Abstract
Background
The surgical resection of colorectal cancer with liver metastases (CLM) has proven to be the most important modality for long-term survival, while effective biomarkers for outcome prediction or postoperative surveillance are still lacking. Currently, circulating biomarkers obtained from a liquid biopsy are widely used to assess the treatment response, disease recurrence and progression. In this study, we analyzed the value of the liquid biopsy, which includes circulating tumor DNA (ctDNA) and cell-free DNA (cfDNA), in patients with CLM.
Methods
Capture-based targeted deep sequencing was performed on matched pre-surgery, post-surgery and liver metastatic tissues of 20 CRC patients who underwent the resection of liver metastases between May and September 2017 using a panel consisting of 41 genes. Mutation landscapes obtained from pre-surgery plasma samples and metastatic tissue samples were compared.
Results
Collectively, we identified 47 mutations from 17 pre-surgery plasma samples (85%), and the remaining 3 patients had no mutation detected from the panel. We revealed a high by-variant concordance rate of 82.14% between pre-surgery plasma samples and liver metastatic tissue samples. We further analyzed the correlation between ctDNA, cfDNA, CEA and tumor burden and revealed a positive correlation between ctDNA and tumor burden (R=0.69, p=0.002). As of the date for data cutoff, 8/20 patients experienced relapse. Our study also demonstrated that pre-surgery ctDNA (p<0.001), cfDNA (p=0.001) and CEA (p=0.012) levels had predictive value for relapse. Patients with low pre-surgery ctDNA (p<0.001), cfDNA (p=0.001) or CEA (p=0.012) levels were more likely to experience prolonged progression-free survival.
Conclusion
Our data demonstrate that the genomic profile obtained from ctDNA is comparable with the genomic profile obtained from metastatic liver tumors. Furthermore, our study also show that pre-surgery ctDNA levels are positively correlated with tumor burden. In addition, pre-surgery ctDNA, cfDNA and CEA levels have predictive value for relapse.
Keywords: colorectal liver metastases, CLM, circulating tumor DNA, ctDNA, tumor burden, prognosis
Introduction
Colorectal cancer (CRC) is currently the third most common cancer worldwide. Approximately 25% of patients with CRC have liver metastases at the initial diagnosis, and another 20–30% of patients will eventually develop liver metastases, contributing to the high mortality rates reported for CRC.1,2 CRC patients with liver metastases (CLM) are thought to have aggressive clinicopathological features, unfavorable cancer biology and poor survival outcomes.3 Although surgical resection of the primary tumor and liver metastases has proven to offer long-term survival, the optimal timing and sequence of chemotherapy and surgery are still undetermined.4 In clinical practice, the involvement of a multidisciplinary team (MDT) in the management of CLM is recommended in clinical guidelines by integrating molecular, clinical and pathological features to customize therapy.5
Due to the advancement of high-throughput sequencing technology, the elucidation of genetic features is more feasible, and a molecular classification with distinct biological behaviors for CRC was proposed by the international consensus.6,7 Although tissue biopsy or surgical resection samples are still regarded as the gold standard for mutation profiling, liquid biopsy is widely used in clinical settings. Obtaining tissue biopsies is not only invasive but also biased because of its temporal and spatial snapshot nature. Furthermore, samples obtained from patients who have progressed on previous treatment options might not be adequate for NGS testing.8–10 Hence, the application of the liquid biopsy, which includes circulating tumor cell (CTC), circulating cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA), has been gaining momentum in clinical practice. cfDNA includes DNA fragments in cell-free components that are released into the bloodstream by cells. The small proportion of cfDNA from neoplastic cells is referred to as ctDNA. Previous studies have reported the value of ctDNA in colorectal cancer management. Several studies confirmed the predictive value of ctDNA in therapeutic response in metastatic colorectal cancer, and prognostic significance of postoperative ctDNA in early stage colon cancer and locally advanced colon and rectal cancer.11–15 ctDNA has also showed potential clinical utility to guide therapeutic decision-making in colorectal cancers.13,14
Considering that the surgical resection of liver metastases is the major choice for CLM treatment, major efforts are needed to elucidate the clinical value of ctDNA in CLM with regard to surgical benefits and optimal treatment strategies.15–17 In the current study, we prospectively analyzed the application of perioperative ctDNA in predicting tumor burden and the risk of recurrence in patients with CLM undergoing the resection of liver metastases.
Materials and Methods
Study Design
This prospective study enrolled 20 CRC patients with liver metastases who underwent surgical resection of their liver metastases at the Department of Colorectal Surgery, Fudan University Shanghai Cancer Center (FUSCC) between May 2017 and September 2017. All patients had stage IV disease. Among them, 9 received simultaneous resection, and 11 received two-stage resection for colorectal cancer and liver metastases. For patients who received two-stage resection, they had surgery on their primary site first and blood collections were carried out before and after metastases resection. Matched pre- and post-surgery blood samples as well as liver tumor tissue samples were obtained from each patient. According to our policy, patients with CLM were all required to attend our institutional multidisciplinary discussion before the surgical resection of primary tumors or metastases. The tumor burden of liver metastases was measured as the sum of the volumes of all ablated tumors by surgery or radiofrequency ablation using the spherical volume formula ([4/3]×π×r3, where r is the maximum tumor radius) and is referred to as the total tumor volume (TTV).18 All patients were followed-up every 3 to 6 months after the surgery. The last follow-up date was November 27, 2018.
This study was reviewed and approved by the Institutional Review Board ofFUSCC. Written informed consent was obtained from all patients to use their cancer tissue and clinical and outcome information for future academic studies during their first hospital admission. This study was carried out in accordance with the Declaration of Helsinki.
Tissue DNA Extraction
DNA of metastatic tumor tissue was extracted using a QIAamp DNA FFPE tissue kit (Qiagen, California, USA) according to the manufacturer’s instructions. DNA concentration was measured with a Qubit dsDNA assay (Life Technologies, California, USA).
DNA Extraction from Blood and cfDNA Extraction
Serial blood samples were collected at two defined time points: pre-surgery (within 7 days before surgery) and post-surgery (within 7 days after surgery). 10 mL of blood were collected in the streck tubes (DNA preserving tubes) and send to be processed immediately within 72 h of collection. Whole blood was centrifuged at 2400×g for 10 min at 4°C. After discarding the red blood cells and buffy coat, we centrifuged the plasma at 16,000×g for another 5 min at 4 °C. Supernatants were subsequently stored at −80 °C until further analysis. cfDNA from plasma was purified using a QIAamp Circulating Nucleic Acid kit according to the manufacturer’s instructions.
Next-Generation Sequencing (NGS) Library Preparation
Tissue DNA was sheared using a Covaris M220 ultrasonicator. The sheared tissue DNA and purified cfDNA underwent end repair, phosphorylation and adaptor ligation. Fragments of 200–400 base pairs (bp) were selected using Agencourt AMPure beads (Beckman Coulter, Brea, CA, USA) followed by hybridization with capture probe baits, hybrid selection with magnetic beads and polymerase chain reaction (PCR) amplification. A bioanalyzer high-sensitivity DNA assay was performed to assess the quality and size of the fragments. A minimum of 50 ng of DNA was used for library construction. Twelve PCR cycles were used for library amplification. The indexed samples were sequenced on a NovaSeq system (Illumina, Inc., San Diego, CA, USA) with paired-end reads with read lengths of 150 bp.
Capture-Based Targeted DNA Sequencing
The genomic profiles of tissue and cfDNA samples were assessed by performing capture-based targeted deep sequencing using the ColonCore panel (Burning Rock Biotech, Ltd.), which covers the whole exons of 41 colon cancer-related genes (Supplemental Table 1) and spans 213 kb of the human genome. DNA quality and size were assessed by a high sensitivity DNA assay using a bioanalyzer. All indexed samples were sequenced on a NextSeq 500 system (Illumina, Inc.) with pair-end reads.
Sequencing Analysis
Sequencing data in FASTQ format were mapped to the human genome (hg19) using BWA 0.7.10. Local alignment optimization, variant calling, and annotation were performed using GATK 3.2, MuTect, and VarScan, respectively. DNA translocation analysis was performed using both TopHat2 and Factera 1.4.3. Gene-level copy number variation was statistically assessed after normalizing the read depth at each region by the total read number and region size and correcting for GC bias using the LOESS algorithm. The amount of ctDNA was calculated as the product of the maximum allelic fraction (MAF) and the cfDNA amount.
Statistics
Normality was assessed with the Shapiro–Wilk test. Continuous variables, such as CEA, cfDNA, and ctDNA, were compared using the Wilcoxon signed-rank test. Correlations between tumor burden and the circulating biomarkers (CEA, cfDNA, and ctDNA) were assessed using Pearson’s correlation after the log base 10 transformation of both variables. The Kaplan-Meier test was used for the survival analysis. The Log rank test was performed to determine statistical significance. Progression-free survival (PFS) was measured from the date of baseline blood collection to the date of progression of liver metastases after surgical resection. The concordance between mutations obtained from tissue and plasma samples was calculated as the ratio of the number of mutations detected in both plasma and tissue samples. All analyses were set at a two-sided p value <0.05 as the threshold for statistical significance. Data are expressed as the mean ± standard deviation (SD). All the analyses and data visualization were performed with RStudio-1.0.143.
Results
Patient Characteristics
Collectively, 20 CRC patients with liver metastases who underwent surgical resection of their liver metastases at the Department of Colorectal Surgery, FUSCC between May 2017 and September 2017 were enrolled. The mean age was 55±13 years. The cohort consisted of 6 females and 14 males. The mean pre-surgery serum CEA level was 31.89 ±47.08 ng/mL; 7 patients fell within the normal range (≤5ng/mL), and 13 patients had elevated levels (>5ng/mL). The primary CRC tumor sites included 9 on the rectum, 6 on the left-sided colon and 5 on the right-sided colon. Nine patients received simultaneous resection, and the remaining 11 patients received two-stage resection for colorectal cancer and liver metastases. For patients who received two-stage resection, they had surgery on their primary site first. Eleven patients received fluorouracil-based neoadjuvant therapy with median time of 3 regimens and 16 patients received fluorouracil-based adjuvant chemotherapy with median time of 5 regimens. Detailed patient characteristics are shown in Table 1.
Table 1.
Demographic and Clinical Characteristics of CLM Patients
| Characteristics | No. |
|---|---|
| Patients Number | 20 |
| Age (years, Mean±SD) | 55±13 |
| Gender (Female/Male) | |
| Female | 6 |
| Male | 14 |
| Smoking (Never/ever) | |
| Never | 4 |
| Ever | 16 |
| BMI (Mean±SD) | 22.5±3.42 |
| Primary tumor sites | |
| Rectums | 9 |
| Left-sided colons | 6 |
| Right-sided colons | 5 |
| Surgery | |
| Simultaneous resection | 9 |
| Two-stage resection | 11 |
| Neoadjuvant therapy | |
| XELOX | 10 |
| FOLFIRI | 1 |
| Therapy after Surgery | |
| XELOX | 9 |
| FOLFOX | 7 |
| Without chemotherapy | 4 |
| Pre-surgery serum CEA (ng/mL, Mean±SD) | 31.89±47.08 |
| Elevated (>5ng/mL) | 13 |
| Normal (≤5ng/mL) | 7 |
| Pre-surgery cfDNA (ng/mL, Mean±SD) | 17.16±15.76 |
| Pre-surgery ctDNA (ng/mL, Mean±SD) | 1.46±3.70 |
| Post-surgery cfDNA (ng/mL, Mean±SD) | 48.89±29.18 |
| Post-surgery ctDNA (ng/mL, Mean±SD) | 0.87±3.31 |
Mutation Landscape in ctDNA of CLM Patients
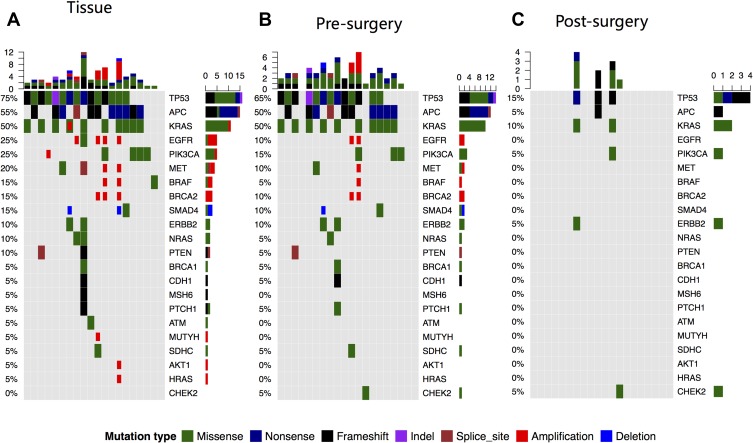
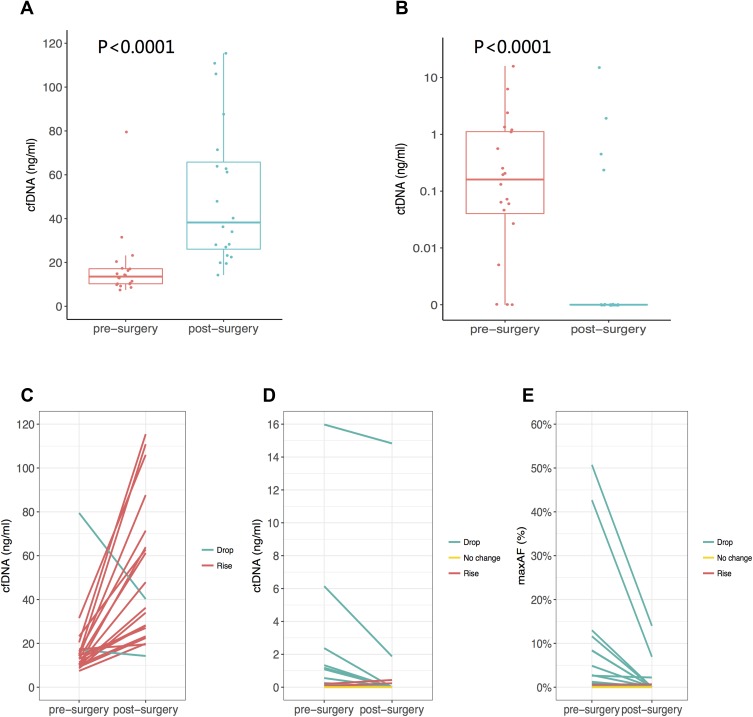
Collectively, we identified 56 mutations spanning 17 genes from the liver metastasis tissue samples of 19 patients, as shown in Figure 1. One patient had no mutation identified from this panel. Their corresponding white blood cells were also sequenced to be used as references to filter out germline mutations. The on-target ratio was 72.5%, with a unique depth of 886x. The most commonly mutated gene was TP53, occurring in 70% (15/20) of patients, followed by APC and KRAS, occurring in 55% (11/20) and 45% (9/20) of patients, respectively. Furthermore, we identified 47 mutations spanning 14 genes from the pre-surgery blood samples, which had a mean cfDNA amount of 17.16±15.76 ng/mL and a mean ctDNA amount of 1.46±3.70 ng/mL, which was calculated by multiplying the cfDNA amount by the maximum allelic fraction (MaxAF). At least one mutation was detected in 17 (85%) patients, and the pre-surgery plasma samples of 3 patients had no mutation detected from this panel. Similar to the liver metastasis tissue samples, the most commonly mutated gene was TP53, occurring in 60% of patients (12/20), followed by KRAS and APC, both occurring in 50% (10/20) of patients. Collectively, tissue and pre-surgical plasma samples achieved a by-variant concordance of 82.14% (46/56) and a by-patient concordance of 70% (14/20). Twenty-four mutations were identified only in the tissue samples, and 1 mutation was identified only in plasma samples. Overall, we identified 10 mutations from the 4 post-surgical plasma samples. All other patients had no mutation detected in their post-surgical samples. For plasma samples, the on-target ratio was 72.2%, with a unique depth of 7803x. The mean cfDNA amount was 48.89 ±29.18 ng/mL. The mean ctDNA amount was 0.87 ±3.31 ng/mL. It is interesting to note that the cfDNA amount was significantly increased (p<0.01) after surgery, but the ctDNA level was significantly reduced (p<0.01) (Figure 2).
Figure 1.
Genomic alterations detected from resected liver metastasis (A), ctDNA from pre-surgery plasma (B) and post-surgery plasma (C). Different mutation types are indicated by different colors. Each column represents a patient and each row represents a gene. Upper and side bars represent the number of mutations a patient had and the number of patients with mutations in a specific gene, respectively.
Figure 2.
The amount of cfDNA (A) and ctDNA (B) obtained from pre-surgery and post-surgery plasma samples. By-patient representations of pre and post-surgery (C) cfDNA (D) ctDNA and (E) maxAF.
Prognostic Value of ctDNA for CLM
We further investigated the impact of the sequence of liver resection on ctDNA. As shown in Table 2, for patients who received two-stage resection, 91% (10/11) had detectable ctDNA prior to resection of the liver tumor; in contrast, only 9% (1/11) of patients had detectable ctDNA after surgery. For patients who received simultaneous resection, 77% (7/9) of patients and 33% (3/9) of patients had detectable ctDNA prior to and after liver resection, respectively.
Table 2.
The Mutated Frequency of ctDNA Between Pre-Surgery and Post-Surgery Phase
| Group | Pre-Surgery Mutation (n) | Post-Surgery Mutation (n) | ||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Simultaneous resection | 7 | 2 | 3 | 6 |
| Two-stage resection | 10 | 1 | 1 | 10 |
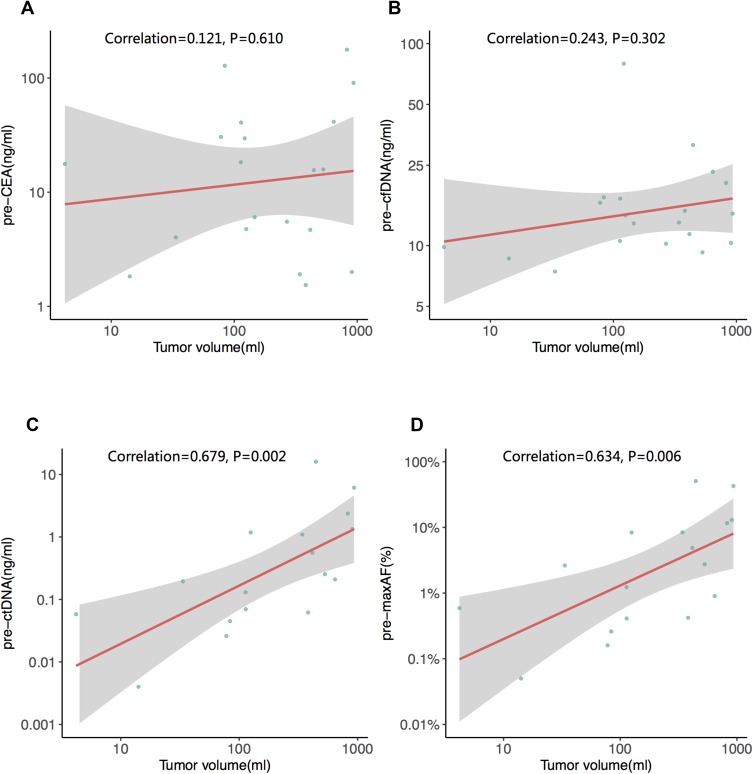
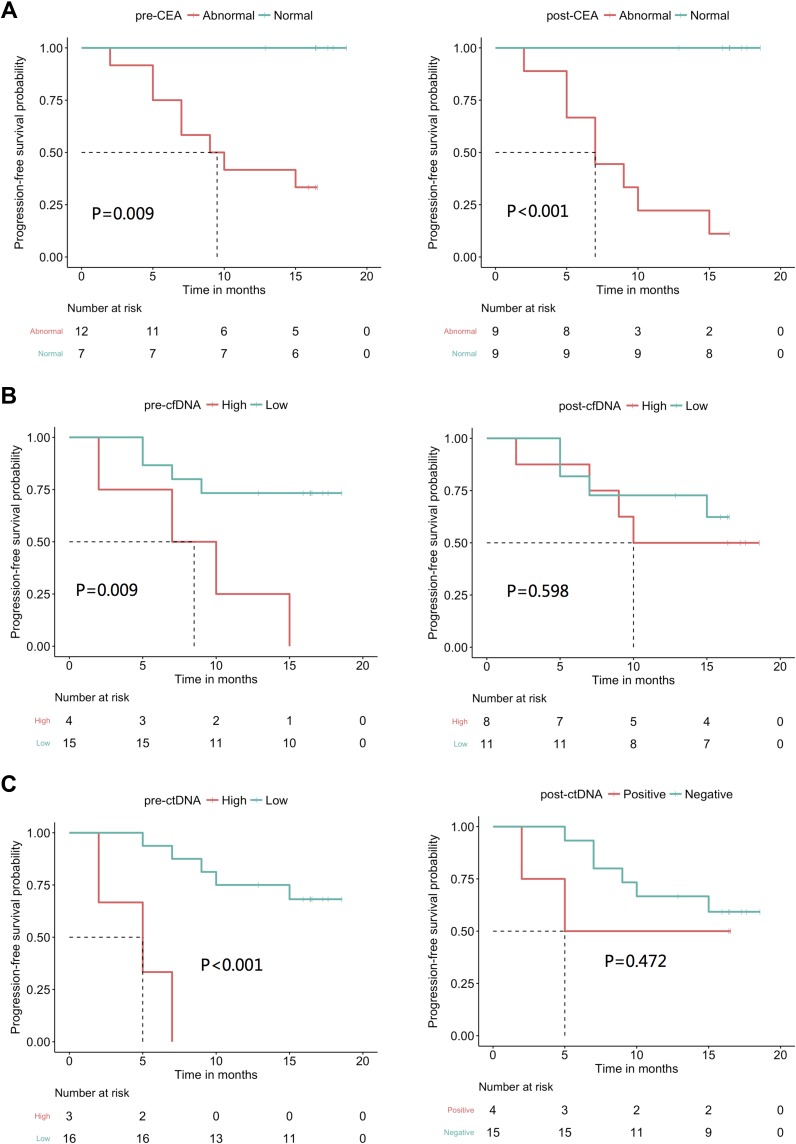
Next, we investigated the association between the tumor volume of liver metastases and CEA, cfDNA and ctDNA. The tumor volume was 326.00 ± 302.19 mL. We determined that only the ctDNA levels were strongly correlated with tumor burden (p=0.002, Figure 3). Furthermore, we also investigated the correlation between PFSand the 3 abovementioned biomarkers (ctDNA, cfDNA and CEA). At the time of analysis (December 15, 2018), the PFS of 19 patients was evaluable; one patient was lost to follow-up. None of the 19 patients were deceased. Eight (42%) of the 19 patients experienced relapse of liver metastases. Our data showed that patients with a normal pre-surgical CEA level experienced longer PFS than patients with an elevated pre-surgical CEA level (p=0.009). Furthermore, dichotomous analyses revealed that patients with low pre-surgical cfDNA (p=0.009) and ctDNA (p<0.001) levels experienced longer PFS (Figure 4). We also evaluated the impact of post-surgical ctDNA on PFS. Only 4 patients had detectable ctDNA after surgery. Although this study did not reveal any difference in PFS between patients with and without post-surgical ctDNA, larger studies are needed to further investigate this finding. Collectively, our results reveal that pre-surgical ctDNA, cfDNA and CEA levels are associated with PFS.
Figure 3.
The correlation between tumor burden and (A) pre-surgery serum CEA (B) pre-surgery cfDNA (C) pre-surgery ctDNA and (D) pre-surgical maximum allelic fraction (MaxAF). The red line is the best fitted line, and the grey area indicates the confidence interval. All the original data are log 10 transformed.
Figure 4.
The predictive value of various markers for PFS. (A) pre and post-surgery serum CEA, (B) pre-and post-surgery cfDNA (C) and pre-and post surgery ctDNA. The dotted lines indicated the median PFS.
Discussion
Hepatic metastasis, often observed in patients with advanced CRC, is a significant clinical problem. For patients with isolated liver metastasis, numerous regional treatments are available, among which surgery remains the gold standard. However, diverse responses in progression-free survival have been observed. CEA is believed to be associated with PFS. The current was a feasibility study aimed at determining the association of CEA, ctDNA and cfDNA with PFS in this patient population. Both ctDNA and cfDNA levels have been suggested to impact the PFS of patients with various cancer types undergoing diverse treatments.19 In the current study, we also compared and contrasted the genomic profiles of pre-surgical and post-surgical plasma samples as well as tissue samples of liver metastases.
Previous studies have reported the concordance of genetic profiles between ctDNA and metastatic tissue. Bachet et al20 demonstrated excellent concordance between RAS status in plasma and tumor tissue from patients with colorectal cancer and liver metastases in a cohort of 412 patients with the κ coefficient of 0.71 (95% CI, 0.64–0.77) and accuracy of 85.2% (95% CI, 81.4% to 88.5%). Consistent with these results, our data indicated high consistency between pre-surgery ctDNA and liver metastasis tissue, achieving 82% by-variant concordance. The observed heterogeneity is due to the nature of a tissue biopsy, which is a snapshot of the tumor. We observed a significant decrease in ctDNA after surgery due to tumor ablation. More interestingly, we observed that patients who underwent simultaneous resection were more likely to have detectable ctDNA after surgery than patients who received two-stage resection (33% vs 9%). What contributes to this difference warrants further investigation.
Previous studies have demonstrated the correlation of ctDNA and tumor burden. The assessment of tumor burden is always critical in the management of metastatic cancer, which might help guide the selection of therapeutic regimens. Radiographical imaging, including computed tomography (CT), magnetic resonance imaging (MRI), and positron-emission tomography coupled with CT (PET-CT), are commonly used to assess tumor burden. However, the sensitivity is limited, especially when the patient is undergoing chemotherapy. Bhangu et al21 indicated that circulating cell-free methylated tumor DNA was correlated with tumor burden, as measured by the CT volume in CLM, and predicted an early response in patients receiving systemic chemotherapy. In the current study, we evaluated the correlation between tumor volume and the total volume of surgically resected and radiofrequency-ablated tumor tissue as the reference to accurately measure tumor burden. Moreover, we studied the association between tumor burden and three tumor markers: CEA, cfDNA, and ctDNA. Our study revealed that elevated CEA and high levels of cfDNA and ctDNA were associated with short PFS. However, only ctDNA was significantly associated with the tumor burden of CLM. We observed a significant increase in the mean cfDNA, from 17.16 ng/mL to 48.89 ng/mL after surgery, potentially due to the relatively short interval between surgery and sampling (within 7 days after surgery) leading to a release of inflammatory factors and necrosis debris of cells under stress.22,23 In contrast, the ctDNA level released from tumor cells was reduced significantly, from 1.47 ng/mL to 0.87 ng/mL, due to ablation and relatively limited chance of finding ctDNA in a large amount of cfDNA. Our finding was consistent with those from previous studies on CRC after primary tumor resection, which indicated that ctDNA was more sensitive than cfDNA in monitoring disease load.24 A recent study in metastatic colorectal cancer demonstrated similar findings, indicating that ctDNA was likely to be more correlated with tumor metastasis and tumor markers than cfDNA.25 Collectively, our results suggest that ctDNA can reflect tumor burden.
This study also had some limitations. First, this prospective study had a limited number of patients. Larger multicenter studies are needed to further validate our findings. The panel used included only 41 genes, which may underestimate the percentage of patients with mutations. Patients with no mutation detected from this panel were regarded as having no ctDNA. Risk adjustment based on known factors that impact progression-free survival was not performed. Due to the relatively short follow-up time, overall survival data were not mature enough to use at the time of data cutoff. Therefore, the prognostic value of ctDNA, cfDNA and CEA could not be evaluated. We are still following all patients for OS. No standard protocol was used to monitor recurrence. Most patients were monitored every 3 months. In conclusion, our data indicate that the mutations obtained from ctDNA are highly consistent with those obtained from resected liver metastases and are associated with tumor burden and PFS, which may be better than serum CEA and cfDNA.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
Dr Jianxing Xiang reports is an employee of Burning Rock Biotech. The authors report no other conflicts of interest in this work.
References
- 1.Chua TC, Saxena A, Chu F, Zhao J, Morris DL. Predictors of cure after hepatic resection of colorectal liver metastases: an analysis of actual 5- and 10-year survivors. J Surg Oncol. 2011;103(8):796–800. doi: 10.1002/jso.21864 [DOI] [PubMed] [Google Scholar]
- 2.Dhir M, Sasson AR. Surgical management of liver metastases from colorectal cancer. J Oncol Pract. 2016;12(1):33–39. doi: 10.1200/JOP.2015.009407 [DOI] [PubMed] [Google Scholar]
- 3.Krell RW, D’Angelica MI. Treatment sequencing for simultaneous colorectal liver metastases. J Surg Oncol. 2019. doi: 10.1002/jso.v119.5 [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–1417. doi: 10.1056/NEJMoa0805019 [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Fan J, Qin X, et al. Chinese guidelines for the diagnosis and comprehensive treatment of colorectal liver metastases (Version 2018). J Cancer Res Clin Oncol. 2019;145(3):725–736. doi: 10.1007/s00432-018-2795-1 [DOI] [PubMed] [Google Scholar]
- 6.Goel G. Molecular characterization and biomarker identification in colorectal cancer: toward realization of the precision medicine dream. Cancer Manag Res. 2018;10:5895–5908. doi: 10.2147/CMAR.S162967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a Phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol. 2011;29(21):2866–2874. doi: 10.1200/JCO.2010.33.4235 [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, Shepherd FA, Hirsh V, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol. 2010;28(5):744–752. doi: 10.1200/JCO.2009.24.3030 [DOI] [PubMed] [Google Scholar]
- 10.Brugger W, Triller N, Blasinska-Morawiec M, et al. Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2011;29(31):4113–4120. doi: 10.1200/JCO.2010.31.8162 [DOI] [PubMed] [Google Scholar]
- 11.Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26(8):1715–1722. doi: 10.1093/annonc/mdv177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Li L, Cohen JD, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol. 2019. doi: 10.1001/jamaoncol.2019.0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68(4):663–671. doi: 10.1136/gutjnl-2017-315852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019. doi: 10.1001/jamaoncol.2019.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra392. doi: 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabriel E, Bagaria SP. Assessing the impact of circulating tumor DNA (ctDNA) in patients with colorectal cancer: separating fact from fiction. Front Oncol. 2018;8:297. doi: 10.3389/fonc.2018.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholer LV, Reinert T, Orntoft MW, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23(18):5437–5445. doi: 10.1158/1078-0432.CCR-17-0510 [DOI] [PubMed] [Google Scholar]
- 18.Toso C, Trotter J, Wei A, et al. Total tumor volume predicts risk of recurrence following liver transplantation in patients with hepatocellular carcinoma. Liver Transpl. 2008;14(8):1107–1115. doi: 10.1002/lt.v14:8 [DOI] [PubMed] [Google Scholar]
- 19.Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst. 2017;109(12). doi: 10.1093/jnci/djx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachet JB, Bouche O, Taieb J, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol. 2018;29(5):1211–1219. doi: 10.1093/annonc/mdy061 [DOI] [PubMed] [Google Scholar]
- 21.Bhangu JS, Beer A, Mittlbock M, et al. Circulating free methylated tumor DNA markers for sensitive assessment of tumor burden and early response monitoring in patients receiving systemic chemotherapy for colorectal cancer liver metastasis. Ann Surg. 2018;268(5):894–902. doi: 10.1097/SLA.0000000000002901 [DOI] [PubMed] [Google Scholar]
- 22.Hummel EM, Hessas E, Muller S, et al. Cell-free DNA release under psychosocial and physical stress conditions. Transl Psychiatry. 2018;8(1):236. doi: 10.1038/s41398-018-0264-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vymetalkova V, Cervena K, Bartu L, Vodicka P. Circulating cell-free DNA and colorectal cancer: a systematic review. Int J Mol Sci. 2018;19(11). doi: 10.3390/ijms19113356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625–634. doi: 10.1136/gutjnl-2014-308859 [DOI] [PubMed] [Google Scholar]
- 25.Osumi H, Shinozaki E, Takeda Y, et al. Clinical relevance of circulating tumor DNA assessed through deep sequencing in patients with metastatic colorectal cancer. Cancer Med. 2019;8(1):408–417. doi: 10.1002/cam4.2019.8.issue-1 [DOI] [PMC free article] [PubMed] [Google Scholar]