Abstract
The topic of self-assembled structures based on elastin-like recombinamers (ELRs, i.e., elastin-like polymers recombinantly bio-produced) has released a noticeable amount of references in the last few years. Most of them are intended for biomedical applications. In this review, a complete revision of the bibliography is carried out. Initially, the self-assembly (SA) concept is considered from a general point of view, and then ELRs are described and characterized based on their intrinsic disorder. A classification of the different self-assembled ELR-based structures is proposed based on their morphologies, paying special attention to their tentative modeling. The impact of the mechanism of SA on these biomaterials is analyzed. Finally, the implications of ELR SA in biological systems are considered.
Keywords: Self-assembled structures, Intrinsically disordered proteins, Biomedical applications
Graphical abstract
1. Introduction
Natural materials in living systems are frequently multifunctional and dynamic. Their functionality and complexity have served as inspiration for researchers to mimic their properties, increasing the number of their potential uses and improving their performance. The increasing knowledge from both fundamental studies and fabrication methods – such as self-assembly (SA) – has rendered sophisticated systems able to mimic some physicochemical properties of such natural components [1].
The SA paradigm in biology has matured scientifically over the past two decades with the emergence of concepts such as intrinsic disorder. The flexibility of these apparently disordered systems implies a lack of constraints on many or all the degrees of freedom involved to form hierarchically structured assemblies or conformational selections that are far from the traditional structure-function concept [2]. The discovery of important protein molecules and domains that, despite being incompletely structured or completely disordered in solution, remained perfectly functional is contributing to the unraveling of one of the most intriguing topics in biology [3]. This breaking theory, contravening what until now was a dogma in biology, can bring new approaches to the field of biomaterials.
Elastic proteins such as tropoelastin (the soluble precursor of elastin), insect resilin, wheat gluten, and spider silks, among others, have provided much inspiration for the design of protein polymers since the sequence origins of disorder were readily identified during early work with these proteins [4].
Following the nature as an inspiration source, in the last 20 years, the topic of self-assembled structures based on elastin-like recombinamers (ELRs) has given rise to a significant amount of works. The goal of this review is to highlight the importance of these structures in the field of biomedical applications and provide a comprehensive overview of the bibliography.
In this review, we will start by considering the concept of SA in a broad sense, paying special attention to its properties and implications on a nanoscale. More specifically, throughout this article, we will focus on self-assembled structures based on ELRs. Along with the description and characteristics of ELRs, the interplay between order and intrinsic disorder in these molecules on a molecular scale is discussed.
Several self-assembled ELR-based morphologies (nanoparticles, fibers, hydrogels, and other exotic shapes and structures) are then discussed, suggesting a modeling of the formation mechanism and relevant parameters that affect the development and stability of the self-assembled structure. Some examples are included to illustrate how the intrinsic disorder in the ELR modulates this kind of structure.
Finally, applications and consequences of ELR SA in biological systems and processes are reviewed. In particular, biomineralization, preformed structural elements, drug delivery, tissue engineering, biosensors, and protein purification are considered.
2. Concept and characteristics of SA
From a general point of view, the concept of SA involves ‘autonomous organization of components into patterns or structures’ [5] that have a higher order than the isolated components [6].
SA processes are ubiquitous in nature and provide a strategy that is, in principle, applicable at all scales: on a planetary scale, weather phenomena; on a macroscopic scale, SA is found in systems consisting of macroscopic building blocks in the fields of, for example, robotics and manufacturing. Even netted systems implemented by computers or sensors interacting only through the flow of information [7], [8], [9] also fall into this category.
With regard to this macroscopic scale, and as an extension of three-dimensional (3D) printing systems, four-dimensional (4D) printing is defined as ‘the ability of an object to change its form and/or function after printing’, with the fourth dimension being represented by the transformation over time. Thus, printed structures become self-assembled materials that transform independently when the construct is dipped in water, heated, or light/electrically activated [10].
When considering SA on a nanoscale, self-assembled structures usually have sizes ranging from a few tens to hundreds of nanometers. This relative size is of the order of magnitude of quantum dots in nanoelectronics, ribosomes, or the length of the tobacco mosaic virus in biology and the ultraviolet wavelength in the electromagnetic spectrum. Obviously, these sizes are significant for the progress of nanotechnology.
The importance of SA on a nanoscale in chemistry and materials science should also be highlighted. As a further step beyond molecular chemistry, from the perspective of supramolecular chemistry, i.e., ‘chemistry beyond the molecule,’ Lehn [11] suggested that the concept of molecular SA corresponds to ‘pattern formation processes tending toward equilibrium.’ Because molecules larger than roughly 1000 atoms or aggregates cannot be synthesized in a bond-by-bond manner, SA provides one solution for arranging matter on larger scales [12], [13].
Atoms, molecules, and a broad range of structures can be used as potential building blocks with different chemical compositions, shapes, and functionalities. Thus, supramolecular SAs with morphologies such as bilayers, micelles, vesicles, tubular structures, liquid crystal phases, and Langmuir monolayers [14], [15], [16] are obtained.
The interactions found in SA structures are typically non-covalent (e.g., van der Waals forces, electrostatic and hydrophobic interactions, capillarity, π-π stacking, and hydrogen bonds) and act at a strictly local level. Thus, the structure builds itself, and its geometry and dimensionality are determined by the interactions between the building blocks [17], [18]. The strength of these non-covalent interactions is usually less energetic than that of covalent interactions by a factor of around 10. Reversibility, significant sensitivity to perturbations arising due to the external environment, and an inherent ability to self-heal and correct errors are some of the dynamic characteristics of self-assembled systems. Thus, these weak interactions rearrange and adapt to the landscape around them [19].
To achieve stability, these interactions must operate and be more favorable energetically (via cooperativity and synergism) than those that would lead to breakup of the structure [20], [21]. This brings thermodynamic aspects into play as it provides some clues about the characteristics that the individual units composing the self-assembled nanostructure should contribute to it.
As SA takes place in the absence of external forces, the process leads to a lower Gibbs free energy, thus meaning that SA structures are thermodynamically more stable than their individual components. Energy minimization drives the process, and the final structure is in thermodynamic equilibrium with its components [22]. As a result, the trade-off between enthalpy and entropy becomes a key aspect of the discussion [23]. Thus, rigidity and the presence of multiple contacts between the interacting surfaces of building blocks are noticeable characteristics of the molecules comprising the SA structure [24].
Whitesides and Grzybowski [5] suggested that two types of SA, namely a ‘dynamic’ and a ‘static’ SA system, can be considered depending on whether energy is dissipated for the SA structure or not. For instance, most folded globular proteins are formed by static SA, and oscillating chemical reactions [25], [26] and biological cells are some examples of dynamic SA. Halley and Winkler [6], however, disagree with this nomenclature and consider this latter dynamic category to correspond to the concept of ‘self-organization’ in biology. Some interesting and subtle differences between ‘SA’ and ‘self-organization’ have been discussed from the viewpoint of complexity [6], [27].
Whitesides and Grzybowski [5] also considered two further variants of SA, namely ‘templated’ SA, where the structures formed adapt to the boundaries or template in the environment, and ‘biological’ SA, where SA allows chemical reactions to be linked to cells. How ordered structures stem from disordered elements at the expense of energy dissipation is a key question in the domain of life [5].
One particularly interesting and novel application of self-assembled systems is the implementation of molecular motors, devices, and machines [28]. Indeed, Professor Feringa was awarded the Nobel Prize in 2016 ‘for the design and synthesis of molecular machines” mainly light and chemically driven [29], [30].
Despite the aforementioned fact, herein, we will focus our interest on ELR-based self-assembled structures. As most of these structures are intended for biomedical applications, most examples included correspond to this field.
3. Intrinsically disordered proteins and ELRs
3.1. Concept of intrinsically disordered proteins
The term intrinsically disordered proteins (IDPs), or intrinsically unstructured proteins, is a generic one used to refer to proteins lacking a cooperatively folded structure under native conditions, as opposed to globular proteins characterized by the presence of structured and well-formed domains. IDPs challenge the traditional paradigm of protein structure in which protein function depends on a fixed 3D structure [31].
Regions of disorder are very common in eukaryotic proteins. Indeed, half of the eukaryotic proteome contains small sequences of 40 or more amino acids that are predicted to be disordered under physiological conditions [32], [33]. The abundance and functional significance of IDPs remained largely unknown until the mid-1990s [32], [34]. This fact can be partly justified by the lack of experimental and computational tools to characterize disorder in this type of protein and intrinsically disordered regions (IDRs). Even today, the characterization of disorder and modeling of assemblies remain a significant challenge and must still be verified by several techniques [32], [35]. The structural characterization of IDPs is usually based on X-ray crystallography, when the interactions of IDPs with their physiological partners result in well-ordered structures, or nuclear magnetic resonance (NMR) spectroscopy, which has the additional advantage of providing insights into dynamic interactions, small-angle X-ray scattering (SAXS), single-molecule fluorescence energy transfer (smFRET), and computer simulations based on polymer physics [32], [36], [37], [38].
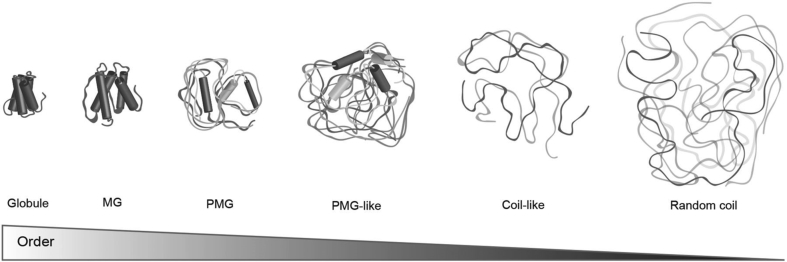
Despite the apparent lack of structured domains, ‘intrinsic disorder’ covers a spectrum of states ranging from fully unstructured or random coil–like to partially structured in the form of (pre-)molten globules [39] (Fig. 1). All these states are vital for fundamental cellular functions, including transcription, translation, cellular signal transduction, and regulatory processes [33]. Based on their functionality, disordered regions are classified into six classes, and a single protein may belong to different functional classes or contain several disordered regions. These classes are as follows: (1) entropic chains, which function without becoming structured but benefit directly from conformational disorder; (2) display sites, where the conformational flexibility of disordered protein regions facilitates posttranslational modifications (PTMs) by allowing easy access and recognition by effectors that mediate downstream outcomes upon binding; (3) chaperones, over half of IDRs are able to adapt and assist RNA and proteins to reach their functionally folded states; (4) effectors, which modify the activity of other proteins upon binding to them by way of competitive interactions or allosteric modulation (this disorder-to-order transition is also known as coupled folding and binding); (5) assemblers, where multiple binding partners bring disordered sequences together to promote the formation of higher order protein complexes; and (6) scavengers, which store and neutralize small ligands [35], [40], [41].
Fig. 1.
Conformational states for proteins range from fully unstructured or random-coil-like structures to partially structured in the form of a premolten globule (PMG) and molten globule (MG) to ordered forms. Proteins can transition between any of these states. Adapted with permission from the study by Staby et al [39]. Copyright Clearance Center, Inc. (‘CCC’).
In performing many of the aforementioned functions, in which IDPs form transient complexes with cellular targets, the interactions are highly specific but with low affinity. Moreover, the resulting complexes are often highly dynamic and short-lived, fluctuating rapidly between conformations [33]. This is because, upon binding, IDRs lose conformational freedom (i.e., entropy), which reduces the overall free energy of binding and leads to a weaker interaction [40]. A large decrease in conformational entropy, which uncouples binding strength from specificity, renders highly specific interactions reversible [42], [43], [44]. However, not all IDPs undergo folding transitions when performing their biological functions, and some disordered regions function as flexible unstructured linkers between globular and disordered interaction domains, whereas others remain disordered even after binding to their targets, forming ‘fuzzy’ complexes [32].
Common functional modules within IDRs include short linear motifs (SLiMs, LMs, or MiniMotifs), which usually have a length of 3–10 amino acids and can occur several times in the same protein [40]. Recent estimates suggest that the human proteome may contain more than 100,000 SLiMs located within IDRs [32]. The limited size of the binding region, and hence low affinity of the SLiM, is compensated by the multiplicity, thereby promoting interactions with greater avidity for partners and/or the recruitment of multiple factors. However, different motifs can overlap, thus resulting in the mutually exclusive binding of interaction partners [33], [40]. Preformed structural elements (PSEs), or molecular recognition features (MoRFs), are another type of peptide motifs within IDRs, which are typically linear in the protein sequence. These sequences typically have a length of 10–70 amino acids, with signs of residual structure before the final binding event, although they are not fully ordered even in their bound form and often contain significant conformational freedom. These MoRFs allow the physiologically effective action of IDPs by way of fast and specific, yet readily reversible, interactions with their partners [45], [46]. Moreover, it has been suggested that SLiMs and MoRFs are different states in the same continuum of binding mechanisms involving disordered regions [40].
The secret to these manifold functions, their inherent flexibility and plasticity (as they can bind to several targets and adopt different structures on different targets), seems to be determined by the presence of multiple well-conserved binding motifs, their low sequence complexity, and biased amino acid composition [32]. The unfolded state is ensured by their low mean hydrophobicity and high net charge, thus weakening the hydrophobic effects that drive folding of peptides into compact tertiary structures [47]. IDPs have a low proportion of the bulky hydrophobic amino acids (W, Y, F, C, I, L, and N) required to drive protein folding and to form a well-organized hydrophobic core but high proportions of charged and hydrophilic amino acids (P, E, K, S, and Q) [32], [35]. The charge balance and distribution of charged residues in the IDP sequence has a marked impact on the dimensions and degree of compaction of IDPs [48], [49]. Indeed, the distribution of these amino acids (proline, glutamine, acidic, basic) is usually not random, and they are often organized into well-discernible domains [35].
Pro-rich repetitive regions are a structural characteristic that is linked to the lack of structure in IDPs. Proline (P) is considered to be the most disorder-promoting residue (and is by far the most soluble amino acid at neutral pH), followed by glutamic acid (E) and serine (S) [50], [51]. One of the reasons why proline promotes disorder is because its cyclic structure renders a more rigid conformation than other amino acids because it does not contain backbone amide hydrogen atoms at physiological pH. As such, it is unable to form stabilizing hydrogen bonds in secondary structures and negatively modulate their propagation, when present [50], [51]. Proline in IDPs and IDRs has a high propensity for adopting non-classical conformations, such as the polyproline type II helix (PPII) (left-handed extended structures that contain three residues per turn and no internal hydrogen bonding) [52].
Proline also exerts pronounced effects on the backbone geometries of the residues preceding it. Thus, while increasing the number of proline in PPII conformations appears to rigidify IDPs, a high abundance of proline combined with favorable glycine contents, or with selective positioning of charged and/or hydrophobic residues, gives rise to different secondary structures [51].
Theillet et al. demonstrated that proline is one is the most disorder-promoting amino acid after comparing the compositions of proteins in four standard data sets, Swiss-Prot, PDB Select 25, surface residues, and DisProt [51].
In another work, Marsh and Forman-Kay [53] compiled Rh measurements and amino acid sequences for a sizeable set of IDPs to investigate the sequence determinants of compaction in these proteins compared with folded and chemically denatured proteins. To assess the sequence dependence of compaction, the Pearson correlation coefficient, raa, was calculated between the fractional content of each type of amino acid and the relative hydrodynamic radius (Rh) for each protein. The results showed that proline had the highest raa value and thus has the strongest association with more highly expanded proteins, suggesting that proline residues strongly correlated with increased hydrodynamic radii and that these are the dominant contributors to compaction of IDPs. The work of Rauscher et al. [54] demonstrates that elastin-like and amyloid-like peptides are separable based on backbone hydration and peptide-peptide hydrogen bonding. The analysis of their sequences revealed that there exists a threshold in proline and glycine composition above which amyloid formation is impeded and elastomeric properties become apparent. The fact that approximately two glycine residues are equivalent to one proline at this threshold confirms the role of proline as the primary determinant of elastin's properties. In the same line, the work of Muiznieks and Keeley [55] says that the maintenance of structural heterogeneity and dynamics of the elastin backbone is achieved in large part by a high proportional composition of both proline (14%) and glycine (35%) residues within hydrophobic sequences.
The previous inherent structural features of IDRs, their biophysical features (multivalency and flexibility), and other in vivo functional regulatory mechanisms (alternative splicing, which modulates the valency of binding sites and posttranslational modifications), as well as environmental factors (temperature, redox potential, and pH), minimize the entropic cost of IDPs from a dynamic equilibrium between different sets of conformational states separated by low energy barriers (flat energy landscape). These principles are extensively exploited in nature but can also be utilized in bioengineering, synthetic biology, and pharmaceutical applications. Conditional transitions between disordered and ordered states triggered by intrinsic and extrinsic factors allow proteins to switch between conformational states, thus enabling diverse protein functions [56].
3.2. ELRs: description and characteristics
Elastin is a key extracellular matrix (ECM) protein that is most abundant in tissues of many vertebrates where elasticity and resilience are of major importance such as blood vessels (50% of dry weight), elastic ligaments (70% of dry weight), lungs (30% of dry weight), and skin (2–4% of dry weight) [57], [58].
Natural elastin is an insoluble polymer synthesized as tropoelastin, its ≈70 kDa soluble precursor, and it is associated with a wide range of elastic peptide and protein sequences that exist in different lengths and with different compositions [58], [59]. The high insolubility of elastin limited its use to researchers for many years who looked for soluble derivatives with similar properties to those present in the natural protein. Recombinantly produced tropoelastin derivatives (ELRs) exhibited many of the properties intrinsic to the mature biopolymer, and they became a vast source of versatile building blocks for the manufacture of biomaterials with potential for diverse applications [60].
ELRs retain the lower critical solution temperature (LCST) phase behavior of natural elastin, undergoing a reversible phase transition (known as an inverse temperature transition [ITT]), above a characteristic transition temperature (Tt) [61], [62]. This ELR transition takes place only in aqueous solution, where the ELR chains are fully hydrated, mainly as a result of hydrophobic hydrations, below a certain Tt. This hydration is ordered into clathrate-like water structures surrounding the apolar moieties of the polymer. Upon heating above Tt, the ELRs undergo a first-order phase transition into polypeptide-rich (37% polymer by weight) and water-rich (63%) phases, with the ELR essentially losing the ordered water structures to form a phase-separated state. The ITT (or coacervation) is a reversible, entropically driven process with an associated latent heat (ΔHt) that combines the increase in solvent entropy by disruption of water structures with concomitant self-association and stabilization as a result of intramolecular hydrophobic contacts [62], [63]. The LCST behavior can be tuned in a precise manner that depends on the mean polarity of the recombinamer (mainly determined by the nature of the guest residue). As such, the Tt increases as the hydrophobicity decreases, and vice versa. In other words, ELRs can be designed to exploit their thermal responsiveness by water-induced chain dynamics alterations [62], [64], [65], [66]. However, other external stimuli, such as temperature, solution ionic strength [67], [68], pH [65], light [69], ELR concentration [70], or the presence of other cosolutes [71] or cosolvents [72], are also known to affect the coacervation process [63], [73].
Thus, ELRs can be defined as rubber-like, protein-based polymers inspired by the sequences present in natural mammalian elastin, which plays a role in the storage of elastic strain energy in a broad range of material properties and functional roles [74], [75]. The term ‘recombinamer’ was coined to highlight both the tailored oligomeric composition and method for producing these materials by recombinant DNA technologies [76].
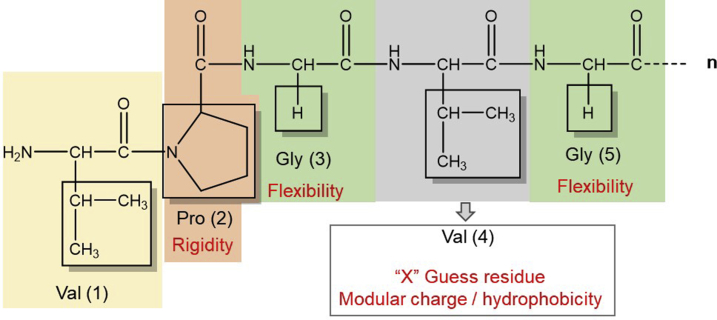
ELRs are based on several common features found in the wide range of elastin-based structures, specifically a pseudo-periodic portion of the hydrophobic domains of elastin, with several types of repeat motifs [77]. These domains mainly comprise the amino acid residues glycine (G), valine (V), and proline (P), commonly arranged as VPG, GV, and GGV [78]. The canonical polypentapeptide sequence (VPGVG)n is the most widely used and studied structural feature for the synthesis of different ELR compositions. Further examples of elastomeric series present in natural elastin that can be incorporated on ELRs include other pentapeptides (e.g., IPGVG), heptapeptides (e.g., LGAGGAG), and nonapeptides (e.g., LGAGGAGVL) [79], [80].
The fourth amino acid valine (Val) in the consensus repeat unit (VPGVG)n is known as the ‘guest residue’ and is represented by an ‘X.’ This position can host any amino acid except proline. Indeed, the choice of a different amino acid in that position can dramatically alter the physicochemical properties of the resulting recombinamer, thus making it a key parameter during ELR design (Fig. 2) [77], [81]. Other substitutions not limited to the guest residue can lead to drastic changes in the macromolecular properties, particularly mechanical behavior. For example, the single substitution of an alanine for a glycine residue in the third position of the VPAVG repeat sequence transforms the mechanical behavior of the ELR from elastomeric to plastic [63], [82].
Fig. 2.
Elastin-like recombinamer (ELR) consensus repeat unit VPGXG and amino acid residues responsible for their main physicochemical properties such as the proline/glycine content, number of tandem (n) repeats, and guest residue composition (‘X’).
The temperature-triggered SA of ELRs can be determined through the use of different physicochemical techniques. The most frequent are turbidity measurements [76], usually with optical density of 350 nm, where the Tt corresponds to the maximum of the first derivative of the turbidity versus temperature, or differential scanning calorimetry (DSC) [64], in which the difference in the amount of heat required to increase the temperature of a sample and reference is measured as a function of temperature. Other techniques can provide additional information about the formation of ordered ELR-based architectures as explained in section 3.1.
Recombinant DNA technology has contributed to obtain ELR-based materials with functions of particular technological and biomedical interest and versatile modifications to introduce complexity that can both mimic and affect in vivo cell-material interactions. But also, major contributions have been made in the well-controlled manipulation of materials structures over multiple length scales and the production of sequentially programmed biomaterial systems for targeted delivery of drugs, genes, and cells via de novo stimuli-responsive strategies [83].
Complex rearrangements (or block copolymers), with independent stimuli responsiveness, allow the construction of complex biomaterials. Some of them in the form of hydrogels, films, particles, fibers, surface coating, and porous scaffolds, with high added value, highlight the utility of ELRs in many biomedical fields [84]. This type of applications has been widely considered in Section 5.
3.3. ELRs as intrinsically disordered proteins
Elastin is an example of a protein for which the interplay between structure and dynamics is crucial to function and is closely related to disorder in its primary structure and solvate [85]. Elastin's essential functional property is the ability to contract reversibly after stretching by an increase in entropy [86], and it is widely accepted that the hydrophobic domains of elastin are responsible for their elastic recoil [87]. The emerging consensus regarding the structural disorder of elastin and its derivatives has been reached after decades of controversial, and seemly contradictory, structural studies [86], [88]. Since the initial theories proposed by Hoeve and Flory [89] and Aaron and Gosline [90], which proposed random coil conformations as the source of elastin's elasticity, the structure-function relationship of elastin has been a matter for ongoing interest and controversy. Given the difficulties encountered when studying the structure of natural elastin (due to problems in isolating pure extracts, its conformational heterogeneity and self-association), pioneering studies on the structural characterization of elastin came from Urry et al. [91], [92] and the study of synthetic elastin-like peptides, based on the most prevalent repeated sequences within the hydrophobic domains, such as (VPG)n, (VPGG)n, (VPGVG)n and (VAPGVG)n [91]. Urry et al. [92] proposed that these sequences adopt a type II β-turn structure, with adjacent proline and glycine residues forming the corners of the turn, stabilized by a hydrogen bond at the end-bond residues between the main chain (C–O) of residue i and the (N–H) of residue i+3. In this model, non-polar side chains are arranged to exclude water molecules [88], [92], [93]. The periodicity of these β-turns in the peptides would predict the formation of an ordered β-spiral consisting of consecutive type II β-turns, as modeled by just under three VPGVG motifs per spiral turn [80], [94]. However, subsequent structural analysis within similar elastomeric sequences of (GVPGV)7 and domains containing VPGVG and VAPGVG repeats determined that the β-spiral model is unstable and that only isolated and fluctuating β-turns are present [88], [95], [96].
The ability to recombinantly produce individual hydrophobic domains, or higher ELRs, has allowed predictions of disorder and conformational flexibility in which the formation of extended secondary structures is restricted in favor of transient and fluctuating local motifs, including β-turns, β-strands, and polyproline II (PPII) structures, to be confirmed [88], [97].
A common aspect that ELRs share with IDPs is tandem repeats with limited hydrophobicity and low sequence complexity. The consensus repeat sequence unit (VPGXG), which stems from the hydrophobic domains present in tropoelastin (the monomeric precursor of natural elastin), complies with this premise [98]. The structural disorder in ELRs is proposed to be a consequence of the high proline and glycine content in the ELR backbone, which has a compositional threshold of (2P+G) ≥ 0.60, thus meaning that the percentage of proline is a stronger determinant than the number of glycine residues [88]. Both amino acids help keep the polypeptide backbone of elastomers disordered and hydrated, although for opposite reasons. Thus, proline is too stiff to form secondary structures and impart structural rigidity on all length scales of ELRs because of its restricted phi dihedral angle, thereby preventing hydrogen-bonded backbone self-interactions. As such, proline is a breaker of common extended secondary structures (α-helix and β-sheet). Similarly, the absence of a side chain on glycine residues allows the ELR to sample a variety of chain conformations in the presence of water, which results in a higher backbone entropy, thereby also disfavoring the formation of ordered assemblies [54], [88], [98].
The entropic force that triggers the conformational shifts in most IDPs occurs as a result of ligand binding, scaffolding, or allosteric regulation after their interaction with one or several partners, whereas different triggers can drive ELRs to adopt a new conformation set of conformations when swollen in water [98].
Upon heating above an LCST higher than a threshold value, the ELR chains separate from the solvent, thus leveraging a combination of the gain in solvent entropy as a result of the release of solvent molecules and the gain in favorable interchain interactions. Changes in intensive solution parameters (typically temperature, but also pH or pressure), and/or in colligative properties, can drive concomitant collapse of the chain into amorphous coacervates (‘fuzzy aggregates’) and break the hydrophobic solvation [77], [78]. The entropic cost for restoring ordered solvent molecules around individual chains increases with increasing temperature. As such, the presence of charged residues and the solvation of proline-rich regions affect the temperature dependence. However, additional sequence features, such as distribution of the polymeric sequences into different block copolymer architectures that modulate the interplay between chain-solvent and chain-chain interactions [64], [78], have also been proposed to modulate ELR behavior.
ELRs remain disordered, even when aggregated, because they retain a high degree of water, thus allowing the hydrophobic domains in the ELR to self-associate and continually interpenetrate with one another to form dynamically heterogeneous structural elements without collapsing into stable, tightly packed structures that exclude solvent and limit conformation entropy [88], [99]. The fundamental balance between the forces underlying the intrinsic disorder of ELRs can result in more stable aggregates when the combined proline and glycine content drops below the compositional threshold. When this happens, the elastomeric properties become less apparent and the formation of amyloid fibrils is possible [54]. Although the proline/glycine content is an important variable as regard separating elastin-forming aggregates from their amyloid-forming counterparts, there are other important factors. For example, zwitterionic charge, the presence of highly hydrophilic or charged residues, the presence of perfect tandem repeats, and the distribution of the polymeric sequences into different block copolymeric architectures can also contribute to the formation of different and more stable polymer assemblies [47], [98]. The way in which these ELR-based sequences are both designed and combined has enormous potential as regard controlling the morphology of SA into supramolecular structures and in the design of novel biomaterials.
In summary, disorder in ELRs is considered at different levels, namely at the sequence level and at the level of aggregation and phase behavior [100]. An example for each level in which SA is modulated by the intrinsic disorder in the ELR is discussed in the following paragraph.
Several parameters are known to control the state of chain disorder at the sequence level. For example, Das and Pappu [101] have shown the importance of the amino acid tandem repeat mixing parameter. Wright and Conticello [102] obtained spherical micelles in a selection of copolymers with sequences (VPGX1G)n and (VPGX2G)n in which the guest residue X1 is significantly more hydrophobic than X2 (subsection 4.1). However, when two guest residues (in particular, alanine and valine) are ‘well mixed’ in the polymer (VPG [X1/X2]G)n, solubility is maintained throughout the entire polymer chain [70], [103].
4. Self-assembled ELR structures: morphologies and modeling
The following morphology-based categories have been considered when grouping the different self-assembled ELRs structures: nanoparticles, fibrillar structures, hydrogels, and, finally, other sophisticated structures.
4.1. Nanoparticles
Herein, we focus our attention on self-assembled nanostructures obtained using just ELRs (single component) or the combination of ELRs with other materials (multicomponent or hybrid nanoparticle).
4.1.1. Single-component nanoparticles
ELR-based corecombinamers comprise the combination of two or more individual and different ELR blocks. The simplest design giving rise to self-assembled nanoparticles is an amphiphilic diblock. Above the transition temperature for the hydrophobic block, hydrophobic and hydrophilic blocks arrange in the core and corona of the micelle, respectively; beyond the transition temperature for the hydrophilic block, micellar structures collapse into phase aggregates. Thus, the hydrophobic block determines the critical micelle temperature, whereas the hydrophilic corona establishes the micelle aggregation temperature [104].
Professor Conticello's group was the first to report [105] a self-assembled ELR nanostructure using an amphiphilic diblock based on a cationic block and a phenylalanine-containing block. The micelles (diameter of around 50 nm) formed initially were found to coalesce into cylindrical micellar aggregates upon increasing the temperature.
The individual block proportion is also a key parameter when designing an ELR-based corecombinamer. Wright and Conticello [102] designed an amphiphilic diblock (AB) or triblock (ABA or BAB) (A and B are the hydrophilic and hydrophobic blocks, respectively) using genetic engineering and studied the impact of the identity and sequence of the individual blocks (macromolecular architecture) on supramolecular SA. At this stage, we will only consider the diblock, as the self-assembled structures obtained with the triblock will be discussed later in subsection 4.3. At ambient temperature and under basic conditions, dilute aqueous solutions of the diblock polymer generate spherical nanoparticles with a hydrodynamic diameter of about 50–90 nm because of collapse of the hydrophobic block. At higher concentrations, different micellar phases are observed (worm-like micelles and micellar clusters) as a result of particle aggregation. Potential medical applications of these materials include drug delivery and soft-tissue augmentation.
Obviously, a deep insight into the effect of the individual blocks comprising the corecombinamer on nanostructure formation, and the different parameters contributing to its control, must be achieved. Dreher et al. [106] designed several linear amphiphilic diblock ELRs with different molecular weights and hydrophilic-to-hydrophobic ratios. When this ratio is between 1:2 and 2:1, the polymer self-assembles into monodisperse spherical micelles (hydrodynamic radius around 30–40 nm) triggered by a clinically relevant temperature (37–42 °C).
Recently, MacEwan et al. [107] introduced subtle changes into the block architecture of a diblock corecombinamer, including gradients between the hydrophilic and hydrophobic blocks. Although the micellar hydrodynamic radius is proportional to the molecular weight of the corecombinamer segments, they concluded that the size and shape of micellar nanostructures is modulated by the incorporation of this interface block.
Professor Rodríguez-Cabello's group has studied the ability to tune the self-assembled morphology using different block arrangements and lengths [108]. Three amphiphilic elastin-like corecombinamers were formed by combining two blocks: a glutamic acid–containing block (E-block) and a fixed-size block containing an alanine residue (A-block). Specifically, three architectures (E50A40, E100A40, and E50A40E50) were designed with a hydrophobic-to-hydrophilic length ratio higher than 1:2. Several thermally driven morphologies were observed at temperatures higher than the transition temperature of the hydrophobic block: spherical micelles for E50A40 and vesicles for E50A40E50 with hydrodynamic radii ranging from 75 to 100 nm.
To complete the previous study, several salts (NaCl, KCl, and Na2HPO4) and concentrations (for NaCl) were used to determine their effect on the morphology of the self-assembled nanostructures [67]. Although the morphology was not significantly modified, a noticeable size decrease was found in all cases along with hydrodynamic diameters ranging from 40 to 80 nm, thus suggesting more compact packed structures in the hydrophobic blocks. However, for a narrow range of low NaCl concentrations, two-state dependence was found, with the nanostructure switching abruptly to a lower diameter. Drug (both hydrophobic and hydrophilic) and gene delivery are some of the medical applications for these vesicles, which remain intact under different conditions.
A quantitative approach has been applied to achieve rational design of the genes encoding the ELR sequence and generate nanoparticles with tailored properties suitable for biomedical applications [104], [109]. The mathematical model proposed by Janib et al. [109] provides a relationship between the critical micelle temperature or the bulk transition temperature and the phase behavior of the hydrophobic and hydrophilic ELR blocks. A comprehensive library of monoblock and diblock ELRs was synthesized and characterized. The hydrophobic-to-hydrophilic ratio was 1:1 in all cases, and the effect of the molecular weight of the individual blocks in the co-ELR on the self-assembled structures was analyzed. This model led to the following conclusions: a minimum molecular weight of the individual ELRs (30 kDa) and a minimum length (48 pentamers) of the hydrophobic block are required for stable SA of nanoparticles; in addition, the amino acid sequence of the hydrophilic block tunes the bulk transition temperature, whereas the critical micelle temperature remains independent of this sequence.
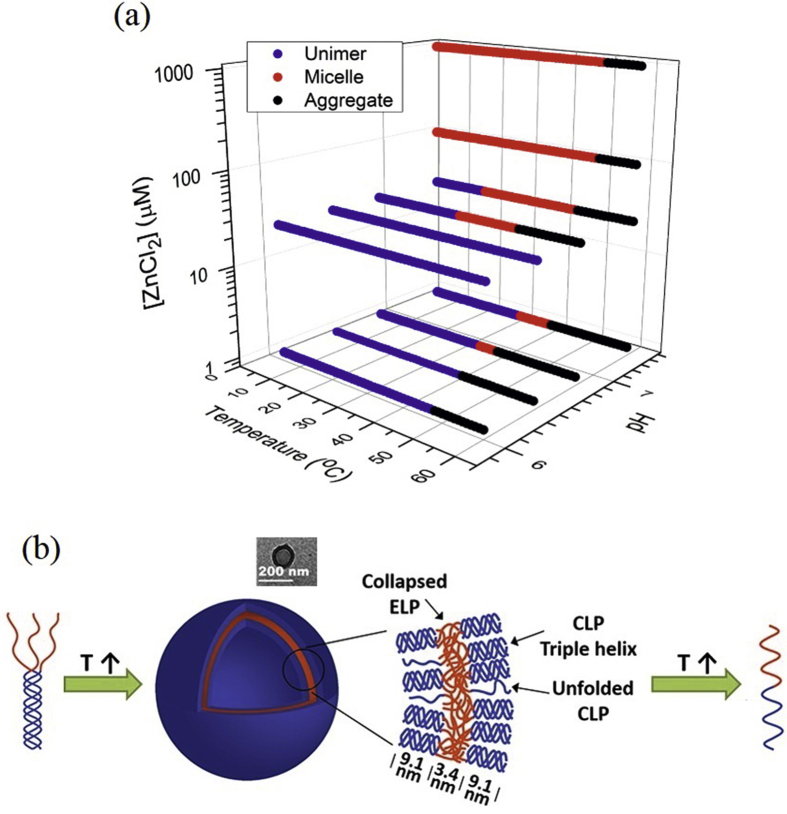
A close control of the main characteristics and properties of the self-assembled nanostructures allows them to be used as chemotherapeutic agents for the treatment of certain diseases. In particular, delivery of anticancer drugs based on thermally or pH-triggered SA has been achieved using the local mild hyperthermia (37–42 °C) and/or abnormal extracellular pH values associated with cancerous tissues [110]. For instance, spherical micelles with a hydrodynamic radius of around 30 nm have been reported by Callahan et al. [111]. In this case, the histidine-rich ELR copolymer was based on two blocks with noticeably different lower critical solution temperatures that self-assemble in response to temperature, pH, and cosolutes (e.g., Zn2+ in physiological concentrations). In Fig. 3 a, the phase diagram includes the combined effect of these three stimuli on the self-assembled structure.
Fig. 3.
Effect of different stimuli on the process of self-assembly in single (a) or hybrid (b) structures. (a) Phase diagram showing the combined effect of temperature, pH, and cosolutes. Data in the absence of ZnCl2 are plotted at 1 μM ZnCl2 to enable presentation on a log scale. Reproduced with permission from Callahan et al. [111]. Copyright American Chemical Society 2012. (b) Schematic of the temperature effect on the ELR-peptide hybrid nanostructure prepared by click chemistry. A cryo-TEM image of the vesicle at 25 °C has been included in the inset. Adapted with permission from Luo and Kiick [112]. Copyright American Chemical Society 2015. CLP, collagen-like peptide; TEM, transmission electron microscopy.
4.1.2. Hybrid or multicomponent nanoparticles
The level of complexity achieved with single-component self-assembled nanostructures still differs markedly from structures found in nature, where multiple-block self-assembled structures are common. An excellent review of multicomponent self-assembled systems comprising protein- and peptide-based building blocks has been published recently by Okesola and Mata [21]. They proposed a classification based on the type of interaction between the building blocks forming the structure. A schematic representation has been included for this quite interesting perspective in Scheme 1. Restricting our interest exclusively to ELRs, three categories are considered in this review for implementing multicomponent SA: systems based on chemical (covalent) conjugation, genetic engineering, and non-specific supramolecular interactions.
-
i)
Chemical conjugation
Scheme 1.
Several self-assembly strategies based on the interactions between the building blocks of the structure for hybrid (consisting of proteins and peptides) self-assembled nanostructures. A and B stand for the intrinsic properties of the building blocks forming the structure, and C is the synergistic property emerging from their interaction. Reproduced from the study by Okesola and Mata [21]. Copyright American Chemical Society 2018.
Luo and Kiick [112] used click chemistry with Cu to join together a short alkyne-functionalized ELR and a short azide-functionalized synthetic collagen-like peptide (CLP). The peptide sequence was selected to exhibit a melting temperature above 37 °C, thus allowing the formation of a stable triple helix at physiological temperature. The ELR-CLP SA gives rise to vesicles (average diameter and wall thickness of around 80–100 and 22 nm, respectively, Fig. 3 b) that show a thermoresponsive behavior at physiological temperatures. However, at elevated temperatures (for instance, 80 °C), the vesicles redissolve. Non-covalent formation of the CLP triple helix anchors the ELR and reduces and tunes its transition temperature.
In an effort to achieve a roadmap for the rational design of drug-loaded nanoparticles to reduce the problems associated with their injection into systemic circulation, Professor Chilkoti's group [113], [114] designed a compound comprising a chimeric polypeptide that includes both a hydrophilic ELR segment and a short, cysteine-rich segment. Maleimide derivatives of small molecules with a wide range of hydrophobicity were subsequently covalently coupled to the ELR via the thiol groups in this short segment. If the ELR and the small molecule have sufficiently different polarities, SA occurs in aqueous solution, thus giving rise to mainly spherical nanoparticles (diameter ranging from 60 to 100 nm). It is, therefore, a side-specific attachment–triggered SA in which several types of hydrophobic small molecules are possible candidates, whose transition temperature occurs between 38 and 42 °C. This strategy was used to conjugate several, mainly anticancer drugs (e.g., doxorubicin) to the polypeptide [113], [115]. Multifunctional nanomedicines with several drug molecules per ELR chain were achieved, thus increasing the effectiveness of the drug.
Thanks to the potential of ELR-based nanodevices, anticancer therapies based on several approaches (such as nanoparticles incorporating cell-penetrating sequences or tumor-homing ligands) are available. The cell-penetrating peptides (CPPs) [116], [117] approach facilitates the incorporation of several anticancer drugs into the cancer cells of solid tumors. These peptides are short molecules that are internalized via mechanisms initiated by electrostatic interactions and hydrogen bonding between the peptide and some components of the cell membrane [118]. Walker et al. [119] conjugated three different CPPs and the anticancer drug doxorubicin to an ELR. The peptides were incorporated by modification of the N-terminus of the ELR, and a terminal cysteine residue on the C-terminus was used to conjugate the drug.
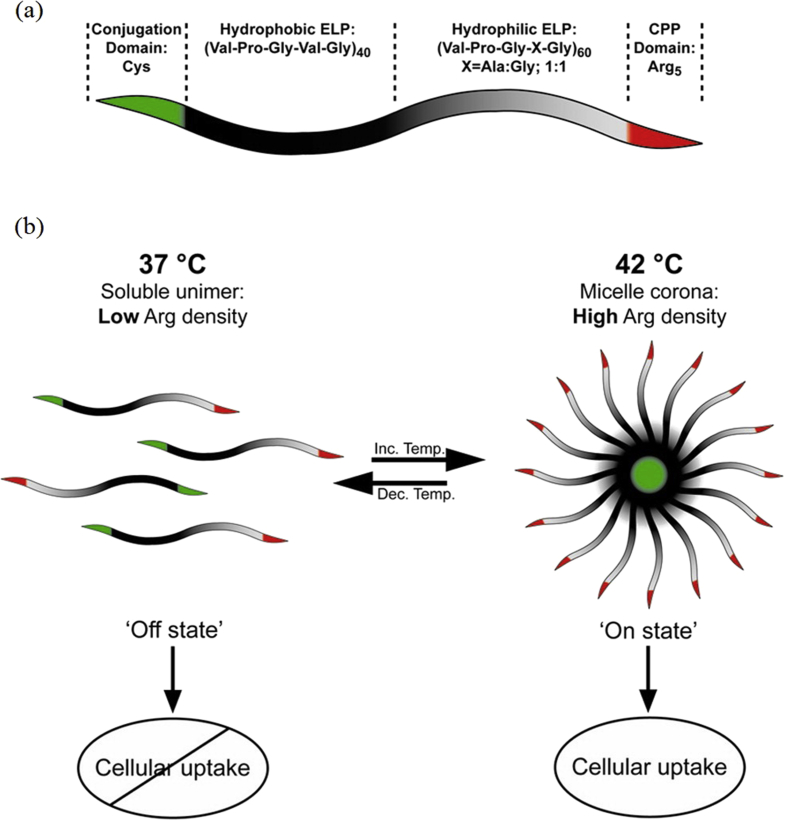
It has been shown that at least six arginine residues are required in the peptide to achieve significant cell uptake. MacEwan and Chilkoti [120] have reported a ‘smart’ dynamic system including a digital ‘off/on’ control switch for the CPP involving modulation of the local density of Arg residues. Assuming the hypothesis that the key factor for internalization is said to be density, five Arg residues have been disposed in the hydrophilic terminus of an amphiphilic corecombinamer (Fig. 4(a)). Special attention was paid to the precise control and tuning of the hydrophobic-to-hydrophilic segment ratio to ensure that this ELR is capable of temperature-triggered SA into micellar nanostructures (hydrodynamic radius of around 20 nm) in the temperature range 39–42 °C. Thus, a thermal stimulus was selected to modulate the local Arg density and allow SA to occur within the tumor. As can be seen in Fig. 4 (b), these ELRs are soluble unimers at 37 °C, exhibiting a low Arg density (‘off’ state). At 42 °C, SA occurs, the interfacial density of Arg residues in the corona increases, and the threshold is exceeded, thus leading to cellular uptake of the micelles (‘on’ state).
Fig. 4.
(a) Scheme of the diblock corecombinamer, including the conjugation and Arg5 domains and (b) implementation of the ‘off’ and ‘on’ state below (37 °C) and above (42 °C) the critical micelle temperature, respectively. Reproduced with permission from MacEwan and Chilkoti [120]. Copyright American Chemical Society 2012. CPP, cell-penetrating peptide.
In the field of tumor targeting, spherical nanocarriers (hydrodynamic radius of around 25–30 nm) have been reported by Hu et al. [121]. In this work, an ELR was genetically fused at its C- and N-termini with a segment including cysteine ((Gly-Gly-Cys)8) and a tumor-targeting peptide F3, respectively. This construct allows for site-specific conjugation of anticancer drugs, for example, doxorubicin.
-
ii)
Genetically encoded hybrid ELRs
This strategy provides a solution at the gene level to obtain hybrid self-assembled nanostructures. Several examples are summarized paragraphs.
Ghoorchian et al. [122] have designed and biosynthesized a novel ELR consisting of a negatively charged peptide of 27 amino acids incorporated at the C-terminal of an ELR with the sequence (GVGVP)40. Above its transition temperature, the foldon domain folds and stable three-armed star-shaped ELR micelles are formed under low salt (NaCl) conditions (diameters in the range 20–30 nm); this size is among the smallest reported for ELR-based particles. The size of the micelles is modulated by salt concentration.
A further example involves the use of the synergetic interaction between an ELR and the viral capsid protein from the cowpea chlorotic mottle virus [123] to form self-assembled nanostructures via two mechanisms: one induced by the thermoresponsive behavior of the ELR (diameter of 18 nm) and the other by pH changes (diameter of 28 nm) under conditions where the ELR does not self-assemble (Fig. 5). While the nanostructure architecture is determined by the virus capsid protein, the ELR phase transition triggers the assembly. One potential application of this system is switchable enzyme encapsulation to control its activity.
Fig. 5.
Two virus-like self-assembled hybrid nanostructures with different sizes triggered by pH and temperature. Reproduced from the study by van Eldijk [123].
McDaniel et al. [124] have obtained cylindrical micelles in highly asymmetric amphiphilic ELRs by incorporating an extremely high hydrophilic weight fraction. Small-angle neutron scattering gave an estimated core radius of 21–23 nm and a length of around 170 nm. All the ELRs consisted of a (VPGAG)n sequence (n = 40, 80, 120) genetically fused to an extremely short, low-molecular-weight domain based on sequences (XGG)8 and (XGy)8, where X represents a hydrophobic amino acid and y is the number of glycine spacers (y = 0, 1, 2) controlling the specific interactions between amino acids. Along with the hydrophobicity of the amino acid X (X = Tyr, Phe, Trp, Gly), the number of glycine spacers is responsible for SA of these ELRs. Because Tyr, Phe, and Trp include aromatic groups, π-π stacking may play a significant role in the hydrophobic interactions in the core of asymmetric amphiphiles.
Nanostructures with different morphologies were obtained when a hybrid protein polymer was produced by fusing a high-molecular-weight ELR (with alanine as a guest residue) with a single-chain antibody (anti-CD20 scFv; Aluri et al. [125]) or an amphipathic peptide (L4F; Pastuszka et al. [126]) at the N-terminus of the ELR. At physiological temperatures, antibody-mediated worm-like nanostructures (length of around 50 nm) are observed [125], whereas unilamellar vesicles (radius and lamellar thickness of 49 and 8 nm, respectively) are found when the peptide is used [126]. In both cases, the transition temperature of the construct is lower than that of the isolated ELR. As therapeutic applications, nanoworms and vesicles can be used for lymphoma therapies [125] and as a potential treatment for hepatic fibrosis [126], respectively.
Finally, the combination of ELRs with fibrous protein domains has also been studied. In an article by Xia et al. [127], silk sequences were alternated with the ELR block with varying silk-to-elastin block ratios in a monomer repeat. Micellar structures were formed in dilute aqueous solution because of the silk domains. Both the assembly ability and average micellar radius (a few tens of nanometers) depend on the silk-to-elastin ratio. When the temperature exceeds the ELR transition temperature, a hierarchical organization of micelles results in spherical nanoparticles (average hydrodynamic radius of around 220 nm).
-
iii)
Non-specific supramolecular interactions
Several interactions, including electrostatic, hydrophobic, π-π stacking, hydrogen bonds, and van der Waals forces, fall within this category [21]. As an example of a dynamic self-assembled system (according to the classification of Whitesides and Grzybowski [5]), these types of interactions between ELRs and peptides have been used by Inostroza-Brito et al. [128] with the following experimental result: if aqueous solutions of the peptide and ELR are mixed above the transition temperature of the ELR, a diffusion reaction mechanism is triggered by coassembly, thus resulting in the formation of a closed, multilayered membrane. The synergistic interaction between the ELR and the peptide modulates the ELR conformational change. Instabilities ‘switch on’ a membrane morphogenesis, thus generating a complex tubular 3D network with dynamic properties, for instance self-healing. Several synthetic and natural exogenous cross-linking agents have been used to stabilize and improve the mechanical properties of these membranes (Inostroza-Brito et al. [129]). Specifically, genipin appears to be a promising alternative that does not compromise biocompatibility.
4.2. Fibrillar structures
ELRs and elastin share the same supramolecular structures [130]. Thus, poly (VPGVG) forms filaments (diameter of around 4–5 nm) above its transition temperature. These fibers are modeled according to the β-spiral model [131], with these spirals combining to form twisted filaments with multiple strands. The Gly-Gly sequence in ELRs such as (VGGVG) [132] or (VGGLG) [133] also gives rise to fibrils organized into filaments.
Assembly into amyloid-like fibrils comprising extended β-sheet structures has also been reported [134]. The amino acid sequence is one of the key factors determining the formation of these fibers. In particular, the (XGGZG) motif, where X and Z can be V, A, or L, has been identified in the sequence [135]. Moreover, fiber formation requires a minimum sequence of 15 amino acids. Thus, while poly (VGGVG) self-assembles into intertwined and elongated fibers, amyloid fibers are found for (VGGVG)3 [136]. Rauscher et al. [54] have shown that the combined ratio of proline-glycine residues is a key factor promoting SA into amyloid fibers. Environmental conditions have also been linked to the formation of amyloid fibrils. For instance, as regard solvent, trifluoroethanol has been shown to be able to induce the formation of this type of fiber [137], [138]. In some cases, the presence of surfaces also affects the growth kinetics of the ELRs nanofibers (both fibrillary and amyloid-like structures) and reinforces their formation because of properties such as surface charge and hydrophobicity [139], [140].
The use of recombinamers based on multiple segments results in an increase in complexity. In the case of diblocks, beaded fibers are obtained in aqueous solutions of a doubly hydrophobic polypeptide including (VPGXG) and (VGGVG) [141].
ELR fibers have been reported recently for biomineralization assisted by boundary SA [142]. In this work, Li et al. [142] provided a deeper insight into the mechanism of biomineralization by way of a detailed study of the SA process used to obtain a bone-like ELR biomaterial. ELRs self-assemble into β-spirals [62], and in the presence of minerals, the interstitial nanocompartments between these β-spirals may serve for mineral deposition, inducing crystal growth. The authors tentatively suggest that capillarity might be the driving force for intrafibrillar mineralization in ELR fibrils. The biological implications of this study will be described in subsection 5.1.
4.3. Hydrogels
Although physical interactions are weak individually, their cooperative and collective action gives rise to ELR hydrogel networks, usually resulting in an enhancement of both the sol-to-gel transition and reversibility in response to environmental stimuli [143]. Ionic interactions, the presence of amphiphilic blocks, and the introduction of SA motifs in the polymer are some of the strategies used to generate physical cross-linking in ELRs.
Electrostatic interactions are known to occur between oppositely charged amino acids located at the guest position of the basic pentamer ELR segment. Specifically, Zhang et al. [144] have reported the formation of complementary ionic bonds between glutamic and lysine side chains. Moreover, when ELRs present periodic cysteine residues, intermolecular disulfide cross-links are formed and a hydrogel is obtained [145].
The folding of the hydrophobic blocks gives rise to aggregation of the amphiphilic blocks in ELRs. The opposite polarity of these hydrophilic and hydrophobic blocks causes repulsion between them, thus resulting in local segregation and the formation of periodic nanostructures [146], [147]. As a design strategy, the incorporation of longer sequences with several blocks of alternating polarity [148] has been proposed to improve hydrogel stability. As an example of this solution, an ELR triblock based on a hydrophobic/hydrophilic/hydrophobic structure was reported by Wu et al. [149]. Above the ELR transition temperature, the hydrophobic segments aggregate, resulting in a hydrogel stabilized by physical cross-links. Interestingly, nanotextured and reversible hydrogels have been also obtained using concentrated aqueous solutions of triblock (BAB) copolymers [102], where B corresponds to the hydrophobic domain that associates into micellar aggregates, thus acting as virtual cross-linking within the matrix.
Properties of the hydrogel can be tuned by the change of the residues in the ELR sequence. For instance, when the fourth residue is changed, transition temperature changes. As for the third position, if the alanine residue is substituted by the consensus glycine residue, mechanical properties are modified to obtain an elastic-mimetic polypeptide, forming an elastic hydrogel [150].
Different self-assembled morphologies can be obtained as a function of the polymer concentration in aqueous solution [151], with the spherical micelles found at low concentration (average hydrodynamic diameter of 100–200 nm) changing into a lyotropic gel as the concentration increases.
An approach based on the incorporation of functional motifs into self-assembled ELRs has recently been suggested. In particular, silk-like [152] or leucine zipper [153] motifs have been used.
Silk elastin–like recombinant polymers consist of tandemly repeated silk-like blocks (GAGAGS) [152] and elastin-like blocks (GVGVP). The first block plays a fundamental role in tuning the SA characteristics of these materials in aqueous solutions [127]. While the crystallization of the silk-like bloks in β-sheets favors the formation of a robust material, elastin-like blocks improve its flexibility and water solubility. The coexistence of silk-like and elastin-like blocks results in a two-step gelation process when heated beyond the transition temperature, with the transition of the ELR occurring first, followed by annealing of the silk sequences [154], with the silk domains crystallizing into β-sheet structures.
As for the leucine zipper motif, it is characterized by a heptad periodicity usually designated ‘abcdefg’ [153], where in ‘a’ and ‘d’ positions are located hydrophobic residues, and in ‘e’ and ‘g’ positions charged residues [155], [156]. Hydrophobic and ionic interactions between charged residues in the zipper motifs give rise to coiled-coil dimers [157], [158]. A triblock architecture in which leucine zippers are embedded at both edges has been typically proposed [153]. Depending on the zipper status (folded or denatured), changes in pH or temperature trigger reversibility in the ELR physical hydrogel. The biocompatibility of these hydrogels has recently been studied [159], thus paving the way for biomedical applications.
4.4. Sophisticated complex architectures
A wide variety of exotic structures were reported, diverse from the previously considered.
-
•
Hollow spheres [160] with diameter sizes from 100 to 1000 nm. On the surface of these spheres, functional groups are available and ready for additional modifications. Controlled release and outstanding cell viability with high pDNA loading efficiency are some of its advantageous applications.
-
•
Multilayer assemblies obtained by the layer-by-layer approach [161] provided nanoscale coatings of surfaces based on the conjugation of an ELR with polyethyleneimine and polyacrylic acid. When cellular cultures are accomplished on these coated surfaces, noticeable differences in some cell properties (such as cytoskeletal organization and focal adhesion) were observed.
-
•
A plasmonic matrix [162] consisting of polypeptides interfaced with gold nanorods, where chemotherapeutics drugs can be incorporated into the matrix. Later, this drug can be selectively released using some external stimuli, such as localized hyperthermic temperatures obtained by laser irradiation.
-
•
An elastin-mimetic dendrimer [163], where a peptide was attached to polyamidoamine dendrimers. Its main advantage is to obtain a unimolecular nanocarrier to implement a drug-delivery system.
-
•
Graphene-ELR nanocomposites [164], including a bioactive sequence (RGD, specifically) at the ELR N-terminus to favor cell binding and spreading.
-
•
ELR-pDNA polyplexes [165], whose size ranges from 100 to 200 nm, that are taken up by cells. As a potential application, intracellular delivery of therapeutic genes with high specific targeting was suggested.
5. ELR SA and biological systems
Nature offers the most complex and sophisticated collection of functional nanostructures that exists and has inspired many ways to derive functional materials with highly ordered hierarchical structures and excellent attributes from the sophisticated biological processes [166]. The structure-function analysis on various length scales of natural biopolymers is an inspiring example of advanced materials because they act as biological machines and biomolecular engines capable of interacting with their environment, adapting and responding to the stimuli that surround them [1].
The potential shown by the biopolymer elastin and its derivatives has been boosted and amplified by the use of recombinant DNA technologies and, nowadays, ELRs reach of even the most advanced polymer chemistry technologies. The recombinant synthesis of ELRs offers a total control of randomness in the polymer sequence and the possibility of incorporating biodegradability and bioactive sequences. These are some central features of these biomaterials besides their biocompatibility that are not found in other materials reported in the literature [167].
The degree of compaction of ELRs, which is related to their primary structure and conformational changes in response to small environmental stimuli, can be exploited for the rational design of a broad variety of useful structures for various in vivo and in vitro applications [168]. Recent manuscripts deal with the intrinsic disorder in ELRs with important applications in the biology of different natural processes and biomedical targets. A general overview of the different self-assembled ELRs structures and their biological/biomedical applications is summarized in Table 1. Some examples are discussed in the following paragraphs.
Table 1.
Summary of self-assembled ELRs structures considered in this review and their applications.
| Morphology | Proposed applications | References |
|---|---|---|
| Nanoparticles | Drug and gene delivery | [67] |
| Encapsulation of small-molecule substrates | [102] | |
| Drug delivery, protein separation, biosensors, and tissue engineering | [104] | |
| Encapsulation, delivery, and release applications | [105] | |
| Drug targeting in clinical applications of hyperthermia | [106] | |
| Biomedical and industrial applications | [107] | |
| Advanced nanocarriers: targeted or intracellular gene or drug delivery | [108] | |
| Potential diagnostic and therapeutic applications: biodegradable multimeric platform for the delivery of payloads, including radiological, chemotherapeutic, or protein-based agents | [109] | |
| Drug delivery (tumor targeting) | [111] | |
| Nanovaccines | [184] | |
| Vaccine carriers | [185] | |
| Drug delivery | [112] | |
| Drugs, imaging agents, and targeting moieties into multifunctional nanomedicines | [113] | |
| In vivo targeted range of hyperthermia | [114] | |
| Hyperthermia targeted chemotherapy of a variety of solid tumors | [115] | |
| Thermal targeting | [119] | |
| Targeting approach for drug delivery in a wide range of cancer types | [120] | |
| Drug nanoparticles for targeted cancer therapy | [121] | |
| Targeted drug delivery | [122] | |
| Switchable enzyme encapsulation | [123] | |
| Drug-loaded nanoparticles thermally targeted to solid tumors | [124] | |
| Lymphoma therapies | [125] | |
| Hepatic fibrosis | [126] | |
| Biomaterials for controlled drug delivery and biomedical engineering | [127] | |
| Delivery of protein therapeutics | [175] | |
| Fibers | Tissue engineering and models for cell studies or drug screening | [133] |
| Tissue engineering and drug-delivery systems | [141] | |
| Biomineralization assisted by boundary self-assembly | [142] | |
| Biomineralization | [172] | |
| Tissue engineering and wound healing | [193] | |
| Hydrogels | Drug delivery and tissue engineering. | [145] |
| Biomineralization | [170], [171] | |
| Robotics, microelectromechanical systems (bioinspired, muscle-like actuators), and tissue engineering | [190] | |
| Biomedicine and nanotechnology | [192] | |
| Smart systems for tissue engineering | [191] | |
| Biomedical devices | [102] | |
| In vivo applications with biomedical devices | [150] | |
| Scaffolds for biomedical uses, in particular, for regenerative medicine | [151] | |
| Tissue engineering (chondrocytic differentiation and cartilage matrix accumulation) | [205] | |
| Functional, biomimetic, artificial extracellular matrix, and cell niches | [154] | |
| Biosensors | [207] | |
| Tissue engineering applications | [159] | |
| Other structures | pDNA loading into hollow spheres | [160] |
| Drug vehicles for targeted therapy (tumor targeting) | [183] | |
| Tissue engineering (myoblast differentiation) | [204] | |
| Tissue engineering | [161], [164] | |
| Biomimetic coatings of biomaterials | [194] | |
| Polypeptide coatings | [195] | |
| Bone tissue engineering | [197] | |
| Protein purification | [208], [209], [210], [211], [212], [213], [214], [215] | |
| Drug delivery | [162], [163] | |
| Biosensors | [202] | |
| Intracellular delivery of therapeutic genes | [165] |
5.1. ELR SA and biomineralization
It has been reported that biomineralization is a highly complex process, even in the simplest organism, and that the proteins involved have very high levels of disorder. The flexible structure and ability of intrinsically disordered proteins to bind to a multitude of surfaces is crucial for biomineralization, either by favoring initial mineral precipitation, thereby reducing the activation energy barrier (prenucleation clusters), or providing surfaces that facilitate this initial mineral deposition (critical nucleus) [169].
Several studies have demonstrated that ELRs can be useful in the biomineralization process. For example, Li et al. [170] used two different microporous cross-linked ELR-based hydrogels (HSS3 and REDV) to template the biomineralization of hydroxyapatite crystals using a biomimetic polymer-induced liquid precursor (PILP) mineralization process. The PILP (polyaspartate in this study) should be well suited to a template with a specific size, morphology, and structure before crystallization and should be able to act as a process-directing agent for calcium and phosphate salts during infiltration and deposition within the matrix. ELR-based hydrogels help to drive the PILP process and provide clear advantages with respect to other macromolecules and synthetic polymers by allowing the formation of controlled morphologies and continuous nucleation on the surface, thus meaning that minerals are specifically deposited within the frameworks, preserving their microstructure with homogeneous but randomly oriented needle-like crystals. In both cases, after 28 days of mineralization, the mechanical properties were found to be of the same order of magnitude as those measured for bovine cortical bone (elastic modulus [E] of 20.3 ± 1.7 GPa), although the hardness (H) was significantly lower than that for bone (0.93 ± 0.07 GPa).
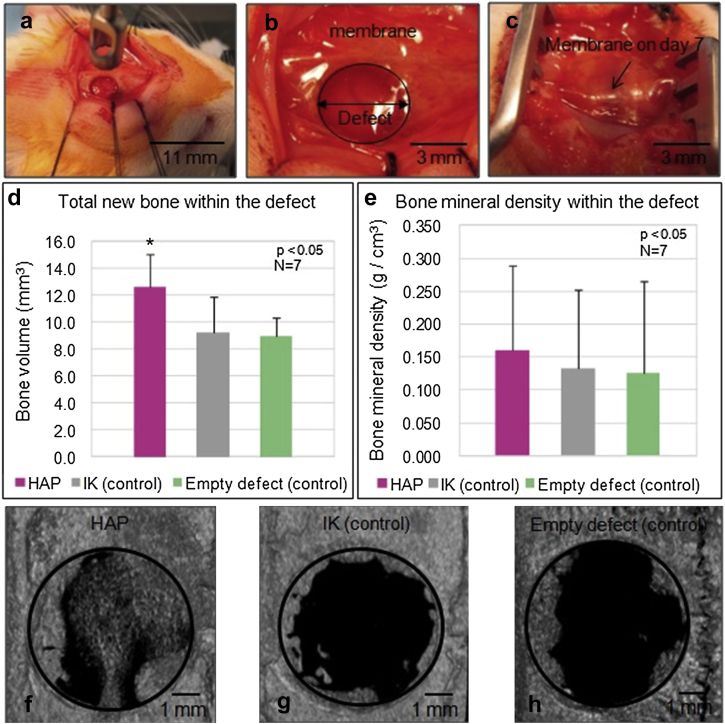
In another study, Tejeda-Montes et al. [171] used smooth ELR-based membranes with high tensile strength (Young's modulus E = 2081 ± 315 kPa), cross-linked under static conditions. The ELRs containing the SNA15 statherin fragment (hydroxyapatite (HAP) sequence) were assayed in vivo using an orthotopic critical size rat calvarial defect model. High-resolution microcomputed tomography (micro-CT) analysis and histological examinations demonstrated a higher mean volume of ossified tissue within the defect than untreated control animals or those treated with ELRs lacking HAP (Fig. 6). The ELR offered a structural stability and provided a nucleus for mineralization and osteoblastic differentiation in a non-cytotoxic and bioactive environment [171].
Fig. 6.
(a) An orthotopic critical-size rat calvarial defect model was used to analyze the bone regeneration capacity of (b) smooth HAP membranes. (c) Membranes were observed to be positioned within the defect site in the same location as they were placed during implantation on day 7. The micro-CT analysis demonstrated that animals implanted with the (d) HAP membranes presented the highest mean volume of ossified tissue (f) within the defect compared with animals receiving the (g) non-bioactive IK membranes and (h) those left untreated. (e) Micro-CT analysis of bone mineral density within the defect revealed no significant differences between the tested groups. Reproduced with permission from Tejeda-Montes et al. [171]. Copyright Elsevier 2014. CT, computed tomography; HAP, hydroxyapatite.
Elsharkawy et al. [172] exploited the disorder-order interactions to facilitate the formation of biomineralized structures. In this work, an amphiphilic co-ELR in the presence of calcium phosphate triggers a hierarchical and controllable framework that operates as a template. A dense network of β-amyloid-like fibers is formed upon cross-linking, and birefringent 3D spherulites (with diameter of a few hundreds of micrometers) are obtained upon drying at room temperature. This hierarchical structure consists of elongated nanocrystals whose aligned and organized growth into prisms generates the spherulite-like structures (Fig. 7 a-d). The thickness and average diameter of these structures are about 85 nm and 38 μm, respectively, and the length of a few tens of micrometers. The dependence of the disordered random coil conformation-to-ordered β-sheet ratio on the amount of cross-linkers was determined, and it was found that no spherulites are observed when this ratio is lower than 0.26, whereas an increase in the amount of cross-linkers induces a corresponding increase in the percentage of random coil conformations and, therefore, the presence of spherulites. If the ELR membrane is incubated in an appropriate solution, a protein-mediated mineralization process takes place inside the membrane bulk, with spherulites acting asnucleating and templating sites for mineralization. The two-stage formation mechanism for the mineralized structure is illustrated schematically in Fig. 7. (e). This tuneable process can also modify the physical properties of the structures to target-specific applications, achieving a Young's modulus (E) of up to 33.0 ± 20.1 GPa and a hardness (H) of up to 1.1 ± 0.8 GPa.
Fig. 7.
SEM images of the mineralized structures: (a) aligned nanocrystals, (b, c) prism-like structures, (d) spherulite-like structures; scale bars: 200 nm, 1 μm, 10 μm, and 20 μm, respectively. (e) Schematic describing the two-stage formation mechanism of the mineralized structures. Reproduced from the study by Elshawkawy et al. [172]. Open Access: http://creativecommons.org/licenses/by/4.0/. ELR, elastin-like recombinamer; SEM, scanning electron microscopy.
5.2. ELR SA and preformed structural elements
The proteins involved in regulatory processes and cellular signaling machinery frequently contain long disordered regions in preformed structural elements. As such, their ability to switch between a variety of conformational ensembles is a key tool for controlling their functionality. Leucine zipper motifs are one good example of this [173]. These structural motifs usually function as transcription factors that play key roles in eukaryotic gene regulation. However, the leucine zipper motif's ability to heterodimerize and change its secondary structure can also be exploited to drive formation of hydrogels with appropriate mechanical properties for use as scaffolds in tissue engineering applications [174]. For example, a study by Fernández-Colino et al. [159] explored the formation of an injectable hydrogel from a thermally responsive amphiphilic tetrablock ELR containing a natural leucine zipper sequence (HLF), with two such sequences per ELR molecule. The presence of leucine for every seven residues on the HLF peptide sequence allows the formation of a preformed coiled coil of parallel alpha helices. The synergistic effect of the ELR's thermoresponsive behavior and the leucine zipper's association results in the formation of a hydrogel with stable mechanical properties above the Tt. The flexibility in the conformational transitions from disordered to α-helical structures determines the reversibility of the process during temperature changes [159]. Although SA of these biomaterials commonly takes the form of hydrogels, other structures, ranging from particles to hollow vesicles to hierarchical supraparticles, have been designed using leucine zipper fusion proteins containing ELRs [175].
Peptide amphiphiles (PAs) present conformational features that are typical of intrinsically disordered molecules. Moreover, many PAs play crucial roles in physiological and pathological events and may assume a precise conformation upon binding to a specific target [176], [177]. PAs can also drive conformational changes in the ELRs, and the advantage of this property has been taken in many different applications. For example, in a study by Ionostroza-Brito [128] (referred to in section 4.1.2), the amphiphilic peptide PAK3 (which self-assembles into nanofibers and can create functional 3D hydrogels), is able to drive the dynamic SA of an ELR (ELP5) in a dynamic system that maintains a controlled non-equilibrium state for substantial periods of time. In addition, this system also enables morphogenesis into tubular structures with high spatiotemporal control. This programmed ordering into cylindrical structures can result in biological signals that allow interaction with cells or many other targets, thus providing these structures with many biomedical functions.
5.3. ELR SA and drug delivery/targeting
Traditional small-molecule drugs target ordered proteins involved in pathological processes via rational ligand-binding designs [178], [179]. However, the natural abundance of functional IDPs and IDRs involved in key cellular processes suggests that these proteins should be seriously considered as drug targets [179]. Given the heterogeneous nature of IDPs, however, this represents a significant challenge. The similarities between ELRs and IDPs suggest that they may prove useful as effectors or receptors for other IDPs or IDRs in complex protein-protein interactions. Their intrinsic disorder, as well as their recombinant synthesis, gives them great potential in such designs. Despite this, most of the literature in this field deals with ELRs as drug-delivery systems, taking advantage of their intrinsic disorder for the construction of smart assemblies that are able to modify the chemical, pharmacokinetic, or pharmacodynamic properties of other active components and for targeting drug loads to specific targets, with ELRs being shown to be powerful vectors. In light of this, the most ambitious drug-delivery systems are designed to achieve the required and sustained levels of administered drugs and also their targeting with spatial control of action to release their cargo at the desired sites. As such, an ideal system should combine sensing of pathological conditions and drug release to correct this adverse condition, thereby avoiding the exposure of healthy tissues to undesirable adverse effects [81].
Several excellent reviews covering the latest therapeutic applications of ELRs in different assemblies have been published [81], [180], [181], [182]. These are mainly based on combining ELR-based block copolymers in the same molecule, but with different stimuli responsiveness in solution. Different factors, such as the chemistry of the guest residue, molecular weight, and processing conditions, allow the formation of recurring patterns in hierarchical structures such as nanoparticulated systems, fibrillar structures, hydrogels, or other insoluble coacervates [63]. These tuned assemblies can be targeted to specific tissues [183] or cellular locations via the multivalent presentation of targeting peptides [165] or by programming ELR molecular SA under physiopathological conditions [111].
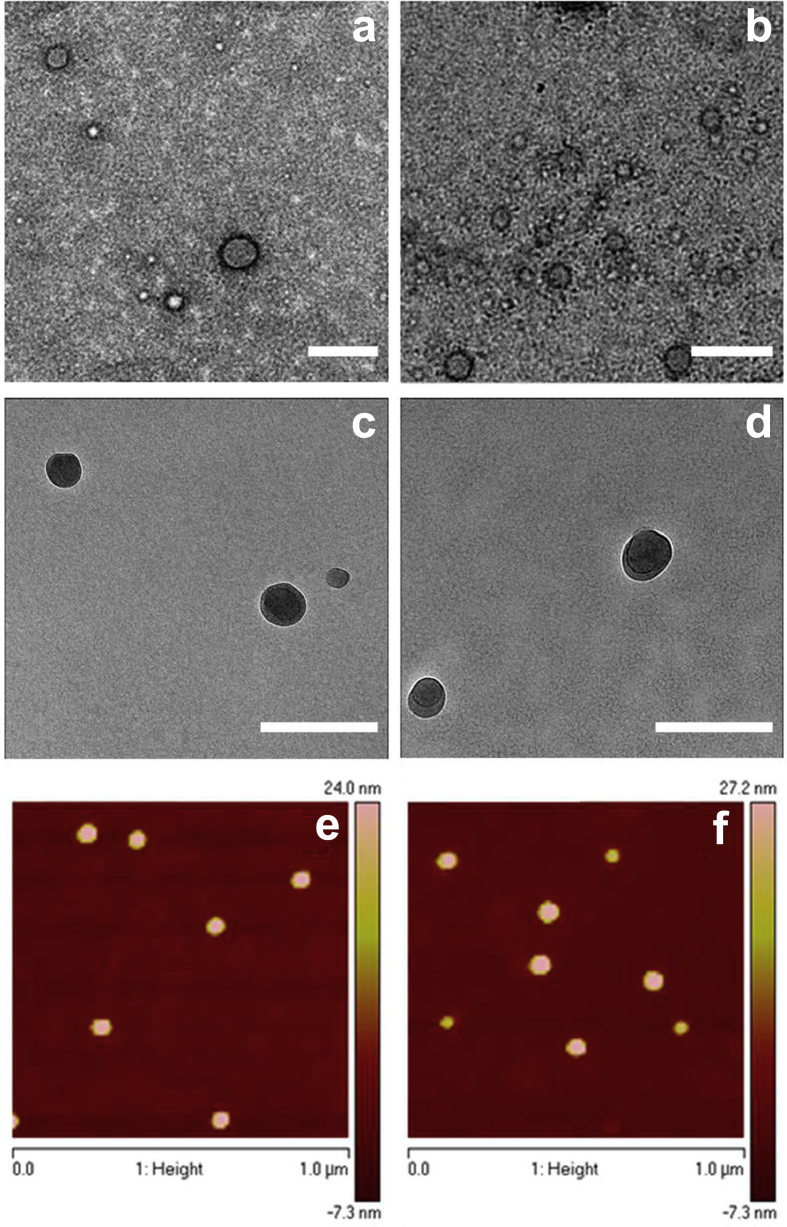
Other therapeutic applications of targeted ELR-based delivery systems displaying multiple peptide copies include those aiming to trigger therapeutic vaccination against low immunogenic antigens (Fig. 8) [184], [185].
Fig. 8.
TEM images of self-assembled nanoparticles stained with 1% uranyl acetate (a, b), cryo-TEM images (c, d), and tapping-mode AFM height images (e, f): E50I60 (left) and dAg-E50I60 (right). Scale bars for TEM and cryo-TEM samples are 200 and 100 nm, respectively. Reproduced with permission from Garcia-Arevalo et al. [184]. Copyright American Chemical Society 2013. TEM, transmission electron microscopy; AFM, atomic force microscopy.
In summary, given their great potential as high-performance delivery systems, the use of ELRs in the drug-delivery field is likely to increase markedly in the future.
5.4. ELR SA and tissue engineering
Elastin is an essential component of many tissues and provides structural integrity, elasticity, resilience, and deformability to large arteries, lungs, ligaments, tendons, skin, and elastic cartilage, among others [57]. Elastin and other components of elastic fibers are examples of essential disordered extracellular proteins involved in tissue organization, forming extensive interactions that organize large molecular assembles while binding multiple interaction partners [186]. As such, several factors make ELRs very attractive in the field of tissue engineering. First, the main proof that entropic elasticity is resilient is that new elastic fibers are synthetized almost exclusively after early development (confined to the fetal and early postnatal periods), with no appreciable turnover in healthy tissues (half-life of around 70 years) [187]. Second, elastin fibers cannot be efficiently repaired after injury without scarring and loss of function. This makes the development of new materials that are able to improve the healing process highly desirable. Over the past few years, the incorporation of ELRs into different assemblies and scaffolds (coacervates, fibers, hydrogels, and films) has been used to design wound healing systems with promising results [188]. Third, ELR-based materials are particularly promising as tissue substitutes because they can be produced with a range of consistencies and stabilities. Matching the elastic properties of the tissue replaced is crucial to integrating the implant with the surrounding living tissue. Moreover, depending on the amino acids in the polymer chains, the result may be gelatinous, rubbery, or as rigid as plastic and can be either stable for long periods or easily degradable. Physically or chemically cross-linked ELR-based hydrogels are common substrates for tissue engineering, and their mechanical properties have been easily tuned by modifying parameters such as sequence and domain arrangements [189], concentration [190], cross-linking degree [191], and porosity [192]. Other supports for cellular growth are available in the form of fibers [193] or films adsorbed onto surfaces [194], thereby changing their inherent properties and allowing the stiffness of these surfaces to be tuned by assembling genetically engineered polypeptides with a tailored amino acid sequence [195]. Innovative 3D printed scaffolds will add a level of complexity even greater than those mentioned previously [196], [197]. Fourth, the success of tissue repair is often limited because of an elicited immune response and/or poor material compliance [168]. ELRs have already been shown to be invisible to the immune system in numerous in vitro and in vivo experiments [198], [199], [200], [201]. Finally, in addition to its structural properties, elastin also plays a cell signaling role in some tissues, thus making elastin mimicry a key property for biomaterial applications and tissue repair. In addition, ELRs can incorporate biologically active amino acid sequences to either guide cell growth for specific cell types or with a specific spatial distribution [202] or regulate its degradation, invasiveness, and duration [203]. But the biologically active ELRs can also trigger the necessary cascade of cellular events that determine cellular differentiation [204], [205].
5.5. ELR SA and biosensors
The flexibility and binding specificity of IDPs allow them to modulate the input-output response of biorecognition systems upon binding to their target molecules. This property can be exploited for the rational design of dynamic IDP-based biosensors [206].
The intrinsic disorder of ELRs and other recombinant proteins appears to be very promising when addressing the design of sensitive biosensors. In this regard, Urry [186] noted the great potential of these elastomeric proteins in the interconversion of energy as these elastic molecules stretch or contract in response to chemical and electrical signals or can generate chemical outputs in response to mechanical stimulation. Indeed, Urry [186] considered ELRs to be ‘elastic biomolecular machines’ because the inverse temperature transition mechanism lies at the heart of most biological energy conversions.
One of the most recent studies concerning the usefulness of ELRs as biological sensors was published by Ibáñez-Fonseca et al. [207]. This study explored the Förster resonance energy transfer (FRET) of two different fluorescent proteins (FPs), namely the green-emitting FP Aequorea coerulescens–enhanced green fluorescent protein (AcEGFP) and the near-infrared-emitting FP (eqFP650), when fused to amphiphilic ELR-containing sequences from silk fibroin protein (SELR). Two techniques were applied for FRET analysis: spectroscopy (donor quenching) and confocal microscopy (acceptor photobleaching). The study was performed at different concentrations of the SELR in solution at which they self-assemble into systems ranging from particles to hydrogels as the concentration increases. The absorption spectrum of SELR-eqFP650 showed a peak mostly overlapping the emission spectrum of the SELR-AcEGFP, hence enabling FRET upon the interaction between two SELR molecules through the silk domains (Fig. 9). These results suggest that peptides/proteins that bind to different targets, such as glucose, lipopolysaccharide, or metal ions, can be included in the SELR sequence, thus increasing the traceability of these biomedical devices [207].
Fig. 9.
The fluorescent proteins (FPs) Aequorea coerulescens–enhanced green fluorescent protein (AcEGFP) and near-infrared-emitting eqFP650 are fused to an amphiphilic SELR based on two types of elastin-like domains, one hydrophilic and the other one hydrophobic, by including glutamic acid and isoleucine as guest residues, respectively. The study of FRET between both SELR-FPsSELR molecules established stacking interactions at middle and high concentrations that are able to minimize the distance between the two FPs, hence enabling FRET. Adapted with permission from Ibanez-Fonseca et al. [207]. Copyright American Chemical Society 2017. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.) SELR, silk elastin-like recombinamer; FRET, Förster resonance energy transfer.
5.6. ELR SA and protein purification
The pioneering work of Meyer and Chilkoti [208] demonstrating that ELRs maintain their thermal transition behavior even after fusion to other tags for protein purification opened up a new field of use for these versatile compounds, namely to replace expensive affinity chromatography methods, which represent a major cost of the final protein product on scale-up [208]. In their work, the genes for two proteins (thioredoxin and tendamistat) were fused to different lengths of ELP genes and purified by both metal affinity chromatography and inverse transition cycling. The ELR was then cleaved using thrombin. Their results confirmed the possibility of purifying proteins with very high yield by exploiting the inverse transition cycling of the tagged ELR, thus suggesting further applications. Subsequent studies corroborated the versatility of this technique, although each target protein or peptide has unique properties that may require adjustment of the inverse temperature cycling purification protocol and selection of the final cleavage of the ELR from the protein [209], [210], [211], [212], [213]. For example, the fusion order, length of the ELR, thermal properties, and stability can make a considerable difference to the expression and activity of the target protein [214], [215].
6. Conclusions
Natural SA of different polymeric molecules has positioned this spontaneous process as a promising tool to create smart and sophisticated systems with challenging applications in the biotechnological and nanotechnological fields.
In this review, the topic of self-assembled structures based on ELRs has been reviewed, and a complete overview of the bibliography has been carried out.
ELRs are recombinantly produced stimuli-responsive polymers inspired by the intrinsically disordered domains of tropoelastin. ELRs and IDPs share many physicochemical properties mainly in terms of low sequence complexity, biased amino acid composition, limited hydrophobicity, and phase behavior, but their relationship to other disordered proteins remains unresolved. The structural disorder in ELRs is proposed to be a consequence of the high proline and glycine content in the ELR backbone in which the formation of extended secondary structures is restricted in favor of transient and fluctuating local motifs.
Owing to the large diversity of designs available at an amino acid level, ELRs have become extremely versatile materials that are able to exhibit a ‘smart’ behavior irrespective of the presence of a particular stimulus.
Depending on the specific ELR and processing conditions, several self-assembled ELR morphologies can be obtained, with stand-alone or hybrid materials being achieved using only an ELR or ELRs combined with another compound, respectively.
As a direct consequence of their excellent biocompatibility, their use in the field of biomedical applications has experienced a noticeable growth in the past and will continue to grow well in the foreseeable future.
Despite the noticeable control and variety of the achieved structures, some limitations and challenges should be indicated, such as the current limited comprehension of the complex SA mechanism or the difficulty in extending the relation between structure and material functionality with practical applications.
Indeed, over the coming years, synergetic effects will provide a range of amazing applications and improvements that we cannot even begin to imagine today.
Data availability
Not applicable.
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
The authors are grateful for the funding from the European Commission (NMP-2014-646075), the Spanish Government (MAT2015-68901-R, PCIN-2015-010, MAT2016-78903-R), Junta de Castilla y León (VA317P18), and Centro en Red de Medicina Regenerativa y Terapia Celular de Castilla y León.
References
- 1.Huebsch N., Mooney D.J. Nature. 2009;462:426. doi: 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boehr D.D. Nat. Chem. Biol. 2009;5(11):789. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dyson H.J. Biophys. J. 2016;110(5):1013. doi: 10.1016/j.bpj.2016.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dzuricky M. Biochemistry. 2018;57(17):2405. doi: 10.1021/acs.biochem.8b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitesides G.M., Grzybowski B. Science. 2002;295(5564):2418. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 6.Halley J.D., Winkler D.A. Complexity. 2008;14(2):10. [Google Scholar]
- 7.Hosokawa K. Artif. Life. 1994;1(4):413. [Google Scholar]
- 8.Cheung K.C. IEEE Trans. Robot. 2011;27(4):718. [Google Scholar]
- 9.Gross R. IEEE Trans. Robot. 2006;22(6):1115. [Google Scholar]
- 10.Tibbits S. Architect Des. 2014;84(1):116. [Google Scholar]
- 11.Lehn J.-M. Angew. Chem. Int. Ed. Engl. 1990;29(11):1304. [Google Scholar]
- 12.Lehn J.-M. Angew. Chem. Int. Ed. Engl. 1988;27(1):89. [Google Scholar]
- 13.Aida T. Science. 2012;335(6070):813. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aronsson C. Sci. Rep. 2015;5:14063. doi: 10.1038/srep14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsuhiko A. Sci. Technol. Adv. Mater. 2008;9(1) 014109. [Google Scholar]
- 16.Discher B.M. Science. 1999;284(5417):1143. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- 17.da Silva R.M.P. Nat. Commun. 2016;7:11561. doi: 10.1038/ncomms11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tantakitti F. Nat. Mater. 2016;15:469. doi: 10.1038/nmat4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S.S. Nat. Nanotechnol. 2017;12:821. doi: 10.1038/nnano.2017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitesides G. Science. 1991;254(5036):1312. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 21.Okesola B.O., Mata A. Chem. Soc. Rev. 2018;47(10):3721. doi: 10.1039/c8cs00121a. [DOI] [PubMed] [Google Scholar]
- 22.Atkinson Ian M., Lindoy Len F. Royal. Soc. Chem.; 2000. Self Assembly in Supramolecular Systems. Chapter 1, pages 1 – 6. [Google Scholar]
- 23.Sharp K. Science. 1991;252(5002):106. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- 24.Smithrud D.B. J. Am. Chem. Soc. 1991;113(14):5420. [Google Scholar]
- 25.Jakubith S. Phys. Rev. Lett. 1990;65(24):3013. doi: 10.1103/PhysRevLett.65.3013. [DOI] [PubMed] [Google Scholar]
- 26.Hess B. Naturwissenschaften. 2000;87(5):199. doi: 10.1007/s001140050704. [DOI] [PubMed] [Google Scholar]
- 27.Goldenfeld N., Kadanoff L.P. Science. 1999;284(5411):87. doi: 10.1126/science.284.5411.87. [DOI] [PubMed] [Google Scholar]
- 28.Kay E.R. Chem. Ind. 2007;38(10) [Google Scholar]
- 29.Koumura N. Nature. 1999;401:152. doi: 10.1038/43646. [DOI] [PubMed] [Google Scholar]
- 30.Fletcher S.P. Science. 2005;310(5745):80. doi: 10.1126/science.1117090. [DOI] [PubMed] [Google Scholar]
- 31.Dyson H.J., Wright P.E. Nat. Rev. Mol. Cell. Biol. 2005;6(3):197. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 32.Wright P.E., Dyson H.J. Nat. Rev. Mol. Cell. Biol. 2014;16:18. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berlow R.B. FEBS. Lett. 2015;589(19 Pt A):2433. doi: 10.1016/j.febslet.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chouard T. Nature. 2011;471(7337):151. doi: 10.1038/471151a. [DOI] [PubMed] [Google Scholar]
- 35.Tompa P. Trends. Biochem. Sci. 2002;27(10):527. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 36.Ferreon A.C. Methods enzymol. 2010;472:179. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 37.Hofmann H. Proc. Natl. Acad. Sci. U. S. A. 2012;109(40):16155. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das R.K., Pappu R.V. Proc. Natl. Acad. Sci. U S A. 2013;110(33):13392. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staby L. Biochem. J. 2017;474(15):2509. doi: 10.1042/BCJ20160631. [DOI] [PubMed] [Google Scholar]
- 40.van der Lee R. Chem. Rev. 2014;114(13):6589. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunker A.K. Biochemistry. 2002;41(21):6573. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 42.Dyson H.J., Wright P.E. Curr. Opin. Struct. Biol. 2002;12(1):54. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 43.Spolar R.S., Record M.T., Jr. Science. 1994;263(5148):777. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 44.Demchenko A.P. J. Mol. Recognit. : JMR. 2001;14(1):42. doi: 10.1002/1099-1352(200101/02)14:1<42::AID-JMR518>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 45.Mohan A. J. Mol. Biol. 2006;362(5):1043. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 46.Fuxreiter M. J. Mol. Biol. 2004;338(5):1015. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 47.Uversky V.N. Proteins: Structure, Function, Bioinformatics. 2000;41(3):415. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 48.Müller-Späth S. Proc. Natl. Acad. Sci. 2010;107(33):14609. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao A.H. Proc. Natl. Acad. Sci. 2010;107(18):8183. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habchi J. Chem. Rev. 2014;114(13):6561. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- 51.Theillet F.X. Intrinsically Disord. Proteins. 2013;1(1) doi: 10.4161/idp.24360. e24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rath A. Biopolymers. 2005;80(2–3):179. doi: 10.1002/bip.20227. [DOI] [PubMed] [Google Scholar]
- 53.Marsh J.A., Forman-Kay J.D. Biophys. J. 2010;98(10):2383. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rauscher S. Structure (London, England : 1993) 2006;14(11):1667. doi: 10.1016/j.str.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 55.Muiznieks L.D., Keeley F.W. J. Biol. Chem. 2010;285(51):39779. doi: 10.1074/jbc.M110.164467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flock T. Curr. Opin. Struct. Biol. 2014;26:62. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 57.Mithieux S.M., Weiss A.S. Adv. Protein. Chem. 2005;70:437. doi: 10.1016/S0065-3233(05)70013-9. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z. Chapter 23 – biodegradable polymers. In: Lanza R., editor. Principles of Tissue Engineering. fourth ed. Academic Press; Boston: 2014. p. 441. [Google Scholar]
- 59.Bellingham C.M. Biopolymers. 2003;70(4):445. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 60.Wise S.G. Engineered tropoelastin and elastin-based biomaterials. In: McPherson A., editor. vol. 78. Academic Press; 2009. p. 1. (Advances in Protein Chemistry and Structural Biology). [DOI] [PubMed] [Google Scholar]
- 61.Urry D.W. Prog. Biophys. Mol. Biol. 1992;57(1):23. doi: 10.1016/0079-6107(92)90003-o. [DOI] [PubMed] [Google Scholar]
- 62.Urry D.W. J. Phys. Chem. B. 1997;101(51):11007. [Google Scholar]
- 63.Rodríguez-Cabello J.C. 4 – elastin-like proteins: molecular design for self-assembling. In: Azevedo H.S., da Silva R.M.P., editors. Self-assembling Biomaterials. Woodhead Publishing; 2018. p. 49. [Google Scholar]
- 64.Ribeiro A. Biophys. J. 2009;97(1):312. doi: 10.1016/j.bpj.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Girotti A. Macromolecules. 2004;37(9):3396. [Google Scholar]
- 66.Rodríguez-Cabello J.C. Chem. Phys. Lett. 2004;388(1):127. [Google Scholar]
- 67.Pinedo-Martín G. Polymer. 2014;55(21):5314. [Google Scholar]
- 68.Cho Y. J. Phys. Chem. B. 2008;112(44):13765. doi: 10.1021/jp8062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alonso M. Macromolecules. 2001;34(23):8072. [Google Scholar]
- 70.Meyer D.E., Chilkoti A. Biomacromolecules. 2004;5(3):846. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 71.Ferreira L.A. Int. J. Mol. Sci. 2015;16(6):13528. doi: 10.3390/ijms160613528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dehghani F. Biomacromolecules. 2008;9(4):1100. doi: 10.1021/bm700891b. [DOI] [PubMed] [Google Scholar]
- 73.Christensen T. Biomacromolecules. 2013;14(5):1514. doi: 10.1021/bm400167h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gosline J. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2002;357(1418):121. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mithieux S.M. Adv. Drug. Deliv. Rev. 2013;65(4):421. doi: 10.1016/j.addr.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 76.Rodríguez-Cabello J.C. Polymer. 2009;50(22):5159. [Google Scholar]
- 77.Li N.K. Biomacromolecules. 2014;15(10):3522. doi: 10.1021/bm500658w. [DOI] [PubMed] [Google Scholar]
- 78.Ruff K.M. J. Mol. Biol. 2018;430(23):4619. doi: 10.1016/j.jmb.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 79.Kowalczyk T. World J. Microbiol. Biotechnol. 2014;30(8):2141. doi: 10.1007/s11274-014-1649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urry D.W. Birkhäuser Boston; Boston (USA): 2006. What Sustains Life? Consilient Mechanisms for Protein-Based Machines and Materials. [Google Scholar]
- 81.Rodriguez-Cabello J.C. Adv. Drug. Deliv. Rev. 2016;97:85. doi: 10.1016/j.addr.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 82.Herrero-Vanrell R. J. Control. Release: Off. J. Control. Release Soc. 2005;102(1):113. doi: 10.1016/j.jconrel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 83.Liang Y. Macromolecules. 2017;50(2):483. doi: 10.1021/acs.macromol.6b02389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carlos Rodríguez-Cabello J. Prog. Polym. Sci. 2005;30(11):1119. [Google Scholar]
- 85.Lee A.L., Wand A.J. Nature. 2001;411(6836):501. doi: 10.1038/35078119. [DOI] [PubMed] [Google Scholar]
- 86.Pometun M.S. J. Biol. Chem. 2004;279(9):7982. doi: 10.1074/jbc.M310948200. [DOI] [PubMed] [Google Scholar]
- 87.Gronau G. Biomaterials. 2012;33(33):8240. doi: 10.1016/j.biomaterials.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muiznieks L.D. Vol. 88. 2010. Biochemistry and cell biology = Biochimie et biologie cellulaire; p. 239. [DOI] [PubMed] [Google Scholar]
- 89.Hoeve C.A., Flory P.J. Biopolymers. 1974;13(4):677. doi: 10.1002/bip.1974.360130404. [DOI] [PubMed] [Google Scholar]
- 90.Aaron B.B., Gosline J.M. Biopolymers. 1981;20(6):1247. [Google Scholar]
- 91.Tjin M.S. Polymer Journal. 2014;46:444. [Google Scholar]
- 92.Urry D.W., Long M.M. CRC Crit. Rev. Biochem. 1976;4(1):1. doi: 10.3109/10409237609102557. [DOI] [PubMed] [Google Scholar]
- 93.McPherson D.T. Biotechnol. Prog. 1992;8(4):347. doi: 10.1021/bp00016a012. [DOI] [PubMed] [Google Scholar]
- 94.Rauscher S., Pomes R. Vol. 725. 2012. Advances in experimental medicine and biology; p. 159. [DOI] [PubMed] [Google Scholar]
- 95.Li B. J. Am. Chem. Soc. 2001;123(48):11991. doi: 10.1021/ja010363e. [DOI] [PubMed] [Google Scholar]
- 96.Tamburro A.M. Biochemistry. 2003;42(45):13347. doi: 10.1021/bi034837t. [DOI] [PubMed] [Google Scholar]
- 97.Wise S.G. Acta Biomater. 2014;10(4):1532. doi: 10.1016/j.actbio.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roberts S. FEBS Lett. 2015;589(19 Pt A):2477. doi: 10.1016/j.febslet.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reichheld S.E. Proc. Natl. Acad. Sci. U. S. A. 2017;114(22):E4408. doi: 10.1073/pnas.1701877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roberts S. FEBS Lett. 2015;589:2477. doi: 10.1016/j.febslet.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Das R.K., Pappu R.V. Proc. Natl. Acad. Sci. 2013;110(33):13392. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wright E.R., Conticello V.P. Adv. Protein. Chem. 2002;54(8):1057. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 103.McDaniel J.R. Biomacromolecules. 2013;14(8):2866. doi: 10.1021/bm4007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hassouneh W. Macromolecules. 2015;48(12):4183. doi: 10.1021/acs.macromol.5b00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee T.A.T. Adv. Mater. 2000;12(15):1105. [Google Scholar]
- 106.Dreher M.R. J. Am. Chem. Soc. 2008;130(2):687. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.MacEwan S.R. Biomacromolecules. 2017;18(2):599. doi: 10.1021/acs.biomac.6b01759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martín L. Biomacromolecules. 2012;13(2):293. doi: 10.1021/bm201436y. [DOI] [PubMed] [Google Scholar]
- 109.Janib S.M. Polymer Chem. 2014;5(5):1614. doi: 10.1039/C3PY00537B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roemer R.B. Annu. Rev. Biomed. Eng. 1999;1(1):347. doi: 10.1146/annurev.bioeng.1.1.347. [DOI] [PubMed] [Google Scholar]
- 111.Callahan D.J. Nano Lett. 2012;12(4):2165. doi: 10.1021/nl300630c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luo T., Kiick K.L. J. Am. Chem. Soc. 2015;137(49):15362. doi: 10.1021/jacs.5b09941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andrew MacKay J. Nature Mater. 2009;8:993. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McDaniel J.R. Angew. Chem., Int. Ed. Engl. 2013;52(6):1683. doi: 10.1002/anie.201200899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McDaniel J.R. J. Control. Release. 2012;159(3):362. doi: 10.1016/j.jconrel.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Derossi D. J. Biol. Chem. 1996;271(30):18188. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 117.Olson E.S. Integr. Biol. 2009;1(5–6):382. doi: 10.1039/b904890a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rothbard J.B. J. Am. Chem. Soc. 2004;126(31):9506. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 119.Walker L. FEBS. Lett. 2012;436(1):825. [Google Scholar]
- 120.MacEwan S.R., Chilkoti A. Nano Lett. 2012;12(6):3322. doi: 10.1021/nl301529p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu J. Chem. Commun. 2015;51(57):11405. doi: 10.1039/c5cc04035c. [DOI] [PubMed] [Google Scholar]
- 122.Ghoorchian A. Macromolecules. 2010;43(9):4340. [Google Scholar]
- 123.van Eldijk M.B. J. Am. Chem. Soc. 2012;134(45):18506. doi: 10.1021/ja308132z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McDaniel J.R. Nano Lett. 2014;14(11):6590. doi: 10.1021/nl503221p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aluri S.R. ACS Nano. 2014;8(3):2064. doi: 10.1021/nn403973g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pastuszka M.K. J. Control. Release. 2014;191:15. doi: 10.1016/j.jconrel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xia X.-X. Biomacromolecules. 2011;12(11):3844. doi: 10.1021/bm201165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Inostroza-Brito K.E. Nat. Chem. 2015;7:897. doi: 10.1038/nchem.2349. [DOI] [PubMed] [Google Scholar]
- 129.Inostroza-Brito K.E. Acta Biomater. 2017;58:80. doi: 10.1016/j.actbio.2017.05.043. [DOI] [PubMed] [Google Scholar]
- 130.Navon Y., Bitton R. Isr. J. Chem. 2016;56(8):581. [Google Scholar]
- 131.Urry D.W., Parker T.M. Mechanics of elastin: molecular mechanism of biological elasticity and its relationship to contraction. In: Linke W.A., editor. Mechanics of Elastic Biomolecules. Springer Netherlands; Dordrecht: 2003. p. 543. [PubMed] [Google Scholar]
- 132.Castiglione Morelli M.A. J. Biomol. Struct. Dyn. 1993;11(1):181. doi: 10.1080/07391102.1993.10508716. [DOI] [PubMed] [Google Scholar]
- 133.Tamburro A.M. J. Biomol. Struct. Dyn. 1992;10(3):441. doi: 10.1080/07391102.1992.10508661. [DOI] [PubMed] [Google Scholar]
- 134.Bochicchio B. Nanomedicine. 2009;4(1):31. doi: 10.2217/17435889.4.1.31. [DOI] [PubMed] [Google Scholar]
- 135.Tamburro A.M. J. Biol. Chem. 2005;280(4):2682. doi: 10.1074/jbc.M411617200. [DOI] [PubMed] [Google Scholar]
- 136.Moscarelli P. Matrix. Biol. 2014;36:15. doi: 10.1016/j.matbio.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 137.Chiti F. Proc. Natl. Acad. Sci. 1999;96(7):3590. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Anderson V.L. Phys. Biol. 2012;9(5) doi: 10.1088/1478-3975/9/5/056005. 056005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhu M. J. Biol. Chem. 2002;277(52):50914. doi: 10.1074/jbc.M207225200. [DOI] [PubMed] [Google Scholar]
- 140.Flamia R. Biomacromolecules. 2004;5(4):1511. doi: 10.1021/bm049930r. [DOI] [PubMed] [Google Scholar]
- 141.Le D.H.T. Biomacromolecules. 2013;14(4):1028. doi: 10.1021/bm301887m. [DOI] [PubMed] [Google Scholar]
- 142.Li Y. ACS Appl. Mater. Interfaces. 2017;9(7):5838. doi: 10.1021/acsami.6b15285. [DOI] [PubMed] [Google Scholar]
- 143.Kopeček J., Yang J. Acta Biomater. 2009;5(3):805. doi: 10.1016/j.actbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang S. Proc. Natl. Acad. Sci. 1993;90(8):3334. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Asai D. Biomaterials. 2012;33(21):5451. doi: 10.1016/j.biomaterials.2012.03.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li M., Ober C.K. Mater. Today. 2006;9(9):30. [Google Scholar]
- 147.Reguera J. J. Am. Chem. Soc. 2004;126(41):13212. doi: 10.1021/ja047417f. [DOI] [PubMed] [Google Scholar]
- 148.Sallach R.E. Biomaterials. 2010;31(4):779. doi: 10.1016/j.biomaterials.2009.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu X. Biomacromolecules. 2005;6(6):3037. doi: 10.1021/bm0503468. [DOI] [PubMed] [Google Scholar]
- 150.Wright E.R. Adv. Funct. Mater. 2002;12(2):149. [Google Scholar]
- 151.Misbah M.H. Polymer. 2015;81:37. [Google Scholar]
- 152.Tanaka K. Insect. Biochem. Mol. Biol. 1999;29(3):269. doi: 10.1016/s0965-1748(98)00135-0. [DOI] [PubMed] [Google Scholar]
- 153.Petka W.A. Science. 1998;281(5375):389. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 154.Fernández-Colino A. Biomacromolecules. 2014;15(10):3781. doi: 10.1021/bm501051t. [DOI] [PubMed] [Google Scholar]
- 155.Moitra J. Biochemistry. 1997;36(41):12567. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- 156.Krylov D. EMBO J. 1994;13(12):2849. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang J. Macromol. Biosci. 2006;6(3):201. doi: 10.1002/mabi.200500208. [DOI] [PubMed] [Google Scholar]
- 158.Tripet B. J. Mol. Biol. 2000;300(2):377. doi: 10.1006/jmbi.2000.3866. [DOI] [PubMed] [Google Scholar]
- 159.Fernández-Colino A. Biomacromolecules. 2015;16(10):3389. doi: 10.1021/acs.biomac.5b01103. [DOI] [PubMed] [Google Scholar]
- 160.Dash B.C. J. Control. Release. 2011;152(3):382. doi: 10.1016/j.jconrel.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 161.Swierczewska M. Acta Biomater. 2008;4(4):827. doi: 10.1016/j.actbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 162.Huang H.-C. Nanomedicine. 2011;6(3):459. doi: 10.2217/nnm.10.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kojima C., Irie K. Peptide Sci. 2013;100(6):714. doi: 10.1002/bip.22276. [DOI] [PubMed] [Google Scholar]
- 164.Wang E. Langmuir. 2014;30(8):2223. doi: 10.1021/la404333b. [DOI] [PubMed] [Google Scholar]
- 165.Piña M.J. Mol. Pharm. 2016;13(3):795. doi: 10.1021/acs.molpharmaceut.5b00712. [DOI] [PubMed] [Google Scholar]
- 166.Zheng X.T. vol. 1253. American Chemical Society; 2017. Bioinspired design and engineering of functional nanostructured materials for biomedical applications; p. 123. (Advances in Bioinspired and Biomedical Materials Volume 2). [Google Scholar]
- 167.Rodriguez-Cabello J.C. J. Biomater. Sci. Poly. Ed. 2007;18(3):269. doi: 10.1163/156856207779996904. [DOI] [PubMed] [Google Scholar]
- 168.Muiznieks L.D., Keeley F.W. ACS Biomater. Sci. Eng. 2017;3(5):661. doi: 10.1021/acsbiomaterials.6b00469. [DOI] [PubMed] [Google Scholar]
- 169.Boskey A.L., Villarreal-Ramirez E. Matrix Biol : J. Int. Soc. Matrix Biol. 2016;52–54:43. doi: 10.1016/j.matbio.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li Y. ACS Appl. Mater. Interfaces. 2015;7(46):25784. doi: 10.1021/acsami.5b07628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tejeda-Montes E. Biomaterials. 2014;35(29):8339. doi: 10.1016/j.biomaterials.2014.05.095. [DOI] [PubMed] [Google Scholar]
- 172.Elsharkawy S. Nat. Commun. 2018;9(1):2145. doi: 10.1038/s41467-018-04319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shammas S.L. Curr. Opin. Struct. Biol. 2017;42:155. doi: 10.1016/j.sbi.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 174.Pastuszka M.K., MacKay J.A. Nanomedicine and nanobiotechnology. Vol. 8. 2016. Wiley interdisciplinary reviews; p. 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jang Y., Champion J.A. Acc. Chem. Res. 2016;49(10):2188. doi: 10.1021/acs.accounts.6b00337. [DOI] [PubMed] [Google Scholar]
- 176.Hendricks M.P. Acc. Chem. Res. 2017;50(10):2440. doi: 10.1021/acs.accounts.7b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vincenzi M. Mol. Biosys. 2015;11(11):2925. doi: 10.1039/c5mb00358j. [DOI] [PubMed] [Google Scholar]
- 178.Hopkins A.L., Groom C.R. Nat. Rev. Drug Discov. 2002;1(9):727. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 179.Uversky V.N. Expert opin. Drug Discov. 2012;7(6):475. doi: 10.1517/17460441.2012.686489. [DOI] [PubMed] [Google Scholar]
- 180.Santos M. Curr. Med. Chem. 2018 [Google Scholar]
- 181.Arias F.J. Curr. Drug Targets. 2018;19(4):360. doi: 10.2174/1389450117666160201114617. [DOI] [PubMed] [Google Scholar]
- 182.Rodríguez-Cabello J.C. Bioconjug. Chem. 2015;26(7):1252. doi: 10.1021/acs.bioconjchem.5b00183. [DOI] [PubMed] [Google Scholar]
- 183.Sarangthem V. Sci. Rep. 2018;8(1):3892. doi: 10.1038/s41598-018-21910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Garcia-Arevalo C. Mol. Pharmacol. 2013;10(2):586. doi: 10.1021/mp300325v. [DOI] [PubMed] [Google Scholar]
- 185.Cho S. J. Drug Targeting. 2016;24(4):328. doi: 10.3109/1061186X.2015.1077847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Urry D.W. Sci. Am. 1995;272(1):64. [Google Scholar]
- 187.Shapiro S.D. J. Clin. Investig. 1991;87(5):1828. doi: 10.1172/JCI115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rodríguez-Cabello J.C. Adv. Drug. Deliv. Rev. 2018;129:118. doi: 10.1016/j.addr.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 189.Miao M. Biopolymers. 2013;99(6):392. doi: 10.1002/bip.22192. [DOI] [PubMed] [Google Scholar]
- 190.Trabbic-Carlson K. Biomacromolecules. 2003;4(3):572. doi: 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]
- 191.Santo V.E. Biomater.Biomed. Eng. 2015;2(1):47. [Google Scholar]
- 192.Gonzalez de Torre I. Acta Biomater. 2014;10(6):2495. doi: 10.1016/j.actbio.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 193.Gonzalez de Torre I. Acta Biomater. 2018;72:137. doi: 10.1016/j.actbio.2018.03.027. [DOI] [PubMed] [Google Scholar]
- 194.Costa R.R. Small (Weinheim an der Bergstrasse, Germany) 2011;7(18):2640. doi: 10.1002/smll.201100875. [DOI] [PubMed] [Google Scholar]
- 195.Costa R.R. Biomacromolecules. 2018;19(8):3401. doi: 10.1021/acs.biomac.8b00723. [DOI] [PubMed] [Google Scholar]
- 196.Do A.-V. Adv. Healthcare Mater. 2015;4(12):1742. doi: 10.1002/adhm.201500168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vila M. Acta Biomater. 2016;45:349. doi: 10.1016/j.actbio.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 198.Ibanez-Fonseca A. J. Tissue Eng. Regenerat. Med. 2018;12(3):e1450. doi: 10.1002/term.2562. [DOI] [PubMed] [Google Scholar]
- 199.Changi K. J. Biomed. Mater. Res. A. 2018;106(4):924. doi: 10.1002/jbm.a.36290. [DOI] [PubMed] [Google Scholar]
- 200.Urry D.W. J. Bioact. Compat. Polym. 1991;6(3):263. [Google Scholar]
- 201.de Torre I.G. Acta Biomater. 2015;12:146. doi: 10.1016/j.actbio.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 202.Flora T. European Poly. J. 2018;106:19. [Google Scholar]
- 203.Coletta D.J. Tissue Eng. Part A. 2017;23(23–24):1361. doi: 10.1089/ten.TEA.2017.0047. [DOI] [PubMed] [Google Scholar]
- 204.Sousa M.P. Mater. Today Chem. 2017;4:150. [Google Scholar]
- 205.Betre H. Biomaterials. 2006;27(1):91. doi: 10.1016/j.biomaterials.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 206.Cissell K.A. Anal. Bioanal. Chem. 2008;391(5):1721. doi: 10.1007/s00216-007-1819-5. [DOI] [PubMed] [Google Scholar]
- 207.Ibanez-Fonseca A. Bioconjug. Chem. 2017;28(3):828. doi: 10.1021/acs.bioconjchem.6b00738. [DOI] [PubMed] [Google Scholar]
- 208.Meyer D.E., Chilkoti A. Nature. 1999;17(11):1112. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 209.da Costa A. Bio Technology. 2018;46:45. [Google Scholar]
- 210.Christensen T. Analy. Biochem. 2007;360(1):166. doi: 10.1016/j.ab.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.VerHeul R. Biomater. Sci. 2018;6(4):863. doi: 10.1039/c8bm00124c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ge X. J. Am. Chem. Soc. 2005;127(32):11228. doi: 10.1021/ja0531125. [DOI] [PubMed] [Google Scholar]
- 213.Hassouneh W. 2010. Current Protocols in Protein Science. (Chapter 6), Unit 6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Trabbic-Carlson K. Protein Eng., Design Selection : PEDS. 2004;17(1):57. doi: 10.1093/protein/gzh006. [DOI] [PubMed] [Google Scholar]
- 215.Fletcher E.E. Protein Exp. Purifi. 2019;153:114. doi: 10.1016/j.pep.2018.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.