Abstract
The process of drug discovery includes individual synthesis and characterization of drug candidates, followed by a biological screening, which is separated from synthesis in space and time. This approach suffers from low throughput and associated high costs, which in turn lead to inefficiency in the field of drug discovery. Here, we present a miniaturized platform combining combinatorial solid-phase synthesis with high-throughput cell screenings. The method is based on the formation of nanoporous poly(2-hydroxyethyl methacrylate-co-ethylene dimethacrylate) layers patterned with hydrophilic spots separated from each other by superhydrophobic liquid-impermeable barriers. The porous polymer inside the hydrophilic spots is used as a support to conduct solid-phase synthesis. The hydrophilic spots can be then filled with droplets containing either reagents for synthesis or live cells. Upon irradiation with UV light, products of solid-phase synthesis are released from the porous polymer because of the photo-cleavable linkers used and diffuse into separate droplets. The light-induced release of the products allows the control of the release spatially, temporally, and quantitatively. To demonstrate the versatility and usability of the platform for various cell lines, we have successfully implemented peptide synthesis to create an exemplary chemical library and demonstrated high cell viability after the UV-triggered small-molecule release.
Keywords: Miniaturization, High-throughput screening, Combinatorial library, Photolytic release
1. Introduction
Drugs play a pivotal role in the history of the humankind. The demand for new drugs is high, not only because of drug-resistant pathogens and the need for better, less-toxic drugs but also because there are just on average only 8 novel first-in-class drugs per year, approved by the Food and Drug Administration [1], [2]. The world is undersupplied with drugs, mainly because of the inefficiency and the high costs of the whole drug-discovery pipeline, which is reflected in averaged 15–22 years and US$ 800 million to US$ 2 billion required for a single drug to enter the market [3], [4], despite enormously high investment in this field and more than 10,000 biotechnology and pharmaceutical companies worldwide.
There are about 18 million purchasable drug-like compounds available in various commercial chemical libraries [5], such as ChemBridge or ChemDiv. These libraries have been accumulated over the last 20–30 years from different sources but usually from individual syntheses performed by different researchers worldwide. Notably, a one-by-one synthesis of compounds that will be included in a chemical library, performed according to standard wet chemistry approaches, requires large amounts of reagents and solvents. Taken with the need of consequent characterization and isolation of new compounds, it makes the efficiency of synthesis of drug-like molecules extremely low and not compatible with the demand for high-throughput (HT) screenings.
To solve this problem, pharmaceutical companies use combinatorial chemistry and HT screening methods to synthesize and test large chemical libraries. Cell screenings are usually performed in 96- or 384-well plates; so, to achieve high throughput, thousands of microtiter plates have to be used. This approach suffers from several drawbacks, such as consumption of large amounts of cells and valuable reagents as well as the need to use robotics to transfer libraries of chemicals into the microtiter plates [6], [7].
Miniaturization and array formats, accompanied by parallelization, can solve some of these problems. The microarray technology enables rapid synthesis of various compounds, including DNA [8], [9], [10], peptides [11], [12], proteins [13], small molecules [11], [14], [15], oligosaccharides [16], [17], and synthetic polymers [18]. Although these methods have provided significant development in the field of HT synthesis in the past years, providing high miniaturization and avoiding transfer step, the scope of the assays that can be performed with surface-bound molecules is limited because the final biological assays are usually performed in bulk solutions [19], [20], [21], [22]. Importantly, cell interaction with soluble cues cannot be assayed in bulk solution because of cross-contamination caused by lateral diffusion of compounds. Therefore, a technology combining miniaturized HT chemical synthesis in confined volumes and cell screening on the same platform is highly anticipated.
Recently, we established a miniaturized screening platform based on the array of nanoliter-sized droplets (droplet microarray [DMA] platform) [23]. DMA has proved itself to serve as a convenient and versatile platform for HT screening of single cells [24], suspension and adherent cells [25], [26], cell spheroids [27], and even single fish-embryos [28]. DMA was also used to perform HT screening on stem cells [29], [30] and is optimized for reversed cell transfection [31].
In the present study, we modify DMA to be used as a miniaturized platform for combinatorial solid-phase synthesis of small molecules and subsequent biological screenings, which resulted in chemBIOS workflow (chemical synthesis is combined with biological readout on the same glass slide); chemBIOS workflow was previously validated for liquid-based synthesis [32]. Owing to compartmentalization, each droplet serves both as a separate reaction compartment and as a microreservoir for culturing cells, rendering library transfer step redundant and enabling high spatial control. Arrayed format enables the combinatorial approach without the need for decoding. The use of a photo-cleavable linker for solid-phase synthesis allowed us to control the concentration of the final drug in separated individual droplet compartments by changing the irradiation time. Finally, a peptide synthesis was established as a model reaction, and HEK293T cells were tested in regard of maintaining cell viability upon UV irradiation to highlight the convenience and future potential of the chemBIOS method.
2. Materials and methods
The glass slides were purchased from Schott Nexterion (Jena, Germany). Ninhydrin and piperidine were purchased from Alfa Aesar (Ward Hill, Massachusetts, USA). 4-Pentynoic acid was purchased from Apollo Scientific (Bredbury, UK). Fmoc-Glu(OtBu)-OH and Fmoc-Gly-OH were purchased from Bachem (Bubendorf, Switzerland). Fmoc-Val-OH, Fmoc-Ala-OH, and Fmoc-Leu-OH were purchased from Iris Biotech (Marktredwitz, Germany) and kindly provided by Dr. Parvesh Wadhwani, Institute of Biological Interfaces, KIT. Hydrochloric acid (37%), ethanol, ethanol absolute, acetone, 4-(dimethylamino)pyridine, 4-[4-(1-hydroxyethyl)-2-methoxy-5-nitrophenoxy]butanoic acid (hydroxyethyl photolinker), pyridine, methanol, N,N-dimethylformamide, acetic acid anhydride, and phenol were purchased from Merck (Darmstadt, Germany). 1-Hydroxybenzotriazole was purchased from Molekula (Newcastle upon Tyne, UK). Sodium hydroxide, 3-(trimethoxysilyl)propyl methacrylate, 2-hydroxyethyl methacrylate, ethylene dimethacrylate, 1-decanol, cyclohexanol, 2,2-dimethoxy-2-phenylacetophenone, 1H,1H,2H,2H-perfluorodecanethiole, cysteaminium chloride, and potassium cyanide were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Calcein AM was purchased from Life Technologies GmbH (Darmstadt, Germany), and propidium iodide (PI), from Invitrogen (Merelbeke, Belgium).
If not stated differently, all chemicals were used without further purification.
2.1. Functionalization of DMA slide with the linker
The DMA slides were prepared according to the previously published procedure [23], [25], [33]. Briefly, a microscope glass slide was coated with a layer of nanoporous poly(2-hydroxyethyl methacrylate-co-ethylene dimethacrylate) (HEMA-co-EDMA) (using fluorinated glass slide during photopolymerization), taped with a sticky film to increase roughness of the surface and subsequently esterified using 4-pentynoic acid. The pattern of repeating superhydrophobic and hydrophilic properties was created by using the corresponding photomask via thiol-yne photoclick reaction using 1H,1H,2H,2H-perfluorodecanethiole and cysteaminium chloride or β-mercaptoethanol, respectively. To functionalize the hydrophilic spots with the linker, (4-[4-(1-hydroxyethyl)-2-methoxy-5-nitrophenoxy]butanoic acid), diisopropyl carbodiimide (DIC), and 1-hydroxybenzotriazole were mixed in dimethylformamide (DMF) to a final concentration of 0.03 M, 0.3 M, and 0.3 M, respectively. In each 2.83-mm spot of the DMA slide functionalized with cysteaminium chloride, 10 μL of solution was pipetted and incubated in dark at room temperature for 18 h (overnight). The DMA slide was then washed with acetone and dried in nitrogen flow. The unreacted amino groups of cysteamine were then capped using a 10% solution of pyridine in acetic acid anhydride. In each spot, 10 μL of capping solution was pipetted. After 5 min, the DMA slide was washed with acetone and dried in nitrogen flow.
2.2. Analytics
The static contact angle was measured using a Drop Shape Analyzer DSA25 (Krüss) by applying 50 μL of deionized water on a non-functionalized hydrophilic surface and on a hydrophilic surface, functionalized with linker and capped with acetic anhydride.
For kinetic experiments (attenuated total reflection infrared [ATR-IR spectroscopy, time-of-flight secondary ion mass spectrometry [ToF-SIMS]), several spots of a DMA slide were functionalized with the linker as described previously, with exposure of the hydrophilic spots to the linker solution at different times (e.g. in the range of 1 h–18 h). After functionalization with the linker, capping of the unreacted amino groups, and thorough washing in acetone and drying in nitrogen flow, the DMA slide was subjected to the respective analytical method. For ToF-SIMS depth profiling, different polymer thicknesses were adjusted during the polymerization process by using Teflon film spacers of 6- and 25-μm thickness. The exact final thickness of the polymer layer was determined by profilometry (Dektak XT Stylus Profiler; Bruker Nano, Karlsruhe, Germany).
X-ray photoelectron spectroscopy (XPS) measurements were performed on the DMA slide, which was exposed to the linker solution for 18 h.
2.2.1. XPS measurements
XPS measurements were performed using a K-Alpha+ XPS spectrometer (ThermoFisher Scientific, East Grinstead, UK). Data acquisition and processing using the Thermo Avantage software is described elsewhere [34]. All samples were analyzed using a microfocused, monochromated Al Kα X-ray source (400-μm spot size). The K-Alpha+ charge compensation system was used during analysis, using electrons of 8-eV energy and low-energy argon ions to prevent any localized charge buildup. The spectra were fitted with one or more Voigt profiles (BE uncertainty: ± 0.2 eV) and a Shirley background. Scofield sensitivity factors were applied for quantification [35]. All spectra were referenced to the C 1s peak (C–C, C–H) at 285.0-eV binding energy controlled by means of the well-known photoelectron peaks of metallic Cu, Ag, and Au, respectively. The K-Alpha + snapmap option was used to image an area of 3 × 3 mm with an X-ray spot of 200 μm (5 iterations were run to reach a better statistic).
2.2.2. Time-of Flight Secondary Ion Mass Spectrometry
ToF-SIMS was performed on a ToF-SIMS-5 instrument (ION-TOF GmbH, Münster, Germany) equipped with a Bi cluster primary ion source, argon cluster source for depth profiling, and a reflectron-type ToF analyzer. Ultra-high vacuum (UHV) base pressure was <10−8 mbar. For high mass resolution, the Bi source was operated in ‘high current bunched’ mode providing short Bi3+ primary ion pulses at 25 keV energy, a lateral resolution of approximately 4 μm, and a target current of 1 pA at 20 kHz repetition rate. The short pulse length of 1.1 ns allowed for high mass resolution. The primary ion beam was rastered across a 500×500 μm2 field of view on the sample, and 128×128 data points were recorded. Larger fields of view were recorded by scanning the primary beam and moving the sample stage. Primary ion doses were kept below 1011 ions/cm2 (static SIMS limit). For charge compensation, an electron flood gun providing electrons of 21 eV was applied, and the secondary ion reflectron, tuned accordingly. Spectra were calibrated on the omnipresent C−, C2-, C3−, or on the C+, CH+, CH2+, and CH3+ peaks. Based on these data sets, the chemical assignments for characteristic fragments were determined.
For depth profiling, a dual beam analysis was performed in non-interlaced mode: The primary ion source was again operated in ‘high current bunched’ mode with a scanned area of 100 × 100 μm2 (3 frames with 64×64 data points), and a sputter gun (operated with Ar1500+ ions, 20 keV, scanned over a concentric field of 300×300 μm, target current 9 nA) was applied to erode the sample followed by a 1.5-s pause for charge compensation. Thus, the sputter ion dose density was >1000 times higher than the Bi ion dose density. This approach allows the recording of larger molecular fragments, such as C4H5O2−, from the base polymer backbone throughout the eroded polymer layer of several μm thickness.
2.2.3. Fourier-transform infrared spectroscopy
A Bruker Tensor 27 Fourier-transform infrared (FTIR) spectrometer (Bruker Optik GmbH, Ettlingen, Germany) was used to obtain the IR spectra of the samples.
All samples were measured in attenuated total reflection geometry without additional preparation on a Bruker Platinum ATR accessory equipped with a diamond crystal, 45° angle of incidence, and one reflection. Spectral range of 4000–370 cm−1 was recorded with a scanner velocity of 10 kHz and a spectral resolution of 4 cm−1 (32 scans). The reference spectra were taken from air. All spectra were evaluated using the Bruker OPUS software.
2.3. Linker loading determination and photorelease kinetic measurements
Two stock solutions were prepared and stored at 8 °C:
-
-
Solution 1: Forty grams of phenol (425 mmol) was dissolved in a mixture of 49 mL of pyridine and 10 mL of absolute ethanol, and 1 mL of a 10 mM aqueous KCN solution was added. Final concentrations were 7.1 M phenol and 0.17 mM KCN.
-
-
Solution 2: Two and a half grams of ninhydrin (14 mmol) was dissolved in 50 mL of absolute ethanol to yield a 281 μM solution
To perform the Kaiser test, the solution 1 was mixed with solution 2 in a 4:1 ratio immediately before the experiment. Ten microliters of the mixture was pipetted manually in each spot. As a negative sample, a DMA slide with the same pattern size but with hydrophilic spots functionalized with β-mercaptoethanol instead of cysteamine was treated in the same way. Both samples were heated on a heating plate at 50 °C for 5 min, and the liquid of both spots was pipetted off and diluted to 1 mL with a methanol/H2O (1:1) solution; the absorbance was measured at 570 nm.
To study the photorelease properties of the linker, glycine was attached to the linker, as described in the following, and then the kinetics of its phototriggered detachment was studied. After UV irradiation (0.5–15 min, 9 data points), the liquid from two spots was pipetted off and added to a mixture of 100-μL solution 1 and 25-μL solution 2 in a vial. For the negative control, deionized water was used instead of the droplet volume. Both were then heated in a water bath at 50 °C for 5 min, diluted to 1 mL with methanol/H2O (1:1), and measured using LAMBDA™ 35 UV spectrometer (PerkinElmer, Waltham, Massachusetts, USA). A molar extinction coefficient of ε = 15700 L∙mol−1 cm−1 was used to calculate the concentration by the Beer-Lamberts law.
2.4. Fmoc-based peptide synthesis
For the overall library scheme, see Fig. 5C and Fig. S7. The DMA slide was functionalized with linker as described previously. For attaching the first amino acid onto the linker 24 μL of a 0.1 M solution of the corresponding Fmoc-protected amino acid in DMF, 8 μL of a 0.1 M solution of 4-dimethylaminopyridine (4-DMAP) in DMF and 8 μL of DIC were premixed in a vial and applied along one row of the DMA slide (4 spots). This was repeated for glycine, valine, alanine, and leucine according to the scheme. During the reaction time of 18 h, the DMA slide was stored in dark at room temperature. Subsequently, the slide was extensively washed with acetone and immersed in dichloromethane (DCM) for 1 h. For coupling the following amino acids, 24 μL of a 0.1 M solution of the corresponding Fmoc-protected amino acid in DMF, 8 μL of 0.3 M solution of 1-hydroxybenzotriazole (1-HOBt) in DMF, and 8 μL of DIC were premixed in a vial, applied in the same manner according to scheme along one row of the DMA slide (4 spots), and incubated at room temperature in dark for 4 h. Then, the slide was extensively washed with acetone and immersed in DCM for 1 h. The coupling step was repeated two times to obtain a library of 16 tripeptides.
Fig. 5.
(A) A schematic representation of the construction of 16-membered combinatorial library on 16 hydrophilic spots of DMA, using 4 different amino acids, arrows indicating the direction of distribution relative to each other. First, amino acids are applied in rows and immobilized on the solid phase. In the second and third steps, amino acids are applied in columns. (B) Reaction scheme utilizing Fmoc chemistry (corresponding combinatorial scheme of tripeptide synthesis, Fig. S7). (C) General reaction scheme of tripeptides (AA1AA2AA3) cleavage from the solid phase; tripeptides bear a free carboxy group at the C-terminus. The photolinker stays attached to the solid phase. After the cleavage, tripeptides diffuse into separate water droplets and can be subjected to ESI-MS analysis. DMA, droplet microarray; HEMA-co-EDMA, 2-hydroxyethyl methacrylate-co-ethylene dimethacrylate.
Before each coupling step, the amino acids have to be deprotected. The deprotection of the amino group was carried out by immersing the whole slide into a solution of 20% piperidine in DMF for 1 h.
2.5. Cell viability assays
Four DMA slides were functionalized with linker and capped with acetic anhydride as described previously, followed by sterilization by immersing in 70% ethanol in water for 1 h in dark. Slides were then air-dried for at least 15 min. While drying, a 2.2% w/v gelatin solution was prepared by adding 3 mL of sterile cell medium to 66 mg of gelatin from bovine skin. To increase the solubility of gelatin, the solution was gently warmed in a water bath at 37 °C. Once gelatin was completely dissolved, the solution was sterilized by filtering through a sterile 0.22-μm filter. The gelatin solution was then applied onto all DMA spots via rolling droplet to produce evenly distributed droplets. The DMA slide was then incubated for 1 h at 37 °C, followed by air-drying of gelatin for 1 h. In each spot, 8 μL of HEK293T cell suspension (4.23∙× 105 cells ml-1) were pipetted, followed by incubation for 3 h at 37 °C. Three slides were illuminated with UV light (364 nm) for 5, 10, and 15 min respectively, and one slide served as a control. Four slides were incubated overnight at 37 °C and stained, using live/dead staining with PI and calcein AM.
3. Results and discussion
DMA comprises a standard microscopic glass slide with a 6-μm thick layer of poly(HEMA-co-EDMA) chemically immobilized to the glass surface (Fig. 1A). The HEMA-co-EDMA polymer surface was functionalized via esterification with 4-pentynoic acid, followed by patterning via the UV-induced thiol-yne click reaction [31] with either cysteamine hydrochloride or 1H,1H,2H,2H-perfluorodecanethiol to form round hydrophilic (HL) spots with a diameter of 2.83 mm (HL, static water contact angle [WCA], θst = 4.4°) [31] surrounded by superhydrophobic (SH; θadv = 173°, θst. = 170°, and θrec = 164°) [31] borders of 0.3 mm (Fig. 1A). Owing to the extreme difference in wettability between SH and HL areas, aqueous solutions, such as cell suspension, applied onto such surface spontaneously form an array of separated microdroplets via discontinuous dewetting [36]. Some organic solvents, such as DMF and dimethyl sulfoxide (DMSO) can also be confined into hydrophilic spots, despite their low surface tensions (37.10 and 43.54 mN/m, respectively).
Fig. 1.
General workflow of solid-phase synthesis using the chemBIOS platform with photocleavable linkers. (A) Superhydrophobic-hydrophilic microarray with hydrophilic spots functionalized with photolinkers L. (B) Synthesis of a combinatorial library of small molecules; (C) Seeding of cells to form an array of aqueous droplets. (D) The screening experiment can be started on demand by irradiating all or selected droplets with 364-nm UV light (1–15 min). (E) Readout step. HEMA-co-EDMA, 2-hydroxyethyl methacrylate-co-ethylene dimethacrylate.
Previously we established two general workflows for screening of compound libraries. The first method involves a library transfer using the ‘sandwiching approach’ (Fig. S1A) [23]. In the ‘reversed drug treatment approach,’ the library of compounds is printed directly onto the spots of a DMA slide (Fig. S1B) [25]. In both approaches, the screening is started as soon as cells get in contact with printed compounds and in all droplets simultaneously, thus limiting the spatiotemporal control. The chemBIOS method was, therefore, designed to give more control over time and position where the screening is performed (Fig. S1C).
Photolytic cleavage of small molecules from the solid phase offers several crucial advantages over the chemical one [37]. Photolabile linkers can only be cleaved upon exposure to the light, which broadens the scope of reactions that can be used in small-molecule synthesis in comparison with chemically cleavable linkers, which can be cleaved, for example, under acidic conditions. Another important factor is the non-contaminant and non-contact nature of light. It is critical if small molecules released from the solid phase are directly used in biological assays [36]. The cleavage upon light irradiation also has higher spatial and temporal resolution than the chemical one, which is essential for screening applications. Furthermore, the intensity and wavelength of light can be adjusted and controlled, which enables facile fine-tuning of releasing properties.
In developing the method of combinatorial solid-phase synthesis on the DMA slide, we chose the [4-(1-hydroxyethyl)-2-methoxy-5-nitrophenoxy]-butanoic acid as the photocleavable linker [38] (represented in Fig. 1A as corresponding amide L). This linker releases acids by UV irradiation at 364 nm, which has been proven not to affect cell viability [39], [40]. The side product of cleavage remains bound to the solid phase. The mechanism of the cleavage is depicted in Fig. S2.
3.1. Loading of linker
The high surface area of the porous polymer layer (9 m2/g) provides numerous accessible reaction sites, while the permeability enables diffusion of chemicals, so the linker can react throughout the polymer layer and not only on the topmost surface. First, we decorated the hydrophilic spots of DMA with amino groups, so linkers carrying carboxylic group could be anchored to the DMA slide as an amide (Fig. 1A). In the next step, we inactivated the unreacted amino groups by formation of an amide with acetic acid anhydride and pyridine as an acid scavenger.
We have determined the linker content on the surface spectrophotometrically by the Kaiser test [41] to be 1.63 nmol/mm2. Spots were functionalized with linkers, and unreacted amino groups were capped with acetic acid anhydride as described previously. Spots functionalized with the linker and capped with acetic acid anhydride stay hydrophilic, with predictably elevated static WCA of 29±1° in comparison with the non-functionalized spot (static WCA = 4.4°). We then determined the kinetics of cleavage by measuring the concentration of a photoreleased compound in the droplet upon exposure to the UV light after distinct time periods (Fig. 4B). Glycine was used as a model compound and was attached and deprotected as described previously. We determined the concentration of glycine photoreleased after UV irradiation spectrophotometrically via the Kaiser test for all irradiation times. We calculated concentration of glycine in the droplet and plotted against the irradiation time, yielding the release curve (Fig. 4B). The linker half-life (time, at which half of the molecules attached to the linker are cleaved) under UV exposure was measured to be around 3 min. The rapid photolytic cleavage of the linker in aqueous environment is important for the application of UV-induced drug release process under biologically relevant conditions. The half-life value is comparable with literature-known values for photolysis in the liquid phase [38]. The DMA slide, therefore, does not suffer from typical obstacles of resin-bound photolabile linkers, such as swelling of resin or light scattering, shadowing and shielding effects, which would increase the half-life of the linker [38]. Complete cleavage was achieved after 15 min of UV irradiation (Fig. 4B), which is six times faster than in a comparable SPOT system, requiring dry cleavage [42].
Fig. 4.
(A) Cell viability as measured by live/dead stainings not affected by the UV light (365 nm, 4 mW/cm2, irradiation time 5–15 min and incubation for 24 h). (B) Phototriggered release of glycine from the surface after different irradiation times, expressed in nmol of glycine per mm2 of the linker-functionalized surface. The quantity per surface area of released glycine was measured by the Kaiser test. The amount of released glycine can be controlled by varying the exposure time.
3.2. Characterization
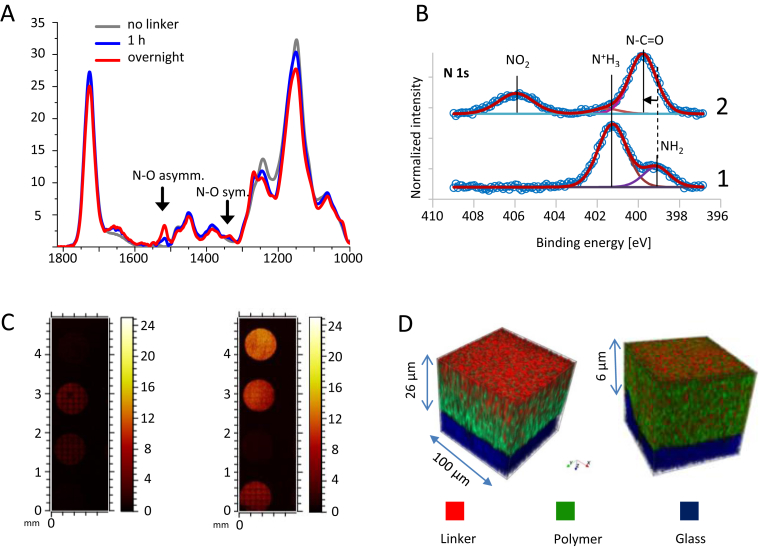
We used ATR-IR measurements for in situ reaction monitoring. ATR-IR spectral data were collected for hydrophilic spots before incubation with linker and after incubation with linker for 1 h and overnight. Incubation of a hydrophilic spot with a 0.03 M linker solution, supplemented with 0.3 M DIC and 0.3 M 1-hydroxybenzotriazole, for 1 h led to the emergence of two new signals in IR spectra that could be assigned to the introduced nitro group (1518 cm−1 for N–O asymmetric vibration and 1336 cm−1 for N–O symmetric stretch) [43]. The intensity of these characteristic signals increased as the reaction proceeds overnight (Fig. 2A).
Fig. 2.
Surface characterization of the DMA. (A) ATR-IR measurements of hydrophilic spots incubated with the linker solution for different periods of time. (B) N 1s XP spectra before (1) and after (2) functionalization with the linker (background subtracted). (C) ToF-SIMS stage scans for NO2− signals of hydrophilic spots reacted with the linker solution for different periods of time (from top to bottom, left: no linker, 2 h, 1 h, no linker; right: 18 h, 6 h, no linker, 3 h). (D) 3D rendering of ToF-SIMS depth profiles; linker, characterized by NO2− (red), polymer characterized by C4H5O2− (green), and glass substrate (SiO2−, SiO3−, and SiO3H−) (blue). XY dimensions are 100x100 μm. DMA, droplet microarray; ATR-IR, attenuated total reflection infrared; ToF-SIMS, time-of-flight secondary ion mass spectrometry; 3D, three-dimensional.
XPS measurements were performed on the spot before and after incubation with 0.03 M linker solution, supplemented with 0.3 M DIC and 0.3 M 1-hydroxybenzotriazole for 18 h (Fig. 2B). Before incubation, the N 1s X-ray photoelectron (XP) spectrum indicated the presence of cysteamine with a component at 399.2 eV and a protonated form with a peak at 401.4 eV. After incubation with linker solution (Fig. 2B), the signal at 399.2 eV is shifted to 399.8 eV, proving the addition of amide groups. An additional signal at 406.0 eV indicates the presence of the linker nitro group, thus revealing the successful attachment of the linker to the solid phase.
To investigate the kinetics of linker immobilization and the loading of the linker as a function of polymer thickness, ToF-SIMS measurements of polymer substrates with different polymer thicknesses and different linker incubation time were conducted. Despite the semiquantitative nature of ToF-SIMS, the relative intensity change of respective ions provides useful information about the kinetics of the reaction. A clear trend of NO2− intensity changing can be observed for different linker incubation times (Fig. 2C). Therefore, to achieve complete loading, it is important to incubate the DMA slide with linker solution overnight. Another important finding was that no NO2− signal was detected on spots without linker. This means that compartmentalization of liquids within the DMA works well and each spot serves as a confined microreactor with no evidence of cross contamination.
The distribution of the linker (loading of the linker as a function of polymer thickness and incubation time) was performed via depth profiling via dynamic SIMS under argon cluster erosion after incubation of hydrophilic spots of different thicknesses (6 and 26 μm) for different times (1–18 h). The distribution of the linker in 6-μm-thick polymer is uniform (Fig. 2D, Fig. S3). However, hydrophilic spots patterned on thicker polymer layers (26 μm), incubated with linker solution overnight, show a gradient of uniformly decreasing amount of linker toward the surface opposite of irradiation according to the NO2− signal in ToF-SIMS. This finding can be attributed to the decreasing amount of the cysteamine (CN− signal in ToF-SIMS), onto which the linker is covalently attached (Fig. S4). The signal of the linker decreased even faster for shorter incubation times (Fig. S5). Despite uneven distribution of the linker in the 26-μm-thick polymer, the depth-integrated signals of linker are the highest for the sample that was produced by incubating the hydrophilic spots with linker solution overnight. The signals decrease simultaneously with the incubation time, which again stressed the importance of incubation of hydrophilic spots with linker solution overnight (Fig. S6).
3.3. Confinement on DMA
The confinement of solution is an essential prerequisite for using hydrophilic spots as distinct microreservoirs without cross contamination between the individual spots. We have proved the confinement of solutions in the bulk of polymer and on top of the slide by XPS (Fig. 3A, area between the spots) and depth profiling via ToF-SIMS (Fig. 3B, cutout of the spot including surrounding). The linker-containing liquid could wet only hydrophilic spots, so the linker was attached only to the cysteamine of the hydrophilic spots, as seen from the intensity of combined N 1s signals of amide and nitro groups (referred to the presence of linker inside the hydrophilic spots; Fig. 3A). XPS image of the area between spots, on the other hand, depicts the high abundance of fluorine (right image, assigned to the fluorine, F 1s, 689.0 eV). The ToF-SIMS–assisted depth profiling of the interface area of the spot and the surrounding has shown NO2− signal only inside the spots and F− signal outside the spots. No NO2− signal was found within fluorinated borders, implying that linker-containing solution could not penetrate the fluorinated boarders, confirming the fluorinated borders seal the spot securely from the surrounding in the bulk of polymer (Fig. 3B). Both XPS and ToF-SIMS measurements show the absence of the linker (and with it lack of active sites for solid-phase synthesis) within the fluorinated boarders, thus rendering them inert and unqualified to act as a solid-phase. These results clearly indicate that compartmentalization of liquids within the DMA works well and each spot serves as a confined microreactor with no evidence of cross contamination.
Fig. 3.
Experiments proving the confinement of liquids inside hydrophilic spots. (A) XPS image of the area between spots depicting the abundance of nitrogen inside the spots (left image, assigned to the amide and nitro group, N 1s, 400.2 and 406.0 eV, respectively) and fluorine outside the spots (right image, assigned to the fluorine, F 1s, 689.0 eV). (B) 3D rendering of ToF-SIMS depth profiles; linker, characterized by NO2− (red), fluorinated borders, characterized by F- (yellow) and glass substrate (SiO2−, SiO3−, and SiO3H−) (blue). XY dimensions are 500x500 μm. XPS, X-ray photoelectron spectroscopy, ToF-SIMS, time-of-flight secondary ion mass spectrometry; 3D, three-dimensional.
3.4. Combinatorial chemistry on DMA
To show the general possibility of solid-phase synthesis on DMA, we performed standard Fmoc chemistry (Fig. 4B) for an exemplary 16-membered library of tripeptides.
For this, the hydrophilic spots were functionalized with the photolinker, as described previously (Fig. 1) and distinct tripeptides were synthesized according to the scheme (Fig. 5A, B and S7). Each pentynoic acid tethered to the HEMA-co-EDMA polymer layer acts as a branching point, bears two linker molecules, and thus doubles the overall number of reaction sites. The combinatorial approach was realized by applying the N-protected Fmoc amino acids glycine (Fmoc-Gly-OH), valine (Fmoc-Val-OH), alanine (Fmoc-Ala-OH), and leucine (Fmoc-Leu-OH) row wise and column wise. We applied amino acids along the rows of the DMA slide, starting with Fmoc-Gly-OH in row 1 and continuing with Fmoc-Val-OH in row 2, Fmoc-Ala-OH in row 3, and Fmoc-Leu-OH in row 4. All amino acids could be deprotected simultaneously by immersing the DMA slide in 20 vol% solution of piperidine in DMF. To conduct the second coupling step, we applied amino acids along the columns, starting with Fmoc-Gly-OH in column 1, continuing with Fmoc-Val-OH in column 2, Fmoc-Ala-OH in column 3, and Fmoc-Leu-OH in column 4. After repeating the deprotection step, we applied amino acids for the third coupling step in the following pattern: Fmoc-Leu-OH in column 1, Fmoc-Ala-OH in column 2, Fmoc-Val-OH in column 3, and Fmoc-Gly-OH in column 4 (Fig. 5A). After the final Fmoc deprotection step, we cleaved the tripeptides from the surface upon UV irradiation for 15 min (Fig. 5C). The tripeptides along the diagonal, GGL, VVA, AAV, and LLG, were analyzed by electrospray ionization mass spectrometry (ESI-MS), confirming the presence of the corresponding [M+H+] peaks (Fig. S8A). This shows that solid-phase synthesis of peptides in confined 10-μL volumes and their release into the corresponding droplets via photo-triggered cleavage can be performed without detectable cross contamination. The fact that the hydrophobic tripeptides could be released from the solid phase in the aqueous solution proves the suitability of the DMA for synthesis and release of drug-like molecules, which are mostly hydrophobic. The 16-membered exemplary library could be constructed within 3 days, therefore requiring <1 h of active participation. Each 2.83 mm in the diameter spot can accommodate up to 10 μL of liquid, so liquid handling was conveniently done by manual pipetting. Smaller spots with varying geometry and less working volumes can be produced by using the corresponding photomask in the slide production process. For example, hydrophilic squares of 333x333 μm accommodate approximately 4 nL. Although conducting chemical synthesis on arrays with smaller features makes automated liquid handling robotics necessary, it allows for a rapid, parallel, and highly miniaturized way to synthesize compound libraries. Being patterned with a 2.83-mm round spot pattern, a standard microscopic glass slide in the size of 75x26 mm can accommodate 80 different reactions, whereas the pattern of 333x333 μm squares amounts to 6048 distinct spots. Thus, a single glass slide can be potentially used for testing 6048 different combinations of starting compounds. On this scale, we assume to face challenges such as precise handling and dispensing of low volumes in nanoliter range, evaporation of liquids, and reaction monitoring, as well as analysis of the reaction yield and product purity. These aspects of the chemBIOS pipeline are currently under investigation.
3.5. Cell compatibility of the photo-triggered release
One of the key features of the combinatorial solid-phase synthesis using the chemBIOS workflow is the possibility to perform cell-based screenings directly on the same DMA slide after the synthesis part is accomplished and without the need to transfer the compounds onto another DMA slide or into the wells of microtiter plates. The 4-[4-(1-hydroxyethyl)-2-methoxy-5-nitrophenoxy]butanoic acid as the UV-triggered linker, used for the solid-phase synthesis, can be cleaved using 364-nm UV light. To prove the harmlessness of the 364-nm UV light for cells cultured on the DMA slide, we have conducted a series of experiments on adherent HEK293T cells. Cells were seeded onto 4 separate DMA slides functionalized with hydroxyethyl photolinker as described previously. Three slides were illuminated with 364-nm UV light for 5, 10, or 15 min. After incubation for 18 h, we assessed the cell viability. These experiments demonstrate that the UV light used for triggering the compound release from the solid phase does not affect viability of HEK293T cells, as seen from life/dead staining on Fig. 4A and Fig. S9.
4. Conclusions
Here, we demonstrated a miniaturized platform combining the possibility to perform combinatorial solid-phase synthesis with HT cell screenings. We used nanoporous poly(HEMA-co-EDMA) layers, which were patterned with hydrophilic spots separated from each other by superhydrophobic liquid-impermeable barriers. The porous polymer inside the hydrophilic spots was functionalized with photolabile linkers, which were used as a support to conduct solid-phase synthesis of a peptide library. We also demonstrated that the hydrophilic spots could be filled with solutions containing either reagent for the synthesis or live cells. Upon irradiation with UV light, products of the solid-phase synthesis could be released from the porous polymer and delivered into the separate droplets. Thus, the light-induced release of the products allowed us to control the release spatially, temporally, and quantitatively. We also showed that the amount of released compounds could be controlled by altering the irradiation time. To demonstrate the versatility and usability of the platform for various cell lines, we have demonstrated high cell viability after the UV-triggered release of the synthesized product. Liquid dispensers can be potentially used to accelerate the construction of chemical libraries and enable higher throughput combinatorial solid-phase synthesis using the chemBIOS pipeline. We believe that the ability not only to synthesize libraries of compounds but also to release them into individual cell microreservoirs with spatiotemporal control will lead to further advancement of miniaturized and HT cell-based assays.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgements
The authors are grateful for support by the ERC Starting Grant (DropCellArray 337077), ERC Proof-of-Concept Grant (CellPrintArray 768929), and the HGF-ERC-0016 Grant from the Helmholtz Associations Initiative and Networking Fund. The authors would also like to thank Udo Geckle for profilometry measurements.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2019.100022.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.U.S. Food&Drug Administration . 2016. Novel Drug Summary.https://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation/ucm534863.htm [Google Scholar]
- 2.Eder J., Sedrani R., Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug Discov. 2014;13(8):577–587. doi: 10.1038/nrd4336. [DOI] [PubMed] [Google Scholar]
- 3.Chong C.R., Sullivan D.J., Jr. New uses for old drugs. Nature. 2007;448:645. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 4.DiMasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Database of commercially-available compounds http://zinc.docking.org/browse/subsets/(accessed 09 January 2018).
- 6.Fernandes T.G., Diogo M.M., Clark D.S., Dordick J.S., Cabral J.M.S. High-throughput cellular microarray platforms: applications in drug discovery, toxicology and stem cell research. Trends Biotechnol. 2009;27(6):342–349. doi: 10.1016/j.tibtech.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith A. Screening for drug discovery: the leading question. Nature. 2002;418:453. doi: 10.1038/418453a. [DOI] [PubMed] [Google Scholar]
- 8.Trevino V., Falciani F., Barrera-Saldaña H.A. DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol. Med. 2007;13(9–10):527–541. doi: 10.2119/2006-00107.Trevino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L., Li P.C.H. Microfluidic DNA microarray analysis: a review. Anal. Chim. Acta. 2011;687:12–27. doi: 10.1016/j.aca.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 10.Heller M.J. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4(1):129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 11.Falsey J.R., Renil M., Park S., Li S., Lam K.S. Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjug. Chem. 2001;12(3):346–353. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- 12.Frank R. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48(42):9217–9232. [Google Scholar]
- 13.MacBeath G., Schreiber S.L. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 14.MacBeath G., Koehler A.N., Schreiber S.L. Printing Small Molecules as Microarrays and Detecting Protein−Ligand Interactions en Masse. J. Am. Chem. Soc. 1999;121(34):7967–7968. [Google Scholar]
- 15.Blackwell H.E. Hitting the SPOT: small-molecule macroarrays advance combinatorial synthesis. Curr. Opin. Chem. Biol. 2006;10(3):203–212. doi: 10.1016/j.cbpa.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 16.Manimala J.C., Roach T.A., Li Z., Gildersleeve J.C. High-throughput carbohydrate microarray analysis of 24 lectins. Angew. Chem. 2006;118(22):3689–3692. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 17.Ban L., Mrksich M. On-Chip synthesis and label-free assays of oligosaccharide arrays. Angew. Chem. Int. Ed. 2008;47(18):3396–3399. doi: 10.1002/anie.200704998. [DOI] [PubMed] [Google Scholar]
- 18.Anderson D.G., Levenberg S., Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004;22:863. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 19.Yang J., Rose F.R.A.J., Gadegaard N., Alexander M.R. A high-throughput assay of cell-surface interactions using topographical and chemical gradients. Adv. Mater. 2009;21(3):300–304. [Google Scholar]
- 20.Ankam S., Teo B.K.K., Kukumberg M., Yim E.K.F. High throughput screening to investigate the interaction of stem cells with their extracellular microenvironment. Organogenesis. 2013;9(3):128–142. doi: 10.4161/org.25425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flaim C.J., Chien S., Bhatia S.N. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2:119. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 22.Hilpert K., Elliott M., Jenssen H., Kindrachuk J., Fjell C.D., Körner J., Winkler D.F.H., Weaver L.L., Henklein P., Ulrich A.S., Chiang S.H.Y., Farmer S.W., Pante N., Volkmer R., Hancock R.E.W. Screening and characterization of surface-tethered cationic peptides for antimicrobial activity. Chem. Biol. 2009;16(1):58–69. doi: 10.1016/j.chembiol.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Popova A.A., Schillo S.M., Demir K., Ueda E., Nesterov-Mueller A., Levkin P.A. Droplet-array (DA) sandwich chip: a versatile platform for high-throughput cell screening based on superhydrophobic–superhydrophilic micropatterning. Adv. Mater. 2015;27(35):5217–5222. doi: 10.1002/adma.201502115. [DOI] [PubMed] [Google Scholar]
- 24.Jogia G.E., Tronser T., Popova A.A., Levkin P.A. Droplet microarray based on superhydrophobic-superhydrophilic patterns for single cell analysis. Microarrays. 2016;5(4):28. doi: 10.3390/microarrays5040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popova A.A., Demir K., Hartanto T.G., Schmitt E., Levkin P.A. Droplet-microarray on superhydrophobic-superhydrophilic patterns for high-throughput live cell screenings. RSC Adv. 2016;6(44):38263–38276. [Google Scholar]
- 26.Popova A.A., Depew C., Permana K.M., Trubitsyn A., Peravali R., Ordiano J.A.G., Reischl M., Levkin P.A. Evaluation of the droplet-microarray platform for high-throughput screening of suspension cells. SLAS Technol. 2017;22(2):163–175. doi: 10.1177/2211068216677204. [DOI] [PubMed] [Google Scholar]
- 27.Popova A.A., Tronser T., Demir K., Haitz P., Kuodyte K., Starkuviene V., Wajda P., Levkin P.A. Facile one step formation and screening of tumor spheroids using droplet-microarray platform. Small. 2019;15:1901299. doi: 10.1002/smll.201901299. [DOI] [PubMed] [Google Scholar]
- 28.Popova A.A., Marcato D., Peravali R., Wehl I., Schepers U., Levkin P.A. Fish-microarray: a miniaturized platform for single-embryo high-throughput screenings. Adv. Funct. Mater. 2018;28:1704386. [Google Scholar]
- 29.Tronser T., Popova A.A., Levkin P.A. Miniaturized platform for high-throughput screening of stem cells. Curr. Opin. Biotechnol. 2017;46(Supplement C):141–149. doi: 10.1016/j.copbio.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Tronser T., Popova A.A., Jaggy M., Bastmeyer M., Levkin P.A. Droplet microarray based on patterned superhydrophobic surfaces prevents stem cell differentiation and enables high-throughput stem cell screening. Adv. Healthc. Mater. 2017;6:1700622. doi: 10.1002/adhm.201700622. [DOI] [PubMed] [Google Scholar]
- 31.Ueda E., Feng W., Levkin P.A. Superhydrophilic–superhydrophobic patterned surfaces as high-density cell microarrays: optimization of reverse transfection. Adv. Healthc. Mater. 2016;5(20):2646–2654. doi: 10.1002/adhm.201600518. [DOI] [PubMed] [Google Scholar]
- 32.Benz M., Molla M.R., Böser A., Rosenfeld A., Levkin P.A. Marrying chemistry with biology by combining on-chip solution-based combinatorial synthesis and cellular screening. Nat. Commun. 2019;10(1):2879. doi: 10.1038/s41467-019-10685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng W., Li L., Ueda E., Li J., Heißler S., Welle A., Trapp O., Levkin P.A. Surface patterning via thiol-yne click chemistry: an extremely fast and versatile approach to superhydrophilic-superhydrophobic micropatterns. Adv. Mater. Interfaces. 2014;1(7):1400269. [Google Scholar]
- 34.Parry K.L., Shard A.G., Short R.D., White R.G., Whittle J.D., Wright A. ARXPS characterisation of plasma polymerised surface chemical gradients. Surf. Interface Anal. 2006;38(11):1497–1504. [Google Scholar]
- 35.Scofield J.H. Hartree-slater subshell photoionization cross-sections at 1254 and 1487 eV, journal of electron spectroscopy and related phenomena. Surf. Interface Anal. 1976;8(2):129–137. [Google Scholar]
- 36.Ueda E., Geyer F.L., Nedashkivska V., Levkin P.A. DropletMicroarray: facile formation of arrays of microdroplets and hydrogel micropads for cell screening applications. Lab Chip. 2012;12(24):5218–5224. doi: 10.1039/c2lc40921f. [DOI] [PubMed] [Google Scholar]
- 37.Klán P., Šolomek T., Bochet C.G., Blanc A., Givens R., Rubina M., Popik V., Kostikov A., Wirz J. Photoremovable protecting groups in chemistry and biology: reaction mechanisms and efficacy. Chem. Rev. 2013;113(1):119–191. doi: 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes C.P. Model studies for new o-nitrobenzyl photolabile Linkers: substituent effects on the rates of photochemical cleavage. J. Org. Chem. 1997;62(8):2370–2380. doi: 10.1021/jo961602x. [DOI] [PubMed] [Google Scholar]
- 39.Usui K., Kikuchi T., Tomizaki K.-y., Kakiyama T., Mihara H. A novel array format for monitoring cellular uptake using a photo-cleavable linker for peptide release. Chem. Commun. 2013;49(57):6394–6396. doi: 10.1039/c3cc41632a. [DOI] [PubMed] [Google Scholar]
- 40.Kakiyama T., Usui K., Tomizaki K.-y., Mie M., Kobatake E., Mihara H. A peptide release system using a photo-cleavable linker in a cell array format for cell-toxicity analysis. Polym. J. 2013;45(5):535–539. [Google Scholar]
- 41.Sarin V.K., Kent S.B.H., Tam J.P., Merrifield R.B. Quantitative monitoring of solid-phase peptide synthesis by the ninhydrin reaction. Anal. Biochem. 1981;117(1):147–157. doi: 10.1016/0003-2697(81)90704-1. [DOI] [PubMed] [Google Scholar]
- 42.Ast T., Heine N., Germeroth L., Schneider-Mergener J., Wenschuh H. Efficient assembly of peptomers on continuous surfaces. Tetrahedron Lett. 1999;40(23):4317–4318. [Google Scholar]
- 43.Hannah R.W. Groups containing N=O bonds, or Si, P, S, or halogen atoms. In: Mayo D.W., Miller F.A., Hannah R.W., editors. Course Notes on the Interpretation of Infrared and Raman Spectra. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2004. pp. 217–246. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.