Abstract
Focal brain lesions disrupt resting-state functional connectivity, but the underlying structural mechanisms are unclear. Here, we examined the direct and indirect effects of structural disconnections on resting-state functional connectivity in a large sample of sub-acute stroke patients with heterogeneous brain lesions. We estimated the impact of each patient’s lesion on the structural connectome by embedding the lesion in a diffusion MRI streamline tractography atlas constructed using data from healthy individuals. We defined direct disconnections as the loss of direct structural connections between two regions, and indirect disconnections as increases in the shortest structural path length between two regions that lack direct structural connections. We then tested the hypothesis that functional connectivity disruptions would be more severe for disconnected regions than for regions with spared connections. On average, nearly 20% of all region pairs were estimated to be either directly or indirectly disconnected by the lesions in our sample, and extensive disconnections were associated primarily with damage to deep white matter locations. Importantly, both directly and indirectly disconnected region pairs showed more severe functional connectivity disruptions than region pairs with spared direct and indirect connections, respectively, although functional connectivity disruptions tended to be most severe between region pairs that sustained direct structural disconnections. Together, these results emphasize the widespread impacts of focal brain lesions on the structural connectome and show that these impacts are reflected by disruptions of the functional connectome. Further, they indicate that in addition to direct structural disconnections, lesion-induced increases in the structural shortest path lengths between indirectly structurally connected region pairs provide information about the remote functional disruptions caused by focal brain lesions.
Keywords: stroke, lesion, functional connectivity, structural connectivity, structural disconnection, shortest path length
1. Introduction
Focal brain lesions that result from stroke and other neurological disorders produce widespread disruptions of brain function that often involve regions remote from the site of injury (Carrera and Tononi, 2014). The distributed functional consequences of focal brain lesions can be measured non-invasively using resting-state fMRI functional connectivity, a measure of the correlation between ongoing low-frequency blood oxygen-level dependent signal fluctuations in different brain regions (Biswal et al., 1995). Lesion-induced functional connectivity disruptions have been strongly associated with clinical deficits in multiple cognitive and behavioral domains (Baldassarre et al., 2016), but it remains unclear how they relate to the structural impact of the underlying lesion. Clarifying the nature of this relationship is thus an important goal that has the potential to inform the development of new therapeutic approaches that aim to restore normal brain function using techniques such as non-invasive neurostimulation (Carrera and Tononi, 2014; Raffin and Siebner, 2014), and beyond clinical relevance, also represents an important step towards understanding how brain structure shapes brain function.
We recently made progress towards this goal by demonstrating that the severity of commonly observed network-level functional connectivity disruptions observed after stroke, namely reductions of interhemispheric integration within networks and ipsilesional segregation between networks, are more strongly related to lesion-induced structural disconnections of inter-regional white matter pathways than to lesion size, location, or damage to putative critical grey matter regions (Griffis et al., 2019). However, our results only hinted at the specific structural features that provide information about when functional connectivity disruptions are sustained between region pairs. For example, we observed significant, albeit weak-to-moderate, connection-level correspondences between the structural disconnection and functional connectivity patterns that co-varied together across patients, suggesting that a lesion that interrupts the direct structural connection between two regions also tends to disrupt their functional connectivity. Here, we explicitly tested whether region pairs that suffer a direct structural disconnection reliably show more severe disruptions of functional connectivity than region pairs with spared direct structural connections.
Most lesion-induced disruptions of functional connectivity, however, occur between regions that can only be connected by traversing intermediate structural links. This suggests that many functional connectivity disruptions may reflect the up/downstream consequences of direct structural disconnections. Supporting this view, a previous study of eleven stroke patients reported that pontine lesions, which should damage intermediary connections along the polysynaptic cortico-pontine-cerebellar pathway, disrupted functional connectivity between somatomotor cortex and the contralateral cerebellar hemisphere (Lu et al., 2011). Therefore, we also sought to identify a structural feature that provides information about the functional connectivity disruptions sustained by indirectly structurally connected region pairs. The report by Lu et al., (2011) suggests that disruptions of the shortest structural paths between indirectly structurally connected region pairs might represent such a feature. Importantly, shortest structural path lengths (SSPLs), which correspond to the minimum number of direct structural links that must be traversed to establish a structural pathway connecting a region pair, have been previously shown to influence functional connectivity in healthy individuals (Goni et al., 2014).
Here, we tested whether damage to the intermediary structural connections along the shortest structural paths linking indirectly structurally connected region pairs represents a general mechanism of lesion-induced functional connectivity disruptions across the cortex. Specifically, we tested whether indirectly structurally connected regions that exhibit lesion-induced increases in SSPLs show disruptions of functional connectivity attributable to this putative indirect structural disconnection (see Fig. 1). Because a lesion that interrupts the direct structural connection between two regions also increases their SSPL (i.e. from one to greater than one), the SSPL measure allowed a unified treatment of both directly and indirectly structurally connected region pairs (Fig. 1). Accordingly, we also compared the magnitudes of the functional connectivity disruptions associated with direct vs. indirect structural disconnections.
Fig. 1.
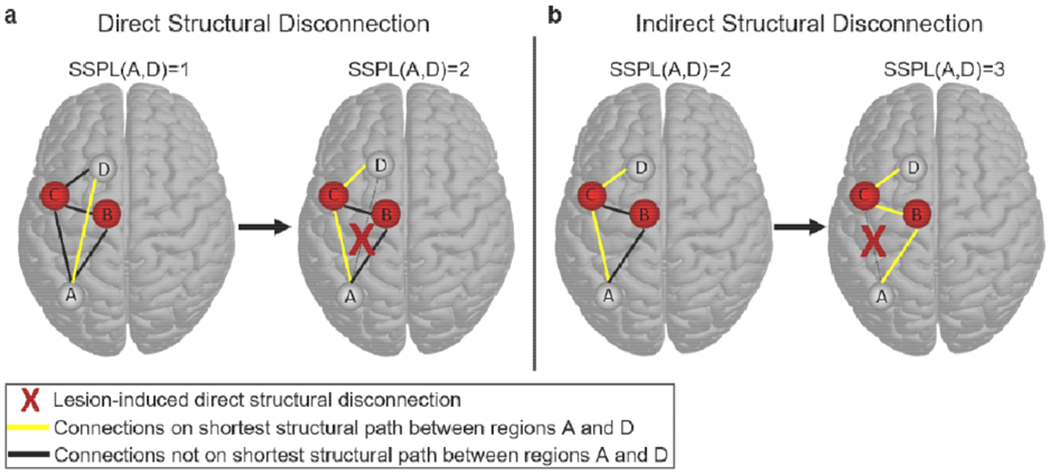
Defining direct and indirect structural disconnections. (a) The brain on the left shows a simple network where regions A and D are directly structurally connected to each other (yellow line), and therefore have an SSPL equal to 1. The brain on the right shows the SSPL (yellow line) between regions A and D after the direct structural connection has been disrupted by a lesion (red X): the SSPL between regions A and D is now 2 because the shortest path passes through region C. This is an example of how direct structural disconnections increase SSPLs between disconnected regions. (b) The brain on the left shows an alternative network configuration where regions A and D are indirectly structurally connected to each other via mutual connections to region C (yellow line), and therefore have an SSPL equal to 2. The brain on the right shows the SSPL (yellow line) between regions A and D after the structural connection between regions A and C has been disrupted by a lesion (red X): the SSPL between regions A and D is now 3 because the shortest path passes through both regions B and C. This is an example of how a direct structural disconnection can increase SSPLs between indirectly structurally connected regions, which we refer to here as an “indirect structural disconnection”.
2. Methods
2.1. Participant information
Written informed consent was obtained prior to enrolling subjects in the study. Study procedures were performed in accordance with the Declaration of Helsinki ethical principles and approved by the Institutional Review Board at Washington University in St. Louis. The complete data collection protocol is described in complete detail elsewhere (Corbetta et al., 2015). Data from 132 first-time stroke patients with clinical evidence of impairment and data from 36 demographically matched healthy controls were considered for inclusion in the analyses. Patients were scanned within the first three weeks of stroke onset (mean time since stroke = 13.09 days, SD = 4.75 days). Patients with previous strokes were not included. After applying quality controls (described in Section 2.7), data from a total of 114 patients and 24 controls were included in the study. Of the included patients, 67% had ischemic strokes, 14% had hemorrhagic strokes, and 19% had other stroke etiologies (e.g. hemorrhagic conversion, vertebral artery dissection, etc.). Additional demographic information is provided in Table 1.
Table 1.
Participant Demographics
| Group | Age (Mean/SD) | Sex | Handedness | Education (Mean/SD) | Lesion Side | Number of Lacunar Infarcts (Mean/SD) | White Matter Disease Severity Score (Mean/SD) |
|---|---|---|---|---|---|---|---|
| Patients (N=114) | 52.4/10.2 | 58 F, 56 M | 106 R, 8 L | 13.3/2.8 | 54 R, 60 L | 1.5/2.6 | 1.14/1.50 |
| Controls (N=24) | 54.5/13.5 | 12 F, 12 M | 23 R, 1 L | 13.5/2.1 | N/A | N/A | N/A |
SD: Standard Deviation, M: Male, F: Female, R: Right, L: Left
Data and code availability statement:
The full set of neuroimaging data (along with behavioral data) are available at http://cnda.wustl.edu/app/template/Login, and specific data and scripts are available upon request to the authors.
2.2. Neuroimaging data collection
Neuroimaging data were collected at the Washington University School of Medicine using a Siemens 3T Tim-Trio scanner with a 12-channel head coil. We obtained sagittal T1-weighted MP-RAGE (TR=1950 msec; TE=2.26 msec, flip angle = 90 degrees; voxel dimensions = 10x1.0x1.0 mm), transverse T2-weighted turbo spin-echo (TR=2500 msec; TE=43 msec; voxel dimensions = 1x1x1), sagittal T2-weighted FLAIR (TR=750 msec; TE=32 msec; voxel dimensions = 1.5x15x1.5 mm) and gradient echo EPI (TR=2000 msec; TE=2 msec; 32 contiguous slices; 4x4 mm in-plane resolution) resting-state functional MRI scans from each subject. Participants were instructed to fixate on a small centrally-located white fixation cross that was presented against a black background on a screen at the back of the magnet bore. An Eyelink 1000 eye-tracking device (SR Research) was used to monitor eye status (i.e. eyes opened/closed) during each functional MRI run. We obtained between six and eight resting-state scans (128 volumes each) from each participant (~30 minutes total).
2.3. Lesion identification
The Analyze software package (Robb and Hanson, 1991) was used to manually segment lesions on structural MRI. Specifically, the T2-weighted and FLAIR scans were co-registered to the T1-weighted scan, and the T1-weighted scan was then linearly registered to atlas space using FSL’s FLIRT tool. The linear transformation was then applied to the co-registered structural scans, and the different structural scans were used in conjunction for lesion segmentation to ensure that lesions were completely segmented. Surrounding vasogenic edema was included in the lesion definition if present. Two board certified neurologists (Maurizio Corbetta and Alexandre Carter) reviewed all segmentations, and segmentations were also reviewed a second time by MC. The final segmentations were converted to binary lesion masks, resampled to have isotropic voxel resolution, and then used to mask out lesioned voxels during non-linear registration of the T1-weighted scan to MNI atlas space via FLS’s FNIRT tool. The resulting non-linear transformation was then applied to the lesion mask to obtain the final MNI-registered lesion segmentation. The group-level lesion overlap map is shown in Figure 2. The number of lacunar infarcts observed on MRI was also recorded for each patient, and the severity of periventricular white matter disease was rated according to the 9-point scale described by Kuller and colleagues (Kuller et al., 2004), as described in a previous publication on this dataset (Corbetta et al., 2015). These data are summarized in Table 1. Full patient-level lesion, lacunar infarct, and white matter disease severity data can be found in Supplementary Table 6 of (Corbetta et al., 2015).
Fig. 2.
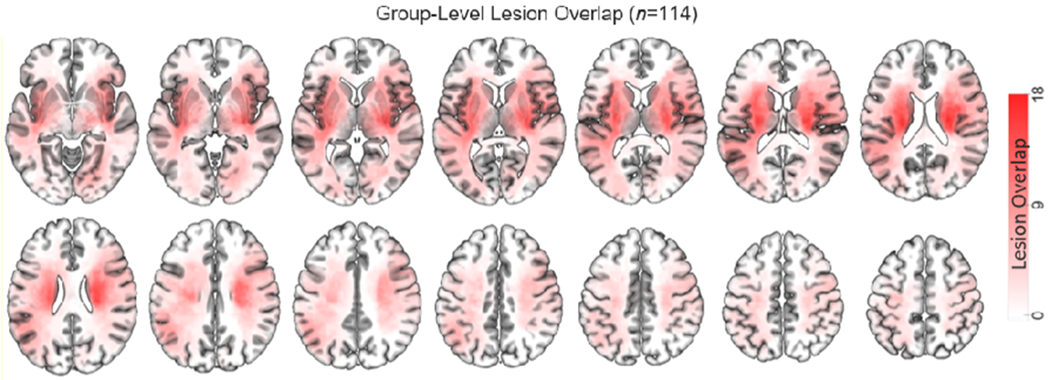
Group-level lesion topography. Color intensities indicate the number of patients with overlapping lesions at each voxel in the brain (max=18).
2.4. Regional parcellation
We parcellated the cortex according to the Gordon333 resting-state functional MRI parcellation (https://sites.wustl.edu/petersenschlaggarlab/resources/). This parcellation contains 333 cortical regions that are each assigned to 1 of 13 large-scale resting-state networks (Gordon et al., 2016). As in previous analyses of this dataset, 9 regions were excluded for having very low numbers of vertices (Griffis et al., 2019; Siegel et al., 2018, 2016). Surface-based functional connectivity analyses were performed using the remaining 324 regional parcels.
A set of 35 sub-cortical and cerebellar regions were also defined to enable a thorough quantification of damage and disconnection. 34 regions corresponding to portions of the cerebellum, thalamus, and basal ganglia were taken from the automatic anatomical labeling (AAL) atlas (Tzourio-Mazoyer et al., 2002). An additional region corresponding to the brainstem was taken from the Harvard-Oxford Subcortical Atlas. The “dilate” command in DSI_studio (http://dsi-studio.labsolver.org/) was used to dilate the cortical regions by 2mm to increase the sensitivity of subsequent region-to-region structural connectivity analyses (Van Den Heuvel et al., 2009; Wilson et al., 2011). This also led to a more liberal definition of cortical damage, since the dilated regions extended slightly beneath the pial surface (Pustina et al., 2017). The volume-based regional parcels are illustrated in Figure 3a (left), and the surface-based cortical parcels are illustrated in Figure 4a.
Fig. 3.
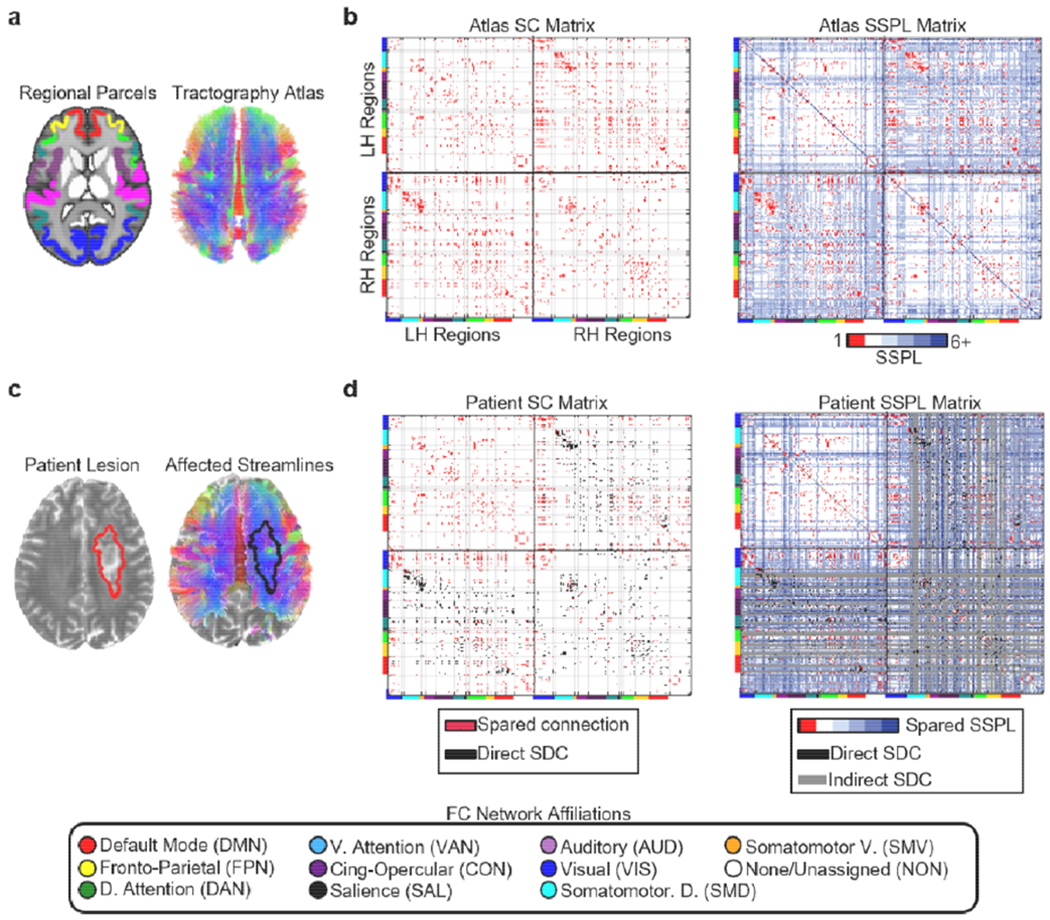
Structural disconnection data. (a) The regional parcels and tractography atlas. (b) Left – Cortical SC matrix derived from the tractography atlas and regional parcels. Right – Cortical SSPL matrix derived from the atlas SC matrix. Atlas SSPL values are integers ranging from 1 to 6. Directly structurally connected regions have SSPLs equal to 1. Cortical regions are organized by hemisphere within each matrix, and the horizontal and vertical black lines on each matrix divide the matrix into quadrants corresponding to intra-LH connections (upper left quadrant), interhemispheric connections (bottom left and upper right quadrants), and intra-RH connections (lower right quadrant). Cortical regions are further organized according to their a priori resting-state network assignments from cortical parcellation as indicated by the colored bars along the edges of each matrix and the legend at the bottom of the figure. The light grey lines extending from the tick marks between these colored bars form “boxes” delineating portions of each matrix corresponding to different sets of within-network (i.e. on-diagonal “boxes” within each quadrant) and between-network (i.e. off-diagonal “boxes” within each quadrant) connections. (c) Lesion segmentation (left) and affected streamlines (right) for a single patient overlaid on that patient’s T2-weighted scan. (d) Left -- spared cortical direct structural connections (red) and direct cortical structural disconnections (black – direct SDC) for the patient shown in (c). Right – spared cortical SSPLs (colored), indirect cortical structural disconnections (gray – indirect SDC), and direct cortical structural disconnections (black– direct SDC) are shown for the same patient shown in (c). Patient SSPLs values are integers ranging from 1 to 6+ (max=infinity, where infinity indicates that no shortest paths exist). The upper left quadrants in the matrices shown in (d) correspond to connections within the contralesional hemisphere, which were spared by the right hemisphere lesion. Note: SSPL calculations also considered cortico-subcortical connections, but these connections are not shown in the matrices above. All brain images are shown in neurological convention (i.e. the left side of the brain is on the left).
Fig. 4.
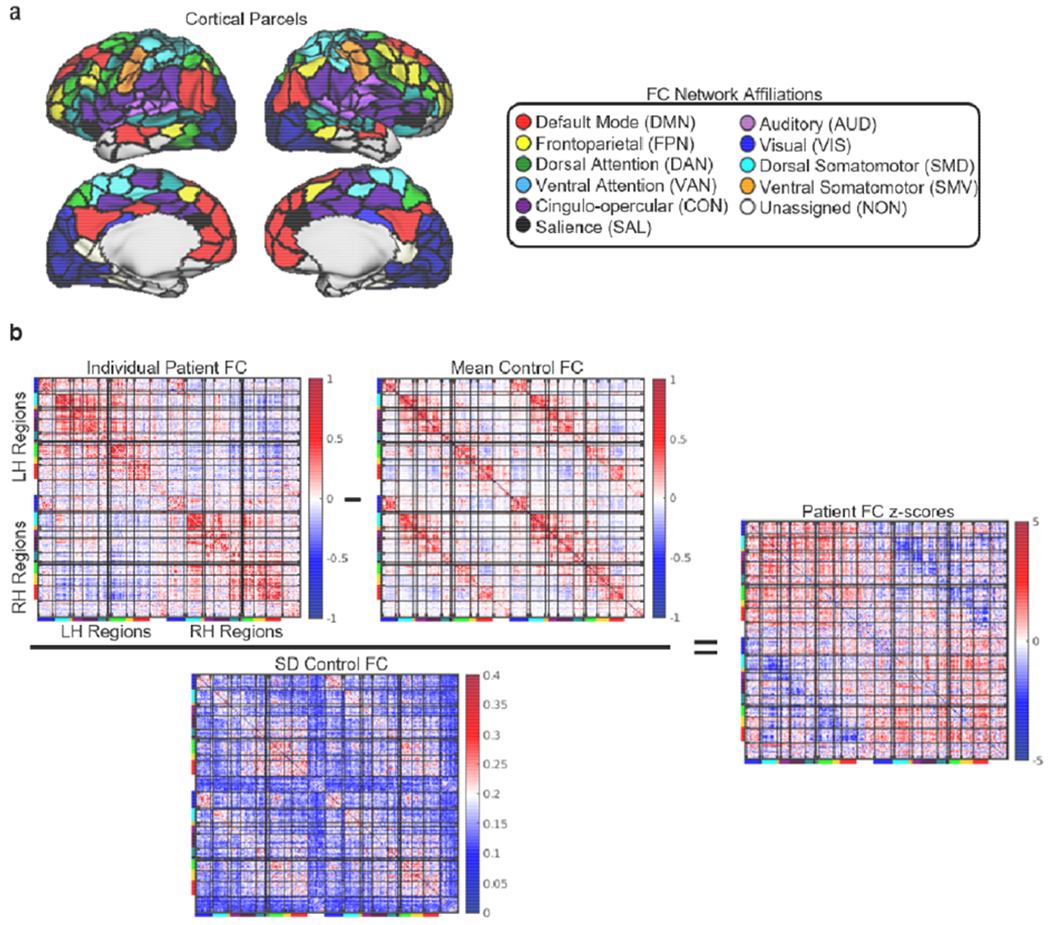
Functional connectivity disruptions. (a) Cortical regions are shown on the cortical surface and are color-coded by network assignment. (b) The transformation of a raw functional connectivity matrix to a functional connectivity z-score matrix is illustrated for the same patient shown in Fig. 3c–d. The bars on matrix axes correspond to the cortical functional connectivity network assignments shown in the legend at the top of the figure.
2.5. Structural connectome atlas
We created a structural connectome atlas using a publicly available diffusion MRI streamline tractography atlas (http://brain.labsolver.org/diffusion-mri-templates/tractography) based on high angular resolution diffusion MRI data collected from 842 healthy Human Connectome Project participants (Yeh et al., 2018) (Fig. 3a, right). These data were accessed under the WU-Minn HCP open access data use term. The tractography atlas contained expert-vetted streamline trajectories in MNI space, and each streamline was assigned to 1 of 66 macroscale white matter pathways (e.g. arcuate fasciculus, cortico-spinal tract, corpus callosum, etc.).
MATLAB scripts implementing functions from the DSI_studio software package was used to define the normative pairwise structural connectome based on the tractography atlas (Fig. 3b, left). We first combined the various streamline bundles from the tractography atlas (e.g. short-range U-fibers, callosal projections, etc.) into a single .trk file that included all streamlines associated with each of the fiber pathways in the atlas. We then extracted all pairwise structural connections, defined as streamlines that bilaterally terminated (i.e. began and ended) within any pair of the 359 volume-based regions, to create a 359x359 structural connectivity adjacency matrix where each cell quantified the number of streamlines connecting a region pair. The close proximity of ventral visual and dorsal cerebellar regions led to a few dorsal cerebellar streamlines terminating within the dilated ventral visual regions, and so any connections between the visual cortex and cerebellum were removed.
A breadth-first search algorithm, implemented in the breadthdist function provided with the Brain Connectivity Toolbox (Rubinov and Sporns, 2010) for MATLAB was then applied to the binarized structural connectivity matrix to obtain a matrix of pairwise SSPLs (Fig. 3b, right), where each cell quantified the minimum number of structural connections that needed to be traversed to establish a structural path between each region pair (Goni et al., 2014). A total of 15 cortical regions had undefined SSPLs, and so they were excluded from further analyses.
2.6. Structural connectivity measures
MATLAB scripts implementing functions from the DSI_studio software package were used to estimate the expected direct structural disconnections for each patient by intersecting the MNI-registered lesion mask with streamline tractography atlas (Fig. 3c). For each patient, all streamlines that intersected the lesion were extracted into a 359×359 direct structural disconnection matrix where each cell quantified the number of streamlines between each region pair that intersected the lesion. The direct structural disconnection matrix was then subtracted from the atlas-derived structural connectivity matrix to obtain a spared structural connection matrix where each cell quantified the number of streamlines between each region pair that were spared by the lesion. The direct structural disconnection and spared structural connection matrices were then normalized by dividing each cell by the corresponding number of streamlines in the atlas structural connectivity matrix so that each cell in the normalized spared structural connection and direct structural disconnection matrices quantified the proportion of streamlines between each region pair that was spared vs. directly disconnected by the lesion, respectively. Each matrix was then binarized by setting all non-zero values to 1. To ensure that our results were not dependent on the binarization threshold, we also performed our main analyses using a binarization threshold of 50% disconnection relative to the atlas (Fig. S4b) Example direct structural disconnections and spared structural connections are shown for an example patient in Figure 3d.
For each patient, a patient-specific SSPL matrix was created by applying the breadthdist function to that patient’s spared connection matrix. The atlas SSPL matrix was then subtracted from the patient-specific SSPL matrix to obtain an SSPL difference matrix where cells corresponding to region pairs with preserved SSPLs had values equal to 0 and cells corresponding to region pairs with increased SSPLs had values greater than 0. The SSPL difference matrix was then binarized so that all cells corresponding to region pairs with increased SSPLs had values of 1. Values for all cells that corresponded to directly structurally connected region pairs were set to 0 so that 1-valued cells in the resulting matrix corresponded to structurally un-connected region pairs that sustained indirect disconnections. A spared indirect connection matrix was then created where all cells that corresponded to structurally un-connected region pairs with spared SSPLs had values equal to 1. All subsequent analyses involving these matrices utilized only the above-diagonal elements (i.e. upper triangles) of each matrix. Example SSPLs and indirect structural disconnections are shown for an example patient in Figure 3d.
2.7. fMRI data processing
The following steps were used to preprocess the fMRI data: slice-timing correction using sinc interpolation, correction of inter-slice intensity differences resulting from interleaved acquisition, normalization of whole-brain intensity values to a mode of 1000, correction for distortion via synthetic field map estimation, and within- and between- scan spatial re-alignment. The fMRI data were re-aligned, co-registered to the corresponding structural scans, registered to atlas space using both linear and nonlinear registration procedures, and resampled to 3mm cubic voxel resolution.
Additional preprocessing was employed to remove contributions from non-neural sources of signal variance. The six head motion parameters obtained from rigid body correction were regressed from the data along with the global whole-brain signal and signals from CSF and white matter tissue compartments that were extracted using FreeSurfer tissue segmentations (Dale et al., 1999; Fischl et al., 1999). Band-pass filtering (0.009 < f < 0.08 Hz) was applied to retain low-frequency fluctuations. Frame censoring was applied using a 0.5mm framewise displacement threshold, and frames that succeeded high-motion frames were also censored to reduce artifacts related to head motion (Power et al., 2014). For each run, the first four frames were discarded to allow for the scanner to reach steady-state magnetization.
Cortical surfaces were generated and fMRI data were further processed according to previously published procedures (Glasser et al., 2013) with some modifications to improve processing of lesioned brains (Siegel et al., 2017). Anatomical surfaces were obtained from the T1-weighted scans using Freesurfer (Dale et al., 1999; Fischl et al., 1999) and were visually inspected to ensure quality. For patients with failed registrations/segmentations, the linearly atlas-registered T1-weighted scans were modified by replacing lesioned voxels with normal tissue voxels from the structural atlas, and the inverse transformation was applied to return the modified volume to native space. The registration/segmentation procedures were then re-run, and the modified voxels were masked out after running the procedures as described in a previous publication on approaches to studying functional connectivity after stroke (Siegel et al., 2017).
Each hemisphere was resampled to 164,000 vertices, and the two hemispheres were registered to each other (Van Essen et al., 2001) before down-sampling to 32,000 vertices per hemisphere. Ribbon-constrained sampling (implemented in Connectome Workbench) was utilized to sample each subject’s fMRI data on the corresponding individual surface. Any voxels with coefficients of variation > 0.5 standard deviations above the mean within a 5 mm sigma Gaussian neighborhood were not included in the mapping from volume to surface (Glasser et al., 2013).
As in our previous work (Griffis et al., 2019), subjects were included if they had at least 180 usable frames of functional MRI data remaining after motion scrubbing. Data from a total of 18 patients and 9 controls were excluded due to failure to meet this criterion, and data from the remaining 114 patients and 24 controls were included in subsequent analyses.
2.8. Functional connectivity
Pairwise functional connectivity matrices were constructed by correlating the mean (i.e. across all within-region vertices) post-processed BOLD timeseries from each surface region with that of each other region in the brain. Fisher z-transformation was then applied to the resulting linear correlation values to obtain 324x324 pairwise functional connectivity matrices for each patient and control. Any vertices that fell within the boundaries of the lesion were excluded from functional connectivity estimation. Any regions with less than 60 vertices remaining after accounting for the lesion were completely excluded by setting the corresponding values to NaN as in previous work (Siegel et al., 2016, 2018; Griffis et al., 2019). All subsequent analyses were performed using only the above-diagonal elements (i.e. upper triangle) of the functional connectivity matrices.
Patient functional connectivity matrices were converted to z-score matrices by subtracting the control mean functional connectivity matrix and dividing the result by the control standard deviation functional connectivity matrix (Fig. 4b). Each cell in the resulting matrices therefore quantified the distances (in standard deviation units) between the observed patient functional connectivity values and the expected values from the control group, with values closer to zero indicating more “normal” functional connectivity and with values farther from zero indicating more “abnormal” functional connectivity. This step was performed to account for the fact that the “normal” magnitudes of functional connectivity values vary considerably across edges in the functional connectome (e.g. see the healthy control matrix in Fig.4b), and to account for the fact that the sets of edges that are directly/indirectly disconnected vary across patients due to the fact that lesion sizes and locations are heterogeneous (Fig. 2).
2.9. Statistical Analyses
We first assessed the relationship between the extents of direct vs. indirect disconnections sustained across patients. This was accomplished by linearly regressing the total number of region pairs that sustained direct disconnections on the number of region pairs that sustained indirect disconnections. Linear regressions were also performed to assess the relationships between direct/indirect disconnection extents and lesion volume.
We also assessed whether particular lesion foci were associated with more extensive disconnections across patients. Since the extent of direct disconnections was strongly related to the extent of indirect disconnections (see Section 3.2, Fig.5c), we defined a combined total disconnection extent measure as the total number of direct and indirect disconnections estimated to be caused by each patient’s lesion. We then assigned this total disconnection extent measure as the response variable in a multivariate partial least squares regression (PLSR) model (Wold et al., 2001) where the predictors were vectorized lesion maps that were stacked on top of each other to form a patient-by-voxel lesion predictor matrix. To adjust for the effects of lesion volume, we employed the direct total lesion volume control (DTLVC) technique, which normalizes each patient’s lesion map to have a unit norm of 1 by dividing the values in each map by the square root of the lesion volume (Zhang et al., 2014). Because PLSR performs a dual decomposition of the predictor and response variables and fits the model using a pre-specified number of predictor components, the number of predictor components is a hyper-parameter of the model that must be determined in a principled way (Abdi et al., 2010a). Here, as in our previous publication employing the PLSR approach (Griffis et al., 2019), we accomplished this by using a leave-one-out (i.e., jack-knife) procedure to identify the optimal number of predictor components and minimize overfitting. This procedure consisted of sequentially adding predictor components until doing so increased the sum of squared prediction errors for the held-out cases, as this is indicative of model overfitting (Abdi, 2010a). After determining the optimal number of predictor components using the leave-one-out procedure, which in this case was determined to be 5, we then fit a final PLSR model to the full dataset using the optimal number of predictor components (Abdi, 2010a; Krishnan et al., 2011). To identify lesion foci that significantly predicted total disconnection extent, we used bootstrap resampling (1000 iterations) to estimate family-wise error (FWE) corrected 95% confidence intervals on the final model regression weights for each voxel in the predictor matrix (Krishnan et al., 2011) according to the bias-corrected and accelerated method (Efron and Tibshirani, 1986). To characterize lesion foci that were most strongly related to lesion volume, this same approach (without the dTLVC procedure) was repeated using lesion volume as the response variable (Fig. S1). A total of 92 patients who suffered at least one direct cortico-cortical structure disconnection were included in these analyses, and analyses were restricted to voxels that were damaged in at least 3 patients. A more detailed description of the PLSR lesion-mapping approach can be found in the Methods section of our previous publication (Griffis et al., 2019), and more detailed descriptions of PLSR within the context of neuroimaging can be found in previous reviews (Krishnan et al., 2011).
Fig. 5.
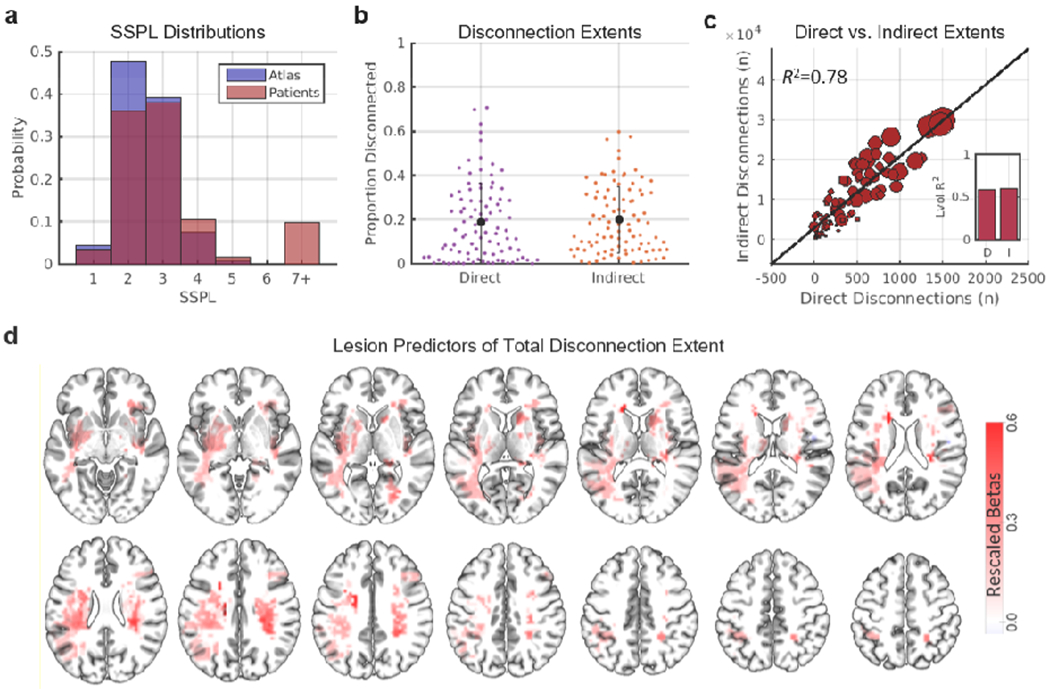
Focal brain lesions disrupt the structural connectome. (a) The red histogram shows the distribution of SSPLs in the atlas-derived structural connectome. The blue histogram shows the distribution of SSPLs in patients. Short SSPLs are decreased and long SSPLs are increased in patients relative to the atlas-derived structural connectome. (b) Distributions and means/standard deviations are shown for the proportion of disconnected direct and indirect cortico-cortical structural connections in the subset of patients that sustained at least one direct cortico-cortical disconnection (n=92). (c) The scatterplot shows the relationship between the total number of direct disconnections (x-axis) and the total number of indirect disconnections (y-axis) in patients. Dots correspond to individual patients, and dot sizes are proportional to lesion volumes. The inset plot shows the amount of variance in direct (D) and indirect (I) disconnection extents explained by lesion volume. (d) Voxels where damage significantly (FWEp<0.05) predicted the total disconnection extent in a multivariate PLSR lesion-mapping analysis with direct total lesion volume control (dTLVC). Color intensities correspond to re-scaled (proportional to the maximum value) regression weight magnitudes for voxels that predicted greater total disconnection extents. See also Fig. S1.
Prior to performing analyses relating patient functional connectivity disruptions to direct and indirect structural disconnections, we aimed to replicate the previously reported dependence of normal functional connectivity on SSPLs using data from the control group (Fig. S2). We first assigned each functional connection in the brain to one of four categories based on its resting-state network (i.e. within-network vs. between-network) and hemispheric (i.e. interhemispheric vs. intrahemispheric) connection types. Within-network connections therefore corresponded to connections between parcels affiliated with the same resting-state network, while between-network connections corresponded to connections between regions affiliated with different resting-state networks. Similarly, intrahemispheric connections corresponded to connections between regions in the same hemisphere, while interhemispheric connections corresponded to connections between regions in different hemispheres. For each control participant, we extracted the mean functional connectivity values for region pairs with SSPLs equal to 1, 2, 3, and 4+ within each connection type category. For each connection type category, we then performed one-way repeated measures ANOVAs with a factor of SSPL to determine whether mean functional connectivity for that connection type category varied systematically across SSPLs in the control group (Fig S1a). Greenhouse-Geisser correction was applied to the degrees of freedom to account for violations of the sphericity assumption (Abdi, 2010b). Because functional connectivity has been shown to depend partially on spatial distance (Goni et al., 2014), these analyses were also repeated after regressing out the effects of log-transformed inter-regional Euclidean distances (computed based on the centroid coordinates for each region pair) on inter-regional functional connectivity strengths (Fig. S2b). The log transformation was applied because this increased the linearity of the relationship between Euclidean distances and mean functional connectivity in the control group (transformed: r=−0.45, untransformed: r=−0.37).
Finally, for each patient, the mean functional connectivity z-scores were separately computed for sets of regions with spared direct structural connections, direct structural disconnections, spared indirect connections, and indirect structural connections with positive vs. negative signs in the control mean functional connectivity matrix. The resulting values were then entered into a three-way repeated measures ANOVA with factors of normative functional connectivity sign, connection type, and connection status. Greenhouse-Geisser correction was again applied to the degrees of freedom to account for violations of the sphericity assumption. Dependent samples t-tests were used to follow up the significant three-way interaction effect. Bonferroni-Holm correction was used to control the family-wise error rate at 0.05 (Aickin and Gensler, 1996). These analyses were also performed after summarizing patient-level functional connectivity z-scores by different network (i.e. within-network and between-network) and hemispheric (i.e. interhemispheric and intrahemispheric) connection types (Fig. S3).
Additional analyses were performed to determine the influence of arbitrary parameter choices on our results (Fig. S4). To ensure that our results were not dependent on the inclusion of partially damaged parcels in the main analyses, we repeated the analyses while excluding all damaged parcels (Fig. S4a). To ensure that our results were not dependent on the threshold for determining direct structural disconnections, we repeated the analyses using a 50% threshold for determining direct structural disconnections (Fig. S4b). To ensure that our results were not dependent on the inclusion of patients with severe hemodynamic lags, we computed voxel-wise lags with respect to the global grey matter signal (Siegel et al., 2015), and we repeated our analyses after excluding patients with mean interhemispheric hemodynamic lag differences greater than 2 standard deviations from the control mean (Fig. S4c). Finally, to ensure that our results were not dependent on the use of healthy controls as the reference group for computing functional connectivity z-scores (i.e., that they did not reflect a general consequence of stroke), we repeated the analyses using a subset of 22 patients who did not sustain any direct cortico-cortical disconnections as the reference group (Fig. S4d). These additional analyses all produced highly similar results to the main analyses reported in the text, indicating that our results did not depend on these analysis choices.
3. Results
3.1. Lesions have widespread impacts on direct and indirect structural connections
Lesion-induced direct structural disconnections should lead to increased SSPLs relative to the normative values derived from the HCP tractography atlas (Fig. 1b), and accordingly, patients showed reductions in short (i.e. < 3) SSPLs and increases in long (i.e. > 4) SSPLs relative to the atlas values (Fig. 5a). To characterize the extents of direct and indirect structural disconnections caused by the lesions in our sample, we measured the proportions of directly and indirectly structurally connected region pairs that sustained direct and indirect structural disconnections, respectively. On average across the subset of patients that sustained at least one direct cortico-cortical structural disconnection (n=92), 19.03% of all region pairs with direct cortico-cortical structural connections were estimated to sustain direct structural disconnections and 20.0% of all region pairs with indirect cortico-cortical structural connections were estimated to sustain indirect structural disconnections (Fig. 5b). These observations indicate that focal brain lesions often have widespread effects on the direct and indirect structural pathways in the brain.
Across patients, the total number of indirect structural disconnections was strongly related to the total number of direct structural disconnections (R2=0.78, p<0.001), indicating that patients with more extensive direct structural disconnections also suffered more extensive indirect structural disconnections (Fig. 5c). Consistent with the expectation that larger lesions would tend to produce more widespread effects on the structural connectome, lesion volume accounted for significant amounts of variance in the extent of both direct (R2=0.59, p<0.001) and indirect (R2=0.61, p<0.001) disconnections (Fig. 5c – inset plot). However, as indicated by the sizeable portions of unexplained variance in direct and indirect disconnection extents (~40%), disconnection extents were only partly determined by lesion size. Accordingly, lesion foci that were identified as significant predictors of total (i.e. direct plus indirect) disconnection extent by the multivariate PLSR lesion-mapping analysis were primarily located within the frontal, temporal, and parietal deep white matter (Fig. 5d – red voxels).
3.2. Direct and indirect structural disconnections disrupt functional connectivity
Previous work has shown that normal functional connectivity strengths vary as a function of SSPLs (Goni et al., 2014). Thus, prior to evaluating the effects of direct and indirect structural disconnections on functional connectivity in the patient group, we replicated the previously reported relationship between functional connectivity strengths and SSPLs using the data from our control group. As shown in Figure S2, average functional connectivity strengths varied reliably as a function of SSPLs for both interhemispheric and intrahemispheric functional connections that were both within and between resting-state networks, and this effect was independent of inter-regional distances.
Next, we proceeded to assess and compare the relative effects of direct and indirect structural disconnections on functional connectivity in the patient group. We performed a three-way repeated-measures analysis of variance (ANOVA) to compare mean functional connectivity z-scores among sets of regions with different structural connection types (i.e. direct vs. indirect), structural connection statuses (i.e. spared vs. disconnected), and normative functional connectivity signs as indicated by the mean control functional connectivity matrix (i.e. positive vs. negative; see Fig. 4b; Fig. 6a,c) in the patient group. Normative functional connectivity signs were included as a factor because prior work has shown that lesion-induced disruptions of positive and negative functional connections tend to be in opposite directions (Baldassarre et al., 2014; Griffis et al., 2019; Lim et al., 2014; Siegel et al., 2016), and this allowed us to avoid mixing effects that we expected a priori to differ in directionality. Within-network functional connections overwhelmingly had positive signs, while the signs of between-network functional connections varied depending on the specific networks in question (Fig. 6a,c).
Fig. 6.
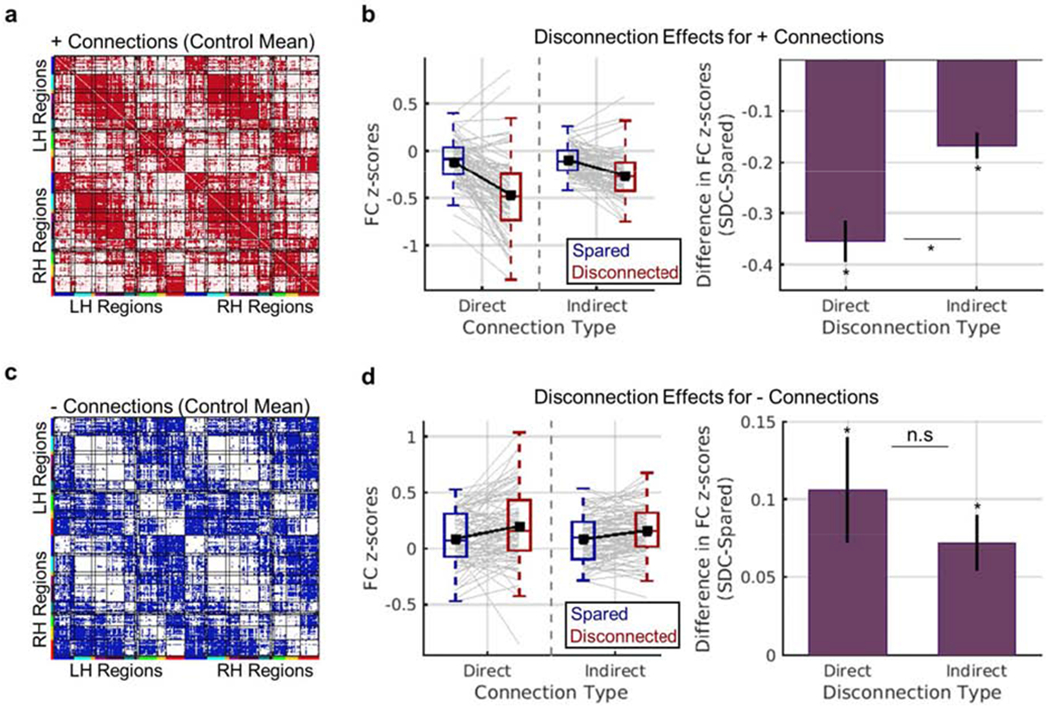
Effects of direct and indirect disconnection on functional connectivity. (a-b) Results for positive functional connections. (a) The matrix shows positive functional connections from the mean control functional connectivity matrix in red. The analyses shown in (b) were restricted to these connections. (b) Left – Distributions of mean patient-level functional connectivity z-scores (y-axis) for each connection status (blue vs. red boxplots) and connection type (x-axis). Grey line plots correspond to individual patient observations, and black line plots show group-level means. Right – Data are summarized to show group-level mean (+/− SEM) differences in functional connectivity z-scores (y-axis) between regions with disconnected vs. spared direct and indirect connections (x-axis). (c-d) Results for negative functional connections. (c) The matrix shows negative functional connections from the mean control functional connectivity matrix in blue. The analyses shown in (d) were restricted to these connections. (d) Same as (b), but for negative functional connections. *dependent samples t-test FWEp<0.004. See also Figs. S3–S4.
The three-way ANOVA, which included 84 patients with valid data for all factor combinations, revealed a significant three-way interaction of connection status, connection type, and normative FC sign (Greenhouse-Geisser corrected F.34,27.54=28.09, p<0.001). We therefore performed follow-up dependent samples t-tests with Bonferroni-Holm correction to further interrogate the effects of interest while controlling the family-wise error rate at 0.05 (Aickin and Gensler, 1996). For positive functional connections (Fig. 6a), mean functional connectivity z-scores were significantly more negative for regions that sustained structural disconnections than for regions with spared structural connections (Fig. 6b). While this effect was observed for both direct and indirect structural disconnection types, the effect was greater for direct structural disconnections than for indirect structural disconnections (Fig. 6b, right plot). For negative functional connections (Fig. 6c), mean functional connectivity z-scores were significantly more positive for regions that sustained structural disconnections than for regions with spared sustained connections (Fig. 6d). While this effect was again observed for both direct and indirect structural disconnection types, the magnitudes of direct and indirect structural disconnection effects on mean functional connectivity z-scores did not significantly differ for negative functional connections (Fig. 6d, right plot). These results support the conclusion that lesion-induced structural disconnections of intermediary structural connections represent a general mechanism underlying lesion-induced functional connectivity disruptions between indirectly structurally connected region pairs across the brain.
Consistent effects of direct and indirect structural disconnections on functional connectivity were also observed when data were further summarized by network (within vs. between) and hemispheric (interhemispheric vs. intrahemispheric) connection types (Fig. S3). Supplemental analyses further indicated that the observed results could not be attributed to regional GM damage, vascular/hemodynamic abnormalities as reflected by large interhemispheric hemodynamic lag differences (Siegel et al., 2015, 2017), or the use of healthy controls as a reference population for defining “normal” functional connectivity (Fig. S4).
4. Discussion
It has long been recognized that focal brain lesions have distributed functional consequences (Carrera and Tononi, 2014). Over the past decade, a number of studies have documented lesion-induced disruptions of resting-state functional connectivity (Eldaief et al., 2016; Gratton et al., 2012; Nomura et al., 2010; Ovadia-Caro et al., 2013; Yuan et al., 2017) and linked them to behavioral deficits in clinical populations (Carter et al., 2010; He et al., 2007; Park et al., 2011; Siegel et al., 2016; Tang et al., 2016). Here, we aimed to clarify how these widespread disruptions of functional connectivity relate to the effects of the lesion on the underlying structural connectome.
Our results show that focal brain lesions have wide-reaching effects on both direct and indirect structural connections, such that roughly one-fifth of all region pairs in the brain were estimated to have suffered either direct or indirect structural connections on average in our patient sample (Fig. 5a–c). The results of our multivariate lesion-mapping analysis further indicate that extensive lesion-induced disconnections are associated with damage to portions of the deep white matter in the frontal, temporal, and parietal lobes (Fig. 5d), including regions that have previously been identified as putative “white matter bottlenecks” where multiple fiber pathways converge in the frontal and temporal deep white matter (Corbetta et al., 2015; Griffis et al., 2017a, 2017b; Mirman et al., 2015a, 2015b; Turken and Dronkers, 2011). Importantly, our results show that these lesion-induced changes in the structural connectome are biologically relevant, as they are reflected by disruptions of resting-state functional connectivity between both directly and indirectly disconnected regions. Altogether, the current results indicate that lesion-induced structural disconnections exert a combination of direct and indirect effects on the structural connectome that disrupt ongoing inter-regional signaling and manifest as disruptions of resting-state functional connectivity.
4.1. Functional connectivity depends on direct and indirect structural connections
Previous work has shown that normal functional connectivity depends on both the direct and indirect structural connections between brain regions (Adachi et al., 2011; Goni et al., 2014; Honey and Sporns, 2009; Mišić et al., 2016). In the healthy brain, it has been observed that functional connectivity tends to be strongest between regions with direct connections (i.e. SSPLs=1) and weakest between regions with long SSPLs (Goni et al., 2014). Importantly, we were able to replicate previously reported SSPL effects on functional connectivity in our healthy control data (Fig. S2), providing additional support for the conclusion that structural connections, both direct and indirect, shape normal functional connectivity patterns. Nonetheless, other factors such as the network embedding of structural paths likely also play important roles in determining normative functional connectivity (Goni et al., 2014; Osmanlioğlu et al., 2019).
We note that common graph theoretical measures that summarize the efficiency of information transfer within local networks (i.e. local efficiency) and at the level of the whole network (i.e. global efficiency) are also based on information about the shortest path lengths between nodes in the network (Rubinov and Sporns, 2010). However, while measures such as local and global efficiency summarize the topological features of local and global networks, respectively, shortest path lengths provide information about the topological distances between each pair of nodes in the network. Here, we were interested in identifying pairs of nodes without direct structural connections that showed (presumed) increases in topological distances due to a lesion’s expected impacts on the structural connectome, which is why we utilized SSPLs in our analyses.
4.2. Disruptions of the structural connectome are reflected in the functional connectome
Despite the clear dependence of functional connectivity on structural connectivity in the healthy brain, most prior empirical work relating lesion-induced disruptions of functional connectivity to the underlying structural damage has focused on the effects of damage to grey matter regions with particular functional network affiliations (Eldaief et al., 2016; Nomura et al., 2010; Ovadia-Caro et al., 2013) or hub-like functional connectivity profiles (Gratton et al., 2012). In contrast, we recently reported that information about direct structural disconnections is superior to information about lesion size, lesion location, or damage to putative hub regions for explaining core network-level functional connectivity disruptions associated with stroke (Griffis et al., 2019). The results reported here extend our previous work by showing that direct structural connections reliably disrupt resting-state functional connectivity between the disconnected region pairs (Fig. 6, see “direct”). Importantly, they also extend previous work on the up/downstream consequences of focal brain lesions (Lu et al., 2011) by showing that damage to intermediary structural connections along the shortest path linking indirectly structurally connected region pairs represents a general mechanism underlying lesion-induced functional connectivity disruptions across the cortex (Fig. 6, see “indirect”).
Direct structural disconnections tended to produce more severe functional connectivity disruptions than indirect structural disconnections for positive functional connections (Fig. 6a). The results of our supplemental analyses suggest that this effect was likely driven primarily by interhemispheric disconnections (Fig. S3b), which are known to rely heavily on direct anatomical projections (Johnston et al., 2008; Roland et al., 2017; Shen et al., 2015). This suggests that the relative magnitudes of functional connectivity disruptions that result from direct and indirect disconnections likely depend on the nature of the affected connections. Determining what factors influence the relative vulnerabilities of different types of functional connections to direct and indirect disconnections is therefore an important goal for future studies on this topic.
Recent studies have shown that lesion-induced direct structural disconnections are associated with neurodegenerative changes in directly disconnected cortical areas (Cheng et al., 2019; Duering et al., 2015; Foulon et al., 2018; Kuceyeski et al., 2014). For example, cortical thinning has been reported to occur in both directly disconnected areas (Duering et al., 2015) and in their contralateral homologues (Cheng et al., 2019) during the first year of recovery. Speculatively, these regional structural changes might be long-term consequences of disconnection-induced functional disruptions such as those reported here, and this should be addressed by future work. Given that we also observed clear effects of indirect disconnection on functional connectivity, future studies on how structural disconnections impact local regional structure might also incorporate measures of indirect structural disconnection to assess the relative contributions of direct and indirect structural disconnections to changes in local regional structure associated with focal brain lesions.
4.3. Potential clinical implications
Our results have potential implications for the development and refinement of techniques that aim to restore brain network function after stroke. For example, there is growing interest in applying non-invasive neuromodulatory techniques such as transcranial magnetic stimulation (TMS) to facilitate stroke recovery (Grefkes and Ward, 2014; Hesse et al., 2011; Naeser et al., 2012), and it has been previously proposed that the putative benefits of techniques such as TMS for stroke recovery may be dependent on the post-stroke structural network architecture (Grefkes and Ward, 2014). The results of our analyses indicate that patient-specific patterns of direct and indirect structural disconnections provide relevant information about the post-stroke structural and functional connectomes. This type of information could potentially be used to both (1) identify potential TMS targets based on their residual post-stroke connectivity patterns and (2) determine how the effects of TMS depend on the attributes of the post-stroke structural connectome.
4.4. Limitations
A primary limitation of the current work is that the inferences are necessarily restricted to the group-level rather than at the level of individual patients. That is, while we show that sets of disconnected regions on average show more severe functional connectivity disruptions than sets of regions with spared connections across patients, we do not show that our disconnection measures are sufficient to allow for the accurate prediction of connection-level functional connectivity disruptions in an individual patient, which will likely be necessary to enable the translation of these findings to clinical applications. Dedicated computational modeling (Adhikari et al., 2017) and/or predictive modeling (Goni et al., 2014) studies that incorporate comprehensive disconnection information are therefore an important next step.
A second limitation is that our structural connectivity and disconnection measures are obtained by embedding patient lesions into a white matter atlas that was constructed based on diffusion MRI data from healthy individuals. Atlas-based approaches cannot account for differences in white matter anatomy across patients, and there is evidence that such differences may contribute to variable outcomes after stroke (Forkel et al., 2014). Nonetheless, the advantages of atlas-based approaches make them an attractive option for studying the effects of brain lesions on the structural connectome. For example, atlas-based measures do not require expensive and time-consuming diffusion MRI data acquisition, and they are less likely than patient-specific diffusion MRI measures to be influenced by inter-individual differences in data quality or reconstruction, which may be particularly problematic for patients with gross structural abnormalities such as lesions caused by stroke (Gleichgerrcht et al., 2017). Atlas-based approaches also provide intuitively interpretable measures of expected structural disconnections that can be consistently employed across independent samples. While the quality of atlas-based measures necessarily depends upon the quality of the atlas used, the streamline tractography atlas used here was constructed using high angular resolution diffusion MRI data obtained from a very large sample of healthy subjects (N=842), and was expert-vetted to ensure correspondence with the known white matter anatomy (Yeh et al., 2018). Nonetheless, dedicated empirical evaluations of atlas-based vs. patient-specific structural disconnection measures are ultimately necessary to determine the relative usefulness of atlas-based vs. patient-specific structural connectivity measures for studying the effects of focal brain lesions on the structural connectome.
A third limitation is that we did not consider other potential brain pathologies that might contribute to disruptions of functional connectivity. For example, recent work indicates that white matter hyperintensities (i.e. leukoaraiosis) resulting from cerebral small vessel disease disrupt long-range structural connections and exacerbate the behavioral consequences of stroke (Wilmskoetter et al., 2019). These and other consequences of cerebral small vessel disease such as lacunar infarcts and microbleeds might be reasonably expected to influence the functional connectivity disruptions observed after stroke in affected patients, as previous work has documented altered structure-function coupling in elderly individuals with widespread white matter hyperintensities (Reijmer et al., 2015). Even so, lacunar infarct load and white matter disease severity in the patient sample studied here was relatively low (Table 1).
Finally, while changes in SSPLs computed based on binary structural connectivity matrices, such as those measured here, provide a simple and straightforward means of estimating the indirect consequences of focal brain lesions, they may be too coarse of a measure to allow for direct practical application. More complex network measures such as search information (Goni et al., 2014) or communicability (Osmanlioğlu et al., 2019) may provide more accurate descriptions of the higher-order structural network topology, which may be necessary to enable clinical translation. Ultimately, further research is necessary to further develop and refine these measures, and dedicated experimental studies are necessary to determine the practical utility of this type of information in the context of non-invasive neuromodulation and other potential clinical applications.
Supplementary Material
Acknowledgments
Funding: R01 NS095741 and R01 HD061117 to M.C.
Data were provided [in part] by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. We thank Alexandre Carter for assisting with lesion segmentation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors do not declare any competing interests.
References
- Abdi H, 2010a. Partial least squares regression and projection on latent structure regression (PLS Regression). Wiley Interdiscip. Rev. Comput. Stat. 2, 97–106. 10.1002/wics.51 [DOI] [Google Scholar]
- Abdi H, 2010b. The greenhouse-geisser correction. Encycl. Res. Des. Sage Publ. 1–10. 10.1007/BF02289823 [DOI] [Google Scholar]
- Adachi Y, Osada T, Sporns O, 2011. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb. cortex 1586–1592. 10.1093/cercor/bhr234 [DOI] [PubMed] [Google Scholar]
- Adhikari MH, Hacker CD, Siegel JS, Griffa A, Hagmann P, Deco G, Corbetta M, 2017. Decreased integration and information capacity in stroke measured by whole brain models of resting state activity. Brain 1068–1085. https://doi.Org/10.1093/brain/awx021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aickin M, Gensler H, 1996. Adjusting for multiple testing when reporting research results: The Bonferroni vs Holm methods. Am. J. Public Health 86, 726–728. https://doi.Org/10.2105/AJPH.86.5.726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre A, Ramsey L, Hacker CL, Callejas A, Astafiev SV, Metcalf NV, Zinn K, Rengachary J, Snyder AZ, Carter AR, Shulman GL, Corbetta M, 2014. Large-scale changes in network interactions as a physiological signature of spatial neglect. Brain 137, 3267–3283. 10.1093/brain/awu297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre A, Ramsey LE, Siegel JS, 2016. Brain connectivity and neurological disorders after stroke. Curr. Opin. Neurol. 29, 706–713. 10.1097/WC0.0000000000000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Zerrin Y, Haughton VM, Hyde JS, 1995. Functional Connectivity in the Motor Cortex of Resting Human Brain Using Echo-Planar MRI. Magn. Reson. Med. 34 [DOI] [PubMed] [Google Scholar]
- Carrera E, Tononi G, 2014. Diaschisis: past, present, future. Brain 137, 2408–2422. 10.1093/brain/awu101 [DOI] [PubMed] [Google Scholar]
- Carter AR, Astafiev SV, Lang CE, Connor LT, Rengachary J, Strube MJ, Pope DLW, Shulman GL, Corbetta M, 2010. Resting interhemispheric functional magnetic resonance imaging connectivity predicts performance after stroke. Ann. Neurol. 67, 365–375. 10.1002/ana.21905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Dietzmann P, Schulz R, Boenstrup M, Krawinkel L, Fiehler J, Gerloff C, Thomalla G, 2019. Cortical atrophy and transcallosal diaschisis following isolated subcortical stroke. J. Cereb. Blood Flow Metab. 10.1177/0271678X19831583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M, Ramsey L, Callejas A, Baldassarre A, Hacker CDD, Siegel JSS, Astafiev SVV, Rengachary J, Zinn K, Lang CEE, Connor LTT, Fucetola R, Strube M, Carter ARR, Shulman GLL, 2015. Common Behavioral Clusters and Subcortical Anatomy in Stroke. Neuron 85, 927–941. 10.1016/j.neuron.2015.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI, 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9, 179–94. 10.1006/nimg.1998.0395 [DOI] [PubMed] [Google Scholar]
- Duering M, Righart R, Wollenweber FA, Zietemann V, Gesierich B, Dichgans M, 2015. Acute infarcts cause focal thinning in remote cortex via degeneration of connecting fiber tracts. Neurology 84, 1685–1692. 10.1212/WNL.0000000000001502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R, 1986. [Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy]: Rejoinder. Stat. Sci. 1, 77–77. 10.1214/ss/1177013817 [DOI] [Google Scholar]
- Eldaief MC, McMains S, Hutchison RM, Halko MA, Pascual-Leone A, 2016. Reconfiguration of Intrinsic Functional Coupling Patterns Following Circumscribed Network Lesions. Cereb. Cortex bhw 139 10.1093/cercor/bhw139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM, 1999. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 9, 195–207. 10.1006/nimg.1998.0396 [DOI] [PubMed] [Google Scholar]
- Forkel SJ, Thiebaut de Schotten M, Dell’Acqua F, Kalra L, Murphy DGM, Williams SCR, Catani M, 2014. Anatomical predictors of aphasia recovery: a tractography study of bilateral perisylvian language networks. Brain 137, 2027–39. 10.1093/brain/awu113 [DOI] [PubMed] [Google Scholar]
- Foulon C, Cerliani L, Kinkingnéhun S, Levy R, Rosso C, Urbanski M, Volle E, Thiebaut de Schotten M, Kinkingnehun S, Levy R, Rosso C, Urbanski M, Volle E, Thiebaut de Schotten M, Kinkingnéhun S, Levy R, Rosso C, Urbanski M, Volle E, de Schotten MT, 2018. Advanced Lesion Symptom Mapping Analyses And Implementation As BCBtoolkit. Gigascience 7, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M., 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleichgerrcht E, Fridriksson J, Rorden C, Bonilha L, 2017. Connectome-based lesion-symptom mapping (CLSM): A novel approach to map neurological function. NeuroImage Clin. 16, 461–467. 10.1016/j.nicl.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goni J, van den Heuvel MP, Avena-Koenigsberger A, Velez de Mendizabal N, Betzel RF, Griffa A, Hagmann P, Corominas-Murtra B, Thiran J-P, Sporns O, 2014. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc. Natl. Acad. Sci. 111, 833–838. 10.1073/pnas.1315529111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE, 2016. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb. Cortex 26, 288–303. 10.1093/cercor/bhu239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C, Nomura EM, Pérez F, D’Esposito M, 2012. Focal brain lesions to critical locations cause widespread disruption of the modular organization of the brain. J. Cogn. Neurosci. 24, 1275–1285. 10.1162/jocn_a_00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefkes C, Ward NS, 2014. Cortical reorganization after stroke: how much and how functional? Neuroscientist 20, 56–70. 10.1177/1073858413491147 [DOI] [PubMed] [Google Scholar]
- Griffis JC, Metcalf NV, Corbetta M, Shulman GL, 2019. Structural disconnections explain brain network dysfunction after stroke. Cell Rep. 28, 1–68. 10.1101/562165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis JC, Nenert R, Allendorfer JB, Szaflarski JP, 2017a. Damage to white matter bottlenecks contributes to language impairments after left hemispheric stroke. NeuroImage Clin. 14, 552–565. 10.1016/j.nicl.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis JC, Nenert R, Allendorfer JB, Szaflarski JP, 2017b. Linking left hemispheric tissue preservation to fMRI language task activation in chronic stroke patients. Cortex 96, 1–18. 10.1016/j.cortex.2017.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BJ, Snyder AZ, Vincent JL, Epstein A, Shulman GL, Corbetta M, 2007. Breakdown of functional connectivity in frontoparietal networks underlies behavioral deficits in spatial neglect. Neuron 53, 905–18. 10.1016/j.neuron.2007.02.013 [DOI] [PubMed] [Google Scholar]
- Hesse MD, Sparing R, Fink GR, 2011. Ameliorating spatial neglect with non-invasive brain stimulation: From pathophysiological concepts to novel treatment strategies. Neuropsychol. Rehabil. 21, 676–702. 10.1080/09602011.2011.573931 [DOI] [PubMed] [Google Scholar]
- Honey C, Sporns O, 2009. Predicting human resting-state functional connectivity from structural connectivity. Proc. … 106, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JM, Vaishnavi SN, Smyth MD, Zhang D, He BJ, Zempel JM, Shimony JS, Snyder AZ, Raichle ME, 2008. Loss of Resting Interhemispheric Functional Connectivity after Complete Section of the Corpus Callosum. J. Neurosci. 28, 6453–6458. 10.1523/JNEUROSCI.0573-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Williams LJ, McIntosh AR, Abdi H, 2011. Partial Least Squares (PLS) methods for neuroimaging: A tutorial and review. Neuroimage 56, 455–475. 10.1016/j.neuroimage.2010.07.034 [DOI] [PubMed] [Google Scholar]
- Kuceyeski A, Kamel H, Navi BB, Raj A, ladecola C, 2014. Predicting future brain tissue loss from white matter connectivity disruption in ischemic stroke. Stroke 45, 717–722. 10.1161/STROKEAHA.113.003645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuller LH, Longstreth WT, Arnold AM, Bernick C, Bryan RN, Beauchamp NJ, 2004. White matter hyperintensity on cranial magnetic resonance imaging: A predictor of stroke. Stroke 35, 1821–1825. 10.1161/01.STR.0000132193.35955.69 [DOI] [PubMed] [Google Scholar]
- Lim DH, LeDue JM, Mohajerani MH, Murphy TH, 2014. Optogenetic Mapping after Stroke Reveals Network-Wide Scaling of Functional Connections and Heterogeneous Recovery of the Peri-Infarct. J. Neurosci. 34, 16455–16466. 10.1523/JNEUROSCI.3384-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Liu H, Zhang M, Wang D, Cao Y, Ma Q, Rong D, Wang X, Buckner RL, Li K, 2011. Focal Pontine Lesions Provide Evidence That Intrinsic Functional Connectivity Reflects Polysynaptic Anatomical Pathways. J. Neurosci. 31, 15065–15071. 10.1523/JNEUROSCI.2364-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman D, Chen Q, Zhang Y, Wang Z, Faseyitan OK, Coslett HB, Schwartz MF, 2015a. Neural organization of spoken language revealed by lesion-symptom mapping. Nat. Commun. 6, 6762 10.1038/ncomms7762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman D, Zhang Y, Wang Z, Coslett HB, Schwartz MF, 2015b. The ins and outs of meaning: Behavioral and neuroanatomical dissociation of semantically-driven word retrieval and multimodal semantic recognition in aphasia. Neuropsychologia 76, 208–219. https://doi.Org/10.1016/j.neuropsychologia.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mišić B, Betzel RF, de Reus MA, van den Heuvel MP, Berman MG, McIntosh AR, Sporns O, 2016. Network-Level Structure-Function Relationships in Human Neocortex. Cereb. Cortex bhw089. 10.1093/cercor/bhw089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeser M. a, Martin PI, Ho M, Treglia E, Kaplan E, Bashir S, Pascual-Leone A, 2012. Transcranial magnetic stimulation and aphasia rehabilitation. Arch. Phys. Med. Rehabil. 93, S26–34. 10.1016/j.apmr.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura EM, Gratton C, Visser RM, Kayser A, Perez F, D’Esposito M, 2010. Double dissociation of two cognitive control networks in patients with focal brain lesions. Proc. Natl. Acad. Sci. 107, 12017–12022. 10.1073/pnas.1002431107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanlioğlu Y, Tunç B, Parker D, Elliott MA, Baum GL, Ciric R, Satterthwaite TD, Gur RE, Gur RC, Verma R, 2019. System-level matching of structural and functional connectomes in the human brain. Neuroimage 199, 93–104. 10.1016/j.neuroimage.2019.05.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovadia-Caro S, Villringer K, Fiebach J, Jungehulsing GJ, van der Meer E, Margulies DS, Villringer A, 2013. Longitudinal effects of lesions on functional networks after stroke. J. Cereb. Blood Flow Metab. 33, 1279–85. 10.1038/jcbfm.2013.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Chang WH, Ohn SH, Kim ST, Bang OY, Pascual-Leone A, Kim Y-H, 2011. Longitudinal changes of resting-state functional connectivity during motor recovery after stroke. Stroke. 42, 1357–62. 10.1161/STROKEAHA.110.596155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE, 2014. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 84, 320–341. 10.1016/j.neuroimage.2013.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustina D, Coslett HB, Ungar L, Faseyitan OK, Medaglia JD, Avants B, Schwartz MF, 2017. Enhanced estimations of post-stroke aphasia severity using stacked multimodal predictions. Hum. Brain Mapp. 00. 10.1002/hbm.23752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffin E, Siebner HR, 2014. Transcranial brain stimulation to promote functional recovery after stroke. Curr. Opin. Neurol. 27, 54–60. 10.1097/WCO.0000000000000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmer YD, Schultz AP, Leemans A, O’Sullivan MJ, Gurol ME, Sperling R, Greenberg SM, Viswanathan A, Hedden T, 2015. Decoupling of structural and functional brain connectivity in older adults with white matter hyperintensities. Neuroimage 117, 222–229. 10.1016/j.neuroimage.2015.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb RA, Hanson DP, 1991. A software system for interactive and quantitative visualization of multidimensional biomedical images. Australas. Phys. Eng. Sci. Med. [PubMed] [Google Scholar]
- Roland JL, Snyder AZ, Hacker CD, Mitra A, Shimony JS, Limbrick DD, Raichle ME, Smyth MD, Leuthardt EC, 2017. On the role of the corpus callosum in interhemispheric functional connectivity in humans. Proc. Natl. Acad. Sci. 114, 201707050 10.1073/pnas.1707050114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O, 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–69. 10.1016/j.neuroimage.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Shen K, Mišić B, Cipollini BN, Bezgin G, Buschkuehl M, Hutchison RM, Jaeggi SM, Kross E, Peltier SJ, Everling S, Jonides J, McIntosh AR, Berman MG, 2015. Stable long-range interhemispheric coordination is supported by direct anatomical projections. Proc. Natl. Acad. Sci. 112, 6473–6478. 10.1073/pnas.1503436112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Ramsey LE, Snyder AZ, Metcalf NV, Chacko RV, Weinberger K, Baldassarre A, Hacker C, Shulman GL, Corbetta M, 2016. Disruptions of network connectivity predict impairment in multiple behavioral domains after stroke. PNAS I, 1–10. 10.1073/pnas.1521083113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Seitzman BA, Ramsey LE, Ortega M, Gordon EM, Dosenbach NUF, Petersen SE, Shulman GL, Corbetta M, 2018. Re-emergence of modular brain networks in stroke recovery. Cortex 101, 44–59. 10.1016/j.cortex.2017.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Shulman GL, Corbetta M, 2017. Measuring functional connectivity in stroke: Approaches and considerations. J. Cereb. Blood Flow Metab. 0271678X1770919. 10.1177/0271678X17709198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Snyder AZ, Ramsey L, Shulman GL, Corbetta M, 2015. The effects of hemodynamic lag on functional connectivity and behavior after stroke. J. Cereb. Blood Flow Metab. 0271678X15614846. 10.1177/0271678X15614846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Zhao Z, Chen C, Zheng X, Sun F, Zhang X, Tian J, Fan M, Wu Y, Jia J, 2016. Decreased Functional Connectivity of Homotopic Brain Regions in Chronic Stroke Patients: A Resting State fMRI Study. PLoS One 11, 1–13. 10.1371/journal.pone.0152875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turken AU, Dronkers NF, 2011. The neural architecture of the language comprehension network: converging evidence from lesion and connectivity analyses. Front. Syst. Neurosci. 5, 1 10.3389/fnsys.2011.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M, 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289. 10.1006/nimg.2001.0978 [DOI] [PubMed] [Google Scholar]
- Van Den Heuvel MP, Mandl RCW, Kahn RS, Hulshoff Pol HE, 2009. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Hum. Brain Mapp. 30, 3127–3141. 10.1002/hbm.20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson C, 2001. An Integrated Software Suite for Surface-based Analyses of Cerebral Cortex. J. Am. Med. Informatics Assoc. 8, 443–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmskoetter J, Marebwa B, Basilakos A, Fridriksson J, Rorden C, Stark BC, Johnson L, Hickok G, Hillis AE, Bonilha L, 2019. Long-range fibre damage in small vessel brain disease affects aphasia severity. Brain 142, 3190–3201. 10.1093/brain/awz251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Galantucci S, Tartaglia MC, Rising K, Patterson DK, Henry ML, Ogar JM, DeLeon J, Miller BL, Gorno-Tempini ML, 2011. Syntactic processing depends on dorsal language tracts. Neuron 72, 397–403. 10.1016/j.neuron.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Sjöström M, Eriksson L, 2001. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 58, 109–130. 10.1016/S0169-7439(01)00155-1 [DOI] [Google Scholar]
- Yeh F-C, Panesar S, Fernandes D, Meola A, Yoshino M, Fernandez-Miranda JC, Vettel JM, Verstynen T, 2018. Population-averaged atlas of the macroscale human structural connectome and its network topology. Neuroimage 178, 57–68. 10.1016/j.neuroimage.2018.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Fang Y, Han Z, Song L, He Y, Bi Y, 2017. Brain hubs in lesion models: Predicting functional network topology with lesion patterns in patients. Sci. Rep. 7, 17908 10.1038/s41598-017-17886-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kimberg DY, Coslett HB, Schwartz MF, Wang Z, Yongsheng Z, Kimberg DY, Coslett HB, Schwartz MF, Wang Z, 2014. Multivariate lesion-symptom mapping using support vector regression. Hum. Brain Mapp. 35, 997 10.1002/hbm.22590 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full set of neuroimaging data (along with behavioral data) are available at http://cnda.wustl.edu/app/template/Login, and specific data and scripts are available upon request to the authors.


