Abstract
Blue fluorescent hexagonal boron nitride quantum dots (h-BNQDs) of ∼10 nm size as an effective enhancer for DNA cleavage activity of anticancer drug doxorubicin (DOX) were synthesized using simple one-step hydrothermal disintegration of exfoliated hexagonal boron nitride at very low temperature ∼ 120 °C. Boron nitride quantum dots (BNQDs) at a concentration of 25 μg/ml enhanced DNA cleavage activity of DOX up to 70% as checked by converting supercoiled fragment into nicked circular PBR322 DNA. The interaction of BNQDs with DOX is proportional to the concentration of BNQDs, with binding constant Kb ∼0.07338 μg/ml. In addition, ab initio theoretical results indicate that DOX is absorbed on BNQDs at the N-terminated edge with binding energy −1.075 eV and prevented the normal replication mechanisms in DNA. BNQDs have been shown to kill the breast cancer cell MCF-7 extensively as compared with the normal human keratinocyte cell HaCaT. The cytotoxicity of BNQDs may be correlated with reduced reactive oxygen species level and increased apoptosis in MCF-7 cells, which may be liable to enhance the anticancerous activity of DOX. The results provide a base to develop BNQD-DOX as a more effective anticancer drug.
Keywords: Hydrothermal, MCF-7, HaCaT, Reduced ROS level
Highlights
-
•
Blue fluorescent hexagonal boron nitride quantum dots (h-BNQDs) were developed at low temperature ∼ 120 °C.
-
•
h-BNQDs as an effective enhancer for DNA cleavage activity of the anticancer drug doxorubicin (DOX).
-
•
Enhanced DNA cleavage activity of DOX checked by converting supercoiled fragment into nicked circular PBR322 DNA.
-
•
BNQDs were shown to kill the breast cancer cell MCF-7 extensively as compared with the normal human keratinocyte cell HaCaT.
-
•
The reduced reactive oxygen species level and increased apoptosis in MCF-7 cells may liable to enhance the anticancerous activity of DOX.
1. Introduction
Besides the unique two-dimensional (2D) single-atomic layered structure [1], [2], 2D boron nitride (BN) nanosheets, ‘white graphene’, show many novel chemical and physical properties such as wide direct energy band gap (5.2 eV), high elastic modulus (36.5 GPa), ultraviolet (UV) photoluminescence, high resistance to oxidation, excellent thermal conductivity (600 W/m.K), and stability [3], [4], [5], [6]. These remarkable properties make it a promising candidate in practical applications such as optoelectronics, electrical insulation, photocatalysis, solid lubrication, substrates for graphene-based electronics, organic pollutant adsorption, and cleanup of oil spillage [3], [4], [5], [6], [7]. More importantly, creating materials in quantum size, comparable to atomic size, can be one of the most effective ways to broaden new physical properties of a material due to quantum confinement [8]. BN quantum dots (BNQDs) have recently been in the spotlight because of their numerous applications, including them being an ideal candidate for use in the biomedical field [9]. However, the synthesis of BNQDs is still challenging due to the high thermal and chemical stability of BN sheets. To the best of our knowledge, only two strategies have been developed by exfoliating and disintegrating of hexagonal boron nitride (h-BN) flakes [9] and by defect engineering [10]. They used a high temperature (>200 °C) and could only synthesize a small amount of BNQDs. Therefore, it is still worthwhile to develop effective synthesis methods of preparation of BNQDs in high yield and at low cost. With this aim, we have developed a simple one-step synthesis method of h-BNQDs of 10 nm size via hydrothermal disintegration of exfoliated h-BN sheets.
Nanostructures offer an unprecedented opportunity for improving drug delivery and targeting because of their unique nanostructure and diverse properties [11], [12], [13], [14], [15], [16], [17]. Breast cancer is one of the leading causes of death among woman worldwide, and treated patients often suffer from disease recurrence and metastasis due to the presence of breast cancer stem cells (BCSCs) [18]. Further increased ATP-binding cassette transporters and slow cycling nature of BCSCs serve as a potential mechanism for chemoresistance. In addition, modulation of Wnt, Hedgehog, Notch, tumor necrosis factor-α/ nuclear factor kappa B (TNF-α/NF-κB), transforming growth factor β (TGF-β), and receptor tyrosine kinase (RTK) signaling pathways might trigger surprising compensatory pathways that could neutralize the treatment effects in breast cancer [18]. However, the side effects of anticancer drugs such as doxorubicin (DOX) used as a single agent remain a significant challenge. Delivery and accumulation of any anticancerous drugs in the nucleus specifically within tumor cells is of great interest because the nucleus is one of the most sensitive targets for drug-induced damage in cancer prevention. Because the nucleus is the site where free DOX is mainly distributed and exerts the effects of the drug through intercalation with DNA and inhibition of macromolecular biosynthesis [19], DOX has been reported to accumulate in the nucleus by the formation of a DOX-proteasome complex [20]. However, a small percentage of drugs are delivered into the nucleus in drug-resistant cells due to overexpression of P-gp involved in drug efflux pumps, drug deactivation and sequestering to acidic compartments, and drug metabolism and detoxification. Therefore, controlled release and better bioavailability of drug are major challenges to increase their therapeutic efficacy in the current scenario [21]. For instance, Wang et al. [11] observed that graphene oxide quantum dots (GQDs) efficiently deliver DOX as conjugate GQDs/DOX with enhanced DNA cleavage activity due to small lateral size with maintenance of the layered structure of graphene. These findings lead us to believe that a conjugate of the anticancer drug with quantum dots (QDs) would improve DNA cleavage activity and reduce side effects of the drug. According to the aforementioned findings and discussion, we have developed a new class of QDs as an enhancer for DNA cleavage of DOX: BNQDs, which have received much more attention due to their non-toxicity, good solubility, strong fluorescence, and strong quantum confinement and are used in non-toxic fluorescent labeling of biological cells [9]. We found that without any premodification, BNQDs show higher anticarcinogenic activity and also enhance DNA cleavage activity remarkably than the previously discussed GQDs in low concentration (20–30 μg/ml) [11]. To further elucidate the nature of the BNQD-DOX-DNA interaction, we performed ab initio computational modeling of the system using density functional theory (DFT) using Vienna Ab initio Simulation Package (VASP). Perhaps to the best of our knowledge, BNQDs would be used first-time time to enhance the DNA cleavage activity of the anticancer drug DOX and explained the interaction of DOX with BNQDs as well as DOX-BNQDs with DNA.
In this work, we report a novel and simple hydrothermal fabrication method of BNQDs (∼10–15 nm size) by cutting exfoliated h-BN sheets, which show increased direct band gap in comparison with the h-BN sheets and exfoliated BN because of strong quantum confinement and some defects. More interestingly, we have demonstrated that BNQDs can be used to enhance the DNA cleavage activity of DOX without any surface modification because of its unique structural and chemical properties along with its effective efficacy in breast cancer chemoprevention.
2. Methods
2.1. Fabrication of BNQDs
The exfoliation of h-BN has been performed using H2SO4 (240 ml) and KMnO4. The exfoliated h-BN dispersion in water (2 mg/ml) has been disintegrated into QDs using the hydrothermal method at 120 °C for 6 h, as described in Fig. 1. A brief experimental process is detailed in the supporting information. A strong polar solvent, deionized water used in the hydrothermal method, possesses appropriate surface energy to cut exfoliated BN flakes under 120 °C temperature. This approach for the fabrication of BNQDs is close to that we have previously demonstrated to create high yield of GQDs [22], owing to the structural similarity of h-BN and graphite. In contrast to graphite, the stacking sequence is AA`; the B atoms in one plane are on top of the N atoms in the other plane. Cyan color fluorescent BNQDs were synthesized by hydrothermal cutting of h-BN sheets followed by an exfoliation of h-BN sheets using acid treatment, as shown in Fig. 1.
Fig. 1.
The schematic for the synthesis of blue fluorescent BNQDs. BNQDs, boron nitride quantum dots; h-BN, hexagonal boron nitride; D.W., distilled water.
2.2. Material characterization
The morphology of the BNQDs was examined by transmission electron microscopy (TEM, Tecnai G2 F30 S-Twin). The height of the BNQD sample was characterized by atomic force microscopy (AFM) (N8 NEOS Senterra) operating in non-contact mode. Raman spectra were obtained with a Renishaw in Via micro-Raman spectrometer with an excitation source 514 nm. The structural measurement was performed using X-ray diffraction (XRD) technique in the Bragg-Brentano parafocussing geometry using PANalytical XPert PRO XRD system. A Fourier-transform infrared (FTIR) spectrometer (FT-IR 4100, Jasco) and a ultraviolet-visible-near-infrared (UV-VIS-NIR) spectrometer (V-570, Jasco) were used for characterization of as-synthesized BNQDs. For photophysical properties of BNQDs, the photoluminenscence (PL) and photoluminescence excitation (PLE ) measurements were carried out using a 325-nm neodymium-doped yttrium aluminium garnet (Nd:YAG) laser and a high-sensitivity photomultiplier tube detector at room temperature.
2.3. DNA cleavage analysis
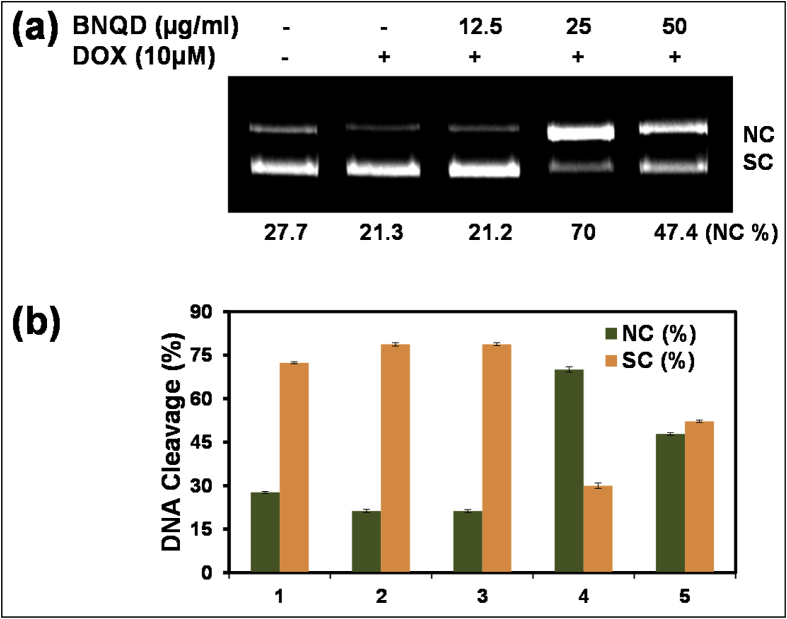
The cleavage efficiency of BNQD with PBR322 DNA was monitored using agarose gel electrophoresis as described previously [21]. PBR322 DNA (300 ng) was treated with different concentrations of BNQDs (12.5–50 μg/ml) in 5% dimethylformanide (DMF)/50 mM Tris-HCl/50 mM NaCl buffer at pH 7.2 for 24 h at 37 °C. NaBH4 solution was used as a reducing agent. Ten micromole of DOX and loading buffer containing 0.25% bromophenol blue, 0.25% xylene cyanol, and 30% glycerol were added, and electrophoresis was performed at 100 V for 1 h in TAE buffer using 1.2% agarose gel containing 0.5 μg/ml of ethidium bromide. The gels were photographed and analyzed by densitometric scanning using AlphaImage Image Analysis System (Alpha Innotech). The cleavage efficiency was measured by determining the ability of the complexes to convert DNA from the supercoiled (SC) to the nicked circular (NC) fragment.
2.4. Cytotoxicity by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay
MCF-7 and HaCaT cells were grown and maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin. Cytotoxicity was analyzed as described in our previous work [23], [24]. Details of the method are described in supporting information.
2.5. Nuclear staining by Hoechst 33258
As described previously [21], MCF-7 cells were seeded and grown on 96-well microtiter plates. Cells were treated with DOX (10 and 20 μM), BNQD (5 and 10 μg/mL), and a combination of both. After incubation with or without the drug for 24 h, cells were processed for Hoechst staining. In brief, cells were washed with PBS and stained with 5 μg/mL of Hoechst 33258 dye in PBS for 20 min at room temperature in a dark chamber. After extensive washing with phosphate-buffered saline (PBS), nuclear staining was examined under a fluorescence microscope (Leica) at 20× magnification. Images were captured digitally.
2.6. Reactive oxygen species measurement
Reactive oxygen species (ROS) level was determined by the oxidative conversion of non-fluorescent 2′, 7′-dichlorofluorescein diacetate (H2DCFDA) to highly fluorescent 2′, 7′-dichlorofluorescein (DCF) as described previously [25], [26]. Details of the method are described in supporting information.
2.7. Statistical analysis
All experiments were repeated three times independently, and one representative image is presented in the Fig. 4, Fig. 6, Fig. 7, Fig. 8. Statistical analysis was performed using SPSS using one-way analysis of variance followed by Tukey's post hoc test. Data represented as mean ± standard error of mean (S.E.M.) *, #, and $ denote significant differences at the level of p < 0.05.
Fig. 4.
Effect of BNQDs on DNA cleavage of PBR322 DNA: (a) Cleavage of DNA (300 ng) by DOX (10 μM) with different concentration of BNQDs (12.5–50 μg/ml) as labeled, (b) densitometric scanning of NC and SC band of DNA. Reaction time was 24 h at 37 °C. Lane 1 is without BNQD and DOX. Lane 2–5 are DNA with 0–50 μg/ml BNQDs. Data represent mean ± S.E.M. of three independent experiments. BNQDs, boron nitride quantum dots; DOX, doxorubicin; NC, nicked circular; SC, supercoiled; S.E.M., standard error of mean.
Fig. 6.
Cytotoxicity of BNQDs and DOX. MCF-7 and HaCaT cells were treated with varying concentrations of (a) BNQDs (0–10 μg/mL) and (b) DOX (0–40 μM) for 24 h and then subjected to MTT dye reduction assay. The percentage of viable cells (relative to control) was plotted against concentration. The data at each point represent mean ± S.E.M. Each experiment was repeated thrice with five replicates each time. Bars indicate standard errors. (c) Upper panel shows apoptotic cells by nucleus staining with Hoechst 33258 (i-iii), and the lower panel shows the level of ROS by green fluorescence H2DCFDA (iv-vi) with 5 and 10 μg/mL of BNQDs after 24 h. *Represents significant difference as compared with control group (*p < 0.05) on MCF-7 cells, and # represents a significant difference as compared with control group (#p < 0.05) on HaCaT cells. BNQD-DOX configuration found. BNQDs, boron nitride quantum dots; DOX, doxorubicin; ROS, reactive oxygen species; H2DCFDA, 2′, 7′-dichlorofluorescein diacetate; S.E.M., standard error of mean.
Fig. 7.
Cytotoxicity of MCF-7 cells. Breast cancer cells were treated with DOX (5, 10, 20, and 40 μM) in the presence of 5 and 10 μg/mL of BNQDs (a) and cells treated with BNQDs (1, 2.5, 5, and 10 μg/mL) in the presence of 10 and 20 μM of DOX (b) for 24 h and then subjected to MTT dye reduction assay. The percentage of viable cells (relative to control) was plotted against concentration. The data at each point represent mean ± S.E.M. Each experiment was repeated thrice with five replicates each time. Bars indicate standard errors. *, #, and $ (* # $ p < 0.05) represent a significant difference as compared with control group. MCF-7 cells treated with 5 μM of DOX and 1 μg/mL of BNQD are considered as the control group. BNQDs, boron nitride quantum dots; DOX, doxorubicin; S.E.M., standard error of mean.
Fig. 8.
Fluorescence image of MCF-7 cells with H2DCFDA and Hoechst 33258. The image from the left panel shows the level of ROS by green fluorescence H2DCFDA with DOX (i, ii) and the combination of DOX and BNQDs (iii-vi); and the right panel shows apoptotic cells by nucleus staining with Hoechst 33258 with DOX (vii, viii) and the combination of DOX and BNQDs (ix-xii). BNQDs, boron nitride quantum dots; DOX, doxorubicin; H2DCFDA, 2′, 7′-dichlorofluorescein diacetate.
2.8. Modeling of BNQDs-DOX interaction with DNA
We performed ab initio computational modeling of the system to elucidate the nature of the BNQDs-DOX-DNA interaction. All relaxations were performed at the DFT level with the Spanish Initiative for Electronic Simulations with Thousands of Atoms (SIESTA) package [27], [28], and core electrons are modeled using Troullier's [29] soft norm-conserving pseudopotentials. The valence electrons are expanded in a basis set of local orbitals using a double-zeta plus polarization orbitals basis set. The Generalised Gradient Approximations (GGA) of Perdew et al. [30] was used for the exchange-correlation functional. All molecules were placed in sufficiently large super cell to avoid nearest-neighbor interactions.
3. Results
3.1. Structural and optical characterization of BNQDs
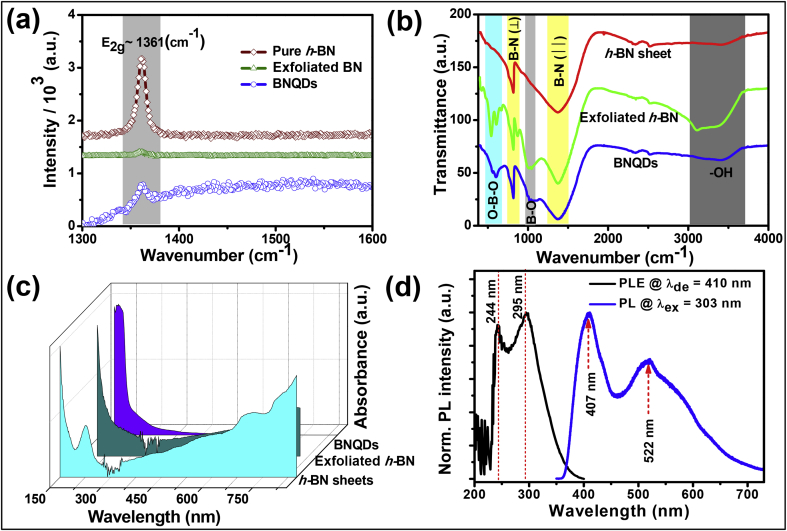
Cyan color fluorescent BNQDs were synthesized by hydrothermal cutting of h-BN sheets. The reaction was initiated by the potassium ions that intercalate and promote the rapid exfoliation of BN sheet under ultrasonication. The formation of BNQDs involved two steps: the potassium ions (K+) intercalate covalently bonded h-BN sheet with weak interlayer van der Waals force followed by disintegration of exfoliated h-BN under hydrothermal condition. The temperature used (120 °C) for disintegration is much less than the previously reported temperature for the fabrication of BNQDs (200 °C) [9]. The morphology of h-BN sheets and synthesized exfoliated h-BN sheets has been characterized by SEM. The starting material was micrometer-sized exfoliated h-BN (Figure S1 and Figure S2b) obtained by the intercalation of the h-BN sheets by K+ in the oxidation process. A more detailed explanation for better understanding is given in supporting information. The exfoliated h-BN disintegrated into QDs under hydrothermal condition. After 15 min of centrifugation at 8000 rpm, impure and unreacted parts get removed, and fluorescent QDs of size ∼10–15 nm in light yellow–colored liquid were collected. The presence of oxygen group may make exfoliated h-BN hydrophilic and helps it disintegrate easily in water solution under hydrothermal treatment. Fig. 2a shows a TEM image of synthesized BNQDs; the average size of BNQDs is around 10 nm. Fig. 2b shows the AFM image of BNQDs. Statistical analysis of Fig. 2b in 2 μm 2 μm region gives an average height of 1.69 nm, which corresponds well to the height distribution curve (in the inset of Fig. 2b) fitted by Gaussian with R2 ∼ 0.9891. Although some of the QDs appeared to be of considerable height as highlighted in the blue circle in the 3D AFM image (Fig. 2c), a strong quantum confinement exists in synthesized BNQDs owing to its small size and average height. XRD patterns of the BNQDs and h-BN sheet are given in Fig. 2d. The peak positions of BNQDs are at 26.24° and 54.01° corresponding to (002) and (004) diffraction planes, respectively [JCPDS, No. 9–12], may be attributed to the hexagonal BNQD formation. The peak at 26.32° corresponds to (002) diffraction peak of h-BN nanostructure [7] and is very intense and well defined. The refinement of XRD pattern was carried out using the Rietveld method using Fullprof software. The standard Bragg positions of h-BN (space group P -6m2) are shown by the vertical bars, and the residue, by the line at the bottom of the XRD patterns.
Fig. 2.
(a) TEM image of BNQDs, (b) AFM topography image of BNQDs and the height distribution curve of 2 μm × 2 μm region in the inset of figure, (c) 3D AFM image of BNQDs, and (d) Rietveld refinement profiles of X-ray diffraction pattern of h-BN sheets and synthesized BNQDs. The circles represent the observed data (Obs), the solid line through the circles is the calculated profile (Calc), and vertical tics below curves represent allowed Bragg reflections for the hexagonal phase. The difference pattern of the observed data and calculated profile (Obs–Calc) is given below the vertical tics. 3D, three-dimensional; AFM, atomic force microscopy; BNQDs, boron nitride quantum dots; TEM, transmission electron microscopy.
The analysis of BNQDs reveals that the samples are of single phase and have no impurities. The average crystallite size for each sample was calculated from the Scherrer equation [31].
| (1) |
λ = 1.5406 Å for this experiment, β is the effective peak width at half maximum after subtraction of instrumental broadening error, and θ is the angular position of the peak.
The lattice parameters (‘a = b’ and ‘c’) and an interlayer spacing (dhkl) between two planes have been measured from Rietveld refinement of the XRD data and also by using the following formula [31]:
| (2) |
where λ is the incident wavelength (λ ≈ 1.5406 Å), θ is the diffraction angle, and h, k, and l are Miller's indices. The volume of the unit cell for a hexagonal system was calculated using the following equation:
| (3) |
The Rietveld refining starts with the crystallographic data of h-BN nanotube [32], and it converges with residual values of conventional Rietveld Rp = 23.9, Rwp = 26.7, Rexp = 8.28, Bragg R-factor = 7.91, chi-squared = 10.42. The goodness of fit of the BNQDs Rietveld final refinement is represented by S (Rwp/Rexp), and it shows S = 3.22 as the best fit result. We found that the BNQDs belong to hexagonal lattice; space groups P -6m2, space group no. 187 and lattice parameter a = 2.367897 Å and c = 6.786186 Å. The XRD pattern of exfoliated h-BN (Figure S3) exhibits a typical crystalline hexagonal structure of BN layers with an interlayer spacing of around 0.40 nm, which is higher than that of the h-BN sheet (0.34 nm), due to intercalation of K atom in the interlayer space. Some more peaks (marked in Figure S3) with low intensity are a manifestation of some impurities and residue of organic compounds in exfoliated h-BN. These impurity peaks correspond to manganese oxide, which is formed after oxidation and reduction process of potassium permanganate.
Raman spectra (Ex∼514 nm) of h-BN, exfoliated h-BN, and BNQDs are shown in Fig. 3a. Only one peak at around 1363 cm−1 is found in the spectrum of BNQDs with lower intensity than that of bulk h-BN. This peak could be attributed to the high-frequency interlayer E2g vibration mode of h-BN which is in good agreement with other reports [33], [34], [35]. In case of BNQDs, full width at half maximum (FWHM) of E2g peak is 15 cm−1, which is higher than that of bulk h-BN (∼11 cm−1) and exfoliated h-BN (∼10 cm−1) but lower than that in earlier reports [35], [36]. No dopant species such as C dopant BN at around 1587 cm−1 were detectable in BNQDs by Raman spectra [37], [38], [39], [40]. The BNQDs and exfoliated h-BN were examined by FTIR spectra in transmittance mode (Fig. 3b) to investigate their bonding nature. The strong IR peaks at 1372 cm−1 and 818 cm−1 owe to B-N vibrations, which are characteristic primary and secondary absorption bands of h-BN corresponding to A2u and E1u IR active modes, respectively. The intense absorption band at 1372 cm−1 is associated with the well-known in-plane vibrational mode (E2g) of the h-BN networks (electron shifting from O to N-B) [9], whereas the absorption band at 818 cm−1 corresponds to out-of-plane vibration [41]. The absorption bands in the regions of 1090–800 cm−1 and 650–400 cm−1 are attributed to the B–O and O–B–O vibrations, respectively [42]. The commonly originated vibration mode at 3397 cm−1 in FTIR spectra of BN samples is the result of the –OH stretching mode due to capturing of ambient moisture.
Fig. 3.
(a) Raman spectrum of h-BN sheets, exfoliated h-BN, and BNQDs, (b) FTIR spectra of h-BN sheets, exfoliated h-BN, and BNQDs, (c) UV–vis absorption spectra of h-BN, exfoliated h-BN, and BNQDs, (d) normalized PL excitation and PL spectra. BNQDs, boron nitride quantum dots; h-BN, hexagonal boron nitride; BN, boron nitride.
The synthesized samples were examined by ultraviolet visible spectroscopy to study the optical energy band gap by Tauc's formulation (Figure S4, see supporting information) [43]. Fig. 3c clearly shows strong absorption edge in the range of 190–500 nm in all samples. The absorption spectrum of h-BN sheets shows absorption edge ca. at 240 nm, which corresponds to an optical band gap of 5.17 eV (Figure S4). The optical band gap of 5.42 eV corresponds to absorption edge at 228 nm in the spectrum of exfoliated h-BN, which is large in comparison with that of the h-BN sheet. This may be due to the presence of some oxygen atoms or boundary effect [43] after intercalation, which is difficult to distinguish. We can also see that after hydrothermal cutting of exfoliated h-BN, the absorption band edge is ca. at 203 nm corresponding to an optical band gap of 6.08 eV (Figure S4). BNQDs show cyan color fluorescence under ultraviolet irradiation (365 nm), as shown in Fig. 1. The peaks of PLE spectra are located at ∼244 nm and ∼295 nm and correspond to higher peak values, and the PL emission spectra were recorded with a laser source of excitation wavelength of 303 nm (Fig. 3d). Asymmetric peaks in the PL spectrum of our sample suggest the presence of multifluorophores or luminescent species. The mechanism of fluorescence in the BN system is presently still a matter of strong debate, varying from case to case. Tang et al. [44] proposed that boron-oxygen species are novel luminescence centers in the boron-nitrogen system. Detailed discussions for the origin of the emission in BN sample are given in supporting information.
3.2. Enhanced DNA cleavage activity of anticancer drug
To further examine the potential applications of BNQDs, we use them in DNA cleavage applications. To investigate the functional mechanism of BNQDs in the enhancement of the nuclease activity of DOX, we varied BNQD concentration as shown in Fig. 4. BNQDs were chosen because of their smaller lateral size, better conjugate state with an enriched oxygen functional group, and well-dispersible nature in water. Cleavage study was analyzed in terms of percentage conversion of SC DNA into NC DNA. Although the interaction mechanism between BNQDs and DNA is not well understood yet, we assume that the BNQDs interact with DOX after intercalating into DNA. Most probably, BNQDs will be a better intercalator to DNA molecules because of their bigger lateral size. The ability to cleave DNA molecule may depend on the generated reactive hydroxyl radicals [45]. Our previous reports have suggested reduction of Cu (II) to Cu (I) or its oxidation to Cu (III) followed by activation of O2-/H2O2-producing reactive oxygen species as the main source of DNA cleavage [46].
Furthermore, complexes with higher electron density around the metal center due to pyridine and methoxy functionalities on the dithioester ligand backbone have been reported to exhibit considerable DNA cleavage capability [46]. The increase of the absorbance at 490 nm has been recognized (Figure S6) to be due to the interaction between BNQDs and DOX with an increase in the concentration of BNQDs with a binding constant (Kb∼ 0.07338 μg/ml) due to electronegative nitrogen and electropositive boron. Our finding includes the enhanced DNA cleavage activity of DOX up to 70% and interaction of BNQDs with DOX with binding constant Kb ∼0.07338 μg/ml. In addition, ab initio theoretical results also indicate the absorption of DOX on BNQD at the N-terminated edge with binding energy −1.075 eV which prevent the normal replication mechanisms in DNA. However, no such direct evidence has been reported yet to explain how BNQDs interact with DOX after intercalating into DNA, although one of the reports of Wang et al. [11] suggested that GQDs efficiently deliver DOX as conjugate GQDs/DOX with enhanced DNA cleavage activity due to small lateral size with maintaining the layered structure of graphene. Perhaps to the best of our knowledge, BNQDs would be used first-time to enhance the DNA cleavage activity of anticancer drug DOX and explained the interaction of DOX with BNQDs and DOX-BNQDs with DNA.
3.3. Modeling for BNQDs-DOX–DNA interaction
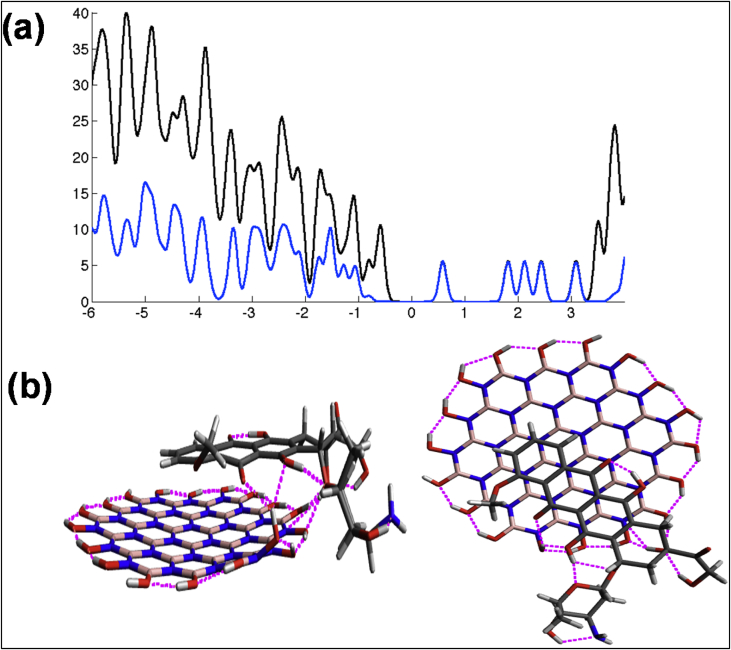
In addition, we investigated theoretically the nature of the BNQD-DOX-DNA interaction using ab initio computational modeling of the system. For the atomic structure of BNQDs, we used geometry proposed by Chigo-Anota et al. [47], and for DOX-DNA interaction, we used the model from the studies by Ane Eizaguirre et al. [48] and Frederick et al. [49] (Fig. 5). The finite range of the orbital was defined by orbital confinement energy of 0.005 Ry. The fineness of the real-space mesh was 250 Ry cutoff, assuring energy and force convergence. The structure was fully optimized with residual forces less than 0.04 eV/Å. Considering BNQDs alone, we can conclude that the molecule is polarized and that consequently B- and N-terminated edges are not equivalent. Molecules are in π stacked configuration. Comparing the binding energy to (probably slightly over bonding) value of 2.749 eV calculated with local density approximation, we conclude that there is the clearly fair amount of wdW interaction and physisorption between BNQDs and DOX. Charge transfer is quite small, ∼0.085e. Another way to appreciate the value of bonding energy is to look at non–π-stacked configurations (Figure S8, S9). The bonding energy in T-shaped configuration drops by two-thirds (to 36%), and with only hydrogen bonding left, it becomes 30% of the π-stacked binding value. Therefore, we can conclude that over two-thirds of the attraction is in stacking of π-conjugated fragments and the rest consists of hydrogen bonding between the BNQD and DOX.
Fig. 5.
(a) The projected density of state (PDOS) (black) and DOX projected states (blue) of the BNQD-DOX complex and (b) side and top views of the minimum-energy BNQD-DOX configuration found. BNQDs, boron nitride quantum dots; DOX, doxorubicin.
3.4. Cytotoxicity of BNQDs and DOX against breast cancer cells (MCF-7) and normal human keratinocyte cells (HaCaT)
Cytotoxicity of BNQDs and DOX on breast cancer MCF-7 and normal human keratinocyte cells HaCaT has been evaluated by MTT assay, which measures the mitochondrial dehydrogenase activity as cell viability. Cytotoxicity of DOX was found much higher in normal human keratinocyte cells HaCaT than in MCF-7 cells. However, BNQDs were less cytotoxic on HaCaT than on MCF-7 cell after 24 h. Approximately 25, 37, 50, and 55% of MCF-7 cells were dead on treating with 1, 2.5, 5, and 10 μg/mL of BNQDs, respectively, whereas ∼37, 49, 45, and 46% of MCF-7 cell death was found after treatment with 5, 10, 20, and 40 μM DOX, respectively (Fig. 6a). However, 8, 13, 33, and 43% cytotoxicity of HaCaT cells was induced by 1, 2.5, 5, and 10 μg/mL of BNQDs, respectively, whereas cell death was approximately 87, 87, 89, and 88% after treatment with 5, 10, 20, and 40 μM DOX, respectively (Fig. 6b). Cancer cells tend to be more susceptible than normal cells to the damaging effects of chemotherapy because of increased glycolytic metabolism, uncontrolled proliferation, inability to arrest the cell cycle, and genetic instability [50]. BNQDs at higher concentration are also toxic to MCF-7 and HaCaT cells because of ROS accumulation. Even at a higher concentration of BNQDs (10 μg/mL), 50% viability of both cells is maintained. But, in the case of DOX even at very low concentration (5 μM), 50% viability of MCF cells is maintained, but that of HaCaT cells was only 20%. Here, even at very low DOX concentration (5 μM), more normal cells are dying than cancer cells.
3.5. BNQDs increase cell apoptosis and reduce ROS accumulation in MCF-7 cells
Chromosome condensation is the most significant change of an apoptotic cell, and the cracking of DNA changes the nuclear morphology. Nuclear fragmentation and condensation is a hallmark of apoptosis. Fluorescent dye Hoechst 33258 binds to the DNA molecule and detects the compacted state of chromatin in apoptotic cells. As Hoechst 33258 is a non-toxic, water-soluble double-benzene imidazole compound, it provides a rapid and convenient assay for apoptosis. The uptake of Hoechst 33258 in the membrane increases during apoptosis of cells, which enhances for highly condensed chromosomes, and it shows strong blue than weak fluorescence in normal cells. Hoechst 33258 staining was used to detect apoptotic cells by fluorescence imaging. Hoechst reagent is readily taken up by cells during the initial stages of apoptosis and selectively stains nuclei of apoptotic cells intense fluorescent blue. The blue fluorescent Hoechst dyes are cell-permeable DNA stains that detect gradations of nuclear damage. In case of the cancer cell MCF-7, BNQD treatment enhances nuclear damage in comparison with control cells after 24 h (Fig. 6c). Our previous report suggests the increased apoptosis by tamoxifen (Tmx)-embedded poly (lactic-co-glycolic acid) (PLGA) nanoparticles (PLGA-Tmx) through nuclear condensation and fragmentation in Hoechst 33258–stained Dalton's lymphoma ascite cells [21]. However, further confirmation of apoptosis could be performed through detection of annexin-V/propidium iodide staining or caspase and Poly (ADP-ribose) polymerase (PARP) proteins [25]. Active caspase 3 causes degradation of lamins, resulting in chromatin condensation and nuclear fragmentation. Previously, we have shown the induction of apoptosis in in vitro and in vivo tumor model after treatment with PI-103, an inhibitor, and quercetin (a natural antioxidant) [25].
ROS level was determined using H2DCFDA dye in MCF-7 cells after 24 h of BNQDs treatment. The H2DCFDA, a cell-permeable, non-fluorescent precursor of DCF, can be used as an intracellular probe for oxidative stress. Intracellular esterases cleave H2DCFDA at the two ester bonds, producing a relatively polar and cell membrane–impermeable product, 2′, 7′-dichlorofluorescein. This non-fluorescent molecule accumulates intracellularly, and subsequent oxidation by hydrogen peroxide (or peroxynitrite) yields the highly fluorescent product DCF. The redox state of the sample can be monitored by detecting the increase or decrease in green fluorescence of DCF. BNQDs reduced green fluorescence of DCF, suggesting a decline in ROS level of MCF-7 cells (Fig. 6c).
3.6. BNQDs enhance the cytotoxicity of DOX in MCF-7 cells
Cytotoxicity of MCF-7 cells after treatment of 5, 10, 20, and 40 μM of DOX for 24 h was determined in the presence of 5 and 10 μg/mL of BNQDs, which was ∼40, 60, 65, and 61% and ∼49, 69, 70, and 66% respectively (Fig. 7a). Similarly, death of MCF-7 cells after treatment with 1, 2.5, 5, and 10 μg/mL of BNQDs in the presence of 10 μM DOX was ∼49, 54, 60, and 66% and in the presence of 20 μM DOX was ∼53, 54, 64, and 69%, respectively (Fig. 7b). The results are in accordance with our previous findings of the induction of cytotoxicity and apoptosis in terms of controlled and regulated delivery of Tmx in breast cancer [21]. In combination, with the synergistic effect of BNQDs and DOX, the cell viability (∼30%) is less than BNQDs (∼60%) and DOX (∼50%) alone in MCF cells. BNQDs also reduced the cell viability of cancer cells as well as normal cells and ROS accumulation. If BNQDs alone are used as a drug, it needs much higher concentration to increase the risk of accumulation of BNQDs in normal cells. However, low-concentration BNQDs in combination with DOX increased the efficiency of DOX-mediated DNA cleavage and consequently reduced cell viability.
3.7. BNQDs improve the activity of DOX in the regression of ROS accumulation and enhance cell apoptosis
Furthermore, DOX (10 and 20 μM) treatment enhances nuclear damage in MCF-7 cells (Fig. 8; vii-viii). The nuclear constriction/damage is vigorously enhanced in the presence of 5 and 10 μg/mL of BNQDs (Fig. 8; ix-xii). Nuclear fragmentation and condensation is a hallmark of apoptosis. The blue fluorescent Hoechst dyes are cell-permeable DNA stains that detect gradations of nuclear damage. The result suggests the improved apoptotic activity of DOX in the presence of BNQDs. Highly ROS such as hydrogen peroxide (H2O2), superoxide anion (O2•−), hydroxyl radicals (OH.), etc. produced in cells are grouped ROS that confer reactivity to different biological targets. ROS has a direct or indirect role in the modulation of signal transduction and transcription factors to regulate cell survival, proliferation, and migration. BNQDs in this regard counterbalanced the enhanced ROS accumulation in MCF-7 cells. Induction of lymphoma cells by external exposure of H2O2 has been reported previously to be counterbalanced by an antioxidant supplement [51].
4. Discussion
The XRD result indicates that the crystallite size tends to decrease from about 35.4 nm to about 15 nm, with the disintegration of h-BN sheets into QDs, which shows that BNQDs have poor crystallinity probably arising from crystal defects due to the cutting of h-BN sheets into small dots and the presence of oxygen functionality. The Rietveld refining calculation gives slightly different lattice parameters of BNQDs from that of h-BN sheets. This is possibly due to the successful disintegration of h-BN sheets.
FTIR spectrum reveals the presence of the oxygen group on as-synthesized BNQDs, which may act similar to a luminescence center. Asymmetric peaks in the PL spectrum of our sample suggest that the presence of multifluorophores or luminescent species. The mechanism of fluorescence in the BN system is presently still a matter of strong debate, varying from case to case. Tang et al. [44] proposed that boron-oxygen species are novel luminescence centers in the boron-nitrogen system. We believe that BOx- (x = 1 and 2) species (Figure S5c) is the effective luminescence center for emission in our sample BNQDs. The detailed discussion for the origin of the emission in BN sample is given in supporting information.
The enhancement of DNA cleavage activity by anticancer drug DOX was performed using synthesized BNQDs. Results show that at low concentration of BNQDs (12.5 μg/ml), the DNA cleavage in the presence of DOX is barely affected. Moreover, the percentage of NC fragment enhanced up to 70% with maximum cleavage activity of DOX with 25 μg/ml of BNQDs compared with 21.3% cleavage by control. Further increasing concentration of BNQDs (50 μg/ml) leads to a decline in NC DNA up to 47.4%. Noticeably, the activity enhancement is critically dependent on the ratio between BNQDs and DOX. This observation is similar to the behavior of the GQDs-DOX [11], suggesting that BNQDs could work together with DOX molecules. The ab initio computational modeling of the system confirms that DOX is adsorbed on BNQDs at the N-terminated edge, as shown in Fig. 5b, with a binding energy of −1.075 eV. The overall binding between DOX and DNA [48] is weakened by the presence of BNQDs, from −1.248 eV to −0.678 eV, which is expected from the BNQD-DOX structure, where some hydrogen bonds needed for DOX-DNA interaction are already occupied. Still, the effect of BNQD-DOX interaction with the DNA can be understood as the impact of a big and massive wedge inserted into the double-stranded DNA. When applied, BNQDs amplify the effect of DOX by introducing a big and bulky unit. That is why both tend to separate the DNA strands and prevent normal replication mechanisms by attaching a massive BNQD-DOX complex to DNA via a number of hydrogen bonds.
Cytotoxicity of DOX was found to be much higher in normal human keratinocyte cells HaCaT than in MCF-7. However, BNQDs were less cytotoxic on HaCaT than on MCF-7 after 24 h. The advantage of any anticancer drug is assessed by much higher potentiality to kill cancer cells with a comparatively basal level of cytotoxicity on normal cells. DOX in this regard shows the major drawback because it kills normal cells to a greater extent than cancer cells. The result suggests BNQDs be a better anticancer agent that may be beneficial in combination with DOX. The ROS level has been considered a significant regulator of cancer progression. The accumulation of ROS due to an imbalance between its production and biological oxidative defense system is described as oxidative stress. Oxidative stress is common through overproduction of reactive oxygen and nitrogen species, either by endogenous or exogenous insult. Unregulated or prolonged production of cellular oxidants has been linked to mutation as well as modification of gene expression leading to carcinogenesis at all the stages. ROS is a group of molecules, including hydrogen peroxide, superoxide anion, and hydroxyl radicals, which confers reactivity to different biological targets. It is produced as a by-product during mitochondrial electron transport of aerobic respiration or by oxidoreductase enzymes and metal-catalyzed oxidation and have the potential to cause some modifications of macromolecules [52]. BNQD treatment on the cancer cell MCF-7 enhances nuclear damage in comparison to control cells after 24 h (Fig. 6c). BNQDs reduced green fluorescence of DCF, suggesting a decline in ROS level of MCF-7 cells (Fig. 6c). These cells show reduced survival with BNQDs. Quercetin has been previously reported to reduce tumorigenesis via regression of ROS and cell survival in murine T-cell lymphoma and HepG2 cells [24], [25]. ROS serves as signaling molecules to regulate biological and physiological processes toward adaptation to changes in environmental nutrients and oxidative environment [53]. It has been suggested to participate in cell survival, proliferation, and migration. ROS is considered a modulator of signaling pathways mediated through extracellular signal regulated kinase (ERK), Mitogen activated protein kinase (MAPK), hypoxia inducible factor (HIF), phosphoinositide 3-kinase (PI3K), protein kinase C (PKC), etc. to take part in carcinogenesis [24], [25], [53], [54].
BNQDs gradually enhance the cytotoxic potential of DOX in MCF-7 cells. Such improved cytotoxicity of DOX in the presence of rotaxane has been reported [55]. The result suggests that BNQDs improve the anticancerous activity of DOX via increasing its cytotoxic potential. DOX treatment showed increased ROS level in MCF-7. The result is supported by earlier findings suggesting that DOX induces ROS accumulation [56]. However, BNQD treatment was found to reduce ROS level in terms of intensity of green fluorescence of DCF in MCF-7 cells (Fig. 8; i-vi) which suggests reduced cell survival. The decrease in ROS level of MCF-7 by DOX (10 and 20 μM) in the presence of 5 and 10 μg/mL of BNQDs was found to be dose-dependent. Thus, BNQDs nullify the toxic effect of DOX. Thus, BNQDs enhance the anticarcinogenic activity of DOX by reducing ROS level and promoting cell death in MCF-7. Some studies established prolonged tissue retention and heavy metals that present the severe concerns for toxicity and clearance of nanoparticles and limit progress toward the clinical use of many of the developing nanoparticles. Also, inclusive variation in the modifiable surface, particle material, size, and surface coating affects the renal or hepatic clearance of nanoparticles including QDs [57]. The studies by Fisher et al. [58] and Schipper et al. [59] also suggested that surface charge affected the renal clearance of QDs in rat and mice system. In addition, renal clearance has been reported to be decreased with the increased size of QD nanoparticles. However, our present study is mainly focused on the synergistic efficacy of BNQDs with DOX in terms of anticancer activity via DNA cleavage, ROS accumulation, and interaction of DOX-BNQDs with DNA; that could be a translational relevance for a clinician but needs to be further investigated in future work.
5. Conclusion
To summarize this work, we propose a simple, ultrafast, economic, ecological, and reproducible hydrothermal route to prepare highly efficient BNQDs from exfoliated h-BN without the drawbacks of other reported methods. The oxidizing agent reacts with h-BN sheets readily under the relevant temperature, and exfoliation of sheet takes place, supported by SEM images. Owing to small size and height, strong quantum confinement exists in synthesized BNQDs. The optical band gap of the BNQDs was estimated to be around 6.08 eV using Tauc's formula, which is smaller than that of h-BN sheets due to quantum confinement. Most interestingly, the h-BNQDs were found to have an application in the clinical field for enhancing the anticarcinogenic activity of DOX. The overall result highlights the impact of BNQDs as an anticancer agent by itself. Furthermore, DOX improves the anticancerous activity in combination with BNQDs with less cytotoxicity. Furthermore, by increasing the concentration of BNQDs, the interaction between BNQDs and DOX was found with the binding constant (Kb ∼ 0.07338 μg/ml). DFT calculations predicted that the BNQD molecule is polarized, and consequently, B- and N-terminated edges are not equivalent. DOX is adsorbed on BNQDs at the N-terminated edge with a binding energy of −1.075 eV. When a massive BNQD-DOX complex attaches to DNA, the normal replication mechanism gets suppressed via formation of hydrogen bonds. The percentage of NC fragment enhanced up to 70% with maximum cleavage activity of DOX with 25 μg/ml of BNQDs compared with 21.3% cleavage by control. The results provide a base to develop BNQD-DOX as a better effective anticancer drug. Our approach may open up a route for the fabrication of high-quality BNQDs, and their exciting properties could pave the way for their use in many biological applications.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
A.S. is thankful to DST, India (project code: M-21-133) for funding. S.U. expresses her gratitude for the U.G.C. financial assistance. A.S. and S.U. acknowledge Biophysics lab, Physics Department, Banaras Hindu University for PL and UV measurement. A.K.M. thanks the Council of Scientific and Industrial Research (CSIR), India, for Senior Research Fellowship. A.G. and R.A. gratefully acknowledge Uppsala University for funding of the U3MEC KoF07-initiative and the Swedish Research Council (Vetenskapsrådet) and Stiftelsen för Strategisk Forskning (SSF (SUSBATT)) for financial support. The calculations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2019.01.001.
Contributor Information
I.-K. Oh, Email: ikoh@kaist.ac.kr.
A. Srivastava, Email: anchalbhu@gmail.com.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Geim A.K., Novoselov K.S. Nat. Mater. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 2.Zhu B.Y., Murali S., Cai W., Li X., Suk J.W., Potts J.R., Ruoff R.S. Adv. Mater. 2010;22:3906–3924. doi: 10.1002/adma.201001068. [DOI] [PubMed] [Google Scholar]
- 3.Lee C., Li Q., Kalb W., Liu X.-Z., Berger H., Carpick R.W., Hone J. Science. 2010;328:76–80. doi: 10.1126/science.1184167. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y. Bando, Yin L., Golberg D. Nano Lett. 2006;6:2982–2986. doi: 10.1021/nl061594s. [DOI] [PubMed] [Google Scholar]
- 5.Jin C., Lin F., Suenaga K., Iijima S. Phys. Rev. Lett. 2009;102:195505–195509. doi: 10.1103/PhysRevLett.102.195505. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z., Zou J., Liu G., Li F., Wang Y., Wang L., Yuan X., Sekiguchi T., Cheng H., Lu G.Q. ACS Nano. 2008;2:2183–2191. doi: 10.1021/nn8004922. [DOI] [PubMed] [Google Scholar]
- 7.Lei W., Portehault D., Liu D., Qin S., Chen Y. Nat. Commun. 2013;4:1–7. doi: 10.1038/ncomms2818. [DOI] [PubMed] [Google Scholar]
- 8.Ithurria S., Tessier M.D., Mahler B., Lobo R.P.S.M., Dubertret B., Efros A.L. Nat. Mater. 2011;10:936–941. doi: 10.1038/nmat3145. [DOI] [PubMed] [Google Scholar]
- 9.Lin L., Xu Y., Zhang S., Ross I.M., Ong A.C.M., Allwood D.A. Small. 2014;10:60–65. doi: 10.1002/smll.201301001. [DOI] [PubMed] [Google Scholar]
- 10.Jung J.-H., Kotal M., Jang M.-H., Lee J., Cho Y.-H., Kim W.-J., Oh I.-K. RSC Adv. 2016;6:73939–73946. [Google Scholar]
- 11.Wang C., Wu C., Zhou X., Han T., Xin X., Wu J., Zhang J., Guo S. Sci. Rep. 2013;3:2852. doi: 10.1038/srep02852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X., Zhang Y., Wang C., Wu X., Yang Y., Zheng B., Wu H., Guo S., Zhang J. ACS Nano. 2012;6:6592–6599. doi: 10.1021/nn301629v. [DOI] [PubMed] [Google Scholar]
- 13.Wu L., Wang J., Ren J., Qu X. Adv. Funct. Mater. 2014;24:2727–2733. [Google Scholar]
- 14.Conde J., Bao C., Tan Y., Cui D., Edelman E.R., Azevedo H.S., Byrne H.J., Artzi N., Tian F. Adv. Funct. Mater. 2015:1–12. doi: 10.1002/adfm.201501283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun C.-Y., Dou S., Du J.-Z., Yang X.-Z., Li Y.-P., Wang J. Adv. Healthc. Mater. 2014;3:261–272. doi: 10.1002/adhm.201300091. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y., Ihme S., Feuring-Buske M., Kuan S.L., Eisele K., Lamla M., Wang Y., Buske C., Weil T. Adv. Healthc. Mater. 2013;2:884–894. doi: 10.1002/adhm.201200296. [DOI] [PubMed] [Google Scholar]
- 17.Ding W., Kameta N., Minamikawa H., Wada M., Shimizu T., Masuda M. Adv. Healthc. Mater. 2012;1:699–706. doi: 10.1002/adhm.201200133. [DOI] [PubMed] [Google Scholar]
- 18.Vinayak M. Int. J. Cancer Oncol. 2016;3:1–7. doi: 10.15436/2377-0902.16.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gewirtz D. Biochem. Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 20.Sahay G., Alakhova D.Y., V Kabanov A., Control J. Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey S.K., Patel D.K., Maurya A.K., Thakur R., Mishra D.P., Vinayak M., Haldar C., Maiti P. Int. J. Biol. Macromol. 2016;89:99–110. doi: 10.1016/j.ijbiomac.2016.04.065. [DOI] [PubMed] [Google Scholar]
- 22.Kumar V., Singh V., Umrao S., Parashar V., Abraham S., Singh A.K., Nath G., Saxena P.S., Srivastava A. RSC Adv. 2014;4:21101–21107. [Google Scholar]
- 23.Maurya A.K., Vinayak M. Mol. Biol. Rep. 2015;42:1419–1429. doi: 10.1007/s11033-015-3921-7. [DOI] [PubMed] [Google Scholar]
- 24.Maurya A.K., Vinayak M. Nutr. Cancer. 2015;67:354–363. doi: 10.1080/01635581.2015.990574. [DOI] [PubMed] [Google Scholar]
- 25.Maurya A.K., Vinayak M. Tumor Biol. 2015;36:8913–8924. doi: 10.1007/s13277-015-3634-5. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh S., Chakraborty P., Saha P., Acharya S., Ray M. RSC Adv. 2014;4:23251–23261. [Google Scholar]
- 27.Soler J.M., Artacho E., Gale J.D., Garcia A., Junquera J., Ordejon P., S. Portal D. J. Phys. Condens. Matter. 2002;14:2745–2779. [Google Scholar]
- 28.Artacho E., Anglada E., Diéguez O., Gale J.D., Garcia A. J. Phys. Condens. Matter. 2008;20:064208–064214. doi: 10.1088/0953-8984/20/6/064208. [DOI] [PubMed] [Google Scholar]
- 29.Troullier N., Martins José Luís. Phys. Rev. B. 1991;43:1993–2006. doi: 10.1103/physrevb.43.1993. [DOI] [PubMed] [Google Scholar]
- 30.Perdew J.P., Burke K., Ernzerhof M. Phys. Rev. Lett. 1996;77:3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 31.Cullity B.D. Addison-Wesley; MA: 1978. Elements of X-ray Diffraction. [Google Scholar]
- 32.Kim Y.-I., Jung J.K., Ryu K.-S., Nahm S.-H., Gregory D.H. J. Phys. D Appl. Phys. 2005;38:1127–1131. [Google Scholar]
- 33.Nemanich R.J., Solin S.A., Martin Richard M. Phys. Rev. B. 1981;23:6348–6356. [Google Scholar]
- 34.Bernard S., Chassagneux F., Berthet M.-P., Vincent H., Bouix J. J. Eur. Ceram. Soc. 2002;22:2047–2059. [Google Scholar]
- 35.Hoffman D.M., Doll G.L., Eklund P.C. Phys. Rev. B. 1984;30:1–6. [Google Scholar]
- 36.Saha S., Muthu D.V.S., Golberg D., Tang C., Zhi C., Bando Y., Sood A.K. Chem. Phys. Lett. 2006;421:86–90. [Google Scholar]
- 37.Zhi C.Y., Bai X.D., Wang E.G. Appl. Phys. Lett. 2002;80:3590–3592. [Google Scholar]
- 38.Umrao S., Jang M.-H., Oh J.-H., Kim G., Sahoo S., Cho Y.-H., Srivastva A., Oh I.-K. Carbon N. Y. 2015;81:514–524. [Google Scholar]
- 39.Srivastava S., Ali M.A., Umrao S., Parashar U.K., Srivastava A., Sumana G., Malhotra B.D., Pandey S.S., Hayase S. Appl. Biochem. Biotechnol. 2014;174:960–970. doi: 10.1007/s12010-014-0965-4. [DOI] [PubMed] [Google Scholar]
- 40.Kumar V., Srivastava S., Umrao S., Kumar R., Nath G., Sumana G., Saxena P.S., Srivastava A. RSC Adv. 2014;4:2267–2273. [Google Scholar]
- 41.Lee C.H., Wang J., Kayatsha V.K., Huang J.Y., Yap Y.K. Nanotechnology. 2008;19:455605–455610. doi: 10.1088/0957-4484/19/45/455605. [DOI] [PubMed] [Google Scholar]
- 42.Holst J.R., Gillan E.G. J. Am. Ceram. Soc. 2008;130:7373–7379. doi: 10.1021/ja709992s. [DOI] [PubMed] [Google Scholar]
- 43.Ci L., Song L., Jin C., Jariwala D., Wu D., Li Y., Srivastava A., Wang Z.F., Storr K., Balicas L., Liu F., Ajayan P.M. Nat. Mater. 2010;9:430–435. doi: 10.1038/nmat2711. [DOI] [PubMed] [Google Scholar]
- 44.Tang C., Bando Y., Zhi C., Golberg D. Chem. Commun. 2007:4599–4601. doi: 10.1039/b711807d. [DOI] [PubMed] [Google Scholar]
- 45.Bin Zheng J.Z., Wang Chong, Xin Xiaozhen, Liu Fei, Zhou Xuejiao, Guo S. J. Phys. Chem. C. 2014;118:7637–7642. [Google Scholar]
- 46.Yadav M.K., Maurya A.K., Rajput G., Manar K.K., Vinayak M., Drew M.G.B., Singh N. J. Coord. Chem. 2017;70:565–583. [Google Scholar]
- 47.Chigo-Anota E., Salazar-Villanueva M., Hernández-Cocoletzi H. J. Nanosci. Nanotechnol. 2011;11:5515–5518. doi: 10.1166/jnn.2011.3441. [DOI] [PubMed] [Google Scholar]
- 48.Ane Eizaguirre L.A.E., Yanez Manuel. Phys. Chem. Chem. Phys. 2012;14:12505–12514. doi: 10.1039/c2cp40931c. [DOI] [PubMed] [Google Scholar]
- 49.Frederick C.A., Williams L.D., Ughetto G., Van Der Mare I.I.G.A., Van Boom J.H., Rich A., Wang A.H. Biochemistry. 1990;29:2538–2549. [PubMed] [Google Scholar]
- 50.Hanahan D., Weinberg R.A. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Maurya A.K., Vinayak M. PLoS One. 2016;11:e0160686. doi: 10.1371/journal.pone.0160686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinberg F., Chandel N.S. Cell. Mol. Life Sci. 2009;66:3663–3673. doi: 10.1007/s00018-009-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkel T. J. Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schieber M., Chandel N.S. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J., Xu Y., Wang X., Zhang L., Zhu J., Pang T., Bao X. Org. Biomol. Chem. 2015;13:7517–7529. doi: 10.1039/c5ob00934k. [DOI] [PubMed] [Google Scholar]
- 56.Fu Z., Guo J., Jing L., Li R., Zhang T., Peng S. Toxicol. Vitr. 2010;24:1584–1591. doi: 10.1016/j.tiv.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Longmire M., Choyke P.L., Kobayashi H. Nanomedicine. 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer H.C., Liu L., Pang K.S., Chan W.C.W. Adv. Funct. Mater. 2006;16:1299–1305. [Google Scholar]
- 59.Schipper M.L., Cheng Z., Lee S.-W., Bentolila L.A., Iyer G., Rao J., Chen X., Wu A.M., Weiss S., Gambhir S.S. J. Nucl. Med. 2007;48:1511–1518. doi: 10.2967/jnumed.107.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.