Abstract
Introduction
This study was conducted to evaluate the microbiomes of endodontic- periodontal lesions (EPL) before and after chemomechanical preparation (CMP).
Methods
Clinical samples were taken from 15 root canals (RC) with necrotic pulp tissues and from their associated periodontal pockets (PP, n=15) of teeth with EPL before and after CMP. The Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) protocol and viable culture were used to analyze samples from RC and PP. The Mann-Whitney test and Benjamini-Hochberg corrections were performed to correlate the clinical and radiographic findings with microbial findings (p<0.05).
Results
Bacteria were detected in 100% of the samples in both sites (15/15) using NGS. Firmicutes was the most predominant phylum in both sites using both methods. The most frequently detected species in the RC before and after CMP by NGS were Enterococcus faecalis, Parvimonas micra, Mogibacterium timidum, Filifactor alocis, and Fretibacterium fastidiosum. Species most frequently detected in the PP before and after CMP by NGS were P. micra, E. faecalis, Streptococcus constellatus, Eubacterium brachy, Tannerella forsythia and F. alocis. Associations were found between periapical lesions ≤ 2 mm and Desulfobulbus sp. oral taxon 041; and with periodontal pockets ≥ 6mm and Dialister invisius and Peptostreptococcus stomatis (all p<0.05, found in the RC before CMP).).
Conclusion
It is concluded that the microbial community present in combined endodontic-periodontal lesions is complex and more diverse than previously reported. It is important to note that bacteria do survive in some root canals after chemomechanical preparation. Finally, the similarity between the microbiota of both sites, before and after chemomechanical preparation, suggests there may be a pathway of infection between the pulp and periodontium.
Keywords: bacteria, endodontic-periodontal lesions, NGS, culture, HOMINGS
Introduction
Endodontic-periodontal lesions, also known as endo-perio lesions, can be defined as pathological changes that reach both the pulp and periodontal tissues. The term endodontic-periodontal, however, does not differentiate itself the source of the injury, which can be initiated either in the pulp or in the periodontium. In these cases, the pulp is always necrotic and the clinical signs of the periodontal disease include the presence of periodontal pockets, loss of attachment and radiographically visible bone loss (39).
Periodontitis is considered the major cause for tooth loss in the adult population, affecting 47% of North Americans (8). Treatment of combined endodontic-periodontal lesions should be well-planned and coordinated, including both endodontic and periodontal therapy in an integrated manner to ensure the success of the combined treatment of these lesions (1). Endodontic infection without treatment may cause a delay in periodontal healing, even in cases where the periodontal treatment has already been initiated (7, 22). Microorganisms and their by-products may escape the infected root canal through several pathways, including the apical foramen and lateral and accessory canals promoting the coalescence with an adjacent periodontal lesion (12).
The best and safest method to decontaminate the root canal is through the careful cleaning of its necrotic content by means of instrumentation, irrigation and aspiration (38). Recently, chlorhexidine has been shown to be an effective endodontic auxiliary chemical substance because of its antimicrobial action and its adsorption to dental hard tissues with gradual and prolonged release at therapeutic levels (16). However, total disinfection of the root canal space is still not fully achieved in clinical practice due to the anatomical complexities of many root canals, and consequent limitations in access by instruments and irrigants (3, 10). Moreover, the efficacy of these measures may also depend upon the susceptibility of the involved species, which may not be uniform.
Traditionally, identification of microorganisms in samples is based on endodontic culture methods involving isolation and subsequent identification using morphological and biochemical tests. However, the prevalence of some oral pathogens may be underestimated using this technique since many bacterial species are difficult to grow or have not-yet been grown in vitro. Recently, the development of next-generation sequencing (NGS) technologies has permitted in-depth sequencing and data analyses to a deeper level than possible with standard molecular biological techniques (42)
Therefore, the aim of this study was to compare the microbial profiles of endodontic-periodontal lesions before and after chemomechanical preparation (CMP) by using NGS, as well as by culture techniques.
Material and Methods
Subject Population
Fifteen subjects presenting combined endodontic-periodontal diseases, from those who came for endodontic treatment to the Piracicaba Dental School, State University of Campinas–UNICAMP, Piracicaba, SP, Brazil, were included in this study.
Endodontic-periodontal lesions were defined as the presence of periodontal pockets > 6 mm, pulpal necrosis and radiographic evidence of radiographic evidence of apical periodontitis. The pulp status was assessed through thermal vitality tests, and the apical condition was determined through the observation of clinical signs, such as tenderness to percussion and pain on palpation.
Exclusion criteria are as follows. Subjects had undergone periodontal treatment within the past year, had received antibiotic treatment within the preceding three months, had teeth with periodontal probing depth < 6 mm, and reported systemic disease. Smoking was not a criterion, but all patients were non-smokers.
The Human Research Ethics Committee of the Piracicaba Dental School approved a protocol describing the sample collection for this investigation, and all patients signed an informed consent for their participation in this research.
Sampling Procedure and clinical data collection
Periodontal plaque sampling
Periodontal plaque samples were collected prior to root canal treatment from the deepest periodontal pocket (PP) of the involved tooth, according to Gonçalves et al. (17), using 3 consecutive sterile paper points, which were kept in place for 60 s and then pooled in a sterile tube containing 1 mL VMGA III transport medium, which was frozen at −70oC until being processed.
The following periodontal clinical parameters were taken by one calibrated examiner at the baseline visit (prior to treatment): pocket depth, bleeding on probing, clinical attachment level, and plaque Index. All measurements were performed using a periodontal probe at six sites per tooth and were recorded. Periapical and interproximal radiographs were taken on all patients at the baseline visit for diagnosis purposes only.
A second periodontal sample was taken in a similar way at the end of the visit, after the rubber dam removal.
Endodontic sample collection and clinical procedures
The method followed for the disinfection of the operative field and sampling procedures had been described previously (9, 12, 28). The teeth were isolated with a rubber dam. The crown and surrounding structures were disinfected with 30% H2O2 [volume/volume (V/V)] for 30s followed by 2.5% NaOCl for the same period of time and then inactivated with 5% sodium thiosulfate. The disinfection of the tooth surface was monitored by taking a swab sample from both external and internal surfaces of the crown and its surrounding structures area around and streaking it on blood agar plates. Plates were incubated aerobically and anaerobically.
A two-stage, access cavity preparation was made under manual irrigation with sterile saline solution without the use of water spray, and by using a sterile high-speed diamond bur. The first stage was performed to promote removal of major contaminants. In the second stage before entering the pulp chamber, the access cavity was disinfected according to the protocol described above. The disinfection of the internal surface of the access cavity was monitored as previously described and all procedures were performed as aseptically as possible. A new sterile bur was used during irrigation with sterile saline to access the canal. In each case, even in multi-rooted teeth, a single root canal was sampled in order to confine the microbial evaluation to a single ecological environment. The criterion used to choose the canal to be investigated in multi-rooted teeth was the canal related to the deepest periodontal pocket.
The first microbial samples were collected with three sterile paper points #20 (Dentsply-Maillefer, Ballaigues, Switzerland), which were consecutively placed for 60 s into the full length of the canal (as determined radiographically). In those cases where a dry canal was identified, an additional sterile paper point moistened in sterile saline was used to ensure sample acquisition. The paper points were pooled in a sterile tube containing 1 mL VMGA III transport medium. When tooth had an atretic root canal interfering with the penetration of the paper point, patency was accomplished through minimal instrumentation with a #10 file (C+ Files, Dentsply, Maillefer, Ballaigues, Switzerland). The file had its cable removed and was transferred to the tube containing 1 mL VMGA III transport medium and subsequently stored at −70oC until being processed.
All root canals (RC) were prepared in the same manner according to Vianna et al. (2007). The coronal two-thirds of each canal were initially prepared using rotary files (NiTi Hero, Taper .06, tip 20, Micromega, Besançon, Françe) instruments at a constant speed of 350 rpm, reaching 4 mm before the total length. Gates-Glidden drills sizes 5, 4, 3 and 2 (DYNA - FFDM, Bourges, France) were used until reaching 2 mm before the length prepared with Hero files. The working length (1 mm from the radiographic apex) was checked with a radiograph after inserting an anatomical file in the canal to the estimated working length and subsequently confirmed using an apical locator (Forum Technologies, Rishon Le-Zion, Israel). The apical stop was established using K-files (DYNA - FFDM, Bourges, France). The apical stop ended after the use of 3 files larger than the initial one. Step-back flaring of the canal was performed using larger files at intervals manipulated in a filing action. The file used to prepare the apical stop was used for recapitulation. Stepping back ended after the use of 3 files larger than the file that prepared the apical stop (42).
The canals were irrigated between each file with 1 mL 2% chlorhexidine gel (Endogel, Itapetininga, SP, Brazil) and then immediately rinsed with 4 mL of sterile physiological saline. The chlorhexidine (CHX) gel consisted of gel base (1% natrosol) and CHX gluconate at pH 7.0. Natrosol gel (hydroxyethyl cellulose) is a non-ionic, highly inert and water-soluble agent. After instrumentation, CHX activity was inactivated with 5 mL of a solution containing 5% Tween 80 and 0.07% (w/v) lecithin for 1 min, which were removed with 5 mL of sterile physiological saline. After the root canal preparation was finished, the canal was irrigated with 17% EDTA (5 mL) for 3 min and then rinsed with 5 mL of sterile physiological saline. Next, the second endodontic sampling was taken. The canals were root-filled if they were dried and asymptomatic. Otherwise, a calcium hydroxide paste was placed for 7 days. The canals were filled with gutta-percha and AH Plus sealer with warm vertical condensation and backfill. The access cavities in all cases were restored with 2 mm of Cavit TM (3M Dental Products, St Paul, MN,USA) and FiltekTM Z250 (3M Dental Products).The same operator performed all procedures in a standard manner.
Microbiological Assessment
I-. Molecular approach
DNA Extraction
Microbial DNA from periodontal and endodontic samples was extracted and purified using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In the last step of extraction, the DNA was eluted in 100 μl of buffer AE (10 mM Tris·Cl; 0.5 mM EDTA, pH 9.0) and subsequently dried in speed vac.
DNA Quantification
10 μL water was added to each one of the 30 Eppendorf tubes containing the dried DNA in order to rehydrate them. DNA concentration and quality was determined using a Nanodrop 8000 (NanoDrop 8000, Thermo Scientific, Wilmington, DE, USA).
16S rDNA profiling
HOMINGS (Human Oral Microbe Identification using Next Generation Sequencing) protocol
Due to the small amount of DNA in the samples, it was found in a pilot study that the two separate PCR reactions would better amplify the 16S rRNA genes of each sample, than in the conventional HOMINGS one-step PCR amplification protocol (5).
Briefly, 10–50 ng of DNA was first amplified using the forward primer NF1: CCA GRG TTY GAT YMT GGC and the reverse primer 1541R (5`-GAAGGAGGTGWTCCADCC-`3) in a total of 25 μL reaction. Two microliters of the DNA template were added to a reaction mixture (25 μL, final volume) containing 1.15 μL NF1 primer (40 μM), 0.85 μL of the 1541R primer (40 μM), 2.5 μL 10X high fidelity PCR buffer, 0.75 μL of 50 mM MgCl2, 2.5 μL deoxynucleotide triphosphate mixture (10 mM), and 0.25 platinum Taq polymerase. The PCR program was carried out in thin-walled tubes with a thermocycler (Perkin-Elmer 9700, Applied Biosystems, Foster City, CA) and included an initial denaturation step at 94°C for 2 min, followed by 32 cycles of denaturation at 94°C for 30s, annealing at 55°C for 30s, elongation at 72°C for 1.5 min with an additional 1 s for each cycle, and a final elongation step at 72°C for 10 min. The PCR products were visualized on a 1.5% agarose gel electrophoresis in Tris-borate-EDTA buffer.
2uL of PCR amplicons from the first assay were amplified using V3-V4 primers and 5 PrimeHotMaster Mix in a 50 μL reaction. The V3-V4 primers used were as follows:
~341F (forward primer)
AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTCCTACGGGAGGCAGCAG and ~806R (reverse primer) CAAGCAGAAGACGGCATACGAGATNNNNNNNNNNNNAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT (Sequences of the primers are in italics)
The cycling conditions were as follows: initial denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 45s, annealing at 50°C for 1min; elongation at 72°C for 1.5 min; and a final extension step at 72°C for 10 min. The amplicon size was approximately 460 bp.
PCR samples were then purified using AMPure beads (Beckman Coulter Genomics, Danvers, MA). One hundred ng of each library was pooled, gel-purified, and quantified using a bioanalyser and subsequently with qPCR (LightCycler® 96 Real-Time PCR System, Roche Diagnostics GmbH, Mannheim, Germany). Twelve pM of the library mixture, spiked with 20% Phix, was run on a MiSeq (Illumina, San Diego, CA).
Sequencing Miseq runs and data analysis
An average of >50,000 sequences of about 441 bp per sequence was obtained in a MiSeq run of 95 samples. Bad reads and chimeric sequences were removed from analyses. Species-specific, 16S rRNA-based oligonucleotide “probes”, many of which originally designed for HOMIM, were used in a BLAST program (called ProbeSeq for HOMINGS) written by Sean Cotton to identify the frequency of oral bacterial targets. It consists of 638 oligonucleotide probes of 17 to 40 bases, recognizing 538 species that target individual oral bacterial species or, in some cases, a few closely-related species. In order to get nearly complete coverage, an additional panel of 129 genus-specific probes was used after species-level determinations. Sequences for this study were analyzed using ProbeSeq version Species-V5 and Genus-V2.
II-. Culture approach
Inside an anaerobic chamber, endodontic samples were serially diluted 10 times in tubes containing Fastidious Anaerobe Broth (Laboratory M, Bury, UK). A 50-mL sample of each serial dilution as well as the undiluted sample was plated onto Fastidious Anaerobe Agar (Laboratory M) with 5% defibrinated sheep blood containing 1 mL/L of hemin and 1 mL/L of vitamin K1. Bacterial plates were incubated at 37oC under anaerobic conditions (10% CO2, 10% H2, and 80% N2) for up to 14 days. Samples were also plated onto brain-heart infusion (BHI) agar with 5% sheep blood (Oxoid, Basingstoke, UK) to allow growth of aerobic and facultative anaerobic microorganisms. The BHI plates were incubated aerobically at 37oC for 2 days. From each bacterial plate, representative colonies of each morphologic type were subcultured. Pure cultures were initially characterized according to their gaseous requirements, Gram-stain characteristic, and ability to produce catalase. The following biochemical identification kits were used for primary speciation of individual isolates: Rapid ID 32 A (BioMérieux, Marcy-l’Etoile, France) for strict anaerobic Gram-negative and Gram-positive rods; RapID ANA II System (Innovative Diagnostic Systems Inc, Atlanta, USA) for strict anaerobic Gram-positive cocci; API Staph (BioMérieux) for staphylococci and micrococci (Gram-positive cocci; catalase-positive cocci); Rapid ID 32 Strep (BioMérieux) for streptococci (Gram-positive cocci; catalase-negative cocci); and the Rapid NH System (Innovative Diagnostic Systems Inc) for Eikenella, Haemophilus, Neisseria and Aggregatibacter.
Statistical analysis
Associations between clinical parameters and microbial profiles were determined using the Mann-Whitney test. Benjamini-Hochberg corrections were used to adjust for multiple comparisons. To compare the relative abundance of bacteria between PPA (periodontal pocket after treatment) and PPB (periodontal pocket before treatment), as well as RCA (root canal after treatment) and RCB (root canal before treatment), percent abundance of the target bacterium after treatment (PPA or RCA) was divided by percent abundance of the same bacterium after the treatment (PPB or RCB) to calculate the abundance ratios of the comparison. The ratio indicates the change in the samples after treatment (PPA percentage/PPB percentage and RCA percentage/RCB percentage). To avoid errors caused by zero denominator, all percentage values were adjusted by adding the minimal non-zero value to avoid the error caused by zero denominator during the ratio calculation.
Results
Clinical features
Of 421 patients with advanced periodontal disease and 306 teeth with deep periodontal pockets, a single tooth from each of 15 adult patients (7 males and 8 females, aged 30 to 72 years; mean age 49.6 years) presented the characteristics needed for this study such as necrotic pulp tissue, presence of periodontal pockets, loss of attachment and radiographically visible bone loss (39). Some teeth used in this study were atretic as the result of periodontal disease upon the pulp. This blockage often causes difficulty in instrumentation. Many teeth were excluded from this study, e.g., had deep pockets up to the apex, and high mobility, but still remained with some pulp vitality
All 15 teeth examined had necrotic pulps and periodontal pockets equal (n=7) or greater (n=8) than 6 mm.
The dental groups involved were incisors (1/15), canines (3/15), premolars (3/15), molars (8/15). Seven upper and 8 lower teeth were included in this work.
The following clinical and radiographic endodontic features were observed: pain (2/15), pain on palpation (7/15), tenderness to percussion (10/15) and periapical lesion (15/15).
Regarding the periodontal characteristics, moderate/severe mobility was recorded in all 15 cases. The average of the clinical parameters was as follows: probe depth: 8; clinical attachment: 10.8 mm; plaque indices: 2. All teeth presented bleeding on probing.
Microbiota of the root canals before and after chemomechanical preparation (CMP) by NGS and culture
NGS
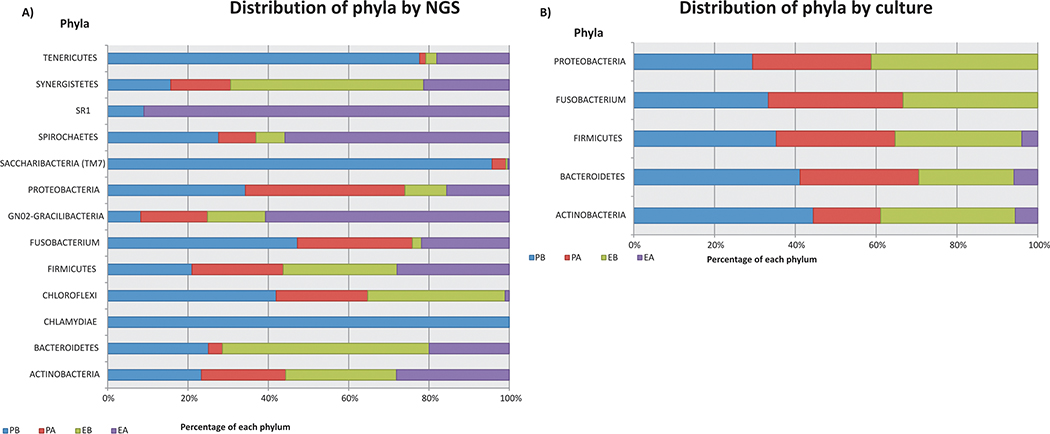
Bacteria belonging to 11 phyla were found before CMP (Fig 1A, Table 1). Firmicutes, (75.09%), followed by Proteobacteria (7.85%), Actinobacteria (7.01%), Bacteroidetes (6.77%) and Synergistetes (2.78%) predominated in the RC investigated before CMP. After CMP, 12 phyla were detected, being Firmicutes (74.01%), Proteobacteria (11.77%), Actinobacteria (7.14%), Bacteroidetes (2.63%) and Synergistetes (1.23%) the most frequently found. Chlamydiae was not detected in any of the root canals, and SR1 was only detected after CMP (Fig 1A, Table 1).
Figure 1 –
Distribution of bacterial phyla in the periodontal pockets (PP) and root canals (RC) of combined endodontic-periodontal lesions before (B) and after (A) chemomechanical preparation in 15 subjects- A) By NGS; B) By culture
Table 1.
Percentage of the bacterial phyla present in the periodontal pockets and root canals of combined endodontic-periodontal lesions before and after chemomechanical preparation in 15 subjects
| Periodontal Pockets |
Root Canals |
|||
|---|---|---|---|---|
| PHYLUM | ||||
| Before Treatment | After Treatment | Before Treatment | After Treatment | |
| Actinobacteria | 5.92920 | 5.33363 | 7.01252 | 7.13658 |
| Bacteroidetes | 3.30395 | 0.46042 | 6.77468 | 2.62611 |
| Chlamydiae | 0.00021 | 0.00000 | 0.00000 | 0.00000 |
| Chloroflexi | 0.14558 | 0.07898 | 0.11873 | 0.00359 |
| Firmicutes | 55.64448 | 60.17821 | 75.08697 | 74.00760 |
| Fusobacteria | 4.68719 | 2.83860 | 0.22696 | 2.16764 |
| Gracilibacteria (GN02) | 0.00088 | 0.00178 | 0.00155 | 0.00650 |
| Proteobacteria | 25.88343 | 29.96746 | 7.84649 | 11.77043 |
| Saccharibacteria (TM7) | 2.96441 | 0.10604 | 0.01504 | 0.01120 |
| Spirochaetes | 0.50762 | 0.17171 | 0.13256 | 1.02706 |
| SR1 | 0.00037 | 0.00000 | 0.00000 | 0.00371 |
| Synergistetes | 0.90935 | 0.86272 | 2.78369 | 1.23418 |
| Tenericutes | 0.02332 | 0.00046 | 0.00082 | 0.00542 |
At the genus level, 82 genera were found in the RC before and 89 after CMP. Dialister, Enterococcus and Streptococcus were the bacterial genera found in all canals investigated (Table 2), while many species were found only in the periodontal site, including the genera Butyrivibrio, Centipeda, Chlamydophila, Delftia, Erythromicrobium, Flavobacteriales, Syntrophomonadaceae[8][G-1], and Turicella (Table 3).
Table 2.
Number and percentage of bacterial genera more frequently detected by NGS in the periodontal pockets and root canals of combined endodontic-periodontal lesions before and after chemomechanical preparation in 15 subjects
| Periodontal Pockets |
Root Canals |
|||
|---|---|---|---|---|
| GENUS | ||||
| Before Treatment | After Treatment | Before Treatment | After Treatment | |
| Actinomyces | 15/100 | 15/100 | 14/93.3 | 15/100 |
| Desulfobulbus | 15/100 | 15/100 | 14/93.3 | 15/100 |
| Dialister | 14/93.3 | 15/100 | 15/100 | 15/100 |
| Enterococcus | 15/100 | 15/100 | 15/100 | 15/100 |
| Eubacterium | 14/93.3 | 15/100 | 14/93.3 | 14/93.3 |
| Filifactor | 13/86.7 | 15/100 | 13/86.7 | 12/80 |
| Fretibacterium | 12/80 | 15/100 | 13/86.7 | 11/73.3 |
| Gemella | 15/100 | 13/86.7 | 8/53.3 | 14/93.3 |
| Granulicatella | 15/100 | 12/80 | 13/86.7 | 14/93.3 |
| Mogibacterium | 14/93.3 | 15/100 | 12/80 | 13/86.7 |
| Parvimonas | 15/100 | 15/100 | 14/93.3 | 15/100 |
| Peptostreptococcaceae[11][G-1]* | 15/100 | 15/100 | 14/93.3 | 15/100 |
| Peptostreptococcus | 14/93.3 | 15/100 | 11/73.3 | 10/66.7 |
| Rothia | 15/100 | 14/93.3 | 14/93.3 | 15/100 |
| Selenomonas | 15/100 | 15/100 | 9/9/60 | 9/9/60 |
| Staphylococcus | 13/86.7 | 13/86.7 | 13/86.7 | 15/100 |
| Streptococcus | 15/100 | 15/100 | 15/100 | 15/100 |
| Veillonella | 15/100 | 14/93.3 | 13/86.7 | 15/100 |
| Veillonellaceae[G-1]* | 13/86.7 | 15/100 | 14/93.3 | 14/93.3 |
phylotype
Table 3.
Number and percentage of bacterial genera less frequently detected by NGS in the periodontal pockets and root canals of combined endodontic-periodontal lesions before and after chemomechanical preparation in 15 subjects
| Periodontal Pockets |
Root Canals |
|||
|---|---|---|---|---|
| GENUS | ||||
| Before Treatment | After Treatment | Before Treatment | After Treatment | |
| Alloscardovia | ND | ND | 2/13.3 | ND |
| Anaerococcus | ND | 1/6.7 | 1/6.7 | ND |
| Anaeroglobus | 1/6.7 | 1/6.7 | 1/6.7 | ND |
| Arsenicicoccus | ND | ND | ND | 1/6.7 |
| Bacillus | 1/6.7 | ND | ND | ND |
| Bacteroidales [G-2]* | 9/60 | 6/40 | ND | 1/6.7 |
| Bacteroides | ND | ND | 3/20 | 1/6.7 |
| Brevundimonas | ND | ND | ND | 1/6.7 |
| Butyrivibrio | ND | 1/6.7 | ND | ND |
| Centipeda | 4/26.7 | 3/20 | ND | ND |
| Chlamydophila | 1/6.7 | ND | ND | ND |
| Delftia | 1/6.7 | ND | ND | ND |
| Desulfomicrobium | 1/6.7 | 1/6.7 | ND | 1/6.7 |
| Desulfovibrio | ND | 1/6.7 | ND | ND |
| Eggerthia | 2/13.3 | ND | 3/20 | 1/6.7 |
| Erythromicrobium | 1/6.7 | ND | ND | ND |
| Flavobacteriales | 1/6.7 | ND | ND | ND |
| Jonquetella | ND | ND | 1/6.7 | 1/6.7 |
| Leptothrix | 1/6.7 | ND | ND | 2/13.3 |
| Megasphaera | 1/6.7 | 1/6.7 | 2/13.3 | ND |
| Mitsuokella | 1/6.7 | 2/13.3 | ND | ND |
| Mollicutes [G-1] | 1/6.7 | 1/6.7 | ND | 1/6.7 |
| Mycoplasma | 6/40 | ND | 1/6.7 | 1/6.7 |
| Ottowia | 1/6.7 | ND | ND | ND |
| Paenibacillus | ND | 1/6.7 | ND | 1/6.7 |
| Parascardovia | 1/6.7 | 2/13.3 | ND | 1/6.7 |
| Peptoniphilus | 3/20 | 2/13.3 | 3/20 | ND |
| Pyramidobacter | ND | 2/13.3 | 1/6.7 | 1/6.7 |
| SR1[G-1]* | 1/6.7 | ND | ND | 2/13.3 |
| Stenotrophomonas | 1/6.7 | 2/13.3 | ND | 4/26.7 |
| Syntrophomonadaceae[8][G-1 ]* | 2/13.3 | 2/13.3 | ND | ND |
| Turicella | ND | 1/6.7 | ND | ND |
ND = not detected
phylotype
Obligate anaerobes and rods predominated in the root canals investigated (Table 4). The number of Gram-positive and Gram-negative taxa detected in the RC was very similar before and after treatment, with a slightly predominance of Gram-negatives after CMP. However it must be pointed out that after treatment, facultative species and cocci tended to predominate (Table 4).
Table 4.
Phenotypic characteristics of the taxa present in the periodontal pockets (PP) and root canals (RC) of combined endodontic-periodontal lesions before (B) and after (A) chemomechanical preparation by NGS (*,**) and culture (***) in 15 subjects
| Periodontal Pockets |
Root Canals |
|||
|---|---|---|---|---|
| BACTERIAL MORPHOLOGY | ||||
| Before Treatment | After Treatment | Before Treatment | After Treatment | |
| Gram-positives* | 167 | 164 | 151 | 146 |
| Gram-positives** | 396387 | 408589 | 521460 | 517549 |
| Gram-positives*** | 26 | 18 | 22 | 3 |
| Gram-negatives* | 166 | 177 | 138 | 171 |
| Gram-negatives** | 248928 | 236726 | 123855 | 127766 |
| Gram-negatives*** | 13 | 11 | 12 | 1 |
| Anaerobes* | 209 | 220 | 193 | 207 |
| Anaerobes** | 416993 | 399815 | 398213.6 | 108455.5 |
| Anaerobes | 22 | 16 | 18 | 1 |
| Facultatives* | 97 | 93 | 70 | 79 |
| Facultatives** | 207033.3 | 212929.9 | 205663.1 | 466686.8 |
| Facultatives | 14 | 11 | 13 | 3 |
| Aerobes* | 27 | 28 | 26 | 31 |
| Aerobes** | 21288.77 | 32570.08 | 41438.22 | 70172.67 |
| Aerobes*** | 3 | 2 | 3 | 0 |
| Cocci* | 74 | 79 | 77 | 69 |
| Cocci** | 296258 | 327843.2 | 368644.5 | 455294.1 |
| Cocci*** | 12 | 12 | 15 | 2 |
| Rods | 259 | 262 | 212 | 248 |
| Rods** | 349057 | 317472 | 276671 | 190021 |
| Rods*** | 27 | 17 | 19 | 2 |
| Number of the detected taxa* | 333 | 341 | 289 | 317 |
| Number of the detected taxa*** | 39 | 29 | 34 | 4 |
NGS = detected taxa
NGS = number of reads
Culture = detected taxa
The species most frequently found before CMP were Enterococcus faecalis, followed by Parvimonas micra, Bacteroidaceae [G-1] sp oral taxon 272, Peptostreptococcaceae [13][G-1] sp oral taxon 113, Mogibacterium timidum, Peptostreptococcus stomatis, Filifactor alocis, and Fretibacterium fastidiosum. The most predominant phylotypes were Stomatobaculum sp oral taxon 373, Peptostreptococcaceae [11][G-1] sp oral taxon 383, Erysipelothrichaceae [G-1] sp oral taxon 905 and Desulfobulbus sp oral taxon 041(Table 5, Supplementary Table 1). After CMP the most detected species were: E. faecalis, Streptococcus salivarius/vestibularis, P. micra, Prevotella nigrescens, Eubacterium brachy, F. alocis and F. fastidiosum. The phylotypes most frequently found were Desulfobulbus sp oral taxon 041, Stomatobaculum sp oral taxon 373, Peptostreptococcaceae [11][G-1] sp oral taxon 383 and Erysipelothrichaceae [G-1] sp oral taxon 905 (Table 5, Supplementary Table 1). Overall, 73 phylotypes were found in the canals before and after CMP.
Table 5.
Microbial diversity and changes in the periodontal pockets (PP) and root canals (RC) of combined endodontic-periodontal lesions before (B) and after (A) chemomechanical preparation in 15 subjects
| MICROORGANISM/GENUS PROBE | PPB | PPA | PPA/PPB | RCB | RCA | RCA/RCB |
|---|---|---|---|---|---|---|
| Actinomyces israelii* | 0.09 | 0.04 | 0.50 | 0.01 | 0.00 | 0.50 |
| Actinomyces israelii** | 13.30 | 13.30 | 1.00 | 0.00 | 0.00 | 1.00 |
| Atopobium sp oral taxon 199* | 0.00 | 0.00 | 1.00 | 1.26 | 0.02 | 0.02 |
| Bacteroidaceae [G-1] sp. oral taxon 272* | 0.06 | 0.04 | 0.71 | 5.13 | 0.16 | 0.03 |
| Desulfobulbus sp oral taxon 041*** | 18.30 | 22.85 | 1.25 | 1.30 | 0.84 | 0.65 |
| Dialister invisus* | 0.08 | 0.10 | 1.22 | 0.68 | 0.37 | 0.55 |
| Dialister pneumosintes* | 0.02 | 0.24 | 8.33 | 0.29 | 0.06 | 0.23 |
| Enterococcus faecalis* | 6.81 | 19.88 | 2.92 | 22.02 | 46.08 | 2.09 |
| Enterococcus faecalis** | 0.00 | 0.00 | 1.00 | 20.00 | 0.00 | 0.00 |
| Erysipelothrichaceae [G-1]sp oral taxon 905*** | 0.00 | 0.00 | 1.00 | 1.92 | 0.00 | 0.01 |
| Eubacterium brachy* | 1.96 | 1.38 | 0.71 | 0.98 | 0.68 | 0.70 |
| Eubacterium nodatum* | 0.14 | 0.18 | 1.27 | 0.10 | 0.06 | 0.64 |
| Eubacterium saphenum* | 1.23 | 1.29 | 1.05 | 0.43 | 0.56 | 1.30 |
| Filifactor alocis* | 1.86 | 2.32 | 1.25 | 2.50 | 0.36 | 0.15 |
| Fretibacerium fastidiosum* | 0.43 | 0.45 | 1.05 | 2.40 | 0.60 | 0.25 |
| Fusobacterium nucl. ssp nucleatum* | 0.29 | 0.17 | 0.60 | 0.01 | 0.00 | 0.50 |
| Fusobacterium nucl. ssp nucleatum** | 53.30 | 0.00 | 0.00 | 53.80 | 73.30 | 1.36 |
| Mogibacterium timidum* | 0.85 | 0.83 | 0.98 | 2.78 | 0.17 | 0.06 |
| Parvimonas micra* | 9.00 | 9.42 | 1.05 | 14.56 | 1.65 | 0.11 |
| Parvimonas micra** | 0.00 | 40.00 | 4001 | 33.30 | 0.00 | 0.00 |
| Parvimonas sp. oral taxon 110* | 2.16 | 0.30 | 0.14 | 0.32 | 0.61 | 1.88 |
| Peptostr.[11][G-1] sp oral taxon 383*** | 0.06 | 0.41 | 6.00 | 2.74 | 0.08 | 0.03 |
| Peptostr. [13][G-1] sp oral taxon 113* | 1.17 | 1.83 | 1.56 | 3.13 | 0.20 | 0.07 |
| Peptostreptococcus stomatis* | 0.97 | 1.05 | 1.08 | 2.66 | 0.30 | 0.12 |
| Porphyromonas endodontalis* | 0.66 | 0.01 | 0.03 | 0.09 | 0.01 | 0.20 |
| Porphyromonas gingivalis* | 0.12 | 0.02 | 0.23 | 0.11 | 0.77 | 6.50 |
| Porphyromonas gingivalis** | 40.00 | 6.70 | 0.17 | 0.00 | 0.00 | 1.00 |
| Prevotella intermedia* | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 1.00 |
| Prevotella intermedia** | 40.00 | 33.30 | 0.83 | 66.70 | 6.70 | 0.10 |
| Prevotella nigrescens* | 0.01 | 0.00 | 0.50 | 0.02 | 0.92 | 31.00 |
| Pseudoramibacter alactolyticus* | 0.36 | 1.00 | 2.73 | 1.09 | 0.12 | 0.12 |
| Rothia mucilaginosa* | 0.95 | 0.61 | 0.65 | 0.16 | 0.61 | 3.65 |
| Stomatobaculum sp oral taxon 373*** | 0.26 | 0.29 | 1.11 | 3.67 | 0.29 | 0.08 |
| Streptococcus constellatus* | 2.54 | 1.93 | 0.76 | 1.27 | 0.32 | 0.26 |
| Streptococcus constellatus** | 40.00 | 20.00 | 0.50 | 13.30 | 0.00 | 0.00 |
| Streptococcus salivarius/vestibularis* | 0.27 | 0.38 | 1.39 | 0.53 | 2.24 | 4.17 |
| Streptococcus salivarius/vestibularis** | 0.00 | 0.00 | 1.00 | 13.30 | 6.70 | 0.50 |
| Tannerella forstythia* | 1.78 | 0.23 | 0.13 | 0.34 | 0.07 | 0.23 |
| Treponema denticola* | 0.06 | 0.00 | 0.14 | 0.01 | 0.14 | 7.50 |
| Treponema maltophilum* | 0.05 | 0.01 | 0.33 | 0.02 | 0.01 | 0.67 |
| Treponema socranskii * | 0.02 | 0.01 | 0.67 | 0.01 | 0.05 | 3.00 |
| Actinomyces Genus probe* | 0.75 | 1.72 | 2.28 | 1.95 | 4.06 | 2.08 |
| Desulfobulbus Genus probe* | 1.28 | 1.27 | 0.99 | 0.06 | 0.03 | 0.57 |
| Dialister Genus probe 1* | 0.00 | 0.01 | 2.00 | 0.01 | 0.00 | 0.50 |
| Dialister Genus probe 2* | 0.00 | 0.00 | 1.00 | 0.02 | 0.01 | 0.67 |
| Enterococccus Genus probe* | 0.12 | 0.29 | 2.31 | 0.24 | 0.87 | 3.52 |
| Erysipelothrichaceae Genus probe* | 0.00 | 0.00 | 1.00 | 0.01 | 0.00 | 0.50 |
| Eubacterium Genus probe 1* | 0.49 | 0.34 | 0.70 | 0.18 | 0.39 | 2.11 |
| Eubacterium Genus probe 2* | 0.02 | 0.04 | 1.67 | 0.03 | 0.01 | 0.50 |
| Filifactor Genus probe* | 0.13 | 0.18 | 1.36 | 0.21 | 0.03 | 0.18 |
| Fretibacerium Genus probe* | 0.09 | 0.14 | 1.50 | 0.23 | 0.09 | 0.42 |
| Fusobacterium Genus probe* | 4.36 | 2.64 | 0.61 | 0.17 | 2.12 | 11.83 |
| Mogibacterium Genus probe* | 0.04 | 0.03 | 0.80 | 0.10 | 0.00 | 0.09 |
| Parvimonas Genus probe* | 1.83 | 0.23 | 0.13 | 2.66 | 0.23 | 0.09 |
| Peptostreptococcus Genus probe* | 0.04 | 0.04 | 1.00 | 0.10 | 0.02 | 0.27 |
| Porphyromonas Genus probe* | 0.02 | 0.00 | 0.33 | 0.01 | 0.00 | 0.50 |
| Prevotella Genus probe* | 0.01 | 0.00 | 0.50 | 0.01 | 0.00 | 0.50 |
| Rothia Genus probe* | 0.05 | 0.80 | 13.50 | 0.02 | 0.06 | 2.33 |
| Staphylococcus Genus probe* | 0.93 | 1.13 | 1.21 | 2.88 | 10.31 | 3.57 |
| Streptococcus Genus probe* | 10.84 | 3.26 | 0.30 | 1.95 | 3.94 | 2.02 |
| Tannerella Genus probe* | 0.04 | 0.00 | 0.20 | 0.01 | 0.00 | 0.50 |
| Treponema Genus probe 3* | 0.00 | 0.00 | 1.00 | 0.00 | 0.06 | 7.00 |
| Treponema Genus probe 4* | 0.05 | 0.02 | 0.50 | 0.03 | 0.01 | 0.50 |
| Treponema Genus probe 5* | 0.04 | 0.01 | 0.40 | 0.01 | 0.06 | 3.50 |
NGS
culture
phylotype
Peptostrep = Peptostreptococcaceae
Before CMP, the number of species per canal varied from 26 to 111 (average 64), and the number of taxa detected by the genus probe per canal varied from 12 to 36 (average 23).
After CMP, the number of taxa per canal varied from 28 to 156 (average 66) and the number of taxa detected by the genus probe per canal varied 14 to 93 (average 32).
There was a considerable variability of the % frequency of RC bacterial taxa after CMP. In some cases, it decreased after treatment (e.g., Dialister invisus, Dialister pneumosintes, E.brachy, Eubacterium nodatum, F.alocis, F. nucleatum sp. nucleatum, Mogibacterium timidum, P. micra, Peptostreptococcus stomatis, Porphyromonas endodontalis, Pseudoramibacter alactolyticus, S. constellatus, T. forsythia). In others, the frequency increased, as for E. faecalis, Eubacterium saphenum, Porphyromonas gingivalis, P. nigrescens, Rothia mucilaginosa, S. salivarius/vestibularis, Treponema denticola and Treponema socranskii) (Table 5, Supplementary Table 1). However, no statistically significant differences were found between the microbiota of the RC before and after CMP (p>0.05).
However, associations were found between periapical lesions ≤ 2 mm and Desulfobulbus sp. oral taxon 041; and with periodontal pocket ≥ 6mm and D. invisius and P. stomatis (all p<0.05).
Culture
Five phyla were found before CMP, including Firmicutes (47.06%), Proteobacteria (20.59%), Actinobacteria (17.65%), Bacteroidetes (11.76%) and Fusobacteria (2.94%). After CMP, 3 phyla were found, corresponding to Firmicutes (50%), Actinobacteria (25%) and Bacteroidetes (25%) (Fig 1B, Table 6).
Table 6.
Percentage of the bacterial phyla presented in the periodontal pockets (PP) and root canals (RC) of combined endodontic-periodontal lesions before (B) and after (A) chemomechanical preparation by culture in 15 subjects
| Periodontal Pockets |
Root Canals |
|||
|---|---|---|---|---|
| PHYLUM | Before Treatment | After Treatment | Before Treatment | After Treatment |
| Actinobacteria | 20.51 | 10.34 | 17.65 | 25.00 |
| Bacteroidetes | 17.95 | 17.24 | 11.76 | 25.00 |
| Firmicutes | 46.15 | 51.72 | 47.06 | 50.00 |
| Fusobacteria | 2.56 | 3.45 | 2.94 | 0.00 |
| Proteobacteria | 12.82 | 17.24 | 20.59 | 0.00 |
Prevotella, Gemella, Streptococus, and Fusobacterium were the most frequently detected genera before CMP. After CMP, the most frequently-detected genera were Gemella, Prevotella, Propionibacterium and Streptococcus (Table 7).
Table 7.
Number and percentage of bacterial genera presented in the periodontal pockets (PP) and root canals (RC) of combined endodontic-periodontal lesions before (B) and after (A) chemomechanical preparation by culture in 15 subjects
| Periodontal Pockets |
Root Canals |
|||
|---|---|---|---|---|
| GENUS | ||||
| Before Treatment | After Treatment | Before Treatment | After Treatment | |
| Actinomyces | 1/6.7 | 1/6.7 | 2/13.3 | ND |
| Aggregatibacter | 2/13.3 | 2/13.3 | 2/13.3 | ND |
| Anaerococcus | 5/33.3 | 6/40 | 3/20 | ND |
| Bifidobacterium | 1/6.7 | 1/6.7 | 1/6.7 | ND |
| Campylobacter | ND | ND | 2/13.3 | ND |
| Capnocytophaga | 1/6.7 | 1/6.7 | ND | ND |
| Enterococcus | ND | ND | 3/20 | ND |
| Eubacterium | ND | ND | 2/13.3 | ND |
| Fusobacterium | 8/53.3 | 11/73.3 | 8/53.3 | ND |
| Gemella | 12/80 | 5/33.3 | 10/66.7 | 1/6.7 |
| Haemophilus | 5/33.3 | 6/40 | 4/26.7 | ND |
| Lactococcus | 2/13.3 | ND | ND | ND |
| Neisseria | 4/26.7 | 1/6.7 | 2/13.3 | ND |
| Parvimonas | 9/60 | 6/40 | 5/33.3 | ND |
| Peptostreptococcus | 1/6.7 | ND | ND | ND |
| Porphyromonas | 8/53.3 | 2/13.3 | ND | ND |
| Prevotella | 6/40 | 5/33.3 | 11/73.3 | 1/6.7 |
| Propionibacterium | 5/33.3 | ND | 2/13.3 | 1/6.7 |
| Staphylococcus | 1/6.7 | 1/6.7 | ND | ND |
| Streptococcus | 1/6.7 | 9/60 | 10/66.7 | 1/6.7 |
ND = not detected
Before CMP, obligate anaerobes and Gram-positive species predominated in the root canals investigated. However after CMP, the number of facultative species increased. The morphological characteristics of the species are shown in Table 4.
Prevotella intermedia or P. nigrescens, F. nucleatum sp. nucleatum, P. micra, S. constellatus and S. salivarius predominated in the canals before CMP. After CMP, microorganisms were cultured from 2 cases, and were identified as P. intermedia or P. nigrescens and Gemella haemolysans in one case; and as Propionibacterium propionicus and S. salivarius for the other canal.
The number of bacterial species found in the canals before CMP varied from 1 to 12 (average 6). Bacteria were isolated in all samples prior to treatment with the average number of colony forming units ranging from 2.5 × 102 to 5×104. However after CMP, CFU varied from zero to 0.4 × 102 and the number of species from 0 to 2.
No statistically significant differences were found between the microbiota found in the root canals before and after CMP (p>0.05).
Microbiota of the periodontal pockets before and after root canal instrumentation by NGS and culture
NGS
Thirteen phyla including Firmicutes (55.44%), Proteobacteria (25.88%), Actinobacteria (5.93%), Fusobacteria (4.69%) and Bacteroidetes (3.30%) were found in the PP before CMP. After CMP, 11 phyla were found, being Firmicutes (60.18%), Proteobacteria (29.97%) and Actinobacteria (5.334%) the most predominant. Chlamydiae and SR1 were only detected before CMP (Fig 1A, Table 1).
At the genus level, 94 genera were found in the PP before and 89 after CMP. Actinomyces, Enterococcus, Parvimonas, Peptostreptococcaceae[11][G-1], Selenomonas and Streptococcus were the bacterial genera found in all PP investigated (Table 2). Some genera were found only in RC, such as Alloscardovia, Bacteroides, Brevundimonas and Jonquetella (Table 3).
Species of obligate anaerobes and rod-shaped species predominated in the PP before and after CMP. Gram-negative anaerobic species (Table 4), tended to be found after CMP.
In terms of number of reads, there was a predominance of rods before CMP and a slight increase in the number of cocci after CMP. Strict anaerobes and Gram-positive species predominated before and after CMP (Table 4).
The most frequently species found in PP before CMP were P. micra, followed by E. faecalis, S.constellatus, Parvimonas sp. oral taxon 110, E. brachy, F. alocis and T. forsythia. Desulfobulbus sp oral taxon 041, Stomatobaculum sp oral taxon 373 and Peptostreptococcaceae [11][G-1] sp. oral taxon 383 were the most predominant phylotypes (Table 5, Supplementary Table 1). After CMP the most frequently species detected in PP were E. faecalis, P. micra, F. alocis, S. constellatus, Peptostreptococcaceae [13][G-1] sp oral taxon 113 and E. brachy. The most frequently detected phylotypes were Desulfobulbus sp oral taxon 041, Peptostreptococcaceae [11][G-1] sp. oral taxon 383 and Stomatobaculum sp oral taxon 373. Seventy three phylotypes were found in the PP before CMP and 82 after.
Before CMP, the number of taxa per PP varied from 59 to 145 (average 110), and the number of taxa detected by a genus probe per PP varied from 18 to 39 (average 31).
After CMP, the number of taxa varied from 53 to 129 (average 92) and the number of taxa detected by the genus probe per canal varied 23 to 36 (average 29).
There was a considerable variability of the % frequency of PP bacterial taxa after CMP (Table 5, Supplementary Table 1). However, in some cases it decreased (e.g., A. israelii, E. brachy, F. nucleatum sp. nucleatum, P. endodontalis, P. gingivalis, R. mucilaginosa, S. constellatus, T. forsythia), and in others increased (e.g. E. faecalis, E. saphenum, Parvimonas micra, Parvimonas sp. oral taxon 110, P. gingivalis, P. nigrescens, R. mucilaginosa, S. salivarius/vestibularis, T. denticola, and T. socranskii).
No statistically significant differences were found between the bacterial taxa detected in PP before and after CMP (p>0.05).
However, statistically significant differences were found between some microorganisms present in the RC and PP before CMP, including Fusobacterium Genus probe, Streptococcus intermedius, TM7 Genus probe, and Gemella morbillorum (p<0.01).
Culture
Five phyla were found in the PP before and after CMP (Fig 1B, Table 6). Before CMP Firmicutes (46.15%), Actinobacteria (20.51%), Bacteroidetes (19.95%), Proteobacteria (12.82%) and Fusobacteria (2.56%) predominated in the PP. After, the same phyla were present but in different percentages such as Firmicutes (51.72%), Proteobacteria (17.24%), Bacteroidetes (17.24%), Actinobacteria (10.34%) and Fusobacteria (3.45%) were found in the PP (Fig 1B, Table 6).
The bacterial genera most frequently found in the PP before CMP were Gemella (80%), Parvimonas (60%), Fusobacterium (53.3%), Porphyromonas (53.3%), Actinomyces (46.7%) and Streptococcus (46.7%). After CMP Fusobacterium (73.3%), Streptococcus (60%), (46.7%), Anaerococcus (40%), Haemophilus (40%) and Parvimonas (40%) (Table 7).
Strict anaerobes and Gram-positive taxa predominated before CMP (Table 4). Most of the Gram-positive taxa were cocci, while the Gram-negatives were rods.
Before CMP, the microorganisms most frequently isolated were Actinomyces israelii, F. nucleatum sp. nucleatum, P. gingivalis, P. intermedia/nigrescens and S. constellatus. The number of taxa found varied from 2 to 11 (average 7) and the average number of CFU was 3.99 × 107.
After CMP, A. israelii, P. micra, P. endodontalis, S. constellatus were still found in the PP. The number of taxa found in the PP varied from 2 to 8 (average 5) and the average number of CFU was 2.73×107.
As described above for the NGS data, there was considerable variability of % frequency of PP bacterial taxa after CMP treatment (Table 5, Supplementary Table 1). In some cases, the % frequency increased (P. micra), in others decreased (F. nucleatum sp. nucleatum, P. endodontalis, P. gingivalis, S. constellatus), and, in others, stayed the same (A. israelii).
No statistically significant differences were found between the taxa found in the periodontal pockets before and after CMP (p>0.05). However, associations were found between Streptococcus constellatus found in the PP after CMP with endodontic lesion > 2 mm (p<0.05).
Discussion
The limited literature on the topic of combined endodontic-periodontal lesions and also the challenges in treatment of the clinical cases with advanced bone loss were motivating factors for the development of this study. It is known that many of the periodontal pathogens are also endodontic pathogens, however there are few studies dedicated to the investigation of combined endodontic-periodontal lesions (6, 24, 25, 27, 29, 34, 44). Only a limited number of studies using Next Generation Sequencing are available, but typically they focused on either the root canal (20, 35) or the periodontal microbiota (4, 37).
The main objective of this work was to investigate the microbial profile of the root canals and their associated periodontal pockets before and after chemomechanical preparation. Therefore, we included the culture technique not only to compare our results with the ones previously reported in the literature, but also to determine whether cells are still viable after chemomechanical procedures.
Microbial profile of the necrotic root canal associated with periodontal disease
The present study confirmed previous findings in the literature regarding the microbial population of root canals, which includes species of the genera Eubacterium, Fusobacterium, Peptostreptococcus, Porphyromonas, Prevotella and Streptococcus (6, 24, 25, 27, 29, 34, 44). However, other species such as Dialister invisus, D. pneumosintes, P. alactolyticus, R. mucilaginosa, T. forsythia, T. maltophilum, T. socranskii and the phylotypes Stomatobaculum sp oral taxon 373, Peptostreptococcaceae sp oral taxon 383and Desulfobulbus sp oral taxon 041, among others, were found in our study. Li et al (27), using polymerase chain reaction-based denaturing gradient gel electrophoresis (PCR-DGGE), cloning, and sequence analysis, reported the presence of Filifactor alocis, Parvimonas micra, Porphyromonas gingivalis, and Tannerella forsythia, in combined endodontic-periodontal lesions, which is in agreement with our work.
An interesting finding of this work was the similarity of the data achieved by NGS and culture, particularly regarding the number of the taxa detected according to the gaseous requirements, Gram stain and cell morphology. It was not surprising that the phylum Firmicutes was most frequently detected, before and after CMP, by both methodologies. Firmicutes includes bacterial taxa such as Parvimonas, Peptostreptococcus, Enterococcus, Eubacterium, Gemella, Dialister, and Streptococcus.
As expected, NGS provided a more comprehensive analysis of the root canal microbiota, revealing species and phylotypes not yet related to the combined endodontic-periodontal lesions. Moreover, the number of taxa per canal was much higher than in previous works involving these combined lesions (6, 24, 25, 27, 29, 34, 44).
Kobayashi et al. (25), using culture technique, reported that obligate anaerobes and coccal forms were by far the predominant types in the root canals of combined endodontic-periodontal lesions compared with the ones found in the associated periodontal pockets. In contrast, we found similar proportions of bacterial taxa at both sites.
Rod-shaped taxa predominated in the root canals investigated before and after CMP, however, according to the number of reads, there was a significant increase of cocci, particularly after CMP. Enterococcus faecalis, a Gram-positive coccus, was found in the root canals in higher numbers before and after CMP, confirming their resistance to the endodontic procedures and their possible role in failure of root canal treatment (13, 14, 15).
Staphylococcus spp., also a Gram-positive coccus, was detected in both sites, particularly in the root canals after CMP by NGS. However, this species was only found in one case by culture and in the periodontal pockets, suggesting possible contamination. Gomes et al. (10) reported the presence of this species before and after CMP, mentioning its resistance to antiseptics and disinfectants commonly used in endodontics.
The efficacy of the CMP with chlorhexidine gel was particularly noted by the culture technique, where only 2/15 of the cases still had viable bacteria. The species recovered by culture were also detected by NGS. There was a large reduction in the count CFU/mL from the root canal immediately after CMP, which corroborates the studies of Berber et al. (2) (in vitro), and Vianna et al. (42, 43) (in vivo), who used the same instrumentation protocol.
On the other hand, NGS showed a predominance of anaerobic taxa after CMP. This might be due that the number of cells may have been insufficient to support bacterial growth or that cells were no longer viable. For culture, strict anaerobic techniques were used to detect as many microorganisms as possible, but the media employed may not have been suitable for the growth of all microorganisms. For example, it was predictable that spirochetes would not survive on the media used and they were not, in fact, isolated, even though they were detected by NGS in the root canals before and after CMP.
Gram-negative anaerobic species were associated with some clinical features such as periapical lesions ≤ 2 mm (Desulfobulbus sp. oral taxon 041, a phylotype) and with periodontal pocket ≥ 6mm (Dialister invisus). Gram-negative bacteria contain LPS (a virulence factor), generally referred to as endotoxin, released during disintegration of bacteria after multiplication and death. A high content of endotoxins in root canals had been associated with endodontic signs and symptoms such as spontaneous pain, pain on palpation and tenderness to percussion. Furthermore, its egression through the apical foramen can cause bone resorption of the periradicular or perpetuate an apical periodontitis (21, 28).
Gram-positive anaerobic species such as Peptostreptococcus stomatis were also associated with periodontal pocket ≥ 6mm. Peptostreptococcus have been recovered from a wide variety of mixed infections involving anaerobic bacteria. The cell walls of Gram-positive bacteria include peptidoglycans and lipoteichoic acids, which can influence inflammatory reactions and enhance pathogenicity of Gram-negative microorganisms. One explanation for the synergy between these species is known enhancement of endotoxin effect by Gram-positive superantigens (13).
Microbial profile of the periodontal pockets associated with endodontic disease
Our results showed that the microbial population of the periodontal pockets in cases of combined endodontic-periodontal lesions is more diverse than previously reported in the literature (6, 24, 25, 27, 29, 34, 44). It comprises not only species of the genera Actinomyces, Enterococcus, Eubacterium, Fusobacterium, Mogibacterium, Parvimonas, Peptostreptococcus, Pseudoramibacter, Porphyromonas, Prevotella, Rothia, Streptococus, Tannerella, and Treponema, but also species such as Dialister invisus (32), D. pneumosintes, Filifactor alocis (31), and Fretibacterium fastidiosum (30), which have been detected by molecular techniques in primarily infected root canals. The microbial profile also includes phylotypes such as Desulfobulbus sp oral taxon 041, Stomatobaculum sp oral taxon 373 and Peptostreptococcaceae sp oral taxon 383.
Not surprisingly, species belonging to the 5 major complexes described by Socransky et al. (38) (i.e., purple, yellow, green, orange and red) were found in the PP. One interesting finding was the abundance of Enterococcus faecalis in the PP, agreeing with the results of Souto & Colombo (40), who reported that this species was frequently detected in the oral microbiota of periodontitis patients suggesting that periodontal infection may favor the colonization by this species. Kipioti et al. (24) also reported the presence of E. faecalis in root canals and adjacent periodontal pockets.
The microbiota of the periodontal pockets before and after CMP was very similar to one found in the root canals, holding a slightly greater number of species The similarity between these microbiomes suggests that there may be a pathway of infection between the pulp and periodontium. The main communication between both sites is the apical foramen, but dentinal tubules and accessory root canals may also be involved (33).
It was not expected to detect a great change in the periodontal community after CMP, as the period of contact of chlorhexidine with the periodontal tissues was relatively short. Moreover, the diameter of the dentinal tubules in the presence of periodontal disease decreases, limiting the diffusion of the intracanal substances to outside the root canals. Further studies involving the use of chlorhexidine based-intracanal medications with different periods of time might prove informative as the time exerts influence in their antimicrobial activity (16, 18). It is important to treat oral infections as associations of periodontal disease have been found with systemic diseases such as cardiovascular disease, type 2 diabetes mellitus, adverse pregnancy outcomes and osteoporosis (23).
It is concluded that the microbial community present in combined endodontic-periodontal lesions is complex and more diverse than previously reported. It is important to note that bacteria do survive in some root canals after chemomechanical preparation. Finally, the similarity between the microbiota of both sites, before and after chemomechanical preparation, suggests there may be a pathway of infection between the pulp and periodontium.
Supplementary Material
References
- 1.Abbott PV, Salgado JC. Strategies for the endodontic management of concurrent endodontic and periodontal diseases. Aust Dent J. 2009; 54 Suppl 1:S70–85. [DOI] [PubMed] [Google Scholar]
- 2.Berber VB, Gomes BP, Sena NT, Vianna ME, Ferraz CC, Zaia AA, Souza-Filho FJ. Efficacy of various concentrations of NaOCl and instrumentation techniques in reducing Enterococcus faecalis within root canals and dentinal tubules. Int Endod J. 2006; 39:10–7. [DOI] [PubMed] [Google Scholar]
- 3.Biffi JC, Rodrigues HH. Ultrasound in endodontics: a quantitative and histological assessment using human teeth. Endod Dent Traumatol. 1989; 5:55–62. [DOI] [PubMed] [Google Scholar]
- 4.Bizzarro S, Loos BG, Laine ML, Crielaard W, Zaura E. Subgingival microbiome in smokers and non-smokers in periodontitis: an exploratory study using traditional targeted techniques and next-generation sequencing. J Clin Periodontol. 2013;40:483–92 [DOI] [PubMed] [Google Scholar]
- 5.Cotton SL, Klepac-Ceraj V, Krishnan K, McCafferty J, Chen T, Kokaras AS, Murphy CM, Toscano ML, Paster BJ. HOMINGS-Species-level Identification of High-throughput Sequencing Data. J Dent Res 93 (Spec Iss B), 400, 2014.24453179 [Google Scholar]
- 6.Didilescu AC, Rusu D, Anghel A, Nica L, Iliescu A, Greabu M, Bancescu G, Stratul SI. Investigation of six selected bacterial species in endo-periodontal lesions. Int Endod J. 2012; 45:282–93. [DOI] [PubMed] [Google Scholar]
- 7.Ehnevid H, Jansson L, Lindskog S, Blomlöf L. Periodontal healing in teeth with periapical lesions. A clinical retrospective study. J Clin Periodontol 1993; 20:254–8. [DOI] [PubMed] [Google Scholar]
- 8.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ; CDC. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012; 91:914–20. [DOI] [PubMed] [Google Scholar]
- 9.Gomes BP, Endo MS, Martinho FC. Comparison of endotoxin levels found in primary and secondary endodontic infections. J Endod. 2012; 38:1082–6. [DOI] [PubMed] [Google Scholar]
- 10.Gomes BP, Lilley JD, Drucker DB. Variations in the susceptibilities of components of the endodontic microflora to biomechanical procedures. Int Endod J. 1996; 29:235–41. [DOI] [PubMed] [Google Scholar]
- 11.Gomes BP, Martinho FC, Vianna ME. Comparison of 2.5% sodium hypochlorite and 2% chlorhexidine gel on oral bacterial lipopolysaccharide reduction from primarily infected root canals. J Endod. 2009; 35:1350–3. [DOI] [PubMed] [Google Scholar]
- 12.Gomes BP, Montagner F, Berber VB, Zaia AA, Ferraz CC, de Almeida JF, Souza-Filho FJ. Antimicrobial action of intracanal medicaments on the external root surface. J Dent. 2009; 37:76–81. [DOI] [PubMed] [Google Scholar]
- 13.Gomes BP, Pinheiro ET, Gadê-Neto CR, Sousa EL, Ferraz CC, Zaia AA, Teixeira FB, Souza-Filho FJ. Microbiological examination of infected dental root canals. Oral Microbiol Immunol. 2004;19:71–6. [DOI] [PubMed] [Google Scholar]
- 14.Gomes BP, Pinheiro ET, Jacinto RC, Zaia AA, Ferraz CC, Souza-Filho FJ. Microbial analysis of canals of root-filled teeth with periapical lesions using polymerase chain reaction. J Endod. 2008; 34:537–40. [DOI] [PubMed] [Google Scholar]
- 15.Gomes BP, Pinheiro ET, Sousa EL, Jacinto RC, Zaia AA, Ferraz CC, de Souza-Filho FJ. Enterococcus faecalis in dental root canals detected by culture and by polymerase chain reaction analysis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006; 102:247–53. [DOI] [PubMed] [Google Scholar]
- 16.Gomes BP, Vianna ME, Zaia AA, Almeida JF, Souza-Filho FJ, Ferraz CC. Chlorhexidine in endodontics. Braz Dent J. 2013; 24:89–102. [DOI] [PubMed] [Google Scholar]
- 17.Gonçalves PF, Klepac-Ceraj V, Huang H, Paster BJ, Aukhil I, Wallet SM, Shaddox LM. Correlation of Aggregatibacter actinomycetemcomitans detection with clinical/immunoinflammatory profile of localized aggressive periodontitis using a 16S rRNA microarray method: a cross-sectional study. PLoS One. 2013; 23 8(12):e85066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S, Tewari S, Tewari S, Mittal S. Effect of time lapse between endodontic and periodontal therapies on the healing of concurrent endodontic-periodontal lesions without communication: A prospective randomized clinical trial. J Endod. 2015; March 25 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Hong BY, Tae-Kwon Lee TK, Lim SM, Chang SW, Park J, Han SH, Zhu Q, Kamran E. Safavi KE, Ashraf F. Fouad A, Kum KY. Microbial analysis in primary and persistent endodontic infections by using pyrosequencing. J Endod. 2013; 39:1136–40. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao WWL, Li KL, Liu ZQ, Jones C, Fraser-Liggett CM, Fouad AF. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genomics. 2012; 28 13:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacinto RC, Gomes BPFA, Shah HN, Ferraz CC, Zaia AA, Souza-Filho FJ. Quantification of endotoxins in necrotic root canals from symptomatic and asymptomatic teeth. J Med Microbiol. 2005; 54(Pt 8):777–83. [DOI] [PubMed] [Google Scholar]
- 22.Jansson L, Ehnevid H, Lindskog S, Blomlöf L. Relationship between periapical and periodontal status: A clinical retrospective study. J Clin Periodontol. 1993; 20:117–23. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Amar S. Periodontal disease and systemic conditions: a bidirectional relationship. Odontology 2006; 94:10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kipioti A, Nakou M, Legakis N, Mitsis F. Microbiological findings of infected root canals and adjacent periodontal pockets in teeth with advanced periodontitis. Oral Surg Oral Med Oral Pathol. 1984; 58:213–20. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T, Hayashi A, Yoshikawa R, Okuda K, Hara K The microbial flora from root canals and periodontal pockets of non-vital teeth associated with advanced periodontitis. Int Endod J. 1990; 23:100–6. [DOI] [PubMed] [Google Scholar]
- 26.Kurihara H, Kobayashi Y, Francisco AI, Isoshima O, Nagai A, Murayama Y. A microbiological and immunological study of endodontic-periodontic lesions. J Endod. 1995; 21:617–21. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Guan R, Sun J, Hou B Bacteria community study of combined periodontal endodontic lesions using denaturing gradient gel electrophoresis and sequencing analysis. J Periodontol. 2014; 85:1442–9. [DOI] [PubMed] [Google Scholar]
- 28.Martinho FC, Leite FR, Nascimento GG, Cirelli JA, Gomes BP. Clinical investigation of bacterial species and endotoxin in endodontic infection and evaluation of root canal content activity against macrophages by cytokine production. Clin Oral Investig. 2014; 18:2095–102. [DOI] [PubMed] [Google Scholar]
- 29.Pereira CV, Stipp RN, Fonseca DC, Pereira LJ, Höfling JF. Detection and clonalanalysis of anaerobic bacteria associated to endodontic-periodontal lesions. J Periodontol. 2011; 82:1767–75. [DOI] [PubMed] [Google Scholar]
- 30.Rôças IN, Neves MA, Provenzano JC, Siqueira JF Jr. Susceptibility of as-yet-uncultivated and difficult-to-culture bacteria to chemomechanical procedures. J Endod. 2014; 40:33–7. [DOI] [PubMed] [Google Scholar]
- 31.Rôças IN, Siqueira JF Jr. Characterization of Dialister species in infected root canals. J Endod. 2006;32:1057–61. [DOI] [PubMed] [Google Scholar]
- 32.Rôças IN, Siqueira JF Jr. Simultaneous detection of Dialister pneumosintes and Filifactor alocis in endodontic infections by 16S rDNA-directed multiplex PCR. J Endod. 2004; 30:851–4. [DOI] [PubMed] [Google Scholar]
- 33.Rubach WC, Mitchell DF. Periodontal disease, accessory canals and pulp pathosis. J Periodontol. 1965; 36:34–8. [DOI] [PubMed] [Google Scholar]
- 34.Rupf S, Kannengiesser S, Merte K, Pfister W, Sigusch B, Eschrich K. Comparison of profiles of key periodontal pathogens in the periodontium and endodontium. Endod Dent Traumatol. 2000;16:269–75. [DOI] [PubMed] [Google Scholar]
- 35.Santos AL, Siqueira JF Jr, Rôças IN, Jesus EC, Rosado AS, Tiedje JM. Comparing the bacterial diversity of acute and chronic dental root canal infections. PLoS One. 2011; 6:e28088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schilder H Cleaning and shaping the root canal. Dent Clin North Am. 1974; 18:269–96. [PubMed] [Google Scholar]
- 37.Schwarzberg K, Le R, Bharti B, Lindsay S, Casaburi G, Salvatore F, Saber MH, Alonaizan F, Slots J, Gottlieb RA, Caporaso JG, Kelley ST. The personal human oral microbiome obscures the effects of treatment on periodontal disease. PLoS One. 2014; 29; 9(1):e86708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solomon C, Chalfin H, Kellert M, Weseley P. The endodontic- periodontal lesion: a rational approach to treatment. J Am Dent Assoc. 1995;126:473–9. [DOI] [PubMed] [Google Scholar]
- 39.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998; 25:134–44. [DOI] [PubMed] [Google Scholar]
- 40.Souto R, Colombo AP. Prevalence of Enterococcus faecalis in subgingival biofilm and saliva of subjects with chronic periodontal infection. Arch Oral Biol. 2008; 53:155–60. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One. 2014; 21; 9(8):e105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vianna ME, Horz HP, Conrads G, Zaia AA, Souza-Filho FJ, Gomes BP. Effect of root canal procedures on endotoxins and endodontic pathogens. Oral Microbiol Immunol. 2007; 22:411–8. [DOI] [PubMed] [Google Scholar]
- 43.Vianna ME, Horz HP, Gomes BP, Conrads G. In vivo evaluation of microbial reduction after chemo-mechanical preparation of human root canals containing necrotic pulp tissue. Int Endod J. 2006; 39:484–92. [DOI] [PubMed] [Google Scholar]
- 44.Xia M, Qi Q. Bacterial analysis of combined periodontal-endodontic lesions by polymerase chain reaction-denaturing gradient gel electrophoresis. J Oral Sci. 2013; 55:287–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.