia in Chinese Han population. Scientific Reports. 2015;4(1) doi: 10.1038/srep05250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pontillo A., Reis E. C., Bricher P. N., et al. NLRP1 L155H polymorphism is a risk factor for preeclampsia development. American Journal of Reproductive Immunology. 2015;73(6):577–581. doi: 10.1111/aji.12353. [DOI] [PubMed] [Google Scholar]
- 51.Lykke J. A., Bare L. A., Olsen J., et al. Vascular associated gene variants in patients with preeclampsia: results from the Danish National Birth Cohort. Acta obstetricia et Gynecologica Scandinavica. 2012;91(9):1053–1060. doi: 10.1111/j.1600-0412.2012.01479.x. [DOI] [PubMed] [Google Scholar]
- 52.Sanchez-Aranguren L. C., Prada C. E., Riano-Medina C. E., Lopez M. Endothelial dysfunction and preeclampsia: role of oxidative stress. Frontiers in Physiology. 2014;5:p. 372. doi: 10.3389/fphys.2014.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Redman C. W., Sargent I. L. Placental debris, oxidative stress and pre-eclampsia. Placenta. 2000;21(7):597–602. doi: 10.1053/plac.2000.0560. [DOI] [PubMed] [Google Scholar]
- 54.Roland-Zejly L., Moisan V., St-Pierre I., Bilodeau J. F. Altered placental glutathione peroxidase mRNA expression in preeclampsia according to the presence or absence of labor. Placenta. 2011;32(2):161–167. doi: 10.1016/j.placenta.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Moreno María U., San José G., Orbe J., et al. Preliminary characterisation of the promoter of the human p22(phox) gene: identification of a new polymorphism associated with hypertension. FEBS Letters. 2003;542(1-3):27–31. doi: 10.1016/s0014-5793(03)00331-4. [DOI] [PubMed] [Google Scholar]
- 56.Siegel D., Ross D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radical Biology & Medicine. 2000;29(3-4):246–253. doi: 10.1016/s0891-5849(00)00310-5. [DOI] [PubMed] [Google Scholar]
- 57.Zhuo P., Diamond A. M. Molecular mechanisms by which selenoproteins affect cancer risk and progression. Biochimica et Biophysica Acta. 2009;1790(11):1546–1554. doi: 10.1016/j.bbagen.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mutze S., Rudnik-Schoneborn S., Zerres K., Rath W. Genes and the preeclampsia syndrome. Journal of Perinatal Medicine. 2008;36(1):38–58. doi: 10.1515/JPM.2008.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used to support the findings of this study are included within the article.


