Abstract
Background and Aims
Inferring the evolutionary relationships of species and their boundaries is critical in order to understand patterns of diversification and their historical drivers. Despite Abies (Pinaceae) being the second most diverse group of conifers, the evolutionary history of Circum-Mediterranean firs (CMFs) remains under debate.
Methods
We used restriction site-associated DNA sequencing (RAD-seq) on all proposed CMF taxa to investigate their phylogenetic relationships and taxonomic status.
Key Results
Based on thousands of genome-wide single nucleotide polymorphisms (SNPs), we present here the first formal test of species delimitation, and the first fully resolved, complete species tree for CMFs. We discovered that all previously recognized taxa in the Mediterranean should be treated as independent species, with the exception of Abies tazaotana and Abies marocana. An unexpectedly early pulse of speciation in the Oligocene–Miocene boundary is here documented for the group, pre-dating previous hypotheses by millions of years, revealing a complex evolutionary history encompassing both ancient and recent gene flow between distant lineages.
Conclusions
Our phylogenomic results contribute to shed light on conifers’ diversification. Our efforts to resolve the CMF phylogenetic relationships help refine their taxonomy and our knowledge of their evolution.
Keywords: Species delimitation, speciation with gene flow, phylogenomics, RAD-seq, conifers, transposable evolution, Abies
INTRODUCTION
Extant gymnosperms have relatively limited species diversity presumably as a result of low speciation rates and high extinction (Crisp and Cook, 2011). However, recent pulses of radiation have been found in some gymnosperm lineages, challenging the perception that gymnosperm taxa are ancient and barely variable (Davis and Schaefer, 2011). Global climate shifts have been suggested as major drivers of diversification in gymnosperm radiations (Nagalingum et al., 2011). However, other powerful contributors to conifer evolution and diversity are the frequent hybridization events (Isoda et al., 2000; Ru et al., 2016) and bursts of transposable elements (TEs; Nystedt et al., 2013), which could facilitate regulatory, karyotypic or other genetic changes (Oliver et al., 2013). Different TE dynamics can contribute to reproductive isolation and promote species radiation (Serrato-Capuchina and Matute, 2018). Delimiting species boundaries and inferring their evolutionary relationships is critical in order to understand the pace and mode of lineage formation in diversifying taxa, as well as their evolutionary drivers (Barley et al., 2013).
Firs (Abies, Pinaceae) are an important component of temperate-cool forests that characterize the boreal regions of the Northern Hemisphere (Farjon and Rushforth, 1989). They represent the second most diverse group of the Pinaceae family and of gymnosperms as a whole (Xiang et al., 2009), consisting of >50 species. Although fir trees have three major areas of distribution, North America, East Asia and the Mediterranean Basin, evolutionary studies have so far primarily focused on the fir trees outside the Mediterranean Basin (Linares, 2011). Based on paleogeographical data (Linares, 2011), colonization and diversification of firs in the Mediterranean could have been associated with climatic and geological changes during the late Miocene and the Pliocene. Numerous investigations have already been conducted on the genus Abies but included only part of the proposed taxa and only single DNA regions or low-resolving molecular markers (see below), hence species delimitation and evolutionary history of all Circum-Mediterranean firs (CMFs) remain debatable.
In the last 200 years, taxonomic classification of firs has undergone numerous revisions, with at least 14 formal classification attempts (Franco, 1950; Liu, 1971; Farjon and Rushforth, 1989). The number of recognized taxa has ranged from six to 12 species and subspecies in different classifications. The most recent taxonomical review (Farjon and Rushforth, 1989) classified CMFs into two groups (section Abies and sect. Piceaster) and eight species. Section Abies comprises Abies alba Mill., the most widely distributed fir in Europe; A. nebrodensis (Lojac.) Mattei, a critically endangered species (conservation status: CR by IUCN) (Nebrodi mountain range, Sicily, Italy); A. cephalonica Loudon (Greece); and A. borisii-regis Mattf. (Northern Balkans and Greece), a hybridogenous taxon of A. alba and A. cephalonica (Bella et al., 2015; Krajmerová et al., 2016). In addition, from the Eastern Mediterranean, Farjon and Rushforth (1989) recognized A. nordmanniana (Steven) Spach (Northeast Caucasus and Turkey) as a single taxon. However, A. nordmanianna is a complex taxon which other authors (Mattfeld et al., 1925; Liepelt et al., 2010; Linares, 2011) have sub-divided into A. equi-trojani (Asch. & Sint. ex Boiss.) Mattf. (Northern Turkey) and A. bornmuelleriana Mattf. (Northern Turkey). This sub-division in only partially supported by molecular evidence (Hrivnák et al., 2017). Finally, A. cilicica (Antoine & Kotschy) Carrière, inhabiting Lebanon, Syria and Southern Turkey, has been included in the next section by some authors (Franco, 1950; Liu, 1971).
Section Piceaster would comprise A. numidica de Lannoy ex Carrière (Northern Algeria) and A. pinsapo Boiss. (Souterhern Spain and Northern Morocco). However, this last taxon has been sub-divided into three different taxa, i.e. A. pinsapo Boiss sensu stricto (Southern Spain), A. marocana Trab. and A. tazaotana Côzar ex Villar (both Northern Morocco), by several authors (Franco, 1950; Arista and Talavera, 1994). Nevertheless, recent studies did not provide molecular support for the Moroccan species distinction (Sánchez-Robles et al., 2014; Terrab et al., 2007).
The inference of macroevolutionary relationships between Abies sections has been shown to be mostly robust to coalescent stochasticity of individual phylogenetic markers (Suyama et al., 2000; Xiang et al., 2009, 2015; Semerikova and Semerikov, 2014). However, the attempts to reconstruct the phylogenetic history of CMFs have yielded strongly conflicting signals between nuclear and plastidial markers (Sánchez-Robles et al., 2014; Semerikova and Semerikov, 2014, 2016; Xiang et al., 2015), resulting in a largely unresolved polytomy; inter-specific relationships and divergence times hence remain to date debated. Although many phylogeographical and phenotypic studies have significantly contributed to clarify species delimitation (e.g. Fady et al., 1992; Scaltsoyiannes et al., 1999; Terrab et al., 2007; Bosela et al., 2016; Liepelt et al., 2010; Hrivnák et al., 2017), it is noteworthy that these previous studies did not include all, or even most CMF taxa. In addition, no genome-wide study has been performed to reconstruct a species tree and determine the causes of the observed molecular discordances. Disagreements among classifications of Abies are largely attributed to insufficient understanding of phylogenetic relationships within the genus, as well as its morphological complexity (Robson et al., 1993; Xiang et al., 2009). This complexity may be further exacerbated by historical gene flow between several species, especially those that are closely related and in geographical proximity (Scaltsoyiannes et al., 1999; Isoda et al., 2000; Semerikova et al., 2011; Sánchez-Robles et al., 2014; Xiang et al., 2015; Krajmerová et al., 2016). Reproductive barriers in the CMFs can be permeable, as suggested by successful artificial crossings, which is in contrast to the strong barriers existing between North American and Mediterranean firs (Mergen et al., 1962; Alizoti et al., 2011; Kormutak et al., 2013). Secondary contacts among several CMF species during climatic cycles in the Quaternary have been previously suggested (Linares, 2011; Sánchez-Robles et al., 2014). However, the existence of ancient gene flow during the evolution of CMFs has never been tested, probably because of the weak phylogenetic signal of the previously used molecular markers.
An accurate CMF species delimitation is fundamental to manage biodiversity and forest genetic resources (Scarascia-Mugnozza et al., 2000; Fady et al., 2016; Kavaliauskas et al., 2018; Potter, 2018). Due to a high degree of endemism, geographically scattered distribution and fragmentation by human activities, four CMF taxa are currently included in the IUCN red list as critically endangered or endangered (A. pinsapo ‘EN’, A. pinsapo var. marocana ‘EN’, A. nebrodensis ‘CR’ and A. numidica ‘CR’; Farjon et al., 1993; Farjon and Page, 1999; Esteban et al., 2010). Hence, the clarification and accurate assessment of species status has important conservation implications (Adams et al., 2014; Melville et al., 2014). New species delimitation analyses on high-throughput data have recently allowed the identification of species boundaries in several cryptic species complexes as well as in rapid adaptative radiations, e.g. in some Reptilia (Barley et al., 2013; Grummer et al., 2014), primates (Hotaling et al., 2016) and plants (Jackson et al., 2017; Brandrud et al., 2019). We applied Illumina-based restriction site-associated DNA sequencing (RAD-seq) on all CMF taxa in order to investigate the evolutionary history of this group.
The main goals in the present study were: (1) to explore the origin and phylogenomic relationships of the modern CMFs; (2) to test taxonomic hypotheses about the current diversity of CMFs; and (3) to investigate the existence of inter-specific hybridization and the role of the TEs on the evolutionary history of the group.
MATERIALS AND METHODS
Taxon sampling and genomic data
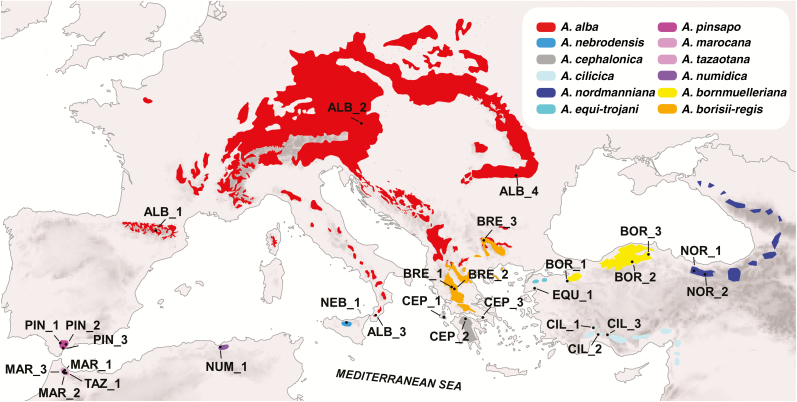
Leaf material from 40 individual trees (2–4 individuals per taxon) of all recognized CMF taxa (Fig. 1), together with one representative of each of the eight remaining sections of the genus Abies were collected at 28 natural sites using the geographic information from the EUFORGEN project (http://www.euforgen.org/) during several field trips in 2015 and 2016 (Supplementary Data Table S1). The samples were preserved in silica gel. Vouchers were deposited in the Herbarium of the University of Seville (SEV). Genomic DNA was extracted using a DNA extraction kit (Qiagen DNeasy Plant Mini Kit) following the manufacturer’s instructions. The RAD library was prepared as in Paun et al. (2016). Genomic DNA was digested by using high-fidelity SbfI (New England Biolabs) and the resulting fragments were double barcoded. The library was sequenced in a separate lane of an Illumina flowcell HiSeq 2500 at the VBCF NGS Unit (www.vbcf.ac.at/ngs) as 100 bp single-end reads.
Fig. 1.
Sampling map for the Mediterranean firs used in this study. Each population is labelled and species ranges are highlighted with different colours (more information is given in Supplementary Data Table S1). Species distributions are based on the EUFORGEN project (http://www.euforgen.org/) and personal observations.
RAD-seq catalogue building and SNP detection
Quality filtering and demultiplexing of the library were performed with deML (Renaud et al., 2015). Additionally, the process_radtags.pl program from STACKS ver. 1.29 (Catchen et al., 2011, 2013) was used to remove reads without a restriction cut site. Different empirical tests were performed with the denovo_map.pl program to optimize the parameters used for catalogue building. We varied the minimum number of identical reads required for a stack to be formed (i.e. the setting ‘m’) from three to 11, the number of differences between alleles in a locus when processing one individual (‘M’) between one and five and the differences between orthologous loci between individuals (‘n’) from zero to four. The final combination of settings (m = 3, M = 2, and n = 1) was chosen to maximize the number of polymorphic RAD loci with a maximum of nine single nucleotide polymorphismes (SNPs; to avoid pooling paralogues in the same locus) which were present in at least 30 individuals (out of 40, i.e. 75 % of the total). For most of the analyses, we used a reduced data set, ‘abies-reduced set’, based on the first SNP of each RAD locus (6090 SNPs in total) to avoid linkage disequilibrium and ensure the independence of the loci. For most analyses, we removed the samples of the hybrid A. borisii-regis (A. alba × A. cephalonica; Krajmerová et al., 2016) from the data set to avoid artefacts in the phylogenomic estimates (hereafter referred to as ‘abies-reduced-nohyb set’). Finally, some analyses focused on the in-group CMFs only and were performed by excluding the outgroup and A. borisii-regis (hereafter the ‘med-abies-reduced set’).
Phylogenomic analyses and tempo of diversification
Phylogenetic reconstruction of the genus Abies, based on the abies-reduced set and the abies-reduced-nohyb set, were conducted by using maximum likelihood (ML) and Bayesian inference (BI). For the ML approach, we used RAxML ver. 8.0 (Stamatakis, 2014) on the CIPRES Web Portal (Miller et al., 2015). The ASC_GTR-GAMMA model as appropriate for a data set of concatenated SNPs and 1000 rapid bootstrap inferences were used to estimate clade confidence. BEAST ver. 2.3.1 (Bouckaert et al., 2014) was used for BI and molecular dating using a GTR mutation model with gamma-distributed mutation rates across sites which provides a good approximation of the marginal likelihood at inter- and intra-specific evolutionary levels using highly variable sequences (Jia et al., 2014). The ucld prior was set to a uniform distribution (minimum 0, maximum 1), the tree prior was speciation Yule process and the chain was 20 million Markov chain Monte Carlo (MCMC) steps, logged every 2000 steps. We ran four runs which were concatenated and used to estimate the posterior distributions of topology and divergence time. In a first analysis, we dated the nodes in the genome-wide SNP-based phylogeny by using a relaxed-clock model in BEAST2 and four calibration points. The age of the oldest Abies fossil (Schorn and Wesley, 1986) was treated as a minimum age of calibration of the root, by applying a log-normal distribution of 47 million years ago (Ma) to the age of the oldest node. In addition, we used three Abies fossils for the calibration of nodes in the ingroup and outgroup. We used the same fossil calibrations as in Xiang et al. (2015). Fossil ages were assigned as a uniform distribution for their stratigraphic range (Supplementary Data Table S2). The minimum age of the stems of Abies and CMFs were assigned using log-normal prior distributions (47 and 23 Ma, respectively); meanwhile uniform prior distributions (a minimum of 16 Ma and a maximum of 47 Ma) were assigned to the crown node of the clade including sects, Grandes and Oiamel, and the crown node of sect. Momi. In a second independent dating analysis, we used two biogeographical calibration points (six calibration points in total; Supplementary Data Table S2). We used the date of the separation of North Africa from Europe (i.e. the end of the Messinian salinity crisis, the opening of the Strait of Gibraltar and Tyrrhenian Sea; approx. 6 Ma, normal prior distribution) as a calibration age of the divergence between A. pinsapo and A. marocana–A. tazaotana but also for divergence between A. nebrodensis and A. alba.
Species delimitation
We compared eight candidate species delimitation models (Fig. 2) by using the Bayesian factor delimitation of species (SNAPP-BFD*; Leaché et al., 2014) as implemented in SNAPP (Bryant et al., 2012). BEAUti v2.3.1 (Bouckaert et al., 2014) was used to set up the analysis file. The null model followed the most recent taxonomic treatment, but considering subspecies as species (11 species in total; delimitation species hypothesis A). Based on previous studies and our phylogenomic results, seven additional hypotheses were tested (Fig. 2). Specifically, we tested that A. marocana and A. tazaotana, which are locally restricted to Mount Tazaot, together form a single species and that their current distributions are just the result of recent fragmentation (hypothesis B; Sánchez-Robles et al., 2014). In addition, we tested the inclusion of the restricted A. nebrodensis in the widespread species A. alba (hypothesis C). We also tested a potential clustering of the Turkish taxa (A. nordmanniana, A. bornmuelleriana and A. equi-trojani) as a single species (hypothesis D). These three taxa have been treated with different taxonomic statuses in several taxonomic revisions (Coode and Cullen, 1965; Farjon and Rushforth, 1989). Finally, we also tested other combinations of these species delimitations (hypothesis E–I). For these analyses, we used the med-abies-reduced set without any missing data. We analysed the alternative tree topologies by using 24 steps of path sampling with 100 000 MCMC steps and 10 000 pre-burn-in steps to estimate marginal likelihoods. Convergence of the parameters (ESS > 200) was checked using Tracer v1.7.1 (Rambaut et al., 2018) after 10 % burn-in removal. Estimated marginal likelihoods for competing models were compared by using Bayes factors (2× loglikelihood difference) to identify the most likely species delimitation. The species tree was subsequently inferred by using the Bayesian multispecies coalescent approach implemented in SNAPP with the same previous parameters (Bryant et al., 2012) and visualized with Densitree 2.01 (Bouckaert, 2010). In addition, we used SVDQuartets (Chifman and Kubatko, 2014) with 100 non-parametric bootstrap replicates in PAUP* version 4.0a147 (Swofford, 2002) to generate a species tree for the best species delimitation.
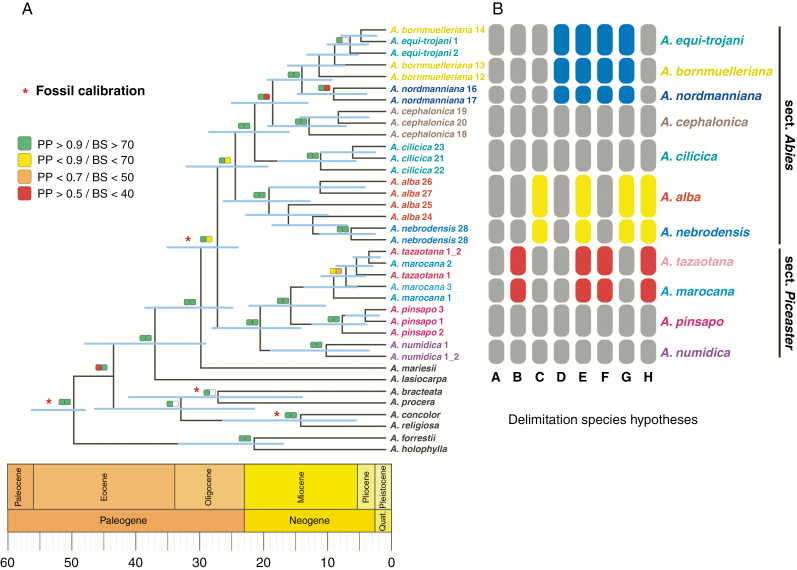
Fig. 2.
Phylogenetic reconstructions for the CMFs based on 6090 SNPs. (A) Chronogram built in BEAST2 using six fossil calibration points (asterisks). Posterior probabilities and bootstrap supports from RAxML are colour marked according to the key. (B) Species delimitation hypotheses tested in the BFD* analysis. Each grey cell represents an independent species. Other colours indicate taxa that are merged into a single species.
We also produced a lineage-through-time (LTT) plot, which displays the number of speciation events through time, taking into account possible extinction events. We calibrated the species trees from SNAPP by enforcing a strict molecular clock using the ‘chronos’ function in the R library ape (Paradis et al., 2004). As calibration point, we used the 95 % highest posterior density (HPD) node age of the CMFs inferred from the previous genome-wide SNP-based BEAST2 analysis. Additionally, we used the gamma statistic (γ) from chronograms (Pybus and Harvey, 2000) to determine whether a signal of decreased diversification was noticeable.
Genetic structure and recent admixture
To assess the genetic structure of CMFs, we performed a Bayesian MCMC approach with STRUCTURE ver. 2.3.4 (Pritchard et al., 2000). Ten independent runs were carried out for the data set for values of K ranging from 1 to 13. The number of iterations was 500 000 with a burn-in of 100 000 steps. STRUCTURE analyses were performed for all the Abies species in the Mediterranean area, as well as separately for each section (sect. Abies and sect. Piceaster, with K ranging from one to ten and one to eight, respectively). STRUCTURE HARVESTER software (Earl and VonHoldt, 2011) was used to determine the optimal number of clusters (K) implementing the Evanno method. The graphical representation of STRUCTURE results was generated by using CLUMPAK (Kopelman et al., 2015). Additionally, fineRADstructure and RADpainter ver. 0.1 (Malinsky et al., 2018) were used to infer the shared ancestry among individuals. The estimated haplotype co-ancestry matrix was used to infer recent admixture and to build a tree by using default parameters. For this analysis, we removed, as recommended, a sample with more missing data (A. equi-trojani_94_1) than the rest.
Divergence with gene flow
As popular approaches such as STRUCTURE (Pritchard et al., 2000) and ADMIXTURE (Alexander et al., 2009) are only appropriate for detecting gene flow over relatively short evolutionary time scales and may have limited power when only a few individuals per population are available (Soraggi et al., 2018), we applied an ABBA–BABA approach (Green et al., 2010) to the SNP matrix. This method distinguishes the contribution of incomplete lineage sorting from introgression by calculating imbalances in alternative tree topology frequencies under a four-taxon scenario (three ingroup taxa and one outgroup). In short, for a particular SNP with an ancestral (‘A’) allele and derived (‘B’) allele, the alternative patterns ‘ABBA’ and ‘BABA’ should be equally frequent under a scenario of incomplete lineage sorting without gene flow. D-statistic and Z-score calculations were performed with the R package evobiR (Blackmon and Adams, 2015), which measures signals of alternative phylogenetic asymmetry and the proportion of the genome that is shared between two taxa due to admixture, respectively (Durand et al., 2011). ABBA/BABA tests were performed on the abies-reduced set including heterozygous sites by generating pseudohaploid sequences (Green et al., 2010). One thousand bootstrap iterations were used to measure the s.d. of the D-statistic (Eaton and Ree, 2013). Based on previous studies (Sánchez-Robles et al., 2014) and our STRUCTURE results, we investigated gene flow among A. alba (sect. Abies) and A. numidica and A. pinsapo (sect. Piceaster). Using the approach proposed by Martin et al., (2013) and the previously estimated species relationships, we performed several ABBA–BABA tests with different candidate species to distinguish between admixture in different time periods. Briefly, if gene flow was only ancient (e.g. prior to A. nebrodensis divergence from A. alba), then A. alba and A. cilicica should both be equally admixed with A. numidica and A. pinsapo. However, if gene flow was more recent (i.e. after A. nebrodensis divergence), then A. alba should be more admixed with A. numidica and A. pinsapo than with A. cilicica. We performed analyses for all combinations of pairs of conspecific individuals from candidate admixers (A. alba, A. nebrodensis, A. cilicica, A. numidica and A. pinsapo) using A. lasiocarpa as outgroup.
Genomic divergence in CMF
Additionally, to infer the contribution of TEs to genomic divergence of CMF species, the presence of TEs was checked in raw reads using the approach proposed by Trucchi et al. (2017). We randomly extracted 100 000 single reads from each individual, which were inspected for TEs by using RepeatMasker ver. 4.0.6 against the RepBase TE database (Kohany et al., 2006) of Viridiplantae ver. 2016-08-29. Each individual sample was analysed independently and the relative abundances (i.e. TEs per 100 000 reads) of different TEs (Gipsy, Copia, LINEs and DNA transposons) were compared between sections by using Poisson-based generalized linear models (GLMs) in the software R.
RESULTS
The 100 bp Illumina RAD-seq for 40 DNA samples (1.7 million reads on average per sample) representing virtually all recognized CMF taxa (Fig. 1; Supplementary Data Text S1; Supplementary Data Table S3) produced in the final STACKS catalogue a total of 1 585 024 RAD loci with an average coverage per sample (± s.d.) of 81.3 ± 25.9 reads per locus. The demultiplexed data have been deposited in the NCBI Short Reads Archive (BioProject ID PRJNA563575). We found a high variability in the percentage of missing RAD loci among samples, ranging from 4.9 to 61.2 % (mean ± s.d. 12.84 ± 10.24 %; Supplementary Data Table S3) but this pattern was not biased towards particular taxa (F19,20 = 1.593; P = 0.155). After retaining only polymorphic RAD loci that were present in at least 30 individuals and had a maximum of nine SNPs per locus, we obtained a final data set with 6090 RAD loci and a total of 23 931 SNPs. However, for further analyses, we retained only one SNP per RAD locus (6090 unlinked SNPs in total).
Genome-based phylogeny and species delimitation
The ML and BI resulted in robust and generally consistent topologies with two highly supported [bootstrap support (BS) = 1.0 and posterior probablity (PP) = 99] lineages within the CMFs (Fig. 2A). These main lineages corresponded to the two previously recognized sections in the Mediterranean, sect. Abies and sect. Piceaster. In the Piceaster lineage, A. numidica (occurring in Algeria) is an early-diverged clade (BS = 1.0 and PP = 99), whereas A. pinsapo (Spain) and A. tazaotana + A. marocana (North-West Morocco) are sister clades. Individuals of A. tazaotana and A. marocana appeared intermixed. In the Abies section, two highly supported clades were shown. The first one included A. alba and A. nebrodensis. The second clade included species from Greece, A. cephalonica, and an inner clade of Turkish taxa, A. cilicica, A. nordmanniana, A. bornmuelleriana and A. equi-trojani. As expected in a bifurcating tree, the individuals of the hybrid taxon A. borisii-regis clustered with either parental species, A. alba and A. cephalonica (Supplementary Data Fig. S1).
For the Bayesian species delimitation analysis in SNAPP, the species hypothesis with the highest likelihood suggested that all previously recognized taxa (either at the species or at the subspecies level) in the Mediterranean should be treated as independent species, with the exception of A. tazaotana and A. marocana [maximum likelihood estimation (MLE) = –6500.7 for distinct, hypothesis A; MLE = –6440.5 for the two taxa grouped as one species, hypothesis B; Bayes factor value = 120.4 representing decisive evidence, Supplementary Data Table S4].
Origin and evolution of Mediterranean firs
The topology of the species tree constructed with SNAPP, based on the best hypothesis according to Bayesian factor delimitation of species (BFD*), was mostly congruent with the concatenated SNP trees (Fig. 3), but the main lineages evolved comparatively early. The Piceaster lineage (including A. numidica, A. pinsapo and A. marocana–A. tazaotana) showed the same internal relationships, whereas the Turkish lineages (including A. nordmanniana, A. bornmuelleriana and A. equi-trojani) diverged rapidly from one another. Additional support for the CMF relationships came from the species tree inferred using SVDQuartets, whose topology was highly supported (Supplementary Data Fig. S2). This species tree, again, supported the Abies and the Piceaster sections as monophyletic, with 100 % BS. Whereas the SVDQuartets tree was largely congruent with inferences from concatenated data sets, the species tree differed in some relationships, especially for deep divergence events.
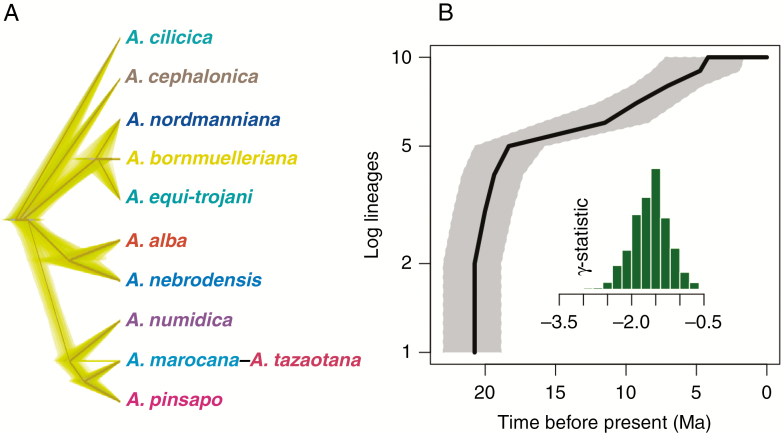
Fig. 3.
SNAPP species cladogram for the best species delimitation hypothesis (A). Lineage through time plot and distribution of γ-statistics for the SNAPP trees (B).
Dating the diversification of CMFs is key to understanding the putative contribution of climatic fluctuations and geological events around the Mediterranean Basin to the evolution of CMF. The estimated divergence times using BEAST and several fossil records are shown in Fig. 2. The chronogram was congruent with an early divergence of the lineages of sect. Abies and sect. Piceaster around the late Oligocene (95 % HPD: 23.1–34.1 Ma). Miocene divergence times for all current CMF lineages are clearly inferred, with speciation times pre-dating by tens of millions of years the Messinian salinity crisis (5.59–5.33 Ma; Krijgsman et al., 1999) and establishment of the the Mediterranean climate (approx. 3.2 Ma; Suc, 1984). For the Piceaster lineage, the split of A. numidica and A. pinsapo–A. marocana–A. tazaotana was estimated at 13.3–27.1 Ma and at 9.4–21.3 Ma for that of A. pinsapo and A. marocana–A. tazaotana. For the lineage of sect. Abies, an Oligocene–Miocene split of A. alba and the eastern lineage was estimated (18.4–31.1 Ma). The eastern lineage diverged in the early Miocene, with 15.1–27.6 Ma for the split of A. cilicica and 12.2–24.0 Ma for that of A. cephalonica. The Turkish species were the last to diverge, around 8.4–18.5 Ma. Additionally, the speciation between A. nebrodensis and A. alba occurred around 9.1–21.8 Ma. These estimates were robust to additional calibration points derived from geological events, such as the opening of the Strait of Gibraltar and Tyrrhenian Sea after the Messinian salinity crisis (Supplementary Data Table S5). Nevertheless, this secondary dating analysis (including geological events) invariably showed shorter ranges for the HPD estimates. Additional dating analyses using other tree prior speciation processes (i.e. birth–death and Bayesian skyline; not shown) provided similar estimates.
We produced an LTT plot, which displays the number of speciation events through time, taking into account extinction events (Fig. 3). The CMF species have accumulated early and quickly in the history of the group. The gamma statistic was further employed to explore the acceleration of speciation rate over the history of CMFs. The distribution of the gamma values on the 2276 SNAPP species trees rejects a constant speciation rate; γ (mean ± s.d.) = –1.61 ± 0.38; P-values < 0.05 (Fig. 3). The negative gamma values revealed an early and rapid accumulation of branching times near the root, indicative of a decelerated diversification. The shift in the number of speciation events occurs quickly in the group, with the highest number of speciation events estimated at the boundary between the Oligocene and the Miocene. The rapid divergence and expansion in the Tertiary and a relative speciation stasis in the Quaternary were congruent with the lack of differences in Abies fossil records along the latitude (t-test, t = –0.384, d.f. = 85.268, P = 0.702) or longitude (t = 1.299, d.f. = 90.376, P = 0.197) between these two epochs (Supplementary Data Fig. S3).
Genomic divergence between fir species and evidence for ancient hybridization during the speciation process
The rapid CMF radiation was congruent with low genomic divergence among species. More than half of the SNPs (51.8 %) represented shared polymorphism between at least two CMF species. Heterozygosity was similar between species, ranging from 0.04 to 0.08 with an average of 0.055 ± 0.002 (χ 211,20 = 16.13, P = 0.134). However, the Piceaster section showed a significantly lower heterozygosity (0.049 ± 0.005) than Abies (0.058 ± 0.003; χ 21,20 = 6.35, P = 0.01; Supplementary Data Fig. S4). The pairwise FST values between species ranged from 0.04 to 0.53 (Supplementary Data Table S6).
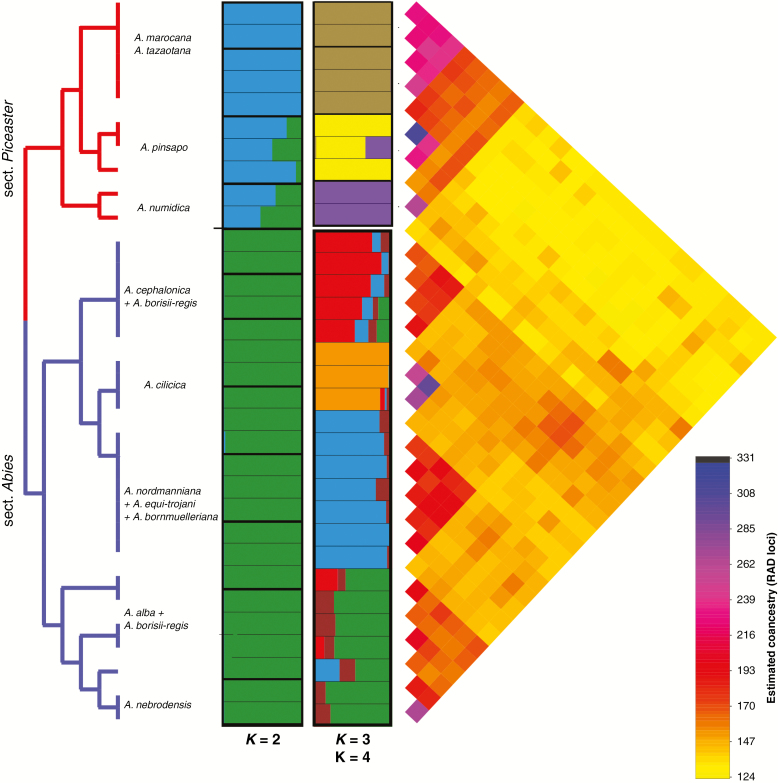
The Bayesian assignment of genetic structure (Fig. 4) showed, again, two main clusters (i.e. best partition K = 2 following the Evanno method; LK = 50984.8, ΔK = 789.2; Supplementary data Fig. S5a) corresponding to the two taxonomic sections. Although most samples showed a high assignment probability to just one section, admixture signals appeared in several individuals (Fig. 4) of A. numidica and A. pinsapo, which showed intermediate probability of belonging to sect. Abies. Additional sub-optimal groupings were K = 5 and K = 7 (Supplementary Data Fig. S5a), which did not show clear taxonomical patterns. Within sections, the genetic structure generally mirrored the species tree (Fig. 4). For sect. Piceaster, K = 3 was the best partition (Supplementary Data Fig. S5b), corresponding to A. numidica, A. pinsapo and A. marocana + A. tazaotana. For sect. Abies, K = 5 partition was selected (Supplementary Data Fig. S5c). The first group corresponded to A. cephalonica individuals and the second one included A. cilicica individuals. Abies alba and A. nebrodensis together supported a third cluster, whereas the last group encompassed A. nordmanniana, A. equi-trojani and A. bornmuelleriana. High haplotype co-ancestry levels among the accessions of Abies section Piceaster were estimated with RADpainter (Fig. 4). In addition, and consistently with recent hybridization between the species of both sections, we observed high levels of haplotype co-ancestry among distantly related species (e.g. A. numidica and A. alba) as estimated with RADpainter (Fig. 4).
Fig. 4.
Different types of evidence of recent gene flow in the CMFs. Hierarchical structure among and between CMF samples estimated by fineRADstructure. Results of the Bayesian assignment analysis (STRUCTURE) for CMFs (and independently for each section). Haplotype co-ancestry matrix based on 6090 SNPs with individual A. equi-trojani_94_1 removed due to excessive missing data. The colour in the heat map corresponds to the number of haplotypes imported from a donor genome to a recipient genome. High levels of haplotype co-ancestry among distantly related species (e.g. A. numidica and A. alba) suggest secondary contacts.
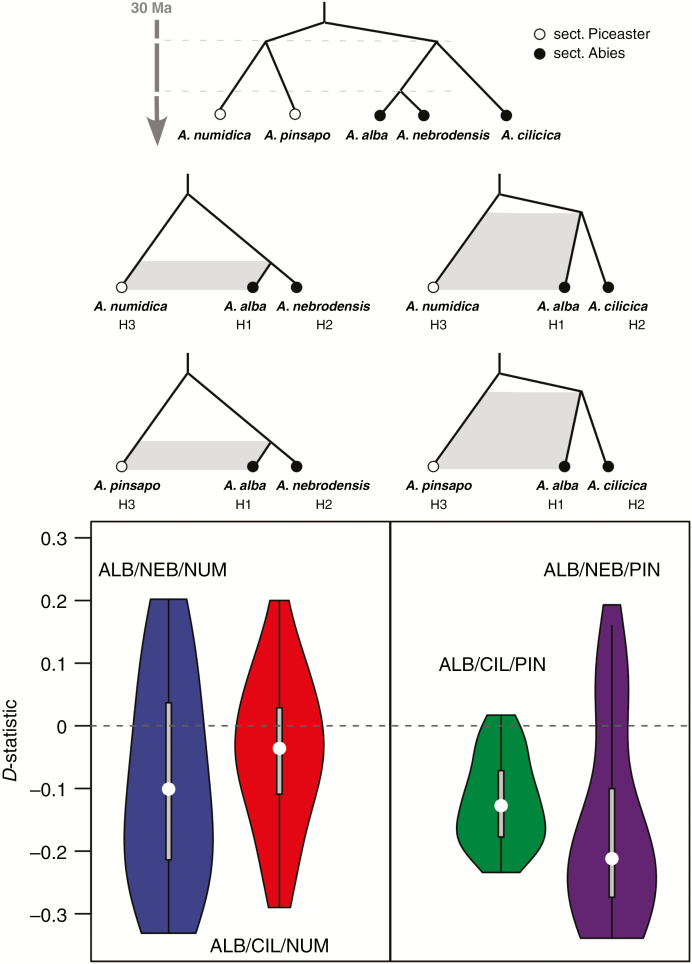
Additionally, we tested if ancestral gene flow was also compatible with these admixture signals. ABBA–BABA tests (Patterson’s D-statistic tests; Fig. 5; Supplementary Data Table S7) supported an ancient divergence with gene flow among species from the two CMF sections. Significant negative D-statistics support a notable ancient gene flow between A. alba and two species of sect. Piceaster, A. numidica and A. pinsapo, but also between A. cilicica and the sect. Piceaster species. In addition, higher absolute admixed values (Z-statistics) were found for comparisons including A. nebrodensis (as opposed to A. cilicica comparisons), indicating additional, recent admixture events. Remarkably, A. alba showed higher admixture with A. pinsapo than with A. numidica.
Fig. 5.
Hypothetical schemes of introgression between species of Abies sect. Abies and A. sect. Piceaster at different time periods. Violin plots show the distribution of the D-statistics (ABBA–BABA tests) resulting from testing different individuals from each species. Detailed information about the significance of the D-statistics for each test is provided in Supplementary Data Table S7. Values different from zero denote introgression between species. ALB, Abies alba; CIL, A. cilicica, NEB, A. nebrodensis; NUM, A. numidica; and PIN, A. pinsapo. Abies lasiocarpa was used as outgroup.
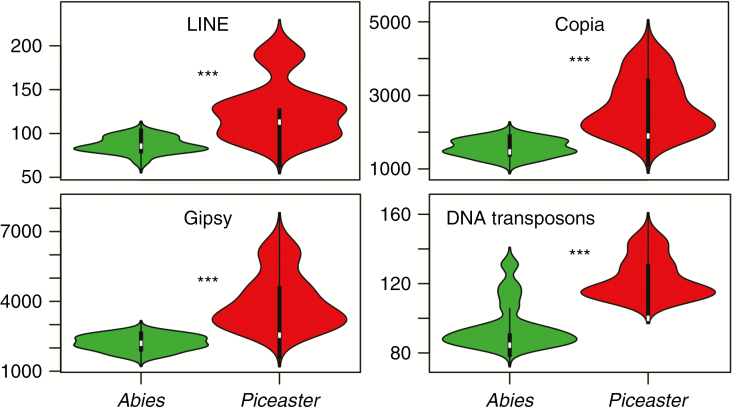
Furthermore, a significant divergence between genomic landscapes of the two Abies sections was also mediated by differences in TE activity. We found significant differences between both Abies sections (Fig. 6) in the relative amount of DNA transposons (χ 2 = 27.5, P < 0.001), Copia (χ 2 = 324.5, P < 0.001), Gipsy (χ 2 = 447.5, P < 0.001) and LINE (χ 2 = 3.3, P < 0.1) within our RAD-seq data. Interestingly, the relative abundances of some TE classes also showed a strong correlation with the latitude of the samples (Supplementary Data Fig. S6).
Fig. 6.
Violin plots of the relative abundance of different transposable element families per individual of the two Mediterranean Abies sections. Asterisks denote statistically significant differences at P < 0.001 (Poisson generalized linear model).
DISCUSSION
To our knowledge, this research represents the first genome-wide evolutionary study in firs and the most intensive sampling including all CMF taxa. Although some limitation of RADseq for outlier scans has been suggested and debated (Tiffin and Ross-Ibarra, 2014; Lowry et al., 2016; but see Catchen et al., 2017), this study highlights the advantages of applying RAD-seq to obtain SNPs for phylogenetic studies in taxa with giant genomes, such as in the genus Abies. Furthermore, the combined use of phylogenetic methods and population genomic methods (such as STRUCTURE) have contributed to reveal the complex evolutionary history of Mediterranean firs, clarifying not just their phylogenetic relationships but also the recent and old secondary contacts.
Phylogeny of the Mediterranean Abies
Our dated phylogenomic tree corroborated the monophyly of all Mediterranean firs, as previously suggested (Suyama et al., 2000; Semerikova and Semerikov, 2014, 2016; Xiang et al., 2015). However, our molecular analyses, for the first time, clearly separated CMFs into the two previously classified sections, Abies and Piceaster (Farjon and Rushforth, 1989). Previous studies using classical markers have been unable to clarify the phylogenetic relationships of these taxa. The monophyly of the Piceaster section, including the south-western species A. numidica, A. marocana, A. tazaotana and A. pinsapo, was additionally supported by other molecular markers (Ziegenhagen et al., 2005; Sánchez-Robles et al., 2014). However, the taxonomic status of the Moroccan species has been controversial; sometimes they were considered as mere varieties of A. pinsapo (Farjon and Rushforth, 1989), sometimes as subspecies (Govaerts, 1995) or distinct species (Maire, 1952). The species delimitation analysis suggested that the Moroccan Abies species must be classified as different species from A. pinsapo, but that they should not be split into independent species. This result is congruent with the genetic distinctiveness of A. pinsapo from A. marocana–A. tazaotana and the lack of genetic structure among populations of the latter by using amplified fragment length polymorphism (AFLP) and plastid markers (Terrab et al., 2007; Sánchez-Robles et al., 2014). Both Moroccan taxa were previously separated (Sánchez Cózar, 1946) on the basis of morphological characters (mainly leaves, cones and bracts) but, because of their close genetic similarity, and based on finer morphological studies (Sekiewicz et al., 2013) as well, we propose to redefine their taxonomic status as a single species. Based on these result, we update the current taxonomy of CMFs (Supplementary Data Text S1).
In the Abies section, the results from the phylogenetic reconstructions and the species delimitation hypothesis test support the identity of A. nebrodensis, endemic to the Madonie range of northern Sicily (Italy), as a species derived from an ancestor of A. alba, the most widely distributed species inhabiting central Europe. This taxonomical status is supported by differences in traditional nuclear and plastid markers (Vicario et al., 1995; Parducci and Szmidt, 1999; Parducci et al., 2001a) and refute its classification as a subspecies or a variety of A. alba, or its alleged hybrid origin from A. alba and A. numidica (Landry, 1984; Parducci et al., 2001b). A close relationship between these two latter species from different sections was previously suggested by Sánchez-Robles et al. (2014) based on AFLP markers, but our results refute this hypothesis. Furthermore, the relationships between A. alba, A. cilicica and A. cephalonica have been resolved here for the first time. The two latter species (plus the A. nordmanniana complex clade) formed a deeply diverged sister lineage to A. alba. In this clade, an early divergence of A. cilicica first occurred, whereas the Greek A. cephalonica and the Eastern Turkish clade diverged later. Although A. cilicica was previously wrongly assigned to the Piceaster section (Franco, 1950; Liu, 1971), our data rather support a phylogenetic relationship with sect. Abies. In addition, the monophyly of the Greek A. cephalonica and the Eastern Turkish Abies species (i.e. the A. nordmanniana complex) was previously highlighted by their shared mitochondrial haplotype (Ziegenhagen et al., 2005). Their genetic distinctiveness was further visible in our STRUCTURE results. The A. nordmanniana complex formed a single cluster, but the BFD* analysis suggested the existence of three different species: A. nordmanniana sensu stricto, A. equi-trojani and A. bormulleriana. A recent study (Hrivnák et al., 2017) on the A. nordmanniana complex resulted in partially congruent phylogenetic relationships with our species delimitation hypothesis. However, the differentiation between the sister species A. equi-trojani and A. bormulleriana was unclear (Hrivnák et al., 2017). Finally, the clarification of CMF taxonomy has predictable positive effects on conservation. New CMF classification could help authorities to focus conservation efforts (especially for the Moroccan and Turkish firs).
Origin and evolution of the Mediterranean firs
The information on the phylogenetic relationships uncovered in this study, combined with our knowledge of the geographical distribution of the species and the fossil record, can provide insights into the origin and evolution of Mediterranean firs. In short, the dated monophyletic origin of the Mediterranean firs and their close relationships with eastern Asian sections Momi and Pseudopicea (Xiang et al., 2015) would suggest a single colonization event by a CMF ancestor from Asia in the late Eocene–Oligocene. This scenario is consistent with the estimated date for the Mediterranean crown group in previous studies on the genus Abies (Aguirre-Planter et al., 2012; Xiang et al., 2015). It is also noteworthy that the global expansion and diversification of firs have been dated during the Eocene as well, in agreement with dispersed fossils all over the Northern Hemisphere (Xiang et al., 2007, 2015). The global climate cooling down through the Eocene–Oligocene (Zachos et al., 2008) period could have favoured the geographical expansion of firs. In a relatively short time, as supported by the fossil records (Cavagnetto and Anadón, 1996), the CMF ancestor could have reached the westernmost region of the Mediterranean Basin (i.e. the Iberian Peninsula).
With regard to the speciation sequence in the Mediterranean firs, a previous hypothesis suggests a slow diversification along the Miocene–Pliocene, culminating with the rise of the modern CMF species after the Messinian climate crisis and the Pliocene marine transgression (Linares, 2011). Contrary to expectations, our conservative molecular dating and the speciation rate analyses suggest a much earlier diversification of the Mediterranean firs in the late Oligocene–Early Miocene, which is congruent with the radiation of the Mesoamerican firs (Aguirre-Planter et al., 2012). The finding of rapid diversification in CMFs is particularly striking given that it concerns one of the most important components of temperate-cool forests in Eurasia and it has been widely studied. Our molecular dating method was carefully designed to incorporate the uncertainty associated with paleontological data and it was robust to different settings (including some biogeographical events). Understanding why CMFs have diversified during the Miocene requires a reconsideration of paradigms concerning patterns of diversification in the Mediterranean basin, as previously suggested by Vargas et al. (2018). Quaternary climatic cycles have been evoked as a main factor of diversification, but the new evidence suggests a secondary role providing secondary contacts and posterior isolation.
Our reconstruction of past evolutionary events suggests that during the late Oligocene–Early Miocene, the archaic Mediterranean Abies would have split into two groups, corresponding to the two sections Piceaster and Abies, perhaps in each extreme of the Mediterranean Basin. In the western region, the ancestor of A. numidica, A. pinsapo and A. marocana (i.e. sect. Piceaster) would remain genetically isolated, and in the central–east region would have differentiated the archaic Abies sect. Abies species. Additionally, the latter species would have quickly split into two lineages, one for proto-A. alba and another one for the ancestor of all eastern species.
Additionally, our LTT plot and gamma statistics analysis showed signals of a rapid speciation burst shortly after the CMF expansion in the early Miocene. These results pre-dated by tens of million years the previous hypothesis about CMF diversification. Interestingly, the divergence between Ephedra and disjunct Cedrus species in the Mediterranean has been estimated to the Miocene (Qiao et al., 2007; Ickert-Bond et al., 2009) as well. Provided CMFs, Cedrus and Ephedra species share a similar biogeographic history in the Mediterranean region, our study supports the idea that living Mediterranean gymnosperms are not relicts and they have originated by an early Neogene pulse of diversification (Davis and Schaefer, 2011) together with many Mediterranean angiosperms (Vargas et al., 2018). The likely reason behind this pulse of speciation in the Mediterranean flora could be attributed to the global climate change during the Miocene. After the temperature and rainfall decreased during the Eocene–Oligocene, the temperature rose slightly through the Neogene until reaching in the Miocene a climatic optimum (15–17 Ma; Zachos et al., 2001). The elevated temperature connected to a global CO2 increase (Zachos et al., 2008) could have aggravated the effects of increased aridity and caused extinctions and geographic isolation in the CMFs (Rundel et al., 2016).
Furthermore, we found differential dynamics in TEs between the two sections, which is in contrast to the slow and steady rate of accumulation of TEs found otherwise in gymnosperms (Nystedt et al., 2013). However, a recent study (Voronova et al., 2017) suggests that some retrotransposon elements have been established, diverged and expanded after separation of the Pinaceae family. In spite of the genome similarity between fir species, differences in TEs could have profound effects on gene expression (Bennetzen and Wang, 2014) and possibly might contribute to adaptative radiation and morphological diversity in CMFs. The role of climatic stress in the activation of TEs has been widely assessed (Capy et al., 2000) and may additionally support the role of the climate in the speciation of CMFs (Kaplan and Guy, 2005; Krasensky and Jonak, 2012; George et al., 2015; Sánchez-Salguero et al., 2017).
Evidence of early divergence with gene flow and secondary contacts in the Mediterranean firs
Generally, our study revealed signs of admixture between CMF species of both sections but also within sections, causing potential conflicts between gene trees, but also when constructing a bifurcating species tree. In spite of the morphological differentiation between CMF species, a lack of reproductive isolation was evidenced by artificial inter-specific crossing experiments (Mergen et al., 1962; Kormutak et al., 2013). The multiple locations of A. borisii-regis in the multiple phylogenomic analysis and the low pairwise FST values support its recent hybridogenous origin due to multiple introgression events between A. alba and A. cephalonica. This pattern of genomic variation is consistent with a previously found genomic cline (Krajmerová et al., 2016) and suggests that A. borisii-regis is an unstabilized hybridogenous taxon. Furthermore, the existence of recent gene flow between other CMF species was also apparent from multiple lines of evidence. The signals of admixture in the STRUCTURE analysis and the high haplotype coancestry are congruent with other previously suggested secondary contacts, most probably during the Pliocene–Pleistocene, between A. alba, A. pinsapo and A. numidica (Parducci et al., 2001a; Sánchez-Robles et al., 2014). Based on chloroplast simple sequence repeat (cpSSR) analysis, Sánchez-Robles et al. (2014) suggested ancient secondary contacts among these species as no recent admixture signals were found using AFLP markers. In our study, we uncovered the importance of this ancient gene flow among species from the two main lineages in the evolution of the CMFs. The process of fast speciation in CMFs involves alternate periods of partial isolation and secondary contacts among divergent species as a likely consequence of climatic oscillations.
Conclusions
In summary, our modern phylogenomic approach sheds light on the diversification of the Circum-Mediterranean firs. Our efforts to resolve the phylogenetic relationships among the different lineages have refined their taxonomy and our knowledge of their evolution. An unexpectedly early pulse of speciation in the Oligocene–Miocene boundary is here documented, pre-dating by millions of years previous hypotheses, revealing a complex evolutionary history encompassing both ancient and more recent gene flow between distant lineages. Different transposable element dynamics could have contributed to the genome divergence of Circum-Mediterranean firs.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Text S1: taxonomic revision. Table S1: population details of the samples included in the study. Table S2: fir fossil calibrations and geological events used in this study. Table S3: SNP screening by using the STACKS software for all the studied individuals. Table S4: marginal likelihoods and Bayes factor values for the alternative species delimitation hypothesis. Table S5: mean ages and 95 % HPD heights for the major nodes of the CMF chronogram using two additional geographic events for calibration. Table S6: Circum-Mediterraneam Abies pairwise FST estimates from 6090 SNPs. Table S7: details of individual ABBA–BABA tests for ancient gene flow among Abies species from sections Abies and Piceaster. Figure S1: maximum likelihood phylogenetic tree of CMFs including the hybrid A. borisii-regis, based on 6090 SNPs. Figure S2: SVDQuartet species tree. Figure S3: geographic pattern of Abies fossil records along latitude and longitude in the Tertiary and the Quaternary periods. Figure S4: violin plots of heterozygosity per individual of the two Mediterranean Abies sections. Figure S5: summarized results from STRUCTURE. Figure S6: relationship between DNA transposons and latitude.
FUNDING
This work was supported by the Spanish Ministerio de Economía y Competitividad (MINECO) [grant no. CGL2013-45463-P] and by the Austrian Science Fund (FWF) [Y661-B16].
ACKNOWLEDGEMENTS
We thank J. Jaramillo-Correa, M. Á. Ortiz, J. A. Mejías, A. Mori, L. Navarro, J. L. Silva and Errol Vela for helping in the sampling. We thank the two anonymous reviewers for their careful reading of our manuscript and their many insightful comments and suggestions. Additionally, we would like to thank the Herbarium and Biology Services (CITIUS2, University of Seville) and the Centro Informático Científico de Andalucía (CICA) for access to the facilities and bioinformatics resources.
LITERATURE CITED
- Adams M, Raadik TA, Burridge CP, Georges A. 2014. Global biodiversity assessment and hyper-cryptic species complexes: more than one species of elephant in the room? Systematic Biology 63: 518–533. [DOI] [PubMed] [Google Scholar]
- Aguirre-Planter É, Jaramillo-Correa JP, Gómez-Acevedo S, Khasa DP, Bousquet J, Eguiarte LE. 2012. Phylogeny, diversification rates and species boundaries of Mesoamerican firs (Abies, Pinaceae) in a genus-wide context. Molecular Phylogenetics and Evolution 62: 263–274. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Research 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizoti G, Fady B, Prada MA, Vendramin GG. 2011. Technical guidelines for genetic conservation and use of Mediterranean firs (Abies spp.). Rome: Bioversity International. [Google Scholar]
- Arista M, Talavera S. 1994. Phenology and anatomy of the reproductive phase of Abies pinsapo Boiss. (Pinaceae). Botanical Journal of the Linnean Society 116: 223–234. [Google Scholar]
- Barley AJ, White J, Diesmos AC, Brown RM. 2013. The challenge of species delimitation at the extremes: diversification without morphological change in philippine sun skinks. Evolution 67: 3556–3572. [DOI] [PubMed] [Google Scholar]
- Bella E, Liepelt S, Parducci L, Drouzas AD. 2015. Genetic insights into the hybrid origin of Abies × borisii-regis Mattf. Plant Systematics and Evolution 301: 749–759. [Google Scholar]
- Bennetzen JL, Wang H. 2014. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annual Review of Plant Biology 65: 505–530. [DOI] [PubMed] [Google Scholar]
- Blackmon H, Adams R. 2015. EvobiR: tools for comparative analyses and teaching evolutionary biology. http://www.uta.edu/karyodb/evobiR/ [Google Scholar]
- Bosela M, Popa I, Gömöry D, et al. . 2016. Effects of post-glacial phylogeny and genetic diversity on the growth variability and climate sensitivity of European silver fir. Journal of Ecology 104: 716–724. [Google Scholar]
- Bouckaert R. 2010. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics 26: 1372–1373. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, et al. . 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandrud MK, Baar J, Lorenzo MT, et al. . 2019. Phylogenomic relationships of diploids and the origin of allotetraploids in Dactylorhiza (Orchidaceae). Systematic Biology. doi: 10.1093/sysbio/syz035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, Roychoudhury A. 2012. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Molecular Biology and Evolution 29: 1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Gasperi G, Biémont C, Bazin C. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85: 101–106. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3 1: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Molecular Ecology 22: 3124–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe P, Bernatchez L, Funk WC, Andrews KR, Allendorf FW. 2017. Unbroken: RADseq remains a powerful tool for understanding the genetics of adaptation in natural populations. Molecular Ecology Resources 17: 362–365. [DOI] [PubMed] [Google Scholar]
- Cavagnetto C, Anadón P. 1996. Preliminary palynological data on floristic and climatic changes during the Middle Eocene–Early Oligocene of the eastern Ebro Basin, northeast Spain. Review of Palaeobotany and Palynology 92: 281–305. [Google Scholar]
- Chifman J, Kubatko L. 2014. Quartet inference from SNP data under the coalescent model. Bioinformatics 30: 3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coode MJE, Cullen J. 1965. Abies Miller. Flora of Turkey 1: 67–70. [Google Scholar]
- Crisp MD, Cook LG. 2011. Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperms. New Phytologist 192: 997–1009. [DOI] [PubMed] [Google Scholar]
- Davis CC, Schaefer H. 2011. Plant evolution: pulses of extinction and speciation in gymnosperm diversity. Current Biology 21: R995–R998. [DOI] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Molecular Biology and Evolution 28: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, VonHoldt BM. 2011. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- Eaton DaR, Ree RH. 2013. Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae). Systematic Biology 62: 689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban LG, de Palacios P, Aguado LR-L. 2010. Abies pinsapo forests in Spain and Morocco: threats and conservation. Oryx 44: 276. [Google Scholar]
- Fady B, Arbez M, Marpeau A. 1992. Geographic variability of terpene composition in Abies cephalonica Loudon and Abies species around the Aegean: hypotheses for their possible phylogeny from the Miocene. Trees 6: 162–171. [Google Scholar]
- Fady B, Aravanopoulos FA, Alizoti P, et al. . 2016. Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. Forest Ecology and Management 375: 66–75. [Google Scholar]
- Farjon A, Page CN. 1999. Conifers: status survey and conservation action plan. Gland, Switzerland and Cambridge, UK: IUCN-SSC Conifer Specialist Group. [Google Scholar]
- Farjon A, Rushforth K. 1989. A classification of Abies Miller (Pinaceae). Notes of the Royal Botanical Garden of Edinburgh 46: 59–79. [Google Scholar]
- Farjon A, Page CN, Schellevis N. 1993. A preliminary world list of threatened conifer taxa. Biodiversity and Conservation 2: 304–326. [Google Scholar]
- Franco JA. 1950. Abetos. Lisboa: Universidade Tecnica de Lisboa. [Google Scholar]
- George JP, Schueler S, Karanitsch-Ackerl S, Mayer K, Klumpp RT, Grabner M. 2015. Inter- and intra-specific variation in drought sensitivity in Abies spec. and its relation to wood density and growth traits. Agricultural and Forest Meteorology 214–215: 430–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts R. 1995. World checklist of seed plants 1(1) Antwerp, Belgium: MIM. [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. . 2010. A draft sequence of the Neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummer JA, Bryson RW, Reeder TW. 2014. Species delimitation using Bayes factors: simulations and application to the Sceloporus scalaris species group (Squamata: Phrynosomatidae). Systematic Biology 63: 119–133. [DOI] [PubMed] [Google Scholar]
- Hotaling S, Foley ME, Lawrence NM, et al. . 2016. Species discovery and validation in a cryptic radiation of endangered primates: coalescent-based species delimitation in Madagascar’s mouse lemurs. Molecular Ecology 25: 2029–2045. [DOI] [PubMed] [Google Scholar]
- Hrivnák M, Paule L, Krajmerová D, et al. . 2017. Genetic variation in Tertiary relics: the case of eastern-Mediterranean Abies (Pinaceae). Ecology and Evolution 7: 10018–10030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickert-Bond SM, Catarina R, Renner SS. 2009. A fossil-calibrated relaxed clock for Ephedra indicates an Oligocene age for the divergence of Asian and New World clades and Miocene dispersal into South America. Journal of Systematics and Evolution 47: 444–456. [Google Scholar]
- Isoda K, Shiraishi S, Watanabe S, Kitamura K. 2000. Molecular evidence of natural hybridization between Abies veitchii and A. homplepis (Pinaceae) revealed by chloroplast, mitochondrial and nuclear DNA markers. Molecular Ecology 9: 1965–1974. [DOI] [PubMed] [Google Scholar]
- Jackson ND, Carstens BC, Morales AE, O’Meara BC. 2017. Species delimitation with gene flow. Systematic Biology 66: 799–812. [DOI] [PubMed] [Google Scholar]
- Jia F, Lo N, Ho SYW. 2014. The impact of modelling rate heterogeneity among sites on phylogenetic estimates of intraspecific evolutionary rates and timescales. PLoS One 9: e95722. doi: 10.1371/journal.pone.0095722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. 2005. RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. The Plant Journal 44: 730–743. [DOI] [PubMed] [Google Scholar]
- Kavaliauskas D, Fussi B, Westergren M, et al. . 2018. The interplay between forest management practices, genetic monitoring, and other long-term monitoring systems. Forests 9: 133. doi: 10.3390/f9030133. [DOI] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources 15: 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormutak A, Vooková B, Čamek V, et al. . 2013. Artificial hybridization of some Abies species. Plant Systematics and Evolution 299: 1175–1184. [Google Scholar]
- Krajmerová D, Paule L, Zhelev P, et al. . 2016. Natural hybridization in eastern-Mediterranean firs: the case of Abies borisii-regis. Plant Biosystems 150: 1189–1199. [Google Scholar]
- Krasensky J, Jonak C. 2012. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. Journal of Experimental Botany 63: 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijgsman W, Hilgen FJ, Raffi I, Sierro FJ, Wilson DS. 1999. Chronology, causes and progression of the Messinian salinity crisis. Nature 400: 652–655. [Google Scholar]
- Landry P. 1984. Synopsis du genre Abies. Bulletin de la Société Botanique de France. Lettres Botaniques 131: 223–229. [Google Scholar]
- Leaché AD, Fujita MK, Minin VN, Bouckaert RR. 2014. Species delimitation using genome-wide SNP data. Systematic Biology 63: 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepelt S, Mayland-Quellhorst E, Lahme M, Ziegenhagen B. 2010. Contrasting geographical patterns of ancient and modern genetic lineages in Mediterranean Abies species. Plant Systematics and Evolution 284: 141–151. [Google Scholar]
- Linares JC. 2011. Biogeography and evolution of Abies (Pinaceae) in the Mediterranean Basin: the roles of long-term climatic change and glacial refugia. Journal of Biogeography 38: 619–630. [Google Scholar]
- Liu T-S. 1971. A monograph of the genus Abies. Taiwan: Department of Forestry, National Taiwan University. [Google Scholar]
- Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, Storfer A. 2016. Breaking RAD: an evaluation of the utility of restriction site associated DNA sequencing for genome scans of adaptation. Molecular Ecology Resources 17: 142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire R. 1952. Flore de l’Afrique du Nord (Maroc, Algérie, Tunisie, Tripolitaine, Cyrénaïque et Sahara), Vol. 1 Paris: Paul Lechevalier. [Google Scholar]
- Malinsky M, Trucchi E, Lawson D, Falush D. 2018. RADpainter and fineRADstructure: population inference from RADseq data. Molecular Biology and Evolution 35: 1284–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Slazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD. 2013. Heliconius and sympatric speciation. Genome Research 23: 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattfeld J, Bornmüller J, von Handel-Mazzetti F. 1925. Zur Kenntnis der Formenkreise der europäischen und kleinasiatischen Tannen. Notizblatt des Botanischen Gartens und Museums zu Berlin-Dahlem 84: 229–246. [Google Scholar]
- Melville J, Smith K, Hobson R, Hunjan S, Shoo L. 2014. The role of integrative taxonomy in the conservation management of cryptic species: the taxonomic status of endangered earless dragons (Agamidae: Tympanocryptis) in the grasslands of Queensland, Australia. PLoS One 9: e101847. doi: 10.1371/journal.pone.0101847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergen F, Burley J, Simpson B. 1962. Artificial hybridization in Abies. Der Züchter 34: 242–251. [Google Scholar]
- Miller MA, Schwartz T, Pickett BE, et al. . 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evolutionary Bioinformatics 11: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingum NS, Marshall CR, Quental TB, Rai HS, Little DP, Mathews S. 2011. Recent synchronous radiation of a living fossil. Science 334: 796–799. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, et al. . 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584. [DOI] [PubMed] [Google Scholar]
- Oliver KR, McComb JA, Greene WK. 2013. Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biology and Evolution 5: 1886–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. Ape: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Parducci L, Szmidt AE. 1999. PCR-RFLP analysis of cpDNA in the genus Abies. Theoretical and Applied Genetics 98: 802–808. [Google Scholar]
- Parducci L, Szmidt AE, Madaghiele A, Anzidei M, Vendramin GG. 2001a Genetic variation at chloroplast microsatellites (cpSSRs) in Abies nebrodensis (Lojac.) Mattei and three neighboring Abies species. Theoretical and Applied Genetics 102: 733–740. [Google Scholar]
- Parducci L, Szmidt A, Ribeiro M, Drouzas AD. 2001b Taxonomic position and origin of the endemic Sicilian fir Abies nebrodensis (Lojac.) Mattei based on allozyme analysis. Forest Genetics 8: 119–127. [Google Scholar]
- Paun O, Turner B, Trucchi E, Munzinger J, Chase MW, Samuel R. 2016. Processes driving the adaptive radiation of a tropical tree (Diospyros, Ebenaceae) in New Caledonia, a biodiversity hotspot. Systematic Biology 65: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter KM. 2018. Do United States protected areas effectively conserve forest tree rarity and evolutionary distinctiveness? Biological Conservation 224: 34–46. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Harvey PH. 2000. Testing macro-evolutionary models using incomplete molecular phylogenies. Proceedings of the Royal Society B: Biological Sciences 267: 2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao CY, Ran JH, Li Y, Wang XQ. 2007. Phylogeny and biogeography of Cedrus (Pinaceae) inferred from sequences of seven paternal chloroplast and maternal mitochondrial DNA regions. Annals of Botany 100: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67: 901–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud G, Stenzel U, Maricic T, Wiebe V, Kelso J. 2015. DeML: robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics 31: 770–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson KA, Maze J, Scagel RK, Banerjee S. 1993. Ontogeny, phylogeny and intraspecific variation in North-American Abies Mill. (Pinaceae): an empirical approach to organization and evolution. Taxon 42: 17–34. [Google Scholar]
- Ru D, Mao K, Zhang L, Wang X, Lu Z, Sun Y. 2016. Genomic evidence for polyphyletic origins and interlineage gene flow within complex taxa: a case study of Picea brachytyla in the Qinghai–Tibet Plateau. Molecular Ecology 25: 2373–2386. [DOI] [PubMed] [Google Scholar]
- Rundel PW, Arroyo MTK, Cowling RM, Keeley JE, Lamont BB, Vargas P. 2016. Mediterranean biomes: evolution of their vegetation, floras and climate. Ecology, Evolution, and Systematics 47: 383–407. [Google Scholar]
- Sánchez Cózar S. 1946. El Abies del Tazaot. Revista de la Real Academia de Ciencias de Madrid XL: 449–468. [Google Scholar]
- Sánchez-Robles JM, Balao F, Terrab A, et al. . 2014. Phylogeography of SW Mediterranean firs: different European origins for the North African Abies species. Molecular Phylogenetics and Evolution 79: 42–53. [DOI] [PubMed] [Google Scholar]
- Sánchez-Salguero R, Camarero JJ, Carrer M, et al. . 2017. Climate extremes and predicted warming threaten Mediterranean Holocene firs forests refugia. Proceedings of the National Academy of Sciences, USA 114: E10142–E10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltsoyiannes A, Tsaktsira M, Drouzas AD. 1999. Allozyme differentiation in the Mediterranean firs (Abies, Pinaceae). A first comparative study with phylogenetic implications. Plant Systematics and Evolution 216: 289–307. [Google Scholar]
- Scarascia-Mugnozza G, Oswald H, Piussi P, Radoglou K. 2000. Forests of the Mediterranean region: gaps in knowledge and research needs. Forest Ecology and Management 132: 97–109. [Google Scholar]
- Schorn HE, Wesley CW. 1986. Abies milleri, sp. nov., from the Middle Eocene Klondike mountain formation, Republic, Ferry county, Washington. Burke Museum Contributions in Anthropology and Natural History 1: 1–7. [Google Scholar]
- Sekiewicz K, Sekiewicz M, Jasińska AK, et al. . 2013. Morphological diversity and structure of West Mediterranean Abies species. Plant Biosystems 147: 125–134. [Google Scholar]
- Semerikova SA, Semerikov VL. 2014. Molecular phylogenetic analysis of the genus Abies (Pinaceae) based on the nucleotide sequence of chloroplast DNA. Russian Journal of Genetics 50: 7–19. [PubMed] [Google Scholar]
- Semerikova SA, Semerikov VL. 2016. Phylogeny of firs (genus Abies, Pinaceae) based on multilocus nuclear markers (AFLP). Russian Journal of Genetics 52: 1164–1175. [PubMed] [Google Scholar]
- Semerikova SA, Semerikov VL, Lascoux M. 2011. Post-glacial history and introgression in Abies (Pinaceae) species of the Russian Far East inferred from both nuclear and cytoplasmic markers. Journal of Biogeography 38: 326–340. [Google Scholar]
- Serrato-Capuchina A, Matute DR. 2018. The role of transposable elements in speciation. Genes 9: pii: E254. doi: 10.3390/genes9050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soraggi S, Wiuf C, Albrechtsen A. 2018. Powerful inference with the D-statistic on low-coverage whole-genome data. G3 8: 551–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suc P. 1984. Origin and evolution of the Mediterranean vegetation and climate in Europe Nature 307: 429–432. [Google Scholar]
- Suyama Y, Yoshimaru H, Tsumura Y. 2000. Molecular phylogenetic position of Japanese Abies (Pinaceae) based on chloroplast DNA sequences. Molecular Phylogenetics and Evolution 16: 271–277. [DOI] [PubMed] [Google Scholar]
- Swofford D. 2002. PAUP* version 4.0. Phylogenetic analysis using parsimony (and other methods). Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Terrab A, Talavera S, Arista M, Paun O, Stuessy TF, Tremetsberger K. 2007. Genetic diversity at chloroplast microsatellites (cpSSRs) and geographic structure in endangered West Mediterranean firs (Abies spp., Pinaceae). Taxon 56: 409–416. [Google Scholar]
- Tiffin P, Ross-Ibarra J. 2014. Advances and limits of using population genetics to understand local adaptation. Trends in Ecology & Evolution 29: 673–680. [DOI] [PubMed] [Google Scholar]
- Trucchi E, Frajman B, Haverkamp THA, Schönswetter P, Paun O. 2017. Genomic analyses suggest parallel ecological divergence in Heliosperma pusillum (Caryophyllaceae). New Phytologist 216: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas P, Fernández-Mazuecos M, Heleno R. 2018. Phylogenetic evidence for a Miocene origin of Mediterranean lineages: species diversity, reproductive traits and geographical isolation. Plant Biology 20: 157–165. [DOI] [PubMed] [Google Scholar]
- Vicario F, Vendramin GG, Rossi P, Liò P, Giannini R. 1995. Allozyme, chloroplast DNA and RAPD markers for determining genetic relationships between Abies alba and the relic population of Abies nebrodensis. Theoretical and Applied Genetics 90: 1012–1018. [DOI] [PubMed] [Google Scholar]
- Voronova A, Belevich V, Korica A, Rungis D. 2017. Retrotransposon distribution and copy number variation in gymnosperm genomes. Tree Genetics and Genomes 13: 88. [Google Scholar]
- Xiang QP, Wei R, Shao YZ, Yang ZY, Wang XQ, Zhang XC. 2015. Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Molecular Phylogenetics and Evolution 82: 1–14. [DOI] [PubMed] [Google Scholar]
- Xiang QQ-Y, Xiang QQ-Y, Guo Y, et al. . 2009. Phylogeny of Abies (Pinaceae) inferred from nrITS sequence data. Taxon 58: 141–152. [Google Scholar]
- Xiang X, Cao M, Zhou Z. 2007. Fossil history and modern distribution of the genus Abies (Pinaceae). Frontiers of Forestry in China 2: 355–365. [Google Scholar]
- Zachos JC, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- Zachos JC, Dickens GR, Zeebe RE. 2008. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451: 279–283. [DOI] [PubMed] [Google Scholar]
- Ziegenhagen B, Fady B, Kuhlenkamp V, Liepelt S. 2005. Differentiating groups of Abies species with a simple molecular marker. Silvae Genetica 54: 123–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.