Abstract
The regulation of human Arf1 GTPase activity by ArfGEFs that stimulate GDP/GTP exchange and ArfGAPs that mediate GTP hydrolysis has attracted attention for the discovery of Arf1 inhibitors as potential anti-cancer agents. The malaria parasite Plasmodium falciparum encodes a Sec7 domain-containing protein - presumably an ArfGEF - and two putative ArfGAPs, as well as an Arf1 homologue (PfArf1) that is essential for blood-stage parasite viability. However, ArfGEF and ArfGAP-mediated activation/deactivation of PfArf1 has not been demonstrated. In this study, we established an in vitro colorimetric microtiter plate-based assay to detect the activation status of truncated human and P. falciparum Arf1 and used it to demonstrate the activation of both proteins by the Sec7 domain of ARNO, their deactivation by the GAP domain of human ArfGAP1 and the inhibition of the respective reactions by the compounds SecinH3 and QS11. In addition, we found that the GAP domains of both P. falciparum ArfGAPs have activities equivalent to that of human ArfGAP1, but are insensitive to QS11. Library screening identified a novel inhibitor which selectively inhibits one of the P. falciparum GAP domains (IC50 4.7 µM), suggesting that the assay format is suitable for screening compound collections for inhibitors of Arf1 regulatory proteins.
Subject terms: Biochemistry, Biological techniques, Cell biology, Drug discovery
Introduction
ADP-ribosylation factor (Arf) GTPases are central regulators of protein trafficking in eukaryotic cells. There are six Arf isoforms, divided into three classes based on sequence homology, of which the most widely studied are Arf1 (Class I) and Arf6 (Class III). Respectively, they principally mediate trafficking in the secretory (Arf1) and endocytic (Arf6) pathways, with additional roles for Arf6 in actin cytoskeleton dynamics1–3. Arf1 is the focus of this study and initiates vesicle formation in the Golgi apparatus by activating lipid modifying enzymes and recruiting coatomer complex I (COPI) coat proteins. The COPI vesicles are responsible for retrograde transport of cargo and trafficking proteins to earlier Golgi compartments and the endoplasmic reticulum4. In addition, Arf1 recruits clathrin adaptor proteins (AP1, AP3 and AP4) and Golgi-localized γ-ear-containing ARF-binding (GGA) proteins to the trans-Golgi network, where they are involved in trapping cargo proteins and the formation of vesicles that deliver secretory proteins to endosomes5.
Presumably, the delivery of newly synthesised secretory proteins to their correct locations places a heavy burden on Arf1 activity in rapidly growing cells. Indeed, Arf1 is upregulated in cancer cell types and plays a role in cancer metastasis phenotypes e.g. cell detachment, migration and invasion, and may additionally be involved in tumour-promoting cell signalling pathways e.g. the phosphatidylinositol 3-kinase (PI3K) and mitogen-activate protein kinase (MAPK) pathways6–9. Moreover, Arf1 inhibitors inhibit cancer cell viability, proliferation and metastatic characteristics10 and tumour growth in mouse models11–13. Like other small GTPases, Arf1 undergoes a cycle of activation and deactivation that is determined by its nucleotide binding status. Exchanging GDP for GTP activates Arf1 through a pronounced conformational change which exposes a myristoylated N-terminal amphipathic α-helix, resulting in membrane association, and enhances effector protein binding. Conversely, hydrolysis of the terminal phosphate of the bound GTP to form GDP deactivates Arf1, returning it to a cytoplasmic pool. Due to the low intrinsic nucleotide exchange and hydrolysis activity of Arf1, Arf1 activation is stimulated by a family of guanine nucleotide exchange factors (GEFs) containing a characteristic Sec7 domain14, while deactivation is promoted by GTPase activating proteins (GAPs) containing GAP domains14,15. The development of Arf1 inhibitors has focused on compounds that disrupt GEF-mediated nucleotide exchange (although the detailed mechanisms may differ) and includes inter alia brefeldin A (BFA) and its analogues, Golgicide A, AMF-26, LM11, Exo2 and SecinH311,16–20. However, Arf GAP inhibitors – QS11 and its derivatives – have been described and reported to inhibit the migration of breast cancer cells21,22.
The genome of the most prevalent and virulent of the malaria parasite species, Plasmodium falciparum, contains six sequences that have been annotated as encoding putative Arf or Arf-like proteins (www.plasmodb.org). One such sequence encodes an Arf1 homologue (PfArf1) that has a very high amino acid sequence conservation (76% identity, 89% similarity) compared to human Arf1. Originally identified by probing a P. falciparum genomic library and PCR from P. falciparum cDNA23–25, the recombinant protein was shown to bind GTP, have ADP-ribosyltransferase and phospholipase D stimulating activity in addition to low intrinsic GTPase activity, all features of Arf GTPases24,25. It is also capable of stimulating P. falciparum phosphatidylinositol 4-phosphate 5-kinase (PIP5K), which is an established role of mammalian Arf1 in the regulation of phosphorylated phosphatidylinositol levels and, consequently, membrane trafficking, signalling and cytoskeleton dynamics26. In blood-stage parasites, PfArf1 fused to GFP was found to co-localise with the Golgi marker GRASP27, while the canonical Arf1 activation inhibitor BFA causes a disruption in Golgi architecture and trafficking of secretory proteins28–32. Taken together, these studies suggest that PfArf1 mimics the key role of mammalian Arf1 in secretory traffic through the Golgi apparatus. As would be expected based on sequence conservation, the crystal structure of GDP-bound PfArf1is very similar to that of human Arf1, with subtle differences in the Switch I and II domains that could affect binding of GEFs and GAPs33. However, direct demonstration of GEF-mediated nucleotide exchange and GAP-mediated GTP hydrolysis by PfArf1 has not been reported.
Interestingly, unlike mammalian cells where the Arf GEF and GAP families contain up to 15 and 27 members respectively14, the P. falciparum genome encodes two putative ArfGAP proteins and a single Sec7 domain-containing putative ArfGEF, responsible for the BFA sensitivity of malaria parasites34,35. The crystal structure of the catalytic GAP domain of one of the GAP isoforms (designated PfArfGAP1 in this study) has been determined and shows an overall similarity of tertiary structure compared to mammalian GAP domains36. However, unlike the highly conserved PfArf1, there is a greater divergence of amino acid sequence homology compared to human ArfGAP1 (39% identity and 52% similarity) and differences in the amino acid residues predicted to interact with Arf136. In this study, using human recombinant proteins as a model, we developed a novel microtiter plate-based assay to detect Arf1 activation (GTP vs. GDP-bound) status and modulation of it by an ArfGEF (ARNO) Sec7 domain and Arf GAP (ArfGAP1) GAP domain. We used the assay to demonstrate and compare the Arf1 GAP activities of the GAP domains of the two putative P. falciparum GAPs, as well as demonstrate ARNO-stimulated nucleotide exchange by PfArf1. Given the interest in Arf1 as a drug target, a further motivation for developing the assay was to introduce an assay format compatible with the screening of compound libraries for Arf1 activity modulators, explored here by detecting the differential inhibition of ARNO and GAP-mediated Arf1 activation/deactivation using standard inhibitors, as well as the identification of a novel, selective PfArf1 GAP inhibitor.
Results
A colorimetric plate-based GST-GGA3 binding assay discriminates between GTP- and GDP-bound Arf1
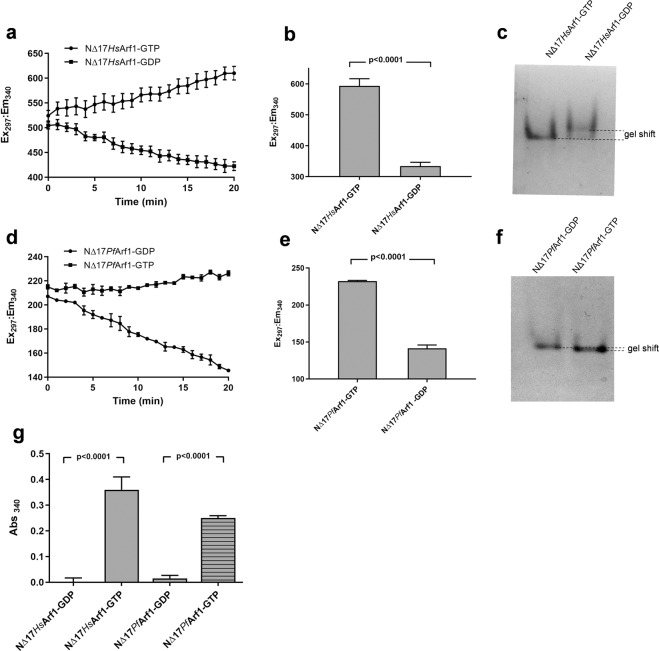
The phenomenon that Arf1only binds to the coat protein GGA3 (via the GAT domain of the latter) when it is in its active GTP-bound vs. inactive GDP-bound conformation has been widely employed as an experimental tool to detect Arf1 activation status in cultured cells using pull-down assays. Typically, glutathione beads coated with a fusion protein consisting of glutathione-S-transferase (GST) and the GAT domain of GGA3 (GST-GGA3GAT) are incubated with cell lysates and bead-bound (active) vs. total Arf1 levels determined by western blotting37. To determine if the selective binding of GST-GGA3GAT to Arf1-GTP could be further exploited to determine the activation status of purified recombinant Arf1 proteins in a microtiter plate format, we conceptualised an assay procedure (Fig. S1) in which Arf1, expressed and purified as a truncated histidine-tagged protein (Fig. S2), is immobilised on nickel-NTA coated 96-well plates, followed by incubation with purified GST-GGA3GAT. The extent of GST-GGA3GAT binding to the plate may be readily determined by the addition of a colorimetric GST enzyme substrate, and should correlate with the level of GTP-bound Arf1. Assessing the viability of this approach required the preparation of GTP-bound and GDP-bound Arf1, respectively, which was achieved by a standard method38. His-tagged human and P. falciparum Arf1, minus the N-terminal 17 amino acids containing the myristoylation site and amphipathic α-helix (NΔ17HsArf1and NΔ17PfArf1, respectively), were incubated with GTP or GDP in the presence of EDTA, followed by the addition of Mg2+ to stabilise the attached nucleotide. The HsArf1 conformational change induced by GTP binding was monitored by kinetic and end-point intrinsic tryptophan fluorescence reads (Fig. 1a,b), as well as by performing native PAGE on the final protein preparations (Fig. 1c). As anticipated by the high level of sequence conservation in PfArf1, the results confirmed that this is a viable approach for preparing and assessing GTP- and GDP-bound NΔ17PfArf1 (Fig. 1d–f), although the native PAGE mobility difference between NΔ17PfArf1-GTP and -GDP was smaller than observed with the human protein. The kinetic tryptophan fluorescence measurements further suggested that the original NΔ17HsArf1preparation was purified from E. coli as a mixture of GDP- and GTP-bound proteins (based on the respective increase and decrease in fluorescence during incubation with GTP and GDP), while NΔ17PfArf1 was predominantly GTP-bound.
Figure 1.
Microtiter plate GST-GGA3GAT binding assay using GTP and GDP preloaded Arf1 proteins. (a,d) Five µM NΔ17HsArf1 (a) and NΔ17PfArf1 (d) were incubated at 25 °C with 50 µM GTP or GDP in the presence of 2 mM (NΔ17HsArf1) or 20 mM (NΔ17PfArf1) EDTA in a black 96-well plate and tryptophan fluorescence (Ex297/Em340) measured at 1 min intervals in a plate reader for 20 min. (b,e) After a further 40 min incubation, MgCl2 was added to a final concentration of 3 mM (NΔ17HsArf1) or 30 mM (NΔ17PfArf1), incubation continued for 10 min and NΔ17HsArf1 (b) and NΔ17PfArf1 (e) tryptophan fluorescence measured as an end-point reading. The nucleotide exchange reactions were conducted in triplicate wells and the data points represent mean fluorescence ± standard deviation. (c,f) After completion of nucleotide exchange, GTP and GDP loaded NΔ17HsArf1 (c) and NΔ17PfArf1(f) were run on 12% native PAGE gels and stained with Coomassie. The gel images were cropped from two separate native PAGE gels, shown in Fig. S6 (Supporting Information). (g) GTP and GDP preloaded NΔ17HsArf1 and NΔ17PfArf1were added to the wells of a Ni-NTA coated clear 96-well plate at a concentration of 1 µM and incubated for 30 min at 4 °C. An equal volume of GST-GGA3GAT was added to a final concentration of 1 µM and incubation continued for 60 min. After washing the wells, GST substrate solution containing reduced L-glutathione and 1-chloro-2,4-dinitrobenzene was added and absorbance measured at 340 nm after a 30 min incubation at room temperature. Mean background absorbance values obtained from empty wells (i.e. lacking immobilised Arf1) incubated with GST-GGA3GAT followed by GST substrate were subtracted from experimental readings. Incubations were carried out in triplicate wells and the bars represent mean Abs340 ± standard deviation. P-values were calculated by two-tailed t-tests.
To determine if GST-GGA3GAT could be used to detect Arf1 activation status using the plate-based colorimetric assay format described above, the nucleotide-loaded Arf1 proteins were incubated in a nickel-NTA coated 96-well plate, followed by sequential incubations with GST-GGA3GAT and a colorimetric GST substrate and absorbance readings performed at 340 nm (Fig. 1g). GTP- vs GDP-bound NΔ17HsArf1could be robustly distinguished by the level of GST enzyme activity captured in the wells and the NΔ17PfArf1 results further confirmed that selective nucleotide-dependent GGA3GAT binding ability is conserved in the malaria protein. To confirm that selective binding of NΔ17PfArf1-GTP to GST-GGA3GAT was due to the recognition of the GGA3GAT portion of the fusion protein, we found that untagged GST failed to bind to NΔ17PfArf1-GTP (or -GDP) (Fig. S3). In addition, GTP- vs. GDP-bound NΔ17PfArf1 was preferentially co-precipitated by GGA3-coated beads (Fig. S3).
Detection of ARNO-mediated nucleotide exchange by human and P. falciparum Arf1
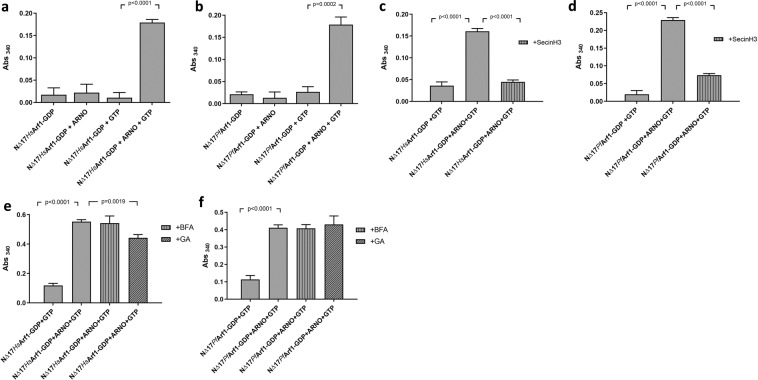
To determine if the assay can be further exploited to detect the activation of Arf1 by an ArfGEF in vitro, GDP-loaded NΔ17HsArf1 and NΔ17PfArf1 were incubated with GTP in the presence of the Sec7 domain of ARNO (ARNOSec7) before adding the reactions to nickel-NTA coated plates and proceeding with the assay described above. ARNOSec7-mediated nucleotide exchange by both NΔ17HsArf1 and NΔ17PfArf1 could be discerned by a marked increase in GST-GGA3GAT binding compared to the respective controls (Fig. 2a,b). The controls consisted of the GDP-bound Arf1 proteins (NΔ17HsArf1-GDP and NΔ17PfArf1-GDP), the GDP-bound Arf1 proteins incubated with ARNOSec7 in the absence of GTP, and the GDP-bound Arf1 proteins incubated with GTP in the absence of ARNOSec7. To confirm that the enhanced GST-GGA3GAT binding was due to an increase in Arf1-GTP levels caused by ARNOSec7 stimulated nucleotide exchange, the reactions were repeated in the presence of 50 µM SecinH3, an inhibitor of the cytohesin family of ArfGEFs to which ARNO belongs20. Inclusion of SecinH3 in the ARNOSec7 exchange reaction reduced GST-GGA3GAT binding by both NΔ17HsArf1 and NΔ17PfArf1 to levels obtained with control reactions lacking ARNOSec7 (Fig. 2c,d), causing a 93% and 74% inhibition of ARNOSec7-mediated NΔ17HsArf1 and NΔ17PfArf1 nucleotide exchange, respectively. The exchange reactions were subsequently repeated in the presence of 50 µM brefeldin A (BFA) or Golgicide A (GA), which are more selective for the BIG and GBF families of ArfGEFs as opposed to cytohesins17,38. Consistent with this bias, neither compound inhibited ARNOSec7-mediated NΔ17PfArf1 activation (Fig. 2f), while Golgicide A caused only a minor 26% inhibition of NΔ17HsArf1 nucleotide exchange (Fig. 2e). In summary, the results confirmed that PfArf1 is susceptible to Sec7-mediated nucleotide exchange in vitro. In addition, it suggested that the assay format can robustly detect the in vitro activation Arf1 by a Sec7 domain, as well as the specific inhibition of the reaction by small compound inhibitors.
Figure 2.
Detection of ARNO-mediated nucleotide exchange using the GST-GGAGAT binding assay. (a,b) One µM GDP preloaded NΔ17HsArf1 (a) or NΔ17PfArf1 (b) was incubated with 0.2 µM ARNOSec7 and 50 µM GTP for 30 min at 37 °C, added to Ni-NTA coated 96-well plates and incubated for a further 30 min at 4 °C. GST-GGA3GAT was added to 1 µM and incubation continued at 4 °C for 60 min, followed by washing, incubation with GST substrate and absorbance readings at 340 nm. Control incubations contained the respective GDP preloaded Arf1 proteins alone, GDP preloaded Arf1 incubated with ARNOSec7 in the absence of GTP and GDP preloaded Arf1 incubated with GTP in the absence of ARNOSec7. (c–f) ARNOSec7 nucleotide exchange reactions were repeated with GDP preloaded NΔ17HsArf1 and NΔ17PfArf1in the presence of 50 µM SecinH3 (c,d) Brefeldin A (BFA) or Golgicide A (GA) (e,f), followed by the GST-GGA3GAT binding assay. Control reactions consisted of the GDP preloaded Arf1 proteins incubated with GTP in the absence of ARNOSec7 and inhibitors. Mean Abs340 values obtained from empty Ni-NTA plate wells incubated with GST-GGA3GAT were subtracted from all other readings. Incubations were carried out in triplicate wells and Abs340 shown as mean ± standard deviation. P-values were derived from two-tailed t-tests.
Detection of GAP-mediated GTP hydrolysis by human and P. falciparum Arf1
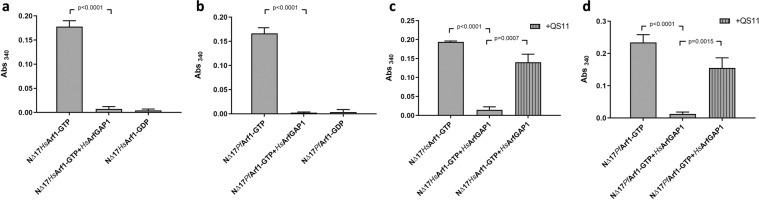
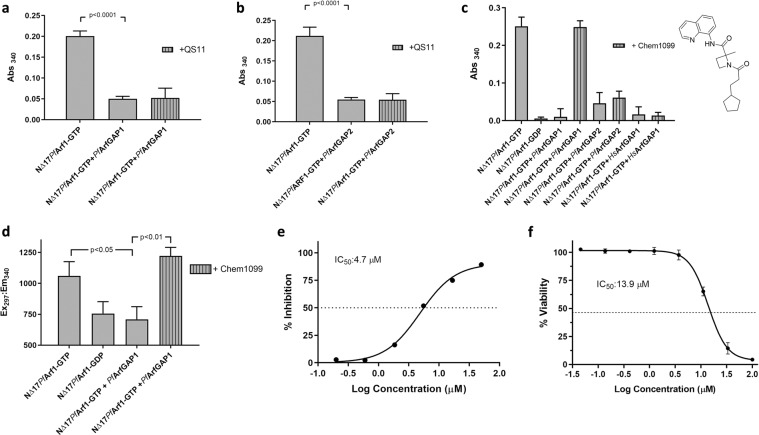
Having demonstrated in vitro Sec7-mediated nucleotide exchange by PfArf1, we next explored whether the assay format could detect GAP-mediated PfArf1 deactivation, using the GAP domain of human ArfGAP1 (HsArfGAP1GAP) as a model GAP. NΔ17HsArf1 and NΔ17PfArf1 preloaded with GTP were incubated with HsArfGAP1GAP, added to a nickel-NTA plate and GST-GGA3GAT binding assessed (Fig. 3a,b). Controls included the GTP-loaded Arf1 proteins incubated in the absence of HsArfGAP1GAP and plate wells containing immobilised GDP-loaded Arf1 proteins. Incubation with the GAP domain completely abrogated the binding of GST-GGA3GAT to both NΔ17HsArf1 and NΔ17PfArf1. To confirm that this was due to GAP-stimulated inactivation (GTP hydrolysis) of the Arf1 proteins, the ArfGAP inhibitor QS1121 was included in the incubations of the GTP-loaded Arf1 proteins with HsArfGAP1GAP at a concentration of 50 µM, which preserved GST-GGA3GAT binding of both NΔ17HsArf1 and NΔ17PfArf1 (Fig. 3c,d). Collectively, the results confirmed that PfArf1 is susceptible to GAP-mediated deactivation and that the assay format can competently detect in vitro ArfGAP activity as well as its inhibition by a small molecule inhibitor.
Figure 3.
Detection of GAP-mediated Arf1 deactivation using the GST-GGAGAT binding assay. (a,b) One µM GTP preloaded NΔ17HsArf1 (a) or NΔ17PfArf1 (b) was incubated with 0.1 µM HsArfGAP1GAP for 30 min at 37 °C, transferred to a Ni-NTA coated 96-well plate and incubation continued at 4 °C for 30 min. GST-GGA3GAT was added to 1 µM and incubation at 4 °C continued for 60 min, followed by washing, incubation with GST substrate and absorbance readings at 340 nm. Control reactions consisted of GTP preloaded Arf1 proteins incubated in the absence of HsArfGAP1GAP and wells incubated with GDP preloaded Arf1 proteins alone. (c,d) The incubations of NΔ17HsArf1 (c) and NΔ17PfArf1 (d) with HsArfGAP1GAP were repeated in the presence of 50 µM QS11. Control reactions consisted of incubations of the GTP preloaded Arf1 proteins in the absence of HsArfGAP1GAP and QS11. Abs340 values obtained from empty Ni-NTA plate wells incubated with GST-GGA3GAT were subtracted from all other readings. Incubations were carried out in triplicate wells and Abs340 shown as mean ± standard deviation. P-values were calculated by two-tailed t-tests.
GAP activity of two putative P. falciparum ArfGAPs
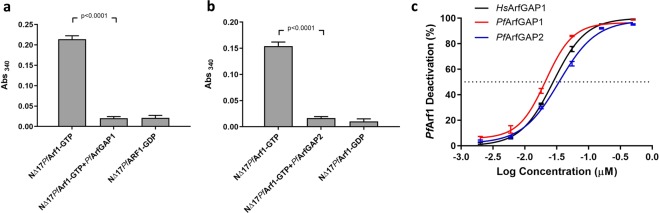
To some extent, stimulation of PfArf1 nucleotide exchange and GTP hydrolysis by human Sec7 and GAP domains (as well as GTP-dependent binding to the human effector protein GGA3) was not unexpected, given the high sequence and structural conservation of PfArf133. However, the question remains to what extent the predicted endogenous P. falciparum GEF and GAPs are capable of acting on PfArf1. In this study, we focused on the two sequences which are annotated as ArfGAPs on the plasmodb.org malaria genome database, which we designated as PfArfGAP1 (Plasmodb entry PF3D7_1244600) and PfArfGAP2 (PF3D7_0526200.1). In contrast to the sequence conservation of PfArf1, the predicted amino acid sequences of the GAP domains of two proteins are considerably less conserved compared to human ArfGAPs (alignments with HsArfGAP1 given in Supplementary Information Fig. S3) and, while the crystal structure of the PfArfGAP1 GAP domain has been published36, neither GAP domain has been reported to have catalytic activity. To demonstrate the latter, we repeated the assays performed with HsArfGAP1GAP. GTP-loaded NΔ17PfArf1 was incubated with the GAP domains of the respective malarial ArfGAPs (PfArfGAP1GAP, PfArfGAP2GAP) and GST-GGA3GAT binding assessed (Fig. 4a,b). As was previously found with HsArfGAP1GAP, both GAP domains reduced GST-GGA3GAT binding to the levels obtained with the GDP-loaded NΔ17PfArf1 controls, suggesting that they had stimulated GTP hydrolysis by the PfArf1 protein. As an end-point assay, the assay format employed here prevented a direct comparison of the GAP activity of the two GAP domains using a kinetic read-out of GTP hydrolysis by NΔ17PfArf1. To address this, a GAP titration assay was performed. NΔ17PfArf1-GTP was incubated at a concentration of 1 µM with serial dilutions of PfArfGAP1GAP, PfArfGAP2GAP and HsArfGAP1GAP, GST-GGA3GAT binding was determined and the dose-response curves compared (Fig. 4c). In this assay format, the GAP activities of the respective GAP domains were not found to be markedly different. Of the two P. falciparum GAP domains, PfArfGAP1GAP was more active, with half-maximal GAP activity (EC50) at 0.021 µM, compared to 0.034 µM for PfArfGAP2GAP (HsArfGAP1GAP was intermediate at 0.028 µM).
Figure 4.
Detection of GAP activity of P. falciparum GAP domains using the GST-GGAGAT binding assay. (a,b) One µM GTP preloaded NΔ17PfArf1 was incubated with 0.1 µM PfArfGAP1GAP (a) or PfArfGAP2GAP (b) for 30 min at 37 °C, transferred to a Ni-NTA coated 96-well plate and incubation continued at 4 °C for 30 min. GST-GGA3GAT was added to 1 µM and incubation at 4 °C continued for 60 min, followed by washing, incubation with GST substrate and absorbance readings at 340 nm. Control reactions consisted of GTP preloaded NΔ17PfArf1 incubated in the absence of the respective GAP domains and wells incubated with GDP preloaded NΔ17PfArf1 alone. Abs340 values obtained from empty Ni-NTA plate wells incubated with GST-GGA3GAT were subtracted from all other readings. Incubations were carried out in triplicate wells and Abs340 is shown as mean ± standard deviation. P-values were calculated using two-tailed t-tests. (c) One µM GTP preloaded NΔ17PfArf1 was incubated with three-fold serial dilutions (0.5–0.002 µM) of PfArfGAP1GAP, PfArfGAP2GAP and HsArfGAP1GAP for 30 min at 37 °C and the GST-GGA3GAT binding assay carried out as described above. Percentage PfArf1 deactivation was calculated from the Abs340 values obtained at the various GAP domain concentrations relative to those obtained with NΔ17PfArf1-GTP (0%) and NΔ17PfArf1-GDP (100%) incubated in the absence of GAP domains. Dose-response curves of percentage PfArf1 deactivation vs. Log[GAP concentration] were generated by non-linear regression analysis using GraphPad Prism.
Identification of a selective small molecule inhibitor of PfArfGAP1GAP activity
To confirm that the reduction in GST-GGA3GAT binding when GTP preloaded NΔ17PfArf1was incubated with 0.1 µM PfArfGAP1GAP and PfArfGAP2GAP was due to GAP activity, the assays were repeated in the presence of 50 µM QS11. In contrast to the results obtained with HsArfGAP1GAP (Fig. 3d), QS11 was unable to restore GST-GGA3GAT binding by NΔ17PfArf1-GTP incubated with either PfArfGAP1GAP or PfArfGAP2GAP (Fig. 5a,b). To identify a potential inhibitor of PfArfGAP1GAP-mediated deactivation of NΔ17PfArf1-GTP, we therefore screened a small BioFocus library of 1120 α-helix mimetics at a concentration of 50 µM (Screening details in Supplementary Information Fig. S5). We focused on the GAP domain of PfArfGAP1 since, in contrast to PfArfGAP2, the coding sequence has been reported to be essential to the survival of blood stage P. falciparum and P. berghei (murine malaria) parasites in genome-wide knockout and transposon mutagenesis studies39,40. This led to the identification of Chem1099 (Fig. 5c) which, at a concentration of 50 µM, preserved the GST-GGA3GAT binding ability of NΔ17PfArf1-GTP incubated with PfArfGAP1GAP, presumably due to inhibition of the GAP activity of the latter (Fig. 5c). Interestingly, the compound was inactive in a parallel screen carried out with PfArf1 and HsArfGAP1GAP (not shown). Indeed, at 50 µM, Chem1099 failed to inhibit the GAP activity of either HsArfGAP1GAP or PfArfGAP2GAP on NΔ17PfArf1-GTP, suggesting GAP selectivity (Fig. 5c). The inhibitory activity of Chem1099 was further confirmed using an alternative assay format. As described earlier, tryptophan fluorescence measurements can be used to assess the conformation of NΔ17PfArf1 which reflects its GTP- vs. GDP-bound status. Incubation of NΔ17PfArf1-GTP with PfArfGAP1GAP reduced its tryptophan fluorescence to levels obtained with a NΔ17PfArf1-GDP control, reflecting stimulation of GTP hydrolysis by the GAP domain (Fig. 5d). By contrast, inclusion of 50 µM Chem1099 in the reaction maintained NΔ17PfArf1-GTP fluorescence levels, suggesting complete inhibition of PfArfGAP1GAP GAP activity. Dose-dependent inhibition of PfArfGAP1GAP activity by Chem1099 was demonstrated by incubating NΔ17PfArf1-GTP and the GAP domain with serial dilutions of the compound followed by the GST-GGA3GAT binding assay and yielded an IC50 value of 4.7 µM (Fig. 5e). To determine if Chem1099 possesses anti-parasitic activity, a dose-response assay was conducted against cultured P. falciparum (3D7) parasites and parasite viability assessed using a plasmodial lactate dehydrogenase assay, which yielded an IC50 of 13.9 µM (Fig. 5f). In conclusion, the results suggest that PfArfGAP1 GAP activity can be inhibited by small compounds in vitro, that inhibitory compounds can discriminate between the GAP domains used in this study and that the assay format can be used to identify GAP inhibitors in compound libraries.
Figure 5.
Selective inhibition of PfArfGAP1GAP activity by a small molecule inhibitor. (a,b) One µM GTP preloaded NΔ17PfArf1 was incubated with 0.1 µM PfArfGAP1GAP (a) or PfArfGAP2GAP (b) for 30 min at 37 °C in the presence of 50 µM QS11, transferred to a Ni-NTA coated 96-well plate and incubation continued at 4 °C for 30 min. GST-GGA3GAT was added to 1 µM and incubation at 4 °C continued for 60 min, followed by washing, incubation with GST substrate and absorbance readings at 340 nm. Control reactions consisted of GTP preloaded NΔ17PfArf1 incubated in the absence of the respective GAP domains, or with the GAP domains in the absence of QS11. Abs340 values obtained from empty Ni-NTA plate wells incubated with GST-GGA3GAT were subtracted from all other readings. Incubations were carried out in triplicate wells and Abs340 is shown as mean ± standard deviation. (c) One µM GTP preloaded NΔ17PfArf1 was incubated respectively with 0.1 µM PfArfGAP1GAP, PfArfGAP2GAP or HsArfGAP1GAP in the absence or presence of 50 µM Chem1099 and the GST-GGA3GAT binding assay repeated as described above. Bars represent mean Abs340 ± standard deviation obtained from triplicate wells. The structure of Chem1099 is shown to the right. (d) Incubation of 1 µM NΔ17PfArf1-GTP with 0.1 µM PfArfGAP1GAP in the presence and absence of 50 µM Chem1099 for 30 min at 37 °C was repeated in a black 96-well plate and tryptophan fluorescence (Ex297/Em340) measured as an end-point reading. Additional wells contained 1 µM NΔ17PfArf1-GDP without PfArfGAP1GAP or without Chem1099. Bars represent mean fluorescence ± standard deviation obtained from triplicate wells. P-values were calculated by two-tailed t-tests. (e) The GST-GGA3GAT binding assay with Chem1099 was repeated with three-fold serial dilutions (50 µM – 0.2 µM) of the compound added to the incubation of NΔ17PfArf1-GTP with PfArfGAP1GAP in triplicate wells. Percentage inhibition of PfArfGAP1GAP activity was calculated from the Abs340 readings obtained at the various compound concentrations relative to the mean Abs340 obtained with NΔ17PfArf1-GTP incubated with PfArfGAP1GAP in the absence of Chem1099 (0%) and wells incubated with NΔ17PfArf1-GTP alone (100%). A dose-response curve was generated from the plot of mean percentage PfArfGAP1GAP inhibition ± standard deviation vs. Log(Chem1099 concentration) and the IC50 value derived by non-linear regression analysis using GraphPad Prism. (f) The antiplasmodial activity of Chem1099 was assessed by incubating P. falciparum (3D7) parasites with a serial dilution of Chem1099 in triplicate wells for 48 h and determining percentage parasite viability (relative to untreated controls) using a plasmodial lactate dehydrogenase assay. The IC50 value was derived by non-linear regression analysis of the % parasite viability vs. Log(Chem1099 concentration) plot using GraphPad Prism.
Discussion
Given the rapid growth rate of the P. falciparum malaria parasite and its reliance on vesicular trafficking to secrete proteins to internal organelles (notably specialised secretory organelles required for erythrocyte invasion), trafficking of proteins to and in the host erythrocyte cytoplasm, as well as extensive endocytosis of erythrocyte cytoplasm41, it is intriguing that, in contrast to mammalian cells, its genome only encodes one predicted Sec7 domain protein (ArfGEF) and two ArfGAPs (according to plasmodb.org annotations) to potentially regulate Arf GTPase function which is central to trafficking in mammalian cells. This is further compounded by the complexity of the parasite life-cycle which, in addition to the blood stages responsible for malaria pathogenesis, includes male and female gametocyte transmission stages, several stages in the Anopheles mosquito vector and human liver stages42. Moreover, although 6 sequences have been annotated as putative ADP-ribosylation factors, four may be Arf-like proteins as opposed to canonical Arf GTPases, one (Plasmodb accession number PF3D7_1034700) appears non-essential for blood-stage parasite survival39,40, and only PfArf1 has been characterised23–27,33. We have focused on PfArf1 and found that it binds to the GAT domain of the human effector protein GGA3 in a nucleotide-dependent manner, which allows it to be characterised in vitro using the plate-based assay format developed with human Arf1 as a model and reported here, as well as potentially allowing an assessment of its activation status in parasites using pull-down assays37.
Like its human counterpart, we confirmed that PfArf1 is susceptible to GDP/GTP nucleotide exchange stimulated by a Sec7 domain. Having used a human cytohesin domain for this purpose, we are currently exploring whether the nucleotide exchange activity extends to the predicted endogenous P. falciparum ArfGEF, despite the unusual secondary structure arrangement of its Sec7 domain34,35. In addition, we confirmed that PfArf1 deactivation can be achieved in vitro using the model GAP domain of human ArfGAP1 and that the GAP domains of the two putative P. falciparum ArfGAPs have equivalent catalytic GAP activities (based on EC50 values obtained in the assay format used here). Interestingly, despite the PfArf1 GAP activity displayed by the GAP domain of PfArfGAP2 and its presence in the parasite blood stages43,44, it has been reported as non-essential for blood-stage parasite survival, in contrast to PfArfGAP1, PfArf1 and the putative ArfGEF39,40. Along with the co-localisation of PfArf1 with the Golgi marker GRASP and the BFA sensitivity of parasite secretion and Golgi structure27–32, this may suggest that the latter trio of proteins form the regulatory network that mediates Arf GTPase-dependent trafficking of secretory proteins through the parasite Golgi apparatus. However, we recognise the caveat that we have performed the assays with truncated PfArf1 and PfArfGAP1 and that interaction in vitro does not necessarily translate into temporal and spatial co-recruitment and interaction on membrane surfaces in vivo. Potentially, this could be interrogated by parasite co-localisation experiments and assessing the effect of specific ArfGEF and ArfGAP1 inhibitors on PfArf1 activation status in parasites.
In addition to exploring the activity of PfArf1 regulatory proteins, the motivation for developing the assay described here was to establish an assay that can robustly detect the inhibition of the Arf1 activation/deactivation cycle and is amenable to screening compound libraries in a microtiter plate-based format. Conceptually, Arf function can be disrupted by inhibiting GTP binding, effector binding, GEF-mediated nucleotide exchange or GAP-mediated GTP hydrolysis. As opposed to inhibiting the binding of substrates/co-factors of traditional metabolic enzymes, protein-protein interactions are extremely challenging to interrupt with drug-like molecules45,46. It is therefore encouraging that this has been achieved with Arf1 (as well as Arf647), with the application of developing potential anti-cancer agents in mind10. The focus of these studies has been on inhibitors of GEF-mediated Arf1 activation, but also includes the discovery of the GAP inhibitors QS11 and its derivatives11,16–22. To support inhibitor discovery, plate-based human Arf1 screening assays that have been reported include a FRET assay for GEF activity48, a fluorescence polarisation assay for GAP activity49, and an additional fluorescence polarisation aptamer displacement assay specific for cytohesins and used to identify SecinH320. Relevant to these efforts, we show that the assay format reported here can competently detect the in vitro inhibition of ARNO Sec7-mediated human and P. falciparum NΔ17Arf1 activation by SecinH3, as opposed to BFA and Golgicide A, as well as inhibition of the deactivation of both proteins by human ArfGAP1 using QS11. In addition, in a preliminary screen of a limited α-helix mimetic library, we identified Chem1099 as a low micromolar in vitro inhibitor of NΔ17PfArf1 deactivation by the GAP domain of PfArfGAP1, further supporting the notion that ArfGAP activity can potentially be inhibited by small chemical compounds and, given the inactivity of Chem1099 against the GAP domains of HsArfGAP1 and PfArfGAP2, that this can be achieved selectively. In light of the reported essentiality of PfArf1 and PfArfGAP1 in blood-stage parasites, it is encouraging that Chem1099 inhibits blood-stage P. falciparum, albeit with a moderate IC50 of 14 µM compared to the low nanomolar activities obtained with standard antimalarials50. However, the assumption that parasite inhibition is due to GAP inhibition is a tenuous one in the absence of extensive mode of action or validation studies. Validation experiments could conceptually include an assessment of the effect of Chem1099 on parasite Golgi structure and function (e.g. through secretion assays), effect of Chem1099 on Arf1 activation status in parasites using pull-down assays on treated parasite lysates, and assessment of Chem1099 IC50 modulation in ArfGAP1 overexpressing or silenced transgenic parasite lines. We are currently expanding our screening of libraries for PfArfGAP1 inhibitors, coupled with biological assays to determine if this avenue of disrupting the PfArf1 activation cycle is detrimental to parasite viability.
Methods
Plasmid constructs and protein expression
For the E. coli expression of the GST-GGA3GAT fusion protein (GST fused to the GAT domain - amino acids 107–286 - of human GGA3), pGEX-4T-2/hGGA3(GAT) (Addgene plasmid #79436, donated by Kazuhisa Nakayama) was used. The other coding sequences were ligated into the NheI/BamHI (Arf1 sequences) or NheI/XhoI sites of pET-28a(+) for expression as His-tagged proteins. The coding sequence of human Arf1 minus the N-terminal 17 amino acids (NΔ17HsArf1) was PCR amplified from pARF1-CFP (Addgene plasmid #11381, donated by Joel Swanson) and the corresponding P. falciparum Arf1 sequence (NΔ17PfArf1) from the full length PfArf1 sequence (PlasmoDB ID PF3D7_1020900) codon-optimised for human expression, synthesised and cloned into pBluescript II by GenScript (Hong Kong). The sequences for the GAP domain of human ArfGAP1 (HsArfGAP1GAP; amino acids 1–140; NCBI sequence NP_060679.1), Sec7 domain of ARNO (ARNOSec7; amino acids 51–253; NP_004219.3) and the putative GAP domain of P. falciparum ArfGAP2 (PfArfGAP2GAP; amino acids 1–161; PF3D7_0526200.1) were codon optimised for E. coli expression and cloned into pET-28a by GenScript. The sequence encoding the putative GAP domain of P. falciparum ArfGAP1 (PfArfGAP1GAP; amino acids 1–161; PF3D7_1244600) was PCR amplified from P. falciparum strain 3D7 genomic DNA. T7 Express lysY E. coli (New England Biolabs) cultured in LB broth was used as expression host for all proteins. Expression was induced after bacterial density had reached OD600 0.5–0.8 with 1 mM IPTG for 3 hours at 37 °C. Bacteria harvested from the induced cultures were lysed by a freeze/thaw cycle, resuspension in buffer containing 2 mg/mL lysozyme and probe sonication. Proteins were purified from the soluble supernatants by nickel-NTA agarose (His-tagged proteins) or glutathione agarose (GST-GGA3GAT) affinity chromatography. Purified proteins were buffer exchanged into assay buffer (25 mM HEPES, 150 mM KCl, 1 mM MgCl2, 1 mM DTT, pH 7.4) using desalting columns and protein concentrations determined using Bradford protein assay. Glycerol was added to a final concentration of 40% (v/v) and the proteins stored at −20 °C until use. More details on protein expression and purification are given in the Supplementary Information (Fig. S2).
Nucleotide loading of Arf1 proteins
To preload NΔ17HsArf1 with GTP or GDP, the protein was diluted to a final concentration of 5 µM in assay buffer (25 mM HEPES, 150 mM KCl, 1 mM MgCl2, 1 mM DTT, pH 7.4) supplemented with 2 mM EDTA and 50 µM GTP or GDP and incubated at 25 °C for 60 minutes. MgCl2 was added to a final concentration of 3 mM and incubation continued for a further 10 min. Nucleotide loading of NΔ17PfArf1 was carried out in the same manner, except that 20 mM EDTA and 30 mM MgCl2 was used. To monitor nucleotide binding, intrinsic tryptophan fluorescence was measured at Ex297/Em340 in a Spectramax M3 plate reader (Molecular Devices). In addition, after completion of nucleotide loading, proteins were analysed in a gel shift (native PAGE) assay. Native PAGE was carried out with a 12% resolving gel and 4% stacking gel using normal SDS-PAGE conditions, except that SDS was omitted from all buffers and reducing agents were omitted from the sample buffer. After electrophoresis, the gel was stained with Coomassie Brilliant Blue.
Plate-based GST-GGA3GAT binding assay
His-tagged NΔ17HsArf1 or NΔ17PfArf1 preloaded with GTP or GDP were diluted to 1 µM in assay buffer supplemented with 1% (w/v) bovine serum albumin (BSA), transferred to a Ni-NTA HisSorb 96-well plate (Qiagen) (50 µL per well) and incubated at 4 °C for 30 min with gentle agitation. GST-GGA3GAT in 50 µL assay buffer was added to a final concentration of 1 µM and incubation continued for an additional 60 min at 4 °C. The protein solutions were aspirated, the wells washed twice in assay buffer containing 0.1% (v/v) Tween-20 followed by four additional washes in assay buffer. GST assay buffer (2 mM reduced L-glutathione and 1 mM 1-chloro-2,4-dinitrobenzene in phosphate-buffered saline, pH 7.4), pre-equilibrated to room temperature, was added to each well (200 µL/well), the plate incubated at room temperature for 30 min and absorbance read at 340 nm in a Spectramax M3 plate reader. Background absorbance readings were obtained from triplicate wells incubated with GST-GGA3GAT in the absence of immobilised Arf1 and the mean absorbance subtracted from the absorbance values of the experimental GST-GGA3GAT wells. Plates were prepared for re-use by rinsing the plate wells in water followed by a 10 min incubation in stripping buffer (20 mM sodium phosphate, 500 mM NaCl, 50 mM EDTA, pH 7.4), an additional wash in water and a 10 min incubation in recharging solution (0.1 M NiSO4). After a final rinse in water, the plates were used immediately.
ARNO-mediated nucleotide exchange and GAP-mediated GTP hydrolysis assays
For nucleotide exchange assays, 1 μM NΔ17HsArf1 or NΔ17PfArf1 preloaded with GDP was incubated with 0.2 µM ARNOSec7 and 50 μM GTP in assay buffer containing 1% BSA in round-bottom plates (50 µL per well) at 37 °C for 30 minutes with continuous agitation. The reactions were transferred to a Ni-NTA plate and the plate-based GST-GGA3GAT binding assay continued as described above. Negative controls included reactions without ARNO, without GTP, or without either. GAP assays were carried out in the same manner, except that Arf1 proteins preloaded with GTP were used, ARNO was replaced with 0.1 µM of the relevant GAP domain (HsArfGAP1GAP, PfArfGAP1GAP, PfArfGAP2GAP) and the addition of GTP was omitted. Negative controls consisted of reactions lacking the GAP domains. To assess the inhibition of nucleotide exchange or GTP hydrolysis, 10 mM stocks of brefeldin A (BFA; Sigma-Aldrich), Golgicide A (GA; Sigma-Aldrich), SecinH3 (Tocris Bioscience) and QS11 (Tocris Bioscience) were prepared in DMSO. The inhibitors were added to the reactions in the round-bottom plate wells to a final concentration of 50 µM [inhibitors were added to the Arf1 solutions immediately before adding ARNO (BFA, GA or SecinH3) or the GAP domains (QS11)]. A corresponding volume of DMSO was added to control reactions lacking the inhibitors (solvent vehicle controls). GAP titration experiments with NΔ17PfArf1 were carried out as described above, except that incubations were carried out with 1 µM NΔ17PfArf1-GTP and 3-fold serial dilutions (0.5–0.002 µM) of the GAP domains. For compound library screening, 50 µL assay buffer containing 1% BSA, 1 µM NΔ17PfArf1-GTP and 0.1 µM PfArfGAP1GAP was incubated in the presence of 50 µM of the test compounds in round-bottom plates for 30 minutes at 37 °C (compounds were added to the reaction mixture before the addition of the GAP domain). The reaction mixtures were transferred to Ni-NTA plates and the GST-GGA3GAT binding assay continued as described above. Dose-dependent inhibition of PfArfGAP1GAP by Chem1099 was determined in the same manner, using 3-fold serial dilutions of the compound. Percentage inhibition of GAP activity at the respective compound concentrations was calculated from Abs340 readings relative to those obtained with NΔ17PfArf1-GTP incubated with PfArfGAP1GAP without Chem1099 (0%) and NΔ17PfArf1-GTP incubated without PfArfGAP1GAP (100%). A dose-response curve of percentage inhibition vs. Log[Chem1099] was generated and the IC50 determined using non-linear regression analysis with GraphPad Prism (v.8.2.0).
Antiplasmodial assay
This was carried out as described previously51. Briefly, cultures of Plasmodium falciparum (3D7) parasites in a 96-well plate were incubated with a 3-fold serial dilution of Chem1099 (100–0.046 µM) for 48 h and parasite levels assessed using a colorimetric plasmodial lactate dehydrogenase (pLDH) assay52. Absorbance readings were converted to percentage parasite viability relative to readings obtained from control wells (parasite cultures without Chem1099) and IC50 derived by non-linear regression analysis of the resulting % viability vs. Log(Chem1099 concentration) using GraphPad Prism.
Supplementary information
Acknowledgements
This study was initiated with funding from the South African Medical Research Council (self-initiated research grant), with additional support from the Deutsche Forschungsgemeinschaft (German-African collaborations in Infectology, PR1099/4-1). This work was further supported through the Grand Challenges Africa programme (GCA/DD/rnd3/032). Grand Challenges Africa is a programme of the African Academy of Sciences (AAS) implemented through the Alliance for Accelerating Excellence in Science in Africa (AESA) platform, an initiative of the AAS and the African Union Development Agency (AUDA-NEPAD). For this work GC Africa is supported by the African Academy of Sciences (AAS), Bill & Melinda Gates Foundation (BMGF), Medicines for Malaria Venture (MMV), and Drug Discovery and Development centre of University of Cape Town (H3D). The views expressed herein are those of the author(s) and not necessarily those of the AAS and her partners. T.S. was supported by a postgraduate bursary from the South African Research Chairs Initiative of the Department of Science and Technology (DST) and National Research Foundation of South Africa (NRF) (Grant No. 98566), F.D.K. by an NRF bursary and A.N. by a Pearson-Young scholarship from Rhodes University.
Author contributions
H.C.H. conceptualised the study and wrote the manuscript, with contributions from all the authors. T.S. performed the experiments, F.D.K., A.N. and D.L. contributed to developing the methodology and reagents used and performed additional experiments reported in the Supporting Information, C.G.L.V. assisted with evaluating the compound screening results and J.M.P., A.L.E. and H.C.H. directed the study.
Data availability
The majority of the data generated or analysed during this study are included in this article and Supplementary Information. Data not shown are available by request from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-61101-3.
References
- 1.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell. Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 2.Donaldson JG, Jackson CL. Arf family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell. Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson CL, Bouvet S. Arfs at a glance. J. Cell Sci. 2014;187:4103–4109. doi: 10.1242/jcs.144899. [DOI] [PubMed] [Google Scholar]
- 4.Beck R, Ravet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 5.Bonifacino JS, Lippincott-Schwartz J. Coat proteins: shaping membrane transport. Nat. Rev. Mol. Cell Biol. 2003;4:409–414. doi: 10.1038/nrm1099. [DOI] [PubMed] [Google Scholar]
- 6.Boulay P-L, et al. ARF1 controls proliferation of breast cancer cells by regulating the retinoblastoma protein. Oncogene. 2011;30:3846–3861. doi: 10.1038/onc.2011.100. [DOI] [PubMed] [Google Scholar]
- 7.Casalou C, Faustino A, Barral DC. Arf proteins in cancer cell migration. Small GTPases. 2016;7:270–282. doi: 10.1080/21541248.2016.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulay P-L, Cotton M, Melançon P, Claing A. ADP-ribosylation factor 1 controls the activation of the Phosphatidylinositol 3-kinase pathway to regulate epidermal growth factor-dependent growth and migration of breast cancer cells. J. Biol. Chem. 2008;283:3642–3634. doi: 10.1074/jbc.M803603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis JE, et al. ARF1 promotes prostrate tumorigenesis via targeting oncogenic MAPK signaling. Oncotarget. 2016;7:39834–39845. doi: 10.18632/oncotarget.9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prieto-Dominguez N, Parnell C, Teng Y. Drugging the small GTPase pathways in cancer treatment: promises and challenges. Cells. 2019;8:255. doi: 10.3390/cells8030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohashi Y, et al. AMF-26, a novel inhibitor of the Golgi system, targeting ADP-ribosylation factor 1 (Arf1) with potential for cancer therapy. J. Biol. Chem. 2012;287:3885–3897. doi: 10.1074/jbc.M111.316125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohashi Y, et al. M-COPA, a Golgi disruptor, inhibits cell surface expression of MET protein and exhibits antitumor activity against MET-addicted gastric cancers. Cancer Res. 2016;76:3895–3903. doi: 10.1158/0008-5472.CAN-15-2220. [DOI] [PubMed] [Google Scholar]
- 13.Sausville EA, et al. Antiproliferative effect in vitro and antitumor activity in vivo of brefeldin A. Cancer J. Sci. Am. 1996;2:52–58. [PubMed] [Google Scholar]
- 14.Sztul E, et al. ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell. 2019;30:1249–1271. doi: 10.1091/mbc.E18-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spang A, Shiba Y, Randazzo PA. ArfGAPs: gatekeepers of vesicle generation. FEBS Lett. 2010;584:2646–2651. doi: 10.1016/j.febslet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seehafer K, et al. Synthesis and biological properties of novel Brefeldin A analogues. J. Med. Chem. 2013;56:5872–5884. doi: 10.1021/jm400615g. [DOI] [PubMed] [Google Scholar]
- 17.Saenz JB, et al. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 2009;5:157–165. doi: 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viaud J, et al. Structure-based discovery of an inhibitor of Arf activation by Sec7 domains through targeting protein-protein complexes. Proc. Natl. Acad. Sci. USA. 2007;104:10370–10375. doi: 10.1073/pnas.0700773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spooner RA, et al. The secretion inhibitor Exo2 perturbs trafficking of Shiga toxin between endosomes and the trans-Golgi network. Biochem. J. 2008;414:471–484. doi: 10.1042/BJ20080149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hafner M, et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, et al. Small-molecule synergist of the Wnt/β-catenin signaling pathway. Proc. Natl. Acad. Sci. USA. 2007;104:7444–7448. doi: 10.1073/pnas.0702136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh M, et al. Structure-activity relationship studies of QS11, a small molecule Wnt synergistic agonist. Bioorg. Med. Chem. Lett. 2015;25:4838–4842. doi: 10.1016/j.bmcl.2015.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truong RM, Francis SE, Chakrabarti D, Goldberg DE. Cloning and characterization of Plasmodium falciparum ADP-ribosylation factor and factor-like genes. Mol. Biochem. Parasitol. 1997;84:247–253. doi: 10.1016/S0166-6851(96)02803-4. [DOI] [PubMed] [Google Scholar]
- 24.Lee F-JS, et al. Identification and characterization of an ADP-ribosylation factor in Plasmodium falciparum. Mol. Biochem. Parasitol. 1997;87:217–223. doi: 10.1016/S0166-6851(97)00061-3. [DOI] [PubMed] [Google Scholar]
- 25.Stafford WH, Stockley RW, Ludbrook SB, Holder AA. Isolation, expression and characterization of the gene for an ADP-ribosylation factor from the human malaria parasite, Plasmodium falciparum. Eur. J. Biochem. 1996;242:104–113. doi: 10.1111/j.1432-1033.1996.0104r.x. [DOI] [PubMed] [Google Scholar]
- 26.Leber W, et al. A unique phosphatidylinositol 4-phosphate 5-kinase is activated by ADP-ribosylation factor in Plasmodium falciparum. Int. J. Parasitol. 2009;39:654–653. doi: 10.1016/j.ijpara.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Thavayogarajah T, et al. Alternative protein secretion in the malaria parasite Plasmodium falciparum. PLoS One. 2015;10:e0125191. doi: 10.1371/journal.pone.0125191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi Mitsuko, Taniguchi Shinya, Ishizuka Yuki, Kim Hye-Sook, Wataya Yusuke, Yamamoto Akitsugu, Moriyama Yoshinori. A Homologue ofN-Ethylmaleimide-sensitive Factor in the Malaria ParasitePlasmodium falciparumIs Exported and Localized in Vesicular Structures in the Cytoplasm of Infected Erythrocytes in the Brefeldin A-sensitive Pathway. Journal of Biological Chemistry. 2001;276(18):15249–15255. doi: 10.1074/jbc.M011709200. [DOI] [PubMed] [Google Scholar]
- 29.Ogun SA, Holder AA. Plasmodium yoelii: brefeldin A-sensitive processing of proteins targeted to the rhoptries. Exp. Parasitol. 1994;79:270–278. doi: 10.1006/expr.1994.1090. [DOI] [PubMed] [Google Scholar]
- 30.Crary JL, Haldar K. Brefeldin A inhibits protein secretion and parasite maturation in the ring stage of Plasmodium falciparum. Mol. Biochem. Parasitol. 1992;53:185–192. doi: 10.1016/0166-6851(92)90020-K. [DOI] [PubMed] [Google Scholar]
- 31.Wickham ME, et al. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 2001;20:5636–5649. doi: 10.1093/emboj/20.20.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benting J, Mattei D, Lingelbach K. Brefeldin A inhibits transport of the glycophorin-binding protein from Plasmodium falciparum into the host erythrocyte. Biochem J. 1994;300:821–826. doi: 10.1042/bj3000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook WJ, Smith CD, Senkovich O, Holder AA, Chattopadhyay D. Structure of Plasmodium falciparum ADP-ribosylation factor 1. Acta Cryst. 2010;F66:1426–1431. doi: 10.1107/S1744309110036997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumgartner F, Wiek S, Paprotka K, Zauner S, Lingelbach K. A point mutation in an unusual Sec7 domain is linked to brefeldin A resistance in a Plasmodium falciparum line generated by drug selection. Mol. Microbiol. 2001;41:1151–1158. doi: 10.1046/j.1365-2958.2001.02572.x. [DOI] [PubMed] [Google Scholar]
- 35.Wiek S, Cowman AF, Lingelbach K. Double cross-over gene replacement within the Sec7 domain of a GDP-GTP exchange factor from Plasmodium falciparum allows the generation of a transgenic brefeldin A-resistant parasite line. Mol. Biochem. Parasitol. 2004;138:51–55. doi: 10.1016/j.molbiopara.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 36.Cook WJ, Senkovich O, Chattopadhyay D. Structure of the catalytic domain of Plasmodium falciparum ARF GTPase-activating protein (ARFGAP) Acta Cryst. 2011;F67:1339–1344. doi: 10.1107/S1744309111032507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen LA, Donaldson JG. Analysis of Arf GTP-binding protein function in cells. Curr. Protoc. Cell Biol. 2010;48:14.12.1–14.12.17. doi: 10.1002/0471143030.cb1412s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell. 2004;15:1487–1505. doi: 10.1091/mbc.e03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, M. et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science360, eaap7847 (2018). [DOI] [PMC free article] [PubMed]
- 40.Bushell E, et al. Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell. 2017;170:260–272. doi: 10.1016/j.cell.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DePonte M, et al. Wherever I may roam: protein and membrane trafficking in the malaria parasite. Mol. Biochem. Parasitol. 2012;186:95–116. doi: 10.1016/j.molbiopara.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Matthews H, Duffy CW, Merrick CJ. Checks and balances? DNA replication and the cell cycle in Plasmodium. Parasit. Vectors. 2018;11:216. doi: 10.1186/s13071-018-2800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto TD, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toenhake CG, et al. Chromatin accessibility-based characterization of the gene regulatory network underlying Plasmodium falciparum blood-stage development. Cell Host Microbe. 2018;23:557–569. doi: 10.1016/j.chom.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing towards the reality. Chem. Biol. 2014;12:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raj M, Bullock BN, Arora PS. Plucking the high hanging fruit: a systematic approach for targeting protein-protein interactions. Bioorg. Med. Chem. 2013;21:4051–4057. doi: 10.1016/j.bmc.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoo J, et al. ARF6 is an actionable node that orchestrates oncogenic GNAQ signaling in uveal melanoma. Cancer Cell. 2016;29:889–904. doi: 10.1016/j.ccell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bill A, et al. A homogeneous fluorescence resonance energy transfer system for monitoring the activation of a protein switch in real time. J. Am. Chem. Soc. 2011;133:8372–8379. doi: 10.1021/ja202513s. [DOI] [PubMed] [Google Scholar]
- 49.Sun W, VanHooke JL, Sondek J, Zhang Q. High-throughput fluorescence polarization assay for the enzymatic activity of GTPase-activating protein of ADP-ribosylation factor (ARFGAP) J. Biomol. Screen. 2011;16:718–723. doi: 10.1177/1087057111408420. [DOI] [PubMed] [Google Scholar]
- 50.Le Manach C, et al. Fast in vitro methods to determine the speed of action and the stage-specificity of anti-malarials in Plasmodium falciparum. Malaria J. 2013;12:424. doi: 10.1186/1475-2875-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lunga MJ, et al. Expanding the SAR of nontoxic antiplasmodial indolyl-3-ethanone ethers and thioethers. ChemMedChem. 2018;13:1353–1362. doi: 10.1002/cmdc.201800235. [DOI] [PubMed] [Google Scholar]
- 52.Makler MT, Hinrichs DJ. Measurement of the lactate dehydrogenase activity of Plasmodium falciparum as an assessment of parasitemia. Am. J. Trop. Med. Hyg. 1993;48:205–210. doi: 10.4269/ajtmh.1993.48.205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The majority of the data generated or analysed during this study are included in this article and Supplementary Information. Data not shown are available by request from the corresponding author.