Abstract
Through fusing CRISPR-Cas9 nickases with cytidine or adenine deaminases, a new paradigm-shifting class of genome-editing technology, termed “base editors,” has recently been developed. Base editors mediate highly efficient, targeted single-base conversion without introducing double-stranded breaks. Analysis of base editing outcomes typically relies on imprecise enzymatic mismatch cleavage assays, time-consuming single-colony sequencing, or expensive next-generation deep sequencing. To overcome these limitations, several groups have recently developed computer programs to measure base-editing efficiency from fluorescence-based Sanger sequencing data such as Edit deconvolution by inference of traces in R (EditR), TIDER, and ICE. These approaches have greatly simplified the quantitation of base-editing experiments. However, the current Sanger sequencing tools lack the capability of batch analysis and producing high-quality images for publication. Here, we provide a base editing analysis tool (BEAT) written in Python to analyze and quantify the base-editing events from Sanger sequencing data in a batch manner, which can also produce intuitive, publication-ready base-editing images.
Introduction
Base editing is a new generation of genome-editing technology that directly installs point mutations into cellular DNA or RNA without making double-stranded DNA breaks, requiring a DNA donor template, or relying on cellular homology-directed repair.1–6 DNA base editors use RNA-programmable Cas9 nickases to target specific DNA locus and nucleobase deaminase enzymes to catalyze the conversion of one base to another. By incorporating different DNA deaminases, two classes of DNA base editor have been developed: cytosine base editors (CBEs), converting a C–G base pair into a T–A base pair,2,4–6 and adenine base editors (ABEs), converting an A–T base pair into a G–C base pair.3 Together, the CBEs and ABEs can mediate four possible transition mutations (C to T, A to G, T to C, and G to A).
Several approaches have been used to analyze base editing events, including imprecise enzymatic mismatch cleavage assays (such as Cel-I, T7E1, Surveyor, or Guide-it Resolvase),7–9 Sanger sequencing of polymerase chain reaction (PCR) amplicons from randomly picked bacterial colonies,4,10 and next-generation deep sequencing (NGS) of the target site.5,6,11–13 However, all these approaches have various disadvantages. For example, the enzymatic cleavage assays cannot resolve the positional effect (i.e., at which positions the bases are mutated), as they only detect the presence of a mismatch bubble formed in heteroduplexes of stochastically annealed PCR amplicons.14 While the Sanger sequencing of PCR amplicons from individually picked bacterial colonies and NGS can resolve the details of the base editing events, they are time-consuming, labor-intensive, and expensive.
Recently, several groups developed fast and simple computer programs to analyze the genome-editing events from the fluorescence-based Sanger sequencing data of PCR amplicons using raw samples such as the web tools Tracking of Indels by Decomposition (TIDE), Poly Peak Parser, Edit deconvolution by inference of traces in R (EditR), and BEEP.15–18 These approaches have greatly simplified the analysis of base-editing studies, although Sanger sequencing as a method to quantify base-editing outcomes has its own drawbacks such as an inability to measure and score specific alleles that are generated from base editing and an inability to detect low-frequency bystander or indel events that occur frequently when using base editors. The current Sanger sequencing tools do not offer the capability of batch analysis, and often they do not allow high-quality image production. Here, we provide a base editing analysis tool (BEAT) written in Python to analyze and quantify the base-editing events from Sanger sequencing data in a batch manner, which can also produce intuitive, publication-ready base-editing images. BEAT is available as a Python script at https://github.com/HanLab-OSU/Beat/, and has been tested under Python 2.7.10 and Python 3.7.1. A stand-alone executable version for Windows 7 is also provided at https://github.com/HanLab-OSU/Beat/.
Materials and Methods
Cell culture and transfection
HEK293 cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 1% penicillin/streptomycin (Gibco). For transient transfection with base editor and guide RNA constructs, 1 × 105 HEK293 cells were seeded per well on six-well plates and transfected with 1 μg gRNA plasmid and 1 μg ABE plasmid using the X-tremeGENE HP DNA Transfection Reagent (Roche) following the manufacturer's instructions.
Constructs
The single A to G point mutant Ano5 and its WT version, as described previously,19 were used as standard controls. The base editors pCMV-ABE7.10 (Addgene; plasmid #102919), pCMV_ABEmax (Addgene; plasmid #112095), and pCMV_AncBE4max (Addgene; plasmid #112094) were obtained. The guide sequences were cloned into pLenti-ogRNA_zeo, which was modified from Lenti-sgRNA(MS2)_zeo backbone (Addgene; plasmid #61427) by using an optimized gRNA scaffold.20,21 The gRNAs were designed using CHOPCHOP.22,23 The gRNA target sequences used are listed in Table 1.
Table 1.
List of gRNA Sequences Used in This Study
| Site 1 | 5′ GAACACAAAGCATAGACTGC GGG |
| Site 2 | 5′ GAGTATGAGGCATAGACTGC AGG |
| Site 5 | 5′ GATGAGATAATGATGAGTCA GGG |
PCR and Sanger sequencing
Genomic DNA (∼1 × 106 cells) was isolated and precipitated by isopropanol 5 days after transfection. PCR reactions were carried out with 100 ng genomic DNA or 10 ng plasmid DNA in the GoTaq Master Mix (Promega) according to the manufacturer's instruction. PCR conditions were 5 min at 95°C (1 × ), followed by 15 s at 95°C, 15 s at 60°C, and 30 s at 72°C (32 cycles). The PCR primers are listed in Table 2. The PCR products were purified using the Wizard SV Gel and PCR Clean-up System (Promega). Purified PCR products (100 ng) were subjected to Sanger sequencing at the Ohio State University Comprehensive Cancer Center Genomics Shared Resource.
Table 2.
List of PCR Primers for the Ano5 Plasmids and gRNA Target Sites
| Ano5 plasmids | F: 5′ GCGATTCAATTTGTTCCTGAG |
| R: 5′ CCTGAATGCAAACTGTGTCAA | |
| Site 1 | F1: 5′ AACCAGTGTCAGGGAGCTGT |
| R1: 5′ ATCCACAGCAACACCCTCTC | |
| F2: 5′ AGGACGTCTGCCCAATATGT | |
| R2: 5′ CAGCCCCATCTGTCAAACTG | |
| Site 2 | F: 5′ CAGGAATATCTGTGTGTGAGCCATA |
| R: 5′ AGGAGTTCGCAGTGAGCCG | |
| Site 5 | F1: 5′ AAGGTTTTGGGCTTCATTCC |
| R1: 5′ CGCCTGGTCACATTGACTTT | |
| F2: 5′ CTCAAAACGGTAGAGCAGGC | |
| R2: 5′ AGGCTGGTCTTGAACTCCTG |
BEAT software
BEAT code was written and tested in Python v2.7.10. For analysis of a single sequence file, BEAT requires as input the folder name, the sequence data file name (.ab1), a spacer sequence of the gRNA, the base change position with the spacer, and the change pattern (e.g., A to G mutation as AG). BEAT can also analyze multiple files in a batch manner. In this case, the file directory, file names, spacer sequences, base change positions, and change patterns could be pre-entered into a CSV file, or all sequencing files within a folder can be analyzed altogether if they have the same spacer sequence.
BEAT first aligns the spacer sequence to the base calls of the Sanger sequencing data to determine the position of the spacer. The BEAT program can handle the case when degenerate bases (e.g., R for A or G) are present in base calls, which is particularly common for base-editing experiments. BEAT then estimates the average background noises for each base from the trace data after trimming the first 100 bases and the last 50 bases. To account for the variability in sequencing, the user can manually select the region to calculate the background noises in case the default trimming does not effectively remove low-quality sequencing. Next, the value of every “N” trace peak value under every non-“N” base call (e.g., G peak value under A, C, or G peaks) is compiled to generate a sample of the noise distribution. We tested several methods, including the z-score, median absolute deviation (MAD), and interquartile ranges (IQR),24 to identify and remove outliers in the noise data, and all produced similar results. By default, we chose the MAD method for outlier identification and removal. After removing the outliers from the noise data, the average noise for each base is then calculated and subtracted from the peak values of each base at each position along the spacer. The percentage of each base at each position is then calculated as the percentage of the background-subtracted peak values of each base over their sum at that position. We found that using the background-subtracted peak values to calculate the percentage of each base yielded similar results as using the peak area as EditR employed. Following the calculation, BEAT then saves the calculated data in a Microsoft Excel file for each sequencing file. It also plots the trace and the table showing the percentage of each base along the spacer, and saves them as a PNG image file.
Statistical analysis
The data are expressed as the mean ± standard error of the mean and were analyzed using GraphPad Prism v5.02 (GraphPad Software). Statistical significance was determined using Student's t-test for two groups or one-way analysis of variance followed by Bonferroni post hoc tests for multiple groups. A p-value of <0.05 was regarded as significant.
Results
In vitro validation and comparison with EditR
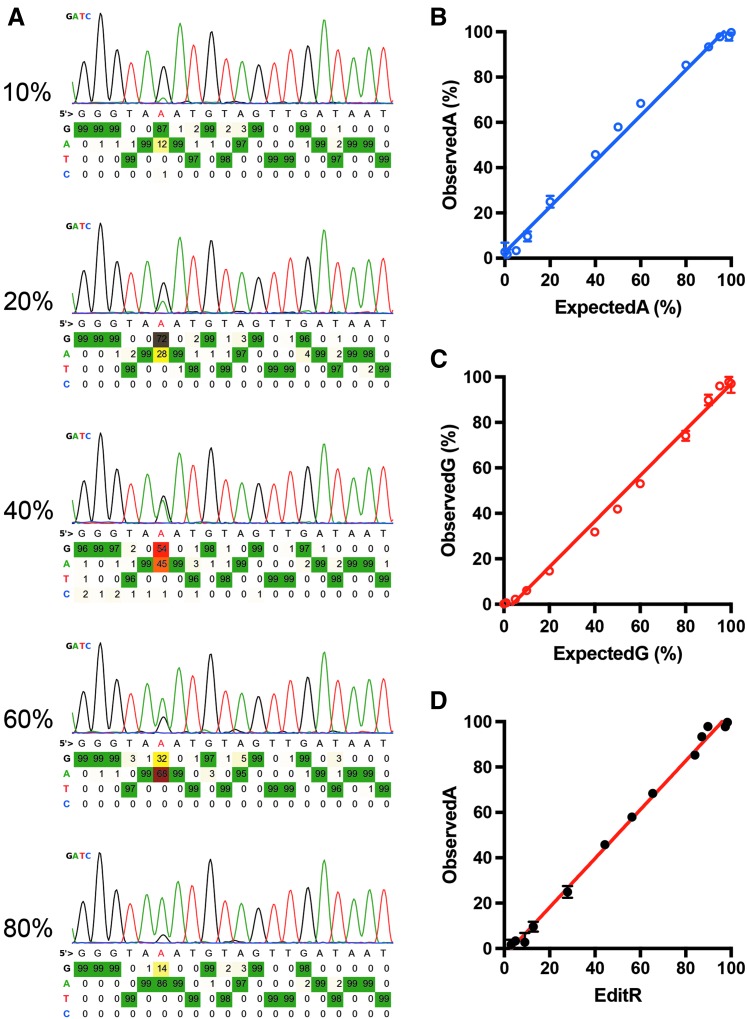
We first tested the sensitivity of BEAT to differentiate PCR amplicons with one defined mutation (A to G) in plasmid DNAs. Plasmids containing wild-type (WT) and mutant intron 6 of the human ANO5 gene were used as a template to amplify a region surrounding the mutation. The PCR products were gel purified, quantified, mixed with different molar ratios, and subjected to Sanger sequencing. The resulting sequencing data were analyzed by BEAT to determine the percentage of A-containing (WT) and G-containing (mutant) products. With an increasing amount of A products, BEAT faithfully detected an increased percentage of A signal (Fig. 1A). Figure 1B shows the correlation of the expected percentage of WT products and the observed percentage by BEAT, and Figure 1C shows the correlation of the expected percentage of mutant products and the corresponding observed values by BEAT. Clearly, when both A and G products were between 5% and 95%, the observed values were highly correlated with the expected values. For example, when these two products were mixed in an equal molar ratio, ∼58% of A products and 42% of G products were identified by the BEAT program. However, when either of these products was out of this range, the detected values deviated significantly from the expected values.
FIG. 1.
In vitro validation of a base editing analysis tool (BEAT). Two polymerase chain reaction (PCR) products carrying a single nucleotide difference were mixed at different ratios—1:0, 0.95:0.05, 0.9:0.1, 0.8:0.2, 0.6:0.4, 0.5:0.5, 0.4:0.6, 0.2:0.8, 0.1:0.9, 0.05:0.95, and 0:1—and subjected to Sanger sequencing. The sequencing trace files were analyzed by BEAT. (A) Sample image output of the sequencing data from the BEAT analysis. The percentages on the left indicate the percentage of A-containing products. (B) Correlation between the percentage of detected A-containing products and the percentage of expected A-containing products. (C) Correlation between the percentage of detected G-containing products and the percentage of expected G-containing products. (D) Correlation of A-containing product percentage as determined by BEAT and Edit deconvolution by inference of traces in R (EditR).
We then compared the performance of BEAT and EditR programs. The detected values by both programs showed a high degree of overlapping (Fig. 1D). They could both detect as low as 5% of G-containing product in the mixture, with this limitation being primarily set by the Sanger sequencing itself.
Application of BEAT to A-to-G edited sequences
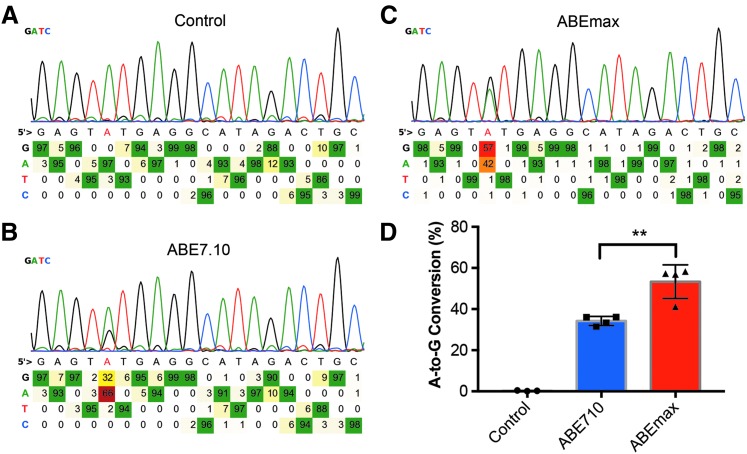
We then tested this approach to quantify the editing efficiency of adenine base editors on a pool of HEK293 cells transfected with ABE7.10 plus or minus a gRNA targeting site 2. BEAT determined that 31% of the A in position 5 was converted G in cell samples co-transfected with ABE7.10 and the gRNA (Fig. 2A), while the control sample with only ABE7.10 transfection did not show an appreciable level of base conversion at this position (Fig. 2B).
FIG. 2.
BEAT detected an increased editing efficiency of codon-optimized ABEmax compared with the original ABE7.10. (A–C) Sanger sequencing data of PCR products from HEK293 cells transfected with irrelevant plasmid control (A), ABE7.10 plus sgRNA targeting Site2 (B), or ABEmax plus sgRNA targeting Site2 (C). (D) A higher rate of A-to-G conversion at position 5 was detected in cells transfected with ABEmax. **p < 0.01.
Previous work demonstrated that genetic codon optimization can boost base-editing efficiency in human cells. We then compared the base-editing efficiency as determined by BEAT in HEK293 cells transfected with codon-optimized ABE7.10 (ABEmax) or regular ABE7.10. Compared to ABE7.10, the ABEmax induced a significantly higher level of A-to-G conversion at position 5 (Fig. 2C and D). Thus, BEAT can be used to quantify the editing efficiencies of different variants of base editors rapidly using Sanger sequencing traces of PCR amplicons.
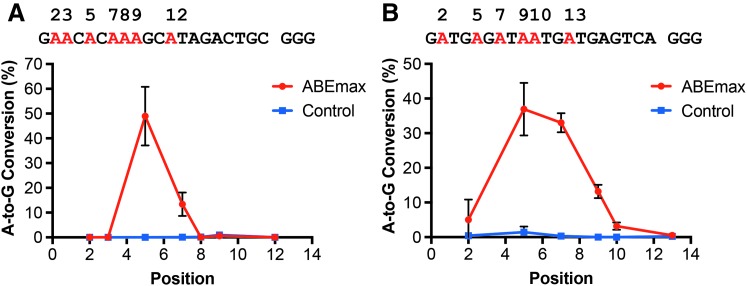
It has been shown that the ABE editors mutate As at a narrow window, ranging from position 4 to position 8 distal from PAM.3 We then chose two A-enriched sites (sites 1 and 5) for testing. Plotting the A-to-G conversion rates enabled us to analyze the editing window quickly and quantitatively. As shown in Figure 3A, ABEmax mutated the As mainly at positions 5 and 7 for site 1, with little to no effects on As at positions 2, 3, 8, and 9 and the other As proximal to the PAM. Similarly, for site 5, we observed that the ABEmax converted A to G mainly at positions 5 and 7, with lower levels at position 9 and no activities at other As (Fig. 3B). These results are consistent with previous reports that ABEmax primarily converts A to G within a narrow window.3
FIG. 3.
Determination of the editing window by ABEmax. (A) The plot of A-to-G conversion rates at multiple positions containing A for site 1 showed that the ABEmax mainly mutated A at positions 5 and 7. (B) For site 5, the ABEmax also converted A to G mainly at positions 5 and 7, with lower levels at position 9.
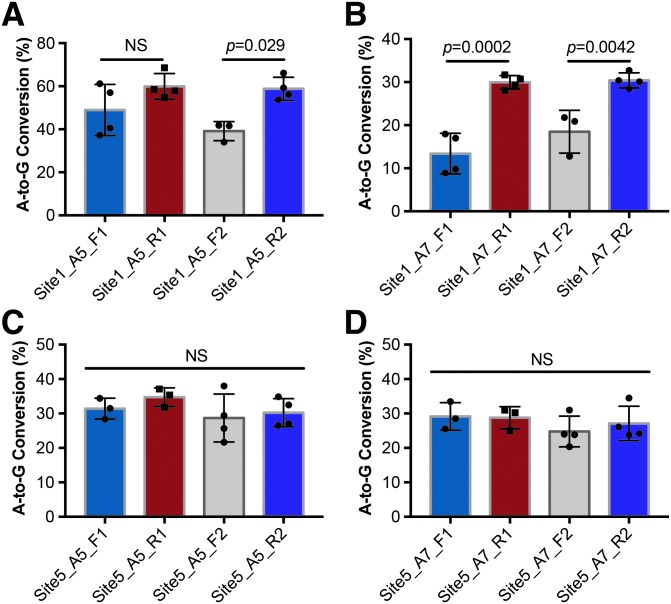
Impact of the amplification and sequencing primers on the outcomes of BEAT quantification
As the quantification of editing frequencies relies on the amplification of the genomic DNA and Sanger sequencing, the outcomes of quantification may be impacted by the choices of different primers for both amplification and sequencing. To test this, we used two different primer pairs to amplify site 1 and site 5 and to sequence each amplicon using both forward and reverse primers. While no significant difference was found when site 5 was analyzed with four different primers in either the forward or reverse direction (Fig. 4C and D), quantification of site 1 was found to be prone to the direction of primers chosen for sequencing (Fig. 4A and B). Sequencing with the reverse primers tended to yield higher editing frequencies (for both A5 and A7) compared to the forward primers. No significant differences were found between the two forward primers or between the two reverse primers. These results suggest that Sanger sequencing can be affected by the choice of sequencing primer directions.
FIG. 4.
The impact of primers on base editing quantification using BEAT. The A-to-G editing frequencies at (A) A5 and (B) A7 of site 1 were measured and quantified using two primer pairs (Site1-F1/R1 and Site1-F2/R2). The A-to-G editing frequencies at (C) A5 and (D) A7 of site 5 were measured and quantified using two primer pairs (Site5-F1/R1 and Site5-F2/R2). NS, not significant.
Discussion
In this work, we provide a Python program, BEAT, for rapid analysis and quantification of base-editing efficiency. The robustness of BEAT has been validated using plasmid DNA PCR products and cellular DNA following transfection of base editors. Analysis of Sanger sequencing traces with BEAT offers us a rapid and low-cost approach to determine editing efficiency at a certain position, as well as at different positions along the entire targeting sequences. The latter would be very useful for rapid analysis of the editing windows for new ABEs or engineered ABE variants to expand or narrow the targeting space. Although we tested ABEs only, the BEAT program should allow analysis of CBE editing events and other targeted genomic DNA modifications such as homology-directed single nucleotide mutations, for which the efficiency can now be improved.25
Compared to other available programs such as EditR, BEAT can analyze not only individual sequencing files but also multiple files in a batch manner. Moreover, publication-ready images are generated in addition to the Microsoft Excel file containing the percentage data of all four bases at each position along the target site, which can be used for further quantitative analysis.
Obviously, our program is limited to analyzing the base-editing events with an efficiency at around 5% or above, as Sanger sequencing is unable to distinguish trace amounts of bases at any position reliably. For application to determine rare editing events such as analysis of low-frequency bystander or indel events, alternative approaches such as deep sequencing would be required. Also, Sanger sequencing of amplicons from pooled samples could not measure or score specific alleles generated from base editing.
As our data showed, quantification of base-editing frequencies using the Sanger sequencing method can be affected by the directions of the sequencing primers at certain site, thus making it unreliable to quantify the absolute editing frequencies. However, for many applications that require only relative quantification of the editing frequencies at the same target sites (e.g., in cases of screening different base-editor variants or determining the editing windows), Sanger sequencing is still the most time- and cost-saving approach.
Acknowledgments
We thank members of the Han lab for helpful discussion and suggestions. R.H. was supported by U.S. National Institutes of Health grants (R01HL116546 and R01AR064241).
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018;19:770–788. DOI: 10.1038/s41576-018-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420–424. DOI: 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551:464–471. DOI: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016;353 DOI: 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- 5. Ma Y, Zhang J, Yin W, et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 2016;13:1029–1035. DOI: 10.1038/nmeth.4027 [DOI] [PubMed] [Google Scholar]
- 6. Hess GT, Fresard L, Han K, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 2016;13:1036–1042. DOI: 10.1038/nmeth.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu L, Zhao P, Mariano A, et al. Targeted myostatin gene editing in multiple mammalian species directed by a single pair of TALE nucleases. Mol Ther Nucleic Acids 2013;2:e112 DOI: 10.1038/mtna.2013.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mariano A, Xu L, Han R. Highly efficient genome editing via 2A-coupled co-expression of two TALEN monomers. BMC Res Notes 2014;7:628 DOI: 10.1186/1756-0500-7-628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu L, Park KH, Zhao L, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther 2016;24:564–569. DOI: 10.1038/mt.2015.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shimatani Z, Kashojiya S, Takayama M, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 2017;35:441–443. DOI: 10.1038/nbt.3833 [DOI] [PubMed] [Google Scholar]
- 11. Komor AC, Zhao KT, Packer MS, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 2017;3:eaao4774 DOI: 10.1126/sciadv.aao4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim YB, Komor AC, Levy JM, et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 2017;35:371–376. DOI: 10.1038/nbt.3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El Refaey M, Xu L, Gao Y, et al. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res 2017;121:923–929. DOI: 10.1161/CIRCRESAHA.117.310996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Till BJ, Burtner C, Comai L, et al. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res 2004;32:2632–2641. DOI: 10.1093/nar/gkh599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brinkman EK, Chen T, Amendola M, et al. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 2014;42:e168 DOI: 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hill JT, Demarest BL, Bisgrove BW, et al. Poly peak parser: method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev Dyn 2014;243:1632–1636. DOI: 10.1002/dvdy.24183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kluesner MG, Nedveck DA, Lahr WS, et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J 2018;1:239–250. DOI: 10.1089/crispr.2018.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv 2018;4:eaau0766 DOI: 10.1126/sciadv.aau0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J, Xu L, Lau YS, et al. A novel ANO5 splicing variant in a LGMD2L patient leads to production of a truncated aggregation-prone Ano5 peptide. J Pathol Clin Res 2018;4:135–145. DOI: 10.1002/cjp2.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma H, Tu LC, Naseri A, et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 2016;34:528–530. DOI: 10.1038/nbt.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen B, Hu J, Almeida R, et al. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res 2016;44:e75 DOI: 10.1093/nar/gkv1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Labun K, Montague TG, Gagnon JA, et al. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 2016;44:W272–276. DOI: 10.1093/nar/gkw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montague TG, Cruz JM, Gagnon JA, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 2014;42:W401–407. DOI: 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rousseeuw PJ, Croux C. Alternatives to the median absolute deviation. J Am Stat Assoc 1993;88:1273–1283. DOI: 10.1080/01621459.1993.10476408 [DOI] [Google Scholar]
- 25. O'Brien AR, Wilson LOW, Burgio G, et al. Unlocking HDR-mediated nucleotide editing by identifying high-efficiency target sites using machine learning. Sci Rep 2019;9:2788 DOI: 10.1038/s41598-019-39142-0 [DOI] [PMC free article] [PubMed] [Google Scholar]